Abstract
Meiotic recombination normally takes place between allelic sequences on homologs. This process can also occur between non-allelic homologous sequences. Such ectopic interaction events can lead to chromosome rearrangements and are normally avoided. However, much remains unknown about how these ectopic interaction events are sensed and eliminated. In this study, using a screen in rice, we characterized a homolog of HUS1 and explored its function in meiotic recombination. In Oshus1 mutants, in conjunction with nearly normal homologous pairing and synapsis, vigorous, aberrant ectopic interactions occurred between nonhomologous chromosomes, leading to multivalent formation and subsequent chromosome fragmentation. These ectopic interactions relied on programed meiotic double strand breaks and were formed in a manner independent of the OsMER3-mediated interference-sensitive crossover pathway. Although early homologous recombination events occurred normally, the number of interference-sensitive crossovers was reduced in the absence of OsHUS1. Together, our results indicate that OsHUS1 might be involved in regulating ectopic interactions during meiosis, probably by forming the canonical RAD9-RAD1-HUS1 (9-1-1) complex.
Author Summary
Meiosis is a special type of cell division that generates gametes for sexual reproduction. During meiosis, recombination not only occurs between allelic sequences on homologs, but also between non-allelic homologous sequences at dispersed loci. Such ectopic recombination is the main cause of chromosomal alterations and accounts for numerous genomic disorders in humans. To ensure genomic integrity, those ectopic recombinations must be quickly resolved. Despite the importance of ectopic recombination suppression, the mechanism underlying this process still remains largely unknown. Here, using rice as a model system, we identified the rice HUS1 homolog, a member of the RAD9-RAD1-HUS1 (9-1-1) complex, and elucidated its roles in meiotic recombination. In Oshus1, vigorous ectopic interactions occur between nonhomologous chromosomes, and the number of crossovers is reduced. We suspect that OsHUS1 participates in regulating ectopic interactions during meiosis, probably by forming the canonical RAD9-RAD1-HUS1 (9-1-1) complex.
Introduction
Meiosis is a highly dynamic process in which chromosomes undergo dramatic structural changes and movements [1],[2]. During the course of meiosis, intimate interactions develop between homologous chromosomes. Among these interactions, homologous recombination (HR) and pairing are the core events that occur during the production of functional gametes [3], [4]. Meiotic recombination is a powerful determinant that creates genetic diversity and provides mechanical stability for the accurate separation of homologous chromosomes. Therefore, meiotic recombination has a strong bias towards homologous chromosomes rather than sister chromatids and is mediated by a complex mechanism [5], [6]. After DNA replication in the premeiotic S phase, a proteinaceous axis is assembled between two chromatids. Homologous recombination then occurs along the chromosomes, beginning with the formation of programmed double strand breaks (DSBs). In conjunction with the initiation of recombination, homologous chromosomes begin to align in pairs.
Studies have shown that for most species, homologous pairing depends on homologous recombination [7]. However, recombination not only occurs between allelic DNA sequences on homologs, but it also frequently occurs between dispersed non-allelic DNA segments that share high sequence similarity [8]. The latter recombination pattern is usually referred to as ectopic recombination (ER, also known as non-allelic homologous recombination). As eukaryotic genomes are rich in repeated DNA sequences, ER can produce chromosomal rearrangements, which in humans result in numerous genomic disorders [9], [10]. Despite the adverse impact of ER on genome integrity, ER occurs relatively frequently; in budding yeast, the frequency of ER is roughly on par with that of allelic recombination [11], [12]. To avoid the deleterious consequences of ER, cells have evolved multiple strategies to suppress ER formation [13]. One strategy is preventing DSB formation in or near DNA repeats. In budding yeast (Saccharomyces cerevisiae), suppression of DSBs in rDNA repeats depends strongly on silent information regulator 2 (Sir2), which encodes a histone deacetylase that promotes the formation of a closed, compact chromatin structure in the rDNA and other regions. Sir2 may suppress DSBs in rDNA in part through the formation of a nucleosomal conformation that is not permissive for SPO11 activity [14]. The second strategy is preventing the use of non-allelic homologous templates for recombination and/or favoring the use of allelic templates. In budding yeast, homologous alignment and synapsis restrict the ability of ectopically located sequences to find each other and recombine [15]. There are also reports on the competition between normal allelic recombination and ER [16]. As both mechanisms involve preventing ectopic interaction intermediate formation, we classified these events as ectopic interaction preventing mechanisms in this study. The frequent occurrence of ER in yeast suggests that these mechanisms cannot totally prevent all ER initiation [17]–[19]. Once a non-allelic partner is used as the template and ER intermediates are built, a mechanism monitors and resolves those ER intermediates into non-crossovers. This ER-eliminating mechanism can be classified as a surveillance mechanism. Studies in S. cerevisiae have shown that the mismatch repair proteins Pms1 and Msh2 are likely to be involved in this mechanism, although direct evidence for this is still lacking. Several DNA helicases, including Sgs1 in yeast and BLM in humans, may also possess anti-crossover activities that are potentially involved in preventing deleterious outcomes of meiotic ER [13].
Rad9, Hus1, and Rad1 (in S. cerevisiae: Ddc1, Mec3, and Rad17) interact in a heterotrimeric complex (dubbed the 9-1-1 complex), which resembles a PCNA-like sliding clamp [20], [21]. In response to genotoxic damage, the toroidal 9-1-1 complex is loaded around damage sites, collaborating with ATM and ATR to carry out its best known function of activating the DNA damage checkpoint [22]. In addition, studies have revealed functional interactions between 9-1-1 and multiple partners, most notably translation polymerases, base excision repair enzymes, and mismatch repair factors [23], [24]. This evidence implies that 9-1-1 also plays a direct role in DNA repair. In addition to its role in the conventional somatic DNA damage response, 9-1-1 also plays roles in meiosis. In S. cerevisiae, it is evident that Ddc1 colocalizes with Rad51 on meiotic chromosomes and is required for the pachytene checkpoint [25]. The rad17 mutants exhibit aberrant synapsis and increased rates of ER during meiosis [26]. In mouse, RAD1 was found to be associated with both synapsed and unsynapsed chromosomes during prophase I [27]. Recently, the HUS1 homologs in Drosophila and mouse were reported to be essential for meiotic DSB repair [28], [29].
The function of the 9-1-1 complex in suppressing meiotic ER was first suggested in yeast [26]. However, this function has not been reported in higher organisms, likely due to a lack of direct cytological evidence. In this study, we aimed to isolate genes that are involved in ectopic interaction suppression. Several mutants showing normal homologous pairing at pachytene and the appearance of ectopic interactions at diakinesis were isolated. Among these, two allelic mutants were characterized in detail. These mutants were found to be mutated in the functional homolog of fission yeast and mammalian HUS1. In the Oshus1 mutants, meiotic homologous pairing took place normally during prophase I, while nonhomologous chromosomes interacted vigorously as well. Multivalents were frequently found to be arranged on the equatorial plate at metaphase I. Chromosome bridges and fragments occurred at anaphase I and telophase I, rendering the mutants completely sterile. These results suggest that OsHUS1 might specifically function in sensing and removing aberrant associations between non-allelic sequences during meiosis, probably via the 9-1-1 complex.
Results
Cloning of OsHUS1
Among our rice sterile mutant libraries, 16 lines with phenotypes meeting the criteria mentioned above were isolated. One of the mutant lines, S7678, which was derived from Nipponbare (a japonica cultivar) tissue culture, was selected for further study. Based on information about its mutation (see below), the mutant was named Oshus1-1. The Oshus1-1 plants did not exhibit defects in vegetative growth under natural growth conditions, except for total male sterility (Figure S1). Fertile plants and sterile plants from the progeny of Oshus1-1+/− produced a 3∶1 segregation ratio (fertile, 214; sterile, 66), which established this mutant as a single recessive mutant (χ2 = 0.30; P>0.05). When we pollinated the mutant flowers with wild-type pollen, the mutant did not set seed, indicating that female fertility is also affected in this mutant.
We isolated OsHUS1 by map-based cloning. A mapping population was constructed by crossing Oshus1-1+/− plants to Nanjing 11 (an indica cultivar) plants. The mutant gene locus was mapped to a physical region of approximately 100 kb on the long arm of chromosome 4. According to information obtained from the public database (Rice Genome Annotation Project, http://rice.plantbiology.msu.edu), we sequenced several genes in this region. As a result, a point mutation (A to T) was found in the gene Os04g44620, which introduced a stop codon (AAG to TAG) in the second exon. We named the mutant Oshus1-1 based on the homology of the protein sequence.
Next, we isolated another mutant from Huanghuazhan (an indica cultivar), which has the same phenotype as that of Oshus1-1. Using map-based cloning and DNA sequencing, we found that this mutant carries a ten-nucleotide deletion in the fourth exon of OsHUS1, causing frame shift and premature stop codon formation. We named this allele Oshus1-2. The chromosome behavior in Oshus1-2 meiocytes was the same as Oshus1-1 (Figure S2A).
We generated a gene-specific p35S OsHUS1-RNAi construct and used it to transform Yandao 8 (a japonica cultivar) rice. Most OsHUS1-RNAi lines showed a severe reduction in fertility (93%, n = 30), and the chromosome behavior in the male meiocytes of these lines mirrored that of Oshus1 (Figure S2B). From these results, we conclude that the mutation in the OsHUS1 gene led to the sterility phenotype.
Characterization of OsHUS1
There are three full-length cDNA sequences of Os04g44620 published in the Rice Genome Annotation Project website, including AK107445, AK101159, and AK064120. Using RT-PCR and RACE (rapid amplification of cDNA ends) on young panicles, we found that AK064120 is the correct sequence for this gene. Alignment of the cDNA sequence with the genomic sequence revealed that OsHUS1 is composed of six exons and five introns (Figure S3). The open reading frame of OsHUS1 has a length of 981 bp, encoding a 326 amino-acid peptide. Using BLASTp, we found that OsHUS1 shares some similarity (approximately 25% identity and 45% similarity) with the HUS1 protein in mammals and fission yeast (Figure S4). Reciprocal BLAST searches further confirmed that the isolated protein is the closest relative of HUS1 in rice (Figure S5).
As shown above, there were several defects during meiosis in Oshus1. We then examined the spatial and temporal expression patterns of OsHUS1. Using quantitative RT-PCR, we found that OsHUS1 could be detected as early as the seedling stage. In adult-stage rice, OsHUS1 was expressed not only in young panicles but also in vegetative organs such as leaves, roots, and internodes (Figure S6), with the highest expression observed in leaf blades.
Chromosome behavior in Oshus1-1
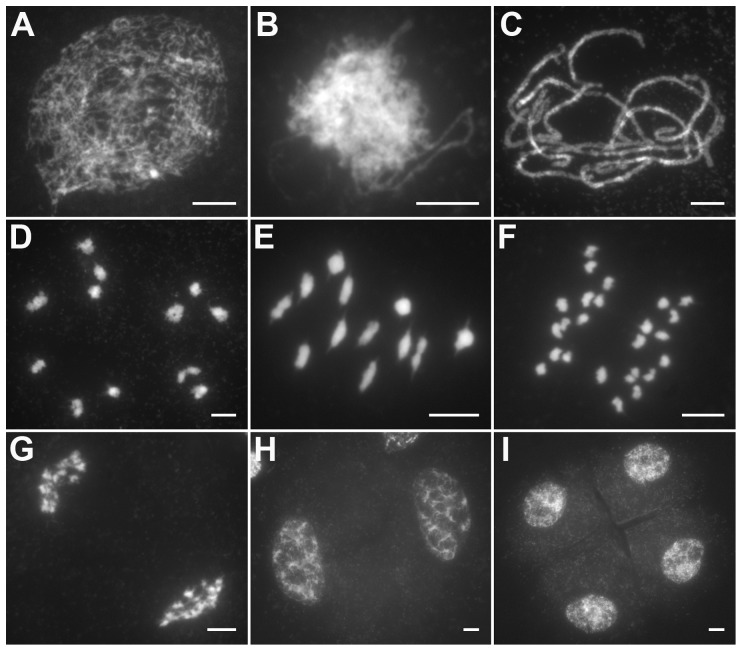
The behavior of meiotic chromosomes was revealed by 4′6-diamidino-2-phenylindole (DAPI) staining. In wild-type pollen mother cells (PMCs), meiosis began with chromosome condensation and the appearance of chromosomes as thin, thread-like structures at leptotene (Figure 1A). As zygotene progressing, homologous chromosomes underwent pairing and synapsis (Figure 1B). During pachytene, homologous pairing culminated in the formation of synaptonemal complexes (SCs; Figure 1C). After the disassembly of the SC at diplotene, the resulting 12 bivalents were further condensed, revealing the presence of chiasmata at diakinesis (Figure 1D). At metaphase I, the bivalents were aligned in the middle of the cell (Figure 1E). Homologous chromosomes separated and migrated toward opposite poles at anaphase I and telophase I (Figure 1F and 1G), generating dyads at the end of meiosis I (Figure 1H). Then, the dyads underwent meiosis II and finally produced tetrads (Figure 1I).
Figure 1. Meiosis in the wild type.
(A) Leptotene. (B) Zygotene. (C) Pachytene. (D) Diakinesis. (E) Metaphase I. (F) Anaphase I. (G) Telophase I. (H) Dyad. (I) Tetrad. Bars, 5 µm.
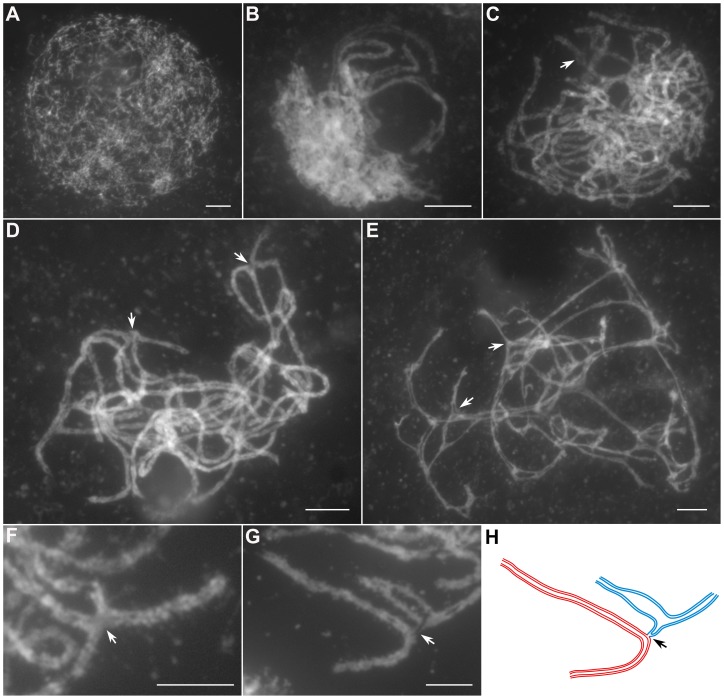
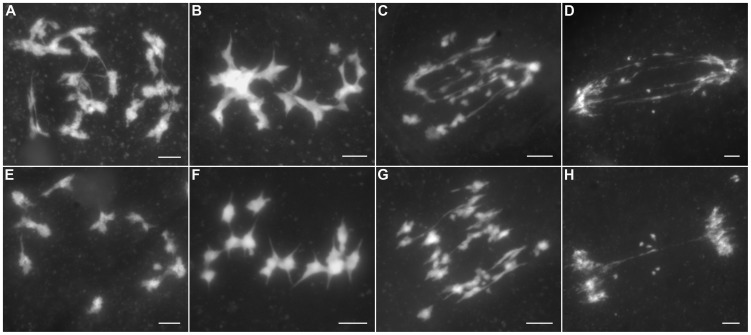
In Oshus1-1 PMCs, the chromosome behaviors appeared the same as those observed in wild-type from leptotene to zygotene (Figure 2A and 2B). However, anomalies began to be manifested at early pachytene. At first glance, almost all homologous chromosomes aligned well. However, upon careful examination, we found that some regions of the chromosomes could not complete close alignment perfectly and exhibited “bubble-like” structures (Figure 2C). During middle pachytene stage, associations between nonhomologous chromosome were observed in all PMCs (n = 252), which caused the chromosomes to stick to each other (Figure 2D). At late pachytene, this type of association became more prominent (Figure 2E–H). At diakinesis, multivalents were detected in all PMCs (n = 521). These multivalents ranged in size from associations of four chromosomes to the extreme case of 24 chromosomes (Figure 3A, 3E); the average number of bivalents per cell was only 1.6. At metaphase I, multivalents and bivalents were located on the equatorial plate due to the drag force exerted on centromeres by spindle fibers (Figure 3B, 3F). During anaphase I, the multivalents and bivalents fell apart, and extensive chromosome bridges and fragments were observed (Figure 3C, 3G). At telophase I, two masses of chromosomes arrived at opposite poles of the nuclei, and several distinct dot-like chromosome fragments still remained on the equatorial plate (Figure 3D, 3H). In a few cells (4%, n = 881), up to 10 or 11 homolog pairs could be individualized at diakinesis and metaphase I. We also found some cells with a small amount of chromosome bridges and fragments at anaphase I and telophase I (Figure 3E, F). These types of defects were maintained during meiosis II, and no normal tetrad was produced.
Figure 2. Meiotic chromosome behaviors of PMCs in the Oshus1-1 mutant.
(A) Leptotene. (B) Zygotene. (C) Early pachytene. (D) Middle pachytene. (E) Late pachytene. (F, G) The enlarged chromosome ectopic association regions. (H) Diagram shows chromosome configuration in (G). Red and blue lines indicate sister chromatids in different bivalents. The arrow indicates “bubble-like” region in (C) and ectopic association site in (D–H). Bars, 5 µm.
Figure 3. The aberrant chromosomal interactions in Oshus1-1 resulted in multivalent associations and chromosome fragments.
(A–D) exhibit PMCs with high frequencies of ectopic chromosomal interactions, while (E–H) show PMCs with low levels of ER. (A, E) Diakinesis. (B, F) Metaphase I. (C, G) Anaphase I. (D, H) Telophase I. Bars, 5 µm.
Synapsis is incomplete in most Oshus1-1 meiocytes
By performing DAPI staining of pachytene chromosomes, we found that in Oshus1-1, homologous chromosomes could pair normally. To validate whether normal SCs were affected by the mutation of OsHUS1, we performed immunofluorescent examination using antibodies against ZEP1 in Oshus1-1 PMCs. ZEP1 is the transverse filament protein of SC in rice and hence, a perfect tool to mark the course of synapsis [30]. In leptotenic and zygotenic Oshus1-1 PMCs, the ZEP1 patterns appeared as dots and short fragments, which were identical to those of the wild-type (Figure 4A and 4B). During pachytene, only approximately 10% (n = 300) of the meiocytes showed full-length ZEP1 signals along the homologous chromosomes (Figure 4C). In the remaining 90% of meiocytes, linear ZEP1 signals extended and could be detected along almost the entire chromosomes, with the exception of a few discontinuities/gaps, some of which exhibited the “bubble-like” structures mentioned above (Figure 4D and 4E). The discontinuities/gaps of ZEP1 signals indicate that the SC integrity might be slightly affected by the mutation of OsHUS1.
Figure 4. Immunolocalization of ZEP1 in the Oshus1-1 mutant.
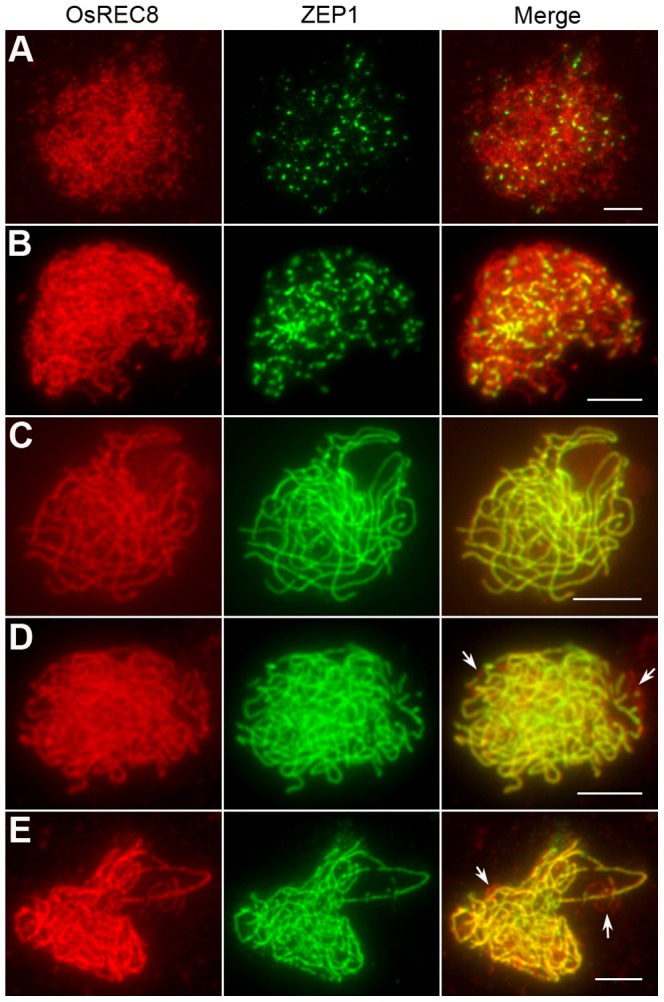
(A) Leptotene. (B) Zygotene. (C–E) Pachytene. Image (C) shows complete ZEP1 signals along the meiotic chromosomes in the pachytene PMC, while (D) and (E) exhibit discontinuities of ZEP1 linear signals. Arrows indicate these gaps. Bars, 5 µm.
Oshus1 mutants show a reduced number of bright HEI10 foci
Many incidents during meiosis are believed to be interdependent, e.g., pairing is recombination-dependent in mammals and higher plants. The nearly normal SC assembly observed in Oshus1 meiocytes is reminiscent of the proper loading of important factors involved in SC assembling and homologous recombination. To further verify the relationship between OsHUS1 and several other meiotic recombination factors, immunodetection was carried out in Oshus1-1 using antibodies against PAIR3, PAIR2, OsZIP4, OsMER3, and HEI10.
PAIR2 is the rice homolog of yeast HOP1 and Arabidopsis ASY1, which associates with unpaired chromosome axes at early meiosis I. PAIR3 is also an axis-associated protein that can bind both unpaired chromosomes and paired chromosomes. Both PAIR2 and PAIR3 are usually utilized to mark the meiotic chromosome axis, and they also play fundamental roles in the recombination process [31]–[33]. OsMER3 and OsZIP4 are members of the ZMM protein family and are essential for early meiotic HR in rice [34], [35]. In the Oshus1-1 mutant, PAIR2 appeared as foci at leptotene and associated with the chromosome axis as linear signals at early zygotene (Figure 5A). PAIR3 signals were first observed as dots at early leptotene and then elongated gradually along the entire lengths of the chromosomes during zygotene (Figure 5B). The appearance of OsMER3 and OsZIP4 commenced at early leptotene, and the number of OsMER3 foci (average 257±15, n = 44, range 221–281) and OsZIP4 foci (average 299±22, n = 35, range 289–328) reached its peak at early zygotene (Figure 5C and 5D); similar results were obtained in the wild-type. At pachytene, both OsMER3 and OsZIP4 decreased rapidly and no signals were found in the later stages in the wild-type and Oshus1-1. The normal loading patterns of these four meiotic factors showed that early HR in Oshus1-1 is not disturbed.
Figure 5. Immunolocalization of several meiotic elements in the Oshus1-1 mutant.
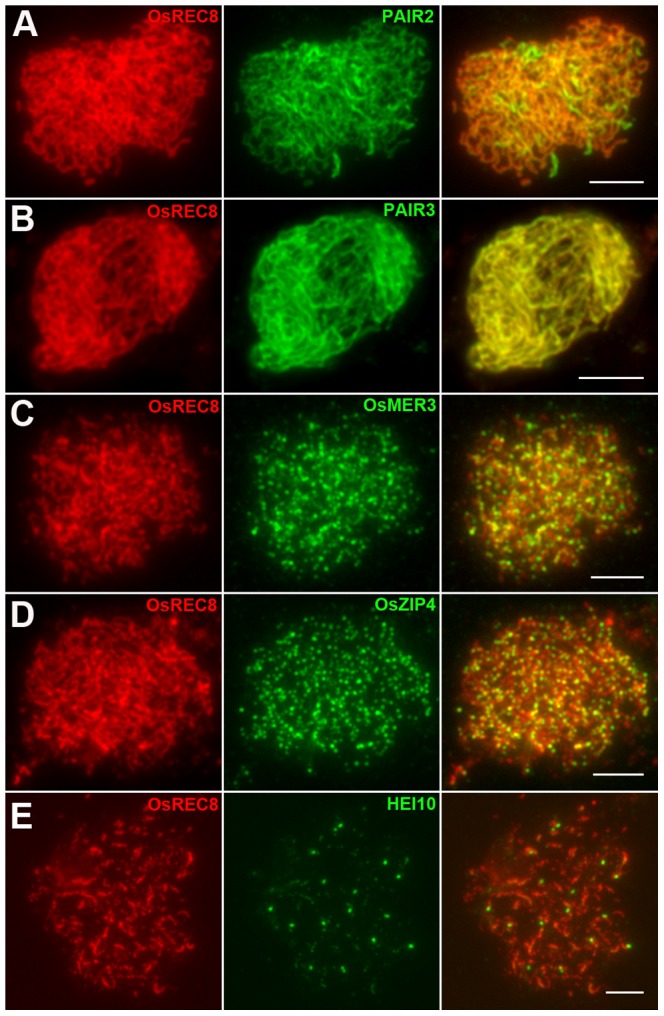
(A) PAIR2 signals at zygotene. (B) PAIR3 signals at zygotene. (C) OsMER3 signals at leptotene. (D) OsZIP4 signals at leptotene. (E) HEI10 bright foci at diplotene. Scale bars, 5 µm.
Previous studies suggest that the interference-sensitive pathway accounts for most of the crossovers (COs) in rice [34]–[36]. We thus wanted to know whether interference-sensitive COs were affected by the mutation of OsHUS1. The HEI10 prominent foci correspond to the interference-sensitive CO sites in rice [36]. We counted the number of HEI10 foci (average 16.9±1.9, n = 17, range 13–20) in Oshus1-1 (Figure 5E) and compared that with the corresponding data for the wild-type (average 24.5±1.8, n = 30, range 22–28). We found that the mean number of HEI10 bright foci of Oshus1-1 was significantly reduced compared with that of the wild-type (t [45] = 13.8, P<0.01). Therefore, the number of interference-sensitive COs is reduced in Oshus1-1 due to the loss of OsHUS1.
Ectopic associations in Oshus1-1 are dependent on PAIR1 but independent of OsRAD51C
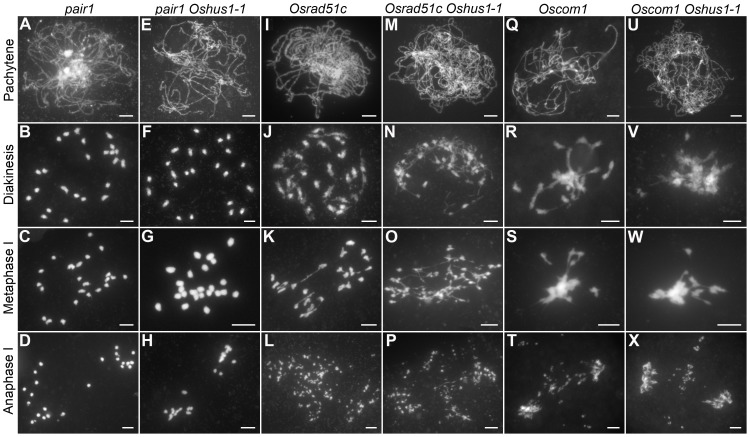
Meiotic recombination is initiated by the formation of DSBs, which is catalyzed by SPO11 proteins; these proteins have been identified in budding yeast, Arabidopsis, and animals [37]. However, to date, no spo11 mutants have been isolated in rice [38]. Recently, three new proteins that are also implicated in DSB formation were reported in Arabidopsis, i.e., PRD1, PRD2, and PRD3 [39], [40]. Among these, PRD3 is thought to be the homolog of rice PAIR1. Furthermore, the phenotype of the pair1 mutant (asynaptic, with no bivalent formation) is reminiscent of the phenotype observed in a mutant lacking DSBs [41]. We isolated an asynaptic mutant (Figure 6A–D), and it was proven to be a new allele of pair1. Then, pair1 Oshus1-1 double mutants were generated using this new pair1 allele. The double mutants showed a typical pair1 phenotype, i.e., an absence of bivalents and lack of chromosome fragments at anaphase I (Figure 6E–H). Therefore, ectopic interactions, as well as chromosome fragmentations in Oshus1-1, require the formation of DSBs.
Figure 6. DAPI staining of Oshus1-1 with other meiotic mutants.
(A–D) pair1. (E–H) The pair1 Oshus1-1 double mutant showing similar chromosome behaviors to those of the pair1 single mutant. (I–L) Osrad51c. (M–P) The Osrad51c Oshus1-1 double mutant displaying a cumulative effect of the two mutations. (Q–T) Oscom1. (U–X) The Oscom1 Oshus1-1 double mutant showing similar chromosome behaviors to those of the Oscom1 single mutant. Bars, 5 µm.
To learn whether OsHUS1 is involved in DSB repair pathway in rice meiosis, we generated Osrad51c Oshus1-1 double mutants. OsRAD51C, like its functional homolog AtRAD51C, is essential for meiotic DSB repair [42]–[44]. In the Osrad51c mutant, homologous pairing and synapsis were defective at zygotene and pachytene, and univalents were observed at diakinesis and metaphase I (Figure 6I–K). In anaphase I, all of the univalents broke into fragments without any chromosome associations and scattered randomly in the nucleus (Figure 6L). These defects are consistent with the role of Osrad51c in meiotic DSB repair. In the Osrad51c Oshus1-1 double mutant, a cumulative effect of the two single mutations was detected; homologous pairing was disrupted, and ectopic chromosome associations were detected in all meiocytes observed. (Figure 6M–O; n = 322). At anaphase I, extensive chromosome fragments were also produced (Figure 6P). Therefore, the occurrence of ectopic interactions in Osrad51c Oshus1-1 suggests that ectopic interactions between nonhomologous chromosomes do not require OsRad51C.
OsCOM1 functions both in promoting homologous recombination and in resolving chromosome entanglements [45]. In the Oscom1 mutant, both homologous pairing and synapsis were abolished at pachytene (Figure 6Q), and aberrant nonhomologous associations were detected. From diakinesis to metaphase I, the most obvious phenotype was an entangled chromosome mass (Figure 6R, S). At anaphase I, chromosome fragments were generated (Figure 6T). We also generated Oscom1 Oshus1-1 double mutants. The phenotype of the Oscom1 Oshus1-1 double mutant could not be distinguished from that of the Oscom1 single mutant (Figure 6U–X), suggesting that OsHUS1 might function after OsCOM1 during meiosis. Of course, we cannot exclude the possibility that the ectopic interaction phenotype of Oshus1 might be hidden by the severe chromosome entanglement of Oscom1.
Ectopic associations in Oshus1-1 are independent of OsMER3 and ZEP1
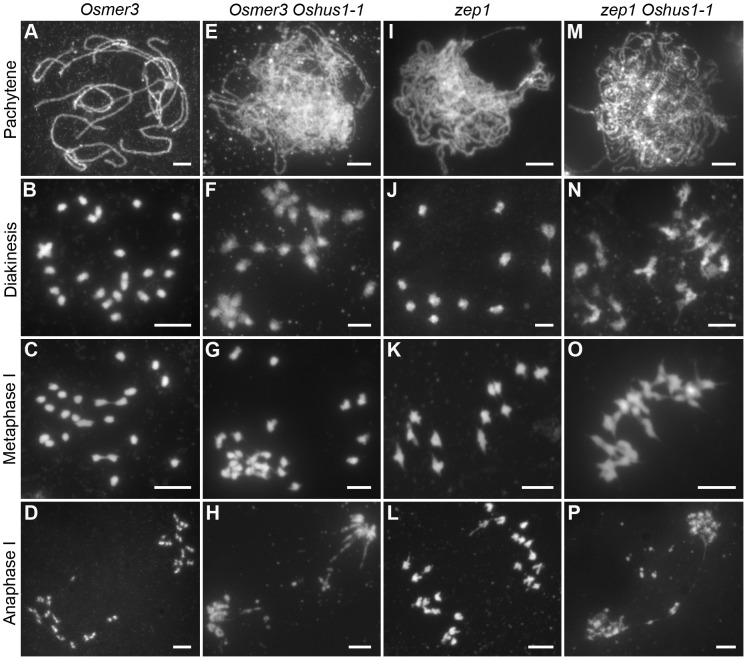
Since most COs in rice are derived from the interference-sensitive pathway, we set out to study the relationship between ectopic interactions and interference-sensitive COs. To this aim, the Osmer3 Oshus1-1 double mutant was generated, and its chromosome behaviors were investigated. In Osmer3, fully aligned chromosomes were detected during pachytene (Figure 7A), indicating the homologous pairing is not affected by the mutation of OsMER3. However, during diakinesis and metaphase I, the mutant cells showed a mixture of both univalent and bivalent chromosomes (Figure 7B, C). In anaphase I, the bivalents separated normally but the scattered univalents segregated randomly (Figure 7D). Intriguingly, in the Osmer3 Oshus1-1 mutant, homologous pairing was not observed at pachytene stage (Figure 7E). FISH experiments further confirmed that homologous pairing was disrupted in Osmer3 Oshus1-1 meiocytes (n = 101, Figure S7). In diakinesis and metaphase I, both multivalents with ectopic interactions and univalents were detected in all meiocytes (n = 122, Figure 7F, G). The multivalents contained an average of 7.0 associated chromosomes (ranging from 2 to 22); the average number of univalents per cell was 8.2 (ranging from 0 to 16). At anaphase I, both univalents and multivalents were pulled toward two poles of the nucleus. Additionally, chromosome bridges and fragments were also found at this stage (Figure 7H). These results suggest that ectopic interactions in Oshus1 arise independently from the OsMER3-mediated pathway.
Figure 7. DAPI staining of Osmer3, Osmer3 Oshus1-1, zep1 and zep1 Oshus1-1 mutant.
(A–D) Osmer3. (E–H) Homologous pairing is disrupted while ectopic associations are remained in Osmer3 Oshus1-1. (I–L) zep1. (M–P) The zep1 Oshus1-1 double mutant exhibiting occurrence of ectopic association. Bars, 5 µm.
To determine whether the defects in Oshus1 are affected by synapsis, we also generated the zep1 Oshus1-1 double mutant. In the zep1 mutant, synapsis was totally disrupted, but 12 bivalents were present at metaphase I and segregated normally at anaphase I (Figure 7I–L). In the zep1 Oshus1-1 double mutant, homologous chromosomes aligned along the entire length of the chromosome, but the SC was not assembled (Figure 7M). However, ectopic interactions were still clearly observed in all meiocytes (n = 298, Figure 7N–P). These results indicate that ectopic interactions are likely independent of synapsis in Oshus1-1.
OsHUS1 localizes to meiotic chromosomes during early prophase I
To further elucidate the role of OsHUS1 in meiosis, we prepared polyclonal antibodies in mice against the entire length of recombinant, His-tagged OsHUS1. Using antibodies against OsREC8 and OsHUS1, we performed dual immunofluorescence staining in rice PMCs. OsREC8, the cohesin protein in rice, was used to indicate the meiotic chromosome axes in this study [34], [46]. During leptotene, OsHUS1 proteins appeared as discrete foci in the nuclei and were loaded on the chromosome axes, as indicated by their full colocalization with OsREC8 (Figure 8A). The intensity of OsHUS1 then reached its peak at early zygotene, but this protein still appeared as foci rather than short lines (Figure 8B). At late zygotene, the number of OsHUS1 foci decreased, and many of them fell off the chromosomes (Figure 8C). At pachytene, the OsHUS1 immunostaining signal was completely absent in the nuclei (Figure 8D). No OsHUS1 signal was observed in male meiocytes of Oshus1-1, which confirmed the specificity of the OsHUS1 antibody (Figure 8E).
Figure 8. OsHUS1 localizes on meiotic chromosomes.
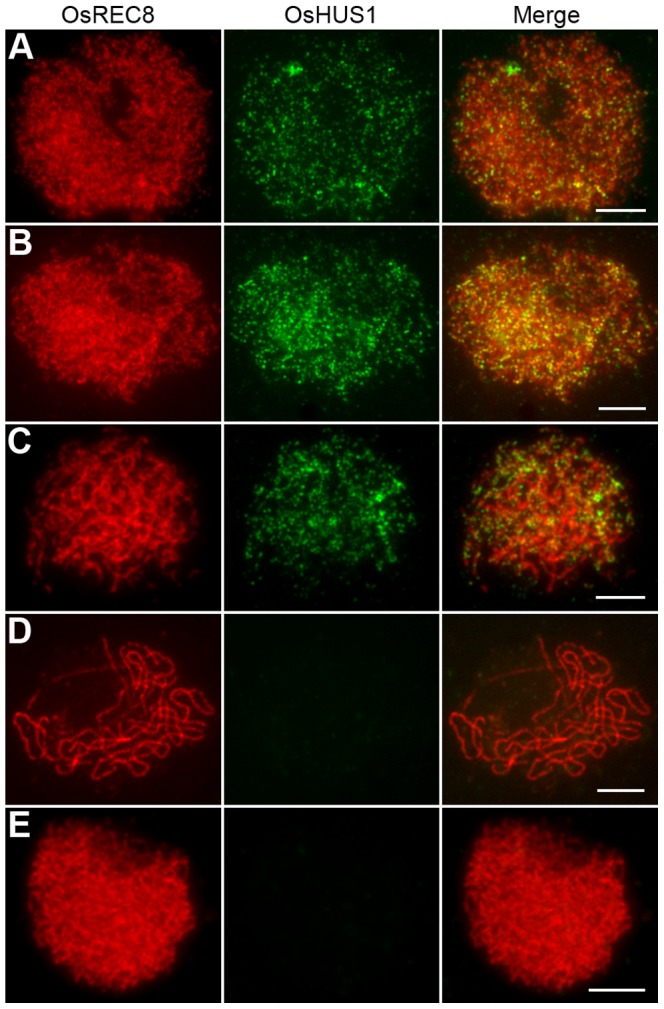
(A) Leptotene. (B) Zygotene. (C) Early pachytene. (D) Late pachytene. (E) No signal was detected in Oshus1-1 meiocytes at zygotene. Bars, 5 µm.
To further investigate the function of OsHUS1 protein, the immunolocalization pattern of OsHUS1 was investigated in Osmer3, zep1, and pair1 mutants. The localization pattern of OsHUS1 was not obviously affected in Osmer3 or zep1 (Figure S8A, B). This result is consistent with the observation that no ectopic interaction was found in either of the mutants. On the contrary, in the pair1 mutant, we failed to detect any OsHUS1 signals (Figure S8C), implying that the function of OsHUS1 depends on the formation of DSBs.
Oshus1-1 seedlings are hypersensitive to mitomycin C
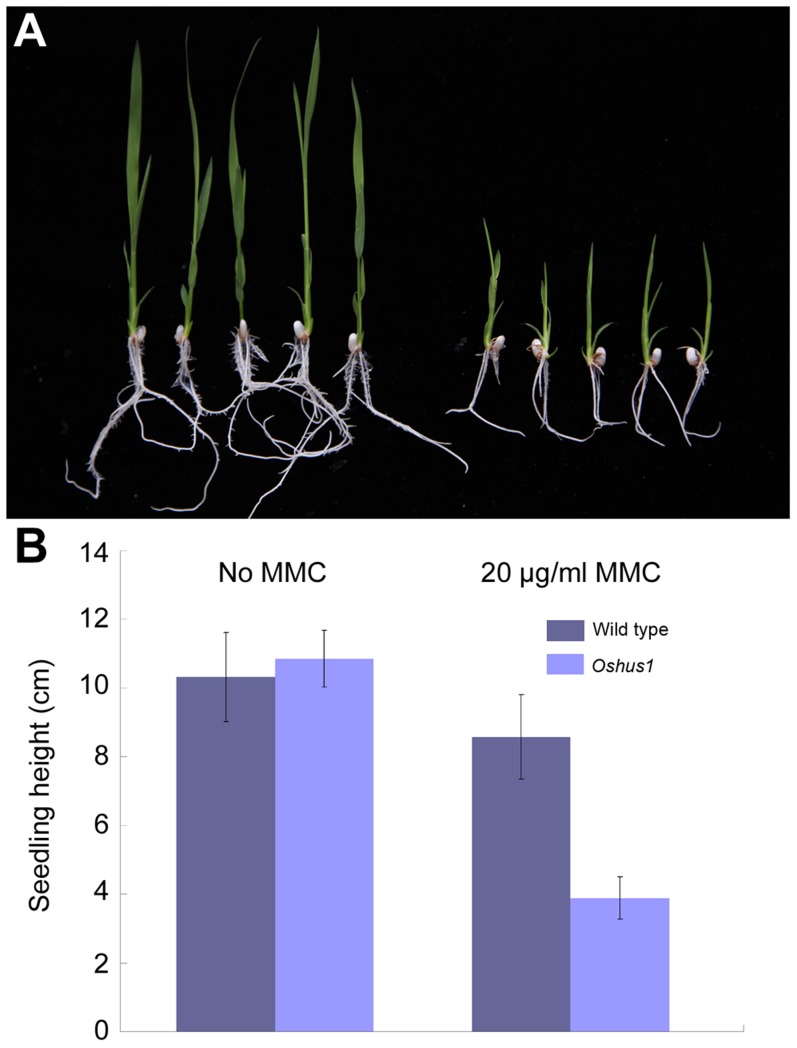
In yeast and mammals, HUS1 protein is implicated in various DNA damage response pathways [47]–[51]. In rice, OsHUS1 has the highest expression abundance in leaves, suggesting that this protein, like its counterparts in yeast and mammals, is potentially involved in the mitotic DNA damage response. To address this possibility, we tested whether Oshus1 plants showed higher sensitivity to mitomycin C (MMC), a DNA cross-link agent, than wild-type plants. Surface-sterilized seeds from wild-type and Oshus1-1 +/− plants were sown on solid 1/2 MS medium containing 0 or 20 µg/ml MMC. When planted on medium lacking MMC, the development of wild-type seedlings was identical to that of progeny derived from an Oshus1-1 +/− plant. However, when treated with MMC, the development of wild-type seedlings was only slightly suppressed, while approximately one-quarter of the progeny derived from the Oshus1-1 +/− plants showed severe growth retardation (Figure 9). Using a PCR genotyping assay, we determined that all of the severely growth-retarded seedlings were Oshus1-1 −/− (n = 20). These data demonstrate that Oshus1-1 rice is hypersensitive to MMC, indicating that OsHUS1 plays an important role in somatic DNA damage repair.
Figure 9. Oshus1-1 is hypersensitive to MMC.
(A) Oshus1-1 seedlings (right) exhibited more growth retardation than that of wild-type seedlings (left) on 1/2 MS medium with MMC. (B) Statistical analysis of WT and Oshus1-1 plant height.
Discussion
OsHUS1 is involved in somatic DNA damage responses
HUS1 is thought to form a PCNA-like complex with its two partners, RAD9 and RAD1 [23]. HUS1 has been intensively investigated in yeast and mammals, with studies primarily focusing on the mitotic DNA damage response. A mutation in MEC3 (the HUS1 counterpart in budding yeast) results in delayed entry into the S phase and slow DNA replication in response to DNA damage-inducing agents [52]. Fission yeast lacking HUS1 also fails to arrest the cell cycle after DNA damage or the blocking of DNA synthesis [53]. Targeted disruption of mouse HUS1 causes embryonic lethality due to the accumulation of chromosome breaks [49].
In this study, we found that rice hus1 seedlings were hypersensitive to the genotoxin MMC, suggesting that OsHUS1 has a conserved function in somatic DNA repair. Expression data for OsHUS1 show high accumulation of its transcript in somatic tissues, which further supports the somatic role of this protein. These findings are also in agreement with the hypothesis that OsHUS1 in rice is the functional homolog of fungal and animal HUS1. By performing a BLASTp search, we found that the homologs of S. pombe RAD9 and RAD1 also exist in rice. In addition, RAD9 is also involved in the regulation of DNA damage repair in the model plant Arabidopsis [54]. Therefore, it is highly possible that OsHUS1 in rice, like its yeast and animal counterparts, also participates in somatic DNA repair responses by forming the 9-1-1 complex.
OsHUS1 may be required for the suppression of ectopic interactions during meiosis
Studies in yeast and humans have revealed parallels between meiotic ER and allelic recombination, such as the observation that both processes occur during prophase I and are initiated by programmed DSBs. ER also results in crossover formation, which can affect genome stability during gametogenesis [12], [13], [55]. Therefore, ER should be inhibited, and/or its intermediates must be quickly eliminated, to ensure accurate homolog segregation during meiosis.
The function of the 9-1-1 complex in suppressing ER was first suggested in yeast [26]. However, to our knowledge, this function has not been reported in higher organisms, likely due to the lack of cytological evidence. Here, in Oshus1 meiocytes, we noticed that at late pachytene, one homolog pair frequently adhered or fused to another homolog pair at several sites, forming cross-like shapes. At the pachytene to diplotene transition (in which homologous pairs began to separate partially due to SC disassembly), the associations became more pronounced. The most remarkable defects observed in Oshus1 meiosis were multivalents at metaphase I and subsequent chromosome fragmentation.
The chromosome behaviors observed in the pair1 Oshus1-1 double mutant indicate that ectopic interactions rely on meiotic DSBs in Oshus1-1, which supports the notion that ectopic and allelic interactions share a common mechanism [56]. DSB formation is essential for homologous chromosome pairing in meiosis [3]. Here, although strong ectopic interactions occurred in Oshus1, homologous pairing took place normally. The nearly perfect ZEP1 signals along the entire lengths of chromosomes at pachytene indicated that synapsis was not severely disturbed in Oshus1. In addition, OsZIP4 and OsMER3 localized normally in Oshus1. It is likely that the early ectopic intermediate-preventing system may function well, and excessive ectopic interaction initiations are prevented in a timely manner in Oshus1. Intriguingly, unlike the Oshus1 and Osmer3 single mutants, the Oshus1 Osmer3 double mutant exhibited disrupted homologous pairing. In light of the competition between allelic and ectopic recombination [57] and the important roles they play during homologous pairing [3], it is attractive to consider that the increase in ectopic interactions and the decrease in allelic associations reduce the chance of homolog recognition and subsequent homolog alignment. Since homolog alignment mainly occurs at zygotene stage, it is reasonable to postulate that ectopic interactions initiate during or prior to zygotene. This hypothesis is consistent with the view that in yeast, ectopic recombination occurs concurrently with allelic recombination during meiosis [13].
Studies in budding yeast have revealed that ER occurs frequently during meiosis [11], [12]. However, it remains unknown whether ER also occurs frequently during plant meiosis. Here, we observed that all meiocytes showed the presence of multivalents in Oshus1. We therefore propose that in wild-type meiocytes, early ectopic interactions, accompanied by allelic interactions, may inevitably occur during homolog searching and homolog recognition. Once homolog recognition is accomplished, those ectopic interaction intermediates might be quickly detected and resolved by the surveillance mechanism. OsHUS1 is likely to be an important component of the surveillance mechanism that specifically eliminates ectopic interaction intermediates during meiosis.
Ectopic associations are independent of the interference-sensitive CO pathway
In budding yeast, a physical assay revealed that levels of ER increase from 1% in wild-type to 3–5% in rad17, rad24, and mec1-1 single mutants. HR is also reduced approximately two-fold in these mutants, from 25–30% in wild-type to 15% in rad17, rad24, and mec1-1. These data indicate that the increase in ER does not quantitatively account for the decrease in HR. Therefore, ER and HR likely occur via different pathways [26]. Here, we demonstrated that the loss of OsMER3 function did not affect ectopic interactions (through characterization of Osmer3 Oshus1-1), implying that these ectopic interactions do not arise from the interference-sensitive crossover formation pathway. In this study, we also observed that the average number of bright HEI10 foci was reduced in the Oshus1-1 mutant, showing that the number of interference-sensitive COs was reduced in the absence of OsHUS1. Thus, the similar alterations in ectopic and allelic interactions between yeast and rice imply that the function of HUS1 may be conserved among different organisms.
Interference plays a role in both controlling and constraining the final distribution of COs. Although the mechanism underlying these processes remains unclear, it has been postulated that spreading interference signals are transmitted along the length of the chromosome axes [58]. Therefore, one possible explanation for the decrease in interference-sensitive CO number is that the spreading interference signals may also be transmitted through associated nonhomologous chromosome axes in Oshus1. Alternatively, it is possible that partial allelic interactions are redirected into ectopic interactions or resolved toward sister chromatids in the absence of OsHUS1.
Possible functions of OsHUS1 during meiosis
Studies in yeast and mammals have shown that the 9-1-1 complex is involved in multiple DNA repair courses by binding to numerous partners, including base excision repair proteins and mismatch repair proteins [23]. Among these, the mismatch repair protein MSH2 is postulated to be involved in the intermediate elimination of ER [13]. In yeast and humans, MSH2-MSH6 heterodimer (MutSα) and MSH2-MSH3 heterodimer (MutSβ) are mismatch recognition factors that function in the mismatch repair pathway. Recent studies have revealed that each subunit of the 9-1-1 complex can interact with both the MSH2/MSH3 and MSH2/MSH6 complexes. In addition, the 9-1-1 complex can also stimulate the DNA binding activity of MutSα [59]. The biochemical properties of the 9-1-1 complex are likely similar during mitosis and meiosis. We therefore postulate that OsHUS1 may also function as a component of the 9-1-1 complex to sense ectopic interaction and further recruit MutS to eliminate ectopic interaction intermediates. The characterization of RAD9, RAD1, and MSH2 homologs in rice will deepen our understanding of the ER-eliminating mechanism.
Materials and Methods
Plant materials
Oshus1-1 was derived from Nipponbare (a japonica cultivar) induced by tissue culture. Oshus1-2 was derived from Huanghuazhan (an indica cultivar) induced by 6°Co∼γ ray radiation. The new pair1 mutant allele was obtained from Nipponbare through tissue culture. In this allele, a Tos17 retrotransposon was inserted in the 7th exon of PAIR1. The new Osrad51c allele was derived from an indica rice variety Zhongxian 3037, induced by 6°Co∼γ ray radiation and found to have a premature stop codon in the 9th exon of OsRAD51C. The Oscom1 and zep1 alleles employed in this study is Oscom1-3 and zep1-1, respectively [30], [45]. Nipponbare was used as the wild type in the related experiments.
Molecular cloning of OsHUS1
STS markers were developed based on sequence differences between japonica variety Nipponbare and indica variety 9311, which were used for map-based cloning of OsHUS1. Primers sequences were listed in Supporting information, Table S1. The cDNA sequence for OsHUS1 was verified by 3′RACE. Total RNA was extracted from rice young panicles (6–8 cm) using TRIZOL reagent (Invitrogen). A measure of 3 µg RNA was reverse-transcribed with Oligo-Adaptor primer (CTGATCTAGAGGTACCGGATCC-d(T)16) using the superscript III RNaseH reverse transcriptase (Invitrogen). Two rounds of PCRs were carried out using Adaptor primer (CTGATCTAGAGGTACCGGATCC), gene specific primers RACE1F (TGTACCTTCTATGGTATTTC) and RACE2F (CTAGACTGACGGACAAGTCC). The product was cloned into pMD19-T vector (TaKaRa) and sequenced.
Generating OsHUS1- RNAi transgenic plants
A 261bp fragment from the exons of OsHUS1 was amplified by PCR with the primer pair OsHUS1RNAiF (AAGGATCCCTGACAGTAGCTGTTACTC) and OsHUS1RNAiR (AGGTCGACACCATAGAAGGTACAGTCGG). The product was introduced into the BamHI-SalI and Bg II-XhoI sites of the pUCCRNAi vector in an inverted repeat orientation. The stem-loop fragment was finally cloned into the pCAMBIA 1300 vector. The OsHUS1-RNAi construct was introduced into Agrobacterium tumefaciens strain EHA105 and transformed the japonica cultivar Yandao 8.
Quantitative RT-PCR analysis
Total RNA was extracted from the internode, leaf, root, panicle and seedling of Nipponbare, and was reverse-transcribed into cDNA. Quantitative RT-PCR analysis was performed using the CFX96 Real Time system (Bio-Rad) and Eva Green (Biotium). The primer pair OsHUS1RTF (CTTGGTGTTCGTGCAACC) and OsHUS1RTR (ACCACCAGGAGAAATACC) was used. The standard control UBIQUITIN gene was examined with the primers UBI-RTF (CAAGATGATCTGCCGCAAATGC) and UBI-RTR (TTTAACCAGTCCATGAACCCG).
Sensitivity test
Husked seeds from the wild-type plants and the heterozygous Oshus1 +/− plants were surface sterilized. Then they were sown on solid 1/2 MS medium containing 20 µg/ml MMC (Solarbio) in a light incubator. Genotype and phenotype assays of the seedlings were assayed 14 days later.
Antibodies
To generate the antibody against OsHUS1, the coding region of it was amplified from Nipponbare leaf cDNA with primer pair OsHUS1PETF (ATGGATCCATGAAGTTCAAGGCCTTC) and OsHUS1PETR (ATCTCGAGACTGCCAGGGTCAAGGAC), and then ligated to the BamHI-XhoI site of the expression vector pET-30a (Novagen). The expression vector was transformed into Escherichia coli strain BL21 (DE3) and was induced for 3 h at 37°C by addition of 0.3 mM IPTG. His-tagged OsHUS1 were accumulated in the inclusion bodies and they were washed and subjected to SDS-PAGE. The main band of His-tagged OsHUS1 on the gel was cut off and powdered and used as an antigen against mice. The OsREC8, PAIR2, PAIR3, OsMER3, OsZIP4, HEI10, and ZEP1 polyclonal antibodies were used as described before [30], [32], [34], [35].
Cytology
Young panicles of at meiosis stage were harvested and fixed in Carnoy's solution (ethanol:glacial acetic acid = 3∶1) for chromosome spreading. Meiotic chromosome preparation and immunofluorescence were performed as previously described [34]. The FISH procedure was performed as described [60]. Microscopy was conducted using a ZEISS A2 fluorescence microscope with a microCCD camera. Image capture and analysis was carried out using IPLab software (BD Biosciences).
Supporting Information
Phenotype of the Oshus1-1 mutant. (A) A wild-type plant; (B) A Oshus1-1 plant; (C) Comparison of a wild-type (left) and a Oshus1-1 panicle (right); (D, E) I2-KI staining of pollen grains in the wild type (D) and Oshus1-1 mutant (E). Bars, 50 µm.
(TIF)
Meiotic chromosomes at Metaphase I in Oshus1-2 and OSHUS1 RNAi plant. (A) Oshus1-2. (B) An OSHUS1 RNAi line. Scale bars, 5 µm.
(TIF)
Structure of the OsHUS1 gene. Exons are represented by black boxes. Gray boxes show the untranslated regions. The position of the Oshus1 mutation is indicated by an arrow.
(TIF)
Alignment of HUS1 homologues. Identical amino acids are shaded in black whereas similar amino acids are shaded in gray.
(TIF)
Phylogenetic tree of the 20 homologs defined by OsHUS1. The tree is constructed using MEGA 4.0 based on the neighbor-joining method. Numbers next to branches are clade credibility values.
(TIF)
Relative expression level of OsHUS1 in different tissues analyzed by quantitative RT-PCR. Values are means ±SEM (standard error of mean) of three independent experiments and value of panicle is set as 1.
(TIF)
Detection of homologous chromosome pairing revealed by FISH in Oshus1-1, Osmer3 and Osmer3 Oshus1-1. (A–C) Pachytene; (D) Diakinesis; FISH signals of 5S rDNA are in green, signals of the BAC clone a0065A15 on the long arm of chromosome 9 are in red, and chromosomes are in blue stained with DAPI. Bars, 5 µm.
(TIF)
Dual immunolocalization of OsREC8 and OsHUS1 in Osmer3, zep1 and pair1 PMCs. (A) Osmer3 shows a normal localization of OsHUS1. (B) zep1 displays a normal localization of OsHUS1. (C) OsHUS1 is absent in pair1. Bars, 5 µm.
(TIF)
Primers used for OsHUS1 map-based cloning.
(DOCX)
Funding Statement
This work was supported by grants from the Ministry of Sciences and Technology of China (2011CB944602 and 2012AA10A301), and the National Natural Science Foundation of China (31230038 and 31170288). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zickler D, Kleckner N (1999) Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33: 603–754. [DOI] [PubMed] [Google Scholar]
- 2. Petronczki M, Siomos MF, Nasmyth K (2003) Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112: 423–440. [DOI] [PubMed] [Google Scholar]
- 3. Hamant O, Ma H, Cande W (2006) Genetics of meiotic prophase I in plants. Annu Rev Plant Biol 57: 267–302. [DOI] [PubMed] [Google Scholar]
- 4. Pawlowski WP, Cande WZ (2005) Coordinating the events of the meiotic prophase. Trends Cell Biol 15: 674–681. [DOI] [PubMed] [Google Scholar]
- 5. Niu H, Wan L, Busygina V, Kwon YH, Allen JA, et al. (2009) Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol Cell 36: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pradillo M, Santos JL (2011) The template choice decision in meiosis: is the sister important? Chromosoma 120: 447–454. [DOI] [PubMed] [Google Scholar]
- 7. Zickler D (2006) From early homologue recognition to synaptonemal complex formation. Chromosoma 115: 158–174. [DOI] [PubMed] [Google Scholar]
- 8. Barzel A, Kupiec M (2008) Finding a match: how do homologous sequences get together for recombination? Nat Rev Genet 9: 27–37. [DOI] [PubMed] [Google Scholar]
- 9. Stankiewicz P, Lupski J (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18: 74–82. [DOI] [PubMed] [Google Scholar]
- 10. Chen J-M, Cooper DN, Férec C, Kehrer-Sawatzki H, Patrinos GP (2010) Genomic rearrangements in inherited disease and cancer. Semin Cancer Biol 20: 222–233. [DOI] [PubMed] [Google Scholar]
- 11. Jinks-Robertson S, Petes TD (1985) High-frequency meiotic gene conversion between repeated genes on nonhomologous chromosomes in yeast. Proc Natl Acad Sci U S A 82: 3350–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goldman ASH, Lichten M (1996) The efficiency of meiotic recombination between dispersed sequences in Saccharomyces cerevisiae depends upon their chromosomal location. Genetics 144: 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sasaki M, Lange J, Keeney S (2010) Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Bio 11: 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mieczkowski PA, Dominska M, Buck MJ, Lieb JD, Petes TD (2007) Loss of a histone deacetylase dramatically alters the genomic distribution of Spo11p-catalyzed DNA breaks in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 104: 3955–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goldman ASH, Lichten M (2000) Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proc Natl Acad Sci U S A 97: 9537–9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoang ML, Tan FJ, Lai DC, Celniker SE, Hoskins RA, et al. (2010) Competitive repair by naturally dispersed repetitive DNA during non-allelic homologous recombination. PLoS Genet 6: e1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lichten M, Haber JE (1989) Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae . Genetics 123: 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kupiec M, Petes TD (1988) Allelic and ectopic recombination between Ty elements in yeast. Genetics 119: 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoang ML, Tan FJ, Lai DC, Celniker SE, Hoskins RA, et al. (2010) Competitive repair by naturally dispersed repetitive DNA during non-allelic homologous recombination. PLoS Genet 6: e1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Venclovas C, Thelen MP (2000) Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res 28: 2481–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu M, Bai L, Gong Y, Xie W, Hang H, et al. (2009) Structure and functional implications of the human Rad9-Hus1-Rad1 cell cycle checkpoint complex. J Biol Chem 284: 20457–20461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harrison JC, Haber JE (2006) Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 40: 209–235. [DOI] [PubMed] [Google Scholar]
- 23. Eichinger CS, Jentsch S (2011) 9-1-1: PCNA's specialized cousin. Trends Biochem Sci 36: 563–568. [DOI] [PubMed] [Google Scholar]
- 24. Helt C, Wang W, Keng P, Bambara R (2005) Evidence that DNA damage detection machinery participates in DNA repair. Cell cycle 4: 529–532. [DOI] [PubMed] [Google Scholar]
- 25. Hong E-JE, Roeder GS (2002) A role for Ddc1 in signaling meiotic double-strand breaks at the pachytene checkpoint. Genes Dev 16: 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grushcow JM, Holzen TM, Park KJ, Weinert T, Lichten M, et al. (1999) Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics 153: 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Freire R, Murguía JR, Tarsounas M, Lowndes NF, Moens PB, et al. (1998) Human and mouse homologs of Schizosaccharomyces pombe rad1 + and Saccharomyces cerevisiae RAD17: linkage to checkpoint control and mammalian meiosis. Genes Dev 12: 2560–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peretz G, Arie LG, Bakhrat A, Abdu U (2009) The Drosophila hus1 gene is required for homologous recombination repair during meiosis. Mech Develop 126: 677–686. [DOI] [PubMed] [Google Scholar]
- 29. Lyndaker A, Lim P, Mleczko J, Diggins C, Holloway J, et al. (2013) Conditional Inactivation of the DNA Damage Response Gene Hus1 in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance. PLoS Genet 9: e1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang M, Wang K, Tang D, Wei C, Li M, et al. (2010) The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22: 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nonomura K, Nakano M, Eiguchi M, Suzuki T, Kurata N (2006) PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J Cell Sci 119: 217–225. [DOI] [PubMed] [Google Scholar]
- 32. Wang K, Wang M, Tang D, Shen Y, Qin B, et al. (2011) PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Mol Biol Cell 22: 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sanchez-Moran E, Santos J-L, Jones GH, Franklin FCH (2007) ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis . Genes Dev 21: 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang K, Tang D, Wang M, Lu J, Yu H, et al. (2009) MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J Cell Sci 122: 2055–2063. [DOI] [PubMed] [Google Scholar]
- 35. Shen Y, Tang D, Wang K, Wang M, Huang J, et al. (2012) ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J Cell Sci 125: 2581–2591. [DOI] [PubMed] [Google Scholar]
- 36. Wang K, Wang M, Tang D, Shen Y, Miao C, et al. (2012) The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet 8: e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murakami H, Keeney S (2008) Regulating the formation of DNA double-strand breaks in meiosis. Genes Dev 22: 286–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luo Q, Li Y, Shen Y, Cheng Z (2014) Ten years of gene discovery for meiotic event control in rice. J Genet Genomics 41: 125–137. [DOI] [PubMed] [Google Scholar]
- 39. De Muyt A, Pereira L, Vezon D, Chelysheva L, Gendrot G, et al. (2009) A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet 5: e1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De Muyt A, Vezon D, Gendrot G, Gallois JL, Stevens R, et al. (2007) AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana . EMBO J 26: 4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nonomura K, Nakano M, Fukuda T, Eiguchi M, Miyao A, et al. (2004) The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16: 1008–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kou Y, Chang Y, Li X, Xiao J, Wang S (2012) The rice RAD51C gene is required for the meiosis of both female and male gametocytes and the DNA repair of somatic cells. J Exp Bot 63: 5323–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li W, Yang X, Lin Z, Timofejeva L, Xiao R, et al. (2005) The AtRAD51C gene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis . Plant Physiol 138: 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Da Ines O, Abe K, Goubely C, Gallego ME, White CI (2012) Differing requirements for RAD51 and DMC1 in meiotic pairing of centromeres and chromosome arms in Arabidopsis thaliana . PLoS Genet 8: e1002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ji J, Tang D, Wang K, Wang M, Che L, et al. (2012) The role of OsCOM1 in homologous chromosome synapsis and recombination in rice meiosis. Plant J 72: 18–30. [DOI] [PubMed] [Google Scholar]
- 46. Shao T, Tang D, Wang K, Wang M, Che L, et al. (2011) OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol 156: 1386–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Enoch T, Carr A, Nurse P (1992) Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev 6: 2035–2046. [DOI] [PubMed] [Google Scholar]
- 48. Longhese MP, Fraschini R, Plevani P, Lucchini G (1996) Yeast pip3/mec3 mutants fail to delay entry into S phase and to slow DNA replication in response to DNA damage, and they define a functional link between Mec3 and DNA primase. Mol Cell Biol 16: 3235–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weiss RS, Enoch T, Leder P (2000) Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev 14: 1886–1898. [PMC free article] [PubMed] [Google Scholar]
- 50. Abdu U, Klovstad M, Butin-Israeli V, Bakhrat A, Schüpbach T (2007) An essential role for Drosophila hus1 in somatic and meiotic DNA damage responses. J Cell Sci 120: 1042–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boulton SJ, Ye M, Hofmann JJ, Stergiou L, Gartner A, et al. (2002) Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol 12: 1908–1918. [DOI] [PubMed] [Google Scholar]
- 52. Weinert TA, Kiser GL, Hartwell LH (1994) Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev 8: 652–665. [DOI] [PubMed] [Google Scholar]
- 53. Kostrub CF, al-Khodairy F, Ghazizadeh H, Carr AM, Enoch T (1997) Molecular analysis of hus1+, a fission yeast gene required for S-M and DNA damage checkpoints. Mol Gen Genet 254: 389–399. [DOI] [PubMed] [Google Scholar]
- 54. Heitzeberg F, Chen IP, Hartung F, Orel N, Angelis KJ, et al. (2004) The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J 38: 954–968. [DOI] [PubMed] [Google Scholar]
- 55. Nag DK, Petes TD (1990) Meiotic recombination between dispersed repeated genes is associated with heteroduplex formation. Mol Cell Biol 10: 4420–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hastings P (2010) Mechanisms of ectopic gene conversion. Genes 1: 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoang ML, Tan FJ, Lai DC, Celniker SE, Hoskins RA, et al. (2010) Competitive repair by naturally dispersed repetitive DNA during non-allelic homologous recombination. PLoS genetics 6: e1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Borner GV, Kleckner N, Hunter N (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- 59. Bai H, Madabushi A, Guan X, Lu A (2010) Interaction between human mismatch repair recognition proteins and checkpoint sensor Rad9-Rad1-Hus1. DNA repair 9: 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang W, Yi C, Bao W, Liu B, Cui J, et al. (2005) The transcribed 165-bp CentO satellite is the major functional centromeric element in the wild rice species Oryza punctata . Plant Physiol 139: 306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phenotype of the Oshus1-1 mutant. (A) A wild-type plant; (B) A Oshus1-1 plant; (C) Comparison of a wild-type (left) and a Oshus1-1 panicle (right); (D, E) I2-KI staining of pollen grains in the wild type (D) and Oshus1-1 mutant (E). Bars, 50 µm.
(TIF)
Meiotic chromosomes at Metaphase I in Oshus1-2 and OSHUS1 RNAi plant. (A) Oshus1-2. (B) An OSHUS1 RNAi line. Scale bars, 5 µm.
(TIF)
Structure of the OsHUS1 gene. Exons are represented by black boxes. Gray boxes show the untranslated regions. The position of the Oshus1 mutation is indicated by an arrow.
(TIF)
Alignment of HUS1 homologues. Identical amino acids are shaded in black whereas similar amino acids are shaded in gray.
(TIF)
Phylogenetic tree of the 20 homologs defined by OsHUS1. The tree is constructed using MEGA 4.0 based on the neighbor-joining method. Numbers next to branches are clade credibility values.
(TIF)
Relative expression level of OsHUS1 in different tissues analyzed by quantitative RT-PCR. Values are means ±SEM (standard error of mean) of three independent experiments and value of panicle is set as 1.
(TIF)
Detection of homologous chromosome pairing revealed by FISH in Oshus1-1, Osmer3 and Osmer3 Oshus1-1. (A–C) Pachytene; (D) Diakinesis; FISH signals of 5S rDNA are in green, signals of the BAC clone a0065A15 on the long arm of chromosome 9 are in red, and chromosomes are in blue stained with DAPI. Bars, 5 µm.
(TIF)
Dual immunolocalization of OsREC8 and OsHUS1 in Osmer3, zep1 and pair1 PMCs. (A) Osmer3 shows a normal localization of OsHUS1. (B) zep1 displays a normal localization of OsHUS1. (C) OsHUS1 is absent in pair1. Bars, 5 µm.
(TIF)
Primers used for OsHUS1 map-based cloning.
(DOCX)