Abstract
Over-expression of the receptor tyrosine kinase HER2/ErbB2 (ERBB2) has been linked to a poor prognosis for breast cancer patients; thus, its activity is a central target for cancer therapy. Likewise, over-expression of heregulin (HRG/NRG1), a growth factor responsible for ErbB2 activation, has also been shown to be a driver of breast cancer progression. Although ErbB2 inhibitors offer a major advancement in the treatment of ErbB2-dependent breast cancers, patients are highly susceptible to developing clinical resistance to these drugs. Therefore, a detailed understanding of the molecular mechanism that underlies HRG/ErbB2-induced tumorigenesis is essential for the development of effective therapeutic strategies for this subset of breast cancer patients. Here, it was demonstrated that HRG promoted anchorage-independent breast cancer cell growth more potently than EGF, and that the HRG-dependent activation of PI3K and mTORC1 are necessary events for cell transformation. Functional evaluation of two distinct mTOR inhibitors, rapamycin and INK-128, on HRG-dependent signaling activities, uncovered a necessary role for mTORC2 in the regulation of the AKT/TSC2/mTORC1 axis by impacting the phosphorylation of AKT at the PDK1-dependent site (T308) as well as at the mTORC2-dependent site (S473). The elimination of Rictor, a critical component of mTORC2, is detrimental to both the activation of mTORC1 and HRG-mediated cellular transformation. Similar results were obtained in multiple breast cancer model systems, highlighting an important role for mTORC2 in HRG/ErbB2-dependent breast cancer.
Implications
These findings suggest the potential benefits of targeting mTORC2 in HRG/ErbB2-induced breast cancer.
Keywords: Heregulin, ErbB2, AKT, mTOR, Rictor
Introduction
ErbB2 over-expression characterizes 20 – 30 percent of all breast cancers and correlates with a poor prognosis for patients presenting with this key biomarker (1,2). Additionally, heregulin (HRG), an EGF-like growth factor that binds to ErbB3 or ErbB4, induces one or the other of these receptors to form a heterodimer with ErbB2, resulting in its activation (3,4). HRG is found to be over-expressed in breast, ovarian and prostate cancers (reviewed in (5)) and can induce cellular transformation by the activation of ErbB2, independent of the expression status of this receptor tyrosine kinase (6). A major advancement in the treatment of ErbB2-positive cancers came with the development of monoclonal antibodies against ErbB2 (trastuzumab/Herceptin) and more recently, ErbB2 kinase inhibitors (e.g. lapatinib, reviewed in (7)). These strategies have provided significant clinical benefits but, as is now being appreciated for many forms of targeted therapy in cancer, patients treated with either trastuzumab or lapatinib are susceptible to the development of resistance to these therapies (8-10). As new treatment options are considered for ErbB2-positive cancers, a molecular understanding of the signaling events that underlie HRG/ErbB2-dependent cellular transformation will be critical.
We have found previously that HRG, but not the closely related growth factor, EGF, signals to the RNA processing machinery to impact cell growth (11). Specifically, the activation of ErbB2 at the cell surface triggers a signaling pathway that leads to the activation of the small GTPase Ran (11). Ran, together with importin α and β, regulates the binding and processing of capped mRNAs by the nuclear cap-binding complex (CBC) to promote mitogenesis (11-13). The over-expression of wild-type Ran or constitutively-active Ran mutants is sufficient to transform NIH-3T3 fibroblasts and non-invasive R37 mammary cells (11,14,15), as well as enhance the transforming potential of the breast cancer cell line, SKBR3 (14), thus underscoring the significance of this signaling endpoint in HRG/ErbB2-dependent transformation.
The ability of HRG to signal to Ran and the CBC is dependent upon the mammalian target of rapamycin (mTOR) (11,12). mTOR is a 280 kDa serine/threonine kinase that forms two functionally distinct complexes in mammalian cells, mTORC1 and mTORC2. The rapamycin-sensitive mTORC1 consists of mTOR, Raptor, mLST8, and PRAS40. mTORC1 controls cell size, proliferation, lipid biogenesis, metabolism, and autophagy by sensing growth factors and the nutrient availability of the cell (reviewed in (16-18)). mTORC2 is insensitive to short-term rapamycin treatment and is comprised of mTOR, Rictor, mSin1, and mLST8 (19-21). Raptor and Rictor are commonly used as markers to discern the two complexes (20,22). Less is understood regarding the functions and regulation of mTORC2, with the exception of its role in cytoskeletal remodeling (20,23). There is, however, emerging evidence for the involvement of mTORC2 in growth factor signaling and tumor progression (24,25).
Many growth factors signal to mTORC1 by activating PI3K, which converts PIP2 to PIP3 at the cell membrane (reviewed in (16-18)). PDK1 (phosphoinositide-dependent kinase 1) is then recruited to the membrane, where it phosphorylates AKT at threonine 308 (reviewed in (26)). AKT achieves maximal activation when it is phosphorylated on both threonine 308 in its activation loop and serine 473 within the hydrophobic motif (27). Once activated, AKT phosphorylates an inhibitory site on TSC2 (tuberous sclerosis complex 2), a GTPase-activating protein (GAP) for the small GTPase Rheb (reviewed in (28)). Rheb binds and activates mTORC1, although the molecular basis for this activation remains poorly defined (29).
In this study, we sought to better understand the cellular signals that underlie the transforming potential of HRG, with an emphasis on HRG signaling to mTORC1. We initially chose SKBR3 breast cancer cells for these studies because based on previous work from our laboratory (14), we felt they would provide an excellent model system for probing the signaling connections between HRG/ErbB2 and mTOR. Here we demonstrate that HRG promotes colony formation more potently than EGF in SKBR3 cells and that the differential activation of mTORC1 is necessary for the enhanced potency. Interestingly, we find that mTORC2 plays a critical role in the ability of HRG to activate mTORC1 and promote cellular transformation. Studies contrasting rapamycin and an ATP-competitive inhibitor of mTOR, INK-128 (30), reveal that the phosphorylation of AKT at serine 473 by mTORC2 is critical for downstream TSC2 phosphorylation and mTORC1 activation in response to HRG. The specific disruption of mTORC2 signaling by the introduction of Rictor shRNAs, not only attenuated the activation of mTORC1 and its upstream signaling activators, but also had a deleterious effect on HRG-mediated colony formation. These initial findings in SKBR3 cells were similarly observed in two other HRG-responsive breast cancer cell lines, MCF7 and ZR-75-1. Taken together, these data highlight mTORC2 as a key signaling intermediate for HRG, demonstrate that mTORC2 is necessary for the HRG-stimulated activation of mTORC1, and provide evidence for an important role for mTORC2 in HRG- and ErbB2-dependent cellular transformation.
Materials and Methods
Antibodies and reagents
The antibodies used for this study were purchased from Cell Signaling Technology with the exception of anti-pan-mTOR (Millipore), and anti-actin (NeoMarker). Rapamycin and LY294002 were purchased from Calbiochem. INK-128 was a generous gift from Dr. Kevan Shokat (UCSF). HRGβ, (residues 178-241) and EGF were obtained from Sigma and Invitrogen, respectively.
shRNAs
The shRNAs targeting Rictor were purchased from Sigma (TRCN0000074288, TRCN0000074290, TRCN0000074289). The lenti-viral constructs expressing Rictor shRNAs were generated according to the manufacturer's protocol (Sigma).
Cell culture conditions
SKBR3 (ATCC® HTB-30™), MCF7 (ATCC® HTB-22™), and ZR-75-1 (ATCC® CRL-1500™) cells were obtained from the American Type Culture Collection (ATCC). Cells were authenticated by the ATCC for viability (prior to freezing and immediately after thawing), growth, morphology, isoenzymology, and short tandem repeat (STR) analysis. Cells were passaged for less than 3 months after resuscitation of frozen aliquots. SKBR3, MCF7, and ZR-75-1 cells were maintained in RPMI 1640 (Invitrogen) containing 10% FBS (Invitrogen) at 37°C, 5% CO2. For growth factor stimulation, SKBR3 cells were seeded at 5-7×105 on 100 mm cell culture plates (Corning), followed by serum-starvation with RPMI for 40-48 h, replenishing with fresh RPMI 24 h after initiation of starvation. SKBR3 cells were then stimulated with HRG at the concentration and times indicated, followed by cell lysis. For growth factor simulation of MCF7 and ZR-75-1 cells, cells were seeded at 1.5×105 on 60 mm plates, followed by serum-starvation with RPMI 18-24 h, and then stimulated with 1 nM HRG at the times indicated. For inhibitor analysis, cells were pre-treated with 50 nM rapamycin, 50 nM INK-128, or 10 μM LY294002 for 30 min followed by the addition of 1 nM HRG. For shRNA knock-down experiments, cells were infected with control or Rictor shRNA expressing lentiviral particles twice, one day apart. Cells were then selected with 2 μg/mL puromycin for 48 hours.
Immunoblot analysis
Cells were lysed with cell lysis buffer (50 mM Hepes, pH 8.0, 150 mM NaCl, 1 mM MgCl2, 25 mM NaF, 1 mM Na3VO4, 50 mM β-glycerophosphate, 10 μg/ml Leupeptin, 10 μg/ml Aprotinin, and 1% Triton X-100). The lysates were resolved by SDS-PAGE, and then the proteins were transferred to polyvinylidene fluoride membranes. The membranes were incubated with the indicated primary antibodies diluted in 20 mM Tris, pH 7.6, 135 mM NaCl, and 0.02% Tween-20. The primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare), followed by exposure to ECL reagent (Perkin Elmer).
Soft-agar assays
SKBR3 cells, or SKBR3 cells infected with the various Rictor shRNAs as described, were seeded at a density of 5×103 cells/well in complete medium (10% FBS, RPMI) containing 0.3% agarose, onto underlays comprised of growth medium containing 0.6% agarose in 6-well dishes. MCF7 and ZR-75-1 cells were treated as described above and seeded at a density of 1-2×104 cells /well. The corresponding growth factors or inhibitors were added in the cell mixture. The cultures were fed with complete medium containing 0.3% agarose along with their respective growth factors or inhibitors every three days. Colonies were counted after 13-18 days.
ImageJ quantification
The Western blots were quantified using ImageJ (http://rsbweb.nih.gov/ij/) under the Gel Analysis Tool. The intensities of the different lanes were taken as a ratio of the phospho-protein over total-protein and then normalized to the control lane, which was set to one. In Figure 4, the difference in intensity was obtained by subtracting the intensity of the control (untreated) samples from the HRG-stimulated samples. The percentage of inhibition is calculated by (1-(Difference in Intensityknock-down)/(Difference in Intensitycontrol)) × 100%.
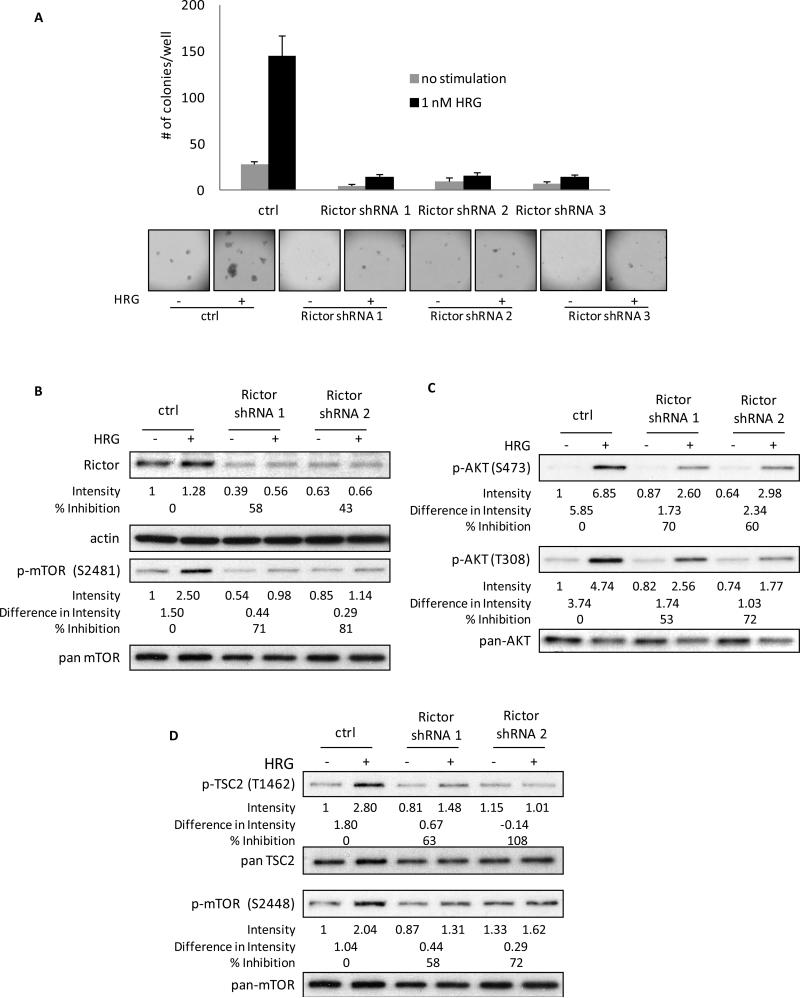
Figure 4.
Rictor is essential for HRG-mediated anchorage-independent growth and HRG-signaling to mTORC1 in SKBR3 cells. A, SKBR3 cells were infected with control virus or viruses containing 3 distinct Rictor shRNAs twice, one day apart, followed by 48 h selection with 2 μg/ml puromycin. Cells were then seeded in 0.3% agarose in complete medium with or without 1 nM HRG. Cells were fed every three days and colonies were counted on day 13. The experiment was done in triplicate and the results were averaged and graphed. B, SKBR3 cells were infected and selected as described above. Cells were then serum-starved for 40-48 h followed by stimulation with 1 nM HRG for 30 minutes. Whole cell lysates were collected and subjected to Western blotting. Blots were probed for Rictor, actin, phospho-mTOR (S2481) and pan-mTOR. Detailed quantification and calculation is described in “Materials and Methods”. C, SKBR3 cells were treated as described in Figure 4B. Blots were probed for phospho-AKT (S473), phospho-AKT (T308), and pan-AKT. D, SKBR3 cells were treated as described in Figure 4B. Blots were probed for phospho-TSC2 (T1462), pan-TSC2, phospho-mTOR (S2448), and pan-mTOR.
Results
To investigate important aspects of HRG/ErbB2-dependent transformation, we started by comparing the relative effectiveness of HRG and EGF to stimulate mitogenesis in breast cancer cells and then attempted to understand what signaling components contribute to any differences observed. The SKBR3 cell line is a low-grade breast cancer cell line that expresses both the EGFR and ErbB2, and as such is a useful model for making comparisons between HRG- and EGF-dependent signaling. We first compared the abilities of HRGβ (herein referred to as HRG) and EGF to enhance the anchorage-independent growth of SKBR3 cells. Cells were seeded in soft agar either in the presence of regular growth media (no treatment), or media supplemented with the addition of 100 nM HRG, or 100 ng/ml EGF, and colonies were then counted after 13 days. As shown in Figure 1A, treating SKBR3 cells with HRG significantly enhances their ability to form colonies in soft agar while EGF does not. Previous studies indicated that mTORC1 is a necessary component for the HRG-specific activation of the Ran GTPase and the CBC in SKBR3 cells (11). Thus, we next examined the differential abilities of HRG and EGF to activate mTORC1. SKBR3 cells were serum-starved for two days and stimulated with HRG or EGF for varying time periods up to 60 minutes. While SKBR3 cells exhibited a high basal level of mTORC1 activity, due both to the relatively high expression of ErbB2 and the presence of amino acids, treatment of the cells with HRG resulted in an additional, albeit modest, time-dependent increase in the phosphorylation of mTOR as determined by Western blotting using a phospho-mTOR (S2448) antibody (Figure 1B, left panel; the right panel shows an example of the 30 minute time-point stimulation by HRG, compared to the background mTOR activity at time zero). In contrast to HRG, EGF was relatively ineffective in its ability to activate mTORC1.
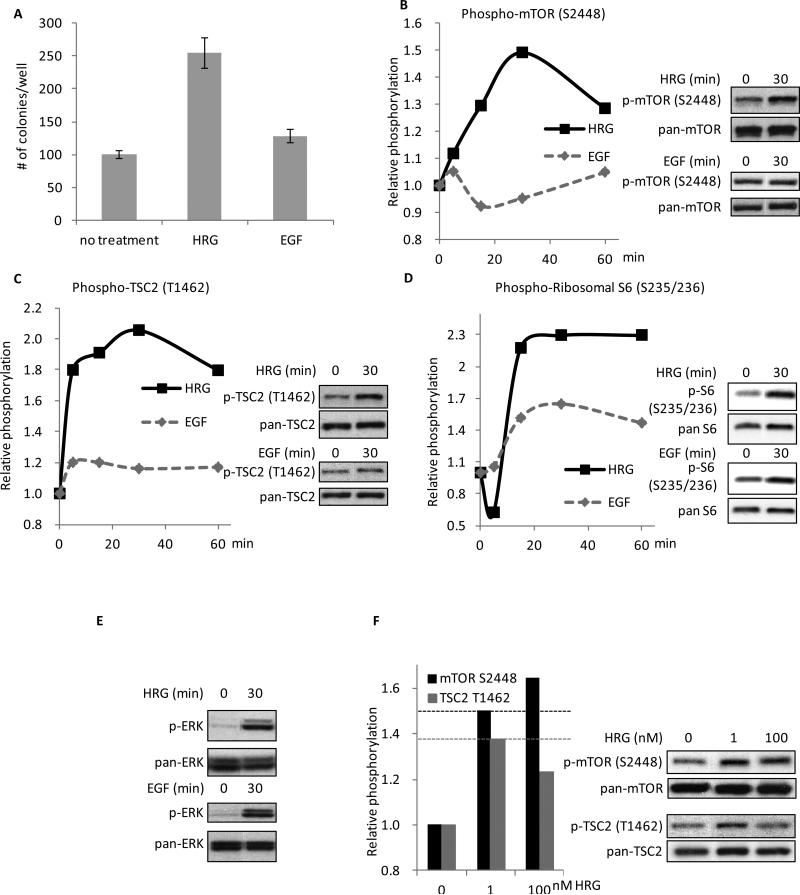
Figure 1.
HRG is more effective than EGF at promoting colony formation in SKBR3 cells and in signaling to mTORC1. A, SKBR3 cells were seeded in 0.3% agarose-containing complete medium with the addition of 100 nM HRG or 100 ng/ml EGF. Cells were fed every three days with the growth factor-containing medium and colonies were counted on day 13. The experiment was done in triplicate and the results were averaged and graphed (p-values: Control vs. HRG = 0.0008; Control vs. EGF = 0.0268; HRG vs. EGF = 0.0021). B, SKBR3 cells were serum-starved for 40-48 h followed by 0-60 minutes of treatment with 100 nM HRG or 100 ng/ml EGF. Whole cell lysates were collected and subjected to Western blotting with phospho-mTOR (S2448) and pan-mTOR antibodies. The experiment was performed in triplicate and one representative blot was quantified using ImageJ. Relative intensities of the bands were taken as a ratio of the phospho-protein over total-protein and then plotted against the zero-minute time-point of each individual blot, which was normalized to one. Two time points, 0 and 30 minutes, are shown on the right as an example of the Western blots. C, SKBR3 cells were treated as stated above. Whole cell lysates were collected and subjected to Western blotting with phospho-TSC2 (T1462) and pan-TSC2 antibodies. The blots were quantified as described above. Two time points (0 and 30 minutes) are shown on the right as an example of the Western blots. D, SKBR3 cells were treated as stated above. Whole cell lysates were collected and subjected to Western blotting with phospho-ribosomal S6 (S235/236) and pan-ribosomal S6 antibodies. The blots were quantified as described above. Two time points (0 and 30 minutes) are shown on the right as an example of the Western blots. E, SKBR3 cells were treated as stated above. Whole cell lysates were collected and subjected to Western blotting with phospho-ERK (T202/Y204) and pan-ERK antibodies. Two time points (0 and 30 minutes) are shown as an example of the Western blots. F, SKBR3 cells were serum-starved for 40-48 h followed by 0, 1, or 100 nM HRG stimulation for 30 minutes. Whole cell lysates were collected and subjected to Western blotting with antibodies against phospho-mTOR (S2448), phospho-TSC2 (T1462), pan-mTOR, and pan-TSC2. The blots were quantified as described above.
We next examined the effects of HRG and EGF on other constituents of the mTORC1 signaling pathway. TSC2 functions upstream of mTORC1 by regulating the GTP-binding activity of the small GTPase, Rheb (reviewed in (28)). Phosphorylation of TSC2 by AKT at T1462 disrupts the ability of TSC2 to regulate Rheb (28), resulting in enhanced mTORC1 function. Probing for the phosphorylation of TSC2 at T1462 indicated that HRG potentiated this phosphorylation to a greater extent than did EGF (Figure 1C), similar to what was observed for phospho-mTOR (S2448). The differential effects of HRG and EGF on this signaling pathway were maintained downstream of mTORC1 as well, as evidenced by the differential phosphorylation of the ribosomal S6 protein (S235/236, Figure 1D), whereas ERK, a downstream target of both HRG and EGF (31), was activated by the two growth factors to similar extents (Figure 1E). A dose response with increasing concentrations of HRG revealed that 1 nM HRG was sufficient to achieve near maximal phosphorylation of mTOR and TSC2 (Figure 1F). Additionally, the activation of these signaling components could be blocked using a selective ErbB2-tyrosine kinase inhibitor (CP-724,714, Supplemental Figure 1) demonstrating that the observed effects were a specific outcome of the activation of ErbB2 by HRG. Like HRGβ, HRGα, a splice variant of HRG associated with the differentiation of normal cells (32), was also able to stimulate the ability of SKBR3 cells to grow in soft agar, as well as activate the mTORC1 pathway (Supplemental Figure 2A and B, respectively).
Given that HRG is more effective than EGF at promoting anchorage-independent growth in SKBR3 cells, and in regulating components of the mTORC1 signaling pathway (i.e., TSC2, mTOR, and S6), we suspected that mTORC1 activity was necessary for HRG-stimulated cellular transformation. To examine this possibility, we utilized inhibitors for mTOR (rapamycin and INK-128), as well as a conventional PI3K inhibitor (LY294002), since the mitogenic activation of mTORC1 is classically described as occurring downstream of PI3K/AKT signaling. Rapamycin is a specific allosteric inhibitor of mTORC1, although prolonged treatment with rapamycin has also been suggested to inhibit mTORC2 (19). INK-128, on the other hand, is a novel ATP-competitive mTOR inhibitor and does not distinguish between mTORC1 and mTORC2 (30).
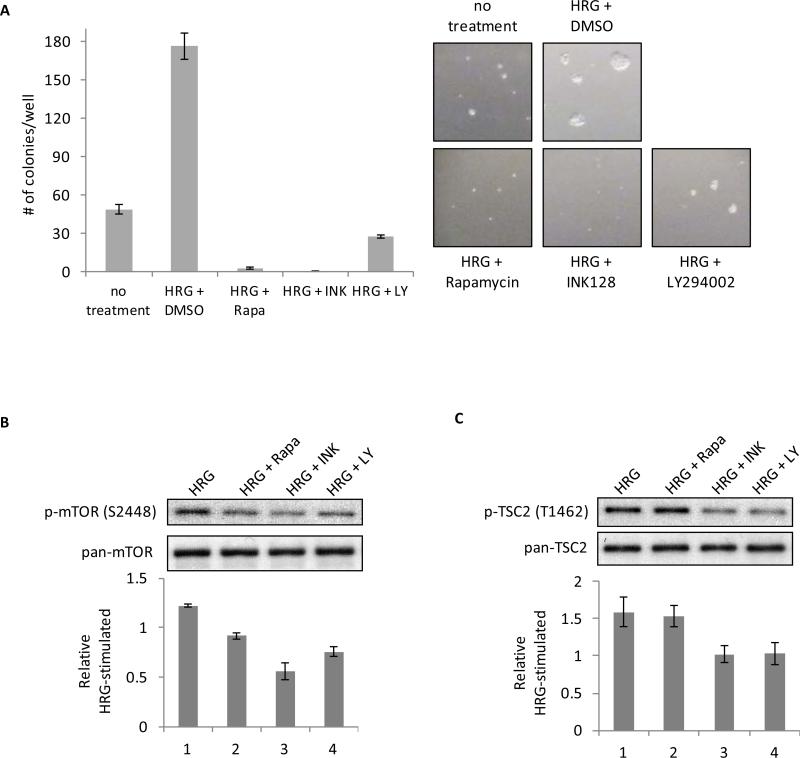
SKBR3 cells were seeded in soft agar in complete medium plus 1 nM HRG, followed by the addition of either DMSO (vehicle), rapamycin, INK-128 or LY294002. These treatments were repeated every three days until colonies were counted on day 13. As shown in Figure 2A, 1 nM HRG markedly augments the ability of SKBR3 cells to form colonies in soft agar, resulting in an increase in colony size as well as number. Both mTOR inhibitors were potent in their ability to block the basal anchorage-independent growth, which is driven by the intrinsic activity of ErbB2 in these breast cancer cells, as well as the HRG-stimulated colony formation in soft agar. INK-128 in particular was striking for its ability to limit the growth of cells beyond the single cell state. The inhibition of PI3K also significantly blocked the basal and HRG-mediated colony formation, albeit to a somewhat lesser extent both in colony number and size.
Figure 2.
Inhibitors of PI3K and mTOR inhibit anchorage-independent growth and HRG-mediated signaling to mTORC1 and TSC2. A, SKBR3 cells were seeded in 0.3% agarose with complete medium containing either DMSO + 1 nM HRG, 50 nM Rapamycin + 1 nM HRG, 50 nM INK-128 + 1 nM HRG, or 10 μM LY294002 + 1 nM HRG. Cells were replenished with the inhibitor + HRG every three days. Colonies were counted on day 13. The experiment was done in triplicate and the results were averaged and graphed. B, SKBR3 cells were serum-starved for 40-48 h. Cells were then pretreated with 50 nM rapamycin, 50 nM INK-128, or 10 μM LY294002 for 30 minutes followed by the addition of the corresponding inhibitor plus 1 nM HRG for 30 minutes. Whole cell lysates were collected and subjected to Western blotting with phospho-mTOR (S2448) and pan-mTOR antibodies. The bar graph was generated by quantifying blots from three independent experiments using ImageJ, as described in “Materials and Methods”, and normalizing the intensity of the bands to the untreated lane. C, SKBR3 cells were treated as described above, followed by blotting with phospho-TSC2 (T1462) and pan-TSC2 antibodies. The bar graph was generated by quantifying blots from three independent experiments using ImageJ and normalizing the intensity of the bands to the untreated lane.
Taken together, these data demonstrate that both mTOR and PI3K activation are required for the HRG- and ErbB2-dependent transformation of SKBR3 cells. They further suggested that the actions of HRG and ErbB2 were dependent upon their ability to stimulate the PI3K/PDK1/AKT pathway. This, in turn, would be expected to result in the inhibitory phosphorylation of TSC2, and the corresponding activation of mTORC1 due to increased Rheb-GTP levels, similar to what has been described for an insulin-stimulated signaling pathway to mTORC1 (16). To test this idea, we examined the effects of rapamycin, INK-128, and LY294002 on phospho-mTOR (S2448) and phospho-TSC2 (T1462). SKBR3 cells were serum-starved for two days and then stimulated with HRG in the presence or absence of these inhibitors. Cell lysates generated from these cells were then analyzed by Western blotting. The expectation was that if the PI3K/PDK1/AKT pathway was the sole signaling event responsible for the HRG-dependent activation of mTORC1, then all three inhibitors should impact the phosphorylation of mTOR at S2448, whereas TSC2, as an up-stream regulator of mTORC1, would be expected to only be sensitive to PI3K inhibition. Indeed, each of the inhibitors was able to reduce the HRG-stimulated phosphorylation of mTOR (Figure 2B), whereas, rapamycin did not affect the ability of HRG to stimulate the phosphorylation of TSC2 at T1462, while the PI3K inhibitor, LY294002, inhibited TSC2 phosphorylation (Figure 2C). However, we also observed that INK-128 was as effective as LY294002 at inhibiting the phosphorylation of TSC2 (T1462) in response to HRG (Figure 2C). It is important to note that the high level of TSC2 phosphorylation under conditions of HRG and rapamycin treatment cannot be attributed to the well-established feedback activation of its upstream activator, AKT, by rapamycin (33), as rapamycin treatment of SKBR3 cells, with or without HRG, results in phosphorylation levels comparable to (but not significantly greater than) the controls (see Supplemental Figure 3).
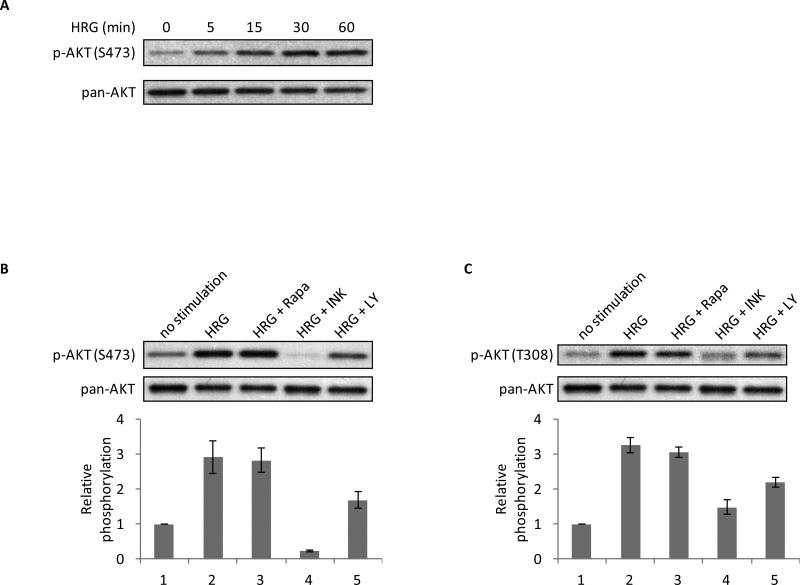
Since the short-term treatment of HRG-stimulated SKBR3 cells with rapamycin did not affect the phosphorylation of TSC2, whereas treatment with INK-128 reduced the phosphorylation, we could only reconcile these data by attributing the effects of INK-128 to the inhibition of mTORC2. The phosphorylation of S473 on AKT by mTORC2 is thought to contribute to the activation of AKT and its downstream signaling targets TSC2 and mTORC1 (28,34,35). Indeed, the treatment of SKBR3 cells with HRG for varying periods of time resulted in an increase in the levels of phospho-AKT (S473) in response to the growth factor (Figure 3A), consistent with the idea that HRG is able to signal to AKT via mTORC2 as well as through PI3K/PDK1.
Figure 3.
mTORC2 is an intermediate in HRG-signaling. A, SKBR3 cells were serum-starved for 40-48 h followed by 0-60 minutes of stimulation with 1 nM HRG. Whole cell lysates were collected and subjected to Western blotting with phospho-AKT (S473) and pan-AKT antibodies. B, SKBR3 cells were serum-starved for 40-48 h. Cells were then pretreated with 50 nM rapamycin, 50 nM INK-128, or 10 μM LY294002 for 30 minutes followed by the addition of the corresponding inhibitor plus 1 nM HRG for 30 minutes. Whole cell lysates were collected and subjected to Western blotting with phospho-AKT (S473) and pan-AKT antibodies. The bar graph was generated by quantifying blots from three independent experiments using ImageJ, as described in “Materials and Methods”, and normalizing the intensity of the bands to the untreated lane. C. SKBR3 cells were treated as described above, followed by blotting with phospho-AKT (T308) and pan-AKT antibodies. The bar graph was generated by quantifying blots from three independent experiments using ImageJ and normalizing the intensity of the bands to the untreated lane.
We next examined the effects of rapamycin, INK-128, and LY294002 on the HRG-stimulated phosphorylation of AKT at S473 as well as at T308. As shown in Figure 3B, rapamycin does not inhibit the phosphorylation of AKT at S473, similar to what we observed for TSC2 (T1462). In contrast, INK-128 causes a dramatic decrease in the phosphorylation of this serine residue. LY294002 causes a partial inhibition of the phosphorylation at S473, consistent with suggestions that PI3K may play a role in signaling upstream of mTORC2 (25). A similar trend was observed for the phosphorylation of AKT at T308 (Figure 3C), suggesting the ability of mTORC2 to impact AKT at both phosphorylation sites.
The results presented in Figures 2 and 3 provide pharmacological evidence to suggest that mTORC2 may be playing an important role in relaying signals arising from the interactions between HRG and ErbB receptors to mTORC1 that contribute to the promotion of the transformed phenotype. While INK-128 appears to be having effects that are distinct from rapamycin, thereby suggesting an involvement of mTORC2 in the HRG/ErbB2-signaling pathway that regulates mTORC1 activity, we wanted to rule out the possibility that INK-128 is simply a more potent inhibitor of mTORC1 under the conditions used.
Rictor is a key component of mTORC2 assembly and function, whereas it is not present within mTORC1 (20). Thus, by targeting Rictor using an shRNA knock-down strategy, we can directly assess the role of mTORC2 in the HRG-stimulated activation of mTORC1 and the resultant transformation of SKBR3 cells. The importance of mTORC2 in the transforming capability of HRG was examined in soft agar assays. SKBR3 cells were infected twice, one day apart, with a Rictor shRNA-carrying virus or a control virus, and cells were then selected with puromycin for 48 hours. Following selection, cells were seeded in soft agar and fed every three days with regular growth medium in the presence or absence of 1 nM HRG until colonies were scored on day 13. As shown in Figure 4A, the cells in the control-infected plates formed colonies in response to HRG. In contrast, colony formation was largely eliminated in cells where Rictor had been knocked down, demonstrating that Rictor, and by extension mTORC2, are necessary for HRG to promote the transformed features of SKBR3 cells.
We next investigated the role of Rictor in relaying HRG-promoted signaling events. Cells were infected and selected as described above and then were serum-starved for 2 days. After serum-starvation, cells were treated with or without 1 nM HRG for 30 minutes. The top panel of Figure 4B shows that the Rictor shRNAs caused an approximately 50% knock-down of Rictor expression as compared to the control samples. The phosphorylation of mTOR at S2481, a rapamycin-insensitive auto-phosphorylation site that is only phosphorylated within an intact mTORC2 (36,37), was next examined to determine how much functional mTORC2 was present in cells following the knock-down of Rictor. There was a 70-80% inhibition of the phosphorylation of mTOR at S2481 between the two different sets of Rictor shRNAs (Figure 4B, middle panel), indicating a significant loss of mTORC2 assembly without affecting the total levels of mTOR protein (Figure 4B, lower panel).
Having confirmed the efficacy of the Rictor knock-down on mTORC2 function in response to HRG, we went on to examine the role of Rictor/mTORC2 in other HRG-stimulated signaling events. AKT phosphorylation was attenuated upon the loss of Rictor from HRG-stimulated cells (Figure 4C). Not only was there a decrease in AKT phosphorylation at the mTORC2 site (i.e., AKT (S473)), but phosphorylation at T308 of AKT (the PDK1 site) was significantly impacted as well. These effects were specific for the elimination of Rictor as the phosphorylation of AKT at both the S473 and T308 sites could be rescued by the concurrent, ectopic expression of an shRNA-insensitive Rictor construct (Supplemental Figure 4). Figure 4D shows the effects of reductions in Rictor expression on the HRG-stimulated phosphorylation of TSC2 (T1462) (top panel) and mTOR (S2448) (lower panel). Destabilization of mTORC2 resulted in the abrogation of mTORC1 function as read out by the decrease in the phosphorylation of both proteins.
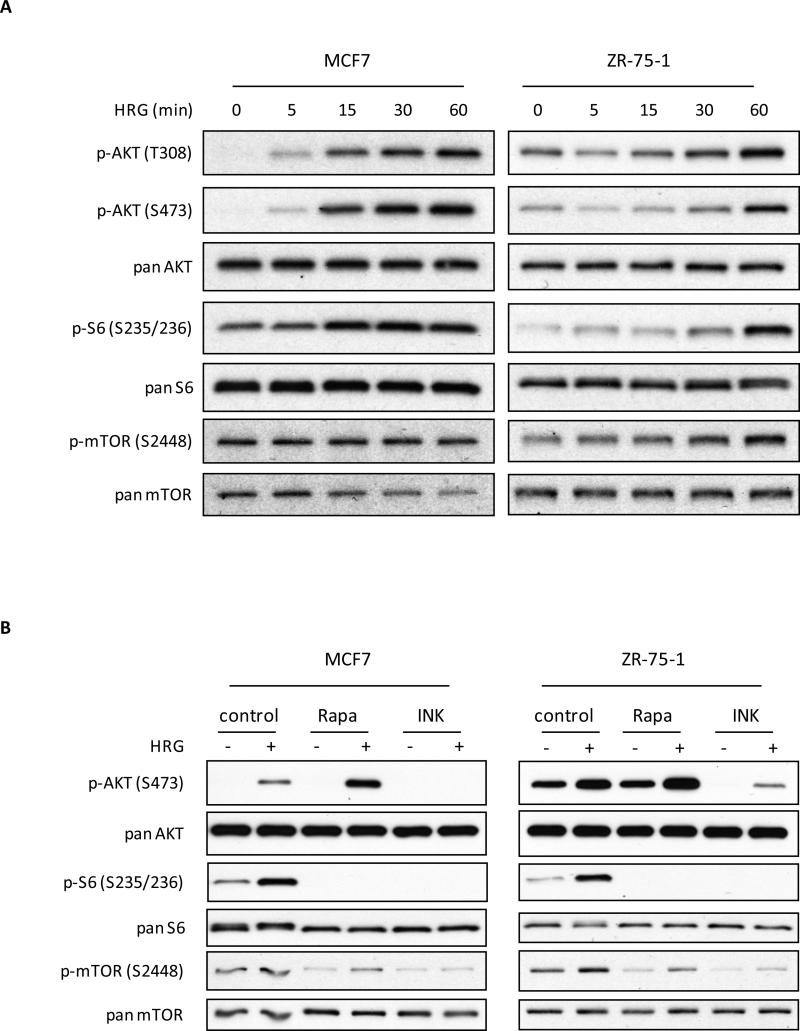
We started this investigation using the SKBR3 cell line as we had previously observed a connection between HRG signaling and mTORC1 activation in this model system. As these new studies uncovered an upstream role for mTORC2 in the HRG-stimulated activation of mTORC1, we questioned whether other HRG-sensitive breast cancer cell lines might also rely on mTORC2 for their HRG-dependent activation of mTORC1. To this end, we examined the generality of these results in two additional ErbB2-expressing breast cancer cell lines (MCF7 and ZR-75-1), which had the ability to respond to the treatment of HRG with a subsequent activation of AKT (T308/S473) and mTORC1, as well as the phosphorylation of the ribosomal S6 protein (Figure 5A). As anticipated, the HRG-dependent phosphorylation of mTOR and ribosomal S6 was effectively blocked by a 30 minute pre-treatment of MCF7 and ZR-75-1 cells with either rapamycin or INK-128 (Figure 5B, lower four panels). Rapamycin treatment did not inhibit AKT phosphorylation, and in contrast to SKBR3 cells, both MCF7 and ZR-75-1 cells showed a relative enhancement of AKT phosphorylation when treated with both HRG and rapamycin suggesting a rapamycin-sensitive feedback loop to AKT (Figure 5B, top panel). INK-128, however, inhibited the ability of HRG to induce the phosphorylation of AKT at S473.
Figure 5.
HRG-dependent signaling in MCF7 and ZR-75-1 breast cancer cell lines. A, MCF7 and ZR-75-1 were serum-starved for 18-24 h, and then challenged with 1 nM HRG for the times indicated. Lysates generated from these cells were then subjected to Western blotting to probe for the HRG-directed activation of components of the mTORC1-related signaling pathway. B, Differential effects of two mTOR inhibitors, rapamycin and INK-128, on MCF7 and ZR-75-1 breast cancer cells. Cells were serum-starved for 18-24 h, and then pre-treated with either 50 nM rapamycin or 50 nM INK-128, followed by the addition of the corresponding inhibitor plus 1 nM HRG for 30 minutes. Whole cell lysates were collected and subjected to Western blotting with phospho-AKT (S473), phospho-ribosomal S6 (S235/236), phospho-mTOR (2448), or the corresponding pan-specific antibodies.
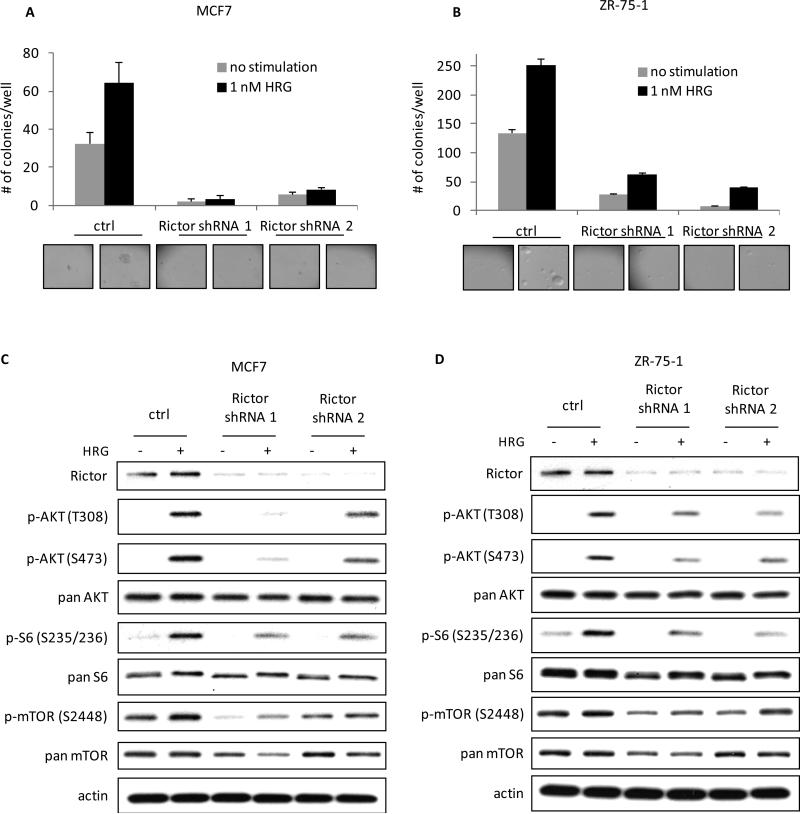
Finally, to confirm that the mTORC2-directed phosphorylation of AKT S473 was important for HRG-dependent transformation and signaling to mTORC1 in MCF7 and ZR-75-1 cells, mTORC2 function was eliminated by the silencing of Rictor, as described above. As was the case for SKBR3 cells, the knock-downs of Rictor in MCF7 and ZR-75-1 cells caused cells to lose their ability to form colonies in soft agar (Figures 6A and B). Concurrent with the loss of transforming potential, the loss of functional mTORC2 blocked the ability of HRG to stimulate AKT phosphorylation and mTORC1 function as read-out by phospho-mTOR and phospho-ribosomal S6 (see Figures 6C and D). Taken together, these results indicate that mTORC2 plays a previously unappreciated role in HRG-promoted transformation via its ability to signal to mTORC1.
Figure 6.
MCF7 and ZR-75-1 breast cancer cells require signaling to mTORC2 for their ability to form colonies in soft agar and to activate mTORC1 in response to HRG. A, mTORC2 was disrupted in MCF7 cells by infecting the cells with viruses containing control or 2 distinct Rictor shRNAs twice, one day apart, followed by 48 h selection with 2 μg/ml puromycin. Cells were then seeded in 0.3% agarose in complete medium with or without 1 nM HRG. Cells were fed every three days and colonies were counted after 18 days. The experiment was done in triplicate and the results were averaged and graphed. B, mTORC2 was disrupted in ZR-75-1 cells as described in Figure 6A and the cells were then analyzed for their ability to form colonies in soft agar in the presence or absence of 1 nM HRG. C, MCF7 cells were infected and selected as described above. Cells were then serum-starved for 18-24 h, followed by stimulation with 1 nM HRG for 30 minutes. Whole cell lysates were collected and subjected to Western blotting. Blots were probed for Rictor, phospho-AKT (T308), phospho-AKT (S473), phospho-ribosomal S6 (S235/236), phospho-mTOR (S2448), the corresponding antibodies for total protein, and actin. D, ZR-75-1 cells were infected with Rictor shRNAs, selected with 2 μg/ml puromycin and then serum-starved for 18-24 h. After a 30 minute treatment with or without HRG, cells were collected and analyzed by Western blotting as described in Figure 6C.
Discussion
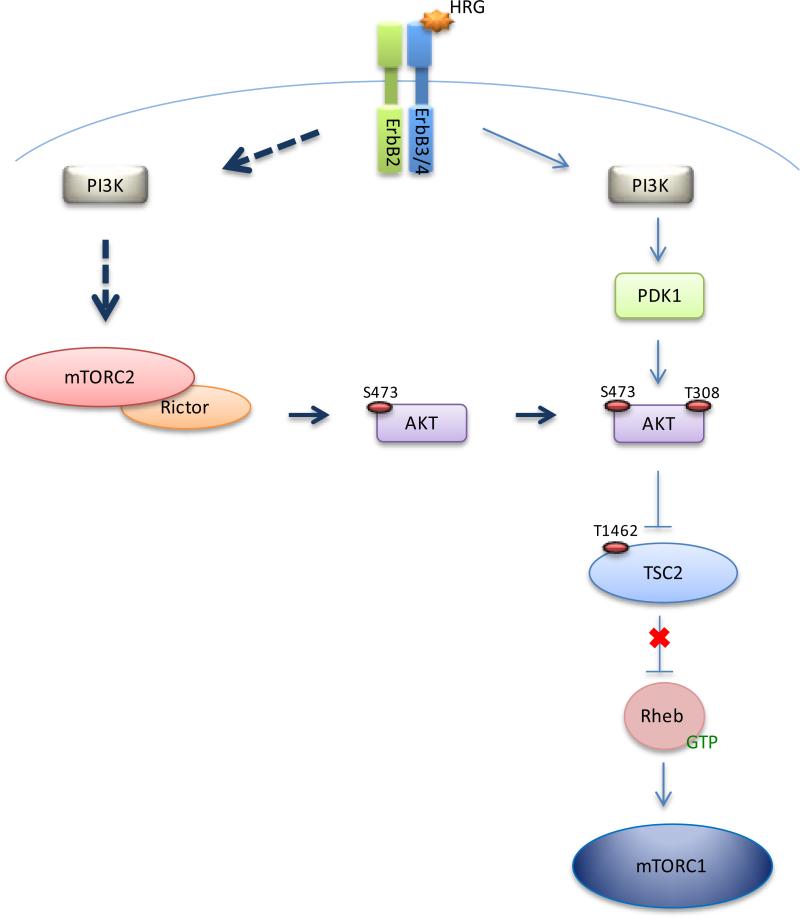
In the present study, we identify mTORC2 as a novel target of HRG/ErbB2 signaling that is necessary for the ability of HRG to promote the enhanced transformation of HRG-sensitive breast cancer cells (i.e., SKBR3, MCF7, and ZR-75-1). Specifically, the use of mechanistically distinct mTOR inhibitors (i.e. rapamycin and INK-128), and a PI3K inhibitor, suggests the ability of mTORC2, as well as PI3K, to feed into the AKT/TSC2/mTORC1 pathway in response to HRG. The model in Figure 7 depicts the HRG signal as bifurcating at PI3K and then converging again at AKT where PI3K/PDK1/AKT (as indicated by the phosphorylation of AKT at T308) represents one branch of the pathway, and PI3K/mTORC2/AKT (i.e., phospho-AKT (S473)) delineates the other. Disruption of the mTORC2 arm of the pathway via the knock-down of Rictor significantly attenuates the ability of HRG to signal to mTORC1 (Figures 4D, 6C, and 6D) as well as to promote oncogenesis, illustrating the necessity of mTORC2 in this context.
Figure 7.
Model for HRG-mediated signaling via mTORC2 to mTORC1. HRG/ErbB2 signals through an mTORC2-dependent pathway to phosphorylate AKT (S473). This phosphorylation precedes the phosphorylation at AKT (T308) by PDK1. Once AKT is fully activated, it phosphorylates TSC2 on multiple sites, sequestering TSC2 away from Rheb, allowing Rheb to activate mTORC1.
The phosphorylation and activation of AKT, which in turn phosphorylates TSC2 and prevents it from negatively regulating Rheb, represents an essential part of the activation of mTORC1 by growth factors (28). It is well known that both T308 and S473 are critical phosphorylation sites for AKT and that having both of these sites phosphorylated exerts a synergistic effect on AKT activation (27). The detection of phosphorylation at these two sites has been routinely used interchangeably to interpret the activation of AKT by PDK1 (38). The discovery by Sabatini and colleagues that AKT (S473) is in fact a preferred mTORC2 substrate (34) raised the possibility for an important role for mTORC2 in mitogenic signaling (25). The phosphorylation of S473 is extremely sensitive to INK-128 but not rapamycin (Figures 3B and 5B), demonstrating the significance of mTORC2 activation by HRG. Also, the effectiveness of INK-128 at inhibiting the phosphorylation of AKT at the PDK1 site (T308) in SKRB3 cells suggests that phosphorylation of S473 may be necessary to allow for the efficient phosphorylation at T308. This raises the possibility that the mTORC2 pathway may in fact be dominant over the PDK1 pathway in the activation of mTORC1 by HRG in SKBR3 cells. This is further underscored by the observation that functionally disabling mTORC2 by the knock-down of Rictor, is sufficient to abolish the HRG-mediated growth of cells in soft agar (Figures 4A, 6A, and 6B).
How do HRG and potentially other growth factors signal to mTORC2? Certainly this is a question that will be garnering acute attention as the appreciation for the role of mTORC2 in mitogenic signaling grows. Thus far, the regulation and function of mTORC2 is less well characterized relative to mTORC1. This most likely stems from the long-standing use of rapamycin to specifically probe mTORC1 function. Prior to the realization that mTORC2 functions as an AKT kinase, mTORC2 was best known for its role in cytoskeletal remodeling (20,23). Our data point to a role for PI3K in the activation of mTORC2, consistent with emerging suggestions that PI3K, as well as Ras, are upstream regulators of mTORC2 (reviewed in (16,25,39)).
We have observed the potential for both mTOR complexes to interact with TSC2 in HEK 293T cells (see Supplemental Figure 5), a cell line which has often been used to characterize the different mTOR complexes (20,22,40). An association of TSC2 with both Raptor and Rictor makes TSC2 an attractive candidate for interfacing mTORC1 and mTORC2, especially given reports of distinct modes of regulation for mTORC1 and mTORC2 by TSC2. While the loss of the TSC1-TSC2 complex from cells gives rise to the activation of mTORC1, consistent with a role for these proteins as negative regulators of mTORC1 (28,41), TSC1-TSC2 deficiency attenuates mTORC2 function, suggesting that the tuberous sclerosis complex positively influences mTORC2 function (40). In addition to our findings, it has also been shown that the N-terminus of TSC2 can interact with the C-terminus of Rictor (42). Thus, TSC2 may serve as a biological bidirectional switch to bring the two complexes in close proximity to achieve signaling and feedback in an efficient manner, both temporally and spatially.
The observation that mTORC2 is necessary for HRG signaling to mTORC1 is underscored by the necessity of mTORC2 for HRG/ErbB2-dependent cellular transformation. Along with an emerging appreciation for the role of mTORC2 in mitogenic signaling is a nascent understanding of its importance in tumorigenesis. mTORC2 was shown to be necessary for prostate cancer development in Pten-deficient mice (24), as well as for the transformation of other cancer cells (i.e. glioma, breast cancer, colorectal cancer), while being less important to normal cells (25). The fact that we find functional mTORC2 to be required for HRG to potentiate the transformation of SKBR3, MCF7, and ZR-75-1 cells raises questions as to whether mTORC2 should be considered as a potential therapeutic target when addressing HRG-mediated and/or ErbB2-positive cancers (see below).
While this study provides evidence for mTORC2 functioning as a signaling intermediary in a pathway from HRG to mTORC1, it also raises the question of whether mTORC2 might play distinct roles that contribute to tumorigenesis. mTORC2 has the potential to promote cell migration and the invasion of SKBR3 and other breast cancer cells in response to HRG, through its function as a cytoskeletal remodeler (43,44). The Rac GTPase, which is well known for its participation in cell migration and cytoskeletal events, has been observed by our laboratory and by other groups to be activated in response to HRG (data not shown, (31,45)). Rac associates with both mTORC1 and mTORC2 and has been suggested to play a role in the cellular localization of these complexes (46). Additionally, a guanine nucleotide exchange factor for Rac, P-Rex1, which can function downstream of mTORC2 (47), has been implicated in breast cancer (48). Future efforts will be directed toward distinguishing the different contributions of mTORC2 to cellular transformation.
The data presented in this study describe a pivotal role for mTORC2, as well as mTORC1, in the ability of HRG/ErbB2 to send signals that drive cellular transformation. Additionally, we show that the mTOR kinase inhibitor, INK-128, is effective not only at inhibiting mTOR (within the context of mTORC1 and mTORC2), but also blocks the ability of PI3K to signal to AKT. Interestingly, the use of mTOR inhibitors (both rapalogs and kinase inhibitors) as a co-therapy with either trastuzumab or lapatinib is currently being investigated for cancers that are refractory to ErbB2-directed monotherapies (8,49,50), as aberrant PI3K/AKT/mTOR activity is one hallmark of resistance to ErbB2 therapy resistance (8,9). Our findings support the rationale of this approach and would point to a greater efficacy with the use of dual mTORC1 and mTORC2 inhibitors.
Supplementary Material
Acknowledgments
We thank Dr. Kevan Shokat (UCSF) for the generous gift of INK-128. We also thank Cindy Westmiller for excellent secretarial assistance.
Financial Support
This work was supported by two grants from the National Institutes of Health to R. Cerione (GM040654, GM047458)
References
- 1.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:07–12. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 3.Carraway KL, 3rd, Sliwkowski MX, Akita R, Platko JV, Guy PM, Nuijens A, et al. The erbB3 gene product is a receptor for heregulin. J Biol Chem. 1994;269:14303–6. [PubMed] [Google Scholar]
- 4.Jones JT, Ballinger MD, Pisacane PI, Lofgren JA, Fitzpatrick VD, Fairbrother WJ, et al. Binding interaction of the heregulinbeta egf domain with ErbB3 and ErbB4 receptors assessed by alanine scanning mutagenesis. J Biol Chem. 1998;273:11667–74. doi: 10.1074/jbc.273.19.11667. [DOI] [PubMed] [Google Scholar]
- 5.Montero JC, Rodriguez-Barrueco R, Ocana A, Diaz-Rodriguez E, Esparis-Ogando A, Pandiella A. Neuregulins and Cancer. Clin Cancer Res. 2008;14:3237–41. doi: 10.1158/1078-0432.CCR-07-5133. [DOI] [PubMed] [Google Scholar]
- 6.Wilson TR, Lee DY, Berry L, Shames DS, Settleman J. Neuregulin-1-mediated autocrine signaling underlies sensitivity to HER2 kinase inhibitors in a subset of human cancers. Cancer Cell. 2011;20:158–72. doi: 10.1016/j.ccr.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–75. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 8.Nahta R, O'Regan RM. Evolving strategies for overcoming resistance to HER2-directed therapy: targeting the PI3K/Akt/mTOR pathway. Clin Breast Cancer. 2010;10:S72–8. doi: 10.3816/CBC.2010.s.015. [DOI] [PubMed] [Google Scholar]
- 9.Chen FL, Xia W, Spector NL. Acquired resistance to small molecule ErbB2 tyrosine kinase inhibitors. Clin Cancer Res. 2008;14:6730–34. doi: 10.1158/1078-0432.CCR-08-0581. [DOI] [PubMed] [Google Scholar]
- 10.Gajria D, Chandarlapaty S. HER2-amplified breast cancer: mechanisms of trastuzumab resistance and novel targeted therapies. Expert Rev. Anticancer Ther. 2011;11:263–75. doi: 10.1586/era.10.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ly TK, Wang J, Pereira R, Rojas KS, Peng X, Feng Q, et al. Activation of the Ran GTPase is subject to growth factor regulation and can give rise to cellular transformation. J Biol Chem. 2010;285:5815–26. doi: 10.1074/jbc.M109.071886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson KF, Fortes P, Singh US, Ohno M, Mattaj IW, Cerione RA. The nuclear cap-binding complex is a novel target of growth factor receptor-coupled signal transduction. J Biol Chem. 1999;274:4166–73. doi: 10.1074/jbc.274.7.4166. [DOI] [PubMed] [Google Scholar]
- 13.Dias SM, Wilson KF, Rojas KS, Ambrosio AL, Cerione RA. The molecular basis for the regulation of the cap-binding complex by the importins. Nat Struct Mol Biol. 2009;16:930–7. doi: 10.1038/nsmb.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milano SK, Kwon W, Pereira R, Antonyak MA, Cerione RA. Characterization of a novel activated Ran GTPase mutant and its ability to induce cellular transformation. J Biol Chem. 2012;287:24955–66. doi: 10.1074/jbc.M111.306514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurisetty VV, Johnston PG, Johnston N, Erwin P, Crowe P, Fernig DG, et al. RAN GTPase is an effector of the invasive/metastatic phenotype induced by osteopontin. Oncogene. 2008;27:71–49. doi: 10.1038/onc.2008.325. [DOI] [PubMed] [Google Scholar]
- 16.Guertin DA, Sabatini DM. Defining the Role of mTOR in Cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Polak P, Hall MN. mTOR and the control of whole body metabolism. Curr Opin Cell Biol. 2009;21:209–18. doi: 10.1016/j.ceb.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Proud CG. mTORC1 signaling: what we still don't know. J Mol Cell Biol. 2010;3:206–20. doi: 10.1093/jmcb/mjq038. [DOI] [PubMed] [Google Scholar]
- 19.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–68. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 20.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 21.Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–32. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 23.Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–8. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 24.Guertin DA, Stevens DM, Saitoh M, Kinkel S, Crosby K, Sheen JH, et al. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell. 2009;15:148–59. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sparks CA, Guertin DA. Targeting mTOR: prospects for mTOR complex 2 inhibitors in cancer therapy. Oncogene. 2010;29:3733–44. doi: 10.1038/onc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanhaesebroeck B, Alessi DR. The PI3K-PDK1 connection: more than just a road to PKB. Biochem J. 2000;346:561–76. [PMC free article] [PubMed] [Google Scholar]
- 27.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–51. [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, Manning BD. The TSC1–TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–90. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–13. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang C, Klein EA, Assoian RK, Kazanietz MG. Heregulin beta1 promotes breast cancer cell proliferation through Rac/ERK-dependent induction of cyclin D1 and p21Cip1. Biochem J. 2008;410:167–75. doi: 10.1042/BJ20070781. [DOI] [PubMed] [Google Scholar]
- 32.Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res. 2003;284:14–30. doi: 10.1016/s0014-4827(02)00102-7. [DOI] [PubMed] [Google Scholar]
- 33.O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996;399:333–8. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 35.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 36.Copp J, Manning G, Hunter T. TORC-specific phosphorylation of mammalian target of rapamycin (mTOR): phospho-Ser2481 is a marker for intact mTOR signaling complex 2. Cancer Res. 2009;69:1821–7. doi: 10.1158/0008-5472.CAN-08-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterson RT, Beal PA, Comb MJ, Schreiber SL. FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J Biol Chem. 2000;275:7416–23. doi: 10.1074/jbc.275.10.7416. [DOI] [PubMed] [Google Scholar]
- 38.Manning BD, Cantley LC. AKT/PKB Signaling: navigating downstream. Cell. 2007;129:1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gan X, Wang J, Su B, Wu D. Evidence for direct activation of mTORC2 kinase activity by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 2011;286:10998–1002. doi: 10.1074/jbc.M110.195016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28:4104–15. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Corradetti MN, Inoki K, Guan KL. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem Sci. 2004;29:32–8. doi: 10.1016/j.tibs.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Huang J, Wu S, Wu CL, Manning BD. Signaling events downstream of mammalian target of rapamycin complex 2 are attenuated in cells and tumors deficient for the tuberous sclerosis complex tumor suppressors. Cancer Res. 2009;69:6107–14. doi: 10.1158/0008-5472.CAN-09-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruan SQ, Wang ZH, Wang SW, Fu ZX, Xu KL, Li DB, et al. Heregulin-beta1-induced GPR30 upregulation promotes the migration and invasion potential of SkBr3 breast cancer cells via ErbB2/ErbB3-MAPK/ERK pathway. Biochem Biophys Res Commun. 2012;420:385–90. doi: 10.1016/j.bbrc.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Ruan SQ, Wang SW, Wang ZH, Zhang SZ. Regulation of HRG-beta1-induced proliferation, migration and invasion of MCF-7 cells by upregulation of GPR30 expression. Mol Med Rep. 2012;6:131–8. doi: 10.3892/mmr.2012.874. [DOI] [PubMed] [Google Scholar]
- 45.Yang C, Liu Y, Lemmon MA, Kazanietz MG. Essential role for Rac in heregulin beta1 mitogenic signaling: a mechanism that involves epidermal growth factor receptor and is independent of ErbB4. Mol Cell Biol. 2006;26:831–42. doi: 10.1128/MCB.26.3.831-842.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saci A, Cantley LC, Carpenter CL. Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol Cell. 2011;42:50–61. doi: 10.1016/j.molcel.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hernandez-Negrete I, Carretero-Ortega J, Rosenfeldt H, Hernández-Garcia R, Calderón-Salinas JV, Reyes-Cruz G, et al. P-Rex1 links mammalian target of rapamycin signaling to Rac activation and cell migration. J Biol Chem. 2007;282:23708–15. doi: 10.1074/jbc.M703771200. [DOI] [PubMed] [Google Scholar]
- 48.Wertheimer E, Gutierrez-Uzquiza A, Rosembilt C, Lopez-Haber C, Sosa MS, Kazanietz MG. Rac signaling in breast cancer: a tale of GEFs and GAPs. Cell Signal. 2012;24:35–62. doi: 10.1016/j.cellsig.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia-Garcia C, Ibrahim YH, Serra V, Calvo MT, Guzmán M, Grueso J, et al. Dual mTORC1/2 and HER2 blockade results in antitumor activity in preclinical models of breast cancer resistant to anti-HER2 therapy. Clin Cancer Res. 2012;18:2603–12. doi: 10.1158/1078-0432.CCR-11-2750. [DOI] [PubMed] [Google Scholar]
- 50.Miller TW, Forbes JT, Shah C, Wyatt SK, Manning HC, Olivares MG, et al. Inhibition of mammalian target of rapamycin is required for optimal antitumor effect of HER2 inhibitors against HER2-overexpressing cancer cells. Clin Cancer Res. 2009;15:7266–76. doi: 10.1158/1078-0432.CCR-09-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.