Abstract
The identification of full length transcripts entirely from short-read RNA sequencing data (RNA-seq) remains a challenge in genome annotation pipelines. Here we describe an automated pipeline for genome annotation that integrates RNA-seq and gene-boundary data sets, which we call generalized RNA integration tool, or GRIT. By applying GRIT to Drosophila melanogaster short-read RNA-seq, cap analysis of gene expression (CAGE) and poly(A)-site-seq data collected for the modENCODE project, we recover the vast majority of previously annotated transcripts and double the total number of transcripts cataloged. We find that 20% of protein coding genes encode multiple protein-localization signals, and that, in 20 day old adult fly heads, genes with multiple poly-adenylation sites are more common than genes with alternate splicing or alternate promoters. When compared to the most widely used transcript assembly tools, GRIT recovers a larger fraction of annotated transcripts at higher precision. GRIT will enable the automated generation of high-quality genome annotations without necessitating extensive manual annotation.
High-throughput sequencing of cDNAs (RNA-seq) yields quantitative information about gene expression, alternative splicing, RNA editing, poly-adenylation sites and other phenomena 1,2,3. The prospect of using RNA-seq data to assemble reads into models of genes and transcripts has motivated the development of algorithms and software 4,5,6,7,8. De novo assembly methods, such as Trinity 5, Oases 7 and Trans-Abyss 8, assemble reads to construct transcript sequences, which are then mapped to a reference genome. Genome-guided approaches, such as Cufflinks 4 and Scripture 6, use reads that are aligned to a reference genome to identify transcript models.
The impact of RNA-seq data on genome annotation has been most substantial for organisms with minimal cDNA resources. For instance, RNA-seq data and Cufflinks were used to produce a de novo annotation for the sea urchin (Strongylocentrotus Purpuratus) but to prevent the inclusion of transcript fragments this study incorporated a stringent filtering system that removed transcript models that lacked ORFs longer than 500aa or that didn’t encode a known protein 9. In contrast, in organisms where substantial annotation efforts are ongoing (e.g., human 10, fruitfly 11, zebrafish 12 and worms 13) the impact of RNA-seq data has largely been by the manual incorporation of elements including new transcription start sites, splice junctions, and poly-adenylation sites. GRIT is designed to assemble a high quality experimentally-driven genome annotation using a reference genome and short read sequencing data, allowing it to be useful for the study both of model and non-model organisms.
It is not surprising that genome annotation has primarily remained in the domain of manual annotation and full-insert cDNA sequencing, because RNA-seq reads are too short to cover full transcripts, typically providing information only about three or four exons at a time 14. This means that it is not always possible to positively identify alternate transcript isoforms, even as the read depth approaches infinity. Furthermore, biases in the RNA-seq assay make positive identification of novel transcript boundaries difficult 15,16,1,17. Other genome annotation tools attempt to circumvent these problems by placing additional restrictions on the space of discoverable transcripts. For instance, Cufflinks only permits the minimal set of transcripts needed to explain the splice junctions, over-simplifying complex loci like Down syndrome cell adhesion molecule 1 (Dscam1) of D. melanogaster. Trinity always extends transcript contigs to the last base, disallowing nested promoters and nested poly(A) sites. As we show, these restrictions can produce annotation sets that are in direct contradiction to observed data from complementary assays.
We use a sparse statistical model amenable to modern optimization techniques, combined with the integration of gene boundary data to analyze RNA-seq data. In principle our approach allows for the construction of any transcript models that can be built by Cufflinks, Trinity, Scripture, Oases or Trans-Abyss, although our requirement that every transcript model be supported by experimental evidence can make GRIT more restrictive in practice. For the purposes of benchmarking, we have used a subset of the modENCODE dataset (1.67B bp, Supplementary Table 1) to compare the performance of GRIT to the most widely used transcript-level RNA-seq analysis tools. GRIT has also been applied to the full set of modENCODE RNA data (over 1 terabases of sequence data from CAGE, rapid amplification of cDNA ends (RACE), expressed sequence tags (EST), cDNA, 454, stranded paired-end RNA-seq and poly(A)-site-seq experiments) to generate a data-driven annotation with unprecedented detail of gene and transcript models for the fruit fly. The full-length transcript models have revealed a number of discoveries, including the findings that >20% of D. melanogaster protein-coding genes encode multiple localization signals and that alternative polyadenylation is more common than alternative splicing in neuronal tissue. These and related insights reported here and in a companion manuscript 18 were not obtainable with other analysis tools, and underscore the importance of integrating multiple types of assays when interpreting RNA sequencing data.
Results
GRIT
A brief overview of the GRIT method is outlined below; for details see the online methods.
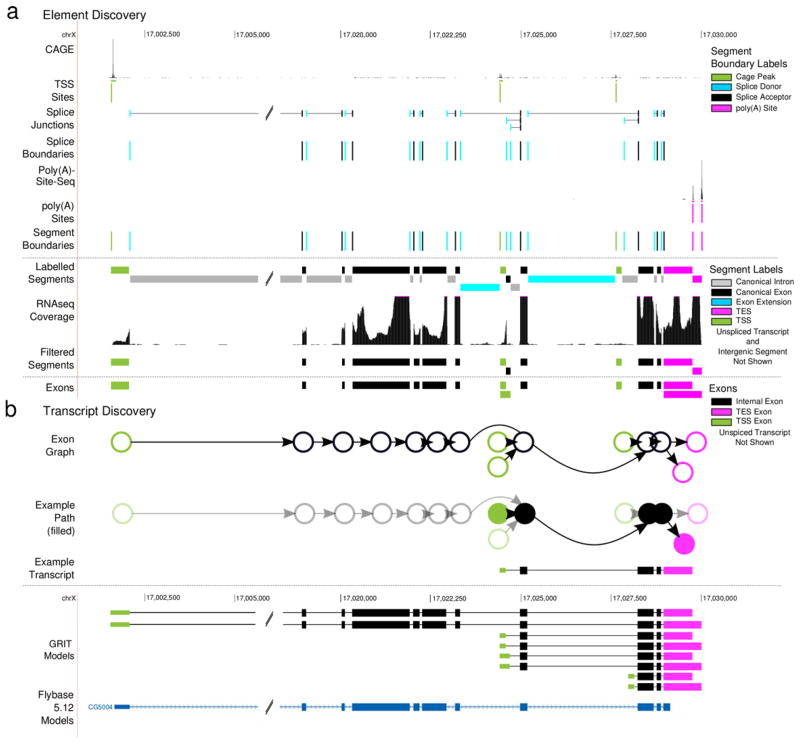
GRIT makes few assumptions about the structure of transcripts; defining them as sets of genomic regions that begin at a transcription start site (TSS), optionally extend through one or more exons connected by splice junctions, and end with a transcription end site (TES). This implies four distinct element types: TSS exons, TES exons, internal exons, and single exon transcripts. TSS exons begin with an experimentally detected promoter (e.g. via the CAGE or RACE assays or 5′ EST sequencing 19), and end with a splice donor site. Similarly, TES exons begin with a splice acceptor site and end with an observed TES (e.g., a poly(A) site). Internal exons begin and end with a verified splice site, and single exon transcripts begin with a TSS and end with a TES. GRIT uses both canonical and non-canonical splice sites.
We identify elements by segmenting the genome into non-overlapping segments with attached labels that describe their segment boundary (Fig. 1a). After removing low coverage segments, groups of adjacent segments are combined and labeled based upon their segment boundaries. For instance, an internal exon’s 5′ boundary is a splice acceptor, and its 3′ boundary is a splice donor. TSS exons’ 5′ boundaries are TSS sites, and their 3′ boundaries are splice donors. Similarly, the 3′ end of a TES exon is a TES, and the 5′ end is a splice acceptor.
Figure 1. Element discovery overview.
(a) Exon discovery. For each gene segment we identify CAGE peaks; segment the gene region using the CAGE peaks, splice boundaries and poly(A) sites; label the segments based upon their boundaries; filter intron segments with low RNA-seq coverage; and build labeled exons from adjacent segments. (b) Transcript discovery. For each gene, we construct a graph where each node is an exon discovered in (b), and each edge is a junction. Then, each candidate transcript is identified with a single path through this directed graph that begins with TSS node, and ends with a TES node.
After the exons are identified, we assemble transcript models. We define the set of candidate transcripts as the union of single exon transcripts and transcripts that begin with a TSS exon, optionally contain splice-junction connected internal exons, and end with a TES exon (Fig. 1b). GRIT differs from other methods, e.g. Cufflinks, in that it considers all possible paths subject to this restriction – rather than some minimal set of covering paths – allowing GRIT to correctly build transcripts in very complex loci like Dscam1 (Supplementary Fig. 1a).
After identifying the possible set of candidate transcripts, we estimate their relative concentrations in the sample of interest, i.e., their expression. This is challenging because reads can not necessarily be assigned to a single transcript. Thus, we first identify transcripts by the non-overlapping exon segments. Then, we group reads into the set of non-overlapping exon segments that they overlap, which we refer to as a “bin” (Supplementary Fig. 2). We define Yi as the number of reads of bin i.
The number of reads that are of a particular bin, when combined with information about the expected distribution of fragments from a particular transcript, provides information about transcript frequencies. Formally, we define an entry Xij in the design matrix X to be the probability of sampling bin j given that the read originated from transcript j. The maximum likelihood estimate of the transcript frequencies, t⃗, is then the vector t̂ that maximizes the log-likelihood, lhd(Y;t⃗) = ΣiYilog[ΣjXijtj], subject to the constraints tj ≥ 0 and Σj tj = 1. Whenever no row of Xij can be constructed from a positively weighted sum of other rows, the statistical model is said to be identifiable given the data, and we can use a convex optimization algorithm to estimate t̂ (Supplementary Note 1). When the model is not identifiable, we use a penalized likelihood which produces a sparse estimate of t⃗; we choose the sparsity parameter so that the sparse estimate achieves approximately the same maximum as the un-penalized likelihood. (Supplementary Note 1)
Forming confidence bounds on a particular transcript’s frequency estimate, t̂j, requires finding the minimum and maximum values that tj can take while still being reasonably likely to produce the observed data set. Given some desired marginal significance level, α, we estimate our lower confidence bound for transcript j by the minimum value that tj can take over all possible values for t⃗ such that . When the model is identifiable, simulations show that this approach produces confidence bounds with the correct rejection rates for realistic sample sizes (Supplementary Fig. 1e). When the model is not identifiable the confidence bounds are conservative, i.e. the lower confidence bounds is zero.
The fact that GRIT produces conservative confidence bounds is a major advantage over other methods. GRIT allows the user to be confident that transcripts with lower confidence bounds greater than zero were likely present in the sample of interest, while unidentifiable regions can be easily detected and targeted for further experimentation. In contrast, the credible bounds that Cufflinks and Rsem 20 produce are strongly dependent on a prior distribution, which can lead to dramatically anti-conservative confidence bounds even in moderately complex genes (Supplementary Fig. 1f).
Comparison to other tools
Current transcript discovery tools make assumptions about the structure of the underlying transcripts, usually restricting them to some identifiable subset. For instance, Cufflinks assumes that the set of possible transcripts is the minimal set of covering paths in the graphical model described in Section 2.1.3. Trinity requires that transcript models extend to the furthest base of an assemblable contig, which disallows transcript models with nested transcription start and termination sites. The GRIT model allows for both of these, but requires gene boundary information. We benchmarked GRIT against Cufflinks, Scripture and Trinity+Rsem, using stranded RNA-seq, CAGE and poly(A)-site-seq data produced from dissected heads of 20 day adult flies (Ad20dHeads) (Supplementary Table 1).
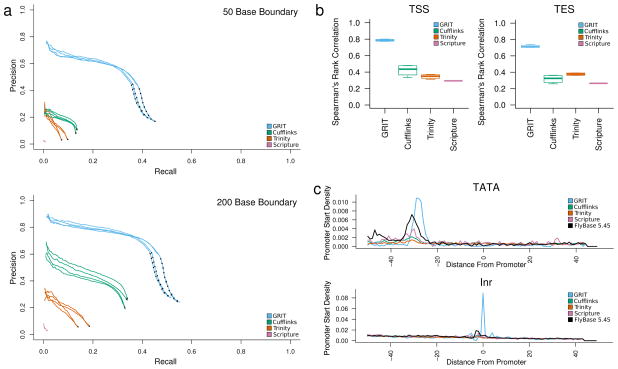
We analyzed the recall and precision of the transcriptomes generated by GRIT and the three other annotation tools by comparing the transcripts predicted by each tool to 13,141 FlyBase 5.45 (Ref. 21) transcripts corresponding to 7,079 genes expressed in Ad20dHeads. Transcripts were considered equivalent when they had the same internal splicing structure and gene boundaries within 50 bp of each other. Under this measure: GRIT recovers 44.2% of transcripts with 17.8% precision; Cufflinks recovers 13.4% of transcripts with 8.8% precision; Trinity+Rsem recovers 8.6% or transcripts with 3.0% precision; Scripture recovers 0.9% of transcripts with 1.4% precision (Fig. 2a). When we filter predicted transcripts with an estimated expression score lower bound less than 1e-6 estimated fragments per kilobase per million reads (FPKMs) at a marginal 99% significance level, then GRIT recovers 39.8% of FlyBase transcripts with 41.3% precision. The Cufflinks, Trinity and Scripture numbers are essentially unchanged.
Figure 2. Comparison with existing tools.
(a) Recall and precision analysis. We compared the set of transcript isoforms discovered by GRIT, Cufflinks, Scripture and Trinity to the FlyBase annotation. A transcript was identified as a match if the internal structure was the same, and the distal boundaries were, variously, within 50 and 200 bp of one-another. (b) FPKM versus CAGE and poly(A)-site-seq counts. For each sample, we calculated the Spearman rank correlation between estimated transcript FPKMs and raw CAGE and poly(A)-site-seq read counts within 50 bp of each annotated promoter/poly(A) site. (c) Motif analysis. For each sample, we considered the sequence within 50 bp of annotated promoters. A position was considered a TATA motif hit if it matched the sequence “T-A-T-A-A”, and an Inr motif match if it matched the sequence “C/T-C/T-A-N-A/T-C/T-C/T”. The plots are aligned with respect to the first base in the annotated promoter, and plot the fraction of promoters that contain a motif match at each position, averaged over replicates.
This substantial rise in GRIT’s precision when low expression transcripts are filtered is largely due to eliminating complex genes. The GRIT annotation is heavily penalized in complex loci, e.g. Dscam1 or Myosin heavy chain (Mhc), because FlyBase includes new transcript models only when they contribute a new exon, intron or gene boundary (FlyBase 5.45 gene notes). The superior performance of GRIT is not purely a result of its increased ability to precisely predict transcript boundaries; when we relax the transcript boundary match distance to 200 bp and even 1,000 bp, GRIT still out-performs competing methods (Fig. 2a, Supplementary Fig. 3).
We studied the consistency of tools’ estimated transcript expression scores by calculating the correlation between estimated FPKMs and both CAGE and poly(A)-site-seq tag counts. GRIT annotated transcripts achieve Spearman rank correlations between 0.71 and 0.80 across replicates, whereas Cufflinks, Trinity and Scripture correlations are all below 0.5 (Fig. 2b).
To study the precision of TSSs, we analyzed the motif enrichment of the two most spatially localized core promoter motifs, TATA 22 and Inr 23, in regions within 50 bp of annotated TSSs (Fig. 2c). The genome sequence surrounding TSSs identified by GRIT and Scripture are significantly enriched (p < 0.01 – see online methods) for the TATA motif 24–32 and 30–35 bp upstream of the TSS, respectively. These correspond to 3.2% and 1.1% of distinct annotated TSSs. Regions identified by Cufflinks and Trinity are not significantly enriched for the TATA motif at any positions. Similarly, regions identified by GRIT are significantly enriched (p < 0.01 – online methods) for the Inr motif enrichment at ±1 bp of the TSS, which corresponds to 12% of annotated TSSs. Neither Cufflinks, Trinity nor Scripture identified regions are significantly enriched (p < 0.01 – online methods) at any bases for the Inr motif. This is expected, because identifying transcript boundaries from RNA-seq data alone is very difficult (Supplementary Result 1, Supplementary Figure 4)
We also analyzed the regions within 50 bp of TSSs annotated in FlyBase 5.45, and found TATA enrichment at 27–34 bp corresponding to 2.9% of distinct TSSs, and Inr enrichment 2–3 bp upstream of annotated TSSs, corresponding to 1.5% of distinct annotated TSSs. Although both GRIT and FlyBase TSS regions show similar TATA enrichment, GRIT more precisely identified the 26–31 bp upstream positioning 23. The GRIT enrichment results are consistent with previous studies19, which report TATA and Inr motifs in 2.1% and 13.8% of peaked promoters identified by RACE 24.
Alternate transcript boundaries are common and functional
Alternate promoters have long been known to serve a regulatory role. The sequence of both 5′ UTRs introns within 5′ UTRs have the potential to alter translational efficiency 25,26,27 and subcellular localization of the mRNA28. Alternative N-terminal protein sequence is known to control the localization of many proteins29.
Genes encoding alternative N-terminal domains, either by alternative promoter usage or splicing, include well-studied examples such as the Prothoracicotropic hormone (Ptth) gene critical for metamorphosis in insects 30,31. Ptth encodes three neural-secreted hormone protein isoforms. The canonical form contains a signal peptide sequence for exportation from the cell. A second isoform with a 25 amino acid N-terminal extension contains a mitochondrial targeting peptide. The third form, first reported by this study, is shorter than the canonical isoform by nine amino acids (Fig. 3a), and is predicted to localize to the cytoplasm or nucleus.
Figure 3. GRIT annotation of D. Melanogaster.
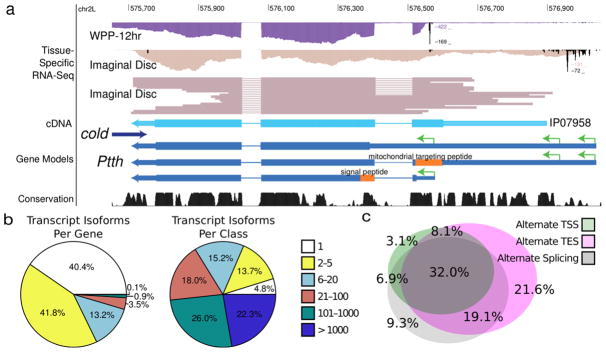
(a) Ptth. The Ptth gene encodes isoforms with multiple proteins due to alternative N-terminal splicing as well as promoter usage. The sample labeled “Imaginal Disc” corresponds to mass isolated tissues enriched more than 50% for imaginal discs. (b) Gene complexity. Although most genes have less than five isoforms, nearly half of transcript isoforms originate in genes that encode 100 or more distinct transcripts. (c) Sources of gene complexity. The Venn digram represents the 59.6% of genes that encode multiple transcript isoforms.
Ptth has the potential to encode multiple localization signals, which appears to be an example of a general phenomenon. Our improved annotation of the Drosophila transcriptome suggests that 19.6% of all protein coding genes encode multiple localization signals – versus 5.7% for FlyBase 5.45 (Supplementary Result 2). We also find substantial complexity at the 3′ ends of transcripts, including neuronal-specific 3′ UTR extensions 32,33. In addition, for 77 genes, we detected polyadenylation sites in canonical CDS exons that result in truncated transcript variants, some of which have been shown to be functional34.
Current tools under-estimate splicing diversity
We identified 47 genes with the capacity to encode >1,000 transcript isoforms 18, 13 of which are only expressed in samples enriched for neuronal tissue. Together, these 13 genes account for nearly 13.5% of the predicted expressed transcript isoforms. In Ad20dHeads, 59.6% of genes expressed encode multiple transcript isoforms (Fig. 3b). Of these, 29.8% exhibit multiple promoters, 48.1% multiple poly(A) events and 40.1% exhibit alternate splicing (Fig. 3c).
Dscam1 has the potential to encode 38,016 distinct protein isoforms35, 3000 of which bind preferentially to themselves, i.e., specific homophilic binding has been observed36. DSCAM1 is known to play a crucial role in axonal tract formation in the developing fly nervous system, and is expressed in neuronal tissue throughout the lifecycle. We observed the highest levels of expression in the central nervous system of pupae (WPP +2day CNS), where we are able to identify a 3′ extension and two novel cassette exons, allowing Dscam1 to produce as many as 228,096 distinct transcripts. In the data collected from Ad20dHeads, GRIT recovers 720 DSCAM isoforms, whereas Cufflinks and Trinity were unable to recover a single full-length transcript.
We used simulated data to study the ability of GRIT, Cufflinks and Trinity to recapitulate known Dscam1 transcripts (Supplementary Fig. 1a). When GRIT was used to analyze 10,000 RNA-seq reads simulated uniformly from the canonical 38,016 isoforms, it recovered every exon, and was thus able to predict every transcript isoforms with perfect precision in 19 of 20 simulations. Trinity was never able to build a full length transcript and Cufflinks recovered one transcript in 1 of 20 simulations, demonstrating the inability of these methods to model complex genes. Running simulations using the 228,096 isoforms identified in WPP +2day CNS produces similar results.
Discussion
The development of tools that enable the accurate interpretation of RNA sequence data is an important challenge. Our tool, GRIT, leverages multiple RNA sequence data types, including CAGE, mRNA-seq, polyA+site-seq, ESTs and cDNAs to discover transcript models. The use of gene boundary data prevents fragmentary transcript models, and models that erroneously merge distinct genes.
Transcript models assembled by GRIT begin with a transcript start site, are connected by intervening mRNA-seq signal, and end in a polyadenylation site. We benchmarked GRIT and three other annotation tools using a subset of the modENCODE Drosophila RNA data sets 18 and found that GRIT performs substantially better than competing methods, both at identifying previously annotated transcript models and discovering new genes and transcripts. We devised a transcript quantification procedure that correctly accounts for model unidentifiability when estimating the confidence bounds, permitting conservative confidence bounds even in gene loci with the potential to produce thousands of transcript isoforms.
In cases where the extant set of transcripts cannot be confidently identified, GRIT could be coupled with other classes of genomic information, including conservation, protein functional data and RNA structure to produce a sparse subset of transcripts that preserve known function. This may aid in generating high-quality transcript annotations. As long read sequencing technologies mature, it may become possible to observe full-length transcripts directly 37. GRIT currently incorporates cDNA sequences into transcript models, providing valuable connectivity information, and will make use of single-molecule data as they become available.
Among the most remarkable findings of our work on the modENCODE Drosophila RNA datasets is the fact that >20% of genes encode proteins with alternative localization signals. Although previous studies have identified individual genes encoding proteins with different subcellular localizations and distinct functional roles 38, our data indicates that this is a ubiquitous function of alternative splicing and promoter usage throughout the genome. This suggests that molecular pleiotropy may be more common than previously thought.
The gene Ptth has been characterized for over a decade. Yet, GRIT discovered a new start codon modulated by an alternative promoter. In addition to emphasizing the importance of accurate gene-boundary information, our studies make evident the need for well-resolved tissue and cell-type transcript maps: the isoform in question is expressed in only two of the 108 modENCODE samples, where it is the dominant form. Future functional studies are needed to determine the biological role of this protein and indeed of the thousands of newly predicted protein isoforms with previously undetected protein localization signals.
GRIT generates full-length transcript models with sample-by-sample expression scores. The accuracy of these automated, purely empirical annotations yield a view of metazoan transcriptomes of unprecedented depth and complexity, which has not been previously obtained through manual annotation or the application of tools that model only a single data type (e.g. RNA-seq without gene boundary information). GRIT alleviates a current analytical bottleneck and will enhance the accessibility and usefulness of RNA sequencing data.
Online Methods
GRIT Method
Below we describe the GRIT methodology including the tuning parameters used for this study; all numerical constants can be changed at the command line. See Supplementary Note 2 for details about the tuning parameters.
GRIT uses reads aligned to a reference genome to build transcript models. We make few assumptions about the structure of a transcript, as follows, and require that every element (e.g. promoter or splice junction) is supported experimentally. We define a transcript as a set of genomic regions that begin at a transcription start site (TSS), optionally extend through one or more exons connected by splice junctions, and end with a transcription end site (TES).
We define four distinct element types: TSS exons, TES exons, internal exons, and single exon transcripts. TSS exons begin with an experimentally detected promoter (e.g. via the CAGE or RACE assays or 5′ EST sequencing 19), and end with a splice donor site. Similarly, TES exons begin with a splice acceptor site and end with an observed TES (e.g. a poly(A) site). Internal exons begin and end with a verified splice site, and single exon transcripts begin with TSS and end with a TES. Our transcript models can use both canonical and non-canonical splice sites. The set of candidate transcripts includes both single exon transcripts and transcripts that begin with a TSS exon, contain splice-junction connected exons, and end with a TES exon (Fig. 1b).
The GRIT annotation pipeline consists of four parts: gene region identification, element discovery, transcript construction and transcript expression estimation.
Gene Region Identification
Segmenting the genome into gene regions involves three distinct steps: identifying exonic regions, identifying intronic regions and merging exonic and intronic regions into gene regions. To build a set of exon regions, we identify all 100 bp regions without any RNA-seq, CAGE, or poly(A)-site-seq reads. These empty regions form boundaries between the different exonic regions. To identify introns, we collect reads that map in a non-contiguous fashion to the reference genome, typically known as junction reads. To avoid junction reads that may be experimental or mapping artifacts, we filter the set of identified junctions using the filtering criterion described as follows.
We require that junctions have an entropy score (defined as − Σipi log2[pi] where ) of at least 2.0 in one biological sample; this filter helps to remove mapping and sequencing artifacts. To remove incorrectly stranded reads, we remove junctions on the strand opposite of canonical acceptor/donor sequences if their frequency is less than 10% of the junction frequency on the canonical strand, and all junctions with a count less than 1% of the count of junctions at the same position but opposite strand. The junction reads that pass this filter are then aggregated into a set of discovered introns.
Finally, we construct gene regions by collecting exon regions that share one or more discovered introns. Note that although 100 bp is too large to properly separate many gene pairs, in practice it provides a good first approximation. During the element discovery stage we use the identified CAGE and poly(A)-site-seq peaks in combination with the read coverage to further segment when necessary (as described below).
Element Discovery
Element discovery proceeds independently in each gene region, and is parallelized for multithreaded processing in GRIT. We split each gene region into non-overlapping segments with attached labels. Segment labels describe the segment boundary (Fig. 1a). For instance, a segment where the 5′ boundary is a splice donor and 3′ boundary is a splice acceptor is a canonical intron; a segment where the 5′ boundary is a splice acceptor and 3′ boundary is a splice donor is a canonical exon. There are four boundary labels: splice acceptor, splice donor, TSS, and TES. Splice donors and acceptors are identified directly from junction reads, as above; TSS and TES are, respectively, identified from CAGE and poly(A)-site-seq data, as follows.
Identifying peaks from transcript boundary data (e.g. CAGE and poly(a)-site-seq) involves both filtering noisy reads and identifying peaks from the filtered data – we use essentially the approach described in Hoskins et al 19. Briefly, we model the data as a mixture of reads sampled from actual gene bounds and from a noise component. Because the dominant source of noise is the selection of RNA fragments that did not originate from true gene bounds (e.g. RNA fragmentation pre-selection, non-specifically bound DNA), we use the RNA-seq data as an estimate of the density of the noise component. For each base i, we estimate the read background density pi as the fraction of RNA-seq reads that start at base i. Then, we model the distribution of the transcript boundary data read count he null as Bin(N,p), where N is the total number of mapped gene boundary reads. If we cannot reject the null hypothesis that the observed transcript boundary data read count originated wholly from the noise component, at significance level 0.01, we zero the count at base i. To identify peaks, we greedily find the set of regions with the smallest combined length that are at least 5 bp long and cover 99% of the gene region’s filtered transcript boundary signal. In the absence of poly(A)-site-seq data, we have successfully applied a machine learning approach to the identification of TESs (Brown et. al. Supplementary Section 12 (Ref. 18)), and expect that a similar approach would work for the identification of TSSs.
There are 16 possible pairwise combinations of the four segment boundary labels, which we group into seven segment labels: TSS segments, canonical introns, canonical exons, exon extensions, TES segments, single exon transcripts and intergenic segments (Fig. 1a). TSS segments are any segments where the 5′ boundary has a TSS label; similarly, a TES segment’s 3′ boundary has a TES label. Canonical introns have a 5′ splice donor label and a 3′ splice acceptor label. Canonical exons have a 5′ splice acceptor label and a 3′ splice donor label. Exon extensions either have two splice donor labels, or two splice acceptor labels. Single exon transcripts have a 5′ TSS label, and a 3′ TES label. Regions that begin with a 5′ TES label and end with a 3′ TSS label are intergenic segments. If intergenic segments are discovered and the average base coverage is sufficiently low, then the gene region is split and the element discovery process is re-started recursively. At this stage, poorly supported segments, meaning those with low read coverage, are removed.
Within a gene region, a low coverage region is defined as a segment where the average read coverage is lower than 1e-2 with high probability; or, the ratio of a segments average read coverage to the highest read coverage segment in the same gene region is less than 1% at a 0.99 significance level. GRIT is relatively robust to changes in these parameters for a given data type, but they may have to be changed when using, for instance, total versus poly(A)+ RNA-seq data (Supplementary Note 2).
The set of candidate exons is all combinations of adjacent segments that start with TSS or splice acceptor, and end with a TES or splice donor. Regions that begin with a TSS label and end with a donor junction are TSS exons; regions that begin with an acceptor junction and end with a TES label are TES exons; regions that begin with a acceptor junction and end with an donor junction are internal exons; regions that start with a TSS label and end with a TES label are single exon transcripts (Fig. 1a).
Transcript Construction
For the purposes of candidate transcript construction, we model a gene as a directed graph in which each exon is a node, and splice junctions are edges (Fig. 1b). Then the set of candidate transcripts is composed of the single exon transcripts and all possible paths through this graph that begin with a TSS exon and end with a TES exon (Fig. 1b). This differs from other methods, e.g. Cufflinks, in that we consider all possible paths subject to this restriction, rather than some minimal set of covering paths.
Transcript Expression Estimation
The primary challenge in estimating transcript expression for a given gene is identifying a vector, t⃗, that corresponds to the relative concentrations of all transcripts in the sample of interest. This is difficult because reads can not necessarily be unambiguously assigned to one transcript. Therefore, the first step in estimating transcript expression levels is further segmenting the transcripts into non-overlapping exon segments, or pseudo exons. It is then possible to unambiguously group reads by the set of pseudo exons that they overlap, which we refer to as a “bin” (Supplementary Fig. 2). Hence, the bins that can be observed unambiguously are a function of gene structure, sequenced read length, and fragment length distribution.
We estimate the fragment length distribution by the 5 base uniform kernel smoothed middle 99% of the empirical distribution of read fragments in the 100 unspliced gene regions with the highest average base coverage that are at least 2000 base pairs long. If there are less than 5000 total fragments that satisfy these criteria, we estimate the fragment distribution by a normal distribution truncated at ±2 standard deviations, with mean and standard deviation estimated from unspliced fragments.
We encode gene structure, read length and type, and fragment length distribution in a design matrix, X, which connects the probability of observing reads of a particular bin to the presence of a particular transcript. Each entry Xij is a conditional probability that applies to individual reads. Formally, Xij is the probability of sampling a read of bin i given that the read originated from transcript j. In practice, we estimate Xij by where fl is the estimated fraction of fragments of length l, Ci,j,l is the count of distinct fragments of length l in transcript j that produce fragments of type i, and Nj is the total number of bins in transcript j. This estimate formalizes the assumption that, within a transcript, all fragments with the same length are equally likely to be observed.
Given a vector of observed bin counts, Y⃗, the maximum likelihood estimate of the transcript frequencies, t⃗, is the vector t̂ that maximizes the log-likelihood, lhd(Y;t⃗) = ΣiYilog[ΣjXijtj], subject to tj ≥ 0 and Σjtj = 1. This is the multinomial log likelihood where the event probabilities, ΣjXijtj, are the bin proportions weighted by the transcript frequencies. The maximum likelihood estimate is unique whenever no row of Xij can be constructed from a positively weighted sum of other rows. In such unique cases, the statistical model is said to be identifiable given the data, and we can use a convex optimization algorithm to estimate t̂ (Supplementary Note 1).
To form confidence bounds on a particular transcript’s frequency estimate, t̂j, our goal is to find the minimum and maximum values that tj can take while still being “reasonably likely” to result in the observed data. We identify a subset ΔR of the probability simplex such that lhd(Y;t⃗) is sufficiently high for every t⃗ ∈ ΔR. Convexity of the likelihood function guarantees this region is simple and convex, which allows us to form our confidence bound for transcript i as the interval [min tj:t⃗ ∈ ΔR, max tj:t⃗ ∈ ΔR,] - a conservative estimate for individual coverage rates.
This interval can be estimated directly by finding the t⃗ on the probability simplex that minimizes tj such that the log likelihood ratio lhd(Y;t⃗mle) − lhd(Y;t⃗) is above some critical value (Supp 1.2.3). Since the asymptotic distribution of lhd(Y;t⃗mle) − lhd(Y;t⃗), a log likelihood ratio statistic39 with one degree of freedom, is we set the critical value to for some desired marginal significance level α. When the model is identifiable, simulations show that this approach produces confidence bounds with the correct rejection rates for realistic sample sizes (Supplementary Fig. 1e).
If the statistical model is not identifiable, then the likelihood solution has no unique maximum even as the read depth approaches infinity. However, for the purposes of visualization or comparative analysis, it may still be useful to quantify a representative set of transcripts, in which case we must make further assumptions. A natural assumption is that the set of transcripts present in solution for a given gene is small. Optimally, we would identify the smallest such subset of transcripts that achieves near the maximum likelihood, but this is not computationally feasible. Instead, we maximize the augmented objective, subject to tj ≥ 0 and Σjtj = 1, where λ is a tuning parameter that determines the sparsity of the resulting solution. Although this optimization problem is not convex, it can be solved by solving Nt convex problems40. We set λ to which guarantees that the estimate for t⃗ lies within the confidence region ΔR (Supplementary Note 1 – Choosing the Sparsity Parameter).
For unidentifiable models, our method produces a lower confidence bound of zero for every transcript in the gene. This allows the user to easily identify regions in which RNA-seq data alone is not sufficient to identify the set of transcripts present. In contrast, Cufflinks and Rsem 20 both use a Bayesian approach, sampling from a posterior distribution to estimate confidence bounds. In complex genes, such as Dscam1 or Mhc, the resulting confidence bounds are strongly dependent on the prior distribution, which typically leads to dramatically anti-conservative confidence bounds (Supplementary Fig. 1f).
Ptth Analysis
Protein sub-cellular localization signals were predicted using the WoLFPSORT41 Command Line Package Version 0.2 in the “animal” setting.
Identifying Flybase Transcripts Expressed in Ad20dHeads
A Flybase transcript was considered expressed in Ad20dHeads if either: it was unspliced; or, it was spliced, and every splice junction was present in at least one RNAseq sample.
TSS Motif Enrichment
To identify motif enrichment in the genome sequence surrounding annotated TSSs, for each tool, we first identified the unique set of transcript start sites. Then, for each TSS, we scanned the genome sequence taken from BDGP5 genome for the TATA motif (TATAAA) and the Inr Motif ([CT][CT]A[ACGT][AT][CT][CT]). A base position was considered a hit if the motif match was exact. Finally, we summed the number of hits at each position, and then divided by the total number of sequences to produce enrichment numbers.
To identify significantly enriched regions, we used a non-parametric approach, performing the above analysis 100000 times from randomly sampled sequence. We sampled chosen randomly from FlyBase 5.45 identified 3′ UTRs, which provides a background with similar sequence composition to 5′ UTRs. A particular position was considered enriched if its value was greater than 99990 of the bootstrapped samples, so that the Type I error rate is expected to be 1%, after correcting for multiple testing (Bonferroni).
Mapping
We used the RNA-seq and CAGE mappings provided by the modENCODE consortium, and distributed by the SRA. (Supplementary Table 1). The poly(A)-site-seq data was mapped with Statmap 19 using the polya assay annotation option, which discards mappings that map to the reference genome before the poly(A) tail is trimmed.
Running Cufflinks
We ran Cufflinks version 2.1.1 on the merged Male and Female RNAseq data using the default configuration options to build the initial transcript sets. Then, we ran Cufflinks in quantification mode (-G option) to provide replicate level quantifications. We did analyze the quantifications produced by running Cufflinks on the replicates independently, but we found them to be of much lower quality than the re-quantified versions (data not shown).
Running Scripture
Due to technical issues, we were not able to run the Scripture version available at http://www.broadinstitute.org/software/scripture/ in house. After some discussion, the authors generously ran alpha version 3.1 on the merged female 20 day adult dissected head data, and provide us the resulting annotation. The parameters used were: 1) premature assembly filter - 0.2; 2) minimum number of spliced reads - 3.0; 3) percentage of total spliced reads - 0.05; 4) alpha for single exon assemblies - 0.01.
Running Trinity
We used the Trinity version 2013-08-14 with the max_number_of_paths_per_node set to 1000 to identify transcript models. We used Rsem to estimate expression (described below) and gmap version 2013-09-11 to map the quantified models to the BDGPv5 reference genome. All other command line options were the packaged defaults.
Running Rsem
We used Rsem version 1.2.7 with the –stranded option to quantify transcripts. We used gmap version 2013-09-11 to map the quantified models to the BDGPv5 reference genome. All other command line options were the packaged defaults.
Simulations
We used the simulation script distributed with GRIT to simulate mapped read data for all simulations. The tool works by first sampling a random transcript from the provided frequency distribution, then sampling a random fragment length from the provided fragment length distribution, and finally choosing a fragment uniformly from the chosen transcript with the chosen fragment length until the desired number of samples is achieved. We do not introduce any sequencing or mapping artifacts into the simulated reads. We note that this simulation is consistent with the GRIT, Cufflinks, and Rsem transcript expression models.
We only compared the performance of GRIT, Cufflinks and Trinity+Rsem in simulations because they were the tools that performed best on real data.
For the synthetic gene simulations (Supplementary Fig. 2 b–f) we sampled from the transcripts uniformly, with a Normal (150, 25) fragment length distribution truncated at ±2 standard deviations. We ran GRIT, Trinity, and Cufflinks in quantification mode. We used GRIT’s compare_annotations.py with a boundary match of ±20 bp to calculate recall and precision numbers. We ran 100 simulations total; 20 simulations with each of 100, 1000, 1e4, and 1e5 simulated reads.
For the Dscam1 simulations (Supplementary Figure 2a), we used the set of DSCAM exons from FlyBase 5.45 to enumerate all possible 38016 DSCAM transcript models. We used a Normal (300,25) fragment length distribution truncated at ± 2 standard deviations. We ran GRIT, Trinity, and Cufflinks in quantification mode. We used GRIT’s compare_annotations.py with a boundary match of ±20 bp to calculate recall and precision numbers.
Supplementary Material
Acknowledgments
We thank the members of the modENCODE transcription consortium for generating the data, Carolyn Cotterman, Erwin Frise, Brenton Garveley, Haiyan Huang and Jingyi Li for discussions. This work was funded by a contract from the National Human Genome Research Institute modENCODE Project, contracts R21 HG006187 to P.J.B.; K99 HG006698 to J.B.B.; U01 HG004271 to S.E.C under Department of Energy contract no. DE-AC02-05CH11231.
Footnotes
Accession Codes
All software associated with this project and the pipelines run to generate these annotations are available for download at http://grit-bio.org/ and as Supplementary Data. All annotation data are available at http://grit-bio.org/nature-biotech-submission.html. SRA accession numbers are provided in Supplementary Table 1.
Author Contributions
N.B. and P.J.B. developed the mathematics and statistics. N.B, J.B.B., and M.S. developed the exon discovery method. N.B. designed the algorithms. N.B. implemented GRIT. B.W.B. aligned the Illumina data. N.B. performed the analysis. P.J.B, J.B.B. and S.E.C. conceived the project. N.B. wrote the paper with input from J.B.B., P.J.B and S.E.C. All authors tested the software, discussed the results and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Bibliography
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10 (1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2010;471 (7339):473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5 (7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 4.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28 (5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011;29 (7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature biotechnology. 2010;28 (5):503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28 (8):1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson G, et al. De novo assembly and analysis of RNA-seq data. Nature methods. 2010;7 (11):909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 9.Tu Q, Cameron RA, Worley KC, Gibbs RA, Davidson EH. Gene structure in the sea urchin Strongylocentrotus purpuratus based on transcriptome analysis. Genome Research. 2012;22 (10):2079–2087. doi: 10.1101/gr.139170.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrow J, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome research. 2012;22 (9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelbart W, et al. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Research. 1999;27 doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins JE, White S, Searle SM, Stemple DL. Incorporating RNA-seq data into the zebrafish Ensembl genebuild. Genome research. 2012;22 (10):2067–2078. doi: 10.1101/gr.137901.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yook K, et al. WormBase 2012: more genomes, more data, new website. Nucleic acids research. 2012;40 (D1):D735–D741. doi: 10.1093/nar/gkr954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang H, Wong WH. Statistical inferences for isoform expression in RNA-Seq. Bioinformatics. 2009;25 (8):1026–1032. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullard J, Purdom E, Hansen K, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC bioinformatics. 2010;11 (1):94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic acids research. 2010;38 (12):e131–e131. doi: 10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Risso D, Schwartz K, Sherlock G, Dudoit S. GC-content normalization for RNA-Seq data. BMC bioinformatics. 2011;12 (1):480. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown JB, et al. Diversity and dynamics of the Drosophila transcriptome. Nature. doi: 10.1038/nature12962. (Submission ID 2012-12-15978A) [DOI] [PMC free article] [PubMed]
- 19.Hoskins RA, et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome research. 2011;21 (2):182–192. doi: 10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B, Dewey C. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12 (1):323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marygold SJ, et al. FlyBase: improvements to the bibliography. Nucleic acids research. 2013;41 (D1):D751–D757. doi: 10.1093/nar/gks1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lifton R, Goldberg M, Karp R, Hogness D. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. presented at Cold Spring Harbor symposia on quantitative biology; 1978. (unpublished) [DOI] [PubMed] [Google Scholar]
- 23.Butler JE, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes \& development. 2002;16 (20):2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins C, Michael D, Mahendroo M, Simpson E. Exon-specific northern analysis and rapid amplification of cDNA ends (RACE) reveal that the proximal promoter II (PII) is responsible for aromatase cytochrome P450 (CYP 19) expression in human ovary. Molecular and cellular endocrinology. 1993;97 (1):R1–R6. doi: 10.1016/0303-7207(93)90227-b. [DOI] [PubMed] [Google Scholar]
- 25.Rojas-Duran MF, Gilbert WV. Alternative transcription start site selection leads to large differences in translation activity in yeast. RNA. 2012;18 (12):2299–2305. doi: 10.1261/rna.035865.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawless C, et al. Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. BMC genomics. 2009;10 (1):7. doi: 10.1186/1471-2164-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penalva LOS. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiology and molecular biology reviews. 2003;67 (3):343–359. doi: 10.1128/MMBR.67.3.343-359.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cenik C, et al. Genome analysis reveals interplay between 5′ UTR introns and nuclear mRNA export for secretory and mitochondrial genes. PLoS genetics. 2011;7 (4):e1001366. doi: 10.1371/journal.pgen.1001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of molecular biology. 2000;300 (4):1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 30.Kawakami A, et al. Molecular cloning of the Bombyx mori prothoracicotropic hormone. Science. 1990;247 (4948):1333–1335. doi: 10.1126/science.2315701. [DOI] [PubMed] [Google Scholar]
- 31.Rewitz KF, Yamanaka N, Gilbert LI, O’Connor MB. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science. 2009;326 (5958):1403–1405. doi: 10.1126/science.1176450. [DOI] [PubMed] [Google Scholar]
- 32.Hilgers V, et al. Neural-specific elongation of 3′ UTRs during Drosophila development. Proceedings of the National Academy of Sciences. 2011;108 (38):15864–15869. doi: 10.1073/pnas.1112672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smibert P, et al. Global Patterns of Tissue-Specific Alternative Polyadenylation in Drosophila. Cell reports. 2012;1 (3):277–289. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Ruscio A, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013 doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Celotto AM, Graveley BR. Alternative splicing of the Drosophila Dscam pre-mRNA is both temporally and spatially regulated. Genetics. 2001;159 (2):599–608. doi: 10.1093/genetics/159.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative Splicing of Drosophila Dscam Generates Axon Guidance Receptors that Exhibit Isoform-Specific Homophilic Binding. Cell. 2004;118 (5):619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharon D, Tilgner H, Grubert F, Snyder M. A single-molecule long-read survey of the human transcriptome. Nature biotechnology. 2013;31 (11):1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juneau K, Nislow C, Davis RW. Alternative splicing of PTC7 in Saccharomyces cerevisiae determines protein localization. Genetics. 2009;183 (1):185–194. doi: 10.1534/genetics.109.105155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bickel PJ, Doksum KA. Mathematical Statistics, volume I. Prentice Hall Englewood; Cliffs, NJ: 2001. [Google Scholar]
- 40.Pilanci M, El Ghaoui L, Chandrasekaran V. Recovery of Sparse Probability Measures via Convex Programming. presented at Advances in Neural Information Processing Systems; 2012. (unpublished) [Google Scholar]
- 41.Horton P, et al. WoLF PSORT: protein localization predictor. Nucleic acids research. 2007;35 (suppl 2):W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.