Abstract
Background
Although they are important disease vectors mosquito biodiversity in Pakistan is poorly known. Recent epidemics of dengue fever have revealed the need for more detailed understanding of the diversity and distributions of mosquito species in this region. DNA barcoding improves the accuracy of mosquito inventories because morphological differences between many species are subtle, leading to misidentifications.
Methodology/Principal Findings
Sequence variation in the barcode region of the mitochondrial COI gene was used to identify mosquito species, reveal genetic diversity, and map the distribution of the dengue-vector species in Pakistan. Analysis of 1684 mosquitoes from 491 sites in Punjab and Khyber Pakhtunkhwa during 2010–2013 revealed 32 species with the assemblage dominated by Culex quinquefasciatus (61% of the collection). The genus Aedes (Stegomyia) comprised 15% of the specimens, and was represented by six taxa with the two dengue vector species, Ae. albopictus and Ae. aegypti, dominant and broadly distributed. Anopheles made up another 6% of the catch with An. subpictus dominating. Barcode sequence divergence in conspecific specimens ranged from 0–2.4%, while congeneric species showed from 2.3–17.8% divergence. A global haplotype analysis of disease-vectors showed the presence of multiple haplotypes, although a single haplotype of each dengue-vector species was dominant in most countries. Geographic distribution of Ae. aegypti and Ae. albopictus showed the later species was dominant and found in both rural and urban environments.
Conclusions
As the first DNA-based analysis of mosquitoes in Pakistan, this study has begun the construction of a barcode reference library for the mosquitoes of this region. Levels of genetic diversity varied among species. Because of its capacity to differentiate species, even those with subtle morphological differences, DNA barcoding aids accurate tracking of vector populations.
Introduction
Mosquitoes are important vectors of animal diseases [1]. Although Pakistan is one of the hotspots for mosquito-vectored diseases [2], [3], mosquito biodiversity in the country is under-explored [4]. However, the recent outbreaks of dengue in Pakistan [5] have generated interest in mosquito distributions in this region [6]–[8]. Among the 3500 mosquito species recorded worldwide (www.mosquito-taxonomic-inventory.info) 104 have been documented from Pakistan and Bangladesh [4], but their morphological identification remains difficult.
Correct vector identification is very important to design strategies for managing vector-borne diseases [9]. Because detailed taxonomic studies have focused on mosquitoes that are vectors of human disease [10], other species have received little attention [11], [12]. Moreover, many closely related species of mosquitoes with differing ecological and host preferences are nearly indistinguishable morphologically [13]. These factors mean that the identification of mosquitoes to a species or sometimes even a genus is often difficult [14]–[16]. As a consequence, DNA-based approaches to mosquito identification [17]–[19], genetic diversity [20], [21], and molecular phylogeny [22], [23] have gained increasing adoption. Although use of nuclear genes is not uncommon [24]–[27], mitochondrial genes have gained primary adoption for analyzing genetic diversity in mosquitoes [28], [29]. DNA barcoding [30] has already seen frequent use for mosquitoes in varied contexts [31]–[37]. As a result, the overall DNA barcode library now includes records for 894 mosquito species among the 320K animal species which have been analyzed (www.boldsystems.org).
Prior studies have monitored mosquito populations using both morphological [38] and molecular approaches [39]. For example, Reisen et al. (1982) [40] used morphological identifications to assess the diversity of mosquitoes in Pakistan, especially those important in the transmission of viral diseases. Mousson et al. (2005) [41] subsequently used mitochondrial DNA variation to study the phylogeography and relationships of Aedes aegypti and Aedes albopictus, while Chen et al. (2002) [25] used 28S rDNA and COII to examine the distribution and vector status of Anopheles minimus in southern China. Correlation between vector genotypes and their capacity to transmit disease pathogens [42] has triggered interest in the genetic diversities of vector species [43]. In a study of three vector species (Ae. aegypti, Ae. albopictus, Aedes vittatus) in India, Angel & Joshi (2008) [44] found that infectivity of dengue virus varied both among species and regionally for a particular species. Studies by Pollitt et al. (2013) [45] and Anderson et al. (2004) [46], correlated mosquito strains/genotypes with their competence to transmit malaria parasites and La Crosse virus.
Studies on species composition and density of local mosquito populations have helped to develop better management strategies for mosquito-borne diseases [47],[48]. Such studies can establish a baseline of mosquito-borne virus activity allowing monitoring of change over time [49]. Ae. aegypti and Ae. albopictus have differing ecological preferences with the former species most prevalent in urbanized areas, while the latter is often common in rural settings [50], [51]. Although native to Southeast Asia, Ae. albopictus has extended its range [52] provoking concerns among disease management strategists [53]. The 2011 dengue epidemic in Pakistan showed considerable regional variation in severity, raising questions about the possible role of shifting distributions of its vector species as the cause.
Since the species checklist by Khan (1971) [4] which included mosquitoes from Pakistan and Bangladesh, only sporadic reports with limited scope have been completed [7], [40], [54]–[56]. Further, information on genetic diversity of regional mosquitoes usable for species assignments or to establish connections between local and the global mosquito fauna is either not available or is insufficient [8]. Although both Ae. aegypti and Ae. albopictus occur in Pakistan [4], there is little information on their relative abundance or distribution [57]. The current study employs DNA barcoding to identify, and analyze genetic diversity in mosquito species making it possible to map the distributions of dengue-vectors in the dengue-affected areas of Pakistan. The study also develops haplotype networks for the important disease vectors at a global scale connecting them in different regions.
Materials and Methods
Ethics statement
No specific permissions were required for this study. Mosquitoes from private residences were collected only after the consent of the respective owners. The study did not involve endangered or protected species.
Collection of mosquitoes
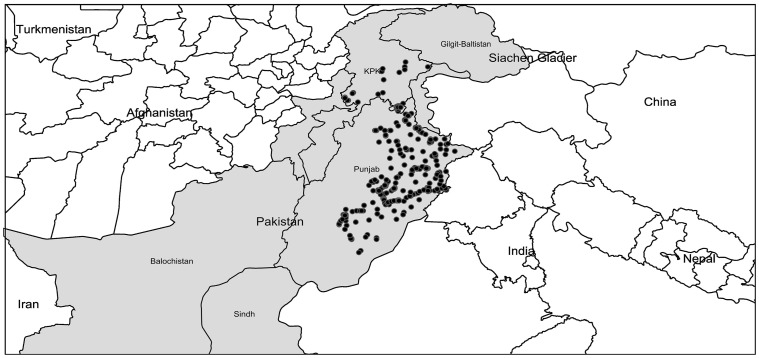
Mosquitoes were collected at 450 urban and rural sites within Punjab, and from 41 locations in Khyber Pakhtunkhwa (KPK) during 2010–2013. These sites ranged in altitude from 92 m - 1004 m in Punjab and from 297 m–2376 m in KPK. Sampling sites included private residences, construction sites, junkyards, water catchments, marshes, ponds, and forests. Adults were collected with nets, aspirators, and light-traps coupled with a CO2 source, while larvae were sampled with pipettes and sieves. GPS coordinates were recorded (Table S1), and collection sites were mapped (Fig. 1) using (http://www.simplemappr.net). A total of 1942 specimens including 190 larvae were randomly chosen for DNA barcode analysis. Each mosquito was assigned a specimen number and photographed, and adults were identified using standard taxonomic keys [4], [40], [55], [58]. Species names follow those employed by the on-line resource (www.mosquito-taxonomic-inventory.info). Specimen data along with the collection information are accessible in the dataset DS-MAMOS (Barcoding Mosquitoes of Pakistan) on BOLD (www.boldsystems.org), the Barcode of Life Data System [59].
Figure 1. Map of collection localities (solid black dots) for mosquitoes in the dengue-affected areas of Punjab and the adjoining Khyber Pakhtunkhwa province.
DNA isolation, PCR amplification and sequencing
A single leg was removed from each adult specimen and transferred to a well pre-loaded with 30µl of 95% ethanol in a 96-well microplate, while larvae were processed using the protocol in Porco et al. (2010) [60]. DNA extraction, PCR amplification, and sequencing were performed at the Canadian Centre for DNA Barcoding (CCDB) following standard protocols [61]. Amplification of the COI-5′ barcode region was performed with primer pair C_LepFo1F (a cocktail of LepF1+LCO1490)/C_LepFo1R (a cocktail of LepR1+HCO2198) (http://www.dnabarcoding.ca/CCDB_DOCS/CCDB_PrimerSets.pdf) using the following PCR conditions: 94°C (1 min); 5 cycles of 94°C (30 s), 45°C (40 s), 72°C (1 min); 35 cycles of 94°C (30 s), 51°C (40 s), 72°C (1 min); and final extension of 72°C (10 min). PCRs were carried out in 12.5 µL reactions with a standard reaction cocktail and 2 µL of DNA template. PCR products were visualized on a 2% agarose E-gel 96 system (Invitrogen Inc.) and successful amplicons were bidirectionally sequenced using BigDye Terminator Cycle Sequencing Kit (v3.1) on an ABI 3730XL DNA Analyzer. The forward and the reverse sequences were assembled and aligned using CodonCode Aligner (CodonCode Corporation, USA). Sequences were subsequently translated in MEGA V5 [62] to verify that they were free of stop codons and gaps and uploaded onto BOLD. All sequences with their GenBank accession numbers (KF406349 to KF407931) are accessible in the dataset DS-MAMOS (Process IDs: MADIP255-10 to MADIP380-10; MADIP458-11 to MADIP473-11; MAMOS001-12 to MAMOS1425-12; MAMOS1521-13 to MAMOS2019-13).
Data analysis
Species discrimination using DNA barcodes
The sequence from each specimen was compared to barcode sequences on GenBank using "Blast", and to sequences for 894 mosquito species on BOLD using the "Identification Request" function. Prior studies have revealed that most different species of Diptera show >2% sequence divergence at COI [63], and researchers have used a 2% distance threshold for species delimitation [64]. Based on reference barcode data, Ratnasingham & Hebert (2013) [65] have established the Barcode Index Number (BIN) system which assigns a unique global identifier to each sequence cluster. In most cases, specimens assigned to different BINs belong to different species, but the BIN system aids the organization of data for records lacking a formal taxonomic assignment. All mosquito sequences obtained in this study were assigned to a BIN.
Genetic diversity and phylogenetic analysis
Nucleotide sequences were aligned using ClustalW [66] in MEGA V5. The FASTA file was submitted to the online version of Automatic Barcode Gap Discovery (ABGD) [67] to generate distance histograms and distance ranks. The presence or absence of a “barcode gap” [68] was also determined for each species as a test of the reliability of its discrimination. Using the barcode gap criterion, a species is distinct from its nearest neighbor (NN) if its maximum intraspecific distance is less than the distance to its NN sequence. The "Barcode Gap Analysis" (BGA) was performed using BOLD. Genetic diversity indices and neutrality tests (Fu's Fs [69] and Tajima's D [70]) were performed in DnaSP v5.10.01 [71]. Calculations of Kimura 2-parameter (K2P) [72] genetic distances and NJ analysis were carried out using MEGA V5. Because most mosquito species were represented by multiple sequences, TAXONDNA was used to generate a consensus barcode for each species, enabling the generation of a compact tree [73]. A tree of all sequences is provided as (Fig. S1). The K2P distance model was used, along with pairwise deletion of missing sites to generate NJ trees, while support for tree nodes was estimated using 500 bootstrap replicates. Consensus sequences were used in Bayesian inference (BI) analysis and BI trees were obtained using MrBayes v3.2.0 [74] and the Markov Chain Monte Carlo (MCMC) technique. The data was partitioned in two ways: a single partition with parameters estimated across all codon positions, and a codon-partition in which each codon position was allowed different parameter estimates. All partitions were allowed a GTR + gamma model and analyses were run for 10 million generations by using four chains with sampling every 1,000 generations. Bayesian posterior probabilities were calculated from the sample points once the MCMC algorithm began to converge. The trees generated through this process were visualized using FigTree v1.4.0.
Haplotype and distribution analysis
Barcode sequences for important disease vectors (Ae. aegypti, Ae. albopictus, Anopheles subpictus, Anopheles stephensi, Anopheles peditaeniatus, Culex tritaeniorhynchus, Cx. quinquefasciatus) from Pakistan, were combined with published records from other countries and aligned in MEGA5 before being exported as MEGA files. Barcode haplotypes were determined using Arlequin v.3.5 [75]. For each species, a minimum spanning tree (MST) based on the number of nucleotide differences between haplotypes was constructed using a distance matrix from Arlequin in Hapstar v. 0.6 [76] to visualize the network of interrelationships between the haplotypes. The distributions of Ae. aegypti and Ae. albopictus were mapped using an online tool (www.simplemappr.net).
Results
Identification of mosquito species and DNA barcode analysis
Morphological study indicated the presence of 21 mosquito species including four species of Aedes (Stegomyia) (Ae. aegypti, Ae. albopictus, Ae. w-albus, Ae. unilineatus), six species of Culex (Cx. quinquefasciatus, Cx. theileri, Cx. tritaeniorhynchus, Cx. bitaeniorhynchus, Cx. mimeticus, Cx. fuscocephala) and seven species of Anopheles (An. subpictus, An. peditaeniatus, An. stephensi, An. splendidus, An. pulcherrimus, An. annularis, An. culicifacies). Barcode sequences were recovered from 1684 of the 1942 specimens (87%). Fig. 2 shows the number of specimens of each species with a barcode sequence. Comparison with records in BOLD and GenBank revealed close sequence matches (<2% divergence) to 11 species which were not recognized morphologically. Anopheles annularis was partitioned into two taxa, An. annularis A and An. annularis B, while An. culicifacies was also found to include two sibling species, An. culicifacies A and a second unidentified taxon, raising the total to 23. Five more species (Culex perexiguus, Phagomyia cogilli, Anopheles sp. nr. dravidicus, Ochlerotatus pulcritarsis, Ochlerotatus caspius) were initially overlooked, but were identified through barcodes, raising the count to 28. Finally, two more species were only identifiable to a generic level (Aedes MA01, Aedes MA02), and two more to a tribe (Aedini 1, Aedini 2). The BIN system assigned these 32 taxa to 31 BINs; the sole case of a shared BIN involved An. annularis A and An. annularis B.
Figure 2. Mosquito species identified from the dengue-affected areas of Pakistan.
The number of specimens of each species in the collection are indicated on the bars.
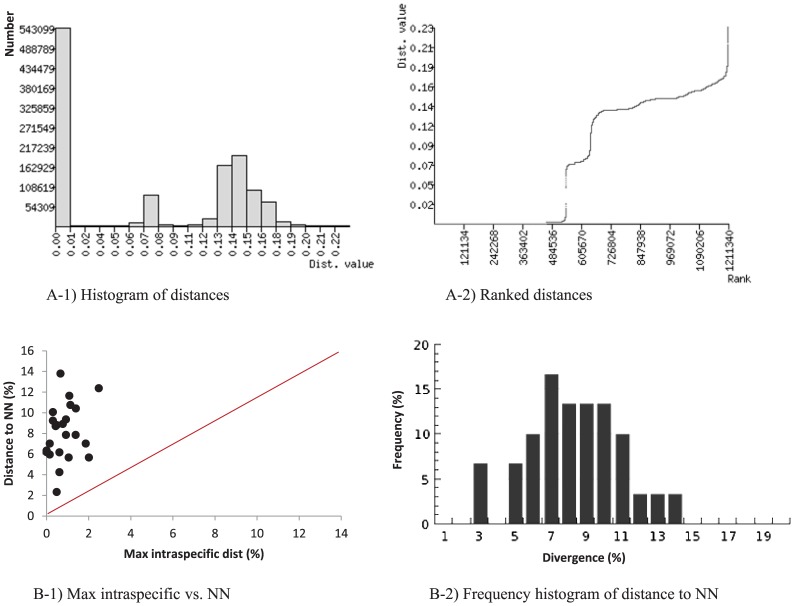
The results of ABGD and BGA analyses revealed a clear gap between intraspecific and interspecific distances (Fig. 3). As well, the minimum distance to the nearest-neighbor (NN) was higher than the maximum intraspecific distance for every species (Fig. 3). In fact, NN distances exceeded 2.3% for all species (Fig. 3 B-1), and most ranged from 4.3–11% (Fig. 3 B-2). An. culicifacies was initially identified as a single species, but the barcode data revealed 4.3% divergence between two taxa; one was An. culicifacies A, but the other could not be identified because of the lack of reference barcode sequences for the other four known taxa in this complex. The barcode data also revealed that An. annularis included representatives of both An. annularis A and An. annularis B, species that were first recognized through polytene chromosome analysis [77]. Intraspecific distances could not be determined for the six species with just a single representative, but all of their NN distances were greater than 2.3%. Sequence divergence increased with taxonomic rank (Table 1) with little overlap between conspecific and congeneric distances. Intraspecific divergences ranged from 0.0–2.4% with a mean of 0.04%, while divergences for the species in a genus ranged from 2.3–17.8% with a mean of 8.2% (Table 1). Genetic diversity indices and results of neutrality tests for the barcodes are shown in Table 2. Average number of pairwise nucleotide differences (k), nucleotide diversity (π) and haplotype diversity (Hd) varied for the species. Fu's Fs and Tajima's D were significant for a majority of the species (Table 2).
Figure 3. Pairwise distance divergence (%) (A) and barcode gap analysis (B) for mosquitoes from Punjab and Khyber Pakhtunkhwa as generated by ABGD [67] and by BOLD [59], respectively.
NN = nearest neighbor.
Table 1. K2P sequence divergence at the COI barcode region among the mosquito species with >2 specimens, among the four genera with two or more species, and in the family Culicidae.
| Distance class | n | Taxa | Comparisons | Min (%) | Mean (%) | Max (%) |
| Intraspecific | 1638 | 24 | 543530 | 0 | 0.04 | 2.4 |
| Congeners | 1529 | 4 | 121341 | 2.3 | 8.1 | 17.8 |
| Confamilial | 1644 | 1 | 685675 | 6.4 | 14.5 | 22.5 |
Table 2. Genetic diversity indices and neutrality tests (Fu's Fs and Tajima's D) in the mtCOI-5′ (barcode) sequences of 21 mosquito species from Pakistan.
| Species | n | S | k | π | h | Hd | Fu's Fs | Tajima's D |
| Ae. aegypti | 48 | 5 | 0.27 | 0.0004 | 5 | 0.22 | −3.95 | −1.90 |
| Ae. albopictus | 182 | 6 | 0.43 | 0.0010 | 7 | 0.38 | −3.88 | −1.22 |
| Ae. unilineatus | 7 | 7 | 2.87 | 0.0055 | 5 | 0.86 | −1.26 | −1.03 |
| Ae. w-albus | 9 | 7 | 1.89 | 0.0031 | 5 | 0.80 | −0.73 | −1.00 |
| Aedes MA01 | 9 | 13 | 4.83 | 0.0073 | 8 | 0.97 | −2.82 | 0.05 |
| Aedes MA02 | 4 | 6 | 3.00 | 0.0046 | 3 | 0.83 | 0.731 | −0.81 |
| Aedini 1 | 5 | 6 | 2.80 | 0.0043 | 4 | 0.90 | −0.445 | −0.19 |
| Ar. subalbatus | 54 | 3 | 0.27 | 0.0004 | 4 | 0.27 | −2.17 | −1.18 |
| An. culicifacies | 5 | 7 | 2.80 | 0.0042 | 5 | 1.00 | −2.37 | −1.16 |
| An. pulcherrimus | 8 | 2 | 0.50 | 0.0008 | 3 | 0.46 | −0.99 | −1.31 |
| An. peditaeniatus | 8 | 7 | 2.25 | 0.0034 | 6 | 0.93 | −2.32 | −0.79 |
| An. stephensi | 28 | 3 | 0.21 | 0.0005 | 4 | 0.21 | −3.27 | −1.73 |
| An. subpictus | 39 | 24 | 3.10 | 0.0052 | 17 | 0.87 | −8.14 | −1.84 |
| Co. pseudotaeniatus | 11 | 23 | 10.1 | 0.0154 | 6 | 0.89 | 1.59 | 0.72 |
| Cx. bitaeniorhynchus | 4 | 1 | 0.50 | 0.0008 | 2 | 0.50 | 0.17 | −0.61 |
| Cx. fuscocephala | 5 | 4 | 2.00 | 0.0030 | 4 | 0.90 | −1.01 | 0.27 |
| Cx. quinquefasciatus | 1018 | 23 | 0.09 | 0.0002 | 24 | 0.09 | −64.4 | −2.31 |
| Cx. tritaeniorhynchus | 89 | 33 | 2.78 | 0.0057 | 33 | 0.90 | −27.08 | −1.88 |
| Ma. uniformis | 4 | 4 | 2.33 | 0.0038 | 4 | 1.00 | −1.62 | 0.65 |
| Oc. pulcritarsis | 72 | 32 | 4.74 | 0.0076 | 37 | 0.96 | −25.58 | −1.04 |
| Ph. cogilli | 48 | 19 | 3.16 | 0.0074 | 18 | 0.89 | −7.36 | −0.86 |
n: number of sequences; S: number of polymorphic sites; k: average number of pairwise nucleotide differences; π: nucleotide diversity; h; number of haplotypes; Hd: haplotype diversity.
Fu's Fs: A negative value of FS is evidence for an excess number of alleles, as would be expected from a recent population expansion or from genetic hitchhiking. Statistical significance: Not significant, P>0.02.
Tajima's D: A negative Tajima's D signifies an excess of low frequency polymorphisms relative to expectation. Statistical significance: Not significant, P>0.10.
Species represented by <3 specimens or barcodes with <500 bp were not included in the analyses.
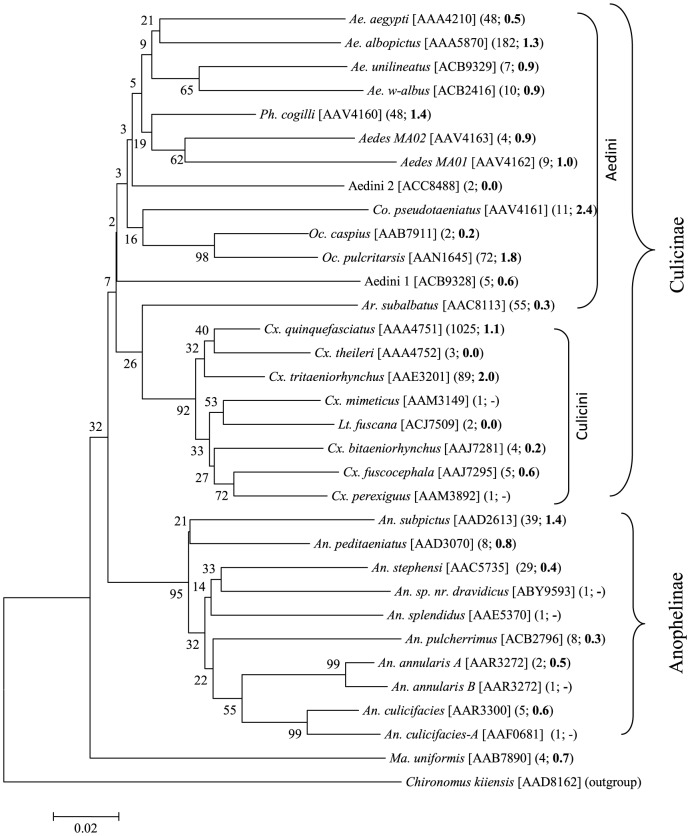
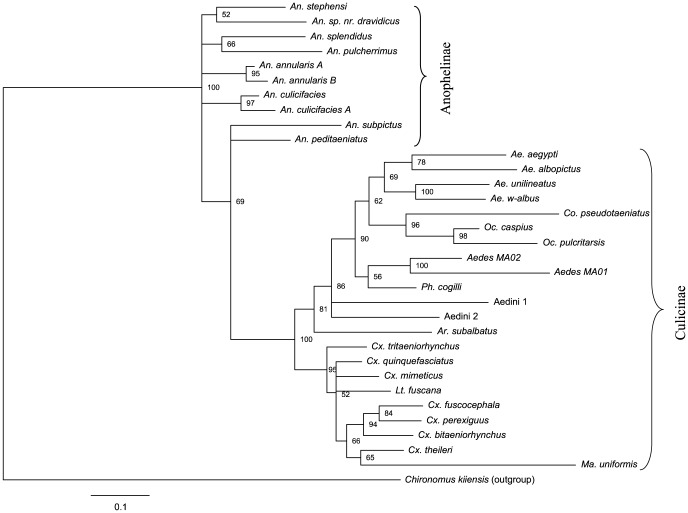
NJ analysis showed that the representatives of each species formed a monophyletic cluster (Fig. 4). The maximum intraspecific distance for Cx. quinquefasciatus (n = 1025) was 1.1%, while the distances for the two dengue vectors, Ae. aegypti (n = 48) and Ae. albopictus (n = 182) were 0.5% and 1.3%, respectively (Fig. 5). Among Anophelini, An. subpictus (n = 39) showed the highest intraspecific distance (1.4%). The only species with >2% intraspecific distance was Collessius pseudotaeniatus (2.4%). Species from the three mosquito tribes (Aedini, Anophelini, Culicini) mostly clustered with other members of their tribe. The cryptic species pair, An. annularis A and An. annularis B shared the same BIN, but their nodes were separated by a 99% bootstrap value in the NJ tree (Fig. 4). A phylogenetic tree of mosquito species estimated using Bayesian inference is presented in Fig. 5. The overall node pattern of the phylogenetic tree was similar to that of NJ tree, other than a close relationship between Aedini and Culicini was more evident and all the species branched with their respective subfamilies and tribes. The posterior probability values for all the nodes were greater than 50%.
Figure 4. NJ analysis of mosquitoes collected from Punjab and Khyber Pakhtunkhwa.
Bootstrap values (500 replicates) are shown above the branches. The scale bar shows K2P distances. Barcode Index Numbers (BINs) follow the species name in square brackets and the number of sequences analyzed and the intraspecific K2P distances (in bold) are included in parenthesis. Analyses were conducted in MEGA5.
Figure 5. Phylogenetic tree of mosquito species estimated using Bayesian inference and the codon partitioned analysis.
Posterior probability shown at nodes.
Global haplotype diversity and distributions of dengue vector species in Pakistan
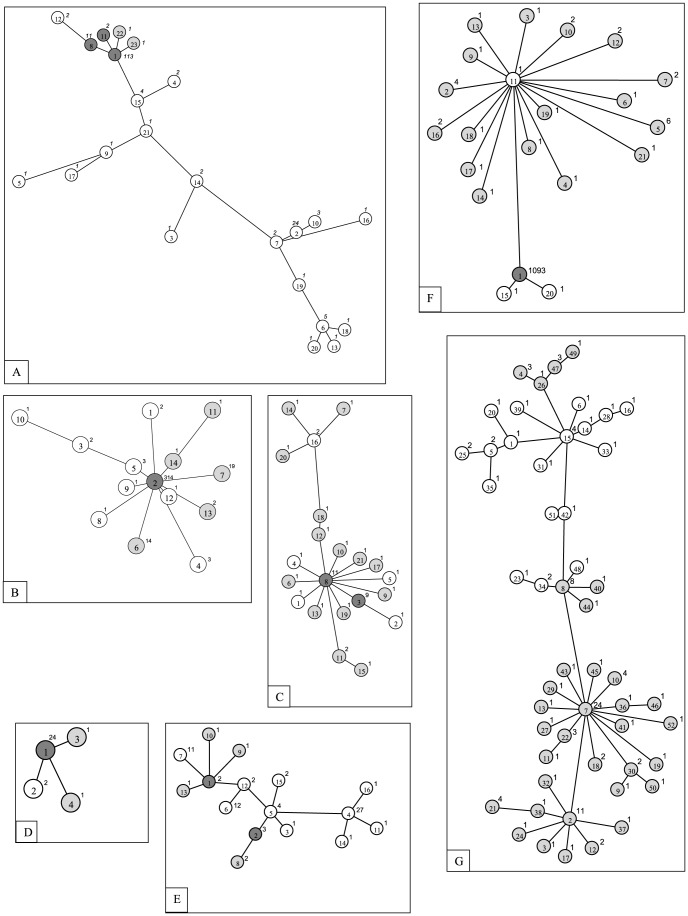
The sequence data for seven disease vector mosquitoes from Pakistan were placed in a broader perspective by including barcode results from other regions. When data (n = 182) from 29 nations was considered for Ae. aegypti, maximum sequence divergence reached 2.2% (mean = 0.5%),while the maximum distance for Ae. albopictus (n = 365) from 21 countries was 1.9% (mean = 0.3%). Twenty three haplotypes were detected in Ae. aegypti although one was dominant (62%), occuring in 20 countries including Pakistan (Fig. 6A). Eighteen of these 23 haplotypes were not detected in Pakistan, while two were only detected there (Fig. 6A). Ae. albopictus showed the presence of 14 haplotypes, one being in abundance (86%) and present in 16 countries (Fig. 6B). Eight of these 14 haplotypes were not detected in Pakistan, while five were only reported from there (Fig. 6B). The malaria vectors, An. subpictus (n = 41; max dist 2.2%) and An. stephensi (n = 28; max dist 0.4%) showed the presence of 21 and 4 haplotypes, respectively (Fig. 6C,D). The most common haplotype of An. stephensi was detected in Pakistan and South Africa (GU908046). There were 16 haplotypes for An. peditaeniatus (n = 72; max dist 1.8%); ten were detected exclusively in Thailand, four in Pakistan and the remaining two were shared between India and Pakistan (Fig. 6E). Barcodes of Cx. quinquefasciatus (n = 1125; max dist 1.2%) from 11 countries revealed 21 haplotypes, 17 of them were found solely in Pakistan, three were not detected in Pakistan, and one most frequent haplotype in Pakistan was also found in 10 other countries (Fig. 6F). Cx. tritaeniorhynchus from four countries (n = 113; max dist 2.3%) showed the presence of 52 barcode haplotypes and 34 were exclusively from Pakistan (Fig. 6G).
Figure 6. Barcode haplotype networks of vector mosquitoes from Pakistan.
Haplotype number and frequency is indicated inside and besides the corresponding circle, respectively. Haplotypes shared between Pakistan and other countries, found solely in Pakistan, and not found in Pakistan are indicated by dark grey, light grey, and blank circles, respectively. Haplotypes (in brackets) and their origin countries follow the species below (except for haplotypes exclusively from Pakistan indicated in light grey). A) Aedes aegypti: (1) Argentina, Australia, Bolivia, Brazil, Cambodia, Canada, Chile, France, French Polynesia, Gabon, Guinea, India, Laos, Pakistan, Russia, Thailand, Uganda, USA, Venezuela, Vietnam; (2) Brazil, Cambodia, Canada, Laos, Martinique, Thailand, USA; (3) South Africa; (4) Canada, USA; (5) (21) Cote d'Ivoire; (6) Australia, Bolivia, Martinique; (7) Martinique, Mexico; (8) Australia, Cambodia, Pakistan, Thailand; (9) Mexico; (10) UK, USA; (11) Bolivia, Pakistan; (12) India, Vietnam; (13) (18) (20) Bolivia; (14) Vietnam; (15) Cameroon, Cote d'Ivoire, Guinea; (16) Tanzania; (17) Australia; (19) Europa Island. B) Aedes albopictus: (1) Thailand; (2) Brazil, France, Germany, Greece, Italy, Japan, Lebanon, Madagascar, Pakistan, Re Union: La possession, Russia, Thailand, Turkey, USA, Hawaii (USA), Vietnam; (3)Romania; (4) Australia, Taiwan; (5) Germany; (8) Cambodia; (9) Madagascar; (10) India; (12) Vietnam. C) Anopheles subpictus: (1) (2) (4) (5) (16) India; (3) (8) India, Pakistan. D) Anopheles stephensi: (1) Pakistan, South Africa; (2) Thailand. E) Anopheles peditaeniatus: (1) (2) India, Pakistan; (3) India; (4) (5) (6) (7) (11) (12) (14) (15) (16) Thailand. F) Culex quinquefasciatus: (1) Brazil, China, India, Iran, Japan, Malaysia, Mexico, Pakistan, Thailand, Uganda, USA; (11) (20) Brazil; (15) Mexico. G) Culex tritaeniorhynchus: (1) (14) (23) (34) (25) (28) (33) (35) (42) (48) (51) Japan; (6) (16) (31) (39) China; (5) (15) China, Japan; (20) Thailand.
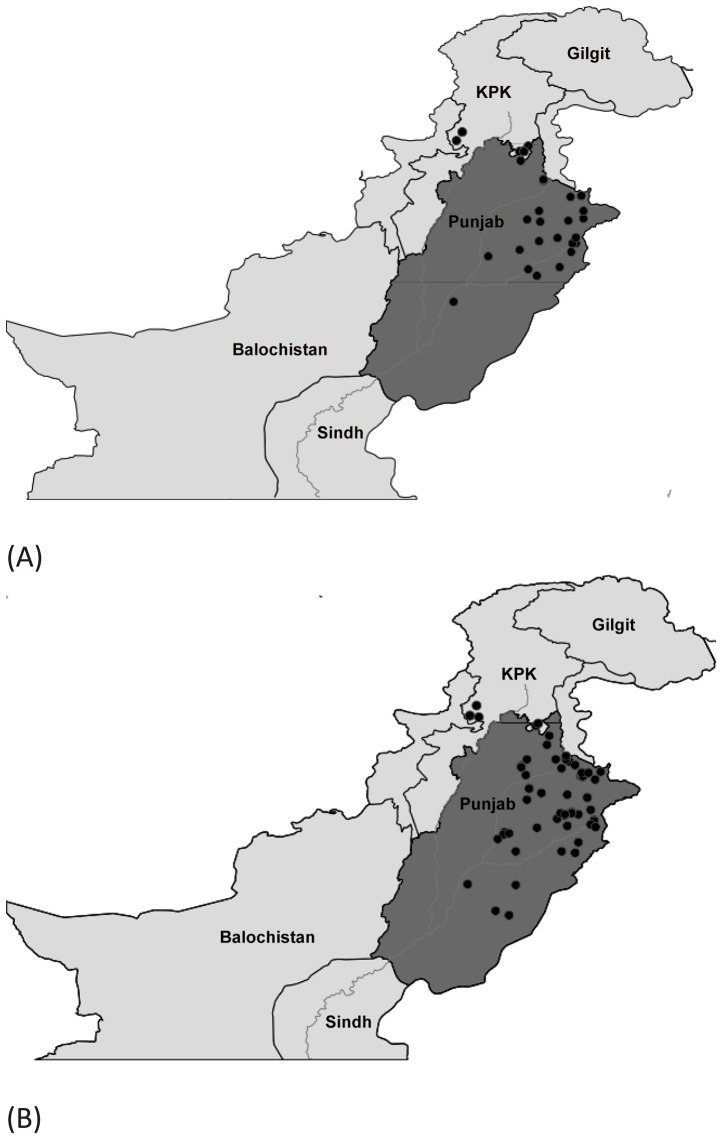
Both Ae. aegypti and Ae. albopictus were present in almost all the dengue affected districts of Punjab (Fig. 7A,B). Although Ae. albopictus was detected from more locations, both species were detected in the urban areas of central Punjab most impacted by dengue. Ae. aegypti was collected at sites ranging in elevation from 112 m–1004 m, while Ae. albopictus had a slightly narrower elevational range (110 m−672 m). Anopheles and Culex species were present in all the areas included in the study with Cx. quinquefasciatus and An. subpictus the most frequent members of their respective genera (distributional data not shown).
Figure 7. Map showing the distribution of Aedes (Stegomyia) aegypti (A) and Aedes (Stegomyia) albopictus (B) in the dengue-affected areas of Punjab, Pakistan.
Discussion
The mosquito fauna of South Asia is known to be diverse; 104 species have been reported from Pakistan and Bangladesh [4]. Local surveys have reported fewer taxa, including 43 species from Punjab [54], 30 species from the Changa Manga National Forest in central Punjab [40], and 21 species from the Swat valley in an adjoining province [7]. Cx. quinquefasciatus was the most abundant species in our study, mirroring results from earlier work in Swat [7], but prior studies in Punjab indicated the dominance of Cx. tritaeniorhynchus [40], [54]. Although Ae. albopictus was the dominant species of Aedes in Punjab, it was not detected at higher elevation sites (1100 m) in Swat [7]. Interestingly, Reisen et al. (1982) [40] found that Aedes lineatopennis was the most abundant species of Aedes, suggesting a shift in species composition through time. Although Ae. albopictus occurred at sites as high as 1200 m on La Reunion [78], it was limited to sites with an elevation less than 700 m in Pakistan. However, other species, such as Oc. pulcritarsis showed a much broader elevational range (111 m−2376 m).
Because of the difficulty of morphology-based identifications in mosquitoes, DNA-based approaches have gained increasing usage. The analysis of sequence variation in the internal transcribed spacer 2 (ITS2) revealed sibling species in both the Anopheles maculipennis [24] and Anopheles crucians [79] complexes. Atrie et al. (1999) [77] detected two sibling species (A and B) in the An. annularis complex through cytogenetic analysis, and subsequent work revealed their discrimination by ribosomal DNA [26], results extended by our analyses which revealed their 2.3% divergence at COI. Kumar et al. (2007) [31] found that DNA barcodes reliably identified 62 of 63 mosquito species from India; Ochlerotatus portonovoensis and Ochlerotatus wardi were the only two species which could not be discriminated. However, Kumar et al. (2013) [35] also concluded that Indian populations of Anopheles fluviatilis previously assigned to a complex of three species (S, T, U) were actually conspecific. Although there are rare cases of failure, the overall success in discrimination of species underscores the importance of constructing a DNA barcode reference library for all Culicidae.
The presence of endemics in South Asia [80], [81] reinforces the importance of developing regional barcode libraries to reveal overlooked species. The mosquito species reported in two major studies from India [31] and China [34] show just 50% overlap with the species assemblage in this study, making it clear that many more species await analysis. The completion of this task is complicated by the difficulty in identifying some taxa, especially those in the tribe Aedini. Species of Aedes are a particular challenge [15], with recurrent taxonomic revisions indicating the ongoing controversy [14]–[16]. This uncertainty in the application of names provides an incentive to consider alternate approaches for species discrimination based on the recognition of DNA sequence clusters. The possible effectiveness of this approach is signaled by the fact that the species examined in this study showed average intraspecific distances ranging from 0−2.4%, while distances between congeneric species varied from 2.3−17.8%. Wang et al. (2012) [34] reported similar levels of conspecific (0−1.67%) and congeneric (2.3−21.8%) divergences in their study of 122 mosquito species in China. The recently established BIN system has created a permanent registry for DNA barcode clusters [65], and the present study provides an opportunity to test the correspondence between the clusters recognized by it and the mosquito species recognized in our study through morphological analysis and examination of the NJ and phylogenetic trees and NN distances. This check indicated near perfect congruence as the 32 species were assigned to 31 BINs; the sole merger involved the sibling species, An. annularis A and An. annularis B. Similarly, all 43 mosquito species analyzed by Kumar et al. (2007) [31] with adequate sequence data for a BIN assignment were placed in a unique BIN.
The importance of sequence analysis is clear in situations where one member of sibling species complex is a vector and the other is not. For example, An. annularis A transmits malaria in some parts of India, but An. annularis B does not [77]. Aside from enabling the discrimination of sibling taxa, DNA barcoding can reveal variation in single species which may have implications for disease transmission [82]. Genetic diversity indices showed a higher genetic divergence in some species and not in others. For example, k and π values were higher in Cx. tritaeniorhynchus and lower in Cx. quinquefasciatus. Intraspecific diversity also varied in the species of Aedes and Anopheles. A recent study on Cx. quinquefasciatus from Malaysia [20] has reported a low COI diversity for this species, and our results are in agreement with that study. But for the same species from India, Sharma et al. (2010) [83] have reported a high diversity in 16S rRNA. This incongruence may support the ongoing debate on mitochondrial and nuclear discordance in animals [84]. Prior work has revealed substantial genetic diversities in both Ae. aegypti and Ae. albopictus, providing important clues on population relationships and origins [85], [86] and insights into their role in the transmission of dengue [41]. Both species showed considerable barcode variation within Pakistan, but Ae. albopictus was less variable than Ae. aegypti when analysis considered the entire range of each species, a result congruent with Mousson et al. (2005) [43]. However, we did not find any evidence of genetic differentiation between specimens from rural and urban habitats or between larval and adult stages although the latter test was weak because larvae comprised just 7% of the specimens (122/1684). Our study revealed a single globally dominant COI haplotype in Ae. aegypti, while Moore et al. (2013) [36] found two major ND4 haplotypes in its African populations. Using exon primed intron crossing and microsatellite markers, Olanratmanee et al. (2013) [87] reported two genetic clusters in one region of Thailand and considered the implications of this diversity for a dengue suppression strategy. Mousson et al. (2005) [41] found little sequence variation in three mitochondrial genes for Ae. albopictus from 15 countries on five continents. Our analysis also revealed one major COI lineage in Ae. albopictus, widely distributed in 16 countries.
The results of our survey indicated that Ae. albopictus was more widely distributed and commoner than Ae. aegypti in Punjab. Akhtar et al. (2012) [57] found that larvae of Ae. aegypti predominated (65%) in collections from water-pots inside houses in Lahore during 2011. Although, the current dominance of Ae. albopictus in Pakistan supports a trend towards expansion of Ae. albopictus and a decline of Ae. aegypti in many areas of the world [88], [89], effect of sampling method on the variation in results cannot be ruled out.
In conclusion, this study has provided baseline data on composition and genetic diversity in the mosquito fauna of Pakistan, information that should be useful in tracking mosquito-borne diseases. The prevalence of Ae. albopictus in urban areas may suggest its importance in the spread of dengue. Because DNA barcoding can resolve cryptic mosquito species and identify their immature stages [90], it provides a valuable tool for large-scale vector identification and disease surveillance programs.
Supporting Information
NJ analysis of mosquitoes collected from dengue-affected areas of Punjab and adjoining Khyber Pakhtunkhwa. Bootstrap values (500 replicates) are shown above the branches. Species names are preceded by the specimen Process IDs (Barcode of Life Data Systems). The scale bar shows K2P distances. Chironomus kiiensis (Diptera: Chironomidae) was included as an outgroup. Analyses were conducted in MEGA5.
(PS)
GPS coordinates of mosquito collection sites in Pakistan.
(XLS)
Acknowledgments
We thank colleagues at the CCDB for aid with sequence analysis, and staff employed with the DNA barcoding project at NIBGE, Faisalabad for their diligence in collecting specimens.
Funding Statement
This research was enabled by grant 106106-001, Engaging Developing Nations in the International Barcode of Life (iBOL) Project, from IDRC. Sequence analysis was enabled by a grant from the Government of Canada through Genome Canada and the Ontario Genomics Institute in support of iBOL. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pant C (1987) Vector borne diseases of man and their socio-economic impact. Insect Sci Applic 8: 655–664. [Google Scholar]
- 2. Chan YC, Salahuddin NI, Khan J, Tan HC, Seah CL, et al. (1995) Dengue haemorrhagic fever outbreak in Karachi, Pakistan, 1994. Trans R Soc Trop Med Hyg 89: 619–620. [DOI] [PubMed] [Google Scholar]
- 3.Stark K, Schoneberg I (2012) Increase in malaria cases imported from Pakistan to Germany in 2012. Euro Surveill 17 : pii = 20320. [DOI] [PubMed] [Google Scholar]
- 4. Khan MA (1971) The mosquitoes of Pakistan. I. A checklist. Mosq Syst Newsletter 3: 147–159. [Google Scholar]
- 5. Shakoor MT, Ayub S, Ayub Z (2012) Dengue fever: Pakistan's worst nightmare. WHO South-East Asia J Public Health 1: 229–231. [DOI] [PubMed] [Google Scholar]
- 6. Mukhtar M, Tahir Z, Baloch TM, Mansoor F, Kamran J (2011) Entomological investigations of dengue vectors in epidemic prone districts of Pakistan during 2006−2010. Dengue Bull 35: 99–115. [Google Scholar]
- 7. Ilahi I, Suleman M (2013) Species composition and relative abundance of mosquitoes in Swat, Pakistan. Intrl J Innov Appl Studies 2: 454–463. [Google Scholar]
- 8. Rasheed SB, Boots M, Frantz AC, Butlin RK (2013) Population structure of the mosquito Aedes aegypti (Stegomyia aegypti) in Pakistan. Med Vet Entomol 27: 430–440. [DOI] [PubMed] [Google Scholar]
- 9. Otranto D, Capelli G, Genchi C (2009) Changing distribution patterns of canine vector borne diseases in Italy: leishmaniosis vs. dirofilariosis. Parasite Vector 2 (suppl 1)S2: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Munstermann LE, Conn JE (1997) Systematics of mosquito disease vectors (Diptera: Culicidae): impact of molecular biology and cladistic analysis. Annu Rev Entomol 42: 351–369. [DOI] [PubMed] [Google Scholar]
- 11. Zavortink TJ (1990) Classical taxonomy of mosquitoes - a memorial to John N. Belkin. J Am Mosq Control Assoc 6: 593–600. [PubMed] [Google Scholar]
- 12. Krzywinski J, Besansky NJ (2003) Molecular systematics of Anopheles: from subgenera to subpopulations. Annu Rev Entomol 48: 111–139. [DOI] [PubMed] [Google Scholar]
- 13. Walton C, Sharpe RG, Pritchard SJ, Thelwell NJ, Butlin RK (2008) Molecular identification of mosquito species. Biol J Linn Soc 68: 241–256. [Google Scholar]
- 14. Reinert JF (2000) New classification for the composite genus Aedes (Diptera: Culicidae: Aedini), elevation of subgenus Ochlerotatus to generic rank, reclassification of the other subgenera, and notes on certain subgenera and species. J Am Mosq Control Assoc 16: 175–188. [PubMed] [Google Scholar]
- 15. Savage HM, Strickman D (2004) The genus and subgenus categories within Culicidae and placement of Ochlerotatus as a subgenus of Aedes . J Am Mosq Control Assoc 20: 208–214. [PubMed] [Google Scholar]
- 16. Reinert JF, Harbach RE, Kitching IJ (2009) Phylogeny and classification of tribe Aedini (Diptera: Culicidae). Zool J Linnean Soc 157: 700–794. [Google Scholar]
- 17. Manonmani A, Townson H, Adeniran T, Jambulingam P, Sahu S, et al. (2001) rDNA-ITS2 polymerase chain reaction assay for the sibling species of Anopheles fluviatilis . Acta Tropica 78: 3–9. [DOI] [PubMed] [Google Scholar]
- 18. Singh OP, Chandra D, Nanda N, Raghavendra K, Sunil S, et al. (2004) Differentiation of members of Anopheles fluviatilis species complex by an allele-specific polymerase chain reaction based on 28S ribosomal DNA sequences. Am J Trop Med Hyg 70: 27–32. [PubMed] [Google Scholar]
- 19. Kang D, Sim C (2013) Identification of Culex complex species using SNP markers based on high-resolution melting analysis. Mol Ecol Res 13: 369–376. [DOI] [PubMed] [Google Scholar]
- 20.Low VL, Lim PE, Chen CD, Lim YAL, Tan TK, et al. (2013) Mitochondrial DNA analysis reveal low genetic diversity in Culex quinquefasciatus from residential areas of Malaysia. Med Vet Entomol. doi: 10.1111/mve.12022 [DOI] [PubMed]
- 21. Pfeiler E, Flores-Lopez CA, Mada-Velez JG, Escalante-Verdugo J, Markow TA (2013) Genetic diversity and population genetics of mosquitoes (Diptera: Culicidae: Culex spp.) from the Sonoran Desert of North America. Sci World J 2013: 724609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shepard JJ, Andreadis TG, Vossbrinck CR (2006) Molecular phylogeny and evolutionary relationships among mosquitoes (Diptera: Culicidae) from the northeastern United States based on small subunit ribosomal DNA (18S ribosomal DNA) sequences. J Med Entomol 43: 443–454. [DOI] [PubMed] [Google Scholar]
- 23. Sharma AK, Chandel K, Tyagi V, Mendki MJ, Tikar SN, et al. (2013) Molecular phylogeny and evolutionary relationship among four mosquito (Diptera: Culicidae) species from India using PCR-RFLP. J Mosquito Res 3: 58–64. [Google Scholar]
- 24. Proft J, Maier WA, Kampen H (1999) Identification of six sibling species of the Anopheles maculipennis complex (Diptera: Culicidae) by a polymerase chain reaction assay. Parasitol Res 85: 837–843. [DOI] [PubMed] [Google Scholar]
- 25. Chen B, Harbach RE, Butlin RK (2002) Molecular and morphological studies on the Anopheles minimus group of mosquitoes in southern China: taxonomic review, distribution and malaria vector status. Med Vet Entomol 16: 253–265. [DOI] [PubMed] [Google Scholar]
- 26. Alam MT, Das MK, Dev V, Ansari MA, Sharma YD (2007) Identification of two cryptic species in the Anopheles (Cellia) annularis complex using ribosomal DNA PCR-RFLP. Parasitol Res 100: 943–948. [DOI] [PubMed] [Google Scholar]
- 27. Surendran SN, Gajapathy K, Kumaran V, Tharmatha T, Jude PJ, et al. (2011) Molecular evidence for the presence of malaria vector species A of the Anopheles annularis complex in Sri Lanka. Parasite Vector 4: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wan Q-H, Wu H, Fujihara T, Fang S-G (2004) Which genetic marker for which conservation genetics issue? Electrophoresis 25: 2165–2176. [DOI] [PubMed] [Google Scholar]
- 29. Galtier N, Nabholz B, Glemin S, Hurst GDD (2009) Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol 18: 4541–4550. [DOI] [PubMed] [Google Scholar]
- 30. Hebert PDN, Cywinska A, Ball S, deWaard J (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumar NP, Rajavel AR, Natarajan R, Jambulingam P (2007) DNA barcodes can distinguish species of Indian mosquitoes (Diptera:Culicidae). J Med Entomol 44: 1–7. [DOI] [PubMed] [Google Scholar]
- 32. Cywinska A, Hunter FF, Hebert PDN (2006) Identifying Canadian mosquito species through DNA barcodes. Med Vet Entomol 20: 413–424. [DOI] [PubMed] [Google Scholar]
- 33. Hemmerter S, Slapeta J, Beebe NW (2009) Resolving genetic diversity in Australian Culex mosquitoes: incongruence between the mitochondrial cytochrome c oxidase I and nuclear acetylcholine esterase 2. Mol Phylogenet Evol 50: 317–325. [DOI] [PubMed] [Google Scholar]
- 34. Wang G, Li C, Guo X, Xing D, Dong Y, et al. (2012) Identifying the main mosquito species in China based on DNA barcoding. PLoS ONE 7: e47051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kumar NP, Krishnamoorthy N, Sahu SS, Rajavel AR, Sabesan S, et al. (2013) DNA barcodes indicate members of the Anopheles fluviatilis (Diptera: Culicidae) species complex to be conspecific in India. Mol Ecol Res 13: 354–361. [DOI] [PubMed] [Google Scholar]
- 36. Moore M, Sylla M, Goss L, Burugu MW, Sang R, et al. (2013) Dual African origin of global Aedes aegypti s.l. populations revealed by mitochondrial DNA. PLoS Negl Trop Dis 7: e2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Werblow A, Bolius S, Dorresteijn AW, Melaun C, Klimpel S (2013) Diversity of Culex torrentium Martini, 1925 - a potential vector of arboviruses and filaria in Europe. Parasitol Res 112: 2495–2501. [DOI] [PubMed] [Google Scholar]
- 38. Coetzee M, Craig M, le Sueur D (2000) Distribution of African malaria mosquitoes belonging to the Anopheles gambiae complex. Parasitol Today 16: 74–77. [DOI] [PubMed] [Google Scholar]
- 39.Bass C, Nikou D, Vontas J, Donnelly MJ, Williamson MS, et al.. (2010) The vector population monitoring tool (VPMT): high-throughput DNA-based diagnostics for the monitoring of mosquito vector populations. Malaria Res Treat doi:10.4061/2010/190434 [DOI] [PMC free article] [PubMed]
- 40. Reisen WK, Hayes CG, Azra K, Niaz S, Mahmood F, et al. (1982) West Nile virus in Pakistan. II. Entomological studies at Changa Manga National Forest, Punjab province. Trans R Soc Trop Med Hyg 76: 437–448. [DOI] [PubMed] [Google Scholar]
- 41. Mousson L, Dauga C, Garrigues T, Schaffner F, Vazeille M, et al. (2005) Phylogeography of Aedes (Stegomyia) aegypti (L.) and Aedes (Stegomyia) albopictus (Skuse) (Diptera: Culicidae) based on mitochondrial DNA variations. Genet Res Camb 86: 1–11. [DOI] [PubMed] [Google Scholar]
- 42. Fansiri T, Fontaine A, Diancourt L, Caro V, Thaisomboonsuk B, et al. (2013) Genetic mapping of specific interactions between Aedes aegypti mosquitoes and dengue viruses. PLoS Genet 9: e1003621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thongsripong P, Green A, Kittayapong P, Kapan D, Wilcox B, et al. (2013) Mosquito vector diversity across habitats in central Thailand endemic for dengue and other arthropod-borne diseases. PLoS Negl Trop Dis 7: e2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Angel B, Joshi V (2008) Distribution and seasonality of vertically transmitted dengue viruses in Aedes mosquitoes in arid and semi-arid areas of Rajasthan, India. J Vector Borne Dis 45: 56–59. [PubMed] [Google Scholar]
- 45. Pollitt LC, Mackinnon M, Mideo N, Read AF (2013) Mosquito transmission, growth phenotypes and the virulence of malaria parasites. Malaria J 12: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anderson JR, Schneider JR, Grimstad PR, Severson DW (2005) Quantitative genetics of vector competence for La Crosse virus and body size in Ochlerotatus hendersoni and Ochlerotatus triseriatus interspecific hybrids. Genetics 169: 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mwangangi JM, Mbogo CM, Orindi BO, Muturi EJ, Midega JT, et al. (2013) Shifts in malaria vector species composition and transmission dynamics along the Kenyan coast over the past 20 years. Malaria J 12: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. LaDeau SL, Leisnham PT, Biehler D, Bodner D (2013) Higher mosquito production in low-income neighborhoods of Baltimore and Washington, DC: understanding ecological drivers and mosquito-borne disease risk in temperate cities. Int J Environ Res Public Health 10: 1505–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Engler O, Savini G, Papa A, Figuerola J, Groschup MH, et al. (2013) European surveillance for West Nile virus in mosquito populations. Int J Environ Res Public Health 10: 4869–4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ishak H, Miyagi I, Toma T, Kamimura K (1997) Breeding habitats of Aedes aegypti (L) and Aedes albopictus (Skuse) in villages of Barru, South Sulawesi, Indonesia. Southeast Asian J Trop Med Public Health 28: 844–850. [PubMed] [Google Scholar]
- 51. Braks MAH, Honorio NA, Lourenco-de-Oliveira R, Juliano SA, Lounibos P (2003) Convergent habitat segregation of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) in Southeastern Brazil and Florida. J Med Entomol 40: 785–794. [DOI] [PubMed] [Google Scholar]
- 52. Lambrechts L, Scott TW, Gubler DJ (2010) Consequences of the expanding global distribution of Aedes albopictus for dengue virus transmission. PLoS Negl Trop Dis 4: e646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Benedict MQ, Levine RS, Hawley WA, Lounibos LP (2007) Spread of the tiger: global risk of invasion by the mosquito Aedes albopictus . Vector Borne Zoonotic Dis 7: 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Reisen WK (1978) A quantitative mosquito survey of 7 villages in Punjab province, Pakistan with notes on bionomics, sampling methodology and the effects of insecticides. Southeast Asian J Trop Med Public Health 9: 587–601. [PubMed] [Google Scholar]
- 55. Amerasinghe FP, Mukhtar M, Herrel N (2002) Keys to the Anopheline mosquitoes (Diptera: Culicidae) of Pakistan. J Med Entomol 39: 28–35. [DOI] [PubMed] [Google Scholar]
- 56. Naeem-Ullah U, Akram W, Suhail A, Rana SA (2010) Grouping of different mosquito species on the bases of larval habitats. Pak J Agri Sci 47: 124–131. [Google Scholar]
- 57. Akhtar MS, Aihetasham A, Saeed M, Abbass G (2012) Aedes survey following a dengue outbreak in Lahore, Pakistan, 2011. Dengue Bull 36: 87–93. [Google Scholar]
- 58. Harbach RE (1988) The mosquitoes of the subgenus Culex in Southwestern Asia and Egypt (Diptera: Culicidae). Contrib Am Entomol Inst 24: 1–246. [Google Scholar]
- 59. Ratnasingham S, Hebert PDN (2007) BOLD: the Barcode of Life Data System (www.barcodinglife.org). Mol Ecol Notes. 7: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Porco D, Rougerie R, Deharveng L, Hebert P (2010) Coupling non-destructive DNA extraction and voucher retrieval for small soft-bodied arthropods in a high throughput context: the example of Collembola. Mol Ecol Res 10: 942–945. [DOI] [PubMed] [Google Scholar]
- 61. Ivanova NV, deWaard JR, Hebert PDN (2006) An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes 6: 998–1002. [Google Scholar]
- 62. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hebert PDN, Ratnasingham S, deWaard J (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergence, among closely related species. Proc R Soc Lond B (Suppl) 270: S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Strutzenberger P, Brehm G, Fiedler K (2011) DNA barcoding-based species delimitation increases species count of Eois (Geometridae) moths in a well-studied tropical mountain forest by up to 50%. Insect Sci 18: 349–362. [Google Scholar]
- 65. Ratnasingham S, Hebert PDN (2013) A DNA-based registry for all animal species: the Barcode Index Number (BIN) System. PLoS ONE 8: e66213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Puillandre N, Lambert A, Brouillet S, Achaz G (2012) ABGD, automated barcode gap discovery for primary species delimitation. Mol Ecol 21: 1864–1877. [DOI] [PubMed] [Google Scholar]
- 68. Meyer CP, Paulay G (2005) DNA barcoding: error rates based on comprehensive sampling. PLoS Biol 3: e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fu Y (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 72. Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 73. Meier R, Shiyang K, Vaidya G, Ng KPL (2006) DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol 55: 715–728. [DOI] [PubMed] [Google Scholar]
- 74. Ronquist F, Teslenko M, van der Mark P (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10: 564–567. [DOI] [PubMed] [Google Scholar]
- 76. Teacher AGF, Griffiths DJ (2011) HapStar: automated haplotype network layout and visualization. Mol Ecol Res 11: 151–153. [DOI] [PubMed] [Google Scholar]
- 77. Atrie B, Subbarao SK, Pillai MKK, Rao SRV, Sharma VP (1999) Population cytogenetic evidence for sibling species in Anopheles annularis (Diptera: Culicidae). Ann Entomol Soc Am 92: 243–249. [Google Scholar]
- 78. Delatte H, Dehecq JS, Thiria J, Domerg C, Paupy C, et al. (2008) Geographic distribution and developmental sites of Aedes albopictus (Diptera: Culicidae) during a Chikungunya epidemic event. Vector Borne Zoonotic Dis 8: 25–34. [DOI] [PubMed] [Google Scholar]
- 79. Wilkerson RC, Reinert JF, Li Cong (2004) Ribosomal DNA ITS2 sequences differentiate six species in the Anopheles crucians complex (Diptera: Culicidae). . J. Med. Entomol. 41: 392–201. [DOI] [PubMed] [Google Scholar]
- 80. Bossuyt F, Meegaskumbura M, Beenaerts N, Gower DJ, Pethiyagoda R, et al. (2004) Local endemism within the Western Ghats - Sri Lanka biodiversity hotspot. Science 306: 479–481. [DOI] [PubMed] [Google Scholar]
- 81. Ashfaq M, Akhtar S, Khan AM, Adamowicz SJ, Hebert PDN (2013) DNA barcode analysis of butterfly species from Pakistan points towards regional endemism. Mol Ecol Res 13: 832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Beebe NW, Whelan PI, van den Hurk A, Ritchie S, Cooper RD (2005) Genetic diversity of the dengue vector Aedes aegypti in Australia and implications for future surveillance and mainland incursion monitoring. Commun Dis Intell Q Rep 29: 299–304. [DOI] [PubMed] [Google Scholar]
- 83. Sharma AK, Mendki MJ, Tikar SN, Kulkarni G, Veer V, et al. (2010) Molecular phylogenetic study of Culex quinquefasciatus mosquito from different geographical regions of India using 16S rRNA gene sequences. Acta Tropica 116: 89–94. [DOI] [PubMed] [Google Scholar]
- 84. Toews DPL, Brelsford A (2012) The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol 21: 3907–3930. [DOI] [PubMed] [Google Scholar]
- 85. Kamgang B, Brengues C, Fontenille D, Njiokou F, Simard F, et al. (2011) Genetic structure of the tiger mosquito, Aedes albopictus, in Cameroon (Central Africa). PLoS ONE 6: e20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mendes dos Santos JM, Fraga EC, Maia JF, Tadei WP (2011) Genetic diversity in dengue mosquito, Aedes aegypti (Diptera: Culicidae) from Amazon region: comparative analysis with isozymes and RAPD loci. Open Trop Med J 4: 11–20. [Google Scholar]
- 87. Olanratmanee P, Kittayapong P, Chansang C, Hoffmann AA, Weeks AR, et al. (2013) Population genetic structure of Aedes (Stegomyia) aegypti (L.) at a micro-spatial scale in Thailand: implications for a dengue suppression strategy. PLoS Negl Trop Dis 7: e1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bagny L, Delatte H, Quilici S, Fontenille D (2009) Progressive decrease in Aedes aegypti distribution in Reunion Island since the 1900s. J Med Entomol 46: 1541–1545. [DOI] [PubMed] [Google Scholar]
- 89. Raharimalala FN, Ravaomanarivo LH, Ravelonandro P, Rafarasoa LS, Zouache K, et al. (2012) Biogeography of the two major arbovirus mosquito vectors, Aedes aegypti and Aedes albopictus (Diptera, Culicidae), in Madagascar. Parasite Vector 5: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dhananjeyan KJ, Paramasivan R, Tewar SC, Rajendran R, Thenmozhi V, et al. (2010) Molecular identification of mosquito vectors using genomic DNA isolated from eggshells, larval and pupal exuvium. Trop Biomed 27: 47–53. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NJ analysis of mosquitoes collected from dengue-affected areas of Punjab and adjoining Khyber Pakhtunkhwa. Bootstrap values (500 replicates) are shown above the branches. Species names are preceded by the specimen Process IDs (Barcode of Life Data Systems). The scale bar shows K2P distances. Chironomus kiiensis (Diptera: Chironomidae) was included as an outgroup. Analyses were conducted in MEGA5.
(PS)
GPS coordinates of mosquito collection sites in Pakistan.
(XLS)