Abstract
The complex chromosomal aberrations found in therapy related acute myeloid leukemia (t-AML) suggest that the DNA double strand break (DSB) response may be altered. In this study we examined the DNA DSB response of primary bone marrow cells from t-AML patients and performed next-generation sequencing of 37 canonical homologous recombination (HR) and non-homologous end-joining (NHEJ) DNA repair genes, and a subset of DNA damage response genes using tumor and paired normal DNA obtained from t-AML patients. Our results suggest that the majority of t-AML patients (11 of 15) have tumor cell-intrinsic, functional dysregulation of their DSB response. Distinct patterns of abnormal DNA damage response in myeloblasts correlated with acquired genetic alterations in TP53 and the presence of inferred chromothripsis. Furthermore, the presence of trisomy 8 in tumor cells was associated with persistently elevated levels of DSBs. Although tumor-acquired point mutations or small indels in canonical HR and NHEJ genes do not appear to be a dominant means by which t-AML leukemogenesis occurs, our functional studies suggest that an abnormal response to DNA damage is a common finding in t-AML.
Keywords: therapy-related AML, DNA damage, DNA repair, Trisomy 8
INTRODUCTION
Therapy-related acute myeloid leukemia/myelodysplastic syndrome (t-AML/MDS) is a well-recognized complication of chemotherapeutic and/or radiation therapy for a primary malignancy.(1) Approximately 10–15 % of AML and MDS cases are therapy-related,(2, 3) although the incidence is expected to rise due to increasing cancer surviviorship. Abnormalities of chromosome 5 (5q−/−5) and 7(7q−/−7) are associated with alkylators and/or radiation, and rearrangements involving the MLL gene on chromosome 11q23 with topoisomerase II inhibitors.(2, 4, 5) The overall prognosis in t-AML is poor,(5, 6) in part due to the presence of adverse risk karyotypes such as abnormalities of chromosomes 5 and 7, MLL gene rearrangements(2, 4, 5) and complex and monosomal karyotypes commonly observed in t-AML/t-MDS. (2, 4, 6)
The chromosomal aberrations frequently found in t-AML cells suggest that the DNA double-strand break (DSB) response may be altered. There is evidence from patients with inherited mutations in key genes of the DSB DNA damage response and DNA DSB repair pathways that dysregulation of these pathways can predispose patients to myeloid malignancies. For example, mutated Fanconi pathway genes result in impaired DNA repair following exposure to DNA damaging agents such as alkylators and are associated with development of AML. Furthermore, patients with germline mutations in TP53 (Li-Fraumeni syndrome) a key regulator of the DNA damage response pathway, can develop leukemias.(7–9) Several groups have reported that polymorphisms in DNA repair genes are associated with t-AML development, most notably in the RAD51 and XRCC3 genes, which are involved in homologous recombination (HR).(10, 11)
Somatic alterations in the TP53 gene are the most commonly reported mutations associated with t-AML.(12, 13) Not surprisingly, acquired TP53 aberrations have been identified in up to 38% patients in t-AML(12–14) and are associated with −5/−5q,(12, 13)complex karyotypes(12, 15, 16) and highly derivative chromosomes.(15) However, the role of acquired alterations in other DSB response and repair genes in t-AML has not been systemically studied.
The major DSB repair pathways in mammalian cells are the homologous recombination (HR) and non-homologous end-joining (NHEJ) pathways (recently reviewed in(17)). We hypothesized that dysregulation of DSB repair by HR or NHEJ exists in t-AML and may result from acquired mutations in HR/NHEJ pathway genes. To test this possibility, we performed functional studies of DSB repair using primary bone marrow cells from t-AML patients. In parallel, we used next-generation sequencing to identify somatic genetic variants in 37 genes, including canonical HR and NHEJ DNA repair genes, and DNA damage response genes using tumor and paired normal DNA obtained from the bone marrow of t-AML patients. In addition, a custom high-resolution array comparative genomic hybridization platform to interrogate copy number alterations of 170 DNA repair genes was performed. We show that a dysfunctional DSB response is present in the majority of t-AML patients tested and that somatic mutations in TP53 and gains on chromosome 8 are associated with distinct and abnormal patterns of DNA DSBs, indicative of abnormal response and/or repair.
METHODS
Patient samples
Samples were obtained from t-AML patients seen at Washington University. All patients provided written informed consent under a protocol approved by the Institutional Review Board of Washington University School of Medicine. A bone marrow sample and a 6-mm punch biopsy of skin (for analysis of unaffected somatic cells) were obtained. Bone marrow samples were frozen as viable cells in fetal calf serum and 10% DMSO in liquid nitrogen. Bone marrow CD34+ cells were obtained from normal healthy volunteers after informed written consent according to institutional guidelines or purchased from Lonza (Walkersville, MD).
Tissue Culture
Cryopreserved samples were cultured in in EGM2 media (Lonza) and 10 ng/mL of the following human cytokines: SCF, TPO, IL-3, IL-6, and FLT-3 ligand in 5% CO2. All cells were “rested” for 24 hours post-thaw prior to experiments, i.e. baseline measurements for all experiments were made 24 hours post-thaw.
γH2AX Assay
Bone marrow cells were mock irradiated or subjected to 2 Gray of irradiation from a Cesium source and cultured for 0.5, 4, 6, 8, and 24 hours, and stained with an antibody cocktail containing CD45, CD3, and CD19 (except for UPN 377512, which had 31% myeloblasts and was stained with CD33). Cells were fixed in BD Cytofix/Cytoperm (BD Biosciences, San Diego, CA), permeabilized in BD Perm/Wash (BD Biosciences) and stained for γH2AX (H2A.X Phosphorylation Assay Kit, Millipore, Temecula, CA) per the manufacturer’s instructions. γH2AX levels were quantified by flow cytometry in myeloblasts (CD45 dim, low side scatter population) and lymphocytes (CD45 bright, low side scatter or CD3/CD19+, low side scatter). Fluorescently labeled beads with known levels of fluorophore (Quantum MESF Microsphere Kit, Bangs Laboratories, Fishers, IN) were included with each experiment to generate a standard curve, and the geometric mean of the fluorescence intensity of the γH2AX signal was converted to mean equivalent soluble fluorophore (MESF) per the manufacturer’s instructions.
Array Comparative Genomic Hybridization Analysis and Validation
A custom high-resolution array comparative genomic hybridization platform (3X 720 K array, NimbleGen, Madison, WI) to interrogate copy number alterations of 170 DNA repair genes (Supplementary Table 2) was generated by dense tiling of probes (80 base pair intervals) spanning each gene and 5 kilobases of flanking genome. This array also contained dense tiling of probes designed to interrogate 835 miRNA genes and 44 miRNA processing genes as previously reported.(18) In addition, probes uniformly spaced throughout the genome at approximately 8600 base pair intervals were included. Two micrograms of genomic DNA from unfractionated bone marrow (tumor) and paired normal tissue (skin) was fragmented, labeled, and hybridized to the array as previously described.(19) (see Supplementary Methods). Full access to the array comparative genomic hybridization data will be made publically available upon publication.
Sequencing
37 candidate genes were selected for mutation screening based on annotation from the Human DNA Repair Genes database (http://sciencepark.mdanderson.org/labs/wood/dna_repair_genes.html, 2010) (21 homologous recombination, 7 non-homologous end-joining, 2 DNA polymerases, and 7 additional regulators of DNA repair).(see Supplementary Methods) (Supplementary Table 3).
Neutral Comet Assay
Bone marrow samples were thawed as above, rested for 24 hours, and subjected to the single cell gel electrophoresis assay using neutral electrophoresis buffer with the Comet Assay Kit (Trevingen, Gaithersburg, MD), per the manufacturer’s instructions. Samples were stained with SYBR green, viewed at 10X on the Nikon Microphot SA microscope, and Comets were scored with CometScore (TriTek, Sumerduck, VA) per the manufacturer’s instructions.
S-Phase and Apoptosis Measurement
Cells were labeled for 16 hours with 10 µM EdU prior to the time point of interest, harvested, and subsequently prepared for flow cytometry analysis of EdU incorporation as a measure of S-phase using the Click-iT EdU Flow Cytometry Assay Kit (Invitrogen). Apoptosis was quantified by flow cytometry by measuring levels of bound, fluorescently labeled fluoromethyl ketone peptide inhibitor of caspase-3 (and 7) using the CaspaTag Caspase-3/7 In Situ Assay Kit (Millipore, Billerica, MA).
Gene Expression Analysis
Microarray analysis was performed using Affymetrix U133plus2 chips. RNA was extracted from bone marrow samples and array data was generated from the same batch analysis for all 14 samples. Affymetrix U133plus2 data is available in GEO (GSE52478). Pathway analysis using Gene Set Enrichment Analysis (GSEA) was used with 1000 phenotype permutations(20) of a set of canonical upregulated human MYC target genes derived from Dang and colleagues.(21)
Retroviral transduction of murine c-kit enriched bone marrow cells
Bone marrow mononuclear cells were harvested from 6–12 week old C57Bl/6 mice and mononuclear cells were stained with MACS CD117+ (c-kit) mouse microbeads (Miltenyi Biotech, Auburn, CA),and subjected to two rounds of positive selection on the autoMACS Separator. The Myc expressing MSCV-Myc-IRES-GFP and the control MSCV-IRES-GFP viruses were generated as previously described.(22) Viral transduction of c-kit enriched cells was performed as previously described.(22) Three or 6 days post-transduction, GFP+/propidium iodide negative cells were sorted (MoFlo) and harvested. DSB quantification was performed by the Neutral Comet assay as above.
Statistical Methods
The γH2AX response of myeloblasts post-IR of individual patients was compared to CD34+ controls and categorized phenotypically based on comparisons of mean γH2AX values at baseline, 0.5 and 4 hours with the Student’s t-test (GraphPad Prism, San Diego, CA) (see Supplementary Methods). The resulting phenotypic groups were further compared to CD34+ controls using a linear mixed model analysis, incorporating a random intercept to account for the correlation between different time points within a subject. Group effect and its interaction with time were included in the model. This model was also used to compare patient lymphocytes in each phenotypic group to normal donor bone marrow lymphocytes. The analysis was generated with SAS 9.2 (SAS Inc, Cary, NC). The p-values generated by the linear mixed model analysis are reported in the results. Differences in the mean percent DNA in tail and Olive moment of Comets were compared using the Mann-Whitney test (GraphPad Prism, San Diego, CA). Means between the groups for S-phase timepoints, apoptosis timepoints, and the γH2AX MESF levels in unirradiated myeloblasts at the 24 hour timepoint were compared using the student’s t-test (Graphpad Prism).
RESULTS
Patient samples
Thirty t-AML patients were chosen for study based on the availability of paired bone marrow (tumor) and skin (normal) DNA samples. Fifteen of these patients had cryopreserved bone marrow specimens available for functional studies. Patient characteristics and assays performed for each patient are shown (Supplementary Table 1). The median time to t-AML from first chemotherapy exposure was 3 years (range, 1.1–13.3 years). Cytogenetic analysis revealed −5/−5q and/or −7 in seven patients (23%), translocations involving chromosome 11q23 (MLL gene rearrangement) in 6 (20.0%), complex cytogenetics (3 or more changes) in eleven patients (36.7 %), and a normal karyotype in 6 (20%) patients. The median myeloblast percentage in the bone marrow was 76 (range, 31–95).
Detection of DNA double-strand breaks in primary t-AML samples
To test the hypothesis that myeloblasts from t-AML patients respond inappropriately to DSBs, we interrogated the kinetics of DSB repair in primary bone marrow cells from 15 t-AML patients and bone marrow obtained from five normal donors as controls. We measured phosphorylated histone H2AX (γH2AX), a well-established marker for DSBs(23, 24) in matched myeloblasts (CD45 dim, side scatter low) (tumor) and lymphocytes (CD3+ or CD19+, side scatter low) (a surrogate for normal cells) from the same patient. Baseline γH2AX measurements in primary cells, coupled with a time course to measure γH2AX induction and resolution after 2 Gray of irradiation (IR) were used to assess the basal DSB burden and the response to acute damage, respectively. The peak induction of γH2AX was evaluated at 30 minutes after IR based on prior reports.(23, 24) An IR dose of 2 Gray optimally induced DSBs in normal donor CD34+ hematopoietic control cells while minimizing changes in cell proliferation and apoptosis compared to higher IR doses (data not shown).
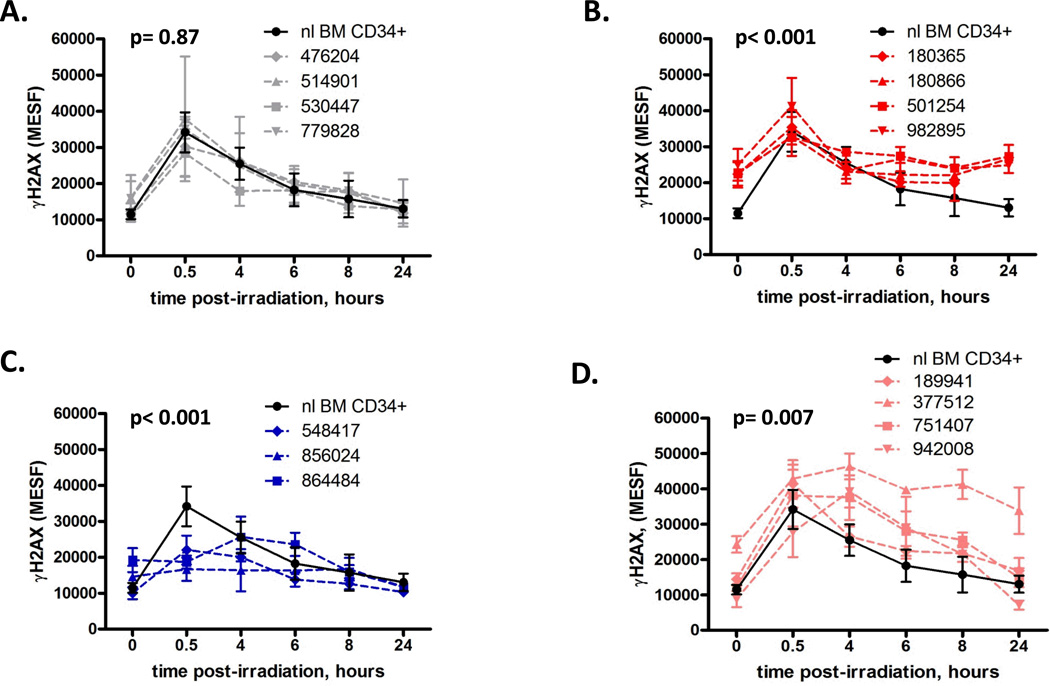
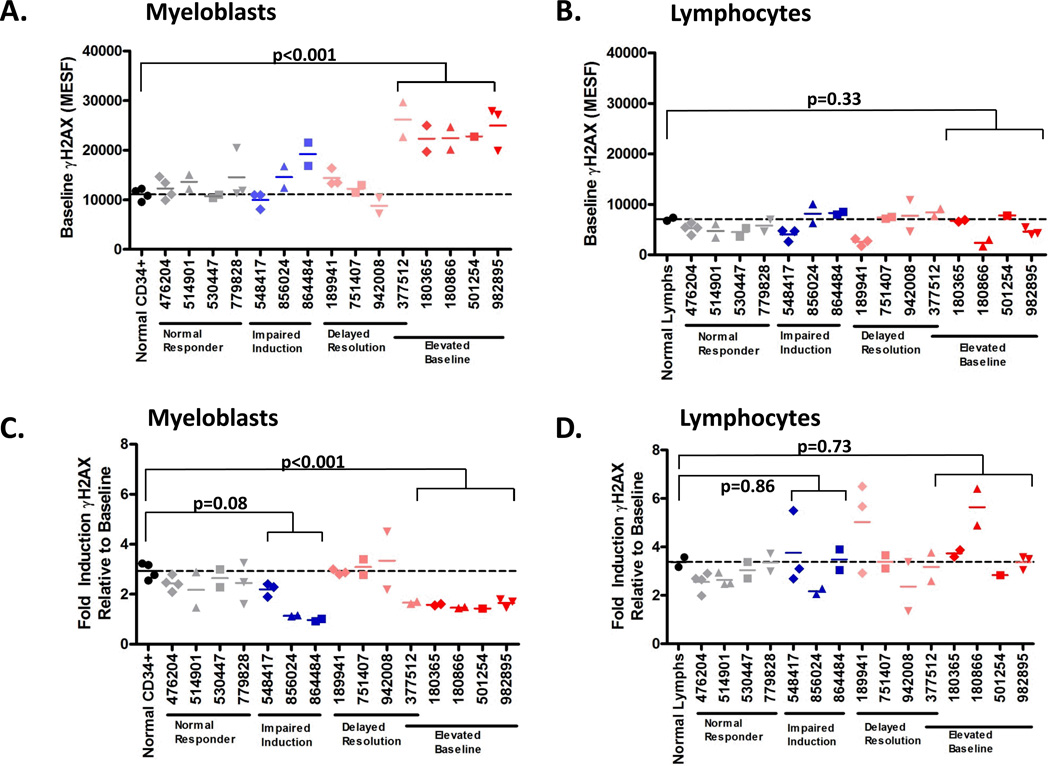
We found that 4 of 15 t-AML patients had myeloblasts that displayed statistically equivalent γH2AX kinetics compared to bone marrow CD34+ cells from normal adults (n=5) (p= 0.867), including baseline and post-damage induction and resolution of γH2AX levels, while 11/15 patients had abnormal γH2AX levels which fell into one of three statistically defined patterns (Figure 1). Myeloblasts from the first subset of patients (n=4) had evidence of higher basal γH2AX levels compared to CD34+ controls (p< 0.001) suggesting an increased basal DSB burden in these cells (Figure 1B). Both at baseline (non-irradiated, defined as time=0) and 24 hours post-irradiation, when γH2AX levels in CD34+ controls have decreased to their baseline levels, the γH2AX levels in this subset of t-AML patients remained elevated (Figure 1B). These myeloblasts also showed impaired γH2AX fold-induction compared to CD34+ controls (mean 1.53 vs. 2.97 fold increase at 30 minutes over baseline, respectively; p<0.001) (Figure 2C), likely due to their elevated basal γH2AX levels. The elevation of basal levels of γH2AX is likely acquired and tumor-cell intrinsic as myeloblasts, but not lymphocytes, from these patients are abnormal (Figure 2).
Figure 1.
Primary myeloblasts from t-AML patients have abnormal γH2AX levels at baseline and post-DNA damage. γH2AX levels were measured in primary myeloblasts from t-AML patients at baseline and post-irradiation (IR) with 2 Gray. The mean fluorescent intensity of the γH2AX signal measured using flow cytometry was converted to mean equivalent soluble fluorophore (MESF) using calibration beads included in each experiment. (A) Myeloblasts from four t-AML patients showed similar baseline, induction γH2AX levels, and γH2AX resolution kinetics as CD34+ cells from normal donors (n=5). (B) Myeloblasts from four t-AML patients showed elevated basal γH2AX levels compared to CD34+ controls at baseline (t=0, unirradiated) (average MESF 23,107 vs. 11,490, respectively; p<0.001) and t=24 (average MESF 25,810 vs. 13,030,repectively; p<0.001). IR-induced DSB formation at t=30 minutes was comparable to CD34+ controls (average MESF 36,580 vs. 34,160, respectively, p=0.29). (C) Myeloblasts from three t-AML patients showed impaired γH2AX induction at 30 minutes post IR compared to CD34+ controls (average MESF 19,520 vs. 34,160, respectively p<0.001). (D) Myeloblasts from four patients showed delayed γH2AX resolution kinetics compared to CD34+ controls (p=0.007). UPN 377512 also had an elevated baseline γH2AX level. Samples were interrogated 2–5 independent times, with the exception of UPN 501254, which was performed once. **p<0.01, *** p<0.001. NL, normal; BM, bone marrow
Figure 2.
Determination of γH2AX levels in myeloblasts and lymphocytes from t-AML patients. Myeloblasts (A), but not lymphocytes, (B) have elevated baseline γH2AX levels compared to CD34+ cells from normal donors (n=5). Similarly, myeloblasts (C), but not lymphocytes (D) (with the exception of UPN 856024) have impaired induction (measured by fold change of γH2AX at 30 min. post-irradiation (IR) over baseline level) of γH2AX compared to CD34+ cells from normal donors. Samples were interrogated 2–5 independent times, with the exception of UPN 501254, which was performed once. MESF was determined as in Fig.1. Dashed line indicates the mean MESF of normal donor bone marrow CD34+ cells or lymphocytes. *p<0.05; ** p<0.01
A second subset of patients (n=3) had impaired γH2AX induction compared to normal donor CD34+ cells (p<0.001) (Figure 1C). Myeloblasts from this group showed approximately two-fold reduction in peak γH2AX levels at 30 minutes post-IR compared to CD34+ controls (p<0.001) (Figure 1C) and a mean 1.44 vs. 2.97 fold increase in γH2AX levels at thirty minutes relative to baseline, respectively (p=0.08) (Figure 2C), suggesting a defect in the ability to detect DSBs. Deficient induction of γH2AX in response to IR was tumor-specific in 2 of these patients (Figure 2 C–D). In one patient, UPN 856024, a slight defect in γH2AX induction was observed in lymphocytes, possibly indicating an inherited defect in the DSB response.
Myeloblasts from the final group of patients (n=4) had delayed resolution of γH2AX levels compared to CD34+ controls post-IR (p=0.007) (Figure 1D). Lymphocytes from these patients did not display a pattern of decreased γH2AX resolution compared to lymphocytes from normal donors (data not shown), suggesting that this finding was also tumor-cell intrinsic.
Collectively, these data suggest the functional integrity of double-strand break repair and/ or response is abnormal in the majority of t-AML patients tested (11 out of 15). The functional alterations were restricted to myeloblasts (with the possible exception of one patient), suggesting that they may be driven by somatic events. To test this possibility, we assessed the role of genetic mutations in the two major pathways of mammalian DSB response, HR and NHEJ.
Genetic Alterations in homologous recombination and non-homologous end-joining genes in t-AML
We sequenced the 21 canonical HR and 7 NHEJ DNA repair genes, as well as a subset of 9 DNA damage response and polymerase genes, using tumor DNA and paired normal DNA obtained from 25 t-AML patients (Supplementary Table 3). We identified 5 heterozygous somatic mutations in 3 genes (EME1, RAD51D and TP53) all with predicted translational consequences (Table 1) (Supplementary Results). In one patient, UPN 189941, whole genome sequencing previously identified a novel heterozygous 3-kilobase deletion spanning exons 7–9 of TP53 in the patient’s skin (normal) DNA, which was homozygous in the tumor DNA as a result of uniparental disomy.(16) In sum, 6 of 25 patients, including UPN 189941, had somatic changes in 3 genes [RAD51D (n=1), EME1 (n=1), TP53, (n=4)] (Table 1).
Table 1.
Somatic Sequence Variants in DNA Repair and Response Genes in t-AML
| UPN | Gene | Amino Acid Change | Pathway |
|---|---|---|---|
| 860923 | EME1 | p.L549V | HR |
| 751407 | RAD51D | p.S144Y | HR |
| 377512 | TP53 | p.R273C | DDR |
| 530447 | TP53 | p.R248Q | DDR |
| 942008 | TP53 | p.R175H | DDR |
| 189941 | TP53 | del of aa 225–3311 | DDR |
3 kb deletion of exons 7–9 homozygous in the tumor secondary to UPD, ref (16)
HR, homologous recombination; DDR, DNA damage response
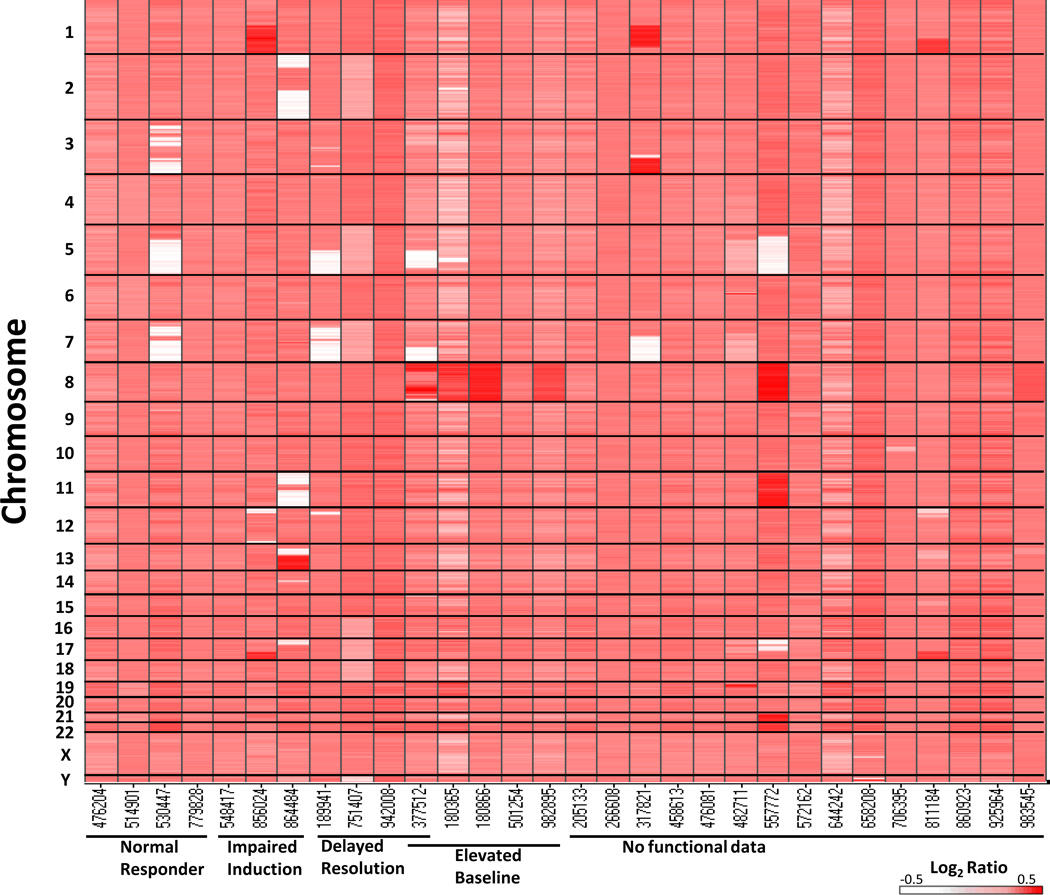
Next, we used a custom, genome-wide array comparative genomic hybridization platform containing high resolution coverage of 170 DNA repair genes (Supplementary Table 2) in order to identify copy-number alterations (amplifications and deletions) involving DNA repair genes. We identified 119 acquired CNAs in 18 of 30 t-AML genomes spanning from 1,367 bp to 243 Mb in size (Figure 3, Supplementary Figure 1, Supplementary Table 4) (Supplementary Results). CNAs involving DNA repair genes were typically found in copy altered regions greater than 5 Mb in size (99.2% of the time). Fifty-eight of the 119 CNAs contained at least one DNA repair gene. Permutation analysis showed no significant enrichment of DNA repair genes in CNAs in the t-AML genomes greater than would be expected by chance alone (p=0.09). Of note, three of the four patients (UPNs 189941, 530447, 377512) with a TP53 mutation also displayed inferred (based on copy number analysis rather than direct sequencing) chromothripsis (chromosomal shattering and reanastomosis)(25, 26) consistent with the recent description of chromothripsis occurring in AML with TP53 mutations.(25) (Supplementary Table 5, Supplementary Figure 2.)
Figure 3.
Copy number alteration heat map for 30 t-AML genomes. The results of copy number analysis of 30 paired tumor and normal DNA samples assayed on a custom genome-wide array CGH platform with high resolution coverage of 170 DNA repair genes. Copy number is represented as the log2 ratio of tumor/normal DNA designated by a color range from white (deletion) to red (amplification). The Y-axis represents the chromosome number, with 1 on the top and Y on the bottom. The X-axis displays samples grouped by functional DSB category.
Correlation of genotype and DSB response phenotype
Tumor cells from UPN 864484 contained a deleted region on chromosome 11 that included several genes known to be involved in the γH2AX response to DSBs, including MRE11A, H2AFX, and ATM. Gene expression analysis revealed that MRE11A (but not H2AFX or ATM) expression in UPN 864484 was the lowest of the 14 t-AML samples tested, which was confirmed by quantitative RT-PCR analysis, consistent with a concordant decrease in gene expression associated with the deletion (Supplementary Figure 3).
We observed TP53 mutations in 3 of 4 patients with myeloblasts showing delayed γH2AX resolution post-acute damage (Figure 1D, Table 1, Supplementary Table 4). However, TP53 mutations were not sufficient to result in this phenotype as UPN 530447 had normal γH2AX kinetics (Figure 1A, Table 1). In addition, we observed that myeloblasts in 5 of 5 patients who harbored trisomy 8 (UPNs 180365, 180866, 501254, 982895) or large copy number gains on chromosome 8 (UPN 377512) (defined by array CGH and/or metaphase cytogenetics), had elevated baseline γH2AX levels compared to normal donor CD34+ cells [Figure 1B and 1D (UPN 377512), Figure 3, Supplementary Figure 4, Supplementary Table 4]. Of note, no somatic (tumor) sequencing variants were identified in the interrogated canonical HR and NHEJ genes in any of these samples. Only UPN 377512, which displayed elevated baseline γH2AX levels and delayed γH2AX resolution after irradiation, (Figure 1D), as well as inferred chromothripsis, (Supplementary Table 4, Supplementary Figure 2) also had a TP53 mutation, as noted above.
Elevated double-strand breaks are present in bone marrow cells isolated from patients with Trisomy 8
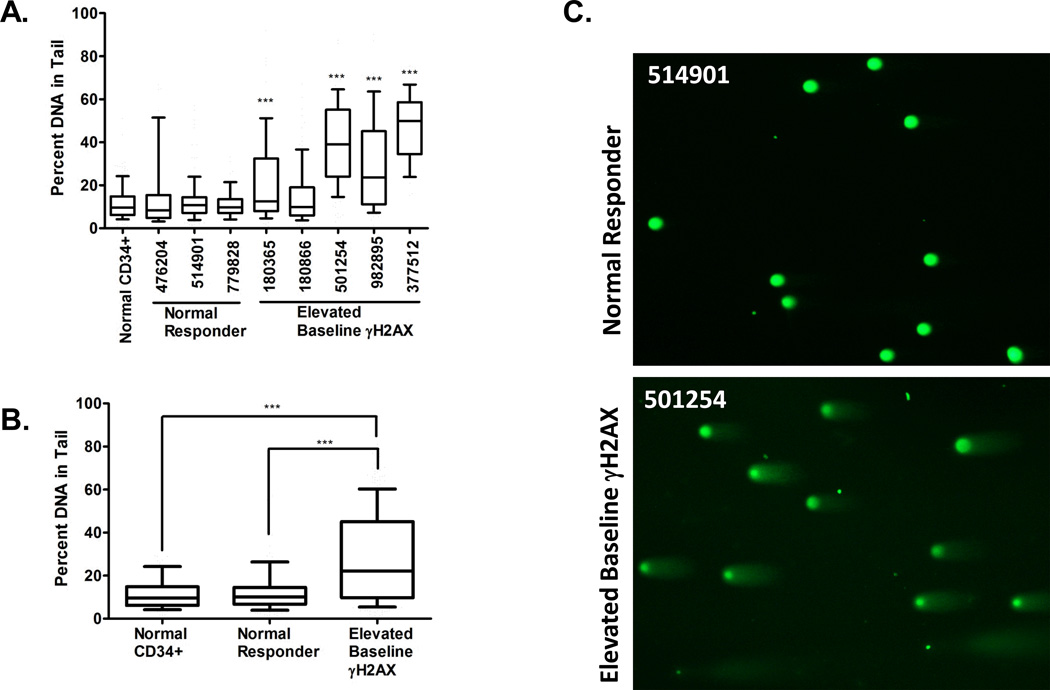
In order to directly measure DSBs in samples with gains on chromosome 8, we performed the Neutral Comet Assay, specific for DSBs(27) using bone marrow cells from patients harboring gains on chromosome 8 and compared them to CD34+ cells from normal donors, and bone marrow cells from t-AML patients in which the γH2AX levels were not different than normal donor CD34+ (Figure 1A), henceforth referred to as normal t-AML responders.
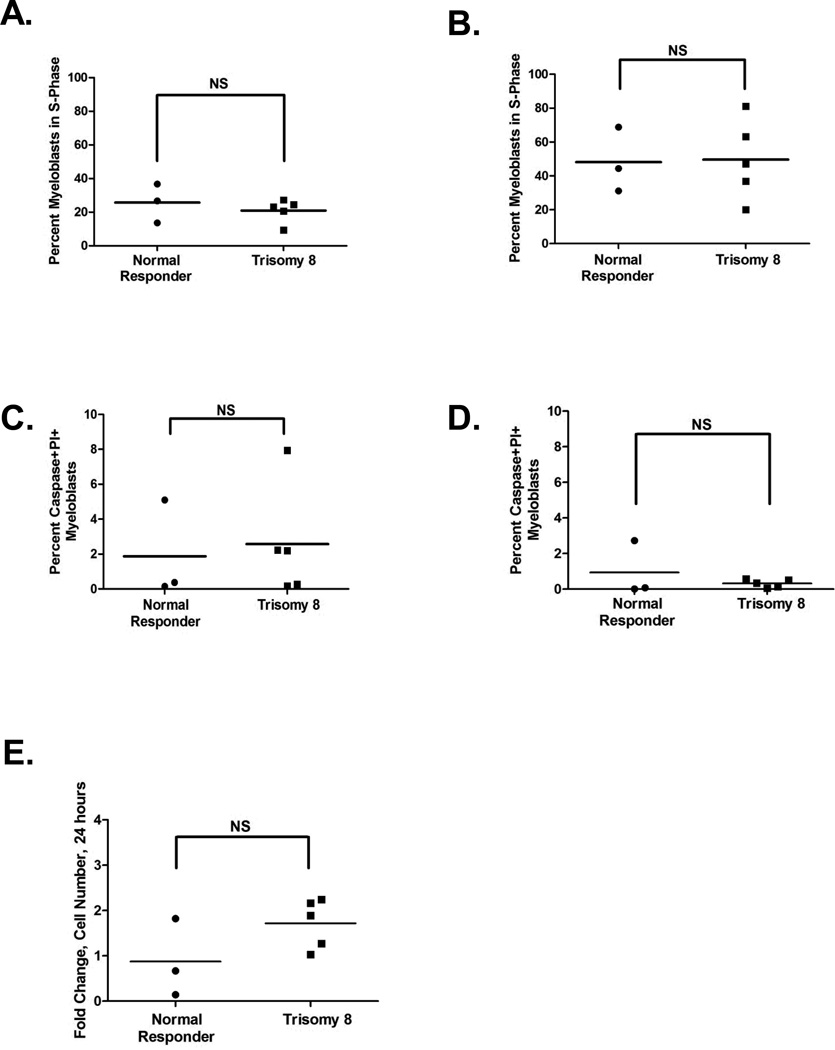
Four of 5 patients with gains on chromosome 8 (UPNs 180365, 501254, 982895, and 377512) had elevated basal DSB compared to normal donor CD34+ cells (Figure 4A). Furthermore, when Comet data were pooled for the normal γH2AX responders, and those with gains on chromosome 8, the mean percent DNA in tail from t-AML patients with chromosomal 8 gains (28.3%) was significantly higher than patients with normal γH2AX (13.2%) (p<0.001) and CD34+ controls (11.8%) (p<0.001). Similar results were observed when Comets were evaluated by Olive moment (p<0.001). Thus, the elevated basal γH2AX levels observed in myeloblasts of patients with gains on chromosome 8 correspond to elevated levels of DSB measured directly by the neutral Comet assay. The elevated levels of DSB in these samples was not associated with alterations in cell cycle parameters or apoptosis (Figure 5), which could induce DSBs, compared to normal t-AML responders, suggesting that myeloblasts harboring gains on chromosome 8 may tolerate persistently elevated levels of DSB burden.
Figure 4.
Elevated double-strand breaks (DSBs) are present in bone marrow cells isolated from t-AML patients harboring gains on chromosome 8. DSB were measured in bone marrow cells by the neutral Comet assay and Comets were scored by Comet Score software. (A) Four of 5 patients with gains on chromosome 8 (UPNs 180365, 501254, 982895, and 377512) had elevated basal DSB compared to normal donor CD34+ cells, while the DSB levels in t-AML patients with normal γH2AX levels (normal t-AML responders, UPNs 476204, 514901, 779828) did not differ from CD34+ controls. Box-whisker plots were generated, with the box showing the 25th to 75th percentiles, the bar representing the median, and the whiskers representing the 10th to the 90th percentiles of the percent of DNA in the tail. The percent of DNA in the tail for CD34+ cells was compared to the percent of DNA in the tail for each UPN. The median number of Comets scored per UPN was 133 (range 44–256). (B) Comet data were pooled for the patients with normal γH2AX kinetics (n=3 patients, 317 total Comets) and those harboring gains on chromosome 8 (n=5 patients, 646 total Comets) and analyzed as above. The mean percent of DNA in the tail from t-AML with trisomy 8 (28.2%) is significantly higher than patients with normal γH2AX (13.2%) and CD34+ controls (11.8%). (C) Representative Comets from a t-AML patient with normal γH2AX kinetics (normal t-AML responder) (UPN 514901) and from a patient with trisomy 8 (UPN 501254) are shown. ***p<0.001.
Figure 5.
Measurements of S-phase, apoptosis, and proliferation in myeloblasts from t-AML patients. Myeloblasts from patients harboring large gains on chromosome 8 (UPNs 180365, 180866, 501254, 982895, 377512) were compared to myeloblasts from normal γH2AX t-AML responders (UPNs 476204, 514901, 779828). (A) The percent of myeloblasts in S-phase from patients with gains on chromosome 8 was not significantly different than those with normal γH2AX kinetics at baseline (mock treated) (mean 21% vs 25.7%, respectively, p=0.42) and (B) after a further 24 hours in culture (mean 49.7% vs 48.1%, respectively, p=0.92). (C) Apoptosis was measured in myeloblasts by the presence of caspase 3+ or 7+ and PI+ cells by flow cytometry and was not significantly different than those with normal γH2AX kinetics at baseline (mean 2.57% vs 1.87%, respectively; p=0.77) and (D) after a further 24 hours in culture (mean 0.32 % vs 0.94 %, respectively; p=0.39). (E) Myeloblasts were enumerated by flow cytometry using Spherotech counting particles in unirradiated cells at baseline and after 24 hours. There was no attrition of myeloblasts from patients with gains on chromosome 8 compared to those with normal γH2AX kinetics after 24 hours in culture (mean fold increase in cells over baseline 1.72 vs 0.88, respectively; p=0.13). The fold change of each sample over the 24 hour period is shown.
MYC expression signature is present in t-AML harboring Trisomy 8
Although multiple genes located on chromosome 8 may contribute to the elevated DSBs observed in trisomy 8 samples, we first examined MYC mRNA expression in t-AML samples with trisomy 8 as MYC is located on chromosome 8 and its overexpression is known to induce DNA damage, including DSB.(28–30) Although the average level of MYC was upregulated in the bone marrow from trisomy 8 subjects compared to normal γH2AX responders by gene expression array analysis (Affymetrix mean signal intensity 30,249 vs. 10,653, respectively; p=0.16) (Supplementary Figure 5) and quantitative RT-PCR analysis (fold change 1.69–4.3 vs. 1.0 –1.43, respectively; p=0.08), the differences were not statistically significant, likely due to our small sample size. UPN 377512, which had elevated baseline γH2AX levels and DSBs (Figures 1D and 4A) harbored large discontinuous gains on chromosome 8 that did not contain MYC. However, MYC expression was elevated compared to normal γH2AX responders in this sample (Affymetrix mean signal intensity 21,314 vs. 10,653, respectively) (data not shown).
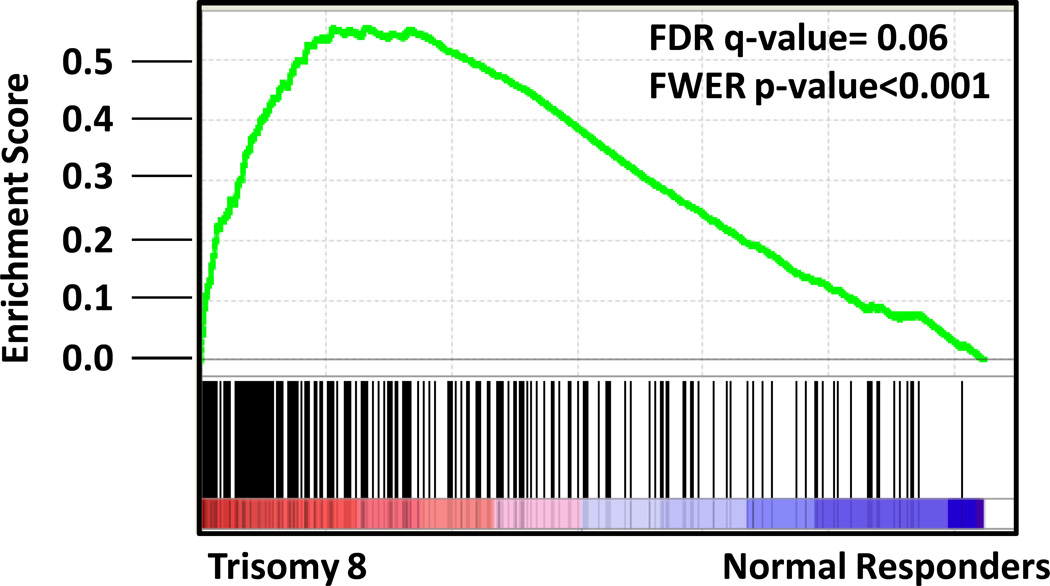
When we examined a larger dataset of de novo AML patients and compared MYC expression between those harboring trisomy 8 (n=22) and those with normal cytogenetics (n=84)(31), MYC was significantly overexpressed in trisomy 8 AML samples (Affymetrix mean signal intensity 33,669 vs. 19,108; p<0.001), in agreement with previously published work.(32) In addition, t-AML patients with trisomy 8 displayed upregulation of a canonical MYC target gene set described by Dang and colleagues(21) compared to normal t-AML γH2AX responders using gene set enrichment analysis (GSEA) (FDR q-value= 0.06; FWER p-value<0.001)(Figure 6, Supplementary Table 6). We also observed elevated reactive oxygen species levels, known to be induced by MYC(28, 30) in patients with trisomy 8, as compared to normal t-AML γH2AX responders (mean 169,300 vs. 46,770 MESF, respectively; p<0.05) (Supplementary Figure 6). Taken together, these data suggest that MYC is upregulated in t-AML patients with trisomy 8 and may partially contribute to elevated DSBs. The gene expression levels of additional putative DNA repair genes located on chromosome 8 were not consistently altered in patients with trisomy 8 (Supplementary Figure 5).
Figure 6.
A MYC gene expression signature is present in trisomy 8 bone marrow. Gene set enrichment analysis (GSEA) of MYC upregulated target genes comparing transcriptional profiles from the bone marrow of t-AML patients with trisomy 8 (UPNs 180365, 180866, 501254, 982895) vs. normal γH2AX t-AML responders (UPNs 476204, 514901, 779828).
Overexpression of Myc in primary murine hematopoietic progenitors results in elevated DSBs
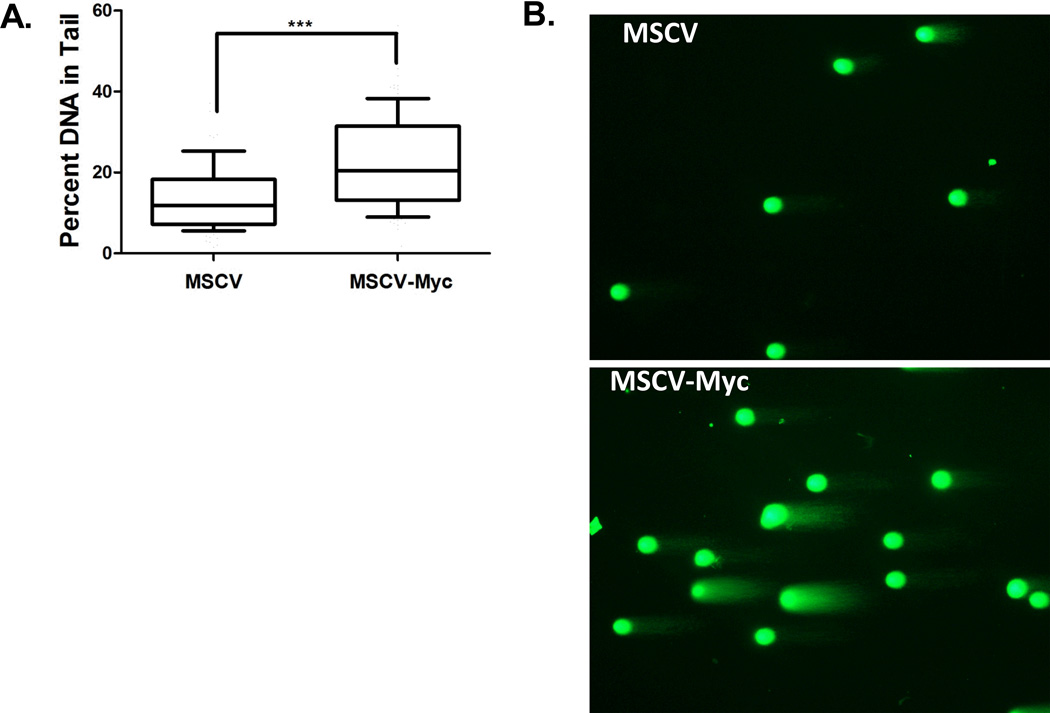
Prior reports have shown that MYC can induce genotoxic stress and DSB formation.(28–30, 33) To directly test if Myc overexpression is sufficient to cause DSB in primary mouse hematopoietic myeloid progenitor cells, we transduced c-kit+ enriched cells with a murine Myc-expressing retrovirus.(22) Murine c-kit+ enriched cells were transduced with MSCV-Ires-GFP or MSCV-Myc-Ires-GFP and transduced cells were sort purified (GFP+/propidium iodide negative) after 3 days. Quantitative RT-PCR analysis confirmed 3-fold higher Myc expression levels in MSCV-Myc-Ires-GFP transduced cells compared to control MSCV-Ires-GFP transduced cells. To directly measure DSBs, purified cells were subjected to the neutral Comet assay. DSBs, as assessed by percent DNA in tail, is significantly higher in Myc-overexpressing cells compared to control cells (22.67% vs. 13.85%, respectively; p<0.001) (Figure 7). Similar results were obtained when Comets were evaluated by the Olive moment (p<0.001) (data not shown).
Figure 7.
Overexpression of Myc in primary mouse hematopoietic progenitors results in elevated DSBs. (A) Murine c-kit+ enriched cells were transduced with MSCV-Ires-GFP control virus or MSCV-Myc-Ires-GFP and virally transduced cells were sort purified (GFP+/propidium iodide negative) after 3 days. DNA was subjected to the neutral Comet assay. The experiment was repeated 2 times, and 100 Comets were scored for each experiment. Box-whisker plots were generated as previously described in Figure 4. (B) Representative Comets from c-kit+ enriched cells transduced with control virus (MSCV) and the Myc overexpressing virus (MSCV-Myc) are shown.
DISCUSSION
In this article, we describe the DNA DSB response of bone marrow cells from t-AML patients. Our results suggest the majority of t-AML patients have tumor cell-intrinsic, functional dysregulation of their DSB response. Distinct patterns of abnormal DNA damage response correlated with genetic alterations in TP53 and were associated with an abnormal response to acute DSB damage, chromothripsis, or both. Furthermore, the presence of trisomy 8 was associated with elevated levels of basal DSB. Although tumor-acquired point mutations or small indels in canonical HR and NHEJ genes do not appear to be a dominant means by which t-AML leukemogenesis occurs, our functional studies suggest that an abnormal response to DNA damage may be a common finding in t-AML.
We observed a subset of patients with reduced H2AX phosphorylation 30 minutes after acute DNA damage compared to normal controls and other t-AML patients. Phosphorylation of H2AX is required for amplification of the DNA damage signal by augmenting the concentration of key DNA damage response proteins at the site of DSBs(34) and haploinsufficiency of H2AX results in genomic instability(35, 36) and increased susceptibility to tumorigenesis.(35) Thus, defective γH2AX signaling may have played a role in promoting leukemogenesis in these patients. Of note, an abnormal karyotype was observed in 2 of the 3 patients with blunted induction of phosphorylated H2AX following DNA damage (UPN 856024 and UPN 864484). We found that in one patient, deficient H2AX phosphorylation was associated with a large acquired deletion spanning MRE11A and concordant low MRE11A expression levels. MRE11A is necessary for optimal signaling of the ATM signaling in response to DSB, which along with the PI3-K like kinase DNA-dependent protein kinase catalytic subunit (DNA-PKcs, official gene symbol PRKDC) functions to phosphorylate H2AX after IR.(37) Interestingly, the γH2AX response pattern displayed by this patient, with defective γH2AX induction followed by a delayed rise in γH2AX levels, is reminiscent of the phenotype displayed by ATM deficient fibroblasts isolated from ataxia-telangiectasia patients(38) and consistent with what would be expected with loss of MRE11A in this patient. The potential basis of the defective γH2AX signaling in response to IR in the other two patients is not known. Nonetheless, defective γH2AX responses to DSB in these patients raises the possibility that traditional cytotoxic chemotherapy, that relies on cell death by inducing and sensing DNA DSB, may be less effective in these patients that do not sense DSB formation normally.
TP53 aberrations are one of the most common mutations in t-AML(12–14) and are associated with complex karyotypes and highly derivative chromosomes.(12, 15) Furthermore, among complex karyotype AML, those with TP53 alterations, accounting for 70% of cases, have a higher degree of genomic complexity, monosomal karyotypes, and inferior overall survival compared to those with unaltered TP53.(39) We noted that all t-AML patients with mutated TP53 had an abnormal DNA DSB response, as evidenced by of delayed γH2AX resolution post-acute damage, and/or inferred (based on copy number analysis rather than direct sequencing) chromothripsis (where catastrophic DNA alterations are tolerated). Previous reports have noted that p53-deficient cell lines show delayed γH2AX resolution post-irradiation,(40, 41) and fibroblasts from patients with Li-Fraumeni syndrome display delayed resolution of DSBs after IR compared to normal fibroblasts, similar to our observations here.(38) Recent reports have implicated p53 in DNA repair after IR through its role in heterochromatin relaxation and induction of WIPI1, the wild-type p53-induced phosphatase that dephosphorylates γH2AX, likely important in homeostasis of the DDR.(41, 42) The mechanism of the dysregulated DSB response in t-AML is likely to be multifactorial and to contribute to the genomic instability and poor outcome to conventional therapies seen in these patients. Further work is needed to elucidate the role(s) of p53 in myeloblasts post DNA damage.
We observed that myeloblasts from t-AML patients that harbored large gains on chromosome 8 had persistently elevated basal levels of DSBs that were not attributable to increased S-phase or apoptosis when compared to normal donor CD34+ control cells and t-AML patients who had normal γH2AX kinetics, suggesting that myeloblasts from these patients may tolerate an elevated DSB burden. Elevated DSBs in these patients could result from ongoing DNA damage, impaired repair of DSBs, or both. MYC expression is increased in MDS and AML from patients with trisomy 8,(32, 43) and we showed t-AML patients with trisomy 8 had upregulation of a MYC expression signature compared to t-AML patients with normal γH2AX kinetics, all of whom lacked trisomy 8. In agreement with previous reports that dysregulated MYC expression can induce DSB and disrupt DSB repair in a variety of contexts,(28–30, 33) we showed that Myc overexpression is sufficient to induce DSB in primary murine hematopoietic precursors. Further work is needed to evaluate the mechanism by which MYC may be leading to DSB persistence and to dissect the potential role of other genes on chromosome 8 in the elevated DSB burden observed in myeloblasts, including studies of UBE2V2, a DNA repair gene that was upregulated in trisomy 8 samples. The ability of myeloblasts to persist despite DNA damage may have implications for chemoresistance with conventional therapies. Recently, BET bromodomain inhibition has been shown to target MYC in experimental models of hematologic malignancies and may provide a novel therapeutic strategy in treating malignancies with pathologic MYC activation.(44, 45)
It is not known whether the dysfunctional DSB response observed in t-AML myeloblasts sensitizes or confers resistance to cell killing by specific chemotherapy. Testing drug sensitivity in vitro is biased because only a subset of AML samples grow adequately to test and there is emerging evidence that selection of leukemic subclones occurs during in vitro culturing that does not represent the clonal architecture observed in the patient (Klco and Ley, unpublished data). An ideal approach to address whether a dysfunctional DSB response confers drug sensitivity would be to screen a large set of t-AML patients enrolled in a clinical trial where the drug treatment, clinical response, and longitudinal follow-up are uniform. Thus innovative therapeutic approaches exploiting dysfunctional DNA response will likely require large clinical trials to test the clinical significance of a dysfunctional DSB response with the long-term goal of identifying more tolerable and efficacious regimens.
Supplementary Material
ACKNOWLEDGEMENTS
Claudia Wiese kindly provided rad51d knockout CHO cells and wild-type human RAD51D construct. Jahangheer Shaik assisted with the copy number alteration permutation analysis. Todd Druley assisted with sequencing design. Gene chip analysis was performed at the Siteman Cancer Center Molecular Genomic Analysis Core Facility. We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, MO., for the use of the Siteman Flow Cytometry Core, which provided the cell sorting service. The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant #P30 CA91842.This work was supported by NIH grants T32-HL007088 and K12 HL087107 (MAJ) and by the MDS Foundation, Washington University Siteman Cancer Center Research Development Award, and the Institute of Clinical and Translational Sciences UL1RR024992 (MJW). We thank Sharon Health for assistance with specimen banking and data management. We thank Dr. Dan Link, Dr. Tim Ley, and Dr. Tom Ellenberger for helpful scientific discussions.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest to disclose.
Supplementary information is available at Leukemia’s website.
REFERENCES
- 1.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009 Jul 30;114(5):937–951. doi: 10.1182/blood-2009-03-209262. PubMed PMID: 19357394. [DOI] [PubMed] [Google Scholar]
- 2.Mauritzson N, Albin M, Rylander L, Billstrom R, Ahlgren T, Mikoczy Z, et al. Pooled analysis of clinical and cytogenetic features in treatment-related and de novo adult acute myeloid leukemia and myelodysplastic syndromes based on a consecutive series of 761 patients analyzed 1976–1993 and on 5098 unselected cases reported in the literature 1974–2001. Leukemia. 2002 Dec;16(12):2366–2378. doi: 10.1038/sj.leu.2402713. PubMed PMID: 12454741. [DOI] [PubMed] [Google Scholar]
- 3.Leone G, Mele L, Pulsoni A, Equitani F, Pagano L. The incidence of secondary leukemias. Haematologica. 1999 Oct;84(10):937–945. PubMed PMID: 10509043. [PubMed] [Google Scholar]
- 4.Kayser S, Dohner K, Krauter J, Kohne CH, Horst HA, Held G, et al. The impact of therapy-related acute myeloid leukemia (AML) on outcome in 2853 adult patients with newly diagnosed AML. Blood. 2011 Feb 17;117(7):2137–2145. doi: 10.1182/blood-2010-08-301713. PubMed PMID: 21127174. [DOI] [PubMed] [Google Scholar]
- 5.Smith SM, Le Beau MM, Huo D, Karrison T, Sobecks RM, Anastasi J, et al. Clinical-cytogenetic associations in 306 patients with therapy-related myelodysplasia and myeloid leukemia: the University of Chicago series. Blood. 2003 Jul 1;102(1):43–52. doi: 10.1182/blood-2002-11-3343. PubMed PMID: 12623843. [DOI] [PubMed] [Google Scholar]
- 6.Schoch C, Kern W, Schnittger S, Hiddemann W, Haferlach T. Karyotype is an independent prognostic parameter in therapy-related acute myeloid leukemia (t-AML): an analysis of 93 patients with t-AML in comparison to 1091 patients with de novo AML. Leukemia. 2004 Jan;18(1):120–125. doi: 10.1038/sj.leu.2403187. PubMed PMID: 14586477. [DOI] [PubMed] [Google Scholar]
- 7.Li FP, Fraumeni JF., Jr Prospective study of a family cancer syndrome. JAMA : the journal of the American Medical Association. 1982 May 21;247(19):2692–2694. PubMed PMID: 7077763. [PubMed] [Google Scholar]
- 8.Birch JM, Alston RD, McNally RJ, Evans DG, Kelsey AM, Harris M, et al. Relative frequency and morphology of cancers in carriers of germline TP53 mutations. Oncogene. 2001 Aug 2;20(34):4621–4628. doi: 10.1038/sj.onc.1204621. PubMed PMID: 11498785. [DOI] [PubMed] [Google Scholar]
- 9.Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nature genetics. 2013 Mar;45(3):242–252. doi: 10.1038/ng.2532. PubMed PMID: 23334668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillem V, Tormo M. Influence of DNA damage and repair upon the risk of treatment related leukemia. Leukemia & lymphoma. 2008 Feb;49(2):204–217. doi: 10.1080/10428190701769657. PubMed PMID: 18231906. [DOI] [PubMed] [Google Scholar]
- 11.Seedhouse C, Faulkner R, Ashraf N, Das-Gupta E, Russell N. Polymorphisms in genes involved in homologous recombination repair interact to increase the risk of developing acute myeloid leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004 Apr 15;10(8):2675–2680. doi: 10.1158/1078-0432.ccr-03-0372. PubMed PMID: 15102670. [DOI] [PubMed] [Google Scholar]
- 12.Shih AH, Chung SS, Dolezal EK, Zhang SJ, Abdel-Wahab OI, Park CY, et al. Mutational analysis of therapy-related myelodysplastic syndromes and acute myelogenous leukemia. Haematologica. 2013 Jun;98(6):908–912. doi: 10.3324/haematol.2012.076729. PubMed PMID: 23349305. Pubmed Central PMCID:3669447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen-Bjergaard J, Andersen MK, Andersen MT, Christiansen DH. Genetics of therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2008 Feb;22(2):240–248. doi: 10.1038/sj.leu.2405078. PubMed PMID: 18200041. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Yehuda D, Krichevsky S, Caspi O, Rund D, Polliack A, Abeliovich D, et al. Microsatellite instability and p53 mutations in therapy-related leukemia suggest mutator phenotype. Blood. 1996 Dec 1;88(11):4296–4303. PubMed PMID: 8943866. [PubMed] [Google Scholar]
- 15.Andersen MK, Christiansen DH, Pedersen-Bjergaard J. Centromeric breakage and highly rearranged chromosome derivatives associated with mutations of TP53 are common in therapy-related MDS and AML after therapy with alkylating agents: an M-FISH study. Genes, chromosomes & cancer. 2005 Apr;42(4):358–371. doi: 10.1002/gcc.20145. PubMed PMID: 15645489. [DOI] [PubMed] [Google Scholar]
- 16.Link DC, Schuettpelz LG, Shen D, Wang J, Walter MJ, Kulkarni S, et al. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA : the journal of the American Medical Association. 2011 Apr 20;305(15):1568–1576. doi: 10.1001/jama.2011.473. PubMed PMID: 21505135. Pubmed Central PMCID: 3170052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodarzi AA, Jeggo PA. The repair and signaling responses to DNA double-strand breaks. Advances in genetics. 2013;82:1–45. doi: 10.1016/B978-0-12-407676-1.00001-9. PubMed PMID: 23721719. [DOI] [PubMed] [Google Scholar]
- 18.Ramsingh G, Jacoby MA, Shao J, De Jesus Pizzaro RE, Shen D, Trissal M, et al. Acquired copy number alterations of miRNA genes in acute myeloid leukemia are uncommon. Blood. 2013 Oct 10;122(15):e44–e51. doi: 10.1182/blood-2013-03-488007. PubMed PMID: 24009227. Pubmed Central PMCID: 3795465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proceedings of the National Academy of Sciences of the United States of America. 2009 Aug 4;106(31):12950–12955. doi: 10.1073/pnas.0903091106. PubMed PMID: 19651600. Pubmed Central PMCID: 2716381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005 Oct 25;102(43):15545–15550. doi: 10.1073/pnas.0506580102. PubMed PMID: 16199517. Pubmed Central PMCID: 1239896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome biology. 2003;4(10):R69. doi: 10.1186/gb-2003-4-10-r69. PubMed PMID: 14519204. Pubmed Central PMCID: 328458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo H, Li Q, O'Neal J, Kreisel F, Le Beau MM, Tomasson MH. c-Myc rapidly induces acute myeloid leukemia in mice without evidence of lymphoma-associated antiapoptotic mutations. Blood. 2005 Oct 1;106(7):2452–2461. doi: 10.1182/blood-2005-02-0734. PubMed PMID: 15972450. [DOI] [PubMed] [Google Scholar]
- 23.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. The Journal of cell biology. 1999 Sep 6;146(5):905–916. doi: 10.1083/jcb.146.5.905. PubMed PMID: 10477747. Pubmed Central PMCID: 2169482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nature reviews Cancer. 2008 Dec;8(12):957–967. doi: 10.1038/nrc2523. PubMed PMID: 19005492. Pubmed Central PMCID: 3094856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012 Jan 20;148(1–2):59–71. doi: 10.1016/j.cell.2011.12.013. PubMed PMID: 22265402. Pubmed Central PMCID: 3332216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011 Jan 7;144(1):27–40. doi: 10.1016/j.cell.2010.11.055. PubMed PMID: 21215367. Pubmed Central PMCID: 3065307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fairbairn DW, Olive PL, O'Neill KL. The comet assay: a comprehensive review. Mutation research. 1995 Feb;339(1):37–59. doi: 10.1016/0165-1110(94)00013-3. PubMed PMID: 7877644. [DOI] [PubMed] [Google Scholar]
- 28.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, et al. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Molecular cell. 2002 May;9(5):1031–1044. doi: 10.1016/s1097-2765(02)00520-8. PubMed PMID: 12049739. [DOI] [PubMed] [Google Scholar]
- 29.Karlsson A, Deb-Basu D, Cherry A, Turner S, Ford J, Felsher DW. Defective double-strand DNA break repair and chromosomal translocations by MYC overexpression. Proceedings of the National Academy of Sciences of the United States of America. 2003 Aug 19;100(17):9974–9979. doi: 10.1073/pnas.1732638100. PubMed PMID: 12909717. Pubmed Central PMCID: 187906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ray S, Atkuri KR, Deb-Basu D, Adler AS, Chang HY, Herzenberg LA, et al. MYC can induce DNA breaks in vivo and in vitro independent of reactive oxygen species. Cancer research. 2006 Jul 1;66(13):6598–6605. doi: 10.1158/0008-5472.CAN-05-3115. PubMed PMID: 16818632. [DOI] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine. 2013 May 30;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. PubMed PMID: 23634996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones L, Wei G, Sevcikova S, Phan V, Jain S, Shieh A, et al. Gain of MYC underlies recurrent trisomy of the MYC chromosome in acute promyelocytic leukemia. The Journal of experimental medicine. 2010 Nov 22;207(12):2581–2594. doi: 10.1084/jem.20091071. PubMed PMID: 21059853. Pubmed Central PMCID: 2989761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Owonikoko TK, Sun SY, Ramalingam SS, Doetsch PW, Xiao ZQ, et al. c-Myc suppression of DNA double-strand break repair. Neoplasia. 2012 Dec;14(12):1190–1202. doi: 10.1593/neo.121258. PubMed PMID: 23308051. Pubmed Central PMCID: 3540944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, et al. Genomic instability in mice lacking histone H2AX. Science (New York, NY. 2002 May 3;296(5569):922–927. doi: 10.1126/science.1069398. PubMed PMID: 11934988. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003 Aug 8;114(3):371–383. doi: 10.1016/s0092-8674(03)00567-1. PubMed PMID: 12914701. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003 Aug 8;114(3):359–370. doi: 10.1016/s0092-8674(03)00566-x. PubMed PMID: 12914700. eng. [DOI] [PubMed] [Google Scholar]
- 37.Riches LC, Lynch AM, Gooderham NJ. Early events in the mammalian response to DNA double-strand breaks. Mutagenesis. 2008 Sep;23(5):331–339. doi: 10.1093/mutage/gen039. PubMed PMID: 18644834. [DOI] [PubMed] [Google Scholar]
- 38.Mirzayans R, Severin D, Murray D. Relationship between DNA double-strand break rejoining and cell survival after exposure to ionizing radiation in human fibroblast strains with differing ATM/p53 status: implications for evaluation of clinical radiosensitivity. International journal of radiation oncology, biology, physics. 2006 Dec 1;66(5):1498–1505. doi: 10.1016/j.ijrobp.2006.08.064. PubMed PMID: 17126209. [DOI] [PubMed] [Google Scholar]
- 39.Rucker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012 Mar 1;119(9):2114–2121. doi: 10.1182/blood-2011-08-375758. PubMed PMID: 22186996. [DOI] [PubMed] [Google Scholar]
- 40.Banath JP, Macphail SH, Olive PL. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer research. 2004 Oct 1;64(19):7144–7149. doi: 10.1158/0008-5472.CAN-04-1433. PubMed PMID: 15466212. [DOI] [PubMed] [Google Scholar]
- 41.Zheng H, Chen L, Pledger WJ, Fang J, Chen J. p53 promotes repair of heterochromatin DNA by regulating JMJD2b and SUV39H1 expression. Oncogene. 2013 Feb 4; doi: 10.1038/onc.2013.6. PubMed PMID: 23376847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moon SH, Nguyen TA, Darlington Y, Lu X, Donehower LA. Dephosphorylation of gamma-H2AX by WIP1: an important homeostatic regulatory event in DNA repair and cell cycle control. Cell cycle. 2010 Jun 1;9(11):2092–2096. doi: 10.4161/cc.9.11.11810. PubMed PMID: 20495376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sloand EM, Pfannes L, Chen G, Shah S, Solomou EE, Barrett J, et al. CD34 cells from patients with trisomy 8 myelodysplastic syndrome (MDS) express early apoptotic markers but avoid programmed cell death by up-regulation of antiapoptotic proteins. Blood. 2007 Mar 15;109(6):2399–2405. doi: 10.1182/blood-2006-01-030643. PubMed PMID: 17090657. Pubmed Central PMCID: 1852203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011 Sep 16;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. PubMed PMID: 21889194. Pubmed Central PMCID: 3187920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011 Oct 27;478(7370):524–528. doi: 10.1038/nature10334. PubMed PMID: 21814200. Pubmed Central PMCID: 3328300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.