Dear Editor,
Exosomes, small membrane vesicles (30-100 nm) of endocytic origin secreted by most cell types, contain functional biomolecules, which can be horizontally transferred to recipient cells1. Exosomes bear a specific protein and lipid composition, and carry a select set of functional mRNAs and microRNAs2. Recently, our group has shown that c-Met shed in exosomes can promote a proangiogenic and prometastatic phenotype in bone marrow-derived progenitor cells during melanoma progression3. In previous research, retrotransposon RNA transcripts, single-stranded DNA (ssDNA), mitochondrial DNA, and oncogene amplifications (i.e., c-myc) have been detected in microvesicles4,5,6. In this report, we provide evidence that tumor-derived exosomes carry double-stranded DNA (dsDNA), as demonstrated through two different approaches, using enzymatic methods (dsDNA-specific shrimp DNase) and physical/structural studies (atomic force microscopy, AFM). Furthermore, we show that exosomal DNA (exoDNA) represents the entire genome and reflects the mutational status of parental tumor cells. We also highlight the translational value of exoDNA in tumor-derived exosomes for its potential usefulness as a circulating biomarker in the early detection of cancer and metastasis.
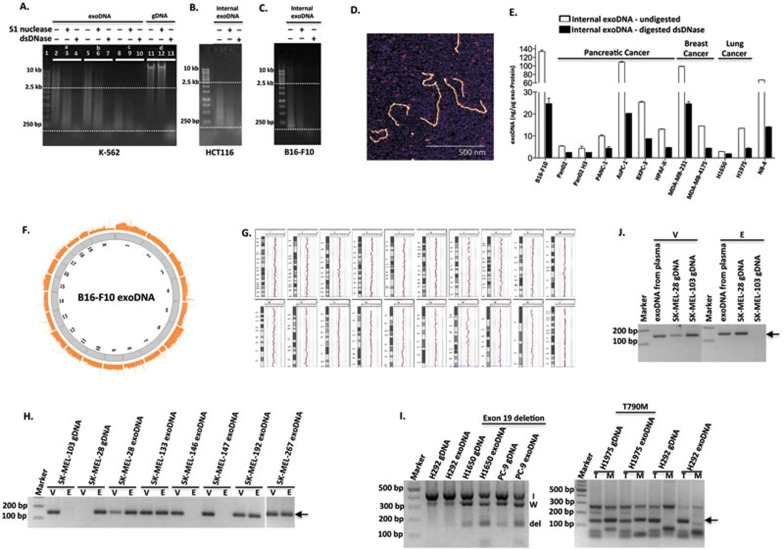
We demonstrate for the first time that the majority of DNA associated with tumor exosomes is double-stranded (Figure 1A-1E). Two types of DNA-specific detection assays were utilized to assess the nature and amount of DNA in three different cancer models including human chronic myeloid leukemia (K-562), human colorectal carcinoma (HCT116), and murine melanoma (B16-F10). One assay is based on the detection of DNA on agarose gels using highly sensitive SYBR Gold nucleic acid staining. The second assay is the QuantiFluor dsDNA Detection System, which uses fluorescent dye that binds specifically to dsDNA (Supplementary information, Data S1). For both assays, we utilized DNases that specifically recognize and digest ssDNA (S1 nuclease) or dsDNA (Shrimp dsDNase). We first verified the specificity of S1 nuclease and dsDNase using purified ssDNA oligonucleotides and Lambda dsDNA as substrates (Supplementary information, Figure S1A). Slight non-specificity of S1 nuclease towards dsDNA was observed. First, to analyze whether DNA is associated with the outer membrane and/or inside exosomes, we extracted DNA from either intact exosomes or exosomes pre-treated with DNases. Remarkably, we observed no change in the pattern of DNA isolated from samples pre-treated with S1 nuclease versus untreated samples (Figure 1A, lane 2 vs lane 5), whereas exosomes pre-treated with dsDNase showed a strong reduction in DNA species greater than 2.5 kb in size and an enrichment of DNA between 100 bp and 2.5 kb (Figure 1A, lane 2 vs lane 8). This indicates that the majority of external exoDNA is dsDNA with a larger size (> 2.5 kb) in comparison to internal exoDNA. Genomic DNA (gDNA) was used as a control for DNase digestion (Figure 1A, lanes 11-13).
Figure 1.
Identification and characterization of exoDNA and its potential use as a diagnostic tool. (A) Equal amounts of DNA extracted from K-562 exosomes, which were untreated (Set a), pre-treated with S1 nuclease (Set b) or dsDNase (Set c), were digested with either S1 nuclease (lanes 3, 6 and 9) or dsDNase (lanes 4, 7 and 10). Digestion of gDNA (Set d) with S1 nuclease (lane 12) or dsDNase (lane 13) serves as controls. (B, C) Analysis of internal exoDNA isolated from HCT116 (B) and B16-F10 (C) after removal of external exoDNA as in A. The results are representative of 2-3 experiments performed independently. (D) AFM image of exoDNA. ExoDNA was extracted from K-562 cells and absorbed on a mica surface in the presence of 5 mM Mg2+. Scale bar, 500 nm. (E) Internal exoDNA was extracted from exosomes secreted by different types of cancer cell lines, including melanoma (B16-F10), pancreatic cancer (Pan02, Pan02 H3, PANC-1, AsPC-1, BXPC-3 and HPAF-II), breast cancer (MDA-MB-231 and MDA-MB-4175), lung cancer (H1650 and H1975), and leukemia (NB-4), and digested with dsDNase. Abundance of dsDNA inside the exosomes, before and after digestion with dsDNase, was expressed as “nanogram of DNA per microgram of exo-Protein”. (F) Circular view of the readings of fragments along each chromosome in the whole-genome sequencing analysis of exoDNA isolated from murine melanoma B16-F10 cell-derived exosomes. (G) ExoDNA represents gDNA shown by comparative genomic hybridization array analysis of B16-F10 exoDNA vs gDNA. (H-J) Mutational analysis of exoDNA. BRAF(V600E) mutation in exoDNA isolated from either cultured melanoma cell lines (H) or circulating exoDNA isolated from SK-MEL-28 melanoma-bearing mice (J) was detected by AS-PCR analysis. gDNA isolated from SK-MEL-28 and SK-MEL-103 cells serves as a positive and negative control, respectively, for V600E mutation (WT (V) and mutant (E) alleles). Arrow indicates the size of expected PCR products. AS-PCR analysis of EGFR mutations in exoDNA isolated from NSCLC cells was shown in I. For del19, “I” indicates internal control; “W”, WT; and “del”, deletion of exon 19. For T790M mutation, “T” indicates WT allele and “M” indicates the mutant allele. The arrow marks the expected size of PCR products.
To further characterize internal exoDNA, first we eradicated external DNA using dsDNase digestion. Then, isolated internal exoDNA was analyzed using both the S1 nuclease/dsDNase digestion method and the QuantiFluor assay. We observed that S1 nuclease digestion resulted in a reduction of DNA to a much lesser extent than dsDNase digestion (Figure 1A, lane 9 vs lane 10 and Figure 1B-1C). The same pattern was observed in internal exoDNA isolated from exosomes untreated or pre-treated with S1 nuclease (Figure 1A, lane 3 vs lane 4 and lane 6 vs lane 7). Consistently, we observed a strong reduction in the binding of dsDNA-specific fluorescent DNA-binding dye in the dsDNase-treated exoDNA samples, compared to those undigested or treated with S1 nuclease (Supplementary information, Figure S1B). Taken together, these results suggest that dsDNA predominates in internal exoDNA. On the other hand, a previous report has demonstrated that tumor microvesicles carry ssDNA4. These conflicting observations suggest that exosomes and microvesicles may vary in DNA cargo composition. However, since both approaches were performed in different biological models, future analyses are required to understand the biological relevance of single-stranded vs double-stranded exoDNA. Using AFM, we further confirmed the presence of dsDNA in exosomes (Figure 1D). The height of the exoDNA was ∼700 pm (Supplementary information, Figure S2), comparable to linear control dsDNA of varying sizes and in line with published values7. Although two other previous studies reported the presence of exoDNA, the conclusions of both studies were based on enzymatic methods that employed digestion of external DNA associated with the surface of exosomes using DNase I, an endonuclease that non-specifically digests both ssDNA and dsDNA6,8. Furthermore, each of these groups used a single enzyme approach employing S1 nuclease to demonstrate the presence of ssDNA6 and DNase I digestion to argue for the presence of dsDNA8 within the exosomes. However, given the lack of specificity for dsDNA of DNase I, our study is the first one to definitively demonstrate the presence of dsDNA in exosomes. We performed a comprehensive analysis of external and internal exoDNA using enzymatic methods specific for dsDNA, and physical studies by imaging exoDNA using AFM.
To further determine whether the association of DNA with exosomes is a common feature of cancer cells, we extended our analysis to a broader panel of cancer cell lines, including melanoma, and breast, lung, prostate and pancreatic cancers. Indeed, we detected exoDNA in all exosomes examined (Supplementary information, Figure S3). We explored the abundance of exoDNA derived from two normal stromal fibroblast lines, namely human dermal and mammary tissue-derived fibroblasts (DF and O97). We found that the amount of fibroblast exoDNA was about 20-fold less than the amount of exoDNA isolated from tumor cells (Supplementary information, Figure S3). Whether this difference is related to the aberrant, rapid replication of tumor cells needs to be further investigated.
Next, we characterized exoDNA by QuantiFluor assay after the eradication of external exoDNA by dsDNase digestion. We showed that dsDNA is a predominant form of internal exoDNA (> 50% reduction in DNA level post dsDNase digestion) in the majority of cell lines (melanoma, leukemia, lung and breast cancers) that we examined (Figure 1E). Notably, exosomes of most pancreatic and lung cancer cell lines studied contained lower amounts of DNA, suggesting differences in exoDNA packaging among different cancer models.
We further employed electron microscopy (EM) to visualize the presence of DNA in exosomes. Exosomes derived from murine B16-F10 melanoma were subjected to staining with anti-DNA antibody followed by gold-conjugated secondary antibody and EM analysis. Interestingly, this study revealed that only a subset of exosomes contained DNA (∼10%; Supplementary information, Figure S4). Our study elicits important questions pertaining to the underlying mechanisms of DNA packaging into exosomes. For example, is this phenomenon due to random selection and limiting capacity inside the exosomes or does it reflect differences in the biogenesis of varying subsets within the heterogeneous populations of exosomes, such as their cellular compartment of origin? Similarly, questions relevant to the shedding of genetic material via exosomes and its consequences within the tumor microenvironment also arise from our work.
Next, using B16-F10 exosomes as a model system, we performed high throughput whole-genome sequencing (Figure 1F) and comparative genomic hybridization analysis (Figure 1G) to further characterize exoDNA. Importantly, our results revealed the entire genome coverage of exoDNA in an unbiased manner. That is, no bias for gene-coding versus intergenic regions and sense versus antisense strands of gene-coding regions were observed in exoDNA. In addition, no specific fragments were highly enriched or depleted in the exoDNA pool compared to the gDNA. However, we did not detect mitochondrial DNA in exoDNA. Interestingly, mitochondrial DNA has been reported in astrocytes and glioblastoma-derived microvesicles5. Again, this inconsistency could be due to differences between cell types and/or composition of membrane vesicles under study. 5′-cytosine methylation is a major modification of nuclear DNA involved in various biological processes, such as transcription and DNA repair. Therefore, we examined the overall level of 5′-cytosine methylation of exoDNA and found that exoDNA is also methylated to a similar level to gDNA (Supplementary information, Figure S5).
Our data prompted us to examine whether exoDNA could be utilized as a surrogate for tumor tissues or cells to detect tumor-specific genetic mutations as recently suggested for other sources of circulating DNA such as cell-free DNA9. To this end, we tested exoDNA isolated from various cancer cell lines for driver mutations known to be present in those cell lines (Figure 1H and 1I). One example is the BRAF(V600E) mutation, present in ∼50% of malignant melanomas10. We performed allele-specific polymerase chain reaction (AS-PCR)11 analysis to evaluate the mutational status of BRAF in exoDNA isolated from several human primary melanoma cell lines, which harbor either wild-type (WT; SK-MEL-146 and SK-MEL-147) or mutated BRAF (SK-MEL-28, SK-MEL-133, SK-MEL-192, and SK-MEL-267). The sensitivity and specificity of the AS-PCR assay was assessed (Supplementary information, Figure S6). By using primers that distinguished WT (“V”) from mutant alleles (“E”) of BRAF, we detected the mutant alleles in exoDNA of all cell lines containing the mutation, whereas only the WT allele was detected in those cell lines with non-mutated BRAF (Figure 1H). A second example of a well-described tumor-associated mutation is the epidermal growth factor receptor (EGFR), which is mutated in several types of cancers, including non-small cell lung cancer (NSCLC)12,13. EGFR mutational status is crucial for selecting patients who will benefit from targeted therapy using tyrosine kinase inhibitors12,13. We employed AS-PCR to assess EGFR mutation in exoDNA from several NSCLC cell lines14,15, including H292 (WT), H1975 (harboring the L858R and T790M gate-keeper mutations), and H1650 and PC-9 (harboring the exon 19 deletion ― del19). As shown in Figure 1I, we detected respective EGFR mutations in 100% of exoDNA isolated from these cell lines. Our findings demonstrate that exoDNA reflects the mutational status of the parental cell lines.
To assess the feasibility of detecting tumor-associated genetic mutations in circulating exoDNA, we employed a pre-clinical animal model of melanoma (SK-MEL-28) harboring the BRAF(V600E) mutation. Cells were subcutaneously implanted in the flank of NOD/SCID mice. Plasma was harvested when the tumor reached the maximum size allowed and circulating exosomes were isolated. ExoDNA was analyzed for the BRAF(V600E) mutation using AS-PCR as described above. We were able to readily detect the V600E mutation in the circulating exoDNA isolated from melanoma-bearing mice (Figure 1J).
In conclusion, we show for the first time the presence of dsDNA in exosomes representing the whole genomic DNA. Our finding that exoDNA can be used to identify mutations present in parental tumor cells illustrates its significant translational potential as a circulating biomarker for cancer in the clinic. ExoDNA is an attractive, potential biomarker candidate in the early detection of cancers and the monitoring of treatment response for several reasons: its protection and thus inherent stability within exosomes; the possibility to isolate or enrich tumor-derived exosomes in complex plasma samples via exosomal surface markers; and its easy and fast preparation.
Acknowledgments
We thank Dr Cyrus Ghajar (Fred Hutchinson Cancer Research Center, Seattle) and members from Dr Bromberg's laboratory (Drs Paul Gao, Qing Chang and Ninhui Mao) for sharing reagents. We thank members of the Genomics Resource Core Facility at WCMC for their high quality service and consultation. We thank Tyler Jacks (Koch Institute for Integrative Cancer Research at MIT) for providing pEGFP-N1-p53 plasmid. We also thank members of our laboratories for helpful discussion. We apologize to colleagues whose work could not be cited due to space constraints. Our work is supported by grants from National Cancer Institute (U01-CA169538, DL), National Institutes of Health (R01-CA169416-01, DL and HP), United States Department of Defense (BC123187P2 (DL), W81XWH-13-1-0425 (DL), W81XWH-12-BCRP-IDEA (DL and JB)), Melanoma Research Alliance (HP), Sohn Conference Foundation (HP), the Children's Cancer and Blood Foundation (HP and DL), The Manning Foundation (DL), The Hartwell Foundation (DL), Champalimaud Foundation (HP and DL), Fundacao para a Ciencia e a Tecnologia (DL), The Nancy C and Daniel P Paduano Foundation (HP and DL), The Mary Kay Foundation (DL), Jose Carreras Leukaemia Foundation (DJCLS R12/06, BKT and KW), Pediatric Oncology Experimental Therapeutic Investigator Consortium (POETIC, DL and HP), James Paduano Foundation (DL and HP), Beth Tortolani Foundation (DL and JB), Malcolm Hewitt Weiner Foundation (DL), Theodore A Rapp Foundation (DL), American Hellenic Educational Progressive Association 5th District Cancer Research Foundation (DL), Charles and Marjorie Holloway Foundation (JB), Sussman Family Fund (JB), Lerner Foundation (JB), Breast Cancer Alliance (JB), and Manhasset Women's Coalition Against Breast Cancer (JB).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
(A) Control digestion of ssDNA oligonucleotide and Lambda dsDNA was performed to verify the specificity of the enzymes.
Comparison of the height of linearized control dsDNA and exosomal DNA using AFM imaging.
DNA is present in tumor cell-derived exosomes.
Detection of DNA in B16-F10 exosomes by immunogold EM.
ExoDNA methylation levels are comparable to those of gDNA.
Evaluation of the sensitivity and specificity of AS-peR assay for BRAF(V600E) mutation detection.
Materials and methods
References
- Peinado H, Lavotshkin S, Lyden D. Semin Cancer Biol. 2011. pp. 139–146. [DOI] [PubMed]
- Valadi H, Ekstrom K, Bossios A, et al. Nat Cell Biol. 2007. pp. 654–659. [DOI] [PubMed]
- Peinado H, Aleckovic M, Lavotshkin S, et al. Nat Med. 2012. pp. 883–891. [DOI] [PMC free article] [PubMed]
- Balaj L, Lessard R, Dai L, et al. Nat Commun. 2011. p. 180. [DOI] [PMC free article] [PubMed]
- Guescini M, Genedani S, Stocchi V, et al. J Neural Transm. 2010. pp. 1–4. [DOI] [PubMed]
- Skog J, Wurdinger T, van Rijn S, et al. Nat Cell Biol. 2008. pp. 1470–1476. [DOI] [PMC free article] [PubMed]
- Adamcik J, Klinov DV, Witz G, et al. FEBS Lett. 2006. pp. 5671–5675. [DOI] [PubMed]
- Kahlert C, Melo SA, Protopopov A, et al. J Biol Chem. 2014. pp. 3869–3875. [DOI] [PMC free article] [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Nat Rev Cancer. 2011. pp. 426–437. [DOI] [PubMed]
- Davies H, Bignell GR, Cox C, et al. Nature. 2002. pp. 949–954. [DOI] [PubMed]
- Jarry A, Masson D, Cassagnau E, et al. Mol Cell Probes. 2004. pp. 349–352. [DOI] [PubMed]
- Paez JG, Janne PA, Lee JC, et al. Science. 2004. pp. 1497–1500. [DOI] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. N Engl J Med. 2004. pp. 2129–2139. [DOI] [PubMed]
- Uhara M, Matsuda K, Taira C, et al. Clin Chim Acta. 2009. pp. 68–72. [DOI] [PubMed]
- Dahse R, Berndt A, Dahse AK, et al. Mol Med Rep. 2008. pp. 45–50. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Control digestion of ssDNA oligonucleotide and Lambda dsDNA was performed to verify the specificity of the enzymes.
Comparison of the height of linearized control dsDNA and exosomal DNA using AFM imaging.
DNA is present in tumor cell-derived exosomes.
Detection of DNA in B16-F10 exosomes by immunogold EM.
ExoDNA methylation levels are comparable to those of gDNA.
Evaluation of the sensitivity and specificity of AS-peR assay for BRAF(V600E) mutation detection.
Materials and methods