Abstract
The expression of certain genes involved in fundamental metabolism is regulated by metabolite-binding “riboswitch” elements embedded within their corresponding mRNAs. We have identified at least six additional elements within the Bacillus subtilis genome that exhibit characteristics of riboswitch function (glmS, gcvT, ydaO/yuaA, ykkC/yxkD, ykoK, and yybP/ykoY). These motifs exhibit extensive sequence and secondary-structure conservation among many bacterial species and occur upstream of related genes. The element located upstream of the glmS gene in Gram-positive organisms functions as a metabolite-dependent ribozyme that responds to glucosamine-6-phosphate. Other motifs form complex folded structures when transcribed as RNA molecules and carry intrinsic terminator structures. These findings indicate that riboswitches serve as a major genetic regulatory mechanism for the control of metabolic genes in many microbial species.
Riboswitches are highly structured domains within mRNAs that precisely sense metabolites and control gene expression (1). These RNA elements are capable of binding to a variety of target compounds and subsequently modulating transcription and translation with performance characteristics that are similar to those of protein genetic factors. Typically, each riboswitch is composed of a conserved metabolite-binding domain (aptamer) located upstream of a variable sequence region (expression platform) that dictates the level of gene expression. Allosteric changes brought about by metabolite binding to the aptamer are harnessed by the expression platform to modulate the expression of the adjacent gene or operon. Riboswitches are versatile genetic control elements. In some instances, both transcription and translation control are used by the same aptamer class in the same prokaryotic organism (e.g., see ref. 2). Evidence also shows that riboswitches can use mRNA-processing events to modulate gene expression (3, 4).
The various metabolites that are detected by known riboswitches are of fundamental importance to living systems (5). On this basis, we have speculated that modern riboswitches might be the remaining representatives of an ancient metabolite-monitoring system that was present in the RNA World (5–9). The wide distribution of some riboswitch classes among microbes (e.g., see refs. 5 and 9–14) and the presence of metabolite-binding RNA domains in eukaryotes (4) support this hypothesis. Each of the seven classes of riboswitches reported (1, 5) was examined for metabolite-binding function because published genetic evidence showed that these elements were important for genetic control. Because the regulation of many metabolism genes has not been characterized in detail, it is possible that numerous other metabolite-binding RNA motifs exist in nature.
The riboswitches known to be present in prokaryotes are typically located in noncoding or intergenic regions (IGRs). Therefore, the examination of unusually long IGRs for indications of conserved sequence and secondary-structure elements should yield new riboswitch candidates. To identify such candidates, we created a database known as the Breaker Laboratory Intergenic Sequence Server (BLISS; http://bliss.biology.yale.edu) that integrates sequence similarity between IGRs from 91 microbial genomes with uniform predictions of gene functions and intrinsic transcription terminators. Examination of BLISS revealed many conserved sequence elements, including six new structural motifs in Bacillus subtilis (Table 1) that are candidates for previously unreported riboswitches.
Table 1. Characteristics of RNA elements that are riboswitch candidates in B. subtilis.
| RNA element | Num | Distribution | Term | Reg | Gene functions |
|---|---|---|---|---|---|
| glmS | 18 | B/C, Fus | — | — | GlcN6P synthetase |
| gcvT | 27 | B/C, α, β, γ, Act | + | + | Glycine cleavage system, Na+/alanine symporter |
| ydaO/yuaA | 15 | B/C, Act | + | — | K+ transporter, membrane metalloproteases |
| ykkC/yxkD | 19 | B/C, Cya, α, β, γ, ε | + | + | Multidrug resistance, N/S/B transport |
| ykoK | 21 | B/C, β, γ, Act | + | — | Divalent metal transporters |
| yybP/ykoY | 56 | B/C, Cya, α, β, γ, Act | + | + | Cation transport ATPase |
Motifs are named for the initial gene of each downstream operon in B. subtilis. The number of sequence representatives (Num), evolutionary distribution, presence of an intrinsic terminator before downstream start codons in Gram-positive bacteria (Term), and predicted or known effect of metabolite binding by the RNA element on gene expression (Reg) are shown for each element. Bacterial classification abbreviations: B/C, Bacillus/Clostridium; α, α-proteobacteria; β, β-proteobacteria; γ, γ-proteobacteria; ε, ε-proteobacteria; Cya, cyanobacteria; Fus, fusobacteria; Act, actinobacteria. N/S/B, nitrate/sulfonate/bicarbonate.
Materials and Methods
Bioinformatics Strategies. The BLISS database and detailed methods are available on the internet (http://bliss.biology.yale.edu). IGRs with a minimum length of 30 nucleotides from 91 complete genomes were analyzed. Conservation between each B. subtilis IGR and other intergenic sequences was identified by blastn searches, and similar IGRs were pair-wise aligned by using the fasta package to highlight additional conservation. For each genome, gene functions were assigned uniformly with the COG database (15), and intrinsic transcription terminators were predicted by a modified version of the program transterm (16). A web interface allows IGR sequence alignments and associated evidence of riboswitch function to be interactively viewed and annotated. Promising secondary-structure models were iteratively refined and extended by motif searching with the program sequencesniffer (J.E.B. and R.R.B., unpublished algorithm) and additional blast searches.
Oligonucleotides and Chemicals. Synthetic DNAs were purchased and purified as described (2). Substrate RNA for the glmS ribozyme assays was chemically synthesized by Dharmacon Research (Lafayette, CO). Radiolabeled [γ-32P]ATP was purchased from Amersham Pharmacia. Glucosamine-6-phosphate (GlcN6P) and glucose-6-phosphate were purchased from Sigma. RNA molecules were prepared by transcription in vitro by using the appropriate PCR-DNA templates and RiboMAX transcription kits (Promega). Templates for transcription were PCR-amplified from chromosomal DNA extracted from B. subtilis strains (Bacillus Genetic Stock Center, Columbus, OH) 1A40 (used for preparation of yybP, ydaO, gcvT, and glmS), 1A210 (yuaA), and 1A234 (ykkC, yxkD, ykoK, and ykvJ). The resulting purified RNAs were 5′-labeled with 32P by using methods similar to those described (17).
Ribozyme Assays and In-line Probing. Ribozyme assays were conducted with ≈5 nM 5′-32P-labeled RNA substrate and were incubated for 5 min at 23°C in the presence of 50 mM Tris·HCl (pH 7.5 at 23°C)/200 mM KCl/10 mM MgCl2/100 nM ribozyme and in the absence or presence of 100 μM effector as indicated for each experiment. Reactions were terminated with an equal volume of 2× gel-loading buffer [90 mM Tris base/90 mM borate/8 M urea/20% sucrose (wt/vol)/1 mM EDTA/0.1% SDS/0.05% xylene cyanol FF/0.05% bromophenol blue), which was supplemented with EDTA to a final concentration of 100 mM. The products were separated by using denaturing 20% PAGE and analyzed by using a PhosphorImager (Molecular Dynamics). In-line probing assays were carried out by using a protocol adapted from those described (2, 18).
Results
Establishing Riboswitch Characteristics. The architectural features of known riboswitches were exploited to identify candidate regulatory motifs. Aptamer domains of riboswitches adopt complex structures that precisely recognize metabolites. Consequently, they tend to reside in unusually large intergenic regions with mixed nucleotide compositions. Indeed, the median length of the 25 B. subtilis IGRs containing known riboswitches is 330 nucleotides compared with only 152 nucleotides for the complete set of 2913 IGRs that were assessed from this organism. Examination of riboswitch sequence alignments generated by BLISS reveals a collection of base-paired stems that in many instances are supported by the presence of nucleotide covariation. Search algorithms that incorporate this secondary structure information and allow the insertion of variable loops can then be used to find new examples of the RNA element.
Riboswitches appear to have a wider evolutionary distribution than most DNA- and RNA-binding protein regulatory factors, which unlike riboswitches are far less conserved between distantly related organisms. Co-occurrence of a sequence motif with orthologous genes in diverse organisms strongly suggests a regulatory role. Similarly, multiple copies of a candidate regulatory motif in a single genome define a genetic regulon, wherein genes are controlled by the same factor in a concerted manner. BLISS aids in assessing these aspects of genetic control systems by displaying both the taxonomic categories of source organisms for each sequence in an IGR alignment and the proposed function (COGs; ref. 15) of the products of downstream genes.
In most instances, the known riboswitches in B. subtilis use expression platforms that use transcription control by means of intrinsic terminator formation (19, 20). Almost invariably, an intrinsic terminator is located within the 5′ untranslated region between the aptamer domain and coding region. It is usually possible to predict the effect of metabolite binding on gene expression from the relative locations of terminators. Metabolite binding stabilizes a specific structure of the aptamer typically by inducing formation of one or more base-paired elements. If the terminator stem overlaps part of the aptamer, then metabolite binding disrupts the terminator and turns “on” gene expression (21). If the terminator is distant from the aptamer, then binding usually disrupts an antiterminator stem and turns “off” expression (1). Nearly all known metabolite-binding riboswitches that control transcription termination operate by the second mechanism. To aid in assessing the possible function of previously uninvestigated elements, BLISS incorporates predictions of intrinsic terminators consisting of a distinctive base-paired stem followed by a run of U residues.
New Riboswitch Candidates. Each IGR from B. subtilis was subjected to sequence homology searching against other IGRs from the same organism and IGRs from 91 different prokaryotic genomes. Individual IGRs that met a specified level of similarity to a given IGR from B. subtilis were declared “hits,” and the sequences of all hits were aligned to form blocks of apparent nucleotide conservation. The resulting data were used to sort IGRs from highest to lowest numbers of hits, and every B. subtilis IGR in the BLISS database with at least five aligned IGRs was manually examined for evidence of regulatory elements.
Short IGRs or those that had extensive homology based on repetitive AT-rich elements were rejected as possible riboswitch candidates. Similarly, IGRs that had short conserved elements that were known or expected to be protein recognition elements also were rejected. In contrast, IGR alignments that had extensive sequence conservation with evidence of secondary structure formation were retained as candidates that required further investigation. In addition to rediscovering six of the seven known types of riboswitches in B. subtilis, this analytical approach provided 14 elements that have considerable sequence similarity and, in certain instances, obvious secondary-structure conservation. These elements were named according to the proposed gene that lies immediately downstream.
Two of the 14 elements (yrvM and yocI) are reportedly small noncoding RNAs (22, 23) and therefore were not examined further in this study. Four of the elements (rplJ, rplM, rplU, and rpsJ) occur immediately upstream of ORFs that encode various ribosomal proteins. We have chosen not to explore these candidates further in this study because precedence exists for protein-mediated feedback control of such mRNAs (24, 25). However, both the noncoding RNAs and the conserved elements associated with ribosomal protein genes have features that are consistent with riboswitches, and further studies to explore possible riboswitch function seem warranted. Two additional elements (ykvJ and ylbH) exhibit weak sequence and structural similarity. Although we have subjected these two elements to further biochemical analysis (see Figs. 4–11, which are published as supporting information on the PNAS web site), they are the poorest candidates for new riboswitch motifs.
The six remaining elements (gcvT, glmS, ydaO, ykkC, ykoK, and yybP) have extensive sequence and apparent structural conservation (Fig. 1). With the exception of the glmS motif, these elements appear multiple times in certain organisms and therefore are likely to be indicative of previously uninvestigated genetic regulons (Table 1). Although some representatives of the six elements reside upstream of genes that encode for proteins with unknown or unconfirmed functions, others encode for biosynthetic or transport proteins. When the gene functions are predicted or confirmed, a striking correspondence occurs between a given element and the types of genes residing downstream. In combination, these factors increase the likelihood that the new motifs serve as genetic control elements that regulate expression of the adjacent genes in response to changing concentrations of specific metabolites. Therefore, we focused our efforts on the RNA versions of these six highly conserved sequence elements.
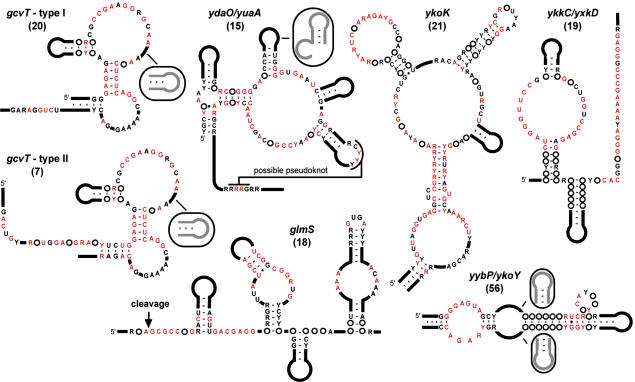
Fig. 1.
Consensus sequences and secondary structure models for riboswitch candidates. Two variations of the gcvT element (types I and II) are depicted. Black and red letters identify nucleotides whose sequence identities are conserved in >80% and 95% of the representatives, respectively. Purine (R) or pyrimidine (Y) designations are given when a single nucleotide does not reach 80% conservation threshold.
In each case, the candidates were examined by using in-line probing (18) to determine whether the proposed secondary structure model corresponds with the actual folded structure of the RNA (2, 4–9). Internucleotide linkages within helical elements undergo reduced spontaneous cleavage relative to unstructured linkages. Therefore, unconstrained loops and bulges can be readily identified on separating and visualizing spontaneous cleavage products by PAGE after a 1- to 2-day incubation of 5′-32P-labeled RNA. Nucleotides that are predicted not to reside in base-paired structures will also exhibit reduced cleavage when they are engaged in tertiary structures that prevent their internucleotide linkages from sampling an in-line orientation.
In certain instances the probing data appear to be inconsistent with details of the secondary structure models. In these cases, the expected function of these motifs as allosteric switches that interact with an expression platform may complicate the interpretation of some cleavage patterns in terms of a static structural model. Models with alternate base-pairings may also be consistent with the phylogenetic data because some putative base-paired stems have highly conserved sequences and could not be supported by covariation. Details of the genetic distributions, sequence alignments, and in-line probing results for each riboswitch candidate are described below or can be found in Figs. 4–11.
The glmS Element Is a Metabolite-Dependent Ribozyme. The glmS element resides upstream of the monocistronic glmS gene in 18 Gram-positive organisms (Fig. 2A). During in-line probing, this RNA exhibits an extraordinarily high level of cleavage at a specific internucleotide linkage. The RNA has subsequently proven to be a metabolite-responsive ribozyme that undergoes rapid self-cleavage activated by GlcN6P (3). The glmS ribozyme also retains its function as a bimolecular construct wherein the RNA has been split into separate “substrate” and “ribozyme” domains (Fig. 2B). The fragmented ribozyme is capable of rapidly cleaving its substrate only when GlcN6P is added to the reaction, whereas the closely related analog glucose-6-phosphate does not induce ribozyme function. The ribozymes do not cleave the substrate molecules to completion in this assay, which is typical of such reactions conducted with synthetic substrates. However, the addition of adequate levels of GlcN6P to transcription assays results in near 100% processing, suggesting that ribozyme processing in vivo is likely to be far more complete.
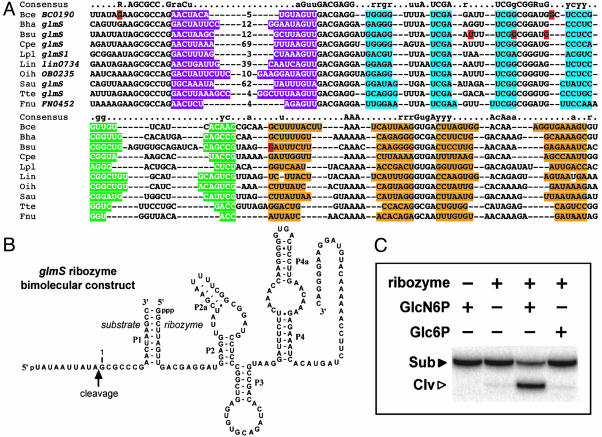
Fig. 2.
The phylogenetic distribution, structural configuration, and function of the glmS RNA element. (A) Sequence alignment of the conserved element residing upstream of glmS gene homologs. Nucleotides shaded with purple, blue, green, and orange identify putative base-paired regions P1 through P4, respectively. Lowercase and uppercase letters of the consensus identify nucleotides that are conserved in 80% and 95% of the representatives, respectively. Nucleotides highlighted in red differ between the database sequence and that determined by the sequencing of clones for Bce and Bsu prepared by our laboratory. The organism and gene name are provided for each representative shown. Organism abbreviations: Bce, Bacillus cereus; Bha, B. halodurans; Bsu, B. subtilis; Cpe, Clostridium perfringens, Lpl, Lactobacillus plantarum; Lin, Listeria innocua; Oih, Oceanobacillus iheyensis; Sau, Staphylococcus aureus; Tte, Thermoanaerobacter tengcongensis; Fnu, Fusobacterium nucleatum. (B) Sequence and predicted secondary structure of the glmS ribozyme that has been engineered to function as a bimolecular construct. (C) Metabolite-dependent ribozyme function of the conserved glmS element. Reactions were conducted in the absence (-) or presence (+) of ribozyme and 100 μM effector as indicated for each lane. Sub and Clv identify the substrate and 5′ cleavage product, respectively. Glc6P, glucose-6-phosphate.
The glmS gene encodes glutamine/fructose-6-phosphate aminotransferase, the enzyme that produces GlcN6P. Deactivating mutations in the ribozyme domain cause proportional derepression of a reporter gene fused to the glmS element (3). Therefore, the glmS ribozyme also appears to serve as a riboswitch that turns off expression of the enzyme responsible for GlcN6P biosynthesis when sufficient amounts of this metabolite are present. The mechanism by which ribozyme cleavage in the 5′ untranslated region of the mRNA leads to reduced gene expression remains to be determined. Overall, these findings demonstrate that the bioinformatics approach used in this study can uncover previously uninvestigated riboswitches. Furthermore, these findings support the hypothesis that some of the remaining elements described herein could be “orphan” riboswitches whose metabolite targets might be established by further biochemical and genetic experimentation.
The gcvT Element. The conserved element associated with the B. subtilis gcvT gene (renamed from yqhI on BLISS) conforms to one of two similar structural types (I and II) with distinctive 5′ and 3′ ends that flank a common central core (Figs. 1 and 3A). The element exhibits evidence of extensive structure formation in the 195-nucleotide RNA construct from B. subtilis (type I) that was subjected to in-line probing (Fig. 3C). Nucleotides 84–87 of gcvT could be paired as depicted (Fig. 3A), but they also are complementary to nucleotides 63–66 and insufficient covariation exists in the sequences to distinguish between these possibilities.
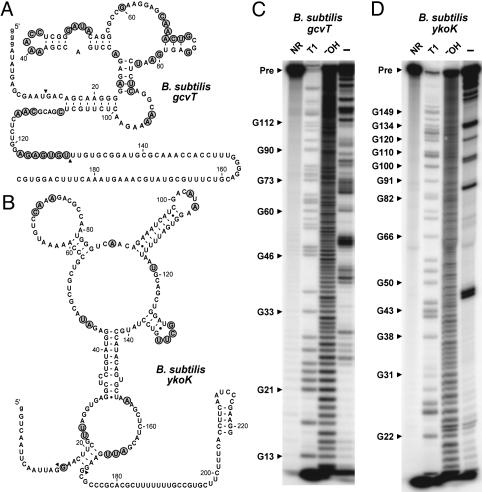
Fig. 3.
In-line probing data for the gcvT and ykoK elements from B. subtilis.(A) Sequence and secondary structure model for the 192 gcvT RNA construct. The nucleotides whose 3′ linkage undergoes a high level of spontaneous cleavage relative to most other linkages are identified by shaded circles. Arrowheads demark the boundaries of the RNA region where spontaneous cleavage data are mapped. (B) Sequence and secondary structure model for the 221 ykoK RNA construct. (C) In-line probing of 5′-32P-labeled 192 gcvT RNA. Products resulting from partial digestion with nuclease T1 (T1), partial digestion with alkali (-OH), and spontaneous cleavage during a 40-h incubation were separated by PAGE. NR, no reaction; Pre, the full-length 192 gcvT RNA. Bands corresponding to certain T1-dependent (G-specific) cleavage products are identified. Nucleotide numbering does not include the unnatural G residues (lowercase letters) inserted to improve transcription in vitro. (D) In-line probing analysis of 5′-32P-labeled 221 ykoK RNA.
The gcvT element occurs most often upstream of the gcvTHP operon encoding the glycine cleavage system, a multisubunit complex that produces 5–10-methylene-tetrahydrofolate, ammonia, and carbon dioxide by breaking down glycine (26, 27). This methylene-carrying form of tetrahydrofolate can be used directly to convert another molecule of glycine to serine or to convert 2′-deoxyuridyl-5′-monophosphate into 2′-deoxythymidyl-5′-monophosphate. It may also be converted into other alkylated tetrahydrofolate derivatives involved in a variety of metabolic pathways, including purine and methionine biosynthesis. The gcvT element often occurs adjacent to genes for putative Na+/alanine symporters, and in one instance it is located upstream of γ-aminobutyrate permease. The element also occurs upstream of a redundant serine hydroxymethyltransferase gene (28) and l-serine deaminase gene in Mycobacterium tuberculosis, and a glycine/d-amino acid oxidase gene in Mesorhizobium loti. Both of these latter systems are used to convert glycine into pyruvate. Despite the many possible target metabolites for this RNA, we have determined that both type I and type II variants of gcvT bind glycine with high selectivity (M.M., M.L., and R.R.B., unpublished data).
The ydaO/yuaA Element. The ydaO/yuaA element occurs upstream of these two genes in B. subtilis and has characteristics of a genetic “off” switch. Structural probing of the corresponding RNA transcript supports the formation of a pseudoknot between a conserved loop and a complementary sequence located downstream in the ydaO RNA, suggesting that this RNA also has a complex tertiary structure (Figs. 1 and 4–11). The ydaO gene product is a predicted amino acid transporter, whereas the protein products of the yuaA-yubG operon constitute a K+ transporter and have been recently renamed KtrA and KtrB (29). The remaining genes appear to encode membrane metalloendopeptidases, cell wall-associated hydrolases, and oligopeptide transporters involved in remodeling the cell wall. B. subtilis strains defective in KtrAB are sensitive to sudden osmotic shock. Therefore, the ydaO/yuaA element could possibly respond to a specific osmoprotectant or might be responsive to a compound whose concentration changes during cell wall remodeling.
The ykkC/yxkD Element. The ykkC/yxkD element is composed of a two-stem junction and a long 3′ conserved region that lacks obvious local secondary structure (Fig. 1). However, the 3′ conserved region always overlaps the GC-rich stem of a transcriptional terminator in Gram-positive organisms. We probed the full ykkC and yxkD leaders and a ykkC construct with the right-hand shoulder of the terminator stem deleted. In all three constructs, the 3′ conserved stretch of nucleotides displays reduced spontaneous cleavage. It is possible that this region is sequestered in the terminator stem when present, but can form an alternative structure when a portion of the stem is deleted.
The ykkCD operon produces a multidrug resistance efflux pump with a broad specificity (30), whereas the yxkD gene encodes a conserved protein of unknown function. In proteobacteria, the ykkC/yxkD RNA element usually occurs upstream of an operon that encodes all three subunits of an ABC-type transporter related to nitrate/sulfonate/bicarbonate transport systems followed by two copies of a different uncharacterized gene. The Synechocystis speB gene affiliated with this element does not have the expected agmatinase or arginase function and appears to be involved in an uncharacterized reaction of secondary metabolism (31). One possible interpretation of these mixed gene functions is that the ykkC/yxkD element switches “on” efflux pumps and detoxification systems in response to harmful environmental molecules.
The ykoK Element. The ykoK motif is the most elaborate of the riboswitch candidates (Figs. 1 and 3B). A 221-nucleotide transcript of this region folds into a highly structured RNA (Fig. 3D). Genes downstream of ykoK elements are similar to those encoding a variety of divalent metal transporters including those specific for Mg2+, Mn2+, Co2+, Ni2+, and Fe2+. Although it is difficult to assign precise functions for transporters based on protein sequence similarity alone, all the regulated gene products probably transport only a single metal. It seems unlikely that such a large RNA would be necessary to sense an easily bound divalent metal, and so a more complex metabolite target might be involved. For comparison, the coenzyme B12 riboswitch carries the most complex of all known natural aptamers and is involved in regulating cobalt transporters in some bacteria (14, 32). This arrangement ensures that cobalt transport matches the demand of coenzyme B12 for its obligatory cobalt ligand. Perhaps a similar genetic logic applies to the ykoK element. It could serve as an “off” switch in response to a cellular compound that requires a metal ligand.
The yybP/ykoY Element. The yybP/ykoY element is the most common and widely distributed of the six riboswitch candidates and the only one at this time that is also detected in Escherichia coli (Table 1). In-line probing confirms the existence of a complex RNA structure that is consistent with the secondary-structure model. The yybP/ykoY element resides upstream of two separate monocistronic transcripts in B. subtilis and E. coli and is found mainly upstream of genes classified into four groups. The first cluster includes E. coli ygjT and B. subtilis ykoY, which are similar to an E. coli gene (terC) that encodes a membrane protein with a poorly defined function related to tellurium resistance. The second group encodes a cation-transport ATPase, whereas the final two clusters are predicted to be membrane proteins (one includes E. coli yebN). No function has been assigned to the annotated B. subtilis yybP reading frame from sequence similarity. One associated gene from Corynebacterium glutamicum is suggestively similar to an arsenite efflux pump. Thus, the possible metabolite target for the yybP/ykoY element remains obscure, although it appears to be a genetic “on” switch in B. subtilis.
Discussion
In this study, we used sequence analysis to identify conserved elements within the IGRs of various microbial species. Similar approaches to identify regulatory motifs in B. subtilis differ from BLISS primarily in their definitions of sequence similarity and the amount of comparative information from other genomes they include. One typical method for predicting transcription-factor-binding sites identifies overrepresented dimer motifs consisting of two sequence words of four to five bases separated by 3–30 bases within B. subtilis intergenic regions (33). A second standard method identifies phylogenetically conserved elements heuristically extended from exact 3 base sequence matches in pairwise alignments of B. subtilis, Bacillus halodurans, and Bacillus stearothermophilus IGRs occurring upstream of orthologous genes (34). BLISS finds matches between all IGRs in 91 complete bacterial genomes to B. subtilis IGRs by using blastn and then assembles pairwise alignments to the B. subtilis IGR into a multiple-sequence alignment. Unlike the other methods, it was designed as a tool for the manual enumeration of promising motifs and does not assign statistical scores to conservation. Instead, the electronic interface displays these predictions of gene functions and intrinsic transcription terminators alongside IGR alignments. An integrated tool allows collaborative annotation of promising intergenic regions as they are examined and as secondary structure models are developed.
Considerable overlap occurs between the regulatory-site predictions of these three methods. In BLISS, riboswitches, T boxes (35), and Fur protein-binding sites (36) dominate B. subtilis IGRs with the most aligned sequences, although some other protein-binding sites are apparent. The predictions of the dimer motif method (33) are largely complementary. It accurately captures sigma- and transcription factor-binding sites but does not detect the 15-bp Fur site and only discovers a single motif within the extended sequences of T boxes. Phylogenetically conserved element predictions (34) are intermediate between these extremes. This approach detects protein-binding sites and covers the conserved portions of several known riboswitches and T boxes with multiple phylogenetically conserved elements. In general, the use of blastn searches to identify similarity introduces a bias toward finding longer sequence motifs that are more typical of riboswitches than protein-binding sites. We have expanded on the sequence homology search results by the iterative application of phylogenetic sequence analysis along with an algorithm that merges sequence and structural constraints to identify additional representatives. This combination of search strategies is particularly effective at identifying larger motifs in their entirety, such as riboswitches and other RNA-mediated genetic control systems like T boxes.
We have confirmed that one motif, the conserved element residing upstream of the glmS gene, is a metabolite-responsive ribozyme whose activity affects gene expression (3). Of the five other top riboswitch candidates examined in greater detail, all exhibit a high level of structure formation, even in regions that are not predicted to form secondary structure. Each motif occurs upstream of related genes in diverse bacterial species. These findings suggest that these elements are expressed as RNA and that the resulting structures are of biological relevance. Riboswitches appear to represent a major form of genetic control once transcription has been initiated. If these new motifs represent previously uninvestigated metabolite-binding RNAs, then the list of different riboswitch classes in modern organisms would expand substantially.
Supplementary Material
Acknowledgments
We thank members of the Breaker laboratory for helpful discussions. This work was supported by National Institutes of Health Grants GM 559343, GM 068819, and NHLBI-N01-HV-28186; National Science Foundation Grants EIA-0129939, EIA-0323510, and EIA-0324045; and the David and Lucile Packard Foundation. J.E.B. is supported by a Howard Hughes Medical Institute predoctoral fellowship.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BLISS, Breaker Laboratory Intergenic Sequence Server; GlcN6P, glucosamine-6-phosphate; IGR, intergenic region.
References
- 1.Winkler, W. C. & Breaker, R. R. (2003) ChemBioChem 4, 1024-1032. [DOI] [PubMed] [Google Scholar]
- 2.Winkler, W. C., Cohen-Chalamish, S. & Breaker, R. R. (2002) Proc. Natl. Acad. Sci. USA 99, 15908-15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winkler, W. C., Nahvi, A., Roth, A., Collins, J. A. & Breaker, R. R. (2004) Nature 428, 281-286. [DOI] [PubMed] [Google Scholar]
- 4.Sudarsan, N., Barrick, J. E. & Breaker, R. R. (2003) RNA 9, 644-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandal, M., Boese, B., Barrick, J. E., Winkler, W. C. & Breaker, R. R. (2003) Cell 113, 577-586. [DOI] [PubMed] [Google Scholar]
- 6.Nahvi, A., Sudarsan, N., Ebert, M. S., Zou, X., Brown, K. L. & Breaker, R. R. (2002) Chem. Biol. 9, 1043-1049. [DOI] [PubMed] [Google Scholar]
- 7.Winkler, W., Nahvi, A. & Breaker, R. R. (2002) Nature 419, 952-956. [DOI] [PubMed] [Google Scholar]
- 8.Winkler, W. C., Nahvi, A., Sudarsan, N., Barrick, J. E. & Breaker, R. R. (2003) Nat. Struct. Biol. 10, 701-707. [DOI] [PubMed] [Google Scholar]
- 9.Sudarsan, N., Wickiser, J. K., Nakamura, S., Ebert, M. S. & Breaker, R. R. (2003) Genes Dev. 17, 2688-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gelfand, M. S., Mironov, A. A., Jomantas, J., Kozlov, Y. I. & Perumov, D. A. (1999) Trends Genet. 15, 439-442. [DOI] [PubMed] [Google Scholar]
- 11.Miranda-Rios, J., Navarro, M. & Soberón, M. (2001) Proc. Natl. Acad. Sci. USA 98, 9736-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stormo, G. D. & Ji, Y. (2001) Proc. Natl. Acad. Sci. USA 98, 9465-9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodionov, D. A., Vitreschak, A. G., Mironov, A. A. & Gelfand, M. S. (2002) J. Biol. Chem. 277, 48949-48959. [DOI] [PubMed] [Google Scholar]
- 14.Vitreschak, A. G., Rodionov, D. A., Mironov, A. A. & Gelfand, M. S. (2003) RNA 9, 1084-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., Kiryutin, B., Galperin, M. Y., Fedorova, N. D. & Koonin, E. V. (2001) Nucleic Acids Res. 29, 22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ermolaeva, M. D., Khalak, H. G., White, O., Smith, H. O. & Salzberg, S. L. (2000) J. Mol. Biol. 301, 27-33. [DOI] [PubMed] [Google Scholar]
- 17.Seetharaman, S., Zivartz, M., Sudarsan, N. & Breaker, R. R. (2001) Nat. Biotechnol. 19, 336-341. [DOI] [PubMed] [Google Scholar]
- 18.Soukup, G. A. & Breaker, R. R. (1999) RNA 5, 1308-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gusarov, I. & Nudler, E. (1999) Mol. Cell 3, 495-504. [DOI] [PubMed] [Google Scholar]
- 20.Yarnell, W. S. & Roberts, J. W. (1999) Science 284, 611-615. [DOI] [PubMed] [Google Scholar]
- 21.Mandal, M. & Breaker, R. R. (2004) Nat. Struct. Mol. Biol. 11, 29-35. [DOI] [PubMed] [Google Scholar]
- 22.Ando, Y., Asari, S., Suzuma, S., Yamane, K. & Nakamura, K. (2002) FEMS Microbiol. Lett. 207, 29-33. [DOI] [PubMed] [Google Scholar]
- 23.Suzuma, S., Asari, S., Bunai, K., Yoshino, K., Ando, Y., Kakeshita, H., Fujita, M., Nakamura, K. & Yamane, K. (2002) Microbiol. 148, 2591-2598. [DOI] [PubMed] [Google Scholar]
- 24.Zengel, J. M. & Lindahl, L. (1994) Prog. Nucleic Acids Res. Mol. Biol. 47, 331-370. [DOI] [PubMed] [Google Scholar]
- 25.Keener, J. & Nomura, M. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (ASM Press, Washington, DC), pp. 1417-1431.
- 26.Fujiwara, K., Okamura-Ikeda, K. & Motokawa, Y. (1984) J. Biol. Chem. 259, 10664-10668. [PubMed] [Google Scholar]
- 27.Okamura-Ikeda, K., Fujiwara, K. & Motokawa, Y. (1987) J. Biol. Chem. 262, 6746-6749. [PubMed] [Google Scholar]
- 28.Chaturvedi, S. & Bhakuni, V. (2003) J. Biol. Chem. 278, 40793-40805. [DOI] [PubMed] [Google Scholar]
- 29.Holtmann, G., Bakker, E. P., Uozumi, N. & Bremer, E. (2003) J. Bacteriol. 185, 1289-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jack, D. L., Storms, M. L., Tchieu, J. H., Paulsen, I. T. & Saier, M. H. (2000) J. Bacteriol. 182, 2311-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekowska, A., Danchin, A. & Risler, J. L. (2000) Microbiology 146, 1815-1828. [DOI] [PubMed] [Google Scholar]
- 32.Nahvi, A., Barrick, J. E. & Breaker R. R. (2004) Nucleic Acids Res. 32, 143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mwangi, M. M. & Siggia, E. D. (2003) BMC Bioinformatics 4, article 18. [DOI] [PMC free article] [PubMed]
- 34.Terai, G., Takagi, T. & Nakai, K. (2001) Genome Biol. 2, research0048.1-research0048.12. [DOI] [PMC free article] [PubMed]
- 35.Grundy, F. J., Winkler, W. C. & Henkin, T. M. (2002) Proc. Natl. Acad. Sci. USA 99, 11121-11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baichoo, N., Wang, T., Ye, R. & Helmann, J. D. (2002) Mol. Microbiol. 45, 1613-1629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.