Abstract
The erythroblastic island provides an important nutritional and survival support niche for efficient erythropoietic differentiation. Island integrity is reliant on adhesive interactions between erythroid and macrophage cells. We show that erythroblastic islands can be formed from single progenitor cells present in differentiating embryoid bodies, and that these correspond to erythro-myeloid progenitors (EMPs) that first appear in the yolk sac of the early developing embryo. Erythroid Krüppel-like factor (EKLF; KLF1), a crucial zinc finger transcription factor, is expressed in the EMPs, and plays an extrinsic role in erythroid maturation by being expressed in the supportive macrophage of the erythroblastic island and regulating relevant genes important for island integrity within these cells. Together with its well-established intrinsic contributions to erythropoiesis, EKLF thus plays a coordinating role between two different cell types whose interaction provides the optimal environment to generate a mature red blood cell.
Keywords: EKLF/KLF1, Erythroblastic island, Erythroid-myeloid progenitor, Mouse
INTRODUCTION
Central to the homeostasis of the hematopoietic system is the correct balance of progenitor cell proliferation versus lineage-committed differentiation (Orkin and Zon, 2008). The first site of hematopoiesis occurs in the yolk sac and provides the primitive erythrocytes essential for survival of the early embryo until the definitive wave of hematopoiesis begins (McGrath and Palis, 2008). Definitive erythropoiesis in the fetal liver and postnatal bone marrow consists of morphologically identifiable, nucleated precursors that progress from the proerythroblast to enucleated reticulocytes (Granick and Levere, 1964). These events occur while the erythroblasts are in close physical contact with a macrophage in structures known as erythroblastic islands (Chasis and Mohandas, 2008; Manwani and Bieker, 2008).
Erythroblastic islands were described over 50 years ago as specialized microenvironmental compartments within which mammalian erythroblasts proliferate and efficiently differentiate (Bessis, 1958). Erythroblastic islands were first observed in bone marrow, although they are also present in the splenic red pulp and in the fetal liver (Chasis and Mohandas, 2008; Manwani and Bieker, 2008). These islands consist of a central macrophage that extends cytoplasmic protrusions to a ring of 5-30 surrounding erythroid cells encompassing proerythroblast to reticulocyte stages (Gifford et al., 2006). Extensive macrophage-erythroblast and erythroblast-erythroblast adhesive interactions are necessary for a thriving definitive erythropoietic community. As a result, structural proteins that mediate these interactions (Fabriek et al., 2007; Lee et al., 2006; Liu et al., 2007; Mankelow et al., 2004; Sadahira et al., 1995; Soni et al., 2006) as well as transcriptional factors (Gutiérrez et al., 2004; Iavarone et al., 2004; Kusakabe et al., 2011) are both crucial for promoting an effective differentiative environment. Erythroblasts can proliferate, mature and enucleate in vitro in the absence of other cell types; however, this process is typically very inefficient at all stages (Hanspal et al., 1998; Rhodes et al., 2008). Macrophages provide not only nutrients, but also proliferative and survival signals to the erythroblasts. Macrophages phagocytose extruded erythroblast nuclei at the conclusion of erythroid maturation (Chasis and Mohandas, 2008; de Back et al., 2014; Manwani and Bieker, 2008), and deoxyribonuclease II alpha (DNASE2A) is required for this process (Kawane et al., 2001; Yoshida et al., 2005).
Chemical ablation of splenic macrophage dramatically impairs erythropoiesis (Sadahira et al., 2000), showing the in vivo importance of macrophage in erythroid biology. Powerful additional validation comes from two recent studies (Chow et al., 2013; Ramos et al., 2013) demonstrating that macrophage play a crucial role in stress erythropoiesis, not only after anemia, when efficient erythroid expansion and enucleation are required, but also under pathological conditions, where they provide a supportive niche for the proliferation of altered erythroid cells, such as is observed in polycythemia vera. These studies also corroborate the role of macrophages in supplying iron to assure effective erythropoiesis.
Despite knowledge of their existence for decades, several gaps remain in our understanding of the molecular controls that are important for coordinating the onset and decline of erythroid-macrophage interactions and how they interface with enucleation events that lead to a mature reticulocyte. This becomes particularly important in the context of aberrant repression or expansion of these final maturation steps under disease conditions (Koury, 2014).
EKLF (erythroid Krüppel-like factor; KLF1) is a red cell-enriched, zinc finger DNA binding protein that interacts with its cognate 5′-CCMCRCCCN-3′ element at target promoters and enhancers (Miller and Bieker, 1993). Its roles in mouse and human β-like globin gene regulation during terminal erythroid differentiation have been well established using genetic, biochemical and molecular approaches (Bauer and Orkin, 2011; Siatecka and Bieker, 2011; Tallack and Perkins, 2010, 2013; Yien and Bieker, 2013). EKLF is also highly expressed in the megakaryocyte/erythroid progenitor, where it may play a determining role in the bipotential decisions that lead to preferential establishment of erythroid progeny (reviewed by Dore and Crispino, 2011; Siatecka and Bieker, 2011).
EKLF's activation target repertoire has expanded beyond the classical β-globin gene to include protein-stabilizing, heme biosynthetic pathway, red cell membrane protein, cell cycle and transcription factor genes in both primitive and definitive erythroid cells (Siatecka and Bieker, 2011; Tallack and Perkins, 2010; Yien and Bieker, 2013). Relatedly, links have been established between mutant or haploinsufficient levels of EKLF and altered human hematology and anemia (Borg et al., 2011; Helias et al., 2013; Siatecka and Bieker, 2011; Singleton et al., 2012).
Comparative analysis of expression arrays between EKLF wild-type and EKLF-null fetal liver cells show that a number of genes involved in execution of the terminal erythroid differentiation program are downregulated in the absence of EKLF (Drissen et al., 2005; Hodge et al., 2006; Pilon et al., 2006, 2011; Tallack et al., 2012, 2010). Our studies, originating from observations of differentiating erythroid cells from embryoid bodies, have uncovered an erythroid-intrinsic role for EKLF in erythroblastic island biology. However, they also quickly converged on an unanticipated molecular regulatory role for EKLF within the macrophage of the island, revealing an important extrinsic role in erythroid/macrophage biology.
RESULTS
Erythroblastic islands from differentiating embryonic stem cells
We made some unanticipated observations during our analyses of embryoid body (EB) differentiation from murine embryonic stem cells (ESCs) (Frontelo et al., 2007; Manwani et al., 2007). Isolated ESCs were differentiated for 4 days to enrich for the presence of primitive erythroid progenitors. The resultant EBs were then dispersed and plated at low density in methylcellulose under conditions optimal for primitive erythroid colony formation (Kennedy and Keller, 2003). Individual erythroid colonies were picked, cytospun onto glass slides and viewed after staining with May-Grünwald Giemsa. Strikingly, all cytospin preparations showed maturing erythroblasts arranged in a ring around a central macrophage (Fig. 1A), a markedly close resemblance to the classic erythroblastic island morphology (e.g. Hanspal et al., 1998). These surprising results suggest that an erythroblastic island can be formed from a primitive erythroid source. These structures will be referred to as embryoid body erythroblastic islands (EBEIs).
Fig. 1.
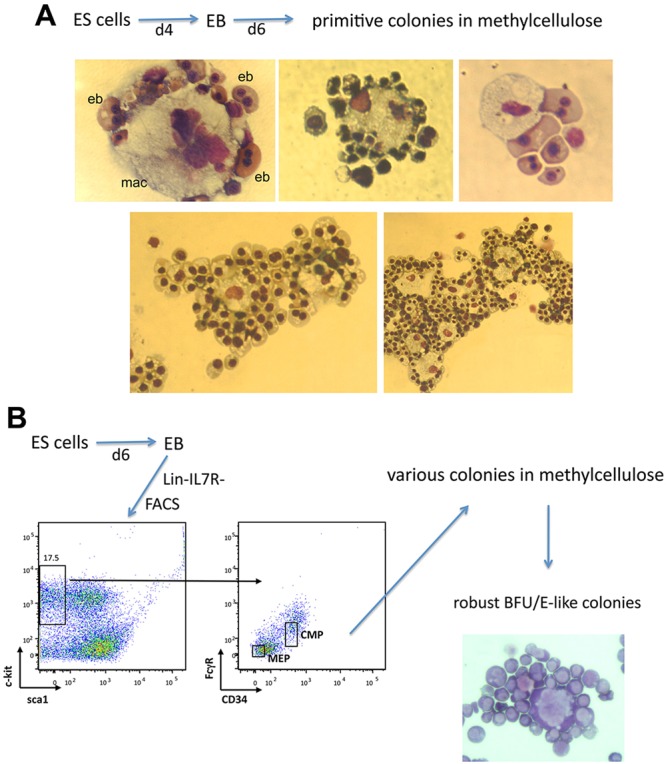
Typical morphology of erythroid colonies derived from differentiating ESCs. Individual methylcellulose erythroid colonies from (A) secondary platings of dispersed EBs or from (B) FACS-sorted dispersed EBs (enriched for MEP or CMP populations as shown) were picked and cytospun onto a glass slide. The slides were air dried and stained with May-Grünwald Giemsa. Note that not all contain a single island (A, bottom); some EB-derived erythroid colonies contain two (left) or more (right) central macrophage. eb, erythoblast; mac, macrophage.
To address whether we could also observe the same results from a definitive erythroid source, we developed a protocol to prospectively sort dispersed cells from differentiating EBs at day 6 (and thus enriched for definitive erythroid cell production) into lin-Kit+sca1-FcRloCD34+ [presumptive common myeloid progenitor (CMP)] and lin-Kit+sca1-FcRloCD34− [presumptive megakaryocyte/erythrocyte progenitor (MEP)] fractions (based on Akashi et al., 2000; Nakorn et al., 2003; Terszowski et al., 2005), and plated these individually sorted cells at low density onto methylcellulose under conditions optimal for definitive colony analysis. It has been difficult to obtain colonies from such prospectively sorted populations when derived from differentiating EBs (Drissen et al., 2010); indeed, we found that use of an ESC-based medium rather than the standard media conditions was absolutely crucial for success (see Materials and Methods). Cytospins of the resulting robust, red colonies again revealed erythroblasts surrounding a central macrophage (Fig. 1B).
These initial observations imply that EBEIs can be formed from single cells; such an idea has not been examined in prior studies of erythroblastic island formation. As a result, to further substantiate the clonal nature of our observations we isolated single Hoechst-/Kit+ cells from EBs at day 6 and plated these in methylcellulose at various concentrations (Fig. 2). The results show a strong linear relationship between cells plated and the resultant colonies (∼9%) that crosses through zero, supporting the idea that the colonies are clonal.
Fig. 2.
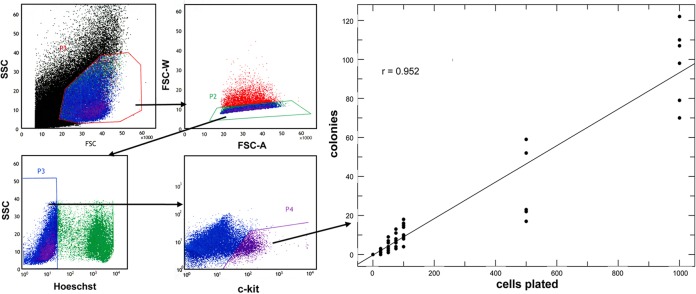
Cell dose response of erythroid colonies derived from single cells. (Left) Day 6 EBs were dispersed, gated for singlets, sorted for viability (Hoechst-) and Kit+ expression before plating in methylcellulose at varying concentrations. (Right) Results from inspection of 60 platings derived from five separate experiments are shown as a regression analysis between cells plated and colony number, leading to a correlation coefficient (r) of 0.952 (P<0.001) and a slope of 0.094.
A number of cell adhesion molecules and their interactions within the erythroblastic islands are crucial for island integrity in definitive populations derived from fetal livers and bone marrow (Chasis and Mohandas, 2008; Manwani and Bieker, 2008). Integrin/actin cytoskeleton interactions may coordinate adhesion and gene expression in the erythroblastic islands by regulating intracellular signaling. We wished to address whether such a mechanism was operant in our primitive EBEI colonies. The erythroid-specific isoform of intercellular adhesion molecule 4 (ICAM4) is expressed on erythroid cells and interacts with αv integrin on macrophage cells (Spring et al., 2001). Blocking ICAM4/αv binding with αv synthetic peptides produces a 70% decrease in islands reconstituted in vitro (Lee et al., 2006), and island formation is predominantly defective in ICAM4-null mice (Lee et al., 2006). As a result, we tested the effects on primitive EBEI formation of a synthetic peptide that has previously been used to block ICAM4/αV adhesion and definitive erythroblastic island formation in reconstitution assays (Lee et al., 2006; Mankelow et al., 2004). One synthetic peptide SVPFWVRMS (FWV) and scrambled peptide (control) were used in increasing concentrations (0.5-2.0 mM) in EBEI assays. The FWV peptide, but not the control, caused a marked, concentration-dependent decrease in the number of colonies per dish (Fig. 3, left). The colonies that did form were smaller and paler (supplementary material Fig. S1). Examination of cytospins from the affected colonies showed complete absence of island-type structures and a reduction in erythroblasts (Fig. 3, right; note that macrophages are still apparent). These data demonstrate that erythroblast ICAM4 binding to macrophage αV integrins is important for proper colony formation even when derived from a primitive erythroid cell.
Fig. 3.
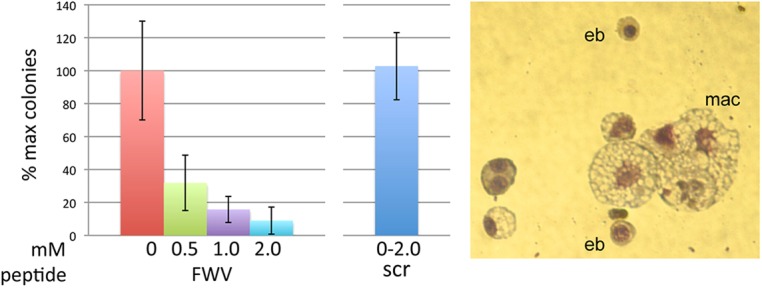
Effect of peptide inhibitor of ICAM4/αV interactions on EBEI formation. (Left) Increasing concentrations of FWV or control peptides were included during the secondary plating for erythroid colony formation, and colonies were counted at day 6. (Right) Colonies were picked from plates treated with 0.5 mM FWV peptide and processed as in Fig. 1 (representative colony is shown; magnification is different from Fig. 1 to show the dispersed nature of the colony).
Collectively, these data strongly suggest that contact between the macrophages and developing erythroblasts is necessary for efficient execution of the primitive and definitive erythroid programs in differentiating EBs. The fact that these arise in a clonal fashion implies that there exists a cell that contains its own intrinsic island-forming capacity.
EKLF regulation of relevant targets in erythroblastic islands
Array and RNA-seq data demonstrate that ICAM4 levels are decreased in EKLF-null primitive and definitive erythroid cells (Hodge et al., 2006; Isern et al., 2010; Pilon et al., 2008; Tallack et al., 2012) and in a congenital dyserythropoietic anemia (CDA) patient harboring the EKLF-E325K mutation that exhibits a defect in red cell enucleation (Arnaud et al., 2010). ICAM4 levels also mirror EKLF levels during primitive and definitive erythroid differentiation (Fig. 4A) (Kingsley et al., 2013). We directly verified the array results by comparative qRT-PCR analysis of RNA from embryonic day 13.5 (E13.5) wild-type or EKLF-het or -null fetal liver definitive cells and found that ICAM4 levels are dramatically reduced in the absence of EKLF (Fig. 4B). Although other molecules, such as EMP (MAEA) or α4β1 integrins (ITGA4/ITGB1) could also be contributory in the context of erythroblastic island formation (Chasis and Mohandas, 2008; Manwani and Bieker, 2008), these are not affected by changes in EKLF levels (Pilon et al., 2011, 2008; Tallack et al., 2012).
Fig. 4.
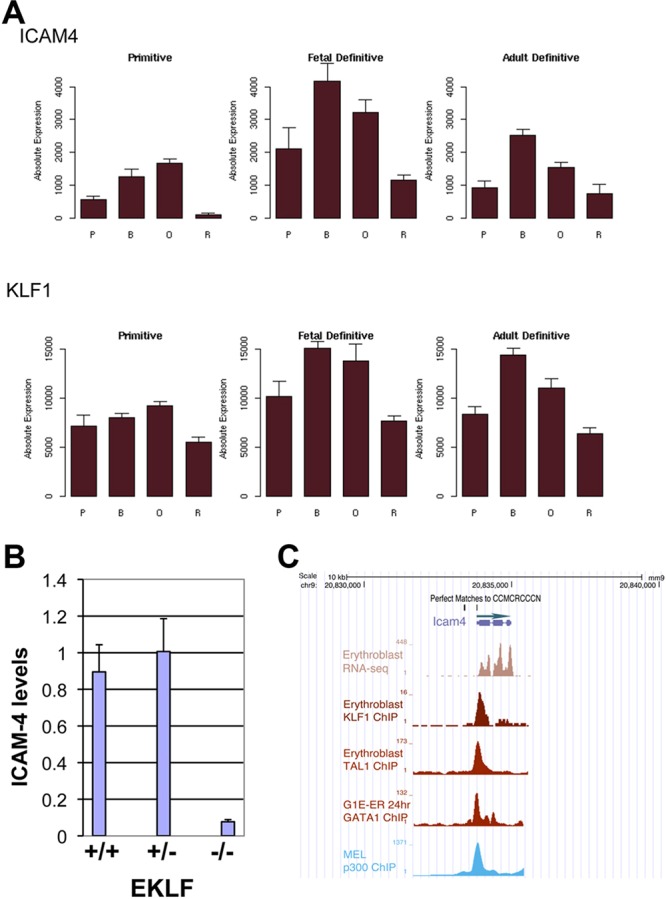
Icam4 is directly regulated by EKLF. (A) Data from the mouse Erythron database (Kingsley et al., 2013) showing changes in ICAM4 and EKLF (KLF1) RNA levels during primitive and definitive (fetal liver or bone marrow) erythroid differentiation. (B) qRT-PCR analysis of ICAM4 levels in E13.5 fetal liver cells. Comparison was made between wild type (+/+), EKLF-het (+/−), or EKLF-null (−/−) as indicated. Normalization is to GAPDH. (C) Genome browser data (http://main.genome-browser.bx.psu.edu/) indicating ICAM4 transcript levels in primary erythroblasts, and overlap between KLF1, TAL1, GATA1 and P300 ChIP data at the 5′ Icam4 promoter. Also indicated are the locations of matches to the KLF1 consensus target site 5′-CCMCRCCCN-3′ (Siatecka et al., 2010; Tallack et al., 2010). B, basophilic erythroblast; O, orthochromatic erythroblast; P, proerythroblast; R, reticulocyte.
Perusal of EKLF chromatin immunoprecipitation sequencing (ChIP-seq) data (Pilon et al., 2011) shows EKLF binding to a region upstream of the Icam4 gene (Fig. 4C). This overlaps with GATA1, TAL1 and p300 binding, consistent with the ‘core’ transcription factor observations that have been noted at a subset of EKLF binding sites (Li et al., 2013; Su et al., 2013; Tallack et al., 2012; Wontakal et al., 2012). Inspection of the EKLF DNA binding cognate sequence shows that it falls into the ‘class I’ site category within 5′-CCMCRCCCN, and thus should bind the neonatal anemia (Nan) EKLF variant based on our analysis of Nan-EKLF biochemical properties (Siatecka and Bieker, 2011; Siatecka et al., 2010). Consistent with this, ICAM4 RNA levels are not affected in Nan mouse fetal livers (data not shown).
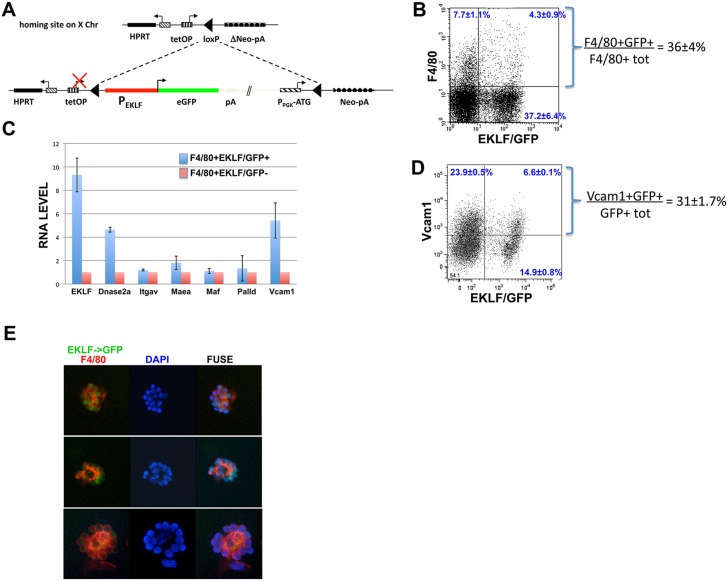
As the colony preparations that gave rise to EBEI were clonal (being from dispersed single cells seeded in methylcellulose), we considered the possibility that EKLF might be expressed in both the erythroid cell and macrophage. There have been hints of EKLF expression in macrophage (Luo et al., 2004), although our analysis of adult bone marrow hematopoietic material had shown no evidence (Frontelo et al., 2007). To address this we used the mouse strain derived from ESCs that contain a single copy of the EKLF promoter directly upstream of a GFP reporter, integrated into the HPRT locus (Fig. 5A) (Lohmann and Bieker, 2008). This EKLF promoter region contains erythroid cell-specific DNase hypersensitive sites and is sufficient to confer tissue-specific expression on a linked reporter in transgenic mice (Chen et al., 1998; Xue et al., 2004; Yien and Bieker, 2013). Our published studies have shown the high cell specificity of GFP expression that results from this pEKLF/GFP construct and, importantly, that GFP onset mirrors EKLF onset (Lohmann and Bieker, 2008). We isolated E13.5 fetal liver cells from pEKLF/GFP mice and monitored, by fluorescence-activated cell sorting (FACS), for overlap of GFP (i.e. EKLF expression) and the F4/80 macrophage marker. Our results show that ∼36% of F4/80+ macrophage cells express EKLF (Fig. 5B). Two analyses were performed to exclude the possibility that the macrophage GFP signal came from adherent erythroid cells or from engulfed erythroid nuclei. First, we included an additional selection for non-clumped cells with single nuclei and obtained similar results (supplementary material Fig. S2A). Second, we cultured the fetal liver cells in the absence of erythropoietin to minimize erythroid expansion, and found that 34-55% of the EKLF/GFP+ population was also F4/80+/Ter119− (supplementary material Fig. S2B).
Fig. 5.
Analysis of the pEKLF/GFP mouse. (A) Schematic of the modified expression system (based on Kyba et al., 2002) after its unidirectional, single-copy integration into the single endogenous modified HPRT locus. The EKLF promoter (950 bp; red) is directly upstream of the GFP reporter (green) (Lohmann and Bieker, 2008). (B) FACS analysis of EKLF expression in macrophage. E13.5 fetal livers (n=8) from pEKLF/GFP mice (Lohmann and Bieker, 2008) were sorted for presence of macrophage (F4/80 marker) and EKLF-expressing cells (GFP marker). The percentage of double-positive F4/80 and GFP cells, compared to total F4/80+ cells, is indicated. (C) qRT-PCR analysis of macrophage genes important for erythroblastic island integrity. F4/80+ macrophage cells were FACS-sorted into EKLF/GFP+ or EKLF/GFP− singlet populations (as in supplementary material Fig. S3). RNA was isolated and used to analyze for the presence of the indicated transcripts. All values are normalized to GAPDH, and these values were then normalized to expression in F4/80+EKLF/GFP− cells (=‘1’). (D) FACS analysis of VCAM1 expression in EKLF-expressing cells (GFP marker) in E13.5 fetal livers from pEKLF/GFP mice (Lohmann and Bieker, 2008). The percentage of double-positive VCAM1 and GFP cells, compared to total GFP+ cells, is indicated. (E) Erythroblastic islands from E13.5 fetal livers were probed for F4/80 (red) and monitored for endogenous EKLF/GFP expression (green). Three typical islands are shown. DAPI (blue) was used as a nuclear marker.
We next investigated whether expression levels of macrophage genes that express proteins important for island integrity are dependent on EKLF. We compared RNA expression levels of αv integrin (Lee et al., 2006; Mankelow et al., 2004), VCAM1 (Sadahira et al., 1995), EMP (MAEA) (Soni et al., 2006), palladin (Liu et al., 2007), MAF (Kusakabe et al., 2011) and DNASE2A (Kawane et al., 2001) between EKLF/GFP+F4/80+ and EKLF/GFP-F4/80+ sorted singlet cells (supplementary material Fig. S3). Other than EMP/MAEA, none are appreciably expressed in the erythroid cell. The controls confirm that EKLF is preferentially expressed in the EKLF/GFP+ population (Fig. 5C). The test samples show that there is no difference in αv integrin, EMP/MAEA, MAF or palladin expression in the two cell populations; on the other hand, DNASE2A [recently shown to be an EKLF target in macrophages (Porcu et al., 2011)] and VCAM1 are more highly expressed in EKLF/GFP+ cells (Fig. 5C). In support of this observation, surface VCAM1 protein is expressed in ∼30% of the EKLF/GFP+ cells (Fig. 5D), and VCAM1 levels are decreased in total fetal liver cell RNA from EKLF-null compared with wild-type embryos (Tallack et al., 2012). These results demonstrate that although αv integrin (the macrophage partner of erythroid ICAM4) is not differentially regulated by the presence of EKLF, a different adhesion molecule (VCAM1, the macrophage partner of erythroid α4β1 integrin) is expressed at a higher level in EKLF/GFP+F4/80+ cells, suggesting that EKLF regulates two erythroid-macrophage adhesive interactions together.
Role for EKLF in erythroblastic island integrity
E13.5 fetal livers from the pEKLF/GFP mouse were then used to enrich for erythroblastic island clusters, which were stained for F4/80 and monitored for GFP. These show a typical island structure, with a central F4/80+ macrophage surrounded by 10-15 GFP+ cells (Fig. 5E). These results demonstrate that EKLF+ cells are associated with the island macrophages when isolated directly from fetal livers, and support the previous observations derived from differentiating ESCs.
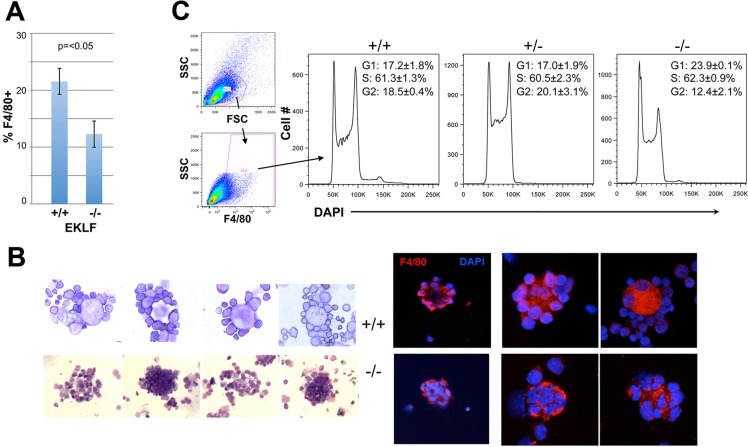
If ICAM4 and VCAM1 are downstream targets of EKLF, then a prediction is that island integrity should be compromised in the absence of EKLF. For this study we compared cells in wild-type and EKLF-null fetal livers (E13.5). FACS analyses show that the percentage (prevalence) of F4/80+ cells is ∼2-fold lower in the EKLF-null fetal liver (Fig. 6A). As it is already known that EKLF is required for erythroid production in vivo, this shows that F4/80 macrophage production is also compromised in the absence of EKLF.
Fig. 6.
Erythroblastic island integrity is dependent on EKLF. Tests used cells derived from wild-type (+/+), EKLF-het (+/−) or EKLF-null (−/−) E13.5 fetal livers as indicated. (A) F4/80+ macrophage cellularity was monitored by FACS and displayed as percentage of total fetal liver cells. (B) Visual analysis of isolated erythroblastic islands (+/+ top row; −/− bottom row). Left, May-Grünwald Giemsa stain of four typical islands from each. Right, macrophage F4/80 stain (red) and DAPI DNA stain (blue) from three typical islands from each (two different magnifications are shown). (C) Cell cycle analysis of dispersed cells is shown.
To address whether definitive erythroblastic islands are affected in the absence of EKLF, we required a globin-independent readout for the red cell component, as adult β-globin levels are virtually nonexistent. This is further complicated by the fact that EKLF is a global regulator of erythroid gene expression, so many standard cell surface markers (e.g. Ter119, CD44) cannot be used. In this context, although CD9 is incompletely affected (∼50%) by the absence of EKLF (Isern et al., 2010), we found that too many (30%) F4/80 macrophage cells also express CD9 (data not shown). As a result we simply stained island clusters from wild-type and EKLF-null E13.5 fetal livers and examined their morphology after May-Grünwald Giemsa stain and after F4/80-DAPI co-stain. We find that clusters from EKLF-null cells are aberrant and consist of macrophages with larger than normal numbers of ingested nuclei (Fig. 6B, left), some of which are grossly misshapen (Fig. 6B, right). These results are reminiscent of those seen with DNASE2A-null fetal liver cells (Kawane et al., 2001). Consistent with observations in EKLF-null erythroid cells (Pilon et al., 2008), analysis of DNA content shows that the cell cycle is dysregulated in EKLF-null F4/80+ cells, as these accumulate at G1 (Fig. 6C); at the same time, there is no evidence for apoptosis (no sub-G1 cells).
Collectively, these studies suggest that in combination with defective red cell interactions following the drop in erythroid ICAM4 and macrophage VCAM1 expression, loss of EKLF also leads to a deficiency in digestion of internalized erythroid nuclei due to the low level of EKLF-dependent DNASE2A in macrophages. The end result is flawed production of mature, enucleated red blood cells from the erythroblastic island, an effect superimposed on the already profound β-thalassemia and global expression defects observed in EKLF-null red cells.
EKLF onset during erythroid-myeloid generation in early development
Waves of hematopoietic progenitors emerge from the developing mouse yolk sac (Bertrand et al., 2005; McGrath et al., 2011; Palis, 2014; Palis et al., 2001, 1999). One arises early (beginning at E7.25) and produces primitive erythroid cells (EryP-CFC). Another arises later (beginning at E8.25) and yields definitive erythroid as well as a varied set of myeloid progeny from a cell termed the erythro-myeloid progenitor (EMP). These transient waves precede the emergence of long-term hematopoietic stem cells during mammalian development from other sites of the embryo (Baron, 2013; Chen et al., 2011; Dzierzak and Speck, 2008; McGrath and Palis, 2005). Our observations on the clonal emergence of erythroid-macrophage islands that arise during primitive or definitive hematopoietic stages of ESC differentiation, the co-expression of EKLF in erythroid and an F4/80 subset of macrophage cells during definitive hematopoiesis, and the availability of the marked GFP mice prompted us to determine the specific cell-surface phenotype of EKLF/GFP+ cells. Previous studies have shown that EKLF is expressed by the neural plate stage (∼E7.5) within the blood islands of the yolk sac (Southwood et al., 1996), and that the transgenic pEKLF construct used here recapitulates this early expression (Xue et al., 2004). However, these studies were focused on the primitive erythroid onset and did not encompass the possibility of a role in EMP onset in the yolk sac.
In the E8.0-E10.0 embryonic yolk sac, the sequential pattern of CD45, Kit and F4/80 expression can be used to establish a temporal developmental sequence (Bertrand et al., 2005; Kierdorf et al., 2013). In particular, CD45-/Kit+/F4/80− cells from the early yolk sac are thought to be EMPs, as they uniquely give rise to cells that express macrophage and erythroid markers. As a result, we isolated GFP+ cells from yolk sacs of E8.5 and E10.5 pEKLF/GFP mice and examined their cell surface expression. Analyses of E8.5 yolk sacs reveal that all stages in the developmental sequence are present at high levels within the EKLF/GFP+ cell population (Fig. 7A). Of particular interest is that ∼20% of the EKLF/GFP+ cells are CD45-/Kit+ EMP progenitors and that this population is significantly enriched ∼4-fold compared with EKLF−/GFP− cells. All subsets are much less apparent in the yolk sac by E10.5 (Fig. 7B), when the increased EKLF/GFP cell numbers retain a small but reproducible high-level of CD45+/Kit− expression (∼2%). These data (along with the F4/80 data of Fig. 4B) suggest that EKLF/GFP is expressed in the EMP progenitor and its more mature progeny before E9.0 (Bertrand et al., 2005; Kierdorf et al., 2013) and before generation of F4/80+ macrophages.
Fig. 7.
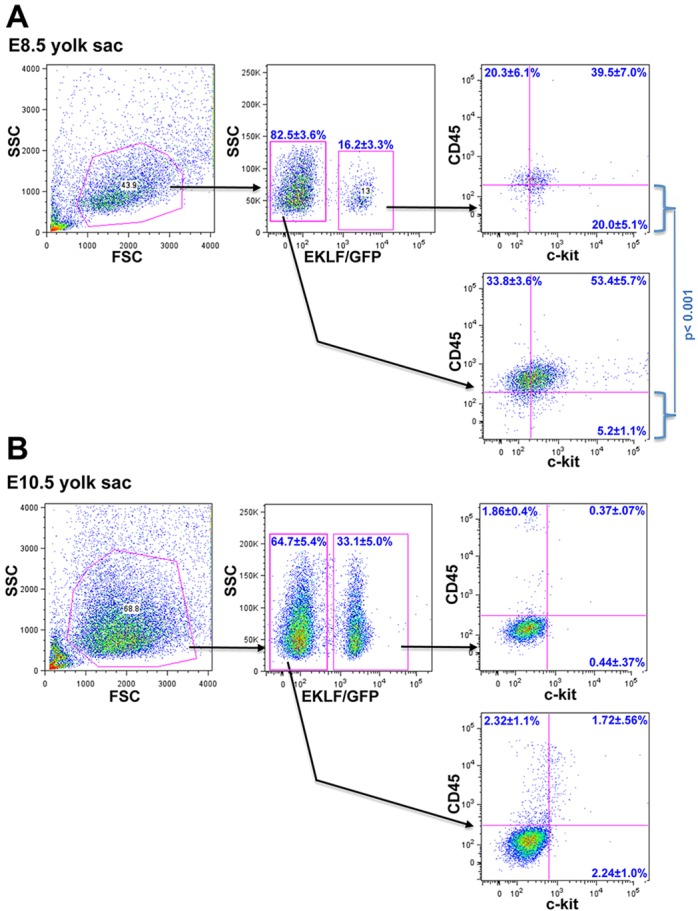
Analysis of EMP and progeny in sorted EKLF/GFP+ or EKLF/GFP− yolk sac cells in early development. Yolk sacs from E8.5 (A; n=5) or E10.5 (B; n=5) embryos were dispersed and analyzed for EKLF/GFP, CD45 and Kit (CD117) expression by FACS. The statistically significant different percentages of CD45–/Kit+ levels between EKLF/GFP+ and EKLF/GFP− cells (i.e. lower right quadrant in each case) are shown for E8.5 (A).
DISCUSSION
Cellular and mechanistic aspects of erythroid/macrophage biology in early development
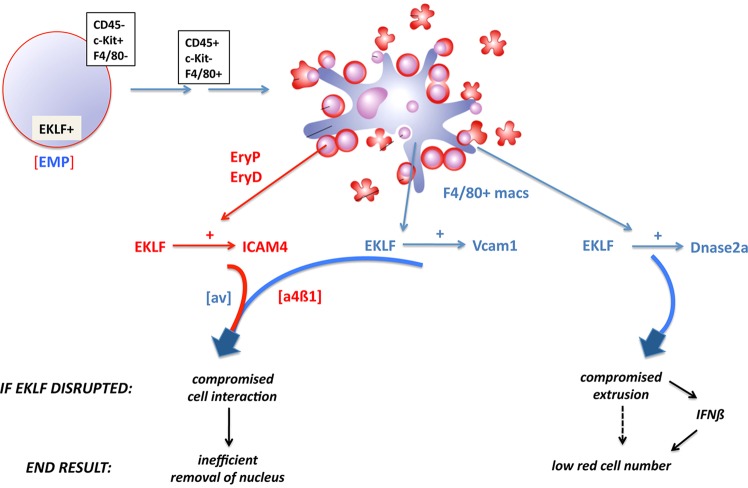
We propose a model that encompasses novel aspects of erythroblastic island biology uncovered by our analyses (summarized in Fig. 8). Surprising is the observation that single cells derived from differentiating EBs can give rise to erythroblastic islands, whether isolated from stages enriched for primitive or definitive erythroid cell production. This progenitor cell is thus able to set up its own island environment. In vivo, these are most likely to arise from the EMP population present at early developmental stages (Fig. 8, top). This EMP population arises in the E8.25 yolk sac then seeds the fetal liver by E11.5, providing the first source of definitive erythroid progenitors prior to or coincident with colonization by distinct hematopoietic stem cell (HSC) sources (Chen et al., 2011; Lux et al., 2008; McGrath et al., 2011; Tober et al., 2013).
Fig. 8.
Model of the role of EKLF in erythroblastic island genetic regulation and integrity. Components (cells and genes) are color-coded: red for erythroid, blue for macrophage. EKLF is expressed in the CD45-/Kit+/F4/80− erythroid-myeloid progenitor (EMP) within the E8.5 yolk sac. This cell sequentially progresses through CD45+/Kit-/F4/80− and CD45+/Kit-/F4/80+ stages (Bertrand et al., 2005; Kierdorf et al., 2013), eventually generating the erythroid and macrophage cells of the erythroblastic island, probably in a clonal manner. EKLF controls expression of cell surface ICAM4 in the erythroid cell (which interacts with macrophage αv integrin), and of cell surface VCAM1 (which interacts with erythroid α4β1 integrin) and nuclear DNASE2A in the macrophage cell. EKLF ablation directly and indirectly leads to compromised cell interactions, ineffective nuclear extrusion, and IFNβ induction, resulting in low red cell numbers that retain their nuclei.
Crucial for integrity of this erythroid/macrophage colony is EKLF. EKLF is expressed at the right time and place in development (the CD45−/Kit+ EMP cell) to exert an effect. Within the island progeny it directly activates ICAM4 in the erythroid compartment, and activates VCAM1 in the macrophage compartment (Fig. 8, bottom left). These enable a two-pronged adhesive intercellular interaction to occur with their respective integrin partners on the opposite cell type. In the absence of EKLF or when the human E325K mutant is expressed, these interactions decrease and contribute to an abundance of nucleated, unprocessed cells in circulation.
In addition, there is a second crucial role for EKLF: activation of DNASE2A in the macrophage (Fig. 8, bottom right). In its absence, macrophages become engorged with undigested erythroid nuclei from any cells that did manage to enter, triggering an inflammatory response such that high levels of IFNβ are released into circulation, leading to anemia and embryonic lethality (Kawane et al., 2001; Nagata, 2007; Yoshida et al., 2005). Circulating erythroid cell numbers in DNASE2A-null embryos are only 1/10th that of wild type (Kawane et al., 2001), and many of these remain nucleated.
The Ristaldi lab has also published data showing that EKLF is expressed in F4/80 macrophage and directly regulates DNASE2A expression (Porcu et al., 2011). Consistent with the downstream effects in DNASE2A-null embryos, IFNβ levels are increased in EKLF-null fetal liver cells (Porcu et al., 2011).
Abnormal numbers of nucleated red cells are observed in EKLF-null embryos (Perkins et al., 1995), and are also a characteristic of human CDA type IV patients who contain a mutation in one allele of KLF1 (Arnaud et al., 2010; Jaffray et al., 2013). This variant is altered within the second zinc finger (E325K), changing its cognate binding specificity and probably converting it to an interfering protein (Singleton et al., 2011). In this context, it is interesting to compare these observations to that of the Nan mouse, which harbors a mutation at the same site but with a different substitution (E339D) (Heruth et al., 2010; Siatecka et al., 2010). These mice, although anemic, do not present with any nucleated circulating erythroid cells, consistent with the idea that its mechanism leading to anemia is different in detail from that of the CDA type IV patients.
EKLF is expressed within CMPs and plays a role in bipotential decisions emanating from MEPs (Frontelo et al., 2007). Our studies further support the concept that EKLF action is important at pre-erythroid stages, in this case within the EMP. During erythropoiesis, EKLF plays a role in all maturation stages; however, our studies now bring in the novel concept that the latter maturational steps are also aided by EKLF regulation of select genes within the macrophage component of the specialized erythroblastic island niche.
Membrane protein dynamics and complexity
Cell surface proteins are dynamically regulated in a temporal fashion during expansion and terminal erythroid differentiation phases, including ICAM4 and β1 integrin, supporting the idea that such changes can directly affect adhesiveness within the island dependent on the maturation stage (An and Mohandas, 2011). The dynamic nature of ICAM4 expression during differentiation parallels that of EKLF, suggesting that changes in EKLF levels exert a finely tuned transcriptional control over these events.
VCAM1 plays a crucial role within the erythroblastic island (Sadahira et al., 1995). In addition, VCAM1 and CD31 are selectively enriched in splenic ‘red pulp’ macrophages compared with other tissue macrophage subsets (e.g. lung, peritoneal, microglia), and F4/80+/VCAM1+ double positivity is coordinately expressed in these cells (Gautier et al., 2012; Kohyama et al., 2009). Red pulp macrophages are closely associated with erythroid cells in the spleen. Our studies suggest that VCAM1 is similarly enriched in fetal liver macrophages that are associated with the erythroblastic island, and is regulated by EKLF.
Definitive red cell interaction with the macrophage is crucial for optimal nutrient access, survival, proliferation and differentiation, but there are additional contributors to effective enucleation (Socolovsky, 2013). Although an abnormal level of circulating nucleated definitive cells are observed in embryos deficient for the DNASE2A, MAF or palladin macrophage proteins (Kawane et al., 2001; Kusakabe et al., 2011; Liu et al., 2007), ablating the majority of macrophage cells does not automatically yield an abundance of nucleated red cells in circulation (Chow et al., 2013; Ramos et al., 2013). In addition, there are a number of structural, enzymatic and chromatin-associated erythroid cell factors that play an intrinsic role in morphologically preparing the cell for proper nuclear extrusion (Ji et al., 2011; Keerthivasan et al., 2011; Konstantinidis et al., 2012; Ney, 2011; Ubukawa et al., 2012; von Lindern, 2006). In the case of EKLF, the abundance of nucleated red cells in circulation seen in its absence or in the CDA KLF1/E325K patients may not only result from defective function in the island macrophage, but also as part of the global erythroid-specific panoply of EKLF targets that include ones important for these final maturation steps.
Regulatory cross-connections
Supplying iron to the maturing erythroid cells is an important in vivo function of island macrophages (Chasis and Mohandas, 2008; Chow et al., 2013; Manwani and Bieker, 2008; Ramos et al., 2013), and can also be demonstrated in co-culture systems [e.g. transport of ferritin (Leimberg et al., 2008)]. In this context, and given the importance of EKLF in controlling a number of steps in the red cell heme biosynthetic pathway (Tallack et al., 2010) (particularly the transferrin receptor and mitoferrin), it is tempting to speculate that EKLF may also play an analogous regulatory role for iron transfer in the macrophage.
EKLF expression is regulated by the BMP4 pathway during early mammalian development, particularly via Smad5 (Adelman et al., 2002; Kang et al., 2012; Lohmann and Bieker, 2008). This same pathway plays a significant role in stress erythropoiesis (Porayette and Paulson, 2008). As macrophage depletion leads to a reduction of BMP4 activity (Chow et al., 2013), part of the explanation for the lower erythroid numbers after stress induction under these conditions is likely to be that EKLF levels are compromised, leading to a global negative effect on red cell expansion and maturation. Conversely, increased EKLF levels lead to precocious erythroid differentiation (Frontelo et al., 2007); thus secreted BMP4 from the adherent macrophages (Millot et al., 2010; Paulson et al., 2011) can use this molecular pathway to support normal or accelerated red cell production. This suggests that EKLF expression levels in the red cell can be dynamically adjusted based on signals from adjacent macrophage both in normal and stress conditions.
Our studies support the notion that an increased understanding of the in vivo steps important for generation of erythroblast islands, accompanied by an understanding of the molecular regulation of the relevant intercellular components within the macrophage and erythroid compartments, will be important for the design of efficient protocols to generate blood cells in vitro, as well as for illuminating how these steps may be tweaked in vivo as needed to alleviate patient morbidity during dysregulated erythropoiesis and anemia.
MATERIALS AND METHODS
Cell isolation and culture
Murine ESC lines were maintained and differentiated to EBs according to established protocols (Adelman et al., 2002; Choi et al., 2005; Kennedy and Keller, 2003; Manwani et al., 2007; Zafonte et al., 2007). ESCs that contain a site-specific integrated GFP driven by the EKLF/KLF1 promoter have been described (Lohmann and Bieker, 2008). Judiciously timed harvesting of EBs enabled enrichment of primitive or definitive erythroid progenitors that were then dispersed and followed with secondary platings for generation of isolated erythroid colonies. Semisolid media (methylcellulose) were used in all ESC and EB colony platings, using a low cell concentration (no more than 50-100 cells per 35 mm dish) to maximize cell separation and EB colony formation (Adelman et al., 2002). Synthetic peptides (BioSynthesis or ElimBio) were premixed into the methylcellulose as needed.
Fetal liver cells harvested from staged embryos were mechanically dispersed to single cells (Lohmann and Bieker, 2008). Yolk sac cells were disaggregated with collagenase (Palis et al., 2001, 1999), and EBs with trypsin. Cytospins were fixed with May-Grünwald solution (Fluka) and stained with Giemsa solution (Sigma-Aldrich), and pictures were taken using a Nikon Microphot-FX fluorescence microscope equipped with a Q-Imaging camera.
FACS analysis
Single-cell suspensions of murine fetal livers, yolk sacs, or EBs were stained for FACS with the following antibodies: Ter119-APC, F4/80-PE, VCAM1-APC and CD45-PE from eBiosciences, Kit-APC from Invitrogen, Ter119-PE from BD Biosciences and F4/80-FITC from Serotec.
Multicolor FACS sorting for CMP or MEP populations was performed as previously described (Frontelo et al., 2007). Dispersed and sorted cells were cultured in MethoCult M3120 (StemCell Technologies) that contained IMDM, 10% PDS or FBS, 5% PFHM-II, 300 µM monothioglycerol, 25 ng/ml ascorbic acid, 2 mM glutamine, 300 µg/ml transferrin, 100 ng/ml SCF, 2 U/ml erythropoietin, 5 ng/ml thrombopoietin and 5 ng/ml IL11 (Kennedy and Keller, 2003). Importantly, colonies from these cell sorts did not form when using culture conditions that typically work well with fetal liver or bone marrow cells (Terszowski et al., 2005).
Flow cytometry data were analyzed using FlowJo (TreeStar). In all FACS analyses, gates are drawn based on negative controls from the same samples within the same experiment; mean±s.d. are shown for each quadrant of interest. Least squares fitting was performed as described (http://www.physics.csbsju.edu/stats/).
Island analysis
Island clusters were enriched and analyzed morphologically or by immunofluorescence as described previously (Fraser et al., 2010; Iavarone et al., 2004; Isern et al., 2008; McGrath et al., 2008). Total RNA was isolated and processed for quantitative analysis as described (Frontelo et al., 2007; Lohmann and Bieker, 2008; Siatecka et al., 2007). Primer sequences used for qPCR analysis are listed in supplementary material Table S1.
Supplementary Material
Acknowledgements
We thank Drs Kyunghee Choi, Jim Palis, Paul Kingsley, Anna Rita Migliaccio, Velia Fowler and Dave Bodine for discussion, and Drs Mirka Siatecka and Shefali Soni for constructive comments on the manuscript. We acknowledge the expertise of the Mount Sinai flow cytometry shared resource facility.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
L.X., M.G., M.N.G. and D.M. performed experiments, D.M. and J.J.B. directed experiments, and J.J.B. wrote the paper with input from the other authors.
Funding
This work was supported by the National Institutes of Health (NIH) [K08 DK002871 to D.M. and R01 DK77822 to J.J.B.], and by a Black Family Stem Cell Institute Exploratory Research Grant to J.J.B. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.103960/-/DC1
References
- Adelman C. A., Chattopadhyay S., Bieker J. J. (2002). The BMP/BMPR/Smad pathway directs expression of the erythroid-specific EKLF and GATA1 transcription factors during embryoid body differentiation in serum-free media. Development 129, 539-549 [DOI] [PubMed] [Google Scholar]
- Akashi K., Traver D., Miyamoto T., Weissman I. L. (2000). A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193-197 10.1038/35004599 [DOI] [PubMed] [Google Scholar]
- An X., Mohandas N. (2011). Erythroblastic islands, terminal erythroid differentiation and reticulocyte maturation. Int. J. Hematol. 93, 139-143 10.1007/s12185-011-0779-x [DOI] [PubMed] [Google Scholar]
- Arnaud L., Saison C., Helias V., Lucien N., Steschenko D., Giarratana M.-C., Prehu C., Foliguet B., Montout L., de Brevern A. G., et al. (2010). A dominant mutation in the gene encoding the erythroid transcription factor KLF1 causes a congenital dyserythropoietic anemia. Am. J. Hum. Genet. 87, 721-727 10.1016/j.ajhg.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M. H. (2013). Concise review: early embryonic erythropoiesis: not so primitive after all. Stem Cells 31, 849-856 10.1002/stem.1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D. E., Orkin S. H. (2011). Update on fetal hemoglobin gene regulation in hemoglobinopathies. Curr. Opin. Pediatr. 23, 1-8 10.1097/MOP.0b013e3283420fd0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J. Y., Jalil A., Klaine M., Jung S., Cumano A., Godin I. (2005). Three pathways to mature macrophages in the early mouse yolk sac. Blood 106, 3004-3011 10.1182/blood-2005-02-0461 [DOI] [PubMed] [Google Scholar]
- Bessis M. (1958). Erythroblastic island, functional unity of bone marrow. Rev. Hematol. 13, 8-11 [PubMed] [Google Scholar]
- Borg J., Patrinos G. P., Felice A. E., Philipsen S. (2011). Erythroid phenotypes associated with KLF1 mutations. Haematologica 96, 635-638 10.3324/haematol.2011.043265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasis J. A., Mohandas N. (2008). Erythroblastic islands: niches for erythropoiesis. Blood 112, 470-478 10.1182/blood-2008-03-077883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Reitman M., Bieker J. J. (1998). Chromatin structure and transcriptional control elements of the erythroid Kruppel-like factor (EKLF) gene. J. Biol. Chem. 273, 25031-25040 10.1074/jbc.273.39.25031 [DOI] [PubMed] [Google Scholar]
- Chen M. J., Li Y., De Obaldia M. E., Yang Q., Yzaguirre A. D., Yamada-Inagawa T., Vink C. S., Bhandoola A., Dzierzak E., Speck N. A. (2011). Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell 9, 541-552 10.1016/j.stem.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Chung Y. S., Zhang W. J. (2005). Hematopoietic and endothelial development of mouse embryonic stem cells in culture. Methods Mol. Med. 105, 359-368 [DOI] [PubMed] [Google Scholar]
- Chow A., Huggins M., Ahmed J., Hashimoto D., Lucas D., Kunisaki Y., Pinho S., Leboeuf M., Noizat C., van Rooijen N., et al. (2013). CD169(+) macrophages provide a niche promoting erythropoiesis under homeostasis and stress. Nat. Med. 19, 429-436 10.1038/nm.3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Back D. Z., Kostova E. B., van Kraaij M., van den Berg T. K., van Bruggen R. (2014). Of macrophages and red blood cells; a complex love story. Front. Physiol. 5, 9 10.3389/fphys.2014.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore L. C., Crispino J. D. (2011). Transcription factor networks in erythroid cell and megakaryocyte development. Blood 118, 231-239 10.1182/blood-2011-04-285981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R., von Lindern M., Kolbus A., Driegen S., Steinlein P., Beug H., Grosveld F., Philipsen S. (2005). The erythroid phenotype of EKLF-null mice: defects in hemoglobin metabolism and membrane stability. Mol. Cell. Biol. 25, 5205-5214 10.1128/MCB.25.12.5205-5214.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R., Guyot B., Zhang L., Atzberger A., Sloane-Stanley J., Wood B., Porcher C., Vyas P. (2010). Lineage-specific combinatorial action of enhancers regulates mouse erythroid Gata1 expression. Blood 115, 3463-3471 10.1182/blood-2009-07-232876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E., Speck N. A. (2008). Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat. Immunol. 9, 129-136 10.1038/ni1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabriek B. O., Polfliet M. M. J., Vloet R. P. M., van der Schors R. C., Ligtenberg A. J. M., Weaver L. K., Geest C., Matsuno K., Moestrup S. K., Dijkstra C. D., et al. (2007). The macrophage CD163 surface glycoprotein is an erythroblast adhesion receptor. Blood 109, 5223-5229 10.1182/blood-2006-08-036467 [DOI] [PubMed] [Google Scholar]
- Fraser S. T., Isern J., Baron M. H. (2010). Use of transgenic fluorescent reporter mouse lines to monitor hematopoietic and erythroid development during embryogenesis. Methods Enzymol. 476, 403-427 10.1016/S0076-6879(10)76022-5 [DOI] [PubMed] [Google Scholar]
- Frontelo P., Manwani D., Galdass M., Karsunky H., Lohmann F., Gallagher P. G., Bieker J. J. (2007). Novel role for EKLF in megakaryocyte lineage commitment. Blood 110, 3871-3880 10.1182/blood-2007-03-082065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier E. L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K. G., Gordonov S., et al. (2012). Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118-1128 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford S. C., Derganc J., Shevkoplyas S. S., Yoshida T., Bitensky M. W. (2006). A detailed study of time-dependent changes in human red blood cells: from reticulocyte maturation to erythrocyte senescence. Br. J. Haematol. 135, 395-404 10.1111/j.1365-2141.2006.06279.x [DOI] [PubMed] [Google Scholar]
- Granick S., Levere R. D. (1964). Heme synthesis in erythroid cells. Prog. Hematol. 27, 1-47 [PubMed] [Google Scholar]
- Gutiérrez L., Lindeboom F., Langeveld A., Grosveld F., Philipsen S., Whyatt D. (2004). Homotypic signalling regulates Gata1 activity in the erythroblastic island. Development 131, 3183-3193 10.1242/dev.01198 [DOI] [PubMed] [Google Scholar]
- Hanspal M., Smockova Y., Uong Q. (1998). Molecular identification and functional characterization of a novel protein that mediates the attachment of erythroblasts to macrophages. Blood 92, 2940-2950 [PubMed] [Google Scholar]
- Helias V., Saison C., Peyrard T., Vera E., Prehu C., Cartron J.-P., Arnaud L. (2013). Molecular analysis of the rare in(Lu) blood type: toward decoding the phenotypic outcome of haploinsufficiency for the transcription factor KLF1. Hum. Mutat. 34, 221-228 10.1002/humu.22218 [DOI] [PubMed] [Google Scholar]
- Heruth D. P., Hawkins T., Logsdon D. P., Gibson M. I., Sokolovsky I. V., Nsumu N. N., Major S. L., Fegley B., Woods G. M., Lewing K. B., et al. (2010). Mutation in erythroid specific transcription factor KLF1 causes Hereditary Spherocytosis in the Nan hemolytic anemia mouse model. Genomics 96, 303-307 10.1016/j.ygeno.2010.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge D., Coghill E., Keys J., Maguire T., Hartmann B., McDowall A., Weiss M., Grimmond S., Perkins A. (2006). A global role for EKLF in definitive and primitive erythropoiesis. Blood 107, 3359-3370 10.1182/blood-2005-07-2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iavarone A., King E. R., Dai X.-M., Leone G., Stanley E. R., Lasorella A. (2004). Retinoblastoma promotes definitive erythropoiesis by repressing Id2 in fetal liver macrophages. Nature 432, 1040-1045 10.1038/nature03068 [DOI] [PubMed] [Google Scholar]
- Isern J., Fraser S. T., He Z., Baron M. H. (2008). The fetal liver is a niche for maturation of primitive erythroid cells. Proc. Natl. Acad. Sci. U.S.A. 105, 6662-6667 10.1073/pnas.0802032105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isern J., Fraser S. T., He Z., Zhang H., Baron M. H. (2010). Dose-dependent regulation of primitive erythroid maturation and identity by the transcription factor Eklf. Blood 116, 3972-3980 10.1182/blood-2010-04-281196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffray J. A., Mitchell W. B., Gnanapragasam M. N., Seshan S. V., Guo X., Westhoff C. M., Bieker J. J., Manwani D. (2013). Erythroid transcription factor EKLF/KLF1 mutation causing congenital dyserythropoietic anemia type IV in a patient of Taiwanese origin: review of all reported cases and development of a clinical diagnostic paradigm. Blood Cells Mol. Dis. 51, 71-75 10.1016/j.bcmd.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P., Murata-Hori M., Lodish H. F. (2011). Formation of mammalian erythrocytes: chromatin condensation and enucleation. Trends Cell Biol. 21, 409-415 10.1016/j.tcb.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.-J., Shin J.-W., Yoon J.-H., Oh I.-H., Lee S.-P., Kim S.-Y., Park S. H., Mamura M. (2012). Inhibition of erythropoiesis by Smad6 in human cord blood hematopoietic stem cells. Biochem. Biophys. Res. Commun. 423, 750-756 10.1016/j.bbrc.2012.06.031 [DOI] [PubMed] [Google Scholar]
- Kawane K., Fukuyama H., Kondoh G., Takeda J., Ohsawa Y., Uchiyama Y., Nagata S. (2001). Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science 292, 1546-1549 10.1126/science.292.5521.1546 [DOI] [PubMed] [Google Scholar]
- Keerthivasan G., Wickrema A., Crispino J. D. (2011). Erythroblast enucleation. Stem Cells Int. 2011, 139851 10.4061/2011/139851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M., Keller G. M. (2003). Hematopoietic commitment of ES cells in culture. Methods Enzymol. 365, 39-59 10.1016/S0076-6879(03)65003-2 [DOI] [PubMed] [Google Scholar]
- Kierdorf K., Erny D., Goldmann T., Sander V., Schulz C., Perdiguero E. G., Wieghofer P., Heinrich A., Riemke P., Hölscher C., et al. (2013). Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16, 273-280 10.1038/nn.3318 [DOI] [PubMed] [Google Scholar]
- Kingsley P. D., Greenfest-Allen E., Frame J. M., Bushnell T. P., Malik J., McGrath K. E., Stoeckert C. J., Palis J. (2013). Ontogeny of erythroid gene expression. Blood 121, e5-e13 10.1182/blood-2012-04-422394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohyama M., Ise W., Edelson B. T., Wilker P. R., Hildner K., Mejia C., Frazier W. A., Murphy T. L., Murphy K. M. (2009). Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature 457, 318-321 10.1038/nature07472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis D. G., Pushkaran S., Johnson J. F., Cancelas J. A., Manganaris S., Harris C. E., Williams D. A., Zheng Y., Kalfa T. A. (2012). Signaling and cytoskeletal requirements in erythroblast enucleation. Blood 119, 6118-6127 10.1182/blood-2011-09-379263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koury M. J. (2014). Abnormal erythropoiesis and the pathophysiology of chronic anemia. Blood Rev. 28, 49-66 10.1016/j.blre.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Kusakabe M., Hasegawa K., Hamada M., Nakamura M., Ohsumi T., Suzuki H., Mai T. T. N., Kudo T., Uchida K., Ninomiya H., et al. (2011). c-Maf plays a crucial role for the definitive erythropoiesis that accompanies erythroblastic island formation in the fetal liver. Blood 118, 1374-1385 10.1182/blood-2010-08-300400 [DOI] [PubMed] [Google Scholar]
- Kyba M., Perlingeiro R. C. R., Daley G. Q. (2002). HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109, 29-37 10.1016/S0092-8674(02)00680-3 [DOI] [PubMed] [Google Scholar]
- Lee G., Lo A., Short S. A., Mankelow T. J., Spring F., Parsons S. F., Yazdanbakhsh K., Mohandas N., Anstee D. J., Chasis J. A. (2006). Targeted gene deletion demonstrates that the cell adhesion molecule ICAM-4 is critical for erythroblastic island formation. Blood 108, 2064-2071 10.1182/blood-2006-03-006759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimberg M. J., Prus E., Konijn A. M., Fibach E. (2008). Macrophages function as a ferritin iron source for cultured human erythroid precursors. J. Cell. Biochem. 103, 1211-1218 10.1002/jcb.21499 [DOI] [PubMed] [Google Scholar]
- Li L., Freudenberg J., Cui K., Dale R., Song S.-H., Dean A., Zhao K., Jothi R., Love P. E. (2013). Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation. Blood 121, 4575-4585 10.1182/blood-2013-01-479451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.-S., Li X.-H., Wang Y., Shu R.-Z., Wang L., Lu S.-Y., Kong H., Jin Y.-E., Zhang L.-J., Fei J., et al. (2007). Disruption of palladin leads to defects in definitive erythropoiesis by interfering with erythroblastic island formation in mouse fetal liver. Blood 110, 870-876 10.1182/blood-2007-01-068528 [DOI] [PubMed] [Google Scholar]
- Lohmann F., Bieker J. J. (2008). Activation of Eklf expression during hematopoiesis by Gata2 and Smad5 prior to erythroid commitment. Development 135, 2071-2082 10.1242/dev.018200 [DOI] [PubMed] [Google Scholar]
- Luo Q., Ma X., Wahl S. M., Bieker J. J., Crossley M., Montaner L. J. (2004). Activation and repression of interleukin-12 p40 transcription by erythroid Kruppel-like factor in macrophages. J. Biol. Chem. 279, 18451-18456 10.1074/jbc.M400320200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux C. T., Yoshimoto M., McGrath K., Conway S. J., Palis J., Yoder M. C. (2008). All primitive and definitive hematopoietic progenitor cells emerging before E10 in the mouse embryo are products of the yolk sac. Blood 111, 3435-3438 10.1182/blood-2007-08-107086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankelow T. J., Spring F. A., Parsons S. F., Brady R. L., Mohandas N., Chasis J. A., Anstee D. J. (2004). Identification of critical amino-acid residues on the erythroid intercellular adhesion molecule-4 (ICAM-4) mediating adhesion to alpha V integrins. Blood 103, 1503-1508 10.1182/blood-2003-08-2792 [DOI] [PubMed] [Google Scholar]
- Manwani D., Bieker J. J. (2008). The erythroblastic island. Curr. Top. Dev. Biol. 82, 23-53 10.1016/S0070-2153(07)00002-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manwani D., Galdass M., Bieker J. J. (2007). Altered regulation of beta-like globin genes by a redesigned erythroid transcription factor. Exp. Hematol. 35, 39-47 10.1016/j.exphem.2006.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath K. E., Palis J. (2005). Hematopoiesis in the yolk sac: more than meets the eye. Exp. Hematol. 33, 1021-1028 10.1016/j.exphem.2005.06.012 [DOI] [PubMed] [Google Scholar]
- McGrath K., Palis J. (2008). Ontogeny of erythropoiesis in the mammalian embryo. Curr. Top. Dev. Biol. 82, 1-22 10.1016/S0070-2153(07)00001-4 [DOI] [PubMed] [Google Scholar]
- McGrath K. E., Kingsley P. D., Koniski A. D., Porter R. L., Bushnell T. P., Palis J. (2008). Enucleation of primitive erythroid cells generates a transient population of “pyrenocytes” in the mammalian fetus. Blood 111, 2409-2417 10.1182/blood-2007-08-107581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath K. E., Frame J. M., Fromm G. J., Koniski A. D., Kingsley P. D., Little J., Bulger M., Palis J. (2011). A transient definitive erythroid lineage with unique regulation of the beta-globin locus in the mammalian embryo. Blood 117, 4600-4608 10.1182/blood-2010-12-325357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller I. J., Bieker J. J. (1993). A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol. Cell. Biol. 13, 2776-2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millot S., Andrieu V., Letteron P., Lyoumi S., Hurtado-Nedelec M., Karim Z., Thibaudeau O., Bennada S., Charrier J.-L., Lasocki S., et al. (2010). Erythropoietin stimulates spleen BMP4-dependent stress erythropoiesis and partially corrects anemia in a mouse model of generalized inflammation. Blood 116, 6072-6081 10.1182/blood-2010-04-281840 [DOI] [PubMed] [Google Scholar]
- Nagata S. (2007). Autoimmune diseases caused by defects in clearing dead cells and nuclei expelled from erythroid precursors. Immunol. Rev. 220, 237-250 10.1111/j.1600-065X.2007.00571.x [DOI] [PubMed] [Google Scholar]
- Nakorn T. N., Miyamoto T., Weissman I. L. (2003). Characterization of mouse clonogenic megakaryocyte progenitors. Proc. Natl. Acad. Sci. U.S.A. 100, 205-210 10.1073/pnas.262655099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ney P. A. (2011). Normal and disordered reticulocyte maturation. Curr. Opin. Hematol. 18, 152-157 10.1097/MOH.0b013e328345213e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S. H., Zon L. I. (2008). Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631-644 10.1016/j.cell.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palis J. (2014). Primitive and definitive erythropoiesis in mammals. Front. Physiol. 5, 3 10.3389/fphys.2014.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palis J., Robertson S., Kennedy M., Wall C., Keller G. (1999). Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126, 5073-5084 [DOI] [PubMed] [Google Scholar]
- Palis J., Chan R. J., Koniski A., Patel R., Starr M., Yoder M. C. (2001). Spatial and temporal emergence of high proliferative potential hematopoietic precursors during murine embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 98, 4528-4533 10.1073/pnas.071002398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson R. F., Shi L., Wu D.-C. (2011). Stress erythropoiesis: new signals and new stress progenitor cells. Curr. Opin. Hematol. 18, 139-145 10.1097/MOH.0b013e32834521c8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A. C., Sharpe A. H., Orkin S. H. (1995). Lethal β-thalassemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375, 318-322 10.1038/375318a0 [DOI] [PubMed] [Google Scholar]
- Pilon A. M., Nilson D. G., Zhou D., Sangerman J., Townes T. M., Bodine D. M., Gallagher P. G. (2006). Alterations in expression and chromatin configuration of the Alpha hemoglobin-stabilizing protein gene in erythroid Kruppel-like factor-deficient mice. Mol. Cell. Biol. 26, 4368-4377 10.1128/MCB.02216-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon A. M., Arcasoy M. O., Dressman H. K., Vayda S. E., Maksimova Y. D., Sangerman J. I., Gallagher P. G., Bodine D. M. (2008). Failure of terminal erythroid differentiation in EKLF-deficient mice is associated with cell cycle perturbation and reduced expression of E2F2. Mol. Cell. Biol. 28, 7394-7401 10.1128/MCB.01087-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon A. M., Ajay S. S., Kumar S. A., Steiner L. A., Cherukuri P. F., Wincovitch S., Anderson S. M., Mullikin J. C., Gallagher P. G., Hardison R. C., et al. (2011). Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood 118, e139-e148 10.1182/blood-2011-05-355107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porayette P., Paulson R. F. (2008). BMP4/Smad5 dependent stress erythropoiesis is required for the expansion of erythroid progenitors during fetal development. Dev. Biol. 317, 24-35 10.1016/j.ydbio.2008.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcu S., Manchinu M. F., Marongiu M. F., Sogos V., Poddie D., Asunis I., Porcu L., Marini M. G., Moi P., Cao A., et al. (2011). Klf1 affects DNase II-alpha expression in the central macrophage of a fetal liver erythroblastic island: a non-cell-autonomous role in definitive erythropoiesis. Mol. Cell. Biol. 31, 4144-4154 10.1128/MCB.05532-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos P., Casu C., Gardenghi S., Breda L., Crielaard B. J., Guy E., Marongiu M. F., Gupta R., Levine R. L., Abdel-Wahab O., et al. (2013). Macrophages support pathological erythropoiesis in polycythemia vera and beta-thalassemia. Nat. Med. 19, 437-445 10.1038/nm.3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes M. M., Kopsombut P., Bondurant M. C., Price J. O., Koury M. J. (2008). Adherence to macrophages in erythroblastic islands enhances erythroblast proliferation and increases erythrocyte production by a different mechanism than erythropoietin. Blood 111, 1700-1708 10.1182/blood-2007-06-098178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadahira Y., Yoshino T., Monobe Y. (1995). Very late activation antigen 4-vascular cell adhesion molecule 1 interaction is involved in the formation of erythroblastic islands. J. Exp. Med. 181, 411-415 10.1084/jem.181.1.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadahira Y., Yasuda T., Yoshino T., Manabe T., Takeishi T., Kobayashi Y., Ebe Y., Naito M. (2000). Impaired splenic erythropoiesis in phlebotomized mice injected with CL2MDP-liposome: an experimental model for studying the role of stromal macrophages in erythropoiesis. J. Leukoc. Biol. 68, 464-470 [PubMed] [Google Scholar]
- Siatecka M., Bieker J. J. (2011). The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood 118, 2044-2054 10.1182/blood-2011-03-331371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siatecka M., Xue L., Bieker J. J. (2007). Sumoylation of EKLF Promotes Transcriptional Repression and Is Involved in Inhibition of Megakaryopoiesis. Mol. Cell. Biol. 27, 8547-8560 10.1128/MCB.00589-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siatecka M., Sahr K. E., Andersen S. G., Mezei M., Bieker J. J., Peters L. L. (2010). Severe anemia in the Nan mutant mouse caused by sequence-selective disruption of erythroid Kruppel-like factor. Proc. Natl. Acad. Sci. U.S.A. 107, 15151-15156 10.1073/pnas.1004996107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton B. K., Lau W., Fairweather V. S. S., Burton N. M., Wilson M. C., Parsons S. F., Richardson B. M., Trakarnsanga K., Brady R. L., Anstee D. J., et al. (2011). Mutations in the second zinc finger of human EKLF reduce promoter affinity but give rise to benign and disease phenotypes. Blood 118, 3137-3145 10.1182/blood-2011-04-349985 [DOI] [PubMed] [Google Scholar]
- Singleton B. K., Frayne J., Anstee D. J. (2012). Blood group phenotypes resulting from mutations in erythroid transcription factors. Curr. Opin. Hematol. 19, 486-493 10.1097/MOH.0b013e328358f92e [DOI] [PubMed] [Google Scholar]
- Socolovsky M. (2013). Exploring the erythroblastic island. Nat. Med. 19, 399-401 10.1038/nm.3156 [DOI] [PubMed] [Google Scholar]
- Soni S., Bala S., Gwynn B., Sahr K. E., Peters L. L., Hanspal M. (2006). Absence of erythroblast macrophage protein (Emp) leads to failure of erythroblast nuclear extrusion. J. Biol. Chem. 281, 20181-20189 10.1074/jbc.M603226200 [DOI] [PubMed] [Google Scholar]
- Southwood C. M., Downs K. M., Bieker J. J. (1996). Erythroid Kruppel-like Factor (EKLF) exhibits an early and sequentially localized pattern of expression during mammalian erythroid ontogeny. Dev. Dyn. 206, 248-259 10.1002/(SICI)1097-0177(199607)206:3%3C248::AID-AJA3%3E3.0.CO;2-I [DOI] [PubMed] [Google Scholar]
- Spring F. A., Parsons S. F., Ortlepp S., Olsson M. L., Sessions R., Brady R. L., Anstee D. J. (2001). Intercellular adhesion molecule-4 binds alpha(4)beta(1) and alpha(V)-family integrins through novel integrin-binding mechanisms. Blood 98, 458-466 10.1182/blood.V98.2.458 [DOI] [PubMed] [Google Scholar]
- Su M. Y., Steiner L. A., Bogardus H., Mishra T., Schulz V. P., Hardison R. C., Gallagher P. G. (2013). Identification of biologically relevant enhancers in human erythroid cells. J. Biol. Chem. 288, 8433-8444 10.1074/jbc.M112.413260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallack M. R., Perkins A. C. (2010). KLF1 directly coordinates almost all aspects of terminal erythroid differentiation. IUBMB Life 62, 886-890 10.1002/iub.404 [DOI] [PubMed] [Google Scholar]
- Tallack M. R., Perkins A. C. (2013). Three fingers on the switch: Kruppel-like factor 1 regulation of gamma-globin to beta-globin gene switching. Curr. Opin. Hematol. 20, 193-200 10.1097/MOH.0b013e32835f59ba [DOI] [PubMed] [Google Scholar]
- Tallack M. R., Whitington T., Yuen W. S., Wainwright E. N., Keys J. R., Gardiner B. B., Nourbakhsh E., Cloonan N., Grimmond S. M., Bailey T. L., et al. (2010). A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 20, 1052-1063 10.1101/gr.106575.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallack M. R., Magor G. W., Dartigues B., Sun L., Huang S., Fittock J. M., Fry S. V., Glazov E. A., Bailey T. L., Perkins A. C. (2012). Novel roles for KLF1 in erythropoiesis revealed by mRNA-seq. Genome Res. 22, 2385-2398 10.1101/gr.135707.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terszowski G., Waskow C., Conradt P., Lenze D., Koenigsmann J., Carstanjen D., Horak I., Rodewald H.-R. (2005). Prospective isolation and global gene expression analysis of the erythrocyte colony-forming unit (CFU-E). Blood 105, 1937-1945 10.1182/blood-2004-09-3459 [DOI] [PubMed] [Google Scholar]
- Tober J., Yzaguirre A. D., Piwarzyk E., Speck N. A. (2013). Distinct temporal requirements for Runx1 in hematopoietic progenitors and stem cells. Development 140, 3765-3776 10.1242/dev.094961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubukawa K., Guo Y.-M., Takahashi M., Hirokawa M., Michishita Y., Nara M., Tagawa H., Takahashi N., Komatsuda A., Nunomura W., et al. (2012). Enucleation of human erythroblasts involves non-muscle myosin IIB. Blood 119, 1036-1044 10.1182/blood-2011-06-361907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lindern M. (2006). Cell-cycle control in erythropoiesis. Blood 108, 781-782 10.1182/blood-2006-05-022368 [DOI] [Google Scholar]
- Wontakal S. N., Guo X., Smith C., MacCarthy T., Bresnick E. H., Bergman A., Snyder M. P., Weissman S. M., Zheng D., Skoultchi A. I. (2012). A core erythroid transcriptional network is repressed by a master regulator of myelo-lymphoid differentiation. Proc. Natl. Acad. Sci. U.S.A. 109, 3832-3837 10.1073/pnas.1121019109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Chen X., Chang Y., Bieker J. J. (2004). Regulatory elements of the EKLF gene that direct erythroid cell-specific expression during mammalian development. Blood 103, 4078-4083 10.1182/blood-2003-09-3231 [DOI] [PubMed] [Google Scholar]
- Yien Y. Y., Bieker J. J. (2013). EKLF/KLF1, a tissue-restricted integrator of transcriptional control, chromatin remodeling, and lineage determination. Mol. Cell. Biol. 33, 4-13 10.1128/MCB.01058-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Okabe Y., Kawane K., Fukuyama H., Nagata S. (2005). Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat. Immunol. 6, 49-56 10.1038/ni1146 [DOI] [PubMed] [Google Scholar]
- Zafonte B. T., Liu S., Lynch-Kattman M., Torregroza I., Benvenuto L., Kennedy M., Keller G., Evans T. (2007). Smad1 expands the hemangioblast population within a limited developmental window. Blood 109, 516-523 10.1182/blood-2006-02-004564 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.