A mutation in the Arabidopsis mRNA polyadenylation factor suppresses the cell death associated with the immunity response mediated by salicylic acid and defective myoinositol biosynthesis.
Abstract
Programmed cell death (PCD) is essential for several aspects of plant life, including development and stress responses. Indeed, incompatible plant-pathogen interactions are well known to induce the hypersensitive response, a localized cell death. Mutational analyses have identified several key PCD components, and we recently identified the mips1 mutant of Arabidopsis (Arabidopsis thaliana), which is deficient for the key enzyme catalyzing the limiting step of myoinositol synthesis. One of the most striking features of mips1 is the light-dependent formation of lesions on leaves due to salicylic acid (SA)-dependent PCD, revealing roles for myoinositol or inositol derivatives in the regulation of PCD. Here, we identified a regulator of plant PCD by screening for mutants that display transcriptomic profiles opposing that of the mips1 mutant. Our screen identified the oxt6 mutant, which has been described previously as being tolerant to oxidative stress. In the oxt6 mutant, a transfer DNA is inserted in the CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30 (CPSF30) gene, which encodes a polyadenylation factor subunit homolog. We show that CPSF30 is required for lesion formation in mips1 via SA-dependent signaling, that the prodeath function of CPSF30 is not mediated by changes in the glutathione status, and that CPSF30 activity is required for Pseudomonas syringae resistance. We also show that the oxt6 mutation suppresses cell death in other lesion-mimic mutants, including lesion-simulating disease1, mitogen-activated protein kinase4, constitutive expressor of pathogenesis-related genes5, and catalase2, suggesting that CPSF30 and, thus, the control of messenger RNA 3′ end processing, through the regulation of SA production, is a key component of plant immune responses.
Programmed cell death (PCD) is essential to several aspects of plant life, including development and defense responses. When confronted by pathogens, plants rely on various induced defenses, such as basal resistance or pathogen-associated molecular pattern-triggered immunity (PTI), a defense response elicited by the recognition of conserved microbial/pathogen-associated molecular patterns. However, because some pathogens are able to suppress PTI, many plants also harbor Resistance (R) genes that, when expressed, produce proteins able to recognize pathogen effectors either directly or indirectly, leading to a strong defense response termed effector-triggered immunity (ETI). Indeed, avirulent pathogens carry Avirulence genes whose protein products or the cellular effects of these products are recognized by plant R proteins.
Many plant-pathogen interactions are well known to induce localized cell death at or around the infection site, a process referred to as the hypersensitive response (HR; Coll et al., 2011). This HR acts to limit pathogen growth to noninfected tissues and is accompanied by ion fluxes, the accumulation of specific signaling molecules such as reactive oxygen species (ROS) or salicylic acid (SA), and the expression of plant immunity markers such as the Pathogenesis Related (PR) genes (Heath, 2000; Mur et al., 2008; Coll et al., 2011). Some of the ETI-associated defense mechanisms, such as the accumulation of SA, are also associated with PTI, thus defining key factors in disease resistance (Vlot et al., 2009).
Although hypersensitive cell death has been extensively investigated, the regulatory mechanisms controlling PCD in plants remains poorly understood. Recently, substantial efforts have been made to unravel the signaling events that lead to cell death and to identify the genes involved. Among the different approaches used to identify the molecular players of plant PCD, mutational analyses were particularly fruitful, as a large number of lesion-mimic mutants (LMMs) exhibiting spontaneous cell death have been isolated in Arabidopsis (Arabidopsis thaliana). Their characterization suggests roles in pathogen defense, responses to environmental stresses, and plant development and has led to numerous studies deciphering signaling pathways that regulate life-or-death decisions (Moeder and Yoshioka, 2008). One of the best characterized LMMs is lesion-simulating disease1 (lsd1), which displays a conditional long-day (LD) runaway cell death phenotype triggered by ROS and SA (Dietrich et al., 1994; Jabs et al., 1996; Mühlenbock et al., 2008; Huang et al., 2010). The cell death phenotype of lsd1 has been shown to require ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) and its interacting partner PHYTOALEXIN DEFICIENT4 (PAD4; Rustérucci et al., 2001). These two proteins constitute a regulatory hub for gene-mediated and basal resistance and are required for the accumulation of SA, but several intermediates in this signaling cascade remain to be identified. We recently characterized the LMM mips1 mutant (also called Arabidopsis thaliana l-myo-Inositol-1-Phosphate Synthase1), which is deficient in d-MYOINOSITOL-3-PHOSPHATE SYNTHASE (MIPS), the key enzyme catalyzing the limiting step of myoinositol (MI) synthesis, using Glc-6-P as a substrate (Meng et al., 2009; Donahue et al., 2010; Luo et al., 2011). The Arabidopsis genome contains three genes encoding MIPS isoforms that share 89% to 93% identity at the amino acid level (GhoshDastidar et al., 2006; Torabinejad and Gillaspy, 2006), but MIPS1 appears to be responsible for most of the MI biosynthesis in leaves. Indeed, MIPS1 is more highly expressed compared with MIPS2 and MIPS3 in wild-type plants, and MI levels are reduced in mips1 but not in mips2 and mips3 (Donahue et al., 2010). Consequently, the mips1 mutant displays pleiotropic developmental defects, such as reduced root growth or altered venation in cotyledons (Meng et al., 2009; Donahue et al., 2010), but one of its most striking features is the light-dependent formation of leaf lesions due to SA-dependent PCD, revealing roles for MI or inositol derivatives in the regulation of these processes. How MI levels can regulate PCD is not clear. It has been shown that peroxisomal hydrogen peroxide (H2O2) induces the formation of SA-dependent lesions in the catalase2 (cat2) Arabidopsis mutant and the expression of disease resistance genes only under lowered MI levels (Chaouch and Noctor, 2010). Hence, the tissue content of MI has been suggested to be a key factor determining whether oxidative stress induces or opposes defense responses, but the underlying molecular mechanisms remain to be unraveled.
To gain further insight into the molecular consequences of reduced MI accumulation, we performed a transcriptome analysis of the mips1 mutant and compared our results with publicly available Arabidopsis expression data. This analysis revealed a strong similarity with plants infected with pathogens, other LMMs such as constitutive expressor of pathogenesis-related genes5 (cpr5/hypersenescence1/onset of leaf death1), and mitogen-activated protein kinase4 (mpk4) mutants (Meng et al., 2009). The CPR5 gene encodes a putative membrane protein of unknown biochemical function and plays highly pleiotropic roles, particularly in pathogen responses, cell proliferation, and cell death (Bowling et al., 1997), whereas MPK4 is a negative regulator belonging to the mitogen-activated protein kinase defense signaling network (Pitzschke et al., 2009). mpk4 mutants display a dwarf phenotype, spontaneous PCD, and constitutive activation of SA and pathogen responses (Colcombet and Hirt, 2008). We showed recently that, upon activation of the flagellin-induced pathogen response, MIPS1 controls its own transcription through chromatin changes induced by the MPK4 pathway (Latrasse et al., 2013). Indeed, the expression of MIPS1 was down-regulated in mpk4 and the accumulation of MIPS1 transcripts was decreased in flagellin-elicited Columbia-0 (Col-0) wild-type plants compared with untreated seedlings, indicating that MIPS1 down-regulation is a component of the mitogen-activated protein kinase-dependent cell death induced by biotic stress and that comparison of transcriptomic profiles can be a useful method to identify the regulators of a common process. To identify negative regulators of PCD, we searched publicly available transcriptomic data for mutants in which genes that are up-regulated in mips1 are down-regulated. Using this criterion, we identified the oxidative stress tolerant6 (oxt6) mutant, which was described originally as being tolerant to oxidative stress in a genetic screen of whole seedling phenotypes (Zhang et al., 2008). In the oxt6 mutant, OXT6 dysfunction is caused by a transfer DNA (T-DNA) insertion in a gene encoding a polyadenylation factor subunit homolog, CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30 (CPSF30). This gene gives rise to two mRNAs and two polypeptides, owing to alternative polyadenylation [poly(A)] site use (Delaney et al., 2006). The smaller of these transcripts is similar to yeast and mammalian polyadenylation factor subunits called Yeast 30 kDa homolog1 (Yth1p) and CPSF30 (for the 30-kD subunit of CPSF), respectively, while the larger transcript encodes a polypeptide consisting of a CPSF30-related domain fused to a second domain that is similar to a mammalian splicing factor-related protein, YTH domain-containing protein1 (YTHDC1 or YT521-B; Stoilov et al., 2002). Previous studies showed that oxt6 mutants lack both polypeptides as well as their encoding mRNAs (Delaney et al., 2006; Zhang et al., 2008).
The poly(A) tail at the 3′ untranslated region (UTR) is an essential feature of virtually all eukaryotic mRNAs that influences stability, nuclear export, and translational efficiency of the mRNA (Eckmann et al., 2011). It is synthesized after RNA polymerase II has transcribed past the cleavage and polyadenylation site and associated signal sequences. These sequences are recognized by two key proteins: CPSF and the Cleavage Stimulation Factor, which associate with the additional cleavage factors CF1 and CF2. These factors cleave the pre-mRNA, which is then immediately polyadenylated by poly(A) polymerase (Mandel et al., 2008). In human, alternative polyadenylation is very widespread, and the broad modulation of alternative polyadenylation has been associated with processes as diverse as cell proliferation and differentiation, neural function, and cancer (Elkon et al., 2013). Likewise in Arabidopsis, alternative polyadenylation may affect 60% to 70% of all genes (Shen et al., 2011; Xing and Li, 2011; Sherstnev et al., 2012). Using a genome-wide approach to assess the role of CPSF30 in alternative polyadenylation in Arabidopsis, poly(A) site choice in the oxt6 mutant was surveyed (Thomas et al., 2012). Interestingly, this study revealed that the set of CPSF30 target genes was enriched for those involved in stress and defense responses, a result consistent with the properties of the oxt6 mutant. In mips1, the onset of PCD is triggered when plants are exposed to high irradiance (Meng et al., 2009; Donahue et al., 2010). In restrictive high-irradiance conditions, many mips1 up-regulated genes are involved in the oxidative stress response, suggesting that PCD in mips1 is induced by increased ROS production, possibly due to photosynthesis or photorespiration. Since oxt6 mutants display enhanced tolerance to oxidative stress, we reasoned that cell death induction in mips1 may involve CPSF30.
Here, we have investigated the role of CPSF30 in the regulation of PCD. We demonstrate that CPSF30 is required for lesion formation in mips1. Furthermore, since a mutation in oxt6 is able to suppress cell death in mips1 and other LMMs such as lsd1, mpk4, cpr5, and cat2, our results suggest that the control of mRNA 3′ end processing contributes to the plant immune response by controlling SA production and signaling.
RESULTS
oxt6 Suppresses Cell Death and Partially Restores a Wild-Type Phenotype in the mips1 Mutant
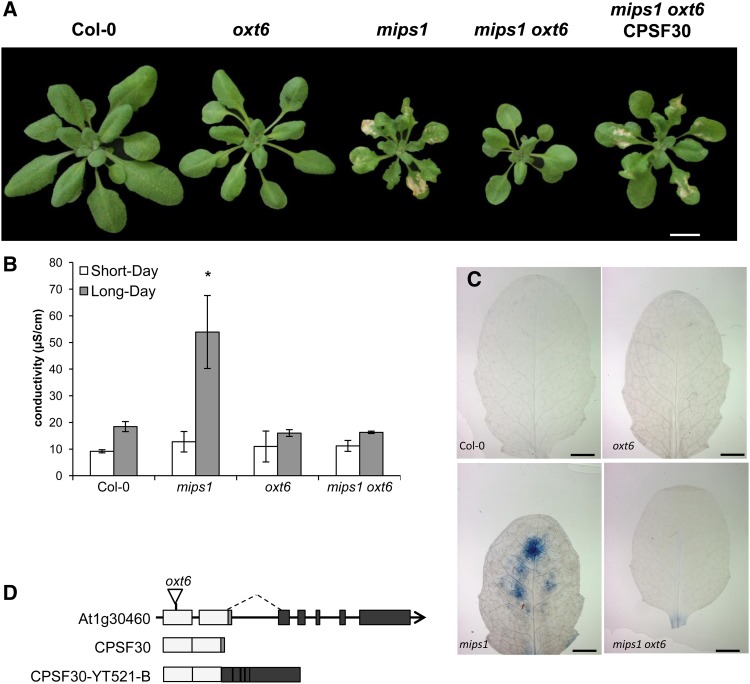
The most striking aspect of the mips1 mutants phenotype is the spontaneous lesion formation observed when plants were transferred in long days, whereas when grown under short-day (SD) conditions, mature mips1 mutants were indistinguishable from the wild type (Meng et al., 2009). To address the role of CPSF30 in the regulation of PCD, a double mips1 oxt6 mutant was generated. Four days after transfer in restrictive LD conditions, cell death was obvious in mips1, whereas the leaves of the mips1 oxt6 double mutant did not show any cell death (Fig. 1A). Ion leakage measurements (Fig. 1B) and trypan blue staining (Fig. 1C) confirmed this observation.
Figure 1.
oxt6-mediated suppression of cell death in mips1. A, Suppression of mips1-mediated cell death in the mips1 oxt6 background and restoration of the cell death phenotype through the complementation of mips1 oxt6 with the smaller of the two OXT6-derived mRNAs. Plants were grown 1 week in vitro and then 14 d in SD conditions in soil and photographed 5 d after transfer to LD conditions. Bar = 1 cm. B, Ion leakage assays, with means and sd calculated from six discs per treatment with three replicates within an experiment in SD and LD conditions. The asterisk denotes a significantly different value according to Student’s t test (P < 0.002). C, Trypan blue staining of wild-type, mips1, oxt6, and mips1 oxt6 leaves. Bars = 2 mm. D, Schematic representation of the OXT6 (AT1G30460) gene structure. Light gray boxes indicate the exons present in the smaller of the two transcripts encoded by this gene, while black boxes represent additional exons present in the larger of the two OXT6-encoded RNAs. The sequence represented by the small dark gray box is absent in the larger RNA (alternative splicing indicated by the dashed line). The structures of the small (CPSF30) and the larger (CPSF30-YT521-B) RNAs are depicted beneath the illustration of the genomic DNA. The position of the T-DNA insertion (Zhang et al., 2008) in the oxt6 mutant is indicated on the genomic DNA.
The oxt6 mutation was located within the first exon of AT1G30460, 147 bp downstream of the translation initiation codon (Zhang et al., 2008), thereby preventing the accumulation of both the short and long CPSF30 mRNAs (Fig. 1D). Because the smaller of the two CPSF30-derived mRNAs is sufficient to restore a wild-type stress-sensitive phenotype to the oxt6 mutant (Zhang et al., 2008), we determined whether this small CPSF30 RNA also could restore lesion formation in the oxt6 mips1 mutant by crossing mips1 with oxt6 plants expressing the smaller CPSF30 mRNA (hereafter referred to as mips1 oxt6 CPSF30). After transfer to LD conditions, cell death was obvious in the mips1 oxt6 CPSF30 plants (Fig. 1A), indicating that the smaller CPSF30 polypeptide is sufficient to promote cell death in the mips1 mutant.
Another striking phenotype of the mips1 mutant was the alterations in cotyledon morphology characterized by irregular margins and altered vascular patterning (Meng et al., 2009; Chen and Xiong, 2010; Donahue et al., 2010; Luo et al., 2011). To investigate whether this abnormality, like lesion formation, also was suppressed in the oxt6 mutant background, we examined cotyledon development in the mips1 oxt6 double mutant and in the mips1 oxt6 CPSF30 complemented line. Cotyledon abnormalities were classified in four classes according to the severity of the phenotype (Fig. 2A). Although the oxt6 mutation reduced the proportion of plants with severe cotyledon defects in the mips1 background, it did not restore wild-type cotyledon development, and the mips1 oxt6 CPSF30 complemented line displayed the same phenotype as the mips1 mutant (Fig. 2B).
Figure 2.
Phenotypic analysis of 7-d-old mips1 oxt6 seedlings. A, Cotyledon phenotypes of mips1 seedlings grown under LD conditions divided into different classes according to the severity of the dimorphism. Bars = 1 mm. B, Relative amounts of each of the cotyledon phenotypic classes in the indicated genotypes. C, Primary root lengths of the indicated genotypes. Asterisks denote significantly different values according to Student’s t test (P < 0.001).
The mips1 mutants also display defects in primary root development (Luo et al., 2011), but the oxt6 mutation did not rescue this mips1 phenotype (Fig. 2C), indicating that CPSF30 is specifically involved in the cell death-related defects of the mips1 mutant but not in other cellular processes requiring MI.
Transcriptional Profiling of mips1 oxt6 and mips1 oxt6 CPSF30
To further explore at the molecular level the suppression of the mips1 phenotype by oxt6, we performed genome-wide transcriptomic analyses using the Complete Arabidopsis Transcriptome MicroArray (CATMA) version 6 on wild-type and mutant plants grown under LD conditions. Three transcriptome comparisons were carried-out, Col-0 versus oxt6, Col-0 versus mips1 oxt6, and Col-0 versus mips1 oxt6 CPSF30, and these data were compared with our previous results from the Col-0 versus mips1 transcriptome performed using CATMA version 2 (Meng et al., 2009).
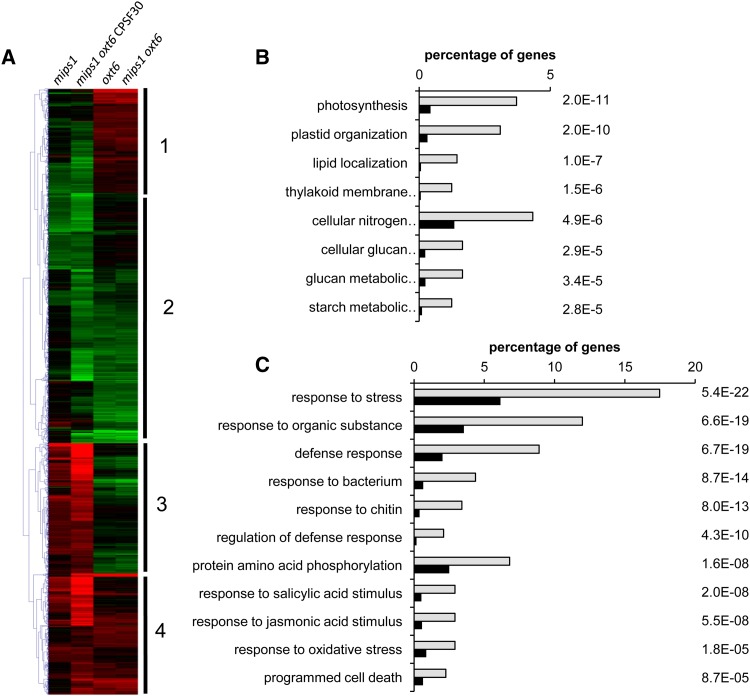
We found that a similar number of genes were differentially expressed in oxt6 single mutants (1,457 down-regulated and 1,206 up-regulated) and mips1 oxt6 double mutants (1,414 down-regulated and 1,162 up-regulated) compared with wild-type Col-0. The most important variation was observed in the mips1 oxt6 CPSF30 line, in which 4,222 transcripts were down-regulated and 5,229 were up-regulated compared with the Col-0 wild type. Explanations for this high variation compared with the mips1 oxt6 double mutant could be due to either misexpression of the CPSF30 transgene, even though its own 5′ regulatory sequences were used, or the absence of the expression of the longer CPSF30 polypeptide in the mips1 oxt6 CPSF30 line. To identify the types of gene networks deregulated by the combination of the mips1 and oxt6 mutations, the data sets were mined with the software tools Genesis (Sturn et al., 2002) and Easy-GO/agriGO (Du et al., 2010). We focused on 2,955 genes that responded differentially in at least two genetic backgrounds and grouped them into hierarchical clusters (Fig. 3A). Overall, the heat map indicates that the transcriptomic profiles of the different mutants can be grouped into two major categories: mips1 and mips1 oxt6 CPSF30 in one group and oxt6 and mips1 oxt6 in the other group. These groupings correlated with the appearance of HR-like phenotypes and also confirmed an epistatic relationship between mips1 and oxt6, with oxt6 being epistatic to mips1.
Figure 3.
Characterization of gene networks deregulated by the mips1 and oxt6 mutations. A, Hierarchical clustering of genes differentially expressed in at least two genetic backgrounds. Red and green correspond to up- and down-regulated expression, respectively. B and C, Enriched GO terms of cluster 1 genes (B) and cluster 3 genes (C). Analyses were performed with the agriGO online software (Du et al., 2010). Light gray bars represent the percentage of genes corresponding to each GO term among genes in the cluster, whereas black bars represent the percentage of genes corresponding to these GO terms in the whole Arabidopsis genome. All of these terms were significantly enriched (P values are indicated to the right of the graphs).
These analyses also revealed the existence of four main clusters with distinct expression profiles (Fig. 3A). Genes belonging to each cluster are listed in Supplemental Table S1. Cluster 1 and 2 genes are underexpressed in mips1 and mips1 oxt6 CPSF30 and overexpressed and underexpressed in oxt6 and oxt6 mips1, respectively. By contrast, cluster 3 and 4 genes are overexpressed in mips1 and mips1 oxt6 CPSF30 and underexpressed and overexpressed in oxt6 and oxt6 mips1, respectively. The identification of clusters 1 and 3 confirmed that the expression of a significant number of genes is inversely controlled according to the presence or absence of CPSF30.
Further analyses to assess overrepresented Gene Ontology (GO) terms indicated a link with photosynthesis and metabolism and the organization of plastids for cluster 1 (Fig. 3B; Supplemental Table S1). These results were consistent with previous mips1 transcriptomic analyses showing that the chloroplastic function was severely impaired in mips1 mutants under restrictive conditions (Meng et al., 2009). Genes involved in photosynthesis also were overrepresented in cluster 2 (Supplemental Fig. S1), but as they were underexpressed in all genotypes, their expression was not dependent on CPSF30 and, thus, not correlated with cell death. This is the case for other genes in cluster 2, including responses to chitin and chemical stimuli, and also for the overrepresented genes in cluster 4 (Supplemental Fig. S1; Supplemental Table S1), which are overexpressed in all conditions and, thus, are neither specific to CPSF30 activity nor to lesion formation.
Finally, GO analysis of cluster 3 revealed an overrepresentation of genes involved in cell death and immune responses (Fig. 3C; Supplemental Table S1), consistent with the phenotype of mips1 and mips1 oxt6 CPSF30 plants. Thus, impaired function of CPSF30 not only prevented the constitutive activation of defense genes in mips1 but actually decreased their expression to levels below the basal values observed in Col-0. Since genes present in cluster 3 are the more likely candidates to account for the cell death phenotype of mips1, we further focused our analysis on this set of genes.
To further understand these deregulations, we compared our transcriptomic data with data on CPSF30-dependent polyadenylation site choice from Thomas et al. (2012). This analysis revealed that more than 66% of the transcripts in cluster 3 displayed poly(A) site choices in their 3′ UTR that were absent in the oxt6 mutant. Knowing that about 38% of the Arabidopsis genes are targeted by CPSF30, it corresponds to a significant enrichment (χ2 > 6.635, P > 0.01), emphasizing a correlation between mRNA polyadenylation mediated by CPSF30 and its potential to contribute to transcriptional regulation.
The Prodeath Function of CPSF30 in the Absence of MI Is Not Mediated by Changes in the Cellular Glutathione Status
PCD is often associated with the concurrent accumulation of ROS (De Pinto et al., 2012), and, as outlined in the introduction, several lines of evidence suggest that the increased sensitivity to oxidative stress in mips1 is the cause of its cell death phenotype.
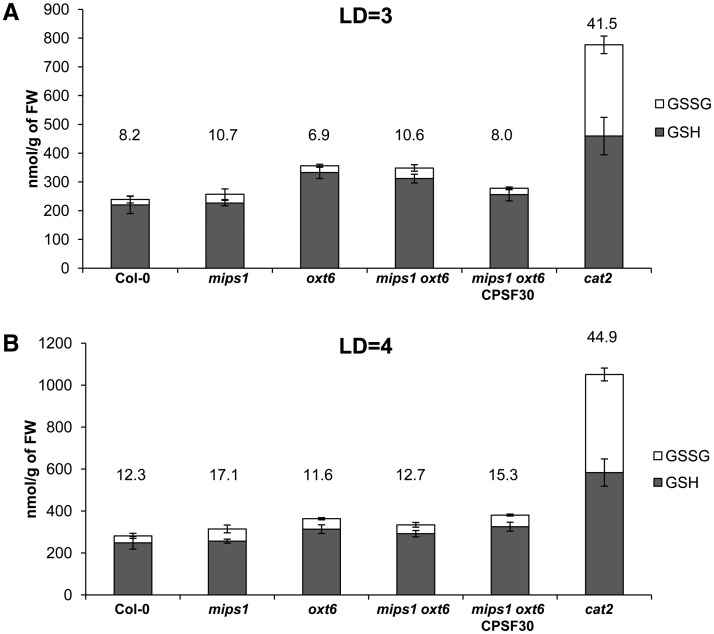
The oxt6 mutant was isolated due to its increased tolerance to oxidative stress, and genes involved in oxidative stress are enriched in cluster 3 defined by our bioinformatic analysis. Thus, we reasoned that the CPSF30-dependent induction of cell death could involve changes in the redox status of the cells. To assess whether the inhibition of cell death in mips1 by the oxt6 mutation was mediated by an antioxidant effect, we quantified the leaf content, 3 or 4 d after transfer in LD conditions, of both the reduced and oxidized forms of glutathione, which are sensitive indicators of intracellular redox status (Noctor et al., 2012). The cat2 mutant, in which ROS signaling is up-regulated (Queval et al., 2007), was used as a positive control. As expected, a large increase in glutathione levels, primarily the oxidized form, was observed in the cat2 mutant (Fig. 4). Although a slight increase in the total glutathione pool was detected 3 d after transfer to LD conditions in oxt6 and mips1 oxt6 mutants compared with the control (Fig. 4A), the ratio of the glutathione oxidized form was similar in the mutants and the control. For the other genotypes at 3 d (Fig. 4A) and 4 d (Fig. 4B) after transfer to LD conditions, the size of the glutathione pool and the percentage of oxidized glutathione remained unchanged, with no significant difference according to Student’s test (P < 0.05). In addition, our transcriptomic analyses revealed that the transcript abundance of some glutathione-related genes that were deregulated in the cat2 mutant (Mhamdi et al., 2010) was not significantly different in mutants lines compared with wild-type Col-0 (genes listed in Supplemental Table S2).
Figure 4.
Cell redox state is not impaired by mips1 and oxt6 mutations. Plants of the indicated genotypes were grown in SD conditions, and leaf glutathione content was measured 3 d (A; LD = 3) or 4 d (B; LD = 4) after transfer to LD conditions. Gray bars represent reduced glutathione (GSH), whereas white bars represent oxidized glutathione (GSSG). The numbers above each bar indicate the percentage of oxidized glutathione. Up error bars are for oxidized glutathione, and down error bars are for reduced glutathione. FW, Fresh weight.
Consistently, in situ detection of H2O2 using 2′,7′-dichlorofluorescein diacetate (data not shown) and 3,3′-diaminobenzidine (DAB; Supplemental Fig. S2) showed no change in ROS accumulation upon transfer of the mips1 mutant to restrictive conditions. DAB staining was detected in dead cells of mips1 and mips1 oxt6 CPSF30 only after 4 d in LD conditions, likely a direct consequence of the ROS damage occurring when cells were dying (Supplemental Fig. S2).
CPSF30 Regulates Cell Death via SA Signaling and Camalexin and Scopoletin Accumulation
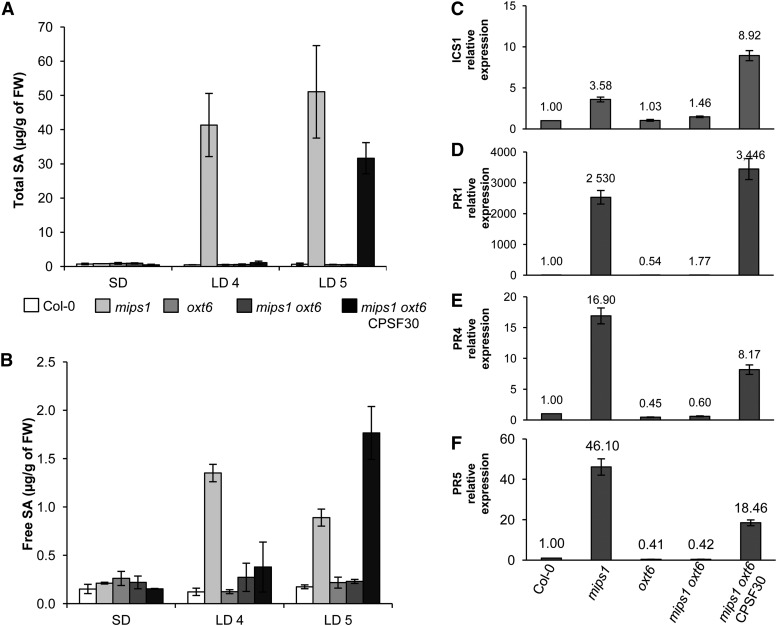
In addition to genes involved in protection against oxidative stress, genes involved in SA-mediated signaling also were overrepresented in cluster 3. Since lesion formation in mips1 is SA dependent, we measured the effect of the oxt6 mutation on SA content in the mips1 background. As reported previously (Meng et al., 2009), free and total SA contents were similar in the different backgrounds under permissive conditions but dramatically increased in the mips1 mutant 4 d after transfer to restrictive conditions (Fig. 5, A and B). This increase in SA was abolished in mips1 oxt6 double mutants, but CPSF30 mRNA complementation reestablished the increase in total and free SA contents, although it occurred 5 d rather than 4 d after transfer to restrictive conditions. This result correlated with a delay in the appearance of lesions observed at 4 d for mips1 compared with 5 d for the mips1 oxt6 CPSF30 line (data not shown).
Figure 5.
CPSF30 acts via SA-dependent cell death and defense response signaling pathways. A and B, Total (A) and free (B) SA levels in the indicated genotypes, with means and sd calculated from four biological replicates grown for 14 d in SD conditions and transferred to LD conditions for 4 d (LD4) or 5 d (LD5). FW, Fresh weight. C to F, Real-time RT-PCR analysis of ICS1 (AT1G74710; C), PR1 (AT2G14610; D), PR4 (AT3G04620; E), and PR5 (AT1G75040; F) expression in the indicated genotypes. The numbers above each bar denote relative transcript abundance compared with wild-type Col-0 and expressed relative to AtUBQ10 (AT4G05320) abundance, which served as a control.
Regulation of SA content may partially occur at the transcriptional level. Indeed, the expression of ISOCHORISMATE SYNTHASE1 (ICS1), which encodes an SA biosynthesis protein, was decreased by MI treatment (Chaouch and Noctor, 2010). Consistently, ICS1 expression levels correlated with the PCD phenotype (Fig. 5C): expression of ICS1 was three times higher in the mips1 mutant than in the wild-type when plants were grown under restrictive conditions, and this induction was abolished in the mips1 oxt6 background. In addition, the oxt6 mutation led to a dramatic down-regulation of the SA pathogenesis-related marker transcripts PR1, PR4, and PR5 (Fig. 5, D–F) in mips1 oxt6 compared with mips1. Because ICS1 was not identified as a CPSF30 target in the genome-wide analysis of polyadenylation sites conducted in the oxt6 mutant (Thomas et al., 2012), we examined whether CPSF30 could target upstream regulators of ICS1. Interestingly, CBP60g (AT5G26920), a key regulator of ICS1 induction and SA synthesis (Zhang et al., 2010), was identified as a target of CPSF30 and was up-regulated in the mips1 mutant but down-regulated in the mips1 oxt6 double mutant, although its expression was not modified in the oxt6 single mutant.
Regarding the SA signaling pathway, among the 18 SA response genes that belong to cluster 3 (Supplemental Table S1), four of them displayed a poly(A) site choice in the 3′ UTR that was specifically dependent on CPSF30 (Thomas et al., 2012). These four genes include a receptor lectin kinase (AT2G37710) induced in response to SA (Blanco et al., 2005), the MYB Domain Protein51 (AT1G18570) also responding to SA stimuli (van de Mortel et al., 2012), GRX480 (AT1G28480), a member of the glutaredoxin protein family, and WRKY70 (AT3G56400), a member of the WRKY transcription factor family, which are also involved in SA-mediated signaling (Ndamukong et al., 2007; Shim et al., 2013).
The indolic compound camalexin (3-thiazol-2′-yl-indole) and the hydroxycoumarin scopoletin (6-methoxy-7-hydroxycoumarin) have been shown to accumulate in Arabidopsis leaves, establishing a hypersensitive cell death to the avirulent bacterium Pseudomonas syringae pv tomato AvrRpm1 (Simon et al., 2010). They have been proposed to act in planta as antimicrobial and antioxidant compounds (Glawischnig, 2007; Simon et al., 2010; Sherstnev et al., 2012). We quantified these compounds in our mutants, and, similar to SA, the increases in both scopoletin and camalexin levels observed in mips1 were not detected in the mips1 oxt6 background (Supplemental Fig. S3). Correlatively, the levels of the PAD3 transcript, which encodes a cytochrome P450 involved in camalexin synthesis (Glawischnig, 2007; Nafisi et al., 2007), were lower in the mips1 oxt6 mutant compared with mips1 (Supplemental Fig. S3).
CPSF30 Activity Is Required for Resistance to P. syringae
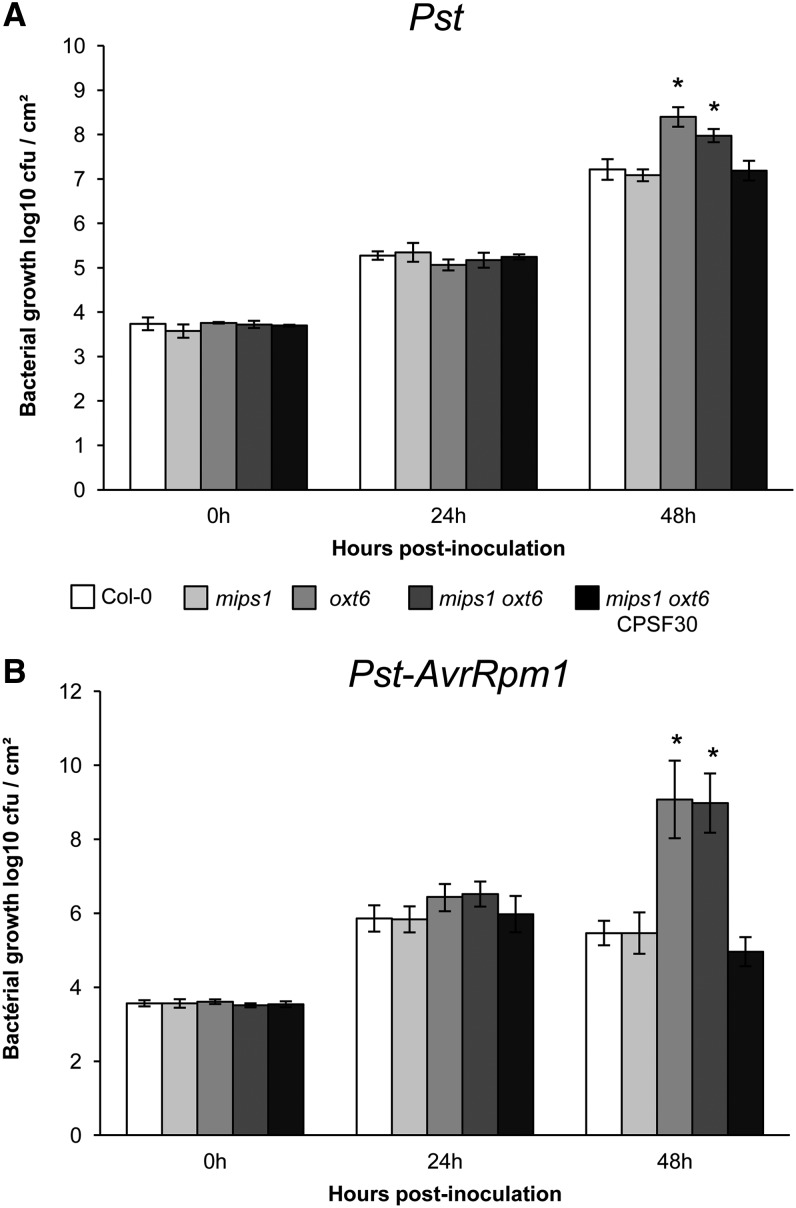
The ability of SA to activate defense genes leads to SA-triggered resistance against several bacterial pathogens. If CPSF30 is required to induce SA production in response to stress, oxt6 mutants are expected to show a decrease in their basal resistance to pathogens. To test this hypothesis, we determined the sensitivity of mips1, oxt6, and mips1 oxt6 mutants and of the CPSF30 complemented line to virulent Pseudomonas syringae pv tomato DC3000 (Pst), which is known to activate SA-mediated resistance in plants. Whereas the mips1 mutant was not compromised in resistance to this bacterial pathogen, as already described by Murphy et al. (2008), oxt6 and mips1 oxt6 mutants showed increased susceptibility to Pst in permissive conditions. In planta growth of the bacteria (measured as yield of colony-forming units [cfu]) was at least 5-fold greater in oxt6 and mips1 oxt6 than in wild-type Col-0 plants (Fig. 6A). However, this increased susceptibility to Pst was no longer detected in the mips1 oxt6 CPSF30 complemented line.
Figure 6.
The oxt6 mutation affects the immune response. The growth of Pst (A) and Pst-AvrRpm1 (B) in the indicated genotypes is shown. Five-week-old plants grown under SD conditions were infiltrated with Pst or Pst-AvrRpm1 at the concentration of 105 cfu mL−1. Samples were taken at 0, 24, and 48 h after inoculation. Error bars represent the sd of five replicates. Asterisks denote significantly different values according to Student’s t test (P < 0.05).
Next, to test whether oxt6 affected R protein-mediated resistance, we inoculated Col-0, mips1, oxt6, mips1 oxt6, and the complemented line with a Pst strain expressing the Avirulence gene avrRpm1 (Pst-AvrRpm1). In Arabidopsis, AvrRpm1 triggers ETI following indirect recognition by the coiled-coil-nucleotide-binding-leucine-rich-repeat receptor RESISTANCE TO PSEUDOMONAS SYRINGAE1 (RPM1) (Boyes et al., 1998). As observed for the basal resistance, in planta bacterial growth was drastically higher in oxt6 and mips1 oxt6 mutants than in wild-type Col-0 plants (Fig. 6B), even though our transcriptomic analyses showed that RPM1 expression was not modified in these lines (data not shown). As seen previously, the decreased resistance to Pst-AvrRpm1 observed in oxt6 and in mips1 oxt6 no longer occurred in the CPSF30 complemented line. Collectively, these data demonstrate that CPSF30 contributed to P. syringae bacterial pathogen resistance in both basal defense and R gene-mediated disease resistance pathways.
In order to correlate these altered levels of resistance to a differential accumulation of SA, as described previously, SA levels were quantified in the Pst-infected leaves of the different genotypes. In all infected genotypes, the total SA content increased 24 h after inoculation with virulent bacteria (Supplemental Fig. S4). However, this accumulation was lower in oxt6 and mips1 oxt6 mutants than in the wild type, mips1, and the complemented line, consistent with the differential Pst susceptibility of these lines.
The oxt6 Mutation Is a General Suppressor of SA-Dependent PCD
To further confirm that CPSF30 is involved in SA production, we asked whether it was required for the onset of PCD in other LMMs. Indeed, several LMMs display both an SA burst and HR responses. To test whether oxt6-mediated cell death inhibition also is involved in the suppression of these HR reactions that have various genetic requirements, we examined the effect of the oxt6 mutation on several LMMs displaying similar autoimmunity phenotypes to mips1.
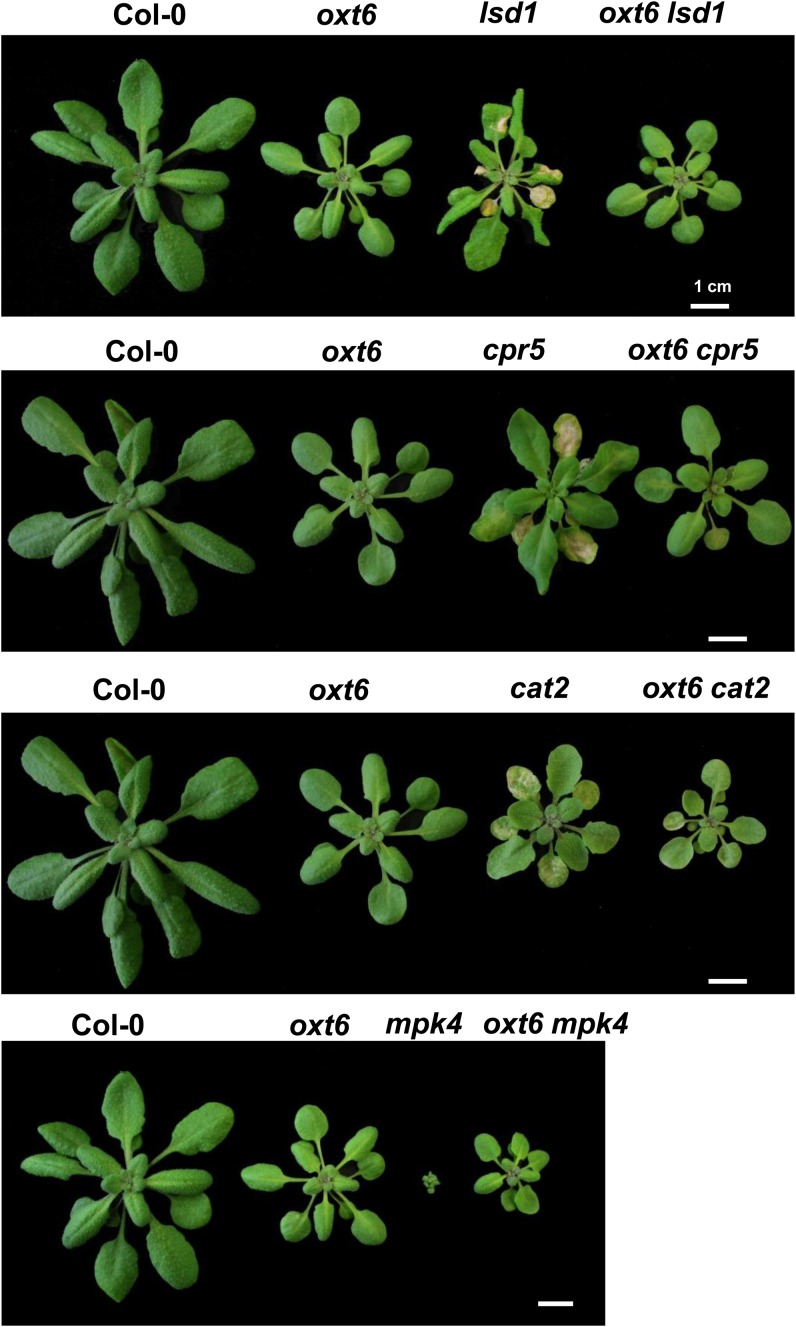
Because a comparison of the mips1 transcriptome and data available from public libraries allowed us to identify two mutants, cpr5 and mpk4, that displayed transcriptomic profiles similar to that of mips1 but opposite to that of oxt6, we selected these LMMs for our initial analysis. Compared with cpr5 and mpk4 single mutants, cpr5 oxt6 and mpk4 oxt6 double mutants did not display visible lesions on their leaves when grown in LD conditions (Fig. 7). However, like mips1 oxt6 double mutants, wild-type development was not totally restored in these double mutants, as they remained smaller than wild-type plants (Fig. 7). The same analysis was conducted on two more LMMs, lsd1 and cat2, which both show light- and SA-dependent PCD (Chaouch et al., 2010; Huang et al., 2010). Like the other LMMs, the oxt6 mutation was able to abolish lesion formation in these two mutants when plants were transferred to LD conditions (Fig. 7), indicating that CPSF30 regulates PCD by modulating SA signaling.
Figure 7.
CPSF30-mediated suppression of cell death in a set of LMMs. Rosette leaf phenotypes of the indicated genotypes are shown. Plants were grown 1 week in vitro and then 14 d in SD conditions in soil and photographed 7 d after transfer to LD conditions. Bars = 1 cm.
DISCUSSION
Due to their sessile nature, plants are confronted continuously with adverse conditions due to both abiotic and biotic stresses. PCD is a vital component of plant immunity, although the molecular mechanisms that control it remain poorly understood. To identify novel regulators of plant PCD, we used a candidate gene approach and found that the polyadenylation factor subunit CPSF30 is required for lesion formation in the mips1 mutant. Our approach revealed that lesions triggered by a decrease in MI content can be genetically reverted in the double mutant mips1 oxt6 and reinduced by complementation with CPSF30. More broadly, loss of CPSF30 function suppressed cell death in a range of LMMs, including lsd1, mpk4, cat2, and cpr5, highlighting the general role of CPSF30 in the control of cell death and plant immune response.
CPSF30 Is a Positive Regulator of the Defense Responses That Acts Downstream of MI Accumulation
As demonstrated by cellular ion leakage and trypan blue staining analysis, both of which measure cell death, oxt6 is epistatic over the mips1 mutation, indicating that CPSF30 contributes to cell death activation in mips1 and acts downstream of MI accumulation. Although the molecular events connecting the reduced MI content with changes in CPSF30 activity remain to be elucidated, one simple explanation would be that MI levels affect the transcriptional regulation of OXT6, leading to OXT6 overexpression in the mips1 background and, subsequently, the constitutive activation of defense genes. However, quantitative reverse transcription (RT)-PCR and CATMA analyses did not detect an overaccumulation of the small or large OXT6 transcript in the mips1 mutant (Supplemental Fig. S5). In fact, their expression was slightly lower in the mips1 mutant compared with the wild type, suggesting that a negative feedback mechanism in the mips1 background could limit OXT6 expression so as to attenuate the autoimmune cell death response. In addition, this result indicates that PCD onset in mips1 is not due to changes in the relative expression levels of the two OXT6-derived mRNAs.
It is worth noting that oxt6 suppresses the autoimmune response but not the root developmental defects of mips1, indicating that these two pathways can be uncoupled. This observation is consistent with plant cell MI derivative diversity, which is involved in a wealth of cellular functions. Due to its position at the starting point of many metabolic pathways, MIPS1 regulates at least two separate biological processes. It negatively controls the autoimmune response and it contributes to proper root development. These broad roles could be due to the role of MI as a precursor of phosphatidylinositol (PtdIns), which is an essential cellular compound involved in various biochemical and physiological processes, including intracellular signal transduction (Gillaspy, 2011), membrane construction and trafficking (Cullen, 2011), and membrane-related protein anchoring (Borner et al., 2005). Indeed, the loss of MIPS1 leads to drastically reduced PtdIns levels and affects endomembrane structure and trafficking and, consequently, pattern formation by regulating auxin distribution (Chen and Xiong, 2010; Zhang et al., 2010; Luo et al., 2011). In addition, overexpression of PHOSPHATIDYLINOSITOL SYNTHASE2, which converts MI to PtdIns, largely restores the mips1 cotyledon defects (Luo et al., 2011). We observed a partial restoration of the mips1 cotyledon defects by the oxt6 mutation, which is consistent with the fact that the severity of cotyledon developmental defects is correlated with the light intensity and, hence, the severity of the cell death phenotype in the previous generation (Luo et al., 2011), possibly due to the negative effect of SA signaling on auxin signaling (Kazan and Manners, 2009). However, although lesion formation was abolished completely in the mips1 oxt6 double mutant, the restoration of the cotyledon phenotype was only partial, suggesting that defects in cotyledon development are only partly dependent on the spontaneous cell death phenotype.
CPSF30 Positively Regulates SA-Mediated Immunity in Arabidopsis
Loss of CPSF30 rescues the cell death phenotype of several LMMs, including cpr5, mpk4, lsd1, and cat2, many of which are thought either to accumulate increased amounts of ROS or to be hypersensitive to ROS production. Indeed, the LSD1 protein is known to function as an integrator of chloroplast-derived signals to regulate PCD in response to excess light (Mühlenbock et al., 2008), the MAPK/ERK KINASE KINASE1-MAP KINASE KINASE1 (MKK1)/MKK2-MPK4 pathway plays a central role in oxidative stress signaling (Pitzschke et al., 2009), and the cat2 mutant is characterized by the production of H2O2 in the peroxisomes, leading to defects in the cell redox state (Chaouch et al., 2010). Since oxt6 was isolated based on its enhanced tolerance to oxidative stress, its ability to restore these mutant phenotypes may be attributed to reduced ROS accumulation. However, our results indicate that ROS production was not enhanced in mips1 mutants and, as such, that CPSF30 likely does not modulate cell death by directly altering ROS production or scavenging. These data are consistent with the results of Chaouch and Noctor (2010) indicating that MI acts downstream of H2O2-dependent cell death.
Despite the importance of the glutathione status in redox signaling, additional complexities of the plant antioxidative system have been recently highlighted within several pathways (Meyer et al., 2012) that were not explored in this work. Thus, we cannot exclude the involvement of CPSF30 in the redox signaling suggested by Zhang et al. (2008).
Strong similarities among the transcriptomic profiles of mips1, cpr5, and mpk4 mutants were identified previously (Meng et al., 2009). Moreover, we have described a regulation of MIPS1 expression by MPK4 (Latrasse et al., 2013); thus, it is logical to find common regulators of MIPS1- and MPK4-dependent cell death. Although all these mutants display a common cell death phenotype, they are impaired in different pathways and not all of their traits are identical. Indeed, unlike in mips1, lsd1, and cat2, in cpr5 and mpk4 mutants, lesion formation is not dependent upon light intensity (Bowling et al., 1997; Brodersen et al., 2006). Yet, one common feature shared by all of the LMMs analyzed in this study is that their cell death phenotypes are dependent on SA accumulation. As such, CPSF30 is a general controller of the SA pathway rather than a specific regulator of MI-mediated PCD. The putative role of CPSF30 as a regulator of SA-mediated signaling is further corroborated by the enhanced susceptibility of oxt6 mutants to both virulent and avirulent P. syringae, which is reminiscent of the increased susceptibility of mutants such as salicylic acid induction deficient2 or plants overexpressing the bacterial salicylate hydroxylase, in which SA signaling is compromised, and invoking CPSF30 as a component of basal immunity as well as of ETI.
The link between CPSF30 and SA-mediated immunity responses is, at first glance, not apparent. Indeed, one could hypothesize that CPSF30 directly affects the expression of a “master” regulator controlling SA accumulation or, alternatively, that the suppression of the mips1 phenotype could be a secondary consequence of a more indirect effect. For example, genes encoding proteins involved in the SA response could be putative targets of CPSF30. Under the first hypothesis, the oxt6 mutation would be expected to prevent the 2-fold increase in ICS1 expression in the mips1 mutant caused by transfer to LD conditions, and this misregulation could account for cell death suppression. Indeed, impairment of ICS1 induction upon pathogen infection in the pad2-1 mutant has been proposed to account for the impaired SA signaling observed in this mutant (Dubreuil-Maurizi et al., 2011). Based on genome-wide analysis (Thomas et al., 2012) and our assays, ICS1 is unlikely to be a direct target of CPSF30. However, the CBP60g transcription factor that regulates ICS1 appears to be differentially polyadenylated in a CPSF30-dependent manner, suggesting that oxt6 could act at this level. Interestingly, genome-wide analysis of polyadenylation sites under various conditions in Arabidopsis revealed that global alternative polyadenylation is reduced upon SA treatment but that 34 Arabidopsis genes use a different polyadenylation site only after SA treatment (Shen et al., 2011). Thus, it is likely that CPSF30 regulates both SA production and response due to its large number of targets.
A Role for mRNA Maturating Complexes in the Regulation of Defense and Cell Death
Gene expression can be regulated via a number of mechanisms impinging on the mRNA 3′ end. In both animal and plant development, alternative 3′ end cleavage sites are widely used to regulate gene expression by modulating the length of the poly(A) tails on mRNAs in the cytoplasm (Eckmann et al., 2011) to influence the stability, nuclear export, and translational efficiency of the mRNAs (Hunt, 2008).
Several lines of evidence indicate that CPSF30 has endonuclease activity, which is consistent with its role in the processing that precedes poly(A) addition and also the modifications in the poly(A) tail lengths of mRNAs in the oxt6 mutant compared with those in wild-type plants (Addepalli and Hunt, 2007; Zhang et al., 2008; Thomas et al., 2012). It also has been shown, on a small set of genes, that alternative poly(A) site choice is a consequence of the absence of CPSF30 (Zhang et al., 2008). However, using a genome-wide strategy, 53% of all of the poly(A) sites seen in the wild type can be processed in the absence of CPSF30 (Thomas et al., 2012), indicating that the polyadenylation complex includes processing endonucleases in addition to CPSF30.
In the context of the recent emerging role of RNA-based regulation in plant immunity, several aspects of nuclear RNA processing recently have been implicated in the response of plants to various stresses (Staiger et al., 2013). Here, we showed that oxt6 and mips1 oxt6 mutants were more susceptible than wild-type control plants to the virulent bacterium Pst. Furthermore, it recently has been shown that the reduced activity of PAPS1, a poly(A) polymerase that is recruited to add the poly(A) tail after the cleavage of the nascent pre-mRNA, leads to a constitutive pathogen response via an EDS1/PAD4-dependent mechanism (Trost et al., 2013; Vi et al., 2013). Likewise, the Arabidopsis protein FPA, which functions independently of the CPSF complex to regulate alternative polyadenylation, has been shown to negatively modulate the flagellin response (Lyons et al., 2013). Combining all of these results, it is tempting to speculate that some pathogen defense responses are at least partly mediated by altering the mRNA polyadenylation status and, thus, the expression of a subset of biotic response factors. This is consistent with our transcriptomic data showing that impaired CPSF30 function not only prevents the constitutive activation of defense pathways but also decreases these pathways to basal values below those observed in wild-type plants. A link with RNA silencing could also be considered, since CPSF100, another member of the polyadenylation complex, has been shown to prevent the formation of aberrant RNAs that would normally enter RNA silencing (Herr et al., 2006).
A role for CPSF30 and other subunits of the CPSF complex as actors in immune responses is relevant outside of the plant kingdom. Indeed, CPSF30 has been shown to be a target of the Nonstructural Protein1 (NS1) of the Influenza A virus. NS1 modifies cellular pre-mRNA processing, including 3′ end formation, by binding to CPSF30, thereby inhibiting interferon-β mRNA processing and the processing of other cellular mRNAs, leading to a blockade of the host innate immune response (Nemeroff et al., 1998; Krug et al., 2003; Noah et al., 2003; Twu et al., 2006). Similarly, the Human immunodeficiency 1 virus recently was shown to require the binding of CPSF6 to its capsid proteins to escape the activation of the innate response in macrophages (Rasaiyaah et al., 2013). Altogether, these results point to intimate relationships between innate immunity and mRNA processing that involve the recruitment of proteins such as CPSF subunits during evolution to govern differential gene expression and activate defense mechanisms.
CONCLUSION
Our work provides, to our knowledge, the first example of CPSF-mediated cell death suppression in a plant. Although OXT6 is the only gene that encodes an obvious CPSF30 factor in the Arabidopsis genome, and genome-wide analyses have revealed that poly(A) site choice in a large majority of Arabidopsis genes is altered in the oxt6 mutant, loss of function of CPSF30 leads to differing but somewhat specific or focused effects, such as the effect on biotic stress responses. Unlike in Arabidopsis, the ortholog of CPSF30 in yeast, Yth1p, is an essential protein (Tacahashi et al., 2003). Thus, it could be argued that, in plants, polyadenylation and the splicing machinery have evolved differentially to fine-tune an apparently wide range of plant-specific responses (e.g. oxidative stress, plant immune responses).
Cell death is strictly regulated to avoid induction during normal growth and prevent unrestricted cell death in response to infection. Our understanding of the SA-mediated defense network has become increasingly more complex with the expanding discovery of additional SA components, not to mention interactions among the components. This complexity is augmented by the fact that SA also cross talks with several other defense responses and hormone signaling pathways. The discovery of RNA-binding proteins involved in the SA-mediated defense signaling network will contribute to our understanding of the posttranscriptional regulation of plant responses during biotic stress and the capacity of this regulation to adapt to and confront pathogens.
MATERIALS AND METHODS
Plant Material and Growth Conditions
T-DNA and other mutant lines of Arabidopsis (Arabidopsis thaliana) were all in the Col-0 background. The oxt6 mutant and the oxt6 CPSF30 complemented line were kind gifts from Deane L. Falcone, cat2 (SALK_059978) was from Graham Noctor, and mpk4 (SALK_056245) was from Heribert Hirt. The lsd1 (SALK_042687) and mips1 (SALK_023626) mutants were obtained from public seed banks (Alonso et al., 2003). Genotyping of these T-DNA lines was performed using the oligonucleotides indicated in Supplemental Table S3. All double mutants were obtained by crossing and were genotyped with the appropriate oligonucleotides listed in Supplemental Table S3.
For primary root length and cotyledon analyses, seeds were sown on commercially available 0.5× Murashige and Skoog medium (Basalt Salt Mixture M0221; Duchefa) solidified with 0.8% (w/v) agar (Phyto-Agar HP696; Kalys) and grown in an LD growth chamber (16-h day/8-h night, 20°C). For all other analyses, plants were grown 1 week as previously and transferred to soil under SD conditions (8-h day/16-h night, 200 µmol photons s−1 m−2, 21°C) for 2 weeks. Plants were then subsequently transferred to LD conditions (16-h day/8-h night, 200 µmol photons s−1 m−2, 21°C) for the times indicated.
Cell Death Assay: Electrolyte Loss and Trypan Blue Staining
For electrolyte loss, leaf discs were removed with a 5-mm punch and washed in distilled water for 10 min. Six leaf discs per genotype were transferred to a tube containing 2 mL of distilled water and agitated 2 h. Conductivity was then measured with the CDM 210 conductivity meter (Radiometer Analytical) and expressed as µS per leaf surface (cm2). Means and sd were from four replicates per genotype per experiment.
For trypan blue staining, rosettes were infiltrated three times under vacuum with lactophenol trypan solution (2.5 mg mL−1 trypan blue, 25% lactic acid, 23% water-saturated phenol, 25% glycerol, and water). Samples were then rinsed with distilled water and heated over boiling water for 2 min. After cooling, samples were destained and cleared with a saturated chloral hydrate solution and observed with a binocular loupe (Zeiss Stemi SV 11).
RNA Extraction and Quantitative RT-PCR
Total RNA was extracted from rosette leaves using the Nucleospin RNA kit (Macherey-Nagel) according to the manufacturer’s instructions. First-strand complementary DNA was synthesized from 2 µg of total RNA using Improm-II reverse transcriptase (A3802; Promega) according to the manufacturer’s instructions. One-twenty-fifth of the synthesized complementary DNA was mixed with 100 nm solution of each oligonucleotide and LightCycler 480 Sybr Green I master mix (Roche Applied Science) for quantitative PCR analysis. Products were amplified and fluorescent signals acquired with the LightCycler 480 detection system. The specificity of amplification products was determined by melting curves. AtUBQ10 was used as an internal control for signal normalization. Exor4 relative quantification software (Roche Applied Science) automatically calculates the relative expression levels of selected genes with algorithms based on the delta delta Cycle threshold method. Data were from duplicates of at least two biological replicates. The oligonucleotides used are described in Supplemental Table S3.
Transcriptome Studies
For the mips1 mutant, the microarray analysis was done previously by Meng et al. (2009) and was performed at the Unité de Recherche en Génomique Végétale using CATMA version 2 containing 24,276 gene-specific tags. In this study, microarray analysis for oxt6, mips1 oxt6, and mips1 oxt6 CPSF30 lines was also performed at the Unité de Recherche en Génomique Végétale using the new CATMA version 6.2 containing 38,511 probes. Wild-type and mutant plants were grown under SD conditions for 3 weeks and then transferred to LD conditions during 4 d to induce lesion formation. Total RNA was obtained by pooling rosette leaves of several plants and was extracted using the Nucleospin RNA kit (Macherey-Nagel); two replicates per line were extracted and used for transcriptome analysis. One dye swap (technical replicate with fluorochrome reversal) was made for each biological repetition. The RT of RNA in the presence of Cy3-dUTP or Cy5-dUTP, the hybridization of labeled samples to the slides, and the scanning of the slides were performed as described previously by Lurin et al. (2004).
For each comparison, one technical replicate with fluorochrome reversal was performed. Microarray data from this article were deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE53247 and at CATdb (http://urgv.evry.inra.fr/CATdb/) with accession number AU12-06_CPSF30.
Microarray Data Analysis and Hierarchical Clustering
For each array, the raw data comprised the logarithm of the median feature pixel intensity at wavelengths 635 nm (red) and 532 nm (green), and no background was subtracted. An array-by-array normalization was performed to remove systematic biases. First, spots considered as badly formed features were excluded. Then, a global intensity-dependent normalization using the Loess procedure was performed to correct the dye bias (Yang et al., 2002). Finally, for each block, the log-ratio median calculated over the values for the entire block was subtracted from each individual log-ratio value to correct print-tip effects. Differential analysis was based on the log ratios averaged on the dye swap: the technical replicates were averaged to get one log ratio per biological replicate, and these values were used to perform a paired Student’s t test. A trimmed variance is calculated from spots that do not display extreme variance (Gagnot et al., 2008). The raw P values were adjusted by the Bonferroni method, which controls the family-wise error rate in order to keep a strong control of the false positives in a multiple-comparison context. We considered probes as being differentially expressed after Bonferroni correction at P < 0.05.
For hierarchical clustering, we selected genes that were differentially expressed in at least two genotypes. Genes were considered as being differentially expressed when the mean of the two biological replicates had a P < 0.05 after Bonferroni correction. Hierarchical clustering was performed using Genesis software (Sturn et al., 2002), with average linkage as agglomeration rule.
GO functional annotations were determined using The Arabidopsis Information Resource version 8.
SA, Scopoletin, and Camalexin Quantifications
Wild-type and mutant plants were grown under SD conditions for 3 weeks and subsequently transferred to LD conditions. About 100 mg of leaves was harvested just before transfer to LD conditions (SD conditions) and 4 and 5 d after transfer to LD conditions. Metabolites were extracted as described by Baillieul et al. (1995) with the following modifications. [14C]SA was added to each sample for extraction loss correction. Samples were dried in a SpeedVac (Savant Instruments). Half of the dried samples were directly used to analyze camalexin and free forms of SA and scopoletin. The other dried samples were subjected to acidic hydrolysis in order to determine total SA (free plus conjugated forms) and scopoletin. HPLC analyses were achieved according to Simon et al. (2010) using the Waters system described previously. Peak purity determination and extraction loss correction were performed as mentioned previously. SA, scopoletin, and camalexin were quantified using a standard curve obtained with their respective standards. Data were analyzed using Empower Pro Software (Waters). Correction for losses was done by quantifying the remaining [14C]SA in each sample, as described previously by Baillieul et al. (1995), using an LS 6500 Multi-Purpose Scintillation Counter (Beckman Coulter). The data presented here are averages of the results obtained from four biological replicates.
Glutathione Quantification and ROS Detection
For glutathione quantification, four biological replicates obtained by pooling rosette leaves from several plants were tested for each genotype. At the time points indicated, approximately 100 mg of tissue was harvested. Extraction and measurement of oxidized and reduced forms of glutathione were measured by plate reader assay as described by Queval and Noctor (2007).
Accumulation of ROS was detected using DAB staining (Dutilleul et al., 2003).
Pathogenic Assays
The virulent strain Pseudomonas syringae pv tomato DC3000 and the avirulent strain Pst-AvrRpm1 were obtained from Jane Glazebrook (University of Minnesota). Bacteria were grown overnight at 30°C in Luria-Bertani medium with appropriate antibiotics (25 µg mL−1 kanamycin and 50 µg mL−1 rifampicin). Bacteria were washed in 10 mm MgCl2 and prepared for inoculations as described by Langlois-Meurinne et al. (2005). Whole leaves of 6- to 7-week-old plants, grown in SD conditions, were infiltrated with bacteria at a concentration of 105 cfu mL−1, using a 1-mL syringe without a needle. For the SA assays after pathogenic infection, a concentration of 107 cfu mL−1 was used. Leaf discs (0.28 cm2 each) were harvested from inoculated leaves at 0, 24, and 48 h post inoculation for the inoculation with Pst and 0 and 48 h post inoculation for the inoculation with Pst-AvrRpm1. For each time point, five samples were made by pooling four leaf discs from different treated plants. The leaves of five different plants were harvested at each time point. Bacterial growth was assessed as described by Tao et al. (2003) with the following modifications. Leaf discs were homogenized in 400 µL of sterile water, and serial dilutions (1:10) were performed for each sample. Several 10-µL aliquots of these 1:10 serial dilutions were spotted on solid Luria-Bertani medium containing kanamycin (25 µg mL−1) and rifampicin (50 µg mL−1), and colony numbers were quantified after 2 to 3 d of incubation at 30°C. Statistical analyses of the differences between two means of log-transformed data were performed according to unpaired Student’s t test.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Enriched GO terms of the genes in clusters 2 and 4.
Supplemental Figure S2. Comparisons of ROS production in the wild type (Col-0), mips1, oxt6, mips1 oxt6, and mips1 oxt6 CPSF30.
Supplemental Figure S3. Camalexin and total scopoletin contents in the indicated genotypes and real-time RT-PCR analysis of PAD3 expression in the indicated lines.
Supplemental Figure S4. Total SA levels in the indicated genotypes after inoculation with Pst.
Supplemental Figure S5. Real-time RT-PCR analysis of the two OXT6-encoded mRNAs in wild-type plants and the mips1 mutant in LD conditions.
Supplemental Table S1. Gene accession numbers of the different clusters with their functional annotations and log ratios averaged from two biological replicates.
Supplemental Table S2. Annotated glutathione-related genes showing no significantly modified transcript levels in mips1, oxt6, mips1 oxt6, and mips1 oxt6 CPSF30.
Supplemental Table S3. Sequences of the oligonucleotides used in this study.
Supplementary Material
Acknowledgments
We thank Deane L. Falcone (University of Massachusetts) for the oxt6 and oxt6 CPSF30 lines, Graham Noctor (Institut de Biologie des Plantes, Orsay) for the cat2 seeds, Heribert Hirt (Unité de Recherche en Génomique Végétale, Institut National de la Recherche Agronomique, Evry) for mpk4 seeds and constructive discussions, Vincent Thareau (Institut de Biologie des Plantes, Orsay) and Marie-Laure Martin-Magniette (Unité de Recherche en Génomique Végétale, Evry) for assistance with bioinformatic and statistical analysis, Mathilde Fagard (Institut National de la Recherche Agronomique, Versailles) for assistance with 2′,7′-dichlorofluorescein diacetate assays, Quentin Hocheux (Institut de Biologie des Plantes, Orsay) for assistance during the phenotypic characterization, and Allison Mallory (Institut de Biologie des Plantes, Orsay) for helpful corrections regarding the manuscript.
Glossary
- PCD
programmed cell death
- PTI
pathogen-associated molecular pattern-triggered immunity
- ETI
effector-triggered immunity
- HR
hypersensitive response
- ROS
reactive oxygen species
- SA
salicylic acid
- LMM
lesion-mimic mutant
- LD
long-day
- MI
myoinositol
- H2O2
hydrogen peroxide
- Col-0
Columbia-0
- T-DNA
transfer DNA
- UTR
untranslated region
- SD
short-day
- GO
Gene Ontology
- DAB
3,3′-diaminobenzidine
- Pst
Pseudomonas syringae pv tomato DC3000
- Pst-AvrRpm1
Pseudomonas syringae pv tomato DC3000 expressing the Avirulence gene avrRpm1
- RT
reverse transcription
- PtdIns
phosphatidylinositol
- CATMA
Complete Arabidopsis Transcriptome MicroArray
- cfu
colony-forming units
Footnotes
This work was supported by the Centre National de la Recherche Scientifique, the Université Paris-Sud, the Agence Nationale de la Recherche (grant no. MAPK-IPS ANR–2010–BLAN–1613–02), and the Labex Saclay Plant Sciences.
The online version of this article contains Web-only data.
References
- Addepalli B, Hunt AG. (2007) A novel endonuclease activity associated with the Arabidopsis ortholog of the 30-kDa subunit of cleavage and polyadenylation specificity factor. Nucleic Acids Res 35: 4453–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Baillieul F, Genetet I, Kopp M, Saindrenan P, Fritig B, Kauffmann S. (1995) A new elicitor of the hypersensitive response in tobacco: a fungal glycoprotein elicits cell death, expression of defence genes, production of salicylic acid, and induction of systemic acquired resistance. Plant J 8: 551–560 [DOI] [PubMed] [Google Scholar]
- Blanco F, Garretón V, Frey N, Dominguez C, Pérez-Acle T, Van der Straeten D, Jordana X, Holuigue L. (2005) Identification of NPR1-dependent and independent genes early induced by salicylic acid treatment in Arabidopsis. Plant Mol Biol 59: 927–944 [DOI] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Weimar T, Michaelson LV, Hawkins ND, Macaskill A, Napier JA, Beale MH, Lilley KS, Dupree P. (2005) Analysis of detergent-resistant membranes in Arabidopsis: evidence for plasma membrane lipid rafts. Plant Physiol 137: 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Nam J, Dangl JL. (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA 95: 15849–15854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Nielsen HB, Zhu S, Newman MA, Shokat KM, Rietz S, Parker J, Mundy J. (2006) Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J 47: 532–546 [DOI] [PubMed] [Google Scholar]
- Chaouch S, Noctor G. (2010) Myo-inositol abolishes salicylic acid-dependent cell death and pathogen defence responses triggered by peroxisomal hydrogen peroxide. New Phytol 188: 711–718 [DOI] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Vanderauwera S, Mhamdi A, Vandorpe M, Langlois-Meurinne M, Van Breusegem F, Saindrenan P, Noctor G. (2010) Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol 153: 1692–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xiong L. (2010) Myo-inositol-1-phosphate synthase is required for polar auxin transport and organ development. J Biol Chem 285: 24238–24247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J, Hirt H. (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL. (2011) Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ. (2011) Phosphoinositides and the regulation of tubular-based endosomal sorting. Biochem Soc Trans 39: 839–850 [DOI] [PubMed] [Google Scholar]
- Delaney KJ, Xu R, Zhang J, Li QQ, Yun KY, Falcone DL, Hunt AG. (2006) Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol 140: 1507–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pinto MC, Locato V, De Gara L. (2012) Redox regulation in plant programmed cell death. Plant Cell Environ 35: 234–244 [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL. (1994) Arabidopsis mutants simulating disease resistance response. Cell 77: 565–577 [DOI] [PubMed] [Google Scholar]
- Donahue JL, Alford SR, Torabinejad J, Kerwin RE, Nourbakhsh A, Ray WK, Hernick M, Huang X, Lyons BM, Hein PP, et al. (2010) The Arabidopsis thaliana myo-inositol 1-phosphate synthase1 gene is required for myo-inositol synthesis and suppression of cell death. Plant Cell 22: 888–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil-Maurizi C, Vitecek J, Marty L, Branciard L, Frettinger P, Wendehenne D, Meyer AJ, Mauch F, Poinssot B. (2011) Glutathione deficiency of the Arabidopsis mutant pad2-1 affects oxidative stress-related events, defense gene expression, and the hypersensitive response. Plant Physiol 157: 2000–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Garmier M, Noctor G, Mathieu C, Chétrit P, Foyer CH, de Paepe R. (2003) Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15: 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR, Rammelt C, Wahle E. (2011) Control of poly(A) tail length. Wiley Interdiscip Rev RNA 2: 348–361 [DOI] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. (2013) Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14: 496–506 [DOI] [PubMed] [Google Scholar]
- Gagnot S, Tamby JP, Martin-Magniette ML, Bitton F, Taconnat L, Balzergue S, Aubourg S, Renou JP, Lecharny A, Brunaud V. (2008) CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res 36: D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GhoshDastidar K, Chatterjee A, Chatterjee A, Majumder AL. (2006) Evolutionary divergence of L-myo-inositol 1-phosphate synthase: significance of a “core catalytic structure.” Subcell Biochem 39: 315–340 [PubMed] [Google Scholar]
- Gillaspy GE. (2011) The cellular language of myo-inositol signaling. New Phytol 192: 823–839 [DOI] [PubMed] [Google Scholar]
- Glawischnig E. (2007) Camalexin. Phytochemistry 68: 401–406 [DOI] [PubMed] [Google Scholar]
- Heath MC. (2000) Hypersensitive response-related death. Plant Mol Biol 44: 321–334 [DOI] [PubMed] [Google Scholar]
- Herr AJ, Molnàr A, Jones A, Baulcombe DC. (2006) Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc Natl Acad Sci USA 103: 14994–15001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Li Y, Zhang X, Zuo J, Yang S. (2010) The Arabidopsis LSD1 gene plays an important role in the regulation of low temperature-dependent cell death. New Phytol 187: 301–312 [DOI] [PubMed] [Google Scholar]
- Hunt AG. (2008) Messenger RNA 3′ end formation in plants. Curr Top Microbiol Immunol 326: 151–177 [DOI] [PubMed] [Google Scholar]
- Jabs T, Dietrich RA, Dangl JL. (1996) Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273: 1853–1856 [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. (2009) Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci 14: 373–382 [DOI] [PubMed] [Google Scholar]
- Krug RM, Yuan W, Noah DL, Latham AG. (2003) Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309: 181–189 [DOI] [PubMed] [Google Scholar]
- Langlois-Meurinne M, Gachon CM, Saindrenan P. (2005) Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol 139: 1890–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D, Jégu T, Meng PH, Mazubert C, Hudik E, Delarue M, Charon C, Crespi M, Hirt H, Raynaud C, et al. (2013) Dual function of MIPS1 as a metabolic enzyme and transcriptional regulator. Nucleic Acids Res 41: 2907–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Qin G, Zhang J, Liang Y, Song Y, Zhao M, Tsuge T, Aoyama T, Liu J, Gu H, et al. (2011) D-myo-inositol-3-phosphate affects phosphatidylinositol-mediated endomembrane function in Arabidopsis and is essential for auxin-regulated embryogenesis. Plant Cell 23: 1352–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons R, Iwase A, Gänsewig T, Sherstnev A, Duc C, Barton GJ, Hanada K, Higuchi-Takeuchi M, Matsui M, Sugimoto K, et al. (2013) The RNA-binding protein FPA regulates flg22-triggered defense responses and transcription factor activity by alternative polyadenylation. Sci Rep 3: 2866–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. (2008) Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci 65: 1099–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng PH, Raynaud C, Tcherkez G, Blanchet S, Massoud K, Domenichini S, Henry Y, Soubigou-Taconnat L, Lelarge-Trouverie C, Saindrenan P, et al. (2009) Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS ONE 4: e7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Belin C, Delorme-Hinoux V, Reichheld JP, Riondet C. (2012) Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxid Redox Signal 17: 1124–1160 [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Hager J, Chaouch S, Queval G, Han Y, Taconnat L, Saindrenan P, Gouia H, Issakidis-Bourguet E, Renou JP, et al. (2010) Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol 153: 1144–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeder W, Yoshioka K. (2008) Lesion mimic mutants: a classical, yet still fundamental approach to study programmed cell death. Plant Signal Behav 3: 764–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenbock P, Szechynska-Hebda M, Plaszczyca M, Baudo M, Mateo A, Mullineaux PM, Parker JE, Karpinska B, Karpinski S. (2008) Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20: 2339–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LA, Kenton P, Lloyd AJ, Ougham H, Prats E. (2008) The hypersensitive response: the centenary is upon us but how much do we know? J Exp Bot 59: 501–520 [DOI] [PubMed] [Google Scholar]
- Murphy AM, Otto B, Brearley CA, Carr JP, Hanke DE. (2008) A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J 56: 638–652 [DOI] [PubMed] [Google Scholar]
- Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, Glazebrook J. (2007) Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C. (2007) SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J 50: 128–139 [DOI] [PubMed] [Google Scholar]
- Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. (1998) Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell 1: 991–1000 [DOI] [PubMed] [Google Scholar]
- Noah DL, Twu KY, Krug RM. (2003) Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAs. Virology 307: 386–395 [DOI] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Chaouch S, Han Y, Neukermans J, Marquez-Garcia B, Queval G, Foyer CH. (2012) Glutathione in plants: an integrated overview. Plant Cell Environ 35: 454–484 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H. (2009) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Queval G, Issakidis-Bourguet E, Hoeberichts FA, Vandorpe M, Gakière B, Vanacker H, Miginiac-Maslow M, Van Breusegem F, Noctor G. (2007) Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J 52: 640–657 [DOI] [PubMed] [Google Scholar]
- Queval G, Noctor G. (2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal Biochem 363: 58–69 [DOI] [PubMed] [Google Scholar]
- Rasaiyaah J, Tan CP, Fletcher AJ, Price AJ, Blondeau C, Hilditch L, Jacques DA, Selwood DL, James LC, Noursadeghi M, et al. (2013) HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503: 402–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci C, Aviv DH, Holt BF, III, Dangl JL, Parker JE. (2001) The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13: 2211–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Venu RC, Nobuta K, Wu X, Notibala V, Demirci C, Meyers BC, Wang GL, Ji G, Li QQ. (2011) Transcriptome dynamics through alternative polyadenylation in developmental and environmental responses in plants revealed by deep sequencing. Genome Res 21: 1478–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherstnev A, Duc C, Cole C, Zacharaki V, Hornyik C, Ozsolak F, Milos PM, Barton GJ, Simpson GG. (2012) Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat Struct Mol Biol 19: 845–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JS, Jung C, Lee S, Min K, Lee YW, Choi Y, Lee JS, Song JT, Kim JK, Choi YD. (2013) AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J 73: 483–495 [DOI] [PubMed] [Google Scholar]
- Simon C, Langlois-Meurinne M, Bellvert F, Garmier M, Didierlaurent L, Massoud K, Chaouch S, Marie A, Bodo B, Kauffmann S, et al. (2010) The differential spatial distribution of secondary metabolites in Arabidopsis leaves reacting hypersensitively to Pseudomonas syringae pv. tomato is dependent on the oxidative burst. J Exp Bot 61: 3355–3370 [DOI] [PubMed] [Google Scholar]
- Staiger D, Korneli C, Lummer M, Navarro L. (2013) Emerging role for RNA-based regulation in plant immunity. New Phytol 197: 394–404 [DOI] [PubMed] [Google Scholar]
- Stoilov P, Rafalska I, Stamm S. (2002) YTH: a new domain in nuclear proteins. Trends Biochem Sci 27: 495–497 [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18: 207–208 [DOI] [PubMed] [Google Scholar]
- Tacahashi Y, Helmling S, Moore CL. (2003) Functional dissection of the zinc finger and flanking domains of the Yth1 cleavage/polyadenylation factor. Nucleic Acids Res 31: 1744–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F. (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PE, Wu X, Liu M, Gaffney B, Ji G, Li QQ, Hunt AG. (2012) Genome-wide control of polyadenylation site choice by CPSF30 in Arabidopsis. Plant Cell 24: 4376–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabinejad J, Gillaspy GE. (2006) Functional genomics of inositol metabolism. Subcell Biochem 39: 47–70 [DOI] [PubMed] [Google Scholar]
- Trost G, Vi SL, Czesnick H, Lange P, Holton N, Giavalisco P, Zipfel C, Kappel C, Lenhard M. (2013) Arabidopsis poly(A) polymerase PAPS1 limits founder-cell recruitment to organ primordia and suppresses the salicylic acid-independent immune response downstream of EDS1/PAD4. Plant J 77: 688–699 [DOI] [PubMed] [Google Scholar]
- Twu KY, Noah DL, Rao P, Kuo RL, Krug RM. (2006) The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J Virol 80: 3957–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Mortel JE, de Vos RC, Dekkers E, Pineda A, Guillod L, Bouwmeester K, van Loon JJ, Dicke M, Raaijmakers JM. (2012) Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol 160: 2173–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vi SL, Trost G, Lange P, Czesnick H, Rao N, Lieber D, Laux T, Gray WM, Manley JL, Groth D, et al. (2013) Target specificity among canonical nuclear poly(A) polymerases in plants modulates organ growth and pathogen response. Proc Natl Acad Sci USA 110: 13994–13999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Xing D, Li QQ. (2011) Alternative polyadenylation and gene expression regulation in plants. Wiley Interdiscip Rev RNA 2: 445–458 [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Addepalli B, Yun KY, Hunt AG, Xu R, Rao S, Li QQ, Falcone DL. (2008) A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS ONE 3: e2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu S, Ding P, Wang D, Cheng YT, He J, Gao M, Xu F, Li Y, Zhu Z, et al. (2010) Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 107: 18220–18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.