Abstract
Objective:
Of the predominant HIV-1 subtypes in Uganda, subtype D infection confers a worse prognosis. HIV-1 infection causes perturbations to natural killer (NK) cells, and yet these cells can exert immune pressure on the virus and influence clinical outcome. Here, we studied NK cell activation and function in Ugandans with chronic untreated HIV-1 subtype D infection in comparison to uninfected community matched controls.
Methods:
Cryopreserved peripheral blood mononuclear cells (PBMCs) from 42 HIV-infected individuals and 28 HIV-negative controls were analysed using eight-colour flow cytometry. NK cell surface expression of CD16, CD56, CD57, HLA-DR and NKG2A were used to investigate activation, maturation and differentiation status. NK cell function was evaluated by measuring interferon-gamma (IFNγ) production in response to K562 cells, or interleukin (IL)-12 and IL-18.
Results:
CD56dim NK cells from HIV-infected individuals produced less IFNγ in response to IL-12 and IL-18 than did CD56dim NK cells from uninfected controls. Infected individuals had lower levels of CD56dim NK cells coexpressing the differentiation markers NKG2A and CD57 than controls. In addition, their NKG2A+CD57+ CD56dim NK cells displayed elevated activation levels as assessed by HLA-DR expression. Cytokine-induced IFNγ production correlated directly with coexpression of CD57 and NKG2A on CD56dim NK cells.
Conclusion:
HIV-1 subtype D infection is associated with impaired NK cell responsiveness to cytokines, decline of the NKG2A+CD57+ CD56dim NK cell subset, as well as elevated activation in this subset. These alterations within the NK cell compartment may contribute to immunopathogenesis of HIV-1 subtype D infection in Ugandans.
Keywords: AIDS, CD57, HIV-1, immune activation, natural killer cells, NKG2A, subtype D
Introduction
The Ugandan HIV-1 epidemic, with a prevalence of 7.3% [1], is predominantly composed of subtype A, D and recombinant viruses [2,3]. HIV-1 subtype affects the rate of disease progression [4], yet it is not clear why. Subtype D is associated with faster progression to AIDS ([5], reviewed in [6,7]), more profound loss of CD4+ T cells [5,8] and decreased frequency of invariant natural killer T cells [9], as compared with subtype A. Further studies of subtype D infection are warranted to understand subtype-dependent differences in HIV-1 immunopathogenesis.
Natural killer (NK) cells are an integral part of the innate immune response to viruses [10]. HIV-1 infection, however, contributes to a range of changes in NK cell phenotype and function as early as primary infection [11–15]. In addition, HIV-1 can acquire escape mutations under NK cell immune pressure [16], although certain killer immunoglobulin-like receptor (KIR) alleles, primarily expressed by NK cells, are associated with slower progression to AIDS [17]. Furthermore, the KIR repertoire of the NK cell compartment adapts and changes during the response to HIV-1 infection [18,19].
NK cells interact with dendritic cells and T cells to shape the immune response to infection. This cross-talk involves the activation of NK cells via interleukin (IL)-12 and IL-18 to release type II interferon, which in turn matures dendritic cells to adequately prime T-cell responses [20,21]. Disruption of the NK–dendritic cell communication may adversely impact immune control of HIV [22,23]. Cytokine-activated NK cells can migrate to lymph nodes [24], and directly activate T cells [25]. Cell-surface receptors such as NKG2A and CD57 can be used to describe CD56dim NK cell differentiation [26]. In the present study, we investigated NK cell activation, differentiation and function in Ugandans with chronic untreated HIV-1 subtype D-infection.
Materials and methods
Study cohort and samples
Study participants were randomly selected from a prospective community-based cohort to characterize HIV-1 infection in Rakai, Uganda, from 1998 until 2004 [5] prior to the availability of antiretroviral therapy in this setting. Cryopreserved peripheral blood mononuclear cells (PBMCs) from 42 treatment-naive individuals infected with HIV-1 subtype D and 28 community-matched HIV-uninfected controls were selected for study (Table 1). Walter Reed Army Institute of Research, Human Subjects Protection Branch (WRAIR#1428), as well as the Uganda National Council of Science and Technology (HS413) approved this study, and all participants provided written consent for participation and for use of stored samples. Plasma viral load was measured using Amplicor HIV-1 Monitor test version 1.5 (Roche Diagnostics, Indianapolis, Indiana, USA). HIV-1 subtypes were determined using the previously described multiregion hybridization assay (MHAacd,[27]). The FACS MultiTEST IMK Kit (BD Biosciences, San Jose, California, USA) was used to enumerate lymphocyte subsets on a dual-laser FACSCalibur (BD Biosciences).
Table 1.
Descriptive statistics for study population.
| Characteristic | HIV-negative (n = 28) | HIV-1 subtype D (n = 42) |
| Age, median (years, range) | 36 (22–48) | 31 (19–54) |
| Sex, no (percentage) | ||
| Female | 14 (50%) | 28 (67%) |
| Male | 14 (50%) | 14 (33%) |
| CD4+ T-cell absolute count, median (cells/μl, range) | 1013 (527–1659) | 475 (85–1336) |
| Viral load, median (copies/ml, range)a | NA | 54 157 (562–1 484 450) |
NA, not applicable.
aViral load measured by Roche Amplicor Monitor v1.5, limit of detection 400 copies/ml.
Flow cytometry
Whole blood collected in acid citrate dextrose anticoagulant was processed within 8 h and PBMC cryopreserved at –130oC in liquid nitrogen, as previously described [28]. PBMCs were thawed and washed in RPMI medium containing 10% foetal bovine serum, 5% l-glutamine, 1% penicillin/streptomycin and 5% HEPES, and cell concentration and viability determined using Guava Viacount reagent and the Guava Personal Cell Analysis System (EMD Millipore, Billerica, Massachusetts, USA) [28]. Samples were stained in 96-well U-bottom plates at 4oC for 30 min in the dark [12]. Aqua Live-Dead stain (Life Technologies Corporation, Carlsbad, California, USA) was used to discriminate viable cells. Commercially available mAbs included anti-CD3, anti-CD14 and anti-CD19 (all on APC-H7), anti-CD56 PE-Cy7, anti-Ki67 PE and anti-HLA-DR V450 (BD Biosciences). Additional mAbs included anti-NKG2A APC (Beckman Coulter, Inc., Brea, California, USA), anti-CD57 FITC (BioLegend, San Diego, California, USA) and anti-perforin PE (eBioscience, San Diego, California, USA). Ki67 and perforin were stained intracellularly after permeabilization using BD Perm/Wash (BD Biosciences). To assess NK cell function, thawed PBMCs were cultured overnight, in the presence of brefeldin A (BD Biosciences), with 10 ng/ml rIL-12 (R&D systems) as well as 10 ng/ml rIL-18 (MBL), or the MHCnull K562 cell line (at an E:T of 5:1), or media alone [12]. Samples were fixed, washed, permeabilized and stained with anti-IFNγ PE (BD Biosciences). All samples were acquired on an eight-colour BD FACSCanto II (BD Biosciences) and analysed using FlowJo version 9.5.3 (Tree Star, Ashland, Oregon, USA).
Statistical analysis
Graph Pad Prism version 6.0a for Mac OSX was used for statistical analysis (GraphPad Software, La Jolla, California, USA). Differences between groups were analysed using the Mann–Whitney test and associations were determined using Spearman's rank correlation. P values less than 0.05 were considered statistically significant.
Results
Impaired CD56dim natural killer cell response to interleukin-12/18 stimulation in HIV-1 subtype D infection
NK cell phenotype and function were studied from the PBMCs of 42 untreated HIV-1 subtype D-infected individuals and 28 community-matched uninfected controls (Table 1). Individuals had a median viral load of 54 147 copies/ml and median CD4+ absolute cell count of 475 cells/μl. NK cells were distinguished on the basis of live, CD3−CD14−CD19− and CD56+ gating (Fig. 1a). After stimulation with IL-12 and IL-18, CD56dim NK cells in HIV-infected individuals produced less IFNγ than those from uninfected controls (P = 0.026, Fig. 1b). The overall trend towards lower absolute counts of CD56dim NK cells (P = 0.07, data not shown) in HIV infection enhanced this pattern in absolute count terms with diminished counts of IFNγ+ CD56dim NK cells in infected individuals (P < 0.001, Fig. 1c). In contrast, IFNγ production in CD56dim NK cells in HIV-infected individuals was unimpaired after K562 stimulation (P = 0.5, Fig. 1d), although the overall trend towards lower absolute counts of CD56dim NK cells led to a lower representation of cells responding to K562 measured as an absolute count (P = 0.036, Fig. 1e). Expression of Ki67 and the cytolytic protein perforin were similar regardless of infection status (data not shown).
Fig. 1.
Characteristics of natural killer cells in HIV-1 subtype D infection.
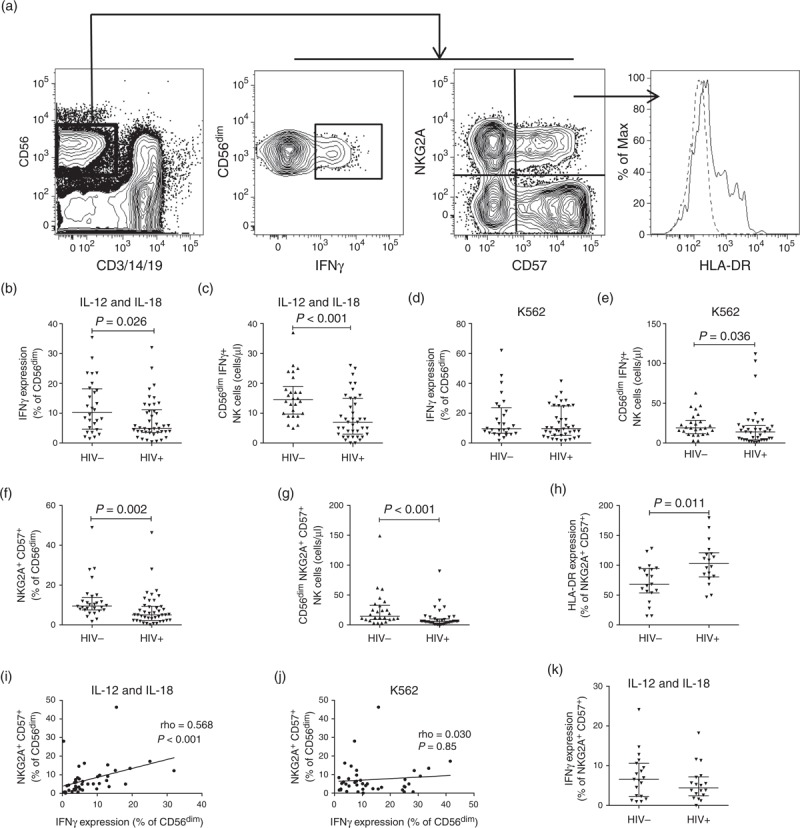
Contour plots, with outliers, illustrating the gating strategy used to identify CD56dim NK cells expressing IFNγ, NKG2A, CD57 and HLA-DR (a). The dashed histogram is the FMO and the continuous line shows HLA-DR expression. Scatter dot plots comparing the median frequencies and interquartile ranges of CD56dim NK cells expressing IFNγ in HIV-infection when stimulated with IL-12 and IL-18, expressed as a percentage (b) or an absolute count (c). Percentage (d) and absolute count (e) responses after stimulation with K562. Levels of NKG2A+CD57+ cells as a percentage (f) and absolute count (g). Activation levels in NKG2A+CD57+ CD56dim NK cells (h). Frequency of the NKG2A+CD57+ CD56dim subset correlated with the frequency of CD56dim NK cells expressing IFNγ when stimulated with IL-12 and IL-18 (i), or K562 (j). IFNγ expression in NKG2A+CD57+CD56dim NK cells after IL-12 and IL-18 stimulation (k).
The NKG2A+CD57+CD56dim natural killer cell subset is activated and declines in subtype D infection
To characterize defects in NK cells in more detail, we studied the phenotype of the CD56dim NK cells using the activation marker HLA-DR, inhibitory receptor NKG2A and the terminal differentiation marker CD57. Individuals had a lower representation of NKG2A+CD57+ CD56dim NK cells than the healthy controls measured both as a percentage of CD56dim NK cells (P = 0.002, Fig. 1f) and as an absolute count of this subset of CD56dim NK cells (P < 0.001, Fig. 1g). The proportions of cells defined by other combinations of these markers (NKG2A+CD57−, NKG2A−CD57+ and NKG2A−CD57−) did not change significantly between HIV-1-infected and uninfected individuals, suggesting that the relative decrease in NKG2A+CD57+ cells was not directly related to an increase in one of the other subsets (data not shown). Furthermore, the NKG2A+CD57+CD56dim NK cell subset was more activated, as determined by HLA-DR expression, in a subset of 18 of the individuals when compared with 19 controls (P = 0.011; Fig. 1h).
NKG2A+CD57+CD56dim natural killer cell frequency correlates with CD56dim interferon-gamma production
We next investigated whether the decline in NKG2A+CD57+ NK cells was associated with the functional impairment of CD56dim NK cells in HIV-1 subtype D-infected individuals. The frequency of NKG2A+CD57+CD56dim NK cells correlated directly with CD56dim IFNγ production in response to IL-12 as well as IL-18 stimulation (rho = 0.568, P < 0.001; Fig. 1i), but not K562 stimulation (rho = 0.030, P = 0.85; Fig. 1j). This association was not observed in controls (rho = –0.080, P = 0.68; data not shown). Surprisingly, cytokine-induced IFNγ production in NKG2A+CD57+ CD56dim NK cells did not differ between cases and controls (Fig. 1k). This suggests that the functional capacity of NKG2A+CD57+CD56dim NK cells may not be directly affected on a per-cell basis, and that their decline may be associated with the functional capacity of the global CD56dim NK cell compartment. Of note, these phenotypic and functional parameters did not correlate with CD4+ cell count or viral load (data not shown).
Discussion
To better understand the innate immunology of HIV-1 subtype D infection in Rakai, Uganda, we studied NK cell responses to IL-12 as well as IL-18 stimulation. CD56dim NK cells from HIV-positive individuals produced less IFNγ in response to cytokine stimulation, as compared with uninfected controls. This is in line with previous findings from a US cohort, despite the geographic, host and viral disparity between these populations [29]. Expression of the inhibitory receptor NKG2A and the terminal differentiation marker CD57 together identified a subset of CD56dim NK cells that was reduced in HIV-1 subtype D infection. These NKG2A+CD57+ cells displayed elevated activation levels in HIV-positive individuals, measured by HLA-DR, and their frequency correlated directly with the proportion of IFNγ expressing CD56dim NK cells in response to cytokine stimulation.
CD56dim NK cells differentiate from high expression of NKG2A in the absence of KIRs and CD57 expression, to a terminally differentiated NKG2A−KIR+CD57+CD56dim subset, with those expressing CD57 losing their ability to respond to cytokine stimulation [26]. Previous studies have indicated that NKG2A expression correlates with the IFNγ response to IL-12 as well as IL-18 cytokine stimulation [30], and that CD57 expression reflects the cytolytic potential of NK cells in the form of perforin and granzyme expression [31]. NKG2A+CD57+CD56dim cells may be in an intermediate differentiation phase, retaining responses to cytokine stimulation and in transition to a more differentiated phenotype [24]. Our finding that the decline of the NKG2A+CD57+CD56dim NK cell subset is associated with impaired NK cell responses to cytokine stimulation suggests a possible important role for this subset in NK cell homeostasis and function.
Pathogen-associated maturational skewing of NK cells has been observed previously. NK cells reconstituting in patients after umbilical cord blood transplantation mature more rapidly in patients infected with cytomegalovirus [32], a pattern also seen in primary HIV infection [33]. Hong et al.[34] observed a decline of less differentiated NK cells in chronic untreated HIV-1 infection. In addition, Marras et al. [35] recently showed that activation, as measured by HLA-DR, of CD56dim NK cells in HIV-1 viremic controllers is associated with differentiation to an NKG2A−CD57+ effector phenotype. Chronic viral infection has also been associated with a defect in miR-155, which is important for IFNγ production [36], maturation and expansion of IL-12 and IL-18 activated NK cells [37]. In this context, the decline in the activated NKG2A+CD57+CD56dim subset could be a consequence of HIV-driven rapid maturation [34] that is not accompanied by expansion of the effector NK cell subset [32,37]. This may reduce the number of NK cells available to produce IFNγ.
Wright et al.[38] found lower plasma IFNγ concentration in ‘late stage’ subtype D-infected patients than in subtype A-infected Ugandans at a similar disease stage. Others have also observed that IFNγ secretion by NK cells declines during HIV-1 infection [39] and is not restored by antiretroviral therapy, despite recovery of phenotypically mature NK cells [29]. These observations, together with the lack of association of the IFNγ ‘defect’ with CD4+ cell count in the present study, may suggest that even relatively healthy HIV-infected individuals have damaged NK cell compartments. In summary, our findings support the notion that HIV-1 infection drives changes in NK cell maturation, with an altered distribution of maturational subsets, elevated expression of activation markers and lower responsiveness to cytokine stimulus.
Acknowledgements
The authors wish to acknowledge the Rakai Community Cohort study volunteers for their participation. The views expressed are those of the authors and should not be construed to represent the positions of the U.S. Army or the Department of Defense.
P.N., M.A.E and J.K.S. designed the study, designed experiments and analysed data. P.N. and M.A.E. performed experiments. O.L., T.C.Q., D.S., N.K.S., R.H.G., N.L.M., F.W.M. and M.L.R. established and developed the cohort, and provided clinical patient data. All authors contributed to manuscript writing.
Primary support was provided by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense. Data collection was supported, in part, by grants R01 AI34826 and R01 AI34265 from the National Institute of Allergy and Infectious Diseases (NIAID); grant 5P30HD06826 from the National Institute of Child and Health Development; grant 5D43TW00010 from the Fogarty Foundation. Additional support was provided by the Swedish Research Council, the Swedish Cancer Society, the Stockholm County Council, Karolinska Institutet and the Division of Intramural Research, NIAID, NIH.
Conflicts of interest
The authors declare no financial conflict of interest.
References
- 1.Ministry of Health U. 2011 Uganda AIDS indicator survey: key findings. Calverton, MD: MOH and ICF International; 2012 [Google Scholar]
- 2.Kapaata A, Lyagoba F, Ssemwanga D, Magambo B, Nanyonjo M, Levin J, et al. HIV-1 subtype distribution trends and evidence of transmission clusters among incident cases in a rural clinical cohort in southwest Uganda, 2004–2010. AIDS Res Hum Retroviruses 2013; 29:520–527 [DOI] [PubMed] [Google Scholar]
- 3.Lihana RW, Ssemwanga D, Abimiku A, Ndembi N. Update on HIV-1 diversity in Africa: a decade in review. AIDS Rev 2012; 14:83–100 [PubMed] [Google Scholar]
- 4.Amornkul PN, Karita E, Kamali A, Rida WN, Sanders EJ, Lakhi S, et al. Disease progression by infecting HIV-1 subtype in a seroconverter cohort in sub-Saharan Africa. AIDS 2013; 27:2775–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiwanuka N, Laeyendecker O, Robb M, Kigozi G, Arroyo M, McCutchan F, et al. Effect of human immunodeficiency virus Type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, with incident HIV-1 infection. J Infect Dis 2008; 197:707–713 [DOI] [PubMed] [Google Scholar]
- 6.Hemelaar J. The origin and diversity of the HIV-1 pandemic. Trends Mol Med 2012; 18:182–192 [DOI] [PubMed] [Google Scholar]
- 7.Pant Pai N, Shivkumar S, Cajas JM. Does genetic diversity of HIV-1 non-B subtypes differentially impact disease progression in treatment-naive HIV-1-infected individuals? A systematic review of evidence: 1996-2010. J Acquir Immune Defic Syndr 2012; 59:382–388 [DOI] [PubMed] [Google Scholar]
- 8.Kiwanuka N, Robb M, Laeyendecker O, Kigozi G, Wabwire-Mangen F, Makumbi FE, et al. HIV-1 viral subtype differences in the rate of CD4+ T-cell decline among HIV seroincident antiretroviral naive persons in Rakai district, Uganda. J Acquir Immune Defic Syndr 2010; 54:180–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flach B, Naluyima P, Blom K, Gonzalez VD, Eller LA, Laeyendecker O, et al. Differential loss of invariant natural killer T cells and FoxP3(+) regulatory T cells in HIV-1 subtype A and subtype D infections. J Acquir Immune Defic Syndr 2013; 63:289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vidal SM, Khakoo SI, Biron CA. Natural killer cell responses during viral infections: flexibility and conditioning of innate immunity by experience. Curr Opin Virol 2011; 1:497–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lichtfuss GF, Cheng WJ, Farsakoglu Y, Paukovics G, Rajasuriar R, Velayudham P, et al. Virologically suppressed HIV patients show activation of NK cells and persistent innate immune activation. J Immunol 2012; 189:1491–1499 [DOI] [PubMed] [Google Scholar]
- 12.Eller MA, Eller LA, Ouma BJ, Thelian D, Gonzalez VD, Guwatudde D, et al. Elevated natural killer cell activity despite altered functional and phenotypic profile in Ugandans with HIV-1 clade A or clade D infection. J Acquir Immune Defic Syndr 2009; 51:380–389 [DOI] [PubMed] [Google Scholar]
- 13.Iannello A, Debbeche O, Samarani S, Ahmad A. Antiviral NK cell responses in HIV infection: II. viral strategies for evasion and lessons for immunotherapy and vaccination. J Leukoc Biol 2008; 84:27–49 [DOI] [PubMed] [Google Scholar]
- 14.Bjorkstrom NK, Ljunggren HG, Sandberg JK. CD56 negative NK cells: origin, function, and role in chronic viral disease. Trends Immunol 2010; 31:401–406 [DOI] [PubMed] [Google Scholar]
- 15.Fauci AS, Mavilio D, Kottilil S. NK cells in HIV infection: paradigm for protection or targets for ambush. Nat Rev Immunol 2005; 5:835–843 [DOI] [PubMed] [Google Scholar]
- 16.Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, et al. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature 2011; 476:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 2007; 39:733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eller MA, Koehler RN, Kijak GH, Eller LA, Guwatudde D, Marovich MA, et al. Human immunodeficiency virus type 1 infection is associated with increased NK cell polyfunctionality and higher levels of KIR3DL1+ NK cells in ugandans carrying the HLA-B Bw4 motif. J Virol 2011; 85:4802–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alter G, Rihn S, Walter K, Nolting A, Martin M, Rosenberg ES, et al. HLA class I subtype-dependent expansion of KIR3DS1+ and KIR3DL1+ NK cells during acute human immunodeficiency virus type 1 infection. J Virol 2009; 83:6798–6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong JL, Berk E, Edwards RP, Kalinski P. IL-18-primed helper NK cells collaborate with dendritic cells to promote recruitment of effector CD8+ T cells to the tumor microenvironment. Cancer Res 2013; 73:4653–4662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long EO, Sik Kim H, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol 2013; 31:227–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mavilio D, Lombardo G, Kinter A, Fogli M, La Sala A, Ortolano S, et al. Characterization of the defective interaction between a subset of natural killer cells and dendritic cells in HIV-1 infection. J Exp Med 2006; 203:2339–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conry SJ, Milkovich KA, Yonkers NL, Rodriguez B, Bernstein HB, Asaad R, et al. Impaired plasmacytoid dendritic cell (PDC)-NK cell activity in viremic human immunodeficiency virus infection attributable to impairments in both PDC and NK cell function. J Virol 2009; 83:11175–11187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juelke K, Killig M, Luetke-Eversloh M, Parente E, Gruen J, Morandi B, et al. CD62L expression identifies a unique subset of polyfunctional CD56dim NK cells. Blood 2010; 116:1299–1307 [DOI] [PubMed] [Google Scholar]
- 25.Zingoni A, Sornasse T, Cocks BG, Tanaka Y, Santoni A, Lanier LL. Cross-talk between activated human NK cells and CD4+ T cells via OX40-OX40 ligand interactions. J Immunol 2004; 173:3716–3724 [DOI] [PubMed] [Google Scholar]
- 26.Bjorkstrom NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood 2010; 116:3853–3864 [DOI] [PubMed] [Google Scholar]
- 27.Arroyo MA, Sateren WB, Serwadda D, Gray RH, Wawer MJ, Sewankambo NK, et al. Higher HIV-1 incidence and genetic complexity along main roads in Rakai District, Uganda. J Acquir Immune Defic Syndr 2006; 43:440–445 [DOI] [PubMed] [Google Scholar]
- 28.Olemukan RE, Eller LA, Ouma BJ, Etonu B, Erima S, Naluyima P, et al. Quality monitoring of HIV-1-infected and uninfected peripheral blood mononuclear cell samples in a resource-limited setting. Clin Vaccine Immunol 2010; 17:910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azzoni L, Papasavvas E, Chehimi J, Kostman JR, Mounzer K, Ondercin J, et al. Sustained impairment of IFN-gamma secretion in suppressed HIV-infected patients despite mature NK cell recovery: evidence for a defective reconstitution of innate immunity. J Immunol 2002; 168:5764–5770 [DOI] [PubMed] [Google Scholar]
- 30.Romee R, Schneider SE, Leong JW, Chase JM, Keppel CR, Sullivan RP, et al. Cytokine activation induces human memory-like NK cells. Blood 2012; 120:4751–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chattopadhyay PK, Betts MR, Price DA, Gostick E, Horton H, Roederer M, et al. The cytolytic enzymes granyzme A, granzyme B, and perforin: expression patterns, cell distribution, and their relationship to cell maturity and bright CD57 expression. J Leukoc Biol 2009; 85:88–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Della Chiesa M, Falco M, Podesta M, Locatelli F, Moretta L, Frassoni F, et al. Phenotypic and functional heterogeneity of human NK cells developing after umbilical cord blood transplantation: a role for human cytomegalovirus?. Blood 2012; 119:399–410 [DOI] [PubMed] [Google Scholar]
- 33.Naranbhai V, Altfeld M, Karim SS, Ndung’u T, Karim QA, Carr WH. Changes in Natural Killer cell activation and function during primary HIV-1 Infection. PLoS One 2013; 8:e53251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong HS, Eberhard JM, Keudel P, Bollmann BA, Ballmaier M, Bhatnagar N, et al. HIV infection is associated with a preferential decline in less-differentiated CD56dim CD16+ NK cells. J Virol 2010; 84:1183–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marras F, Nicco E, Bozzano F, Di Biagio A, Dentone C, Pontali E, et al. Natural killer cells in HIV controller patients express an activated effector phenotype and do not up-regulate NKp44 on IL-2 stimulation. Proc Natl Acad Sci U S A 2013; 110:11970–11975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trotta R, Chen L, Ciarlariello D, Josyula S, Mao C, Costinean S, et al. miR-155 regulates IFN-gamma production in natural killer cells. Blood 2012; 119:3478–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zawislak CL, Beaulieu AM, Loeb GB, Karo J, Canner D, Bezman NA, et al. Stage-specific regulation of natural killer cell homeostasis and response against viral infection by microRNA-155. Proc Natl Acad Sci U S A 2013; 110:6967–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright E, Mugaba S, Grant P, Parkes-Ratanshi R, Van der Paal L, Grosskurth H, et al. Coreceptor and cytokine concentrations may not explain differences in disease progression observed in HIV-1 clade A and D infected Ugandans. PLoS One 2011; 6:e19902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alter G, Teigen N, Davis BT, Addo MM, Suscovich TJ, Waring MT, et al. Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood 2005; 106:3366–3369 [DOI] [PubMed] [Google Scholar]


