Abstract
Proteins are very sensitive to their solvent environments. Urea is a common chemical denaturant of proteins, yet some animals contain high concentrations of urea. These animals have evolved an interesting mechanism to counteract the effects of urea by using trimethylamine N-oxide (TMAO). The molecular basis for the ability of TMAO to act as a chemical chaperone remains unknown. Here, we describe molecular dynamics simulations of a small globular protein, chymotrypsin inhibitor 2, in 8 M urea and 4 M TMAO/8 M urea solutions, in addition to other control simulations, to investigate this effect at the atomic level. In 8 M urea, the protein unfolds, and urea acts in both a direct and indirect manner to achieve this effect. In contrast, introduction of 4 M TMAO counteracts the effect of urea and the protein remains well structured. TMAO makes few direct interactions with the protein. Instead, it prevents unfolding of the protein by structuring the solvent. In particular, TMAO orders the solvent and discourages it from competing with intraprotein H bonds and breaking up the hydrophobic core of the protein.
Mechanisms have evolved in nature to allow living organisms to compensate for extreme conditions. For example, certain marine creatures have adapted to life at high pressures and salinity by using osmolytes to maintain cellular volume and buoyancy (1, 2). However, certain osmolytes, like urea, can degrade protein function and disrupt their structures at the high concentrations found in some animals and marine life (1–3), although elevated pressures (≈70 MPa) could mitigate some of the deleterious effects of urea (4–6). Nevertheless, the answer to this paradox was the discovery of protective osmolytes such as betaine and trimethylamine N-oxide (TMAO) in certain elasmobranchs by Yancey et al. (1, 7–8) and later in other marine organisms and mammals (1, 6, 9). In marine animals, the TMAO concentration varies with habitat depth, presumably as a response to pressure (4–5). Furthermore, in organisms that concentrate urea as an osmolyte (7) and buoyancy factor (2), TMAO has been found in urea at ratios of 3:1 and 2:1 (7, 10).
TMAO can restore enzyme function that has been lost because of the presence of urea (6, 10–12) by restoring the protein to its native structure (13–15). The mechanism of action of these protective osmolytes is not understood fully; both direct (16–19) and indirect (13, 19–21) interactions have been proposed, and the mechanism may be molecule-specific (14). Our understanding of the mechanism of action of chemical denaturants, such as urea and guanidinium chloride, is in a similar state (19, 20, 22–33). Consequently, we are pursuing molecular dynamics (MD) simulations of such compounds in an attempt to characterize these mechanisms at the molecular level.
Here, we investigate the ability of TMAO to overcome the effect of urea on protein structure at the atomic level. Chymotrypsin inhibitor 2 (CI2) was chosen for this study because of the extensive amount of information available regarding its folding and unfolding behavior from both theoretical and experimental studies (34). Simulation studies in our laboratory have focused on its unfolding pathway by means of thermal denaturation (35–40) and chemical denaturation by urea (22). Here, we describe simulations of CI2 in a ternary solution of 8 M urea/4 M TMAO at 60°C, which is below the Tm of the protein (41), focusing on the properties of the protein and solvent environment. The results are compared with the behavior of the protein at high temperature (125°C) in water (40) and in 8 M urea at elevated temperature (60°C) (22). Control simulations of CI2 in 4 M TMAO at 60°C, 1 M TMAO at 25°C, and in pure water at 25°C and 60°C were performed also. For the simulations presented here, we found that a small fraction of the total TMAO molecules interacted specifically with Lys and Arg side chains. The dominant mode of stabilization of the native state was indirect: TMAO molecules ordered and strengthened water structure, thereby discouraging unfolding of the protein. Furthermore, TMAO decreased urea–protein interactions and strengthened urea–water interactions, thereby mitigating the denaturant action of urea.
Methods
MD simulations were performed by using encad (42). The protocols, as well as the potential functions of protein (43), urea (20, 22), TMAO (20), and water (44), have been described. The simulations began with the crystal structure of CI2 (1ypc, ref. 45). The simulations of CI2 in water at 125°C and in 8 M urea have been described (22, 40). The control trajectory of CI2 in pure water at 60°C was prepared for MD as described (22), yielding 2,596 water molecules.
We constructed 8 M-urea systems (mole fraction of 0.186) as described (22). Briefly, water molecules were replaced randomly with urea, resulting in 2,103 water molecules and 493 urea molecules. The box volume was adjusted to reproduce the experimental density for 60°C, 1.103 g/ml (46). The 8 M-urea system described here is UR1 from ref. 22.
Ternary systems of 8 M urea and 4 M TMAO were constructed in a similar fashion. Water molecules were randomly replaced first with TMAO molecules and then with urea molecules to achieve approximate mole fractions of 0.10 and 0.17 for TMAO and urea, respectively, resulting in a total of 1,893 water, 259 TMAO, and 444 urea molecules. The box volume was adjusted to set the final density of 1.129, which was estimated based on predicted partial molal volumes of TMAO, urea, and water at 60°C (2). Several additional steps were performed to prepare the system for MD. First, the potential energy of the system was minimized for 1,500 steps. Next, water alone was minimized for 1,001 steps, followed by 1,001 steps of MD and another 1,001 steps of minimization. The protein was then subjected to 1,001 steps of minimization. Finally, the entire system was minimized for 1,001 steps. A duplicate ternary simulation was prepared by increasing the final energy minimization to 1,101 steps.
The 4 M and 1 M TMAO systems were prepared as described above resulting in simulation boxes with TMAO mole fractions of 0.10 and 0.02. The simulation boxes included 2,446 water and 271 TMAO molecules in the 4 M TMAO system and 2,545 water molecules and 51 TMAO molecules in the 1-M system. The box volume was adjusted to set the densities for 4 M at 60°C (1.015 g/ml) and 1 M at 25°C (1.0 g/ml) solutions of TMAO (22). The 4-M system was prepared in several steps. First, the water potential energy was minimized for 1,001 steps, followed by 1,001 steps of MD, and it was then minimized for an additional 1,001 steps. Next, protein and TMAO potential energies were minimized for 1,001 steps, followed by a final 1,001 steps for the whole system.
Production simulations were then performed for each system. A nonbonded cutoff of 8 Å with smooth, force-shifted truncation was used (43, 44), and the nonbonded list was updated every 2–5 steps. Periodic boundary conditions and the minimum image convention were used to reduce edge effects within the microcanonical (NVE, i.e., the number of particles, volume, and energy are constant) ensemble. The simulations were then performed for 10 ns by using a 2-fs time step. It took up to 400 ps for some of the mixed solvent boxes to equilibrate. Structures were saved every 0.2 ps for analysis, resulting in 50,000 structures for each 10-ns simulation.
Results
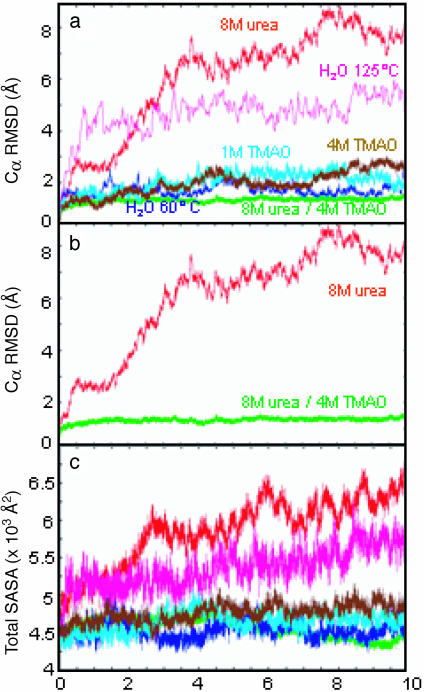
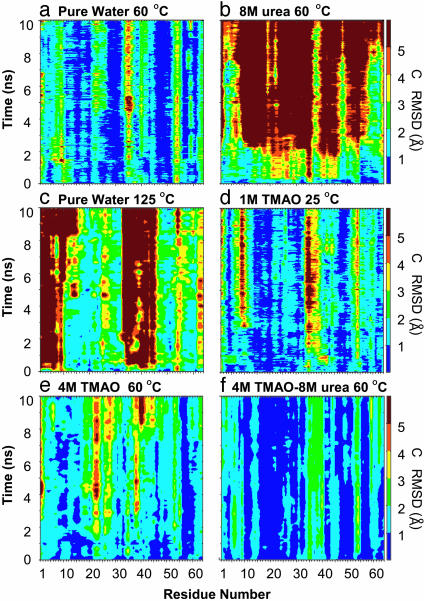
Protein Properties. General properties of the protein were monitored for each simulation. The Cα RMS deviation (RMSD) for each simulation containing TMAO was well below the 125°C and 8 M-urea simulations (Fig. 1). In fact, the TMAO trajectories were most similar to the simulation of CI2 in pure water at 60°C (Fig. 1). Comparison of the 8 M-urea and 4 M TMAO/8 M urea ternary simulations is striking (Fig. 1b). A more detailed view of the Cα RMSD is shown on a per-residue basis in Fig. 2. In the 8 M-urea denaturation simulations, contributions to the high-Cα RMSD came from residues throughout the protein, whereas the N terminus moved away from the β-strands and allowed the hydrophobic core to become solvated with water in high-temperature simulations (125°C). The deviations were quite low throughout the structure in the presence of 4 M TMAO/8 M urea (Fig. 2f).
Fig. 1.
Cα RMSD from the crystal structure and the total SASA of CI2 in various solvent environments. (a and c) CI2 in pure water at 60°C and 125°C, in 8 M urea at 60°C, in 1 M TMAO at 25°C, in 4 M TMAO at 60°C, and in a ternary mixture of 8 M urea and 4 M TMAO at 60°C. (b) Simulations at 60°C of 8 M urea and 4 M TMAO/8 M urea, which are displayed also in a.
Fig. 2.
Cα RMSD from the starting structure mapped per residue of CI2 in various solvent environments.
The simulation in 1 M TMAO shows a similar pattern of backbone motion in the active-site loop (residues 34–45) as the simulation in pure water at 60°C (Fig. 2 a and d). The deviations were also low in 4 M TMAO except in the active-site loop and around residues 23–30. The CI2 simulation in 4 M TMAO/8 M urea displayed low Cα RMSD values compared with all other simulations (Figs. 1 and 2). Furthermore, the pattern of Cα deviation for each residue in the ternary simulation was similar to the pure-water control simulation, in which the N-terminal and active-site loop residues exhibited the highest values.
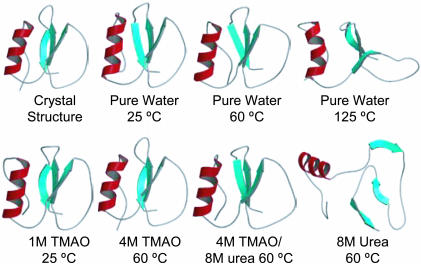
There were no significant changes in the secondary-structure content in the pure water, 1 M TMAO, and 4 M TMAO simulations at ≤60°C (Fig. 3). In contrast, the native β-structure dissolved in the 8 M-urea and high-temperature water simulations, although a dynamic helix was present in both simulations (Fig. 3). Interestingly, 4 M TMAO mitigated the effects of urea, and the native helix and β-structure were maintained (Fig. 3).
Fig. 3.
The CI2 crystal structure and final 10-ns structures from MD simulations in different solvent environments.
We also monitored each simulation for tertiary contacts and solvent-accessible surface area (SASA) to explore how CI2 behaved in various solvent environments. The number of tertiary contacts in the TMAO and pure-water simulations ranged from 210–250. The number of contacts dropped 30–40% in 8 M urea and at 125°C in water. The total SASA for CI2 in each solvent environment is shown in Fig. 1c. In 8 M urea and at 125°C in water, exposure of nonpolar side chains was the major contribution to the overall increase in surface area. The solvent exposure of CI2 was similar in the TMAO, pure-water control, and ternary simulations. The fluctuations in total SASA in the TMAO and control simulations were due to periodic exposure of nonpolar side chains; no significant changes in main-chain or polar side-chain exposure were observed.
Solvent Properties and Interactions with CI2. The bulk water–water H-bond lifetimes increased from 0.8 ps in pure water to 3.0 ps in 1 M TMAO and 3.8 ps in 4 M TMAO. In contrast, the values in 8 M urea were 1.3 ps for water–protein H bonds and 1.01 ps for water–water H bonds. Urea–protein H-bond lifetimes were 1.0 ps. In the ternary simulation, water–protein H-bonding lifetimes were 1.8 ps and average bulk-water lifetimes were 5.2 ps. Interestingly, in the ternary simulation, water–water H-bond lifetimes in the hydration layer remained constant at 0.41 ps, whereas the urea–protein H-bonding lifetimes decreased 20% (from 1.0 to 0.8 ps).
We observed roughly the same number of urea molecules in the hydration shell [≤3.5 Å between any protein and solvent heavy atoms; see Beck et al. (47) for the reasons for using this distance cutoff] in the ternary simulation (the average was 45 molecules over the last nanosecond) as in the 8 M-urea simulation (44 ureas) (Table 1). Thus, addition of TMAO did not stabilize the protein by occluding urea from the surface of the protein (see Fig. 5). Interestingly, fewer of the urea molecules in the hydration shell made H bonds with the protein in the presence of TMAO (36 H bonds on average in 8 M urea and 30 H bonds on average in 8 M urea/4 M TMAO). There were also fewer TMAO-NO—H-protein H bonds in the ternary simulation compared with the 4 M-TMAO simulation (9 versus 17, on average). In contrast to urea, this decrease was a result of having less TMAO in the hydration shell in the presence of urea (27 TMAO molecules in 4 M TMAO versus 18 TMAO molecules in the ternary system). Considering the folded protein, where there is little difference in the SASA between simulations, the average number of hydration water molecules decreased in the presence of TMAO, dropping from an average of 159 water molecules in the pure-water simulation to 110 water molecules in 4 M urea and 123 water molecules in the ternary solvent.
Table 1. Properties of the hydration shell of Cl2 with different solvent environments at 60°C.
| No. of H bonds
|
||||||
|---|---|---|---|---|---|---|
| Simulation | No. of waters | No. of ureas | No. of TMAOs | Water—protein | Urea—protein | TMAO—protein |
| Pure water | 159 | — | — | 127 | — | — |
| 8 M urea | 157 | 44 | — | 113 | 36 | — |
| 4 M TMAO | 110 | — | 27 | 103 | — | 17 |
| 8 M urea/4 M TMAO | 123 | 45 | 18 | 84 | 30 | 9 |
The hydration shell is defined as any solvent molecules with heavy-atom distances ≤3.5 Å from the protein. H bonds required a donor—acceptor distance of ≤2.6 Å and a H bonding angle within 35° of linearity. All values are averages over the last 2 ns of the simulation.
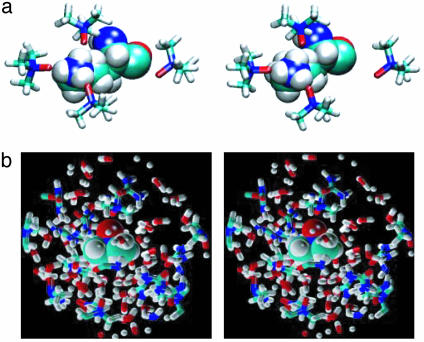
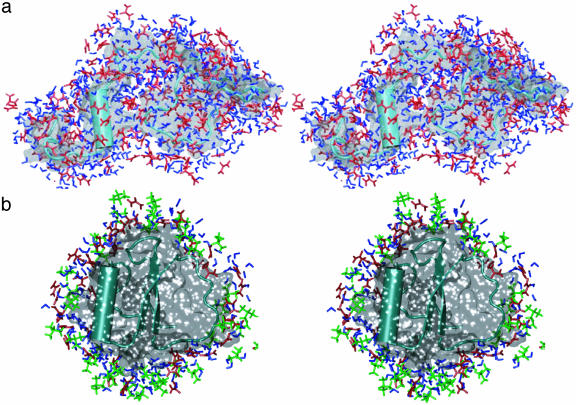
Fig. 5.
Stereoviews of solvent structure in the hydration shell and in bulk solvent. (a) TMAO H bonding with Lys-11 at 10 ns in the 4 M TMAO simulation. (b) Hydration shell of a TMAO molecule in the bulk solvent from the 10-ns snapshot of the 8 M urea/4 M TMAO simulation.
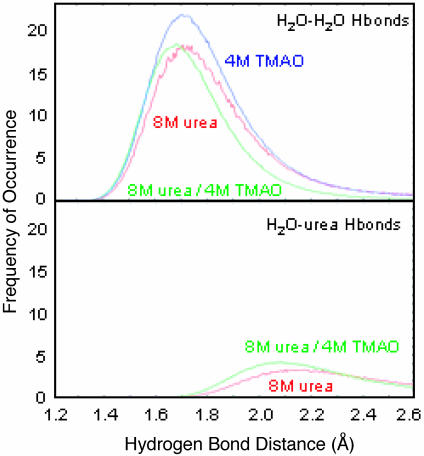
In previous work, we found that the strength of water–water H bonds was sensitive to the solvent environment (20, 22). The average lengths of water–urea, water–water, and water–TMAO H bonds were 1.90, 1.80, and 1.51, respectively. In these CI2 simulations, we also found that TMAO shifted water–water H-bond lengths to shorter distances in 4 M TMAO and in the ternary simulation compared with the 8 M-urea simulation (Fig. 4). There was an increase in the number of water–urea H bonds as well as a shift toward shorter, stronger H bonds upon addition of TMAO (Fig. 4).
Fig. 4.
H-bond length distributions in bulk solvent for water–water and water–urea H bonds. All data were normalized by the average number of water molecules over the analysis interval (5,000 snapshots) for nonhydration water molecules (>3.5 Å from the protein).
TMAO can only be a H-bond acceptor, and specific TMAO interactions with the positively charged side chains of Lys and Arg were observed. In 1 M TMAO, Lys residues formed two H bonds, at most, with TMAO, with an average lifetime of ≈1 ns. However, in 4 M TMAO, TMAO formed H bonds with all Lys residues until each Lys side chain made at least two TMAO H bonds. For example, Lys-11, Lys-17, and Lys-18 participated in three TMAO H bonds by 7 ns. Fig. 6a shows several TMAO molecules close to Lys-11, with three of them involved in very tight (1.5–1.6 Å) H bonds at 10 ns. Arg-43 and Arg-62 each formed only single H bonds with TMAO. Arg-46 and Arg-48 formed intraprotein H bonds throughout the simulation. No other polar side chains made significant H bonds with TMAO. The number of TMAO molecules within the hydration layer was roughly linear with respect to concentration.
Fig. 6.
The hydration shell around 10-ns structures of CI2 in 8 M urea (a) and 4 M TMAO/8 M urea (b). Green, TMAO; blue, water; red, urea.
In the ternary simulation, there were an average of 65 H bonds between urea and TMAO. H bonding between urea and TMAO consisted of only one urea molecule per TMAO molecule. However, bifurcated H bonds between a single urea and TMAO molecule were observed. Other urea molecules within a 9-Å radius of the TMAO nitrogen atom were sandwiched between water molecules surrounding TMAO methyl groups (see Fig. 6b).
Discussion
The intriguing discovery of the ability of TMAO to stabilize proteins and counteract urea-induced denaturation (4–6, 11, 12) prompted our MD simulations of the system, with the aim of providing detailed structural information for these water–cosolvent–protein systems. An additional aim of these simulations was to develop cosolvent models that are robust and display the correct physical properties, so as to make MD simulations more realistic. Previous MD simulations of model peptides (cyclic di-Gly and di-Ala) in solutions of TMAO and urea showed that TMAO rarely interacted with the peptides, whereas urea interacted with both of them (20). Also, as a control for the work presented here, we recently described the unfolding of CI2 in 8 M urea. Urea denatured the protein both by means of direct attack on the main chain and indirectly by altering water structure, facilitating the exposure of nonpolar side chains to the solvent (22). Here, we take this work a step further to investigate how TMAO affects protein and solvent at the atomic level.
Our results show that CI2 maintained native structure in the presence of TMAO at room temperature, at 60°C and in 8 M urea (Figs. 1, 2, 3 and 5). The Cα RMSD values for CI2 in TMAO were similar to CI2 in pure water (Fig. 1). Deviations in the backbone occurred mainly in areas that are normally susceptible to motion, such as the active-site loop and the N terminus (Fig. 2). Other sections of the backbone had higher deviations as a result of Lys and Arg H bonds with TMAO (Fig. 6a). For example, the high-Cα RMSDs for residues 23–30 in the 4 M-TMAO simulation were a result of Lys-24 H bonding with TMAO instead of the main-chain carbonyl oxygen of residue 42.
The overall topology and SASA were preserved in the presence of TMAO, in contrast to high-temperature and urea simulations (Figs. 1, 3, and 5). Furthermore, native secondary structure was preserved, and no nonnative secondary structure was imposed on the protein by TMAO, although some minor fluctuations in the active-site loop were observed. As was the case with the cyclic dipeptides (20), TMAO rarely interacted with the main-chain atoms of the protein. Studies have shown that TMAO is somewhat excluded from the surface of the protein (20, 21, 24, 26). Water-binding sites on the denatured protein outnumbered those for urea, as determined by H bonding (22) and shown qualitatively in Fig. 5. This finding is in agreement with recent magnetic-relaxation dispersion experiments addressing solvation of a urea-denatured protein (48).
Experimental work has posited that urea interactions with the protein surface are not perturbed significantly by the presence of TMAO (10). There were a comparable number of urea molecules within the hydration shell of CI2 in the ternary and 8 M-urea simulations. This finding is surprising because the SASA of CI2 is ≈50% greater in 8 M urea than in the ternary solution, but it shows that TMAO does not act by preventing urea–water interactions. However, TMAO did disrupt urea–protein H bonds. In our earlier urea-denaturation simulations, water molecules were the first to solvate the hydrophobic core of CI2. Urea then followed when the core had opened sufficiently (22). That is, water was the first denaturant. TMAO appears to prevent this initial water attack.
Bulk water was affected also by the cosolvents, and the effects were different for urea and TMAO. Simulations of CI2 in 8 M urea have shown that many solvent H bonds were lost or lengthened (22). In contrast, TMAO shortened and strengthened water–water H bonds even in the presence of 8 M urea (Fig. 4). These high concentrations of cosolvent also affected the dynamics of water in the simulations. The presence of 8 M urea reduced the water self-diffusion coefficient by 50% (22). Water diffusion in solutions of TMAO was also reduced, but, at present, no experimental data exist for comparison.
Direct interactions between TMAO and urea in the ternary simulations were characterized by single urea–TMAO H bonds. Generally, urea was observed to be sandwiched between water molecules that formed the hydration layer of the TMAO methyl groups (Fig. 6b). TMAO dominated the structure and dynamics of the ternary simulations by increasing the number of tight water–water H bonds in the bulk solvent when compared with simulations of 8 M urea (Fig. 4). Moreover, water–urea H bonds increased and were shorter in the presence of TMAO (Fig. 4). Specific interactions of TMAO with Lys and Arg side chains also aided in ordering of the hydration layer, thereby discouraging water–protein and urea–protein interactions further. Thus, TMAO-induced ordering of both the hydration layer and bulk solvent contributed to stabilization of the protein.
Conclusions
We present results from the first set of MD simulations to explore the ability of TMAO to counteract the effects of urea on the structure and dynamics of protein and solvent: that is, how a chemical chaperone prevents protein unfolding. We found that the native state of CI2 was maintained in the presence of a 2:1 urea/TMAO molar ratio, compared with simulations of CI2 in 8 M urea in which the protein unfolded. There were a comparable number of urea molecules in the hydration shell in the ternary and 8 M-urea simulations, and the relative water content in the hydration shell dropped upon addition of TMAO, suggesting that TMAO does not stabilize the native state by occluding urea from the hydration shell. Instead, analysis of the solvent environments in the 8 M urea, 4 M TMAO, and 8 M urea/4 M TMAO simulations showed that TMAO enhanced water H bonding, even in the presence of urea. In fact, TMAO also strengthened urea–water interactions and led to a decrease in urea–protein H bonding. This enhancement of solvent structure in the hydration shell prevented the initial attack of the protein by water and subsequently urea, as observed in the 8 M-urea simulations. TMAO also enabled better water–water and water–cosolvent H bonding in bulk solvent, thereby further discouraging solvent–protein interactions and the subsequent unfolding of the protein.
Acknowledgments
We thank Dr. Kenneth Murphy for first suggesting that we should simulate TMAO. This work was supported by National Institutes of Health Grants R01GM50789 (to V.D.) and T32GM07750 (to B.J.B.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CI2, chymotrypsin inhibitor 2; MD, molecular dynamics; RMSD, RMS deviation; SASA, solvent-accessible surface area; TMAO, trimethylamine N-oxide.
References
- 1.Yancey, P. H., Clark, M. E., Hand, S. C., Bowlus, R. D. & Somero, G. N. (1982) Science 217, 1214-1222. [DOI] [PubMed] [Google Scholar]
- 2.Withers, P. C., Morrison, G. & Guppy, M. (1994) Physiol. Zool. 67, 693-705. [Google Scholar]
- 3.Barton, K. N., Buhr, M. M. & Ballantyne, J. S. (1999) Am. J. Physiol. 276, R397-R406. [DOI] [PubMed] [Google Scholar]
- 4.Yancey, P. H., Blake, W. R. & Conley, J. (2002) Comp. Biochem. Physiol. A Mol. Integr. Physiol. 133, 667-676. [DOI] [PubMed] [Google Scholar]
- 5.Yancey, P. H., Fyfe-Johnson, A. L., Kelly, R. H., Walker, V. P. & Aunon, M. T. (2001) J. Exp. Zool. 289, 172-176. [DOI] [PubMed] [Google Scholar]
- 6.Yancey, P. H. & Siebenaller, J. F. (1999) J. Exp. Biol. 202, 3597-3603. [DOI] [PubMed] [Google Scholar]
- 7.Yancey, P. H. & Somero, G. N. (1980) J. Exp. Zool. 212, 205-213. [Google Scholar]
- 8.Yancey, P. H. & Somero, G. N. (1979) Biochem. J. 183, 317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yancey, P. H. & Burg, M. B. (1990) Am. J. Physiol. 258, 198-204. [DOI] [PubMed] [Google Scholar]
- 10.Lin, T. Y. & Timasheff, S. N. (1994) Biochemistry 33, 12695-12701. [DOI] [PubMed] [Google Scholar]
- 11.Baskakov, I. & Bolen, D. W. (1998) Biophys. J. 74, 2658-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baskakov, I., Wang, A. J. & Bolen, D. W. (1998) Biophys. J. 74, 2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, A. J. & Bolen, D. W. (1997) Biochemistry 36, 9101-9108. [DOI] [PubMed] [Google Scholar]
- 14.Tseng, H.-C. & Graves, D. J. (1998) Biochem. Biophys. Res. Commun. 250, 726-730. [DOI] [PubMed] [Google Scholar]
- 15.Tatzelt, J., Prusiner, S. B. & Welch, W. J. (1996) EMBO J. 15, 6363-6373. [PMC free article] [PubMed] [Google Scholar]
- 16.Xie, G. F. & Timasheff, S. N. (1997) Protein Sci. 6, 222-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie, G. F. & Timasheff, S. N. (1997) Protein Sci. 6, 211-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie, G. F. & Timasheff, S. N. (1997) Biophys. Chem. 64, 25-43. [DOI] [PubMed] [Google Scholar]
- 19.Lin, T. Y. & Timasheff, S. N. (1996) Protein Sci. 5, 372-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou, Q., Bennion, B. J., Daggett, V. & Murphy, K. (2002) J. Am. Chem. Soc. 124, 1192-1202. [DOI] [PubMed] [Google Scholar]
- 21.Wang, A. J. & Bolen, D. W. (1996) Biophys. J. 71, 2117-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennion, B. J. & Daggett, V. (2003) Proc. Natl. Acad. Sci. USA, 100, 5142-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felitsky, D. J. & Record, M. T. (2003) Biochemistry 42, 2202-2217. [DOI] [PubMed] [Google Scholar]
- 24.Anderson, C. F., Felitsky, D. J., Hong, J. & Record, M. T. (2002) Biophys. Chem. 101, 497-511. [DOI] [PubMed] [Google Scholar]
- 25.Courtenay, E. S., Capp, M. W. & Record, M. T. (2001) Protein Sci. 10, 2485-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schellman, J. A. (2002) Biophys. Chem. 96, 91-101. [DOI] [PubMed] [Google Scholar]
- 27.Schellman, J. A. (1978) Biopolymers 17, 1305-1322. [DOI] [PubMed] [Google Scholar]
- 28.Schellman, J. A. (1987) Biopolymers 26, 549-559. [DOI] [PubMed] [Google Scholar]
- 29.Schellman, J. A. (1990) Biophys. Chem. 37, 121-140. [DOI] [PubMed] [Google Scholar]
- 30.Schellman, J. A. & Gassner, N. C. (1996) Biophys. Chem. 59, 259-275. [DOI] [PubMed] [Google Scholar]
- 31.Timasheff, S. N. (2002) Biophys. Chem. 101–102, 99-111. [DOI] [PubMed] [Google Scholar]
- 32.Timasheff, S. N. (2002) Proc. Natl. Acad. Sci. USA 99, 9721-9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timasheff, S. N. (1998) Adv. Protein Chem., Vol 51, 355-432. [DOI] [PubMed] [Google Scholar]
- 34.Fersht, A. R. & Daggett, V. (2002) Cell 108, 573-582. [DOI] [PubMed] [Google Scholar]
- 35.Daggett, V., Li, A., Itzhaki, L. S., Otzen, D. E. & Fersht, A. R. (1996) J. Mol. Biol. 257, 430-440. [DOI] [PubMed] [Google Scholar]
- 36.Ladurner, A. G., Itzhaki, L. S., Daggett, V. & Fersht, A. R. (1998) Proc. Natl. Acad. Sci. USA 95, 8473-8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, A. & Daggett, V. (1994) Proc. Natl. Acad. Sci. USA 91, 10430-10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, A. & Daggett, V. (1996) J. Mol. Biol. 257, 412-429. [DOI] [PubMed] [Google Scholar]
- 39.Kazmirski, S. L., Wong, K.-B., Freund, S. M. V., Tan, Y.-J., Fersht, A. R. & Daggett, V. (2001) Proc. Natl. Acad. Sci. USA 98, 4349-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Day, R., Bennion, B. J., Ham, S. & Daggett, V. (2002) J. Mol. Biol. 322, 189-203. [DOI] [PubMed] [Google Scholar]
- 41.Jackson, S. E. & Fersht, A. R. (1991) Biochemistry 30, 10428-10435. [DOI] [PubMed] [Google Scholar]
- 42.Levitt, M. (1990) encad, Energy Calculations and Dynamics (Yeda, Rehovot, Israel).
- 43.Levitt, M., Hirshberg, M., Sharon, R. & Daggett, V. (1995) Comput. Phys. Commun. 91, 215-231. [Google Scholar]
- 44.Levitt, M., Hirshberg, M., Sharon, R., Laidig, K. E. & Daggett, V. (1997) J. Phys. Chem. B 101, 5051-5061. [Google Scholar]
- 45.Harpaz, Y., Elmasry, N., Fersht, A. R. & Henrick, K. (1994) Proc. Natl. Acad. Sci. USA 91, 311-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weast, R. & Astle, M. J. (1980) Handbook of Chemistry and Physics (CRC, Boca Raton, FL).
- 47.Beck, D. A. C., Alonso, D. O. V. & Daggett, V. (2003) Biophys. Chem. 100, 221-237. [DOI] [PubMed] [Google Scholar]
- 48.Modig, K., Kurian, E., Prendergast, F. G. & Halle, B. (2003) Protein Sci. 12, 2768-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]