Significance
Spontaneous mutations are rare and difficult to observe in large numbers experimentally. By sequencing the genomes of 145 diploid mutation accumulation (MA) lines of the budding yeast Saccharomyces cerevisiae, we identified nearly 1,000 mutations, a larger number than in any prior eukaryotic MA experiment as far as we are aware. For the first time, to our knowledge, in MA data, we were able to estimate rates of context-dependent single-nucleotide mutations. We were also able to observe mutational classes not seen in earlier yeast MA experiments and infer the rate of strongly deleterious mutations from patterns of missing mutations in each mutational class. Our findings both answer outstanding questions in the field, as well as highlight the need for more studies of spontaneous mutation.
Keywords: neighbor-dependent mutation rate, strongly deleterious mutation
Abstract
Mutation is the ultimate source of genetic variation. The most direct and unbiased method of studying spontaneous mutations is via mutation accumulation (MA) lines. Until recently, MA experiments were limited by the cost of sequencing and thus provided us with small numbers of mutational events and therefore imprecise estimates of rates and patterns of mutation. We used whole-genome sequencing to identify nearly 1,000 spontaneous mutation events accumulated over ∼311,000 generations in 145 diploid MA lines of the budding yeast Saccharomyces cerevisiae. MA experiments are usually assumed to have negligible levels of selection, but even mild selection will remove strongly deleterious events. We take advantage of such patterns of selection and show that mutation classes such as indels and aneuploidies (especially monosomies) are proportionately much more likely to contribute mutations of large effect. We also provide conservative estimates of indel, aneuploidy, environment-dependent dominant lethal, and recessive lethal mutation rates. To our knowledge, for the first time in yeast MA data, we identified a sufficiently large number of single-nucleotide mutations to measure context-dependent mutation rates and were able to (i) confirm strong AT bias of mutation in yeast driven by high rate of mutations from C/G to T/A and (ii) detect a higher rate of mutation at C/G nucleotides in two specific contexts consistent with cytosine methylation in S. cerevisiae.
Spontaneous mutations are the source of all genetic variation in nature. The rate of emergence of new mutations and the relative proportions of advantageous, neutral, and deleterious mutations are key determinants in how species evolve and adapt to new selective challenges. Unfortunately, our knowledge of the properties of spontaneous mutations remains incomplete primarily due to the difficulty of observing large enough numbers of mutational events in an unbiased way.
Analyzing patterns of divergence in nonfunctional sequences is a statistically powerful method used to study relative rates of different mutation classes. This method is applicable to most organisms and now can generally be carried out on a genome-wide scale. However, this approach relies crucially on the assumption that mutations in certain regions, such as pseudogenes or fourfold degenerate codon positions, are not affected by selection and are thus reliable approximations of true mutation rate. It is now becoming apparent that selection or selection-like processes, such as biased gene conversion, are acting even at these sequences and can substantially bias the observed patterns (1–5).
Studies focusing on mutations in reporter genes use a more restrictive method that can be applied only in model organisms. In some cases, such reporter genes can be placed genome-wide and thus provide estimates of genomic variation in mutation rates. However, this approach is limited by the inability to detect mutations without a visible phenotype and thus also gives us a biased picture of the mutational process (6–12).
A more unbiased approach for the study of mutations is to directly compare genomes of parents and offspring (13–17). Unfortunately, this approach is currently experimentally too expensive and laborious to generate sufficient numbers of mutational events for a systematic analysis of mutations. A conceptually similar method is to carry out a mutation accumulation (MA) experiment for a larger number of generations by passaging the population through sharp bottlenecks. Here, only the most strongly deleterious mutations will be missed from the final tally. Although until recently MA experiments were limited by the rarity of mutations, advances in next-generation sequencing are making large-scale MA experiments feasible.
Previous MA experiments have been conducted in a number of eukaryotic species, including Chlamydomonas reinhardtii, Arabidopsis thaliana, Caenorhabditis elegans, Saccharomyces cerevisiae and Drosophila melanogaster (18–25). In the budding yeast S. cerevisiae, MA experiments have been conducted in both haploids (4 lines, ∼4,800 divisions each, 33 single nucleotide changes) (26) and diploids (20 vegetative MA lines, ∼1,740 divisions each, 29 mutations) (27), providing estimates for overall mutation rates and insight into general mutational patterns. Even so, the recovery of tens of mutations is not sufficient to study either the rate of strongly deleterious mutations or context-dependent mutation rates.
Here, we present whole-genome sequencing results from 145 diploid MA lines, propagated asexually for an average of 2,062 generations each. We identified 867 single nucleotide mutations (SNMs), three double mutations (two SNMs occurring next to each other), 26 small indels under 50 bp, three copy number variants (CNVs), and 31 whole-chromosome copy-number changes. The last four mutation classes were observed only very rarely if at all in previous experiments. In total, we identified almost 1,000 new mutation events, an order of magnitude more than previous experiments in yeast and more than in any other MA experiment with wild-type strains as far as we are aware. The sufficiently large number of SNMs allowed us for the first time, to our knowledge, to detect context-dependent variation in mutation rates.
Results
The 145 mutation accumulation (MA) lines in this analysis were described previously (28, 29). Briefly, an ancestor strain for the experiment was generated from a diploidized haploid strain of genotype ade2, lys2–801, his3–D200, leu2–3.112, ura3–52, and ho. An initial set of 152 clonal lineages were established for mutation accumulation and propagated independently for ∼2,062 generations, each with a single-cell bottleneck every ∼20 generations (28, 29). Only the colony closest to a location marked in advance on each plate was propagated at each transfer, thereby eliminating inadvertent selection on colony size. The effective population size of the MA lines during the experiment was estimated to be ∼10 (29), leading to the expectation that only mutations of large heterozygous deleterious fitness effects conferring >10% selective disadvantage (sh > 0.1) would be missed (26).
For 145 lines, 100-bp paired-end Illumina libraries were constructed using high-throughput Nextera-based technology and sequenced to a total depth of 1,500× (∼10× per line). Coverage appears uniform across the genome, with the exception of telomeric and centromeric regions (Fig. S1). The remaining 7 lines did not grow when attempts were made to revive them from frozen stocks. Sequencing reads were mapped using Burrows–Wheeler aligner (BWA), and variants were called with genome analysis toolkit (GATK), after which customized post UnifiedGenotyper filters were applied to minimize false positives (Materials and Methods). Calls were made based upon the prior expectation that, unless the majority of new mutations occur in strong hotspots, the probability of an identical mutation independently occurring in two lines over ∼2,062 generations should be on the order of 10−13. Thus, despite the relatively low 10× sequencing depth per diploid line, we were able to call mutations with high fidelity in part because we had a strong prior expectation that all true spontaneous mutations should be present in a single line in a heterozygous state and that all fixed differences between the ancestral strain and the reference genome should be present in all lines in a homozygous state.
We identified a total of 3,137 single-nucleotide differences and 63 small indel differences (<50 bp) (Dataset S1) between the ancestral strain in our experiment and the reference S288C genome and used these to build the MA ancestral reference genome. The mapping and variant-calling pipeline was repeated on this new MA reference to obtain single nucleotide mutations (SNMs) and small indels that arose during the MA experiment. Whole-chromosome aneuploidies and large copy number variants (CNVs) were identified using sequencing depth traces (Materials and Methods).
Aneuploidy and Large CNVs.
Thirty-one of the 145 sequenced lines had whole-chromosome copy number changes, 29 of which were whole-chromosome duplications (9.7 ± 1.8 × 10−5 events per diploid genome per generation), and only two losses of entire chromosomes (0.7 ± 0.04 × 10−5 events per diploid genome per generation) (Table 1).
Table 1.
The line IDs of strains carrying an extra (3n) or a lost (1n) copy of each chromosome compared with the starting diploid (2n) state
| Chromosome | Length, kb | 3n strains | 1n strains |
| 1 | 231 | 152 | |
| 2 | 814 | 43,71,77 | |
| 3 | 317 | 43,49 | |
| 4 | 1,532 | 10,48,80 | |
| 5 | 577 | 50,117,146 | |
| 6 | 271 | ||
| 7 | 1,091 | 115 | |
| 8 | 563 | 83,108,111,152 | |
| 9 | 440 | 15,88,119 | 29,108 |
| 10 | 746 | 31 | |
| 11 | 667 | 30 | |
| 12 | 1,079 | 123 | |
| 13 | 925 | ||
| 14 | 785 | 63,73,124 | |
| 15 | 1,092 | ||
| 16 | 949 | 10,112,141 |
Both observed chromosomal losses were of chromosome IX, one of the smallest chromosomes in S. cerevisiae, and likely led to a loss of sporulation ability as successful sporulation was never observed in these two lines. Because our lines faced selection against strongly deleterious mutations, it is possible that loss of chromosome IX is better tolerated than losses of other chromosomes, despite strong phenotypic effects.
Given that mechanistically chromosomal losses and gains are likely to occur at the same time during mitosis, the much higher number of observed chromosomal gains suggests that chromosomal losses were strongly and dominantly deleterious. The observed rate of aneuploidies is thus a conservative estimate especially for the chromosomal losses.
Recent studies of aneuploidy in yeast showed that the rate of chromosomal loss correlates negatively with chromosomal length, with particularly high rates at chromosomes III and XII (30). Chromosomes also vary in stability when in aneuploid states (31). With our limited number of observed events, the distribution of whole-chromosomal copy number gains appeared random across chromosomes (Poisson distribution, P = 0.17, G test). As mentioned, however, the chromosome losses were observed for one of the smallest chromosomes.
No other forms of aneuploidy, such as tetrasomy or whole-genome duplication, were observed. Three large copy number variants were also observed in coverage traces (duplications of chromosomal segments 650–750 kb on chromosome XII in MA27, 0–40 kb on chromosome XV in MA64, and 888–988 kb on chromosome IV in MA84), but the events were too few in number to allow further analysis.
Small Indels.
We identified 26 high-confidence small indels (<50 bp), with a slight bias toward deletions (18 deletions vs. 8 insertions, P = 0.05, χ2 test) and a net loss of 58 bp, yielding an estimated indel mutation rate of 5.03 ± 0.99 × 10−12 per base per generation. The ratio of indels to SNMs observed was ∼0.03, which is consistent with previous yeast MA experiments that observed one indel for 33 SNMs (26) and zero indels for 19 SNMs (27). Observed indels included one tandem-repeat insertion and one tandem-repeat deletion. All but one occurred near simple repeats (Table 2).
Table 2.
Genomic location and affected bases of small indel mutations
| Chrm | Position | Type | Ref seq | Alt seq | Upstream seq | Downstream seq | |
| 2 | 8294 | 8304 | DEL | AGGGGTGCCGG | A | TGCCTATTAT | AAAAACCCTT |
| 4 | 49408 | 49408 | IN | G | GA | AGGGAAAAAT | AAAAAAAGGA |
| 4* | 67487 | 67487 | IN | A | ACTTTTT | CCCTTCACAT | CTTTTTCTTTTTCTTTTTCTTTT |
| 4 | 271299 | 271301 | DEL | TGA | T | CCACAGTAAT | AAATGTCAAAAAAA |
| 4 | 806439 | 806440 | DEL | AG | A | GGGGCGGCCTTGGCGGC | GGGGAGGCCTCTG |
| 4 | 914652 | 914682 | DEL | CGGCTGGTTTCTTTTCAGCTGGGGCTTTGGA | C | GTCTTTTTAG | TGTATGTGTGTATG |
| 5 | 384833 | 384833 | IN | A | ATGT | ATTCATGATG | TGTTGTTGTTGTTGTT |
| 6 | 162303 | 162304 | DEL | CT | C | TGCGCAGTTT | TTTTTTCTGATTTTTATTTTTTT |
| 7 | 728202 | 728203 | DEL | GT | G | ATGCTGTCTTG | TTTTGTATCGTCGTT |
| 7 | 904867 | 904867 | IN | T | TA | GGAATGGGTA | AAAAAATACAAGAA |
| 8 | 275558 | 275559 | DEL | CA | C | GGATACTACC | AAATGCCGTAT |
| 9 | 248934 | 248935 | DEL | AT | A | TGGTGTCGTT | TTTTTATTTTTATTTTTTTTTT |
| 9 | 351612 | 351613 | DEL | TA | T | TAAACGGATA | TTTTTTTTTGCGTCC |
| 10* | 121756 | 121759 | DEL | AAAG | A | GAAAGGAAAA | AAGTGTCCTTTT |
| 10 | 741950 | 741950 | IN | A | AC | CGACTCCAGCT | ACTGAGCGCATGGT |
| 11 | 364868 | 364870 | DEL | GCT | G | TTTTTTTGTCAAAC | GAGTAATAGAATATA |
| 11 | 409353 | 409354 | DEL | GA | G | ATTCAAACCT | CCCGGCTATAAGTTCTTTT |
| 12 | 35617 | 35626 | DEL | AATCCAGTAG | A | AAAGTGGGCT | TAATGAGGGA |
| 12 | 199746 | 199750 | DEL | GTCTT | G | AACCATTCTA | CTTTGGTGAAA |
| 12 | 311700 | 311700 | IN | A | AG | GCGACAGTGC | GGGGGACGATC |
| 13 | 444769 | 444769 | IN | C | CA | CACCCAAGGC | AAAAAAAAATT |
| 13 | 511778 | 511780 | DEL | TAA | T | TATAATATATTTTAATA | ATTTATTTATTAATA |
| 13 | 565730 | 565732 | DEL | CAG | C | TTTTTGAGAAA | AGGGAAGATCCACA |
| 13 | 838018 | 838019 | DEL | AT | A | CCCGGGAGAT | TTTTTTACTTTTGA |
| 13 | 910069 | 910070 | DEL | TA | T | CAACCACACT | TTACTATAACAGAT |
| 15 | 107864 | 107864 | IN | G | GT | GCGAAAGCGA | TTTTTGGAGA |
Indels are often found next to simple repeats.
Two of the indels involve tandem repeat sequences.
Although 75% of the yeast genome is genic, only 14 of the spontaneous indels (53%) were found in genic regions (P = 0.013, χ2 test). The paucity of genic indels was even more pronounced among the 63 indels that distinguish S288C reference genome from that of the MA ancestor strain, only 19 of which (30%) were located in genic regions (P < 0.001, χ2 test). Although indels within genic sequences appeared to be more tolerated under an MA regime, a significant number remained significantly deleterious and could not be observed.
Single Nucleotide Mutations.
We identified a total of 867 SNM events after excluding strains with complex mutations, genomic regions prone to mapping errors, and double mutations. From raw SNM calls, visual inspection revealed five lines (MA4, MA8, MA60, MA61, and MA70) with runs of SNMs called consecutively in a short genomic region. Reads mapped to such regions suggested complex mutations that resulted in faulty variant calling. Because it was difficult to determine the exact number of mutational events that occurred at these regions, these five lines were removed from analysis and did not contribute to the final SNM set. In the remaining 140 lines, we excluded 600 kb of the genome that comprise annotated repeats, bases that were too low in coverage (<8×) for SNP calling (15% of the total), and six SNMs that appeared to be double mutations. [The likelihood of two SNMs occurring next to each other by chance in our dataset was extremely low (P < 0.001).]
The final set of 867 SNMs formed the largest category of mutations by far and allowed us to estimate the genome-wide single-nucleotide mutation rate at 1.67 ± 0.04 × 10−10 per base per generation (Dataset S1). This estimate is lower than previous haploid yeast mutation rate estimates from 33 genome-wide events (26) and gene-specific estimates (11, 32). However, the estimate is close to previous vegetative diploid yeast estimates from 19 genome-wide events, in line with the expectation that vegetative diploid yeast is genetically more stable than other states (27).
The three pairs of adjacent mutations made up 0.35% of all SNMs. Although there were only three instances observed, to our knowledge, this was the first time such double mutations were observed in eukaryotic MA experiments. Our double-mutation rate is similar to a previous estimate of 0.3% from primate sequences (33) but lower than another estimate of 1–10% based on ancient, conserved coding sequences (34). We also found two SNMs that appeared to be homozygous derived with high sequencing depth (37× and 46×, respectively), but no reads supporting the reference allele.
We compared the distribution patterns for the 867 MA SNMs with the 3,137 fixed differences between the ancestor of the MA lines and the S288C reference genome. We did not observe a difference in the proportion of SNMs occurring within genic or nongenic regions in the two datasets (proportion of fixed changes in coding sequences = 75.5 ± 1.8%, proportion of SNMs in coding sequences = 74.0 ± 3.4%, P = 0.29, χ2 test). However, within coding sequences, where ∼75% of all changes should be nonsynonymous if the mutations occurred randomly and were not subject to selection, the two datasets behaved differently. We found that, whereas fixed differences between the MA ancestor and S288C reference did show a clear deviation from this expectation with a significant deficit of nonsynonymous changes at 40.2 ± 2.1% (P < 0.001, χ2 test), SNMs acquired during MA showed no signals of selection and were equally likely to be synonymous or nonsynonymous (proportion of nonsynonymous SNMs = 75.4 ± 3.9%, P = 0.514, χ2 test). It appeared that the MA SNM dataset, unlike indels and aneuploidy, was not affected by selection in the MA regime and was an accurate reflection of the true SNM spontaneous mutation rate and spectrum.
Low False-Positive and False-Negative Rates in Calling SNMs.
SNM calling in low-coverage diploids can be difficult because one allele may never be sampled by chance, or sampled at such low frequency that naive variant callers might generally classify them as sequencing errors. We used a stringent SNM calling procedure (Materials and Methods), but it was essential to quantify both false-positive and false-negative rates experimentally.
To assess the rate of false positives, we first used Sanger resequencing to verify 53/56 SNMs that were called in five MA lines. PCR products were not obtainable for the remaining 3 SNMs. These lines were chosen because they contained recessive-lethal mutations. However, in other aspects, these 53 SNMs should be representative of SNMs in other strains. We did not find any inaccurately called SNMs, suggesting that the false-positive rate was low in our SNM set.
To estimate both the false-negative rate and further refine our estimate of the false-positive rate, we further sequenced complete tetrads from 19 of the MA lines (76 haploid lines, 1,520× coverage total, 20× coverage on average per line). Our reasoning was that it is both much easier to call SNMs in haploids, and true SNMs should be present in all reads from two of the spores in a tetrad while being completely absent in all reads from the other two. Only positions covered to >8× sequencing depth in diploids and >4× sequencing depth in all four haploid spores were used for error-rate estimation. In such positions, 126 SNMs were called in both diploids and tetrads (with 2:2 segregation), six SNMs were called only in diploids, and nine SNMs were called only in tetrads. All nine false-negative SNMs had at least one read supporting the alternate allele in diploid sequences but did not pass the postvariant calling filter for various reasons. These nine SNMs were added to our list of identified SNMs. The six SNMs that were called only in diploids were completely absent in all of the haploid spores in the tetrad, despite high-confidence calling and multiple reads supporting the alternate allele in diploid-sequencing data. Thus, these may represent either false-positive SNMs or additional changes that occurred after the final generation of the MA experiment in the diploid lineages used for sequencing. To be conservative, these six SNMs were removed from the list of SNMs. The estimated false-negative rate of diploid SNM calls was 6.8% whereas the estimated false-positive rate was conservatively estimated at 4.8% (assuming SNMs called in diploids but not tetrads were indeed false positives rather than post-MA mutations).
SNM Mutation Spectrum and Biases.
We tested whether SNM rates varied across lines or chromosomes. Because genome coverage varied by line and was correlated with the number of discovered SNMs, the raw number of SNMs in a line did not reflect the true mutation rate in that line. To obtain an unbiased distribution of SNMs in a line, we limited this portion of the analysis to the 97 lines with >60% of the genome covered to at least 8× read depth. Within these lines, only positions covered to at least 8× in all 97 lines were considered. For the 256 SNMs that fall within these regions, we then tabulated SNM counts for each line. The distribution of this adjusted number of SNMs per line fit both Poisson (P = 0.07, G test) and negative binomial (P = 0.1, G test) distributions but was clearly not binomial (P < 0.001, G test) (Fig. 1). In addition, the number of SNMs per chromosome followed a strong linear correlation with chromosome size (adjusted R2 = 0.94, P < 0.001) (Fig. 2). Both results suggested that per-base-pair mutation rate did not vary substantially across lines or chromosomes on a genome-wide scale. However, note that the five lines excluded from the analysis for carrying regions with complex mutations suggest that mutation rates may vary greatly within specific regions, and different techniques would be required to clarify the chronological order and total number of events in such regions.
Fig. 1.
Histogram of SNM counts per line in 97 MA lines with >60% of the genome covered to >8× read depth. Only positions sequenced to >8× read depth in all 97 lines were considered. A total of 256 SNMs were called in such regions in the 97 lines. Histogram shape may be Poisson (P = 0.07, G test) or negative binomial (P = 0.1, G test), but not binomial (P < 0.001, G test).
Fig. 2.
(x axis) Number of mutations observed per chromosome. (y axis) Length of chromosome. Number of mutations observed on a chromosome is strongly correlated with chromosomal length (adjusted R2 = 0.94, P < 0.001).
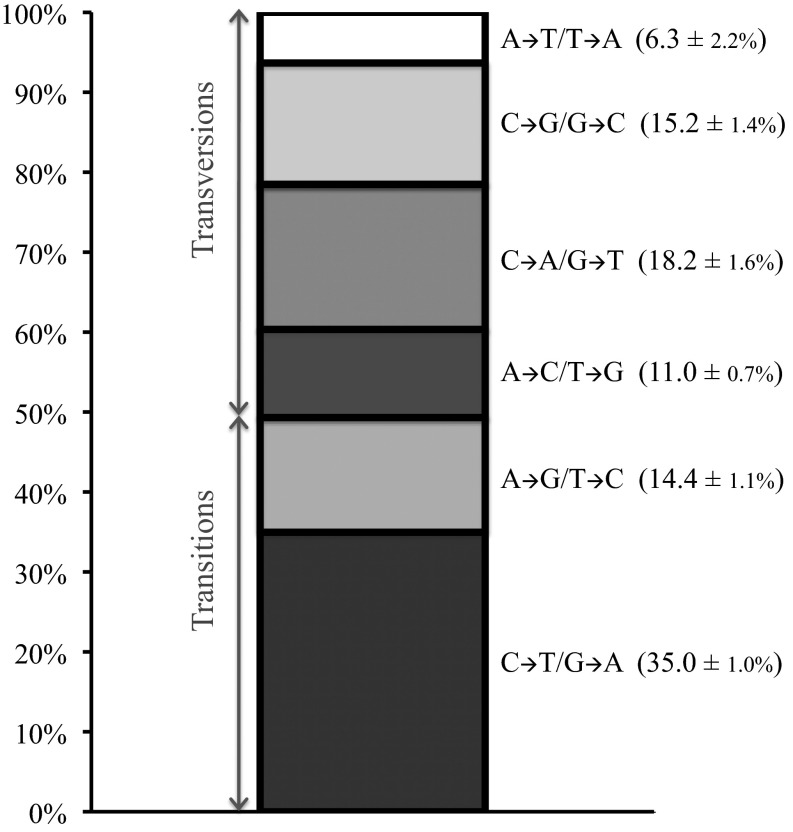
In all species assayed to date, two patterns appear to be universal—a high transition per transversion ratio (Ts/Tv) and a GC-to-AT mutation bias that is true within both transitions and transversions (14, 35). If all six base mutation types are equally frequent, relative mutation rates should be 0.17 each. In our data, we estimated the Ts/Tv bias of the mutational process alone to be 0.95, higher than previous estimates of 0.6–0.7 from a much smaller number of events (26, 27), but lower than the Ts/Tv of 2.96 among the fixed mutations between the ancestral and reference strains. This difference suggested that much of the Ts/Tv bias in polymorphism and divergence was driven by natural selection and not mutation (Fig. 3).
Fig. 3.
Relative mutation rates of each of six possible nucleotide changes.
The remaining deviation from an expected Ts/Tv of 0.5 if all mutation types occurred with equal probabilities was entirely driven by C-to-T transitions, which occurred at twice the rate of the average mutation (relative rate 0.35 ± 0.01, P < 0.001, two-tailed Z test). The other transition, T to C, did not occur at a particularly high rate (relative mutation rate 0.144 ± 0.011, P = 0.238, two-tailed Z test).
A strong GC-to-AT bias was also observed, driven by both C-to-T transitions and C-to-A changes within transversions (relative mutation rate 0.182 ± 0.016, P < 0.001, two-tailed Z test). From observed rates, we expect a mutation-driven equilibrium genomic GC content of 32%. This percentage is lower than the observed genome-wide GC content of 38% and is consistent with previous claims (26) that the yeast genome is unlikely to be in GC-content equilibrium from mutational bias alone.
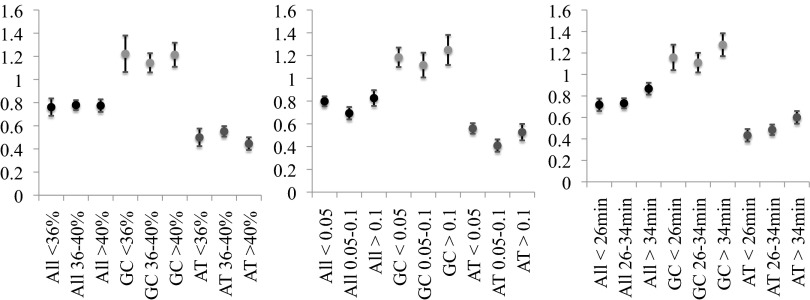
We further tested whether GC content (per 1-kb window), transcription rate (36), and replication time during the cell cycle (37) affected local mutation rate. SNM rates were calculated for genomic regions falling into three categories for each tested variable (low, medium, or high for transcription rate; early, midcycle, or late for replication time). We found no correlation in mutation rate with GC content after controlling for local coverage (P = 0.999, Pearson’s correlation test), a variable that is correlated with GC content and with the probability of identifying a mutation (Fig. 4, Left). We also did not observe changes in mutation rate with transcription rate (P = 0.8707, Pearson’s correlation test) (Fig. 4, Center). We found a possible weak trend where late-replicating sites had ∼20% higher mutation rates than early-replicating sites, but this difference was not statistically significant (P = 0.123, two-tailed Z test) (Fig. 4, Right).
Fig. 4.
Relative mutation rates at all bases, only G/C bases, and only A/T bases with respect to local (Left) 1-kb GC content, (Center) transcription rate, and (Right) replication time during cell cycle. Categories were defined by obtaining bins that contain roughly equal numbers of SNMs.
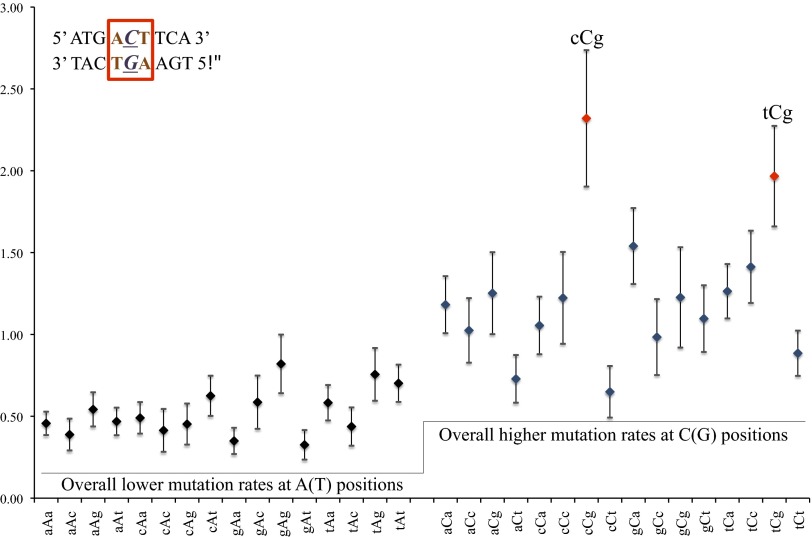
With the large number of SNMs identified, we were able to explore the possible influence of neighboring bases on mutation rate. Neighboring bases were defined as the ones immediately preceding and following the position of interest. Every position in the genome, in conjunction with its neighboring bases, was assigned to one of 64 possible triplets. We ignored strand orientation and folded the spectrum such that complementary triplets belong to the same category (for example, GCT and its complement AGC both contribute to mutation rate at the central C/G position under category GCT) and estimated relative mutation rates in each neighbor context. As expected, all GC bases had a higher mutation rate than AT bases. Mutation rates at AT bases in all environments were uniform (P = 0.123, G test), but mutation rates at GC bases were not (P < 0.001, G test). Two GC environments in particular appeared to have mutation rates twice as high as at other GC environments: CCG (P = 0.028, two-tailed Z test) and TCG (P = 0.015, two-tailed Z test) (Fig. 5).
Fig. 5.
(y axis) Relative mutation rate. (x axis) Neighbor environment. Neighbor-dependent mutation rate is defined as the effects of immediate flanking nucleotides (e.g., bases a and t in an aCt environment) on mutation rate at base of interest (base C in an aCt environment). Environment classes represent mutation rates regardless of strand orientation (aCt class includes overall mutation rate at aCt and aGt positions). Average mutation rate of 1.24 (1.11 when excluding cCg and tCg contexts) at C/G bases shows clear overall elevation over a corresponding rate of 0.52 at A/T bases. In addition, two environments (cCg and tCg) show additional elevations in mutation rate.
Strongly Deleterious Mutations.
Although seven MA lines could not be revived after a standard laboratory freeze–thaw cycle and were necessarily excluded from analyses, they provided an insight into the dominant-lethal mutation rate—in this context referring to all strongly deleterious mutations of sh > 0.1 that would be missed in the final dataset even under a vegetative MA growth regime. Although such dominant-lethal mutations can never be observed in the environment (environment A) from which the organism is sampled, by transferring lines carrying mutations accumulated in environment A to a second environment (environment B), the percentage of lines that did not survive (and thus carry dominant-lethal mutations in the new environment) is a conservative estimate of the rate at which environment-specific dominant-lethal mutations arise, not including mutations impeding critical functions that would have rendered the organism inviable in any environment. The seven MA lines that accumulated mutations during culturing at 30 °C on rich, solid medium (environment A) that were lethal post freeze-thaw (environment B) place the environment-dependent dominant-lethal mutation rate at 2.2 ± 0.8 × 10−5 events per diploid genome per generation.
Similarly, through segregation patterns of tetrads from diploid lines, we were able to obtain an estimate of the recessive lethal mutation rate—in this context referring to all mutations that result in inviable haploid spores. All 145 surviving lines were sporulated according to the protocol previously described in ref. 38. At least 10 lines carried recessive-lethal mutations and consistently produced only two surviving spores per tetrad. Although it was impossible to identify dominant-lethal mutations, it was possible to do so for recessive lethals. For eight of the lines that consistently produced only two visible colonies per tetrad, a pooled library of 96 surviving spores from 48 tetrads was sequenced to an average depth of 60× (Materials and Methods). Recessive-lethal mutations should be the only mutations present in the diploid parent but absent in surviving spores. In five of the lines (MA18, MA27, MA75, MA97, and MA98), we identified a single nonsense mutation in an essential gene (Table 3). MA36 had a 30-bp deletion within an essential gene that most likely led to a nonfunctional protein (Table 3). All six of the putative lethal mutations were verified by Sanger sequencing. In MA70 and MA105, no candidates were found because causal mutations either were not covered by sufficient sequencing reads or were from classes of mutations not assayed. MA70 was one of the lines we later identified as having complex mutations, and it is possible that these difficult-to-map mutations were responsible for the recessive lethality in this strain. The 10 lines carrying such mutations place recessive-lethal mutations at an estimated rate of 3.2 ± 1 × 10−5 per diploid genome per generation.
Table 3.
The genomic positions and affected genes of recessive-lethal mutations in six MA diploid lines, ordered by line ID number
| Line | Chrm | Position | Gene | Amino acid | Post mutation |
| 18 | II | 87066 | YBL074C | Q | Stop |
| 36 | IV | 914652 | YDR224C | — | 30-bp deletion |
| 27 | V | 192081 | YER018C | G | Stop |
| 75 | XV | 260436 | YOL034W | Q | Stop |
| 97 | IV | 438662 | YDL007W | E | Stop |
| 98 | XV | 927326 | YOR326W | Q | Stop |
In addition, a conservative strongly deleterious mutation rate, including all mutations that exhibit large fitness costs within the yeast life cycle, could be estimated from missing mutations in mutation classes that showed signals of selection, such as aneuploidy and small indels, as well as lines showing unusual segregation patterns during sporulation. From aneuploidy, ∼27 whole-chromosome deletions were missing compared with whole-chromosome duplications. From small indels, ∼22 indels were missing from genic regions compared with rates within nongenic regions. These are both conservative estimates, with the assumption that all chromosomal gains and noncoding indels had mild fitness effects and were retained in the MA lines. In addition, four MA lines were unable to produce any spores entirely, and another 36 showed either low viability or otherwise unusual segregation patterns in their spores. These 40 lines likely carry mutations of large deleterious effects. Together with previously identified dominant and recessive lethal mutations, the final estimate for strongly deleterious mutation rate is 3.3 ± 0.3 × 10−4 per diploid genome per generation, a tenth of the total observed mutation rate, which is 3.3 ± 0.1 × 10−3 per diploid genome per generation.
Discussion
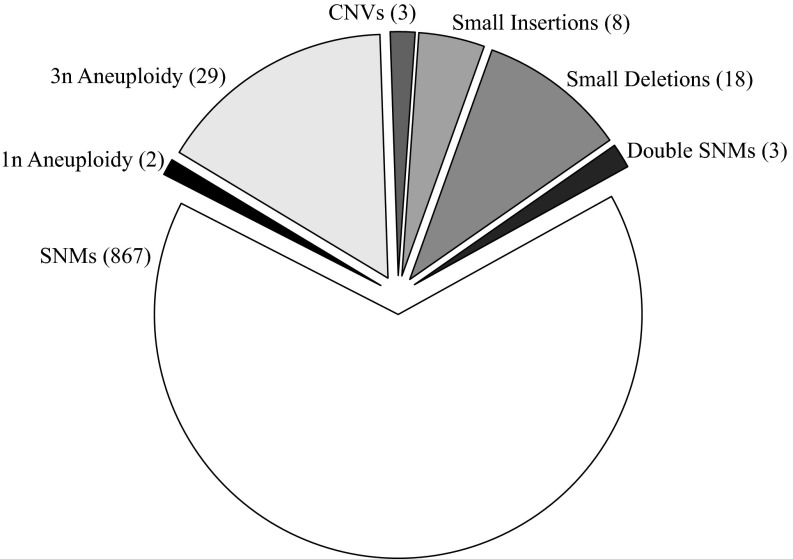
We observed 924 high-confidence spontaneous mutations including 867 single-nucleotide changes, 3 double mutations, 26 small indels under 50 bp (8 insertions, 18 deletions), 31 whole-chromosome copy-number changes (29 chromosome gains, 2 chromosome losses), and 3 large copy-number changes >30 kb (Fig. 6). This dataset, accumulated over a total of ∼311,000 mitotic generations, is the largest set of spontaneous mutations identified for any wild-type organism to our knowledge.
Fig. 6.
Summary of all mutations identified. Numbers in parentheses represent numbers of events called in each mutation class.
We chose to analyze vegetative lines from a diploid ancestral strain because such lines shield all recessive deleterious mutations including recessive lethals from selection. The advantages of using diploid lines outweighed the greater difficulty of analyzing diploid genome sequences. The most serious issue with analyzing diploids is that mutation calls must be more conservative than when analyzing haploids, but we were largely able to overcome this problem by taking advantage of the large number of MA lines to distinguish between true SNMs and sequencing errors.
In one mutation class—SNMs—we found no clear signals of selection and were able to identify enough events that an analysis of context-dependent effects on mutation rate was possible. We analyzed the effect of neighboring bases on mutation rate and found two classes of neighbor contexts, CCG and TCG, with mutation rates twice as high as mutation rates at GC base pairs in other neighbor contexts. The best-known example of neighbor-dependent mutation rate elevation at CG nucleotides is in 5-methylcytosine CG positions in mammals. Budding yeast is generally assumed to have no methylation, given its lack of a gene encoding DNA methyltransferase and difficulties in chemically detecting methylated molecules (39–41). However, a recent study using air chromatography found ∼0.364% methylation at cytosines in S. cerevisiae (42). If methylated cytosines in yeast have similar 10×–50× elevations in mutation rate as in humans (43–45), 1/5–1/25 of all CCG and TCG sites would have to be methylated for the observed ∼2× overall increase in mutation rate at these sites. Taking 0.364% to be the methylation rate in S. cerevisiae and assuming all methylation takes place within CCG and TCG contexts where we found elevated mutation rates, which correspond to ∼3% of the analyzed sequences, this value leads to an estimate of ∼1/22 of CCG and TCG sites being methylated. These calculations suggest that methylation at CCG and TTG sites is a parsimonious explanation for the observed elevation of mutation rate in these contexts and that methylation in S. cerevisiae might be confined to two specific contexts.
We further explored three potential factors that have been found to affect SNM mutation rate in non-MA experiments. First, local GC content is known to be associated with gene density, codon use, substitution rate, and mutation rate at CG sites (43, 46–48). We found no genome-wide correlation in mutation rate with local GC content in yeast, suggesting that GC content-related effects on substitution rate are largely due to postmutation selection or selection-like processes such as biased gene conversion. Next, we looked at highly transcribed genes that are thought to have elevated mutation rates due to transcription-associated mutagenesis (49–52). In our data, we found no genome-wide correlation in mutation rate with transcription rate. Finally, replication time during the cell cycle determines exposure to different repair mechanisms of varying fidelity (12, 53–56). We observed a weak and statistically nonsignificant trend where late-replicating sites have ∼20% higher mutation rates than early-replicating sites, close to the 30% increase seen previously in ref. 55, but more modest than the sixfold increase found in ref. 12. To our knowledge, this is the first time that such analyses have been carried out on yeast MA data.
We also observed patterns that suggest that different mutation classes may contribute strongly deleterious mutations at comparable counts. In our analysis of dominant-lethal mutations, we found at least 27 aneuploidy events (42% of all aneuploidy) and 22 small indels (46% of all indels) that were selected out of the lines even under the low-selection MA regime (Fig. S2). Because the total number of SNMs observed was much larger than aneuploidies and indels, and strongly deleterious events were detected through their absence from the final dataset, a similar absolute number of missing SNMs (∼3%) would not have been detectable. However, a similar trend was found in recessive-lethal mutations that consisted of two whole chromosome losses, one small indel, and five SNMs. The fact that aneuploidy, small indels, and SNMs were all observed at similar frequencies among dominant and recessive lethal mutations, despite SNMs being 30 times as frequent as aneuploidies and indels, holds interesting implications, for the evolution of individual and compounded mutation rates of mutation classes (57, 58), that would require more theoretical and experimental exploration.
Although the large number of mutations identified in this dataset allowed precise estimates of mutational biases (such as Ts/Tv ratio and AT bias, context-specific SNM rates, mutation rates in relatively rare mutation classes such as aneuploidy and small indels, and the relative contributions of each class to strongly deleterious mutations), there are remaining questions that would benefit greatly from even more data. For example, the yeast genome consists mostly of coding sequences, and dynamics unique to such regions may drive the observed elevation of mutation rates in CCG and TCG contexts. Larger numbers of SNMs in noncoding regions would reveal whether a different pattern exists within noncoding regions whereas more events per context would also clarify whether rates within individual contexts are driven by specific nucleotide changes, some subset, or overall elevation of mutation rate. In addition, more power is required to accurately estimate the fine scale effects of replication time on local mutation rate, in addition to other possible factors. Although we observed more aneuploidy and small indel events than ever before, there were too few total events to perform the analyses possible in SNMs. Specifically, small indels showed a potential bias toward deletions and net loss of genic material, but it was impossible to clarify whether this bias was due to selection against strongly deleterious mutations or a true bias in the mechanisms that generate indels (59–64). Lastly, measuring the individual fitness effects of each new mutation is also critical (65). A larger dataset of mutations and their fitness effects, not infeasible in the near future, would allow us to better answer these outstanding questions.
Materials and Methods
Mutation Accumulation Lines.
The MA lines were previously described in refs. 28 and 29. In brief, the ancestral strain for the experiment was created from a haploid strain of genotype ade2, lys2-801, his3-∆D200, leu2–3.112, ura3–52, ho. The haploid strain was transformed with an HO-expressing plasmid to create the diploid ancestor strain, after which the plasmid was removed. The ancestor is homozygous at all except the mating-type locus. For mutation accumulation, 151 lines descended from the ancestor were propagated independently on YPD solid medium (1% yeast extract, 2% peptone, 2% dextrose, 2% agar) and put through a single-cell bottleneck every ∼20 generations (48 h) by streaking to single colonies, for a total of 100 bottlenecks, or ∼2,062 generations (200 d). Mitochondrial petite mutations that often accumulate in such experiments and cause problematic results were screened out using the color assay made possible by the presence of the ade2 mutation in the propagating strain. In addition, picking the colony closest to a location marked on the plate in advance eliminated any preferential selection for colony morphology during bottlenecks. For 19 of the MA lines, complete tetrads, in which all four spores were viable, were also obtained (38).
Sequencing.
All lines were cultured from frozen stock on supplemented YPD solid medium (1% yeast extract, 2% peptone, 2% dextrose, 2% agar, 0.05 mg/mL Ade, 0.05 mg/mL Trp) for 2 d. When visible, colonies were inoculated into 3-mL liquid cultures (same recipe without agar) overnight on a shaking 30 °C incubator until saturation. Cells were then pelleted for DNA extraction.
MA diploids and tetrads.
MA diploid DNA extractions were carried out using the YeaSTAR Kit (Zymo Research), following steps in the protocol with chloroform. For the 19 complete tetrads, DNA extractions were carried out using the ZR-96 Fungal/Bacterial DNA Kit (Zymo Research). Library making for both datasets was outsourced to Moleculo, and resulting Illumina 100-bp paired-end libraries (with unique barcodes for each line) were pooled and run on four lanes (145 MA diploid lines, ∼10× coverage per line) and 1/2 lane (19 MA complete tetrads, 76 lines, ∼20× coverage per line) on HiSeq. 2000 machines.
MA surviving spore pools.
For the pools of 96 surviving spores from each of eight MA lines carrying recessive lethals, cultures of haploid lines were pooled before DNA extraction. Cells in the final 288-mL liquid cultures were pelleted for DNA extraction. DNA extractions were carried out using the Qiagen Genomic Q-Tip 100 following standard protocols. Then, 2 µg of DNA from each pool was used for 100-bp paired-end library construction following standard protocols. The eight pools were individually barcoded and sequenced on a single lane of Illumina HiSeq. 2000 to an average of ∼60× sequencing depth per pool.
Mapping and SNM/Small Indel Identification.
Mapping of sequence reads from each library was carried out in two stages. Fastq files were first mapped to the reference genome with BWA v0.5.9 (66), sorted and indexed with SAMtools v0.1.18 (67), and assigned MA line ID with Picard Tools v1.55. Duplicated read pairs were removed, and remaining reads were locally realigned with GATK v2.1–8 (68). UnifiedGenotyper was used to call candidate variants across all samples simultaneously. The resulting VCF file was filtered for variants called as derived homozygous across all sequenced lines. These variants are fixed differences between the MA ancestral line and the S288C reference genome and were subsequently incorporated into the reference genome to generate an ancestral MA reference genome. The same mapping process was repeated on this ancestral MA reference to fully eliminate confounding influences from these fixed differences that may affect mapping accuracy. SNM and indel calling was carried out independently on the 145 MA diploid lines, the 19 MA tetrads, and the eight pools of spores. The final spontaneous point mutation and small indel calls were filtered with the strong prior expectation that each would be present only in a single MA line. In the 145 MA diploids and eight pools of spores, a minimum of eight reads covering the position and at least two reads supporting the alternative allele was required for variant calling. In the 19 full tetrads, because lines were haploid, a minimum of four reads covering the position and >90% of all reads supporting the same allele was required for variant calling. Around 600 kb of the genome—these regions were annotated in the SGD database as simple repeats, centromeric regions, telomeric regions, or LTRs (SGD project; http://downloads.yeastgenome.org/curation/chromosomal_feature/SGD_features.tab, downloaded August 4, 2012)—were excluded from analysis due to their susceptibility to mismapping and associated miscalls. All parameters and used commands are available in Dataset S1.
Sanger Verification.
Five recessive-lethal mutation candidates from eight MA lines carrying recessive lethals were verified by Sanger sequencing. In addition, five of the eight lines were randomly picked, and all mutations but three were verified by Sanger sequencing (for those three SNMs, we were unable to obtain PCR products with two different sets of PCR primers). All 53 SNMs verified were true mutations (for SNPs and corresponding primer sequences, see Supporting Information).
Aneuploidy Identification.
Average coverage was computed for each chromosome per line, excluding an 18-kb region consisting of two consecutive 9.1-kb rDNA locus repeats on Chromosome XII associated with ERCs (extrachromsomal rDNA circles) that show consistently excessive coverage. Whole-chromosome aneuploidies were called if average coverage of a chromosome differs more than 35% from other chromsomes in the same line (likelihood P < 0.001, χ2 test). Whole-genome duplications that would create lines with 3N chromosome content were ruled out as all lines were sporulated and produced viable spores with 2:2 segregation of alpha and a mating types. Subsequent back crossing and sporulation of F1 spores were also conducted for a large number of the lines and did not suggest the presence any 4N lines.
Supplementary Material
Acknowledgments
We thank the reviewers for their immensely valuable feedback. We thank Gavin Sherlock, Carlos Bustamante, Hua Tang, Anna-Sophie Fiston-Lavier, Sasha Levy, Dan Kvitek, Katja Schwartz, Barbara Dunn, Way Sung, Ziming Weng, Sandeep Venkataram, and Diamantis Sellis for their scientific expertise and invaluable input. We thank members of the D.A.P. laboratory for helpful discussions. We thank Ruth Hershberg, Nandita Garud, Alan Bergland, Zoe Assaf, David Lawrie and Phillip Messer for comments that helped improve an earlier draft. We thank Stanford Bio-X for computational support, and Stanford Center for Genomics and Personalized Medicine (SCGPM) for sequencing resources. Y.O.Z. was supported by the A*STAR National Science Scholarship PhD. M.L.S. was supported by National Institutes of Health (NIH) Grant R01GM097415. D.W.H. was supported by NIH Grant R01GM097415. D.A.P. was supported by NIH Grants R01GM100366 and R01GM097415.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323011111/-/DCSupplemental.
References
- 1.Ellegren H, Smith NG, Webster MT. Mutation rate variation in the mammalian genome. Curr Opin Genet Dev. 2003;13(6):562–568. doi: 10.1016/j.gde.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Ochman H. Neutral mutations and neutral substitutions in bacterial genomes. Mol Biol Evol. 2003;20(12):2091–2096. doi: 10.1093/molbev/msg229. [DOI] [PubMed] [Google Scholar]
- 3.Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 4.Lawrie DS, Petrov DA, Messer PW. Faster than neutral evolution of constrained sequences: The complex interplay of mutational biases and weak selection. Genome Biol Evol. 2011;3:383–395. doi: 10.1093/gbe/evr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kousathanas A, Oliver F, Halligan DL, Keightley PD. Positive and negative selection on noncoding DNA close to protein-coding genes in wild house mice. Mol Biol Evol. 2011;28(3):1183–1191. doi: 10.1093/molbev/msq299. [DOI] [PubMed] [Google Scholar]
- 6.Mukai T, Cockerham CC. Spontaneous mutation rates at enzyme loci in Drosophila melanogaster. Proc Natl Acad Sci USA. 1977;74(6):2514–2517. doi: 10.1073/pnas.74.6.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell LB, Russell WL. Spontaneous mutations recovered as mosaics in the mouse specific-locus test. Proc Natl Acad Sci USA. 1996;93(23):13072–13077. doi: 10.1073/pnas.93.23.13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148(4):1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat. 2003;21(1):12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- 10.Baer CF, Miyamoto MM, Denver DR. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat Rev Genet. 2007;8(8):619–631. doi: 10.1038/nrg2158. [DOI] [PubMed] [Google Scholar]
- 11.Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178(1):67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang GI, Murray AW. Mutation rates across budding yeast chromosome VI are correlated with replication timing. Genome Biol Evol. 2011;3:799–811. doi: 10.1093/gbe/evr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itsara A, et al. De novo rates and selection of large copy number variation. Genome Res. 2010;20(11):1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci USA. 2010;107(3):961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conrad DF, et al. 1000 Genomes Project Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43(7):712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell CD, Eichler EE. Properties and rates of germline mutations in humans. Trends Genet. 2013;29(10):575–584. doi: 10.1016/j.tig.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baer CF, et al. Comparative evolutionary genetics of spontaneous mutations affecting fitness in rhabditid nematodes. Proc Natl Acad Sci USA. 2005;102(16):5785–5790. doi: 10.1073/pnas.0406056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haag-Liautard C, et al. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature. 2007;445(7123):82–85. doi: 10.1038/nature05388. [DOI] [PubMed] [Google Scholar]
- 20.Keightley PD, et al. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 2009;19(7):1195–1201. doi: 10.1101/gr.091231.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327(5961):92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denver DR, et al. Variation in base-substitution mutation in experimental and natural lineages of Caenorhabditis nematodes. Genome Biol Evol. 2012;4(4):513–522. doi: 10.1093/gbe/evs028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ness RW, Morgan AD, Colegrave N, Keightley PD. Estimate of the spontaneous mutation rate in Chlamydomonas reinhardtii. Genetics. 2012;192(4):1447–1454. doi: 10.1534/genetics.112.145078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rutter MT, et al. Fitness of Arabidopsis thaliana mutation accumulation lines whose spontaneous mutations are known. Evolution. 2012;66(7):2335–2339. doi: 10.1111/j.1558-5646.2012.01583.x. [DOI] [PubMed] [Google Scholar]
- 25.Schrider DR, Houle D, Lynch M, Hahn MW. Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster. Genetics. 2013;194(4):937–954. doi: 10.1534/genetics.113.151670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch M, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci USA. 2008;105(27):9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishant KT, et al. The baker’s yeast diploid genome is remarkably stable in vegetative growth and meiosis. PLoS Genet. 2010;6(9):e1001109. doi: 10.1371/journal.pgen.1001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joseph SB, Hall DW. Spontaneous mutations in diploid Saccharomyces cerevisiae: More beneficial than expected. Genetics. 2004;168(4):1817–1825. doi: 10.1534/genetics.104.033761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall DW, Mahmoudizad R, Hurd AW, Joseph SB. Spontaneous mutations in diploid Saccharomyces cerevisiae: Another thousand cell generations. Genet Res. 2008;90(3):229–241. doi: 10.1017/S0016672308009324. [DOI] [PubMed] [Google Scholar]
- 30.Kumaran R, Yang SY, Leu JY. Characterization of chromosome stability in diploid, polyploid and hybrid yeast cells. PLoS ONE. 2013;8(7):e68094. doi: 10.1371/journal.pone.0068094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu J, Pavelka N, Bradford WD, Rancati G, Li R. Karyotypic determinants of chromosome instability in aneuploid budding yeast. PLoS Genet. 2012;8(5):e1002719. doi: 10.1371/journal.pgen.1002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunz BA, Ramachandran K, Vonarx EJ. DNA sequence analysis of spontaneous mutagenesis in Saccharomyces cerevisiae. Genetics. 1998;148(4):1491–1505. doi: 10.1093/genetics/148.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith NGC, Webster MT, Ellegren H. A low rate of simultaneous double-nucleotide mutations in primates. Mol Biol Evol. 2003;20(1):47–53. doi: 10.1093/molbev/msg003. [DOI] [PubMed] [Google Scholar]
- 34.Averof M, Rokas A, Wolfe KH, Sharp PM. Evidence for a high frequency of simultaneous double-nucleotide substitutions. Science. 2000;287(5456):1283–1286. doi: 10.1126/science.287.5456.1283. [DOI] [PubMed] [Google Scholar]
- 35.Hershberg R, Petrov DA. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 2010;6(9):e1001115. doi: 10.1371/journal.pgen.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelechano V, Chávez S, Pérez-Ortín JE. A complete set of nascent transcription rates for yeast genes. PLoS ONE. 2010;5(11):e15442. doi: 10.1371/journal.pone.0015442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raghuraman MK, et al. Replication dynamics of the yeast genome. Science. 2001;294(5540):115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- 38.Hall DW, Joseph SB. A high frequency of beneficial mutations across multiple fitness components in Saccharomyces cerevisiae. Genetics. 2010;185(4):1397–1409. doi: 10.1534/genetics.110.118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antequera F, Tamame M, Villanueva JR, Santos T. DNA methylation in the fungi. J Biol Chem. 1984;259(13):8033–8036. [PubMed] [Google Scholar]
- 40.Proffitt JH, Davie JR, Swinton D, Hattman S. 5-Methylcytosine is not detectable in Saccharomyces cerevisiae DNA. Mol Cell Biol. 1984;4(5):985–988. doi: 10.1128/mcb.4.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkinson CR, Bartlett R, Nurse P, Bird AP. The fission yeast gene pmt1+ encodes a DNA methyltransferase homologue. Nucleic Acids Res. 1995;23(2):203–210. doi: 10.1093/nar/23.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang Y, Gao XD, Wang Y, Yuan BF, Feng YQ. Widespread existence of cytosine methylation in yeast DNA measured by gas chromatography/mass spectrometry. Anal Chem. 2012;84(16):7249–7255. doi: 10.1021/ac301727c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fryxell KJ, Moon WJ. CpG mutation rates in the human genome are highly dependent on local GC content. Mol Biol Evol. 2005;22(3):650–658. doi: 10.1093/molbev/msi043. [DOI] [PubMed] [Google Scholar]
- 44.Hodgkinson A, Ladoukakis E, Eyre-Walker A. Cryptic variation in the human mutation rate. PLoS Biol. 2009;7(2):e1000027. doi: 10.1371/journal.pbio.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernando-Herraez I, et al. Dynamics of DNA methylation in recent human and great ape evolution. PLoS Genet. 2013;9(9):e1003763. doi: 10.1371/journal.pgen.1003763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharp PM, Lloyd AT. Regional base composition variation along yeast chromosome III: Evolution of chromosome primary structure. Nucleic Acids Res. 1993;21(2):179–183. doi: 10.1093/nar/21.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murakami Y, et al. Analysis of the nucleotide sequence of chromosome VI from Saccharomyces cerevisiae. Nat Genet. 1995;10(3):261–268. doi: 10.1038/ng0795-261. [DOI] [PubMed] [Google Scholar]
- 48.Mugal CF, Arndt PF, Ellegren H. Twisted signatures of GC-biased gene conversion embedded in an evolutionary stable karyotype. Mol Biol Evol. 2013;30(7):1700–1712. doi: 10.1093/molbev/mst067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair (Amst) 2007;6(9):1285–1296. doi: 10.1016/j.dnarep.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nat Rev Genet. 2012;13(3):204–214. doi: 10.1038/nrg3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park C, Qian W, Zhang J. Genomic evidence for elevated mutation rates in highly expressed genes. EMBO Rep. 2012;13(12):1123–1129. doi: 10.1038/embor.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X, Zhang J. No gene-specific optimization of mutation rate in Escherichia coli. Mol Biol Evol. 2013;30(7):1559–1562. doi: 10.1093/molbev/mst060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hawk JD, Stefanovic L, Boyer JC, Petes TD, Farber RA. Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proc Natl Acad Sci USA. 2005;102(24):8639–8643. doi: 10.1073/pnas.0503415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamatoyannopoulos JA, et al. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41(4):393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agier N, Fischer G. The mutational profile of the yeast genome is shaped by replication. Mol Biol Evol. 2012;29(3):905–913. doi: 10.1093/molbev/msr280. [DOI] [PubMed] [Google Scholar]
- 56.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26(8):345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sung W, Ackerman MS, Miller SF, Doak TG, Lynch M. Drift-barrier hypothesis and mutation-rate evolution. Proc Natl Acad Sci USA. 2012;109(45):18488–18492. doi: 10.1073/pnas.1216223109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petrov DA, Hartl DL. Pseudogene evolution and natural selection for a compact genome. J Hered. 2000;91(3):221–227. doi: 10.1093/jhered/91.3.221. [DOI] [PubMed] [Google Scholar]
- 60.Petrov DA, Sangster TA, Johnston JS, Hartl DL, Shaw KL. Evidence for DNA loss as a determinant of genome size. Science. 2000;287(5455):1060–1062. doi: 10.1126/science.287.5455.1060. [DOI] [PubMed] [Google Scholar]
- 61.Petrov DA. Mutational equilibrium model of genome size evolution. Theor Popul Biol. 2002;61(4):531–544. doi: 10.1006/tpbi.2002.1605. [DOI] [PubMed] [Google Scholar]
- 62.Messer PW, Arndt PF. The majority of recent short DNA insertions in the human genome are tandem duplications. Mol Biol Evol. 2007;24(5):1190–1197. doi: 10.1093/molbev/msm035. [DOI] [PubMed] [Google Scholar]
- 63.Kvikstad EM, Chiaromonte F, Makova KD. Ride the wavelet: A multiscale analysis of genomic contexts flanking small insertions and deletions. Genome Res. 2009;19(7):1153–1164. doi: 10.1101/gr.088922.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun C, López Arriaza JR, Mueller RL. Slow DNA loss in the gigantic genomes of salamanders. Genome Biol Evol. 2012;4(12):1340–1348. doi: 10.1093/gbe/evs103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Halligan DL, Keightley PD. Spontaneous mutation accumulation studies in evolutionary genetics. Annu Rev Ecol Evol Syst. 2009;40:151–172. [Google Scholar]
- 66.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li R, et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19(6):1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.