Abstract
IκBα inhibits the transcription factor, NFκB, by forming a very tightly bound complex in which the ankyrin repeat domain (ARD) of IκBα interacts primarily with the dimerization domain of NFκB. The first four ankyrin repeats (ARs) of the IκBα ARD are well-folded, but the AR5-6 region is intrinsically disordered according to amide H/D exchange and protein folding/unfolding experiments. We previously showed that mutations towards the consensus sequence for stable ankyrin repeats resulted in a “prefolded” mutant. To investigate whether the consensus mutations were uniquely able to order the AR5-6 region, we used a predictor of protein disordered regions PONDR VL-XT to select mutations that would alter the intrinsic disorder towards a more ordered structure (D→O mutants). The algorithm predicted two mutations, E282W and P261F, neither of which correspond to the consensus sequence for ankyrin repeats. Amide exchange and CD were used to assess ordering. Although only the E282W was predicted to be more ordered by CD and amide exchange, stopped-flow fluorescence studies showed that both of the D→O mutants were less efficient at dissociating NFκB from DNA.
Introduction
Intrinsically disordered proteins have disordered or weakly folded regions and comprise a large portion of the proteome 1. Often the disordered regions fold upon association to their binding partners, and this coupled folding and binding provides several advantages. Flexibility can allow proteins to conform to the shape of a binding partner thereby increasing complementarily, such as with colicin and TolB. 1 The flexibility can allow the protein to adopt different structures for different binding partners, as is the case with p53, which has been observed in helical, β-strand, or extended structures with different binding partners.2 Coupled folding and binding can influence the binding kinetics; for example, in the fly-casting mechanism, an unfolded protein binds faster than a well-structured protein because the unfolded protein has a larger capture radius than the folded protein.3 Simulations on pKID showed that the binding rate could be reduced by increasing the amount of structure in unbound pKID, supporting a fly-casting process.4
According to CD and amide exchange studies, the last two ankyrin repeats (AR) of IκBα are disordered and are only fully folded when IκBα is bound to NFκB.5 The weakly folded AR5-6 region serves as a switch between degradation mechanisms. When IκBα is free, AR5-6 and the C-terminal PEST sequence exposes a degron that targets the protein for a rapid signal- and ubiquitin-independent proteosomal degradation process.6, 7 This degron is masked when IκBα is bound to NFκB, and the bound protein requires phosphorylation and ubiquitylation at the N-terminal signal response element for targeting to proteosomal degradation.8 The switch between degradation mechanisms is accompanied by a large difference in the intracellular half-life of IκBα; free IκBα has a half-life of 10 min, whereas an NFκB-bound IκBα has a half-life of 12 h.9 The short half-life of free IκBα keeps cellular levels of the inhibitor low, which is essential for robust activation of NFκB. IκBα has a high affinity for NFκB, resulting from an extremely slow dissociation rate.10 This has been shown to be due in part to the coupled folding and binding of the AR(5-6) region. IκBα inhibits the transcriptional activity of NFκB, and we recently showed that it actually accelerates the dissociation of NFκB from DNA—a process that requires the disordered AR5-6 region of IκBα.11
Alignment of the many hundreds of AR sequences helped to define a consensus sequence, and AR domains that are designed with this consensus in mind are highly stable.12 We noticed that the more consensus-like ARs of IκBα were AR(1-4), and these had the least amide exchange, whereas AR(5-6) conform less to the consensus and undergo rapid exchange13 in agreement with lower sequence conservation of the disordered regions. 14 Introduction of two mutations to the consensus sequence in AR6, Y254L and T257A resulted in “pre-folding” of AR6 so that all six ARs became part of the cooperatively folding AR domain (ARD). 15 This mutant had impaired binding affinity and was also less efficient at dissociating NFκB from the DNA.11, 15
Here, we searched for another way to find mutations in the weakly folded AR6 of IκBα that might alter folding and function. We used an in-house “mutator” software (see Materials and Methods for details) to predict disorder-to-order (D→O) transition mutations in AR6 of IκBα. The algorithm is based on a bioinformatic approach that analyzes disordered regions of proteins based on sequence information. “mutator” predicted two D→O mutations, P261F and E282W, neither of which introduced consensus residues. These mutants were prepared and analyzed for the “foldedness” using amide H/D exchange and CD, for binding to NFκB using stopped-flow fluorescence following the single tryptophan in AR6, W258, as a reporter, and for the ability to dissociate NFκB from DNA using stopped-flow fluorescence following pyrene-labeled DNA. We show that both D→O mutants have slower rate constants for dissociating NFκB from the DNA, emphasizing the functional role for intrinsic disorder in IκBα.
Materials and Methods
Prediction of the disorder-to-order transition mutations
The in-house “mutator” software was used to predict the D→O transition mutations in the mouse IκBα protein. This software was designed based on the observation that some naturally occurring disease-associated mutations cause a large decrease in the predicted disorder score.16, 17 The effect of one D→O mutation was verified using accelerated molecular dynamics simulations. In agreement with our predictions, an increased α-helical propensity of the region harboring the mutation was observed.17
The “mutator” is based on the PONDR VL-XT disorder predictor,18 which assigns the disorder score to each amino acid residue in the protein. The “mutator” automatically replaces each residue within a user-defined window with each of the 19 remaining residues and recalculates PONDR VL-XT scores after each change. Subsequently, to suggest the mutation for mutagenesis, the “mutator” calculates the area under the prediction curve (AUC) and suggests the mutation that causes the largest decrease of the AUC. Since the disorder score is calculated based on the sliding window, a change of one amino acid influences the disorder score of the entire region. According to VL-XT prediction, full-length human IκBα has two long disordered regions: residues 1-68 and 253-298, with the latter interspersed by a short, ordered segment. It also has two short disordered regions, 81-92 and 254-262. The region 253-298, containing AR5-AR6 and the PEST was selected for mutagenesis in this study. Two mutations in IκBα which caused the largest difference in the score, P261F and E282W, were selected as candidates for experimental confirmation.
Protein Expression and Purification
Human wild-type IκBα67-287 referred to simply as IκBα and mutants (P261F and E282W) were expressed in E. coli BL21 DE3 cells and purified using a Hi-Load Q Sepharose (GE Healthcare, Pittsburgh, PA, USA) followed by a Superdex 75 column (GE Healthcare) as described previously.10, 13 The protein concentrations were determined by spectrophotometry (ε IκBα WT and P261F = 12,950 M−1cm−1, and ε IκBα E282W = 18,450 M−1cm−1).
The N-terminal hexahistidine-NFκB (His6-p5039-350/RelA19-321) heterodimer was co-expressed using the method described previously19 and purified by nickel affinity chromatography (Ni-NTA Agarose, Qiagen, Valencia, CA, USA), cation exchange chromatography (Mono S column, GE Healthcare), and size exclusion chromatography (Superdex 200, GE Healthcare). The protein concentration was determined by spectrophotometry (ε NFκB = 43,760 M−1cm−1).
Murine RelA190-321 with an N-terminal cysteine, and murine p50248-350, were expressed, purified, and quantified as described.10 The RelA dimerization domain, RelA190-321 in 25 mM Tris pH 7.2, 150 mM NaCl, 1 mM EDTA was biotinylated by incubation with a 1:1 molar ratio of biotin PEO maleimide (Pierce Chemicals), neutation at room temperature for 1 h, and purification immediately by size exclusion chromatography on a S75 Superdex 16/60 column at 4 °C in SPR Immobilization Buffer (SPR-IB) (500 mM NaCl, 10 mM Tris pH 7.5, 0.5 mM EDTA, 0.5 mM sodium azide, 0.005% v/v P20). Murine p50248-350 was purified by size exclusion chromatography as above. To form the (p50248-350/RelA190-321) heterodimer, referred to as biotinylated NFκB, a 50-fold excess of unbiotinylated p50248-350 was incubated with biotin- RelA190-321 at room temperature for 1 h and subsequently at 4 °C overnight.
Hydrogen/deuterium exchange mass spectrometry
Hydrogen/deuterium exchange mass spectrometry (HDXMS) was performed using Waters nanoACQUITY UPLC system with H/DX technology (at Lilly, Inc. San Diego). For each different deuteration time, 5 μL of purified protein ([WT] = 84 μM, [E282W] = 82 μM and [P261F] = 90 μM) was mixed with 55 μL of D2O buffer (containing 0.1× PBS) for a specified amount of time (10 s to 10 min) at 15 °C. The exchange was quenched for 2 min at 1 °C with an equal volume of 100 mM phosphate with 320 mM TCEP (pH 2.4). The quenched sample was injected into a 50 μL sample loop, followed by on-line pepsin column digestion (Applied Biosystems, Poroszyme Immobilized Pepsin cartridge). The resulting peptic peptides were captured on a Vanguard trap column, separated by analytical column (Waters, Acquity UPLC BEH C18, 1.7 μm, 1.0×50 mm) with a gradient of 3%–85% acetonitrile in 12 min where both mobile phases contained 0.2% formic acid, and directed into a Waters SYNAPT G2 quadrupole time-of-flight mass spectrometer. The mass spectrometer was set to collect data in the MSE, ESI+ mode; mass acquisition range of 255.00 – 1950.00 (m/z); scan time 0.4 s. Continuous lock mass correction was accomplished with infusion of a peptide standard every 30 s (mass accuracy of 1 ppm for calibration standard). The peptides were initially identified by using PLGS 2.5 (Waters, Inc.) with a mass accuracy of 3–5 ppm and fragmental ions, and the relative deuterium uptake for each peptide was calculated by comparing the centroids of the mass envelopes of the deuterated samples with the undeuterated controls using DynamX 2.0 (Waters Corporation). Details of this methodology have been previously reported.20
Circular Dichroism Spectroscopy
CD measurements were performed with an Aviv 202 spectropolarimeter (Aviv Biomedical, Lakewood, NJ). The proteins were dissolved in 10 mM PO4 pH 7.5, 150 mM NaCl, 1mM DTT, 0.5 mM EDTA at a concentration of 60 μM except for the E282W which was at 55 μM. All the spectra were collected at a constant temperature of 25 °C. The uncorrected ellipticity is reported.
SPR Experiments
Sensorgrams were recorded on a Biacore 3000 instrument using streptavidin (SA) chips (GE Healthcare). Biotinylated NFκB was immobilized in SPR-IB on flow cell (FC) 2, FC 3, and FC 4 with 50, 100, and 150 RU, respectively, leaving FC 1 unmodified. Data were collected at the high collection rate on FCs 2, 3, and 4 automatically subtracting FC 1. Binding experiments were conducted on IκBα, IκBα P261F, IκBα E282W, and IκBα E282Q in SPR Running Buffer (SPR-RB) (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.5 mM, sodium azide, 10% w/v glycerol, 0.005% P20) at 50 μL/min using the kinject function with a 5 min contact time and a 20 min dissociation. The surface was regenerated by a 1 min pulse of 1.5 M urea diluted from a 6 M stock in SPR-RB. The data were analyzed as previously described.21
Stopped-flow fluorescence
To measure the association of IκBα to NFκB or to the NFκB-DNA complex, we took advantage of the native Trp fluorescence of IκBα W258. Six different concentrations of NFκB or of the NFκB-DNA complex (0.3, 0.4, 0.5, 0.6, 0.7, 0.8 μM) were mixed with IκBα (0.1 μM). All stopped-flow kinetic experiments were performed at 25 °C using an SX-20 stop-flow apparatus (Applied Photophysics, Leatherhead, UK) set to collect 2000 pt linearly with a final mixing volume of 200 μL. Twenty traces were collected for each concentration, and 10 traces that overlaid well were selected and averaged. The data were processed using pro Fit 6.1.14 and were fit to a single exponential. The observed association rates were plotted against the IκBα concentration and fit using a linear model to obtain the association rate constant. We did not observe any difference in the fluorescence change for the E282W as compared to wild type IκBα even though this mutation introduces a second Trp. We believe this is because position 282 is at the very C-terminus of the protein in a relatively solvent-exposed region.
To measure the IκBα-mediated dissociation of DNA from NFκB we used a hairpin DNA sequence corresponding to the IFN-κB site, 5′-AmMC6/GGGAAATTCCTCCCCCAGGAATTTCCC-3′ (IDT Technologies, Coranville, CT, USA), which was labeled with pyrene (N-hydroxyl succinimide ester) and is referred to as DNA*, as described.22 The pyrene was excited at 343 nm, and the fluorescence emission was monitored at 376 nm with a cut-off filter at 350 nm. Intrinsic fluorescence of the naturally-occurring IκBα W258 was followed by exciting at 280 nm and monitoring the emission at 345-355 nm with a cut-off filter at 320 nm. The rate of IκBα-mediated dissociation of the DNA from NFκB was measured by adding different concentrations of IκBα (0.25, 0.5, 0.75, 1, 1.25, 1.5, 17.5, and 2 μM) to a 1 μM NFκB:1.2 μM DNA* complex. The curves were fitted with pro Fit 6.1.14 using a single exponential dissociation model. The observed dissociation rates were plotted against the IκBα concentration and fit using a linear model to obtain the rate constant for IκBα-mediated dissociation of the NFκB-DNA complex.
All of the experiments reported above were repeated using IκBα P261F and IκBα E282W (and for some E282Q) in place of the wild type IκBα.
Results and Discussion
Predicting disorder to order transitions in IκBα using the “mutator” algorithm
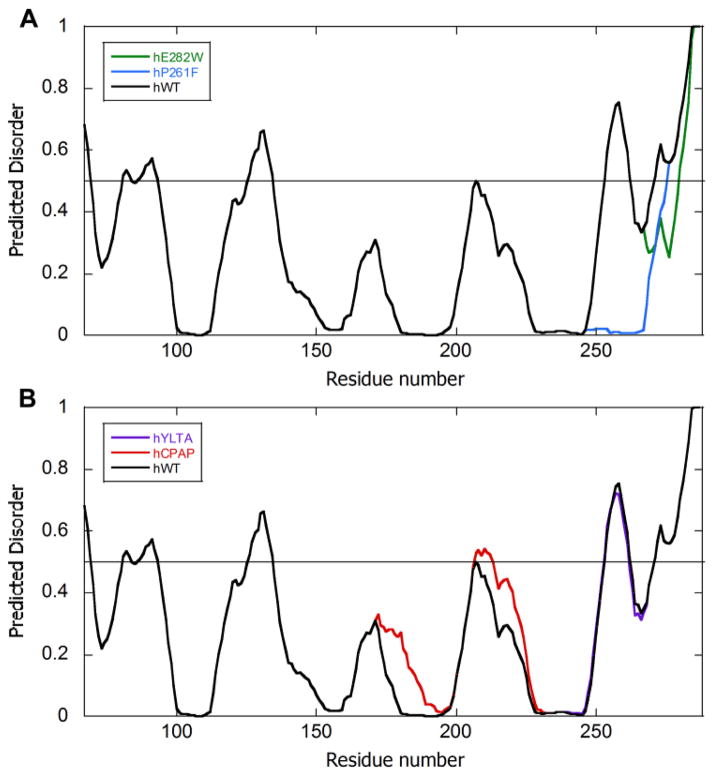
IκBα67-287 has increased intrinsic disorder towards the C-terminus of the protein (Figure 1). According to VL-XT prediction, the IκBα ARD has a long disordered region extending from residue 253 to residue 298 interspersed by a short ordered segment, and this region was selected for mutagenesis in this study. Two mutations in IκBα, P261F and E282W, were predicted to increase order of the C-terminal disordered region of the protein (Figure 1A). It is interesting to note that when we analyzed the mutations that were known to stabilize IκBα based on mutation to the stable ankyrin repeat consensus sequence, these mutations did NOT result in predicted ordering of the 253-298 region according to the VL-XT prediction (Figure 1B). These results demonstrate that the disorder predictor accesses a different realm of protein folding space than the consensus sequence, which is what we were aiming for in using this approach.
Figure 1.
Plot of the PONDR VL-XT predicted disorder for the ARD (residues 67-287) of (A) wild type human IκBα (black), the P261F mutant (blue), and. the E282W mutant (green), (B) wild type IκBα (black), the C186P/A220P mutant (red) and,. the Y254L/T257A mutant (purple). The P261F mutant is predicted to be more ordered in residues 253-275, whereas the E282W mutant is predicted to be more ordered in residues 268-287. The C186P/A220P and the Y254L/T257A mutations do not influence the disorder score of the region 253-298, except for a slight increase in disorder in the 207-212 region for the C186PA220P mutant. The score of ≥ 0.5 signifies predicted disorder, whereas the score of <0.5 signifies predicted order (depicted by the horizontal line in each figure).
Assessment of the “foldedness” of the D→O mutants
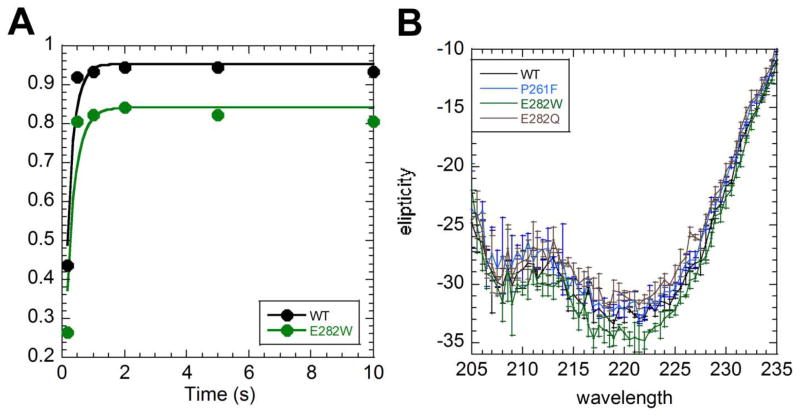
We previously showed that all of the amides in AR5 and AR6 of IκBα exchange rapidly, whereas the amides in AR(1-4) exchange much more slowly.13 We interpreted the differential exchange behavior in light of folding simulations that indicated AR(1-4) formed a stable, folded, domain whereas AR(5-6) folded later and more weakly.23 In fact, the consensus YLTA mutant showed significantly reduced exchange in AR5 and AR6. 15 A similar approach was used to assess whether the D→O mutant proteins were more well-folded. The wild type and each mutant protein were incubated in deuterated buffer for 0–10 min, and the amide exchange was assessed after quench and pepsin digestion by mass spectrometry.13 We were only able to accurately assess the foldedness of the E282W mutant, however, because the P261F mutation introduced a pepsin cleavage site that resulted in a different distribution of peptides that could not be directly compared to the wild type. Three peptides showed slightly reduced exchange in the E282W mutant, but the only one with significantly reduced exchange was the peptide that covered the site of the mutation (Figure 2A). Thus, amide exchange indicates that at least for the E282W mutant, introduction of this single, non-consensus mutation, which was predicted by a bioinformatic algorithm, results in a mor folded cornormation.
Figure 2.
Amide H/D exchange and CD results for the E282W D→O mutant. (A) The C-terminal residues showed reduced exchange in the E282W mutant (green) compared to wild type IκBα (black). (B) Circular dichroism spectroscopy showed a statistically significant increase in helicity for the E282W mutant, but not for the P261F mutant (error bars are the standard deviation from three independent experiments).
To further explore the possible ordering introduced by the D→O mutations, we measured the ellipticity by circular dichroism (CD) of equal concentrations of wild type, P261F, E282W, and a control mutant, E282Q. The E282W mutant showed a significant increase in helical signal, whereas the P261F and E282Q mutants had the same helical content as wild type IκBα (Figure 2B). Thus, both CD and amide exchange indicated a slight increase in ordered structure for the E282W mutant.
Effect of D→O mutations in IκBα on binding kinetics
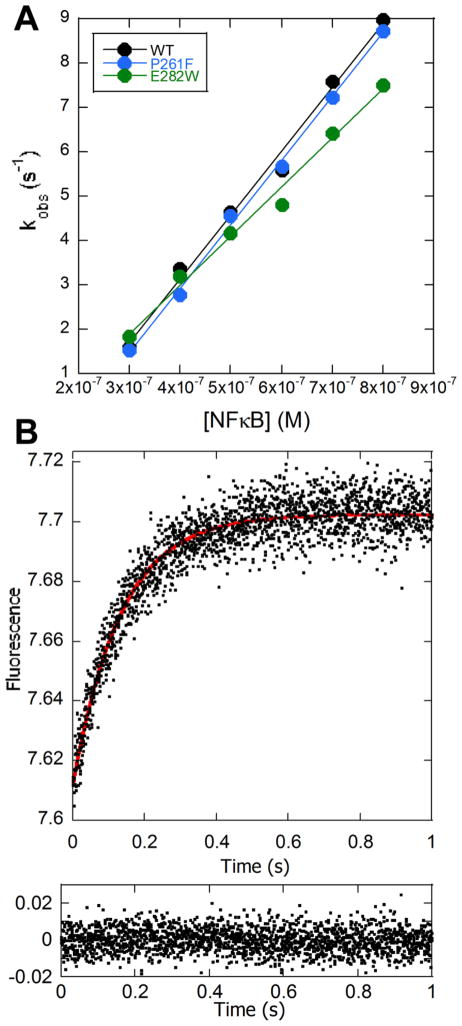
To assess whether the mutations affected the kinetics of binding of the mutant IκBα proteins to NFκB (RelA/p50), we used both SPR and stopped-flow fluorescence. For the stopped-flow experiments, we monitored the change in fluorescence of the naturally-occurring Trp258 in AR6 of IκBα that increases fluorescence upon binding 24. The P261F mutant bound with the same association rate constant as the wild type (1.4 (± 0.1) × 107 M−1 s−1), and the E282W mutant showed a slight decrease in binding rate (1.1 (± 0.1) × 107 M−1 s−1) (Figure 3). We could not measure dissociation by stopped-flow because the NFκB-IκBα complex binds extremely tightly; however the SPR results showed similar dissociation rates and relatively similar KD’s of 172 ± 4 pM for WT, 462 ± 8 pM for P261F and 155 ± 14 pM for the E282W mutant (Suppl. Figure 1 and Suppl. Table 1). Although P261F binds to NFκB slightly more weakly than WT IκBα, its KD still exhibits the strong binding associated with the IκBα-NFκB complex. Furthermore, we found no correlation between binding affinity and the rate at which the IκBα variants dissociated NFκB from DNA.
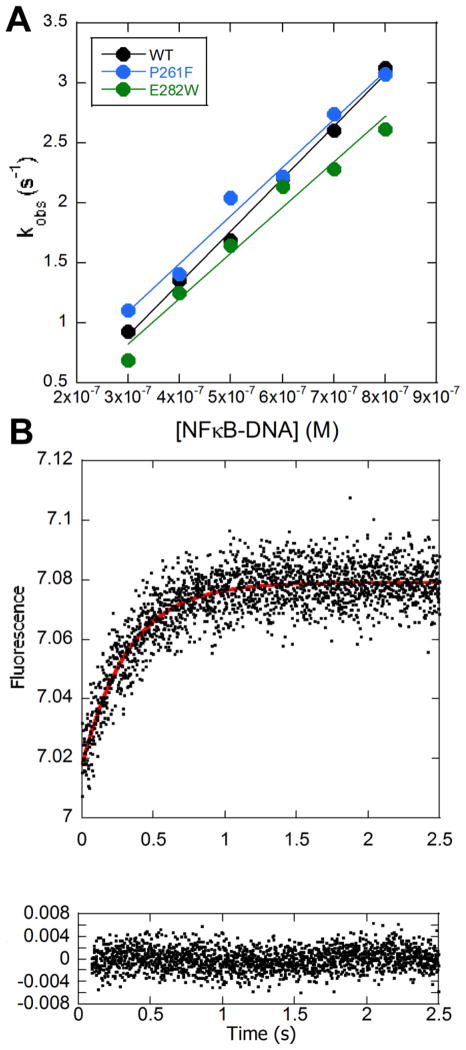
Figure 3.
Stopped-flow fluorescence kinetic experiments in which IκBα (0.1 μM) was mixed with varying concentrations of NFκB, and the change in intensity of the Trp fluorescence from the native W258 in IκBα was monitored. (A) Plot of the fluorescence relaxation times determined at constant IκBα concentration and varying NFκB concentrations, which could be linearly fit yielding the association rate constant. (B) A representative stopped-flow fluorescence trace obtained by mixing 0.1 μM IκBα with 0.8 μM NFκB is shown with the corresponding residuals for the single exponential fit (red line in trace) plotted below the trace.
Effect of disorder to order mutations in IκBα-mediated dissociation of NFκB from DNA
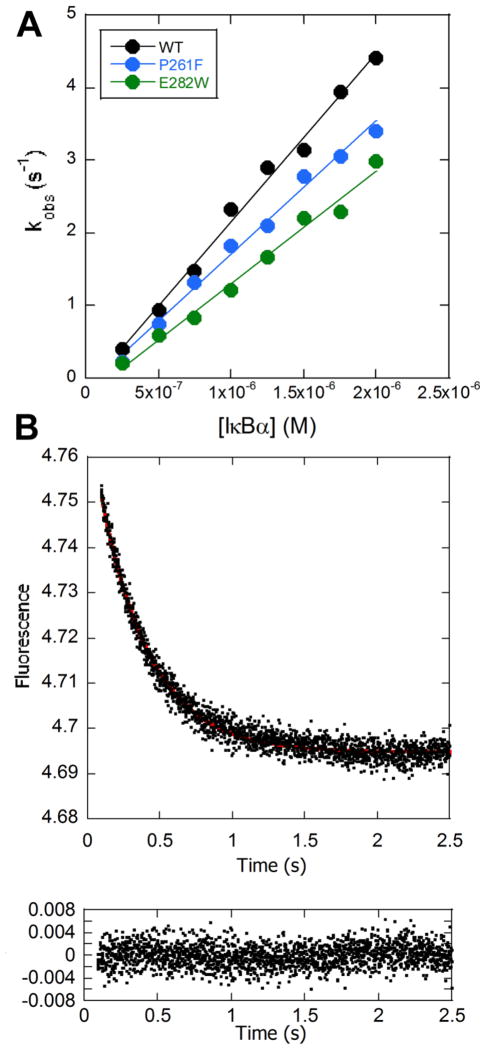
We previously showed that introduction of consensus mutations that stabilize the ARD of IκBα decreases the rate constant for IκBα-mediated dissociation of NFκB from DNA.11 We used stopped-flow fluorescence to follow how well the non-consensus D→O mutant IκBαs mediated dissociation of DNA* from the NFκB-DNA* complex as described previously 24. Both mutants showed decreased rate constants for IκBα mediated dissociation of NFκB from DNA*. Wild type IκBα facilitated dissociation with a second order rate constant of 2.3 (±0.1) × 106 M−1 s−1, whereas the rate constant for the P261F mutant was 1.8 (±0.1) × 106 M−1 s−1, a decrease of 22%, and the rate constant for the E282W mutant was 1.6 (±0.1) × 106 M−1 s−1, a decrease of 30% (Figure 4). While these differences were small, they were statistically significant and reproducible across several experiments and with different protein preparations. To control for the change in charge of the E282W mutant, we also measured the rate constant for the E282Q mutant. Compared to wild type IκBα, this mutant had the same predicted disorder and a rate constant of 2.2 (±0.1) × 106 M−1 s−1 that is nearly identical to the wild type protein (Supplementary Figure 2).
Figure 4.
Stopped-flow fluorescence was used to measure the dissociation of DNA* from the NFκB- DNA* complex in the presence of varying concentrations of IκBα. Upon binding of IκBα to the NFκB-DNA complex, the fluorescence increased in the first 100 ms and then decreased to the values expected for free DNA*. The IκBα-concentration-dependent dissociation rate was measured from the second part of the traces. (A) The dissociation became more rapid with increasing IκBα concentration, and the plot of the fluorescence relaxation times vs. IκBα concentrations could be fit linearly yielding the second order rate constant for IκBα mediated dissociation of the NFκB-DNA* complex. (B) A representative stopped flow trace obtained by mixing 0.1 μM NFκB-DNA* complex with 1.25 μM IκBα is shown with the corresponding residuals for the single exponential fit (red line in trace) plotted below the trace.
To determine whether the decreased second order rate constant for dissociating NFκB from the DNA was due to decreased association of the mutant IκBαs with the NFκB-DNA complex, the first step in the facilitated dissociation, we measured the association of each IκBα with the NFκB-DNA complex, again by following the change in fluorescence of the Trp258 in IκBα. The wild type IκBα associated with the NFκB-DNA complex with a rate constant of 4.4 (±0.1) × 106 M−1 s−1, the P261F mutant rate constant was 4.0 (±0.3) × 106 M−1 s−1, a decrease of 9%, and the E282W rate constant was 3.8 (±0.3) × 106 M−1 s−1, a decrease of 14%. These results show that association accounts for some, but not all of the decrease (Figure 5). Thus, the mutants appear to be less able to actually facilitate dissociation of the DNA after the transient ternary complex is formed.
Figure 5.
Stopped-flow fluorescence kinetic experiments in which IκBα (0.1μM) was mixed with varying concentrations of NFκB-DNA complex and the change in intensity of the Trp fluorescence from the native W258 in IκBα was monitored. (A) Plot of the fluorescence relaxation times determined at constant IκBα concentration and varying NFκB-DNA complex concentrations. The linear fit yielded the association rate constant. (B) A representative stopped-flow fluorescence trace of IκBα (0.1 μM) mixed with the NFκB-DNA complex (1.25 μM) and the single exponential fit (red line in trace). The corresponding residuals for the single exponential fit are shown below the trace.
In conclusion, we have used a sequence-based algorithm to predict D→O mutations in the intrinsically disordered C-terminal region of IκBα. Two mutations were predicted to cause the largest D→O transition in this region, and both were tested. To examine changes in “foldedness,” amide H/D exchange and CD spectroscopy were used. Only the E282W mutant showed decreased amide exchange that was significant in residues 278-287, precisely the residues that were predicted to transition to a more ordered state by the “mutator” algorithm. This mutant also showed a significant increase in helicity by CD spectroscopy, further corroborating that this mutant is more well-folded.
We had previously shown that mutations towards the consensus for stable ankyrin repeats decreased the rate constant for IκBα-mediated dissociation of NFκB from DNA.11 Here we attempted to access more ordered conformations by an alternative approach; the mutator algorithm. Interestingly, this algorithm does not predict a significant ordering for the consensus mutations (i.e., IκBα C186P/A220P and IκBα Y254L/T257A), suggesting that consensus mutation and purely sequence-based predictions provide alternative approaches to introducing order or foldedness, vaguely defined, in ankyrin repeat domains. Both D→O mutations decreased the rate constant for IκBα-mediated dissociation of NFκB from DNA; however, the E282W mutation had a stronger effect. This observation emphasizes the requirement for disorder in IκBα to efficiently dissociate NFκB from DNA and suggests that the defective dissociation of the consensus mutations was indeed due to their ordering of the disordered AR5-AR6 of IκBα. While the D→O mutants affected the association of the mutant IκBαs to the NFκB-DNA complex, these mutations more strongly affected the rate of IκBα–mediated dissociation. The results emphasize how perfectly tuned the IκBα energy landscape is for the function of dissociating NFκB from the DNA.
Supplementary Material
Acknowledgments
This work was supported by P01-GM071862. We would like to thank Molecular Kinetics Inc. for the access to the PONDR VL-XT predictor. LMI was supported by the grants NIH RO1 HD065288 and NIH RO1 MH091350.
References
- 1.Wright PE, Dyson HJ. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. BMC Genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoemaker BA, Portman JJ, Wolynes PG. Proc Natl Acad Sci U S A. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turjanski AG, Gutkind JS, Best RB, Hummer G. PLoS Comput Biol. 2008;4:e1000060. doi: 10.1371/journal.pcbi.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Truhlar SM, Torpey JW, Komives EA. Proc Natl Acad Sci U S A. 2006;103:18951–18956. doi: 10.1073/pnas.0605794103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathes E, O’Dea EL, Hoffmann A, Ghosh G. EMBO J. 2008;27:1357–1367. doi: 10.1038/emboj.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathes E, Wang L, Komives E, Ghosh G. J Biol Chem. 2010;285:32927–32936. doi: 10.1074/jbc.M110.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh S, May MJ, Kopp EB. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 9.O’Dea EL, Barken D, Peralta RQ, Tran KT, Werner SL, Kearns JD, Levchenko A, Hoffmann A. Mol Syst Biol. 2007;3:111. doi: 10.1038/msb4100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergqvist S, Croy CH, Kjaergaard M, Huxford T, Ghosh G, Komives EA. J Mol Biol. 2006;360:421–434. doi: 10.1016/j.jmb.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergqvist S, Alverdi V, Mengel B, Hoffmann A, Ghosh G, Komives EA. Proc Natl Acad Sci U S A. 2009;106:19328–333. doi: 10.1073/pnas.0908797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binz HK, Stumpp MT, Forrer P, Amstutz P, Pluckthun A. J Mol Biol. 2003;332:489–503. doi: 10.1016/s0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 13.Croy CH, Bergqvist S, Huxford T, Ghosh G, Komives EA. Protein Sci. 2004;13:1767–1777. doi: 10.1110/ps.04731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown CJ, Johnson AK, Daughdrill GW. Mol Biol Evol. 2010;27:609–621. doi: 10.1093/molbev/msp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Truhlar SM, Mathes E, Cervantes CF, Ghosh G, Komives EA. J Mol Biol. 2008;380:67–82. doi: 10.1016/j.jmb.2008.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vacic V, Iakoucheva LM. Mol Biosyst. 2012;8:27–32. doi: 10.1039/c1mb05251a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vacic V, Markwick PR, Oldfield CJ, Zhao X, Haynes C, Uversky VN, Iakoucheva LM. PLoS Comput Biol. 2012;8:e1002709. doi: 10.1371/journal.pcbi.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Genome Informatics Series Workshop. 1999;10:30–40. [PubMed] [Google Scholar]
- 19.Sue SC, Cervantes C, Komives EA, Dyson HJ. J Mol Biol. 2008;380:917–931. doi: 10.1016/j.jmb.2008.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergqvist S, Ghosh G, Komives EA. Protein Sci. 2008;17:2051–2058. doi: 10.1110/ps.037481.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Studer SM, Joseph S. Meth Enzymol. 2007;430:31–44. doi: 10.1016/S0076-6879(07)30002-5. [DOI] [PubMed] [Google Scholar]
- 23.Ferreiro DU, Cho SS, Komives EA, Wolynes PG. J Mol Biol. 2005;354:679–692. doi: 10.1016/j.jmb.2005.09.078. [DOI] [PubMed] [Google Scholar]
- 24.Alverdi V, Hetrick B, Joseph S, Komives EA. Proc Natl Acad Sci U S A. 2014;111:225–230. doi: 10.1073/pnas.1318115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.