Abstract
The Lyz endolysin of bacteriophage P1 was found to cause lysis of the host without a holin. Induction of a plasmid-cloned lyz resulted in lysis, and the lytic event could be triggered prematurely by treatments that dissipate the proton-motive force. Instead of requiring a holin, export was mediated by an N-terminal transmembrane domain (TMD) and required host sec function. Exported Lyz of identical SDS/PAGE mobility was found in both the membrane and periplasmic compartments, indicating that periplasmic Lyz was not generated by the proteolytic cleavage of the membrane-associated form. In gene fusion experiments, the Lyz TMD directed PhoA to both the membrane and periplasmic compartments, whereas the TMD of the integral membrane protein FtsI restricts Lyz to the membrane. Thus, the N-terminal domain of Lyz is both necessary and sufficient not only for export of this endolysin to the membrane but also for its release into the periplasm. The unusual N-terminal domain, rich in residues that are weakly hydrophobic, thus functions as a signal-arrest-release sequence, which first acts as a normal signal-arrest domain to direct the endolysin to the periplasm in membrane-tethered form and then allows it to be released as a soluble active enzyme in the periplasm. Examination of the protein sequences of related bacteriophage endolysins suggests that the presence of an N-terminal signal-arrest-release sequence is not unique to Lyz. These observations are discussed in relation to the role of holins in the control of host lysis by bacteriophage encoding a secretory endolysin.
Double-stranded DNA phage typically use a holin-endolysin system to achieve lysis of their bacterial hosts (1, 2). Holins are small proteins with no known enzymatic function. They are localized to the cytoplasmic membrane and function to control the timing of lysis. Endolysins are proteins with one of several muralytic activities responsible for the destruction of the peptidoglycan. For example, in the phage λ infection cycle, the holin, a product of the S gene, accumulates in the inner membrane throughout the period of late gene expression, whereas the active endolysin, R, accumulates in the cytoplasm without deleterious effects on the host (3). Suddenly, at a genetically determined time, the holin disrupts the inner membrane, allowing R to attack the peptidoglycan of the infected cell; lysis occurs within seconds. The λ paradigm, where the holin is required for the export of the endolysin from the cytoplasm to the periplasm, was long thought to be universal. However, Sao Jose et al. (4) reported that the endolysin (Lys-44) from oenococcal phage fOg44 carries a cleavable, N-terminal signal sequence that functions in both Escherichia coli and Oenococcus oeni. Thus, the catalytic domain of Lys-44 is exported by the sec translocon, and its signal sequence is proteolytically removed by leader peptidase. Many other phages of Gram-positive bacteria have similar N-terminal signals and are thus likely to be similarly exported. Despite the presence of a secretory endolysin, however, phage fOg44 appears to have a canonical holin gene. Moreover, in fOg44 infections, significant secretion of the endolysin, as monitored by the appearance of the processed product, occurs long before lysis is achieved, indicating that the sec-mediated export of the endolysin was not sufficient for lysis. These findings give rise to a number of questions about the role and mode of action of holins in phages where the endolysin is sec-exported. What is the role of the holin, if not to release the endolysin to the periplasm? How is lysis timing achieved if the endolysin is already secreted across the cytoplasmic membrane? Confounding the resolution of these issues is the limited understanding of the nature of the murein envelope in Gram-positive bacteria, where little is known about the chemical environment, and where, unexpectedly, high levels of a wide range cytosolic enzymes have been found (5).
An investigation of the lysis system of bacteriophage P1, one of the classic coliphages, warrants further study. P1 is unusual in that its endolysin gene, lyz, is not clustered with the holin and antiholin, as is found in all lambdoid and many other phage genomes (6). Moreover, unlike phage λ, which requires both its holin and endolysin to effect host lysis, P1 mutants deleted for the putative holin gene lydA are plaque formers, although lysis is somewhat delayed when compared with the wild-type phage (7, 8). Here, we report the results of experiments to determine the mechanism of the apparent holin independence of P1 Lyz-mediated host lysis. These results are discussed in terms of a type of subcellular localization signal found in a number of endolysins from phages of Gram-negative bacteria, and how this localization is integral to the control of lysis.
Materials and Methods
Bacterial Strains, Growth Media, and Culture Conditions. All bacterial cultures were grown in standard LB medium, supplemented with various antibiotics when appropriate: 100 μg/ml for ampicillin, 10 μg/ml for chloramphenicol, 40 μg/ml for kanamycin, and 10 μg/ml for tetracycline. When indicated, isopropyl β-d-thiogalactoside, dinitrophenol (DNP), NaN3, or CHCl3 were added at final concentrations of 1 mM, 10 mM, 1 mM and 1%, respectively.
The E. coli strains MC4100 and XL1-Blue have been described (9). An azide-resistant mutant of XL1-Blue was selected by plating on LB media containing 1 mM NaN3. Some experiments used RY8653 (MC4100 phoR dsbA::kan1 zih12::Tn10), kindly provided by T. J. Silhavy, Princeton University, Princeton; RY1531 (MC4100 secAts), kindly provided by J. Beckwith (Harvard Medical School, Boston) (10); or TG1 [F′ traD36 proAB lacIq Δ(lacZ)M15/supF hsdD5 thi Δ(lac - proAB)], a phenotypically PhoA- host (11). Standard conditions for the growth of cultures and the monitoring of lysis kinetics have been described (9, 12). When appropriate, the presence of active, cytosolic endolysin in nonlysing cultures was tested for by the addition of CHCl3.
Standard DNA Manipulation, PCR, and DNA Sequencing. Procedures for the isolation of plasmid DNA, DNA amplification by PCR, PCR product purification, DNA transformation, and DNA sequencing have been described (13–15). Oligonucleotides were obtained from Integrated DNA Technologies, Coralville, IA, and were used without further purification. Ligation reactions were performed by using the Rapid DNA ligation kit from Roche Molecular Biochemicals according to the manufacturer's instructions. All other enzymes were purchased from Promega, with the exception of Pfu polymerase, which was from Stratagene. Automated fluorescent sequencing was performed at the Gene Technologies Laboratory in the Department of Biology at Texas A&M University.
Plasmid Construction. The various endolysins, endolysin chimeras, and genes encoding FtsI and PhoA were placed under the control of the lac promoter of pJF118 (16). The DNA inserts for these constructs were PCR-amplified from the following sources: for pJFLyz, the P1 gene lyz was from P1vir DNA; for pJFR, the λ R gene was from pS105 (13); for pJF19, the P22 gene 19 was from P22 DNA; for pJFR21, the bacteriophage 21 R gene was from pBR121 (17); and for pJFFtsI and pJFPhoA, the ftsI and phoA genes were from E. coli chromosomal DNA. To construct pZAdsbA, the dsbA gene was PCR-amplified from E. coli chromosomal DNA. The PCR product was digested with and cloned into unique KpnI and XbaI restriction sites in the chloramphenicol resistance plasmid pZA-31, under control of the pL/tetO-1 promoter (18). The plasmid pFtsIΦLyz, in which the transmembrane domain (TMD) of FtsI replaced the N-terminal hydrophobic domain of Lyz was constructed by first amplifying the DNA encoding the sequence FALLCGCILLALAFLLG from FtsI. The upstream primer had, at its 5′ end, 15 nucleotides of homology to positions -3 to +12 of the gene lyz in pJFLyz. The downstream primer had, at its 5′ end, 15 nucleotides of homology to positions +70 to +85 of lyz. The purified PCR product was then used to conduct a modified site-directed mutagenesis reaction by using the QuikChange kit from Stratagene with pJFLyz as the template. The resultant PCR product was digested with DpnI and was transformed into XL1-Blue. The plasmids pR21ΦLyz, in which the sequence encoding the N-terminal hydrophobic domain of the phage 21 endolysin R21 replaced that of Lyz, pLyzΦ19, in which sequence encoding the N-terminal hydrophobic domain of Lyz was inserted between the first two codons of gene 19 from phage P22; and pLyzΦPhoA, in which the signal sequence of PhoA was replaced with that of the N-terminal hydrophobic domain of Lyz, were constructed in a similar way. A cmyc-tagged allele of the R21 gene was generated by using primers encoding the epitope flanked either with 15 nucleotides of homology 5′ to the insertion site in R21 or 15 nucleotides 3′ to the insertion site. These primers were used for site-directed mutagenesis by using pJFR21 as described above to give pJFR21cmyc. The plasmid pcmycLyzΦ19, in which the cmyc epitope was inserted between the first two codons of the chimeric endolysin found in pLyzΦ19, was constructed in a similar way. All constructs were verified by DNA sequencing.
Subcellular Fractionation and Alkaline Phosphatase Assay. Cell pellets from 40-ml cultures were resuspended in 4 ml of FP buffer (0.1 M sodium phosphate/0.1 M KCl/5 mM EDTA/1 mM DTT/1 mM phenylmethylsulfonyl fluoride, pH 7.0) and were then disrupted by passage through a French pressure cell (Spectronic Instruments, Rochester, N.Y.) at 16,000 psi (1 psi = 6.89 kPa). The membrane and soluble fractions were separated by centrifugation at 100,000 × g for 60 min at 16°C. To isolate the periplasmic fraction, cell pellets from 25-ml cultures were resuspended in 500 μl of 25% sucrose/30 mM Tris·HCl, pH 8.0. Next, 10 μl of 0.25 M EDTA, 10 μl of lysozyme (20 mg/ml), and 500 μl of distilled water were added in sequence. After 5 min at room temperature, microscopic examination showed that ≈95% of the cells had formed spheroplasts. The samples were centrifuged at 8,000 × g for 30 min to separate the released periplasm from the spheroplasts (membrane and cytosol).
PhoA activity assays was determined by using p-nitrophenyl phosphate as the substrate and a millimolar extinction coefficient of 18.3 for p-nitrophenol at 420 nm. One milliunit of activity is the amount of enzyme needed to form 1 μm of product per minute under standard assay conditions (19). TG1 cells carrying the empty vector, pJF, contained <5% of the activity detected in cells carrying pJFLyzΦPhoA.
SDS/PAGE and Western Blotting. SDS/PAGE, Western blotting, and immunodetection were performed as described (13). Antibodies against the purified His6-tagged Lyz and λ R endolysins were prepared in chickens by Aves Labs, (Tigard, OR). Proteins tagged with the cmyc epitope were detected by using a mouse monoclonal antibody from Babco (Richmond, CA). For detection of PhoA and its derivatives, a rabbit polyclonal antibody from 5 Prime → 3 Prime was used. Horseradish peroxidase-conjugated secondary antibodies against chicken IgY, mouse IgG, and rabbit IgG were from Aves Labs, Pierce, and Pierce, respectively. Generally, primary antibodies were used at a 1:1,000 dilution, whereas secondary antibodies were used at a 1:3,000 dilution. Blots were developed by using the chromogenic substrate 4-chloro-1-naphthol (Sigma) or with the SuperSignal chemiluminescence kit (Pierce). Equivalent sample loadings were used whenever multiple fractions obtained from the same culture were analyzed.
Results
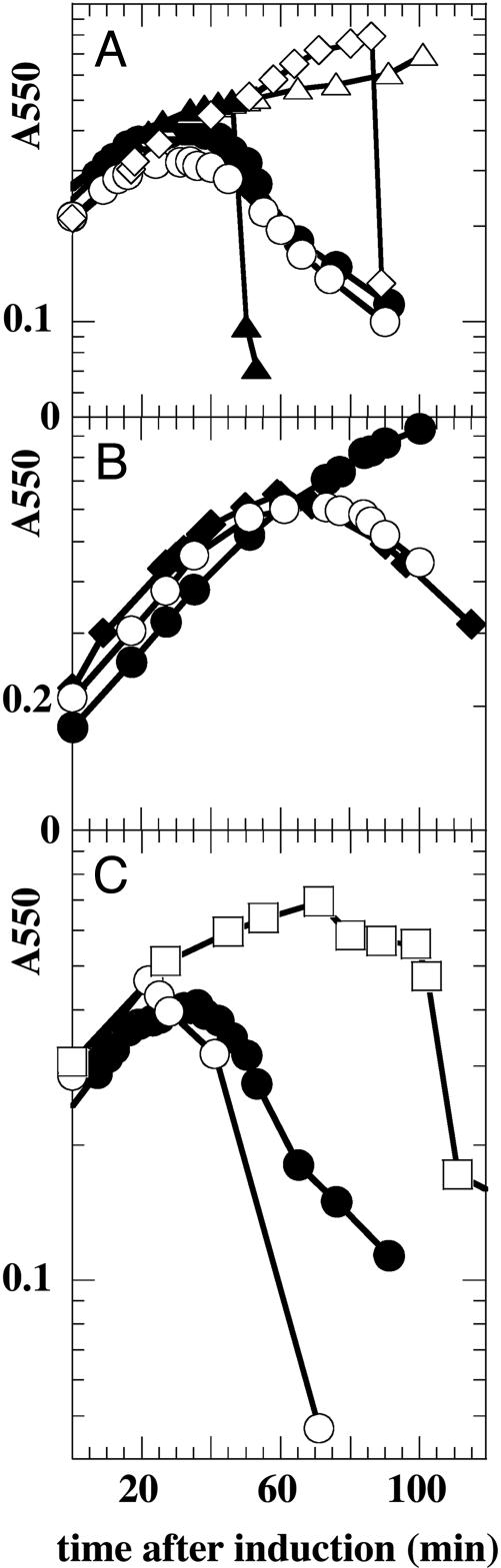
The P1 Endolysin Causes Lysis of E. coli in the Absence of Holin Function. The lyz gene encodes the P1 endolysin of 185 residues, which is homologous to the T4 gpe lysozyme. Unexpectedly, when lyz was cloned under an inducible promoter and expressed in logarithmically growing cells in the absence of a holin gene, overt lysis was observed beginning within 35 min (Fig. 1A). Inspection of the culture before lysis revealed that the cells began adopting a spherical morphology ≈25 min after induction. Both of these observations suggest that Lyz can gain access to the periplasm and degrade the host peptidoglycan in the absence of its cognate holin. Inspection of the predicted Lyz sequence reveals a hydrophobic domain potentially capable of serving as a signal sequence (Fig. 2). The possibility that this sequence allows export of Lyz by using the sec system was tested by examining the effect of the SecA inhibitor, azide, on the holin-independent lysis observed after induction of the cloned lyz gene. Azide was found to inhibit Lyz-mediated lysis in a host carrying the wild-type secA locus, but not in an isogenic strain carrying an azide-resistant allele of secA. The addition of CHCl3 to permeabilize the membrane of the azide-treated culture resulted in its immediate lysis, which is consistent with the azide effect being at the level of membrane translocation (Fig. 1 A). Similar results were obtained by using a secAts allele at the nonpermissive temperature. Again, lysis was blocked until CHCl3 was added to permeabilize the membrane (Fig. 1 A).
Fig. 1.
Induction of the Lyz endolysin results in holin-independent lysis. (A) Lyz-mediated lysis requires SecA. Azide-sensitive (▴, ▵, and •) or resistant (○) XL1-Blue cells carrying pJFLyz were induced at time 0, and culture turbidity was followed as a function of time. To two of the azide-sensitive cultures (▴ and ▵), 1 mM sodium azide was added 10 min after induction, and to one of these cultures (▴), CHCl3 was added at 50 min. In another culture of MM52 secAts pJFLyz (⋄), the cells were shifted from 30°C to 42°C at 90 min before induction, and CHCl3 was added at 85 min after induction. Cells retained at 30°C and then induced underwent lysis beginning at 50 min after induction (data not shown). (B) Lyz function requires DsbA activity. MC4100dsbA+pJFLyz (♦), RY8653 dsbA::kan pJFLyz pZA-31 (•), and RY8653 dsbA::kan pJFLyz pZA-dsbA (○) were induced at time 0 and were monitored for turbidity as a function of time. (C) Lyz-mediated lysis is triggered by energy poisons unless SecA-mediated secretion is inhibited. Cultures of XL1-Blue carrying pJFLyz were induced at time 0 and were monitored for turbidity as a function of time. To one culture (•), no further additions were made. To a second culture, 10 mM DNP was added 20 min after induction (○), and to a third, 1 mM sodium azide was added 10 min after induction, followed by 10 mM DNP at 70 min and CHCl3 at 100 min (□).
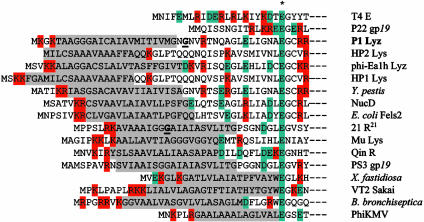
Fig. 2.
N-terminal sequences of P1 Lyz and other homologs of T4 gpe lysozyme. The N-terminal regions of P1 Lyz and other phage or prophage-encoded homologs of T4 lysozyme are displayed, aligned by the presumptive catalytic Glu residue (indicated by an asterisk), and compared with the soluble lysozyme sequences of the canonical lysozyme T4 gpe (GI126605) and with the P22 endolysin gp19 (GI963553). The putative TMD of the SAR signal is high-lighted in gray, and basic and acidic residues are highlighted in red and teal, respectively. The residue after which there is a potential signal sequence cleavage site in the P1 Lyz sequence is in bold and is underlined. In addition to the endolysins from P1, T4, and P22, endolysins shown from functional bacteriophages include: Lys of phage Mu (GI9633512), Lys of Haemophilus influenzae phage HP1 (GI1708889), Lyz of Erwinia amylovora phage phiEA1H (GI11342495), gp45 of Pseudomonas aeruginosa phage φKMV, R21 of lambdoid phage 21 (GI126600), and gp19 of Salmonella typhimurium phage PS34 (GI3676081). In the R21 sequence, the Gly-16 residue altered to Cys in the R21ΦLyz fusion is underlined and is bold. Included in this list are putative endolysins from: a Yersinia pestis prophage (GI16122337), from a Fels-2-like prophage of uropathogenic E. coli (GI26246847), a prophage of Xylella fastidiosa (GI15837115), a prophage of Bordetella bronchiseptica (GI33602455), and the VT2 Sakai prophage of O157:H7 E. coli (15834216). Also included is the chromosomal endolysin NucD, encoded by a prophage remnant in Serratia marcescens, and the endolysin R from Qin, a cryptic prophage segment from E. coli K-12 (GI26249022), both of which have been demonstrated to have lytic function (32, 33). (Only a representative set of the putative SAR endolysins is shown.) Accession nos. are for the GenBank database.
Additional genetic evidence that Lyz is exported to the periplasm was obtained by using a dsbA host. There are seven cysteine residues in Lyz, six of which reside in its hydrophilic, catalytically active C-terminal domain and might form up to three disulfide bonds if this domain is externalized to the periplasm. These disulfides could be necessary for either the stability or activity of Lyz. As can be seen in Fig. 1B, Lyzmediated lysis is not observed in a dsbA host but is recovered when dsbA gene function is provided from a compatible plasmid. Significantly, no Lyz could be detected by Western blot in dsbA cells, suggesting that in the absence of periplasmic DsbA activity all of the Lyz protein misfolds and is degraded. However, if dsbA cells are treated with 1 mM azide before induction, Lyz does accumulate and can be detected by Western blot (Fig. 6, which is published as supporting information on the PNAS web site). This finding suggests that normally there is no cytoplasmic pool of Lyz.
In comparison with the saltatory and rapid lysis seen in a P1-infected culture (20), the lysis observed in cells expressing P1 gene lyz alone is gradual. However, the addition of DNP to cultures 20 min after the induction of Lyz expression dramatically accelerates lysis (Fig. 1C). Cyanide has a similar effect (data not shown), suggesting that the activity of the exported Lyz remains largely cryptic until the proton-motive force (pmf) across the cytoplasmic membrane is dissipated. Significantly, the addition of DNP to azide-inhibited cultures did not result in lysis, even though the cells contained sufficient, active Lyz to cause lysis after the addition of CHCl3 (Fig. 1C). We conclude that Lyz is externalized by the sec system, requires DsbA to catalyze the formation of stabilizing disulfide bonds in the periplasm, and can be activated to induce lysis by collapse of the pmf.
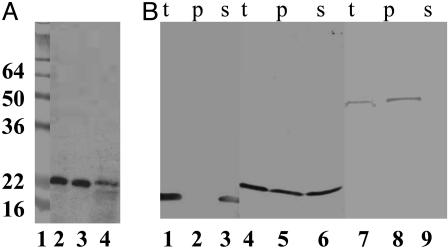
Subcellular Localization of Lyz: The Signal-Arrest-Release (SAR) Sequence. When cells expressing the lyz gene were disrupted and fractionated, the Lyz protein was found in both soluble and membrane fractions (Fig. 3A). This unexpected dual localization was found to be insensitive to the level of expression of lyz (Fig. 7, which is published as supporting information on the PNAS web site). Moreover, induction of a P1 lysogen and the plasmid-borne lyz clone resulted in the production of comparable levels of Lyz protein which exhibited identical mobility after SDS/PAGE (Fig. 7B). Thus, the dual localization of Lyz in the absence of proteolytic processing is not an artifact of overexpression from a plasmid clone. To assess the localization of the soluble fraction, cells expressing lyz were converted into spheroplasts. Approximately half of the Lyz fractionated with the soluble periplasmic contents (Fig. 3B). As controls, the bacteriophage λ endolysin, R, and alkaline phosphatase were found almost exclusively in the spheroplast (cytosol and membranes) and periplasmic fractions, respectively. Analysis of the Lyz sequence by algorithms designed to predict the presence of secretory signals gave conflicting results; the N-terminal hydrophobic domain was predicted to be either a cleavable signal sequence or an N-terminal signalarrest domain. Eliminating the putative signal sequence cleavage site (Fig. 2) by site-directed mutagenesis was without effect on either inducible lysis or the presence of the enzyme in the soluble and membrane fractions (data not shown). Moreover, the membrane and soluble forms of Lyz had identical mobilities in SDS/PAGE (Fig. 3A). Finally, when a c-myc epitope was added at the N terminus of the gene product, both soluble and membrane forms were immunoreactive with anti-cmyc antibody (data not shown). Thus, although periplasmic, the soluble form of Lyz is not generated from the membrane-associated form by proteolytic removal of the N-terminal hydrophobic domain. Attempts to demonstrate proteinase K sensitivity of spheroplast-associated Lyz failed, presumably due to the resistance of the membrane-bound form to proteolysis. Nevertheless, all of the Lyz that fractionated with the membranes could be extracted with detergent (data not shown), indicating that the insoluble fraction did not represent inclusion bodies that cosedimented with the membranes but instead consisted of Lyz protein embedded in the bilayer by its N-terminal TMD.
Fig. 3.
Subcellular localization of Lyz. (A) Lyz is found in both the membrane and soluble fractions. Total membrane and soluble fractions were prepared from an induced culture of XL1-Blue cells carrying pJFLyz and were analyzed by SDS/PAGE and immunoblotting as described in Materials and Methods. Lane 1, molecular mass standards; lane 2, total culture lysate; lane 3, soluble fraction; and lane 4, membrane fraction. The masses of the prestained standards are given in kDa on the left. (B) Soluble Lyz is found in the periplasm. Periplasmic and spheroplast fractions were prepared from induced cultures of MC4100 carrying either pJFR (lanes 1–3), pJFLyz (lanes 4–6), or pJFPhoA (lanes 7–9), and were analyzed by SDS/PAGE and immunoblotting as described in Materials and Methods. t, total culture; p, periplasmic fraction; s, spheroplasts.
Taken together, these results strongly indicate that the N-terminal hydrophobic domain of Lyz first serves as a signal-arrest domain, in directing externalization of the enzyme by the sec translocon without leader peptidase cleavage, leaving the protein tethered to the membrane, but then also allows release into the periplasm. Consequently we propose to designate this domain as a SAR domain, another class of secretion signal.
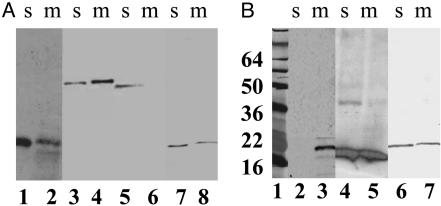
The SAR Domain of Lyz Is Necessary and Sufficient for Localization to Two Cell Compartments. To test whether the SAR domain of Lyz is sufficient to localize a protein to both the cytoplasmic membrane and the periplasm, this sequence was either used to replace the normal, cleavable signal sequence of the periplasmic enzyme, PhoA, or fused to the N terminus of the soluble, cytoplasmic endolysin from bacteriophage P22, gp19. The LyzΦgp19 protein was functional, and ≈20% of it was membrane-bound (Fig. 4A). The LyzΦPhoA fusion protein was also found to be distributed in both the membrane and soluble fractions (Fig. 4A). Moreover, the membrane and soluble forms of LyzΦPhoA had identical mobilities in SDS/PAGE, indicating that the soluble form is not generated from the membrane-associated form by proteolysis of the fusion protein. The specific activity of the two forms is approximately the same, based on the relative signal in the Western blot (Fig. 4A) and on PhoA enzyme assays, which showed 8 milliunits/ml culture and 32 milliunits/ml culture in the soluble and membrane fractions, respectively. Because PhoA is active only in the periplasm, this result indicates that the catalytic domains of both the membrane and soluble forms are externalized and that there is no significant proportion in the cytosol.
Fig. 4.
The SAR domain of Lyz is sufficient for localization in both membrane and soluble compartments. (A) The N-terminal domain of Lyz confers partial membrane localization on soluble proteins. Total membrane and soluble fractions were prepared from induced cultures of XL1-Blue cells carrying pJFLyz (lanes 1–2), TG1 cells carrying pJFLyzΦPhoA (lanes 3–4), pJFPhoA (lanes 5–6), or pcmycLyzΦ19 (lanes 7–8), and were analyzed by SDS/PAGE and immunoblotting as described in Materials and Methods. s, soluble fractions; m, membrane fractions. (B) The N terminus of the phage 21 endolysin is a functional SAR sequence. Total membrane and soluble fractions were prepared from induced cultures of XL1-Blue cells carrying pJFFtsIΦLyz (lanes 2–3), pJFR21cmyc (lanes 4–5), or pJFR21ΦLyz (lanes 6–7), and were analyzed by SDS/PAGE and immunoblotting as described in Materials and Methods. The masses of the prestained standards (lane 1) are given in kDa on the left.
Next, we replaced the N-terminal domain of Lyz with the well characterized signal-arrest domain of FtsI (21). The FtsIΦLyz chimera was recovered exclusively in the membrane fraction (Fig. 4B). We were unable to localize the reciprocal LyzΦFtsI fusion protein, presumably because its instability prevented detection by Western blotting. Finally, we examined the distribution of another endolysin with a potential SAR domain, the endolysin R21 from bacteriophage 21 (Fig. 2). When synthesized in E. coli, R21, like Lyz, causes lysis in the absence of its cognate holin (data not shown). As with Lyz, R21 was found in both soluble and membrane-bound forms (Fig. 4B). When the putative SAR domain of R21 was used to replace the SAR sequence of Lyz, the chimera R21ΦLyz, while enzymatically inactive, was localized similarly to wild-type R21 (Fig. 4B). The lack of activity of R21ΦLyz was surprising, considering the similarity of the sequences of these two endolysins (Fig. 2). The most obvious difference between the R21 and Lyz SAR sequences was the lack of a cysteine residue in the former. When Gly-13 in R21ΦLyz (corresponding to Gly-16 in the R21 sequence; see Fig. 2) was changed to a Cys residue, the chimera became lytically active (data not shown). Thus, the SAR domains of Lyz and R21 are essentially interchangeable, except for the requirement of a cysteine residue in the former, a finding that will be considered elsewhere (M.X., A. Arelandu, D.K.S., S. Swanson, J. Sacchettini, and R.Y., unpublished work).
Discussion
A Subcellular Localization Signal: The SAR Sequence. The results presented here demonstrate that the phage P1 endolysin, Lyz, is secreted by the sec translocon of the host, unlike the well studied endolysins of other phages of Gram-negative bacteria, all of which have been shown to accumulate in the cytosol and to be released by holin-mediated membrane disruption (1, 2). The N-terminal secretory signal of Lyz is not removed during export, suggesting that it constitutes a signal-arrest domain, which, on completion of translocon function, tethers the exported protein to the membrane. Surprisingly, however, a significant portion of the Lyz is found in the periplasm. This finding is independent of the level of expression of lyz, because identical results were found with lyz mounted on a low copy number plasmid (Fig. 7) and also when lyz, with its cognate translation signals, was used to replace the SR genes of phage λ (see Supporting Materials and Methods and Fig. 8, which are published as supporting information on the PNAS web site). Because the sec translocon will initially localize an uncleaved, signal-arrest domain to the membrane, the presence of a significant fraction of the protein in the periplasm indicates that some of the initially membrane-bound protein was subsequently released into the periplasm. Consequently, the N-terminal domain of the P1 endolysin represents another type of subcellular localization signal, the SAR sequence. The unusual feature of the SAR sequence is that it endows a protein with the ability to convert from a membrane-integrated state to a freely soluble state, without proteolytic cleavage. The SAR sequence from Lyz was found to confer the two-compartment disposition on fusions with the P22 lysozyme, gp19 and with PhoA (Fig. 4A). Thus, the Lyz SAR sequence is both necessary and sufficient for the sec-mediated export to a membranetethered state and subsequent release to the periplasm.
The endolysin gene R21 of lambdoid phage 21 also encodes a T4 gpe homolog with a functional SAR sequence. The SAR sequences of Lyz and R21 do not share significant sequence similarity but do share the characteristic of having 40–60% of the residues as Gly or Ala, which contribute very little to the hydrophobic character of TMDs (ref. 22 and Fig. 2). A survey of T4 lysozyme homologs in phage, prophage, and putative prophage sequences reveals that many have N-terminal domains that resemble the SAR sequences defined functionally in Lyz and R21 (Fig. 2). Overall, 57% of the residues in these SAR sequences are either the weakly hydrophobic residues, Gly and Ala, or uncharged polar residues, Ser, Thr, Gln, and Tyr (Fig. 2). In contrast, the average TMD normally has only 36% of these types of residues (23). How much this contributes to the unusual character of the SAR sequence to allow release of the N-terminal TMD is an open question; other less obvious factors, such as the lack of TMD oligomerization motifs (24), may also be important. One feature that may be important for the function of the SAR sequence is the relative paucity of basic residues in their cytoplasmically disposed flanking sequences. All of the SAR endolysins for which there is experimental evidence of lytic function have 0–2 basic residues in a very short predicted cytoplasmic domain, and most of these are Lys residues, which have considerable hydrophobic character due to the C4H8 component of its side chain. In addition, most SAR endolysins have a short turn-predicted sequence between the bulk of the hydrophobic domain and the periplasmic domain (Fig. 2). This feature may facilitate the folding up of the SAR helix against the globular mass of the catalytic domain when the protein is released from the membrane. It will be of interest to see whether proteins other than phage lysozymes have SAR N-terminal domains.
Interestingly, a precedent exists for the release of a tethered protein from the membrane in certain signal-sequence mutants of lamB. A double mutation near the cleavage site (A23D and A25Y) abolishes leader peptidase cleavage, leaving the LamB tethered to the membrane by its uncleaved signal sequence, where it is rapidly degraded. However, a suppressor R6L allows LamB to reach the outer membrane and fold into functional porin and λ receptor, with the mutant signal sequence intact (25). Inspection of the mutant sequences suggests that the suppressor may be creating a SAR sequence from the LamB signal sequence (Fig. 9, which is published as supporting information on the PNAS web site). The triple mutant has a signal-arrest domain of 19 uncharged residues because the four-carbon aliphatic segment of the Lys side chain allows its α carbon to be buried a full helical turn within the bilayer (26). This domain resembles the SAR sequences in having 10 of 19 residues either negligibly hydrophobic (Ala or Gly) or overtly hydrophilic (Lys, Ser, Thr, and Gln), with only a single basic side chain to serve as a membrane anchor. Duguay and Silhavy (25) speculated that this mutant protein might be assisted in exiting the membrane by periplasmic folding factors, a notion that could also apply to the endolysins described here.
Host Lysis by Bacteriophage P1. Phage P1 is one of the most intensively studied phages, which was established as a major experimental system the same year as phage λ (27). P1, like other classical phages, was subjected to thorough amber mutant screening, by which all essential genes were identified. Two essential genes with primary lysis phenotypes were found: lyz, which encodes the endolysin of P1 and has a lysis-negative null phenotype; and lydB, encoding the antiholin, which has a null phenotype of early lysis, such that no plaque-forming units are generated (20). Strikingly absent was a lysis defect that could be attributed to a holin. The results presented here provide an rationale for this long-term mystery. We have shown conclusively that lysis can be mediated by the endolysin, Lyz, which is capable of attacking the host murein without requiring a holin to disrupt the membrane. Instead, Lyz is exported by the host sec machinery. Thus, the lydA amber mutant was never isolated because it does not have a lysis-negative phenotype detectable in simple plate tests.
To our knowledge, this report is the first of its kind of a sec-exported endolysin in phages of Gram-negative bacteria and serves to both confirm and generalize the results reported by Santos and colleagues (4), who have shown that endolysins encoded by phages of Gram-positive hosts can have secretory signal sequences. In the well studied cases of phage λ and T4, where the endolysins absolutely require holin function for escape from the cytoplasm, it has been demonstrated that the timing of lysis, and thus the yield of virions for the infection, is an exquisitely sensitive function of the primary structure of the holins (28–31). Thus, the finding of the endolysins with cleavable signal sequences was a surprise and posed a challenge to the presumed central regulatory role of holins. Nevertheless, the available physiological data and also genomic analysis suggest that even in phages with secretory endolysins, in both Gram-negative and Gram-positive systems, a holin is present.
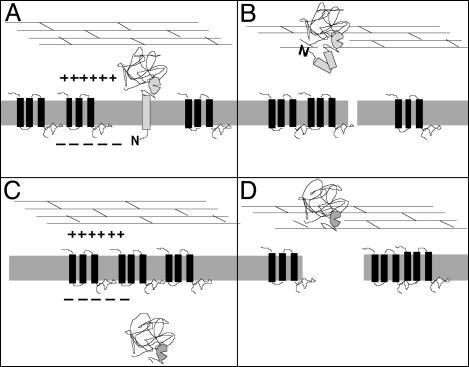
Role of the Holin in the Regulation of the SAR Endolysin. With the properties of the SAR endolysins in mind, it is proposed that there are two different modes by which holins can control lysis timing; both can be viewed as activation of the endolysin (Fig. 5). At the programmed time, the canonical holins of λ and T4 disrupt the membrane and allow escape of the cytoplasmically located, active endolysin to the periplasm, after which degradation of the murein and lysis follow within seconds (Fig. 5 A and B). With the SAR endolysins, we suggest that the endolysin is first localized to the periplasm in its membrane tethered form, where it is either inactive or its activity is cryptic (i.e., restrained from access to the peptidoglycan). Triggering of the holin at the programmed lysis time will facilitate the instantaneous and quantitative release of the SAR endolysin from the membrane. This result might simply be due to the collapse of the pmf that occurs after holin triggering. Indeed, energy poisons were found to trigger Lyz-mediated lysis unless Lyz secretion is prevented by the inhibition of SecA (Fig. 1C). The observation that loss of the pmf alone is sufficient to release Lyz from the membrane might explain why much of this endolysin is found in the soluble/periplasmic fractions when cells are subjected to subcellular fractionation well before overt lysis occurs. In this instance, the pmf is transiently depressed by the manipulations necessary to obtain the subcellular fractions allowing release of the membrane-tethered endolysin. This finding would imply that holin-independent lysis mediated by Lyz is due to the slow, spontaneous release of a small fraction of the membrane-tethered Lyz, accounting for its gradual nature when compared with natural P1 infections.
Fig. 5.
Model for triggering of lysis with SAR endolysins. (A) A SAR endolysin is initially tethered in an inactive form to the energized membrane, in which the holin protein accumulates without affecting the pmf. (B) At the programmed lysis time, the holin triggers, disrupting the membrane sufficiently to abolish the pmf, and perhaps also to assist the liberation of the endolysin from the membrane, which results in activation of the endolysin. In contrast, with the canonical lysozymes of T4, λ, and T7, the endolysin accumulates in its active form in the cytosol (C) and, when the holin triggers, is released to the cell wall by membrane disruption sufficient to allow passage of large proteins (D).
It should be noted that a more active role for the holin in facilitating the release of Lyz is not precluded by this analysis. In vivo, the holin may directly facilitate the exit of the SAR sequences by causing a more profound disruption of the membrane than just ablation of the pmf. In any case, it seems that with both canonical endolysins, retained within the cytosol, and SAR endolysins, prepositioned to the periplasm, holin action may be considered to be the programmed activation of the muralytic enzyme.
Supplementary Material
Acknowledgments
We thank Michael Yarmolinsky for providing strains; Tram Anh Tran for providing plasmid pJFPhoA; other members of the R.Y. group for critical discussions; and Mario Santos and colleagues for providing unstinting communication of unpublished work, which was essential to the concepts discussed here. This work was supported by Public Health Service Grant GM27099 and Welch Foundation Award A1384 (to R.Y.), and by the office of the Vice President for Research at Texas A&M University.
Abbreviations: DNP, dinitrophenol; pmf, proton-motive force; TMD, transmembrane domain; SAR, signal-arrest-release.
References
- 1.Wang, I.-N., Smith, D. L. & Young, R. (2000) Annu. Rev. Microbiol. 54, 799-825. [DOI] [PubMed] [Google Scholar]
- 2.Young, R., Wang, I.-N. & Roof, W. D. (2000) Trends Microbiol. 8, 120-128. [DOI] [PubMed] [Google Scholar]
- 3.Gründling, A., Manson, M. D. & Young, R. (2001) Proc. Natl. Acad. Sci. USA 98, 9348-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.São-José, C., Parreira, R., Vieira, G. & Santos, M. A. (2000) J. Bacteriol. 182, 5823-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkins, J. C., Beighton, D. & Homer, K. A. (2003) Appl. Environ. Microbiol. 69, 5290-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt, C., Velleman, M. & Arber, W. (1996) J. Bacteriol. 178, 1099-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iida, S. & Arber, W. (1977) Mol. Gen. Genet. 153, 259-269. [DOI] [PubMed] [Google Scholar]
- 8.Yarmolinsky, M. B. & Sternberg, N. (1988) in The Bacteriophages, ed. Calendar, R. (Plenum, New York), pp. 291-438.
- 9.Smith, D. L., Struck, D. K., Scholtz, J. M. & Young, R. (1998) J. Bacteriol. 180, 2531-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliver, D. B. & Beckwith, J. (1981) Cell 25, 765-772. [DOI] [PubMed] [Google Scholar]
- 11.Nørholm, M. H. & Dandanell, G. (2001) J. Bacteriol. 183, 4900-4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, C.-Y., Nam, K. & Young, R. (1995) J. Bacteriol. 177, 3283-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gründling, A., Bläsi, U. & Young, R. (2000) J. Biol. Chem. 275, 769-776. [DOI] [PubMed] [Google Scholar]
- 14.Smith, D. L. & Young, R. (1998) J. Bacteriol. 180, 4199-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith, D. L., Chang, C.-Y. & Young, R. (1998) Gene Expression 7, 39-52. [PMC free article] [PubMed] [Google Scholar]
- 16.Fürste, J. P., Pansegrau, W., Frank, R., Blöcker, H., Scholz, P., Bagdasarian, M. & Lanka, E. (1986) Gene 48, 119-131. [DOI] [PubMed] [Google Scholar]
- 17.Barenboim, M., Chang, C.-Y., dib Hajj, F. & Young, R. (1999) Mol. Microbiol. 32, 715-727. [DOI] [PubMed] [Google Scholar]
- 18.Lutz, R. & Bujard, H. (1997) Nucleic Acids Res. 25, 1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulein, R., Rutz, C. & Rosenthal, W. (1997) Protein Eng. 10, 707-713. [DOI] [PubMed] [Google Scholar]
- 20.Walker, J. T. & Walker, D. H., Jr. (1980) J. Virol. 35, 519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guzman, L. M., Weiss, D. S. & Beckwith, J. (1997) J. Bacteriol. 179, 5094-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson, I., Johnson, A. E. & von Heijne, G. (2003) J. Biol. Chem. 278, 29389-29393. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Y., Engelman, D. M. & Gerstein, M. (2002) Genome Biol. 3, 54.1-54.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curran, A. R. & Engelman, D. M. (2003) Curr. Opin. Struct. Biol. 13, 412-417. [DOI] [PubMed] [Google Scholar]
- 25.Duguay, A. R. & Silhavy, T. J. (2002) J. Bacteriol. 184, 6918-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Killian, J. A. & von Heijne, G. (2000) Trends Biochem. Sci. 25, 429-434. [DOI] [PubMed] [Google Scholar]
- 27.Bertani, G. (1951) J. Bacteriol. 62, 293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, I.-N., Deaton, J. F. & Young, R. (2003) J. Bacteriol. 185, 779-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramanculov, E. R. & Young, R. (2001) Gene 265, 25-36. [DOI] [PubMed] [Google Scholar]
- 30.Gründling, A., Bläsi, U. & Young, R. (2000) J. Bacteriol. 182, 6082-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raab, R., Neal, G., Sohaskey, C., Smith, J. & Young, R. (1988) J. Mol. Biol. 199, 95-105. [DOI] [PubMed] [Google Scholar]
- 32.Berkmen, M., Benedik, M. J. & Bläsi, U. (1997) J. Bacteriol. 179, 6522-6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaiser, K. (1980) Mol. Gen. Genet. 179, 547-554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.