Abstract
Staphylococcus aureus is an important pathogen, with methicillin-resistant (MRSA) and multi-drug resistant strains becoming increasingly prevalent in both human and veterinary clinics. S. aureus causing bovine mastitis yields high annual losses to the dairy industry. Conventional treatment of mastitis by broad range antibiotics is often not successful and may contribute to development of antibiotic resistance. Bacteriophage endolysins present a promising new source of antimicrobials. The endolysin of prophage ΦSH2 of Staphylococcus haemolyticus strain JCSC1435 (ΦSH2 lysin) is a peptidoglycan hydrolase consisting of two catalytic domains (CHAP and amidase) and an SH3b cell wall binding domain. In this work, we demonstrated its lytic activity against live staphylococcal cells and investigated the contribution of each functional module to bacterial lysis by testing a series of deletion constructs in zymograms and turbidity reduction assays. The CHAP domain exhibited three-fold higher activity than the full length protein and optimum activity in physiological saline. This activity was further enhanced by the presence of bivalent calcium ions. The SH3b domain was shown to be required for full activity of the complete ΦSH2 lysin. The full length enzyme and the CHAP domain showed activity against multiple staphylococcal strains, including MRSA strains, mastitis isolates, and CoNS.
Keywords: mastitis, phage endolysin, coagulase negative staphylococci
1. Introduction
Staphylococcus is a genus of Gram-positive cocci that includes both human and animal pathogens. Multiple drug resistant strains of Staphylococcus aureus (e.g., methicillin-resistant S. aureus, MRSA) pose a major threat to human health. A recent study reports clinical estimates indicating that MRSA strains caused more than 94 000 serious infections and more than 18 000 deaths in the United States in 2005 (Klevens et al., 2007).
Also in the dairy industry, staphylococci are a persistent problem. Bovine mastitis, an infection of the mammary gland, results in annual losses between $1.7 billion and $2 billion in the United States alone (Sordillo and Streicher, 2002). S. aureus is among the most relevant causative agents of this disease, accounting for 18% of mastitis cases in a study carried out on dairy herds in the states of New York and Pennsylvania (Wilson et al., 1997). The conventional method of treatment by antibiotics is less than 50% effective and often leads to premature culling (Deluyker et al., 2005). Furthermore, the use of broad range antibiotics such as pirlimycin and penicillin, which are commonly applied for treatment of mastitis (Cattell et al., 2001), can contribute to the development of resistance in mastitis pathogens and nonrelated bacteria (Fischetti, 2005; Lee, 2003; Vanderhaeghen et al., 2010). In an international study including several European countries and the United States, 57% of 811 S. aureus isolates from bovine mastitis were shown to be β-lactamase positive (De Oliveira et al., 2000). Moreover, there is a debate on whether antibiotic resistance can be transferred from farm animals to humans (Ferber, 2002, 2003).
In contrast to antibiotics, the use of pathogen-specific antimicrobials such as bacteriophage endolysins is expected to reduce the risk of resistance development (Walsh, 2003). Bacteriophage endolysins are proteins which are produced inside an infected host cell at the end of the lytic multiplication cycle of the phage in order to lyse the bacterial cell from within, thereby releasing the phage progeny. In most cases, the lysis event is triggered by the action of a holin, another phage encoded protein which creates pores in the cytoplasmic membrane, enabling the endolysin to gain access to the host cell wall and degrade the peptidoglycan (Young and Blasi,1995). When exposed externally to Gram positive bacteria, in the absence of an outer membrane, these enzymes can also degrade the peptidoglycan, and lyse the cells. This makes them potential antimicrobials against Gram-positive pathogens such as Staphylococcus aureus (Fischetti, 2005; Loessner, 2005). Furthermore, development of resistance in Gram-positive organisms against the highly specific action of phage endolysins is believed unlikely due to coevolution of phage and host, and up to now no resistant strains have been reported despite repeated efforts to find them (Fischetti, 2005; Loeffler et al., 2001; Schuch et al., 2002). It should be noted, though, that bacterial strains resistant against other (non-endolysin) peptidoglycan hydrolases such as the bacteriocin Lysostaphin (Dehart et al., 1995; Gründling et al. 2006; Sugai et al. 1997) or human lysozyme (Guariglia-Oropeza and Helmann, 2011; Vollmer, 2008) have been described. Resistance against Lysostaphin has been ascribed in most cases to changes within the pentaglycine bridge (Dehart et al., 1995; Rohrer et al., 1999; Stranden et al., 1997; Sugai et al., 1997; Thumm AND Götz, 1997), which is the target of Lysostaphin (Schindler and Schuhardt, 1964) and presumably constitutes the most variable part of staphylococcal peptidoglycan (Schleifer and Kandler, 1972).
Endolysins from a Gram-positive background show a modular architecture, consisting of one or more enzymatically active domains (EADs), which cleave certain bonds within the bacterial peptidoglycan, and often a cell wall binding domain (CBD), which directs the enzyme to its substrate and confers specificity for the target cells. The latter is usually located at the C-terminus of the protein (Borysowski et al., 2006; Fischetti, 2005; Loessner, 2005). According to the bonds cleaved by the EADs, these domains can be classified into five different groups: (i) muramidases (also known as lysozymes) and (ii) glucosaminidases, which are both glycosidases and cleave one of the two β-1,4 glycosidic bonds within the glycan strand of the peptidoglycan each; (iii) lytic transglycosylases, which cleave the same bond as muramidases, but by a different mechanism; (iv) amidases, which cut between the glycan and the peptide moieties; and (v) endopeptidases, which cleave within the peptide moiety. The latter can be further divided into those enzymes cutting within the stem peptide, those cutting within the inter-peptide bridge, and those cleaving between the stem peptide and the inter-peptide bridge (Borysowski et al., 2006; Hermoso et al., 2007; Loessner, 2005). As demonstrated by several studies, the modular organization of phage endolysins allows artificial rearrangement of their functional domains, yielding protein chimeras with new and potentially optimized properties for control of pathogens (Becker et al., 2009b; Croux et al., 1993a; Croux et al., 1993b; Diaz et al., 1990; Donovan et al., 2006; Schmelcher et al., 2011). Therefore, bacteriophage endolysins do not only represent a promising alternative to antibiotics in their native form, but also a source of functional modules for the construction of tailor-made antimicrobials (Donovan et al., 2009).
The genomic sequence of Staphylococcus haemolyticus strain JCSC1435 was published recently, and two prophages were identified within the genome (Takeuchi et al., 2005). One of the prophages, ΦSH2, contains a gene (SH2333) coding for a putative endolysin, which was annotated as N-acetylmuramoyl-L-alanine amidase (BAE05642.1). Bioinformatic analysis suggests that this protein consists of two enzymatically active domains and one C-terminal SH3b CBD. A conserved domain database (www.ncbi.nlm.nhi.gov) search identified a CHAP (Cysteine, Histidine-dependent Amidohydrolases/Peptidases) domain (pfam # PF05257; Bateman and Rawlings, 2003; Rigden et al., 2003) at the N-terminus, an Amidase_2 domain (N-acetylmuramoyl-L-alanine amidase; pfam # PF01510) in the center, and a bacterial SH3 domain (SH3b domain), which is associated with cell wall binding, at the C-terminus of the protein (pfam # PF08460; Whisstock and Lesk, 1999). The crystal structure of one representative of the Amidase_2 family, the AmiE domain of the major S. epidermidis autolysin AtlE has recently been reported as the first protein structure with an amidase-like fold from a Gram-positive bacterial background, and also the structure of its binding domain and the phylogenetic relationship of the protein have been analyzed (Albrecht et al., 2012; Zoll et al., 2010; Zoll et al., 2012). The ΦSH2 lysin shares its domain architecture with a number of other staphylococcal lysins described so far, such as the Φ11 prophage endolysin LytA (Wang et al., 1991), the phage K endolysin LysK (O'Flaherty et al. 2005), the phage Twort endolysin plyTW (Loessner et al., 1998), and the S. warneri phage WMY endolysin lysWMY (Yokoi et al., 2005). When more than 50 SH3b-containing staphylococcal peptidoglycan hydrolases (including the aforementioned phage endolysins) were classified based on overall sequence homology, the majority of the proteins fell into five groups featuring mostly >90% within-group but mostly <50% between-group sequence identity. The ΦSH2 lysin (previously referred to as “haemolyticus JCSC1435” ) was one of six “stand-alone” proteins that maintained the domain architecture but shared less than 50% identity with any of the groups (Becker et al., 2009b).
Peptidoglycan structure varies only slightly between different species and strains of the genus Staphylococcus, comprising a highly conserved stem peptide and a glycine-rich inter-peptide bridge (usually a penta- or hexaglycine bridge) in which single residues can be replaced by L-serine or L-alanine (Schleifer and Kandler,1972). Therefore, it is not uncommon that an endolysin from a phage specific for a certain staphylococcal species also shows activity against other species of the genus, as demonstrated for lysWMY (Yokoi et al., 2005) and LysK (Becker et al., 2009a). S. haemolyticus peptidoglycan differs from that of S. aureus (pentaglycine bridge) by variations of the inter-peptide bridge, the most predominant cross bridges being COOH-Gly-Gly-Ser-Gly-Gly-NH2 and COOH-Ala-Gly-Ser-Gly-Gly-NH2 (Billot-Klein et al., 1996).
In this work, we analyzed the staphylolytic potential of the ΦSH2 lysin by creating a series of truncations of the enzyme and examining their lytic activities against Staphylococcus aureus cells in comparison with the full-length protein.
2. Material and Methods
2.1 Plasmids, constructs, and strains
The S. haemolyticus JCSC1435 lysin (GenBank accession no. BAE05642.1) nucleotide sequence was obtained from the genomic sequence of S. haemolyticus JCSC1435 (Takeuchi et al., 2005) (GenBank accession no.NC_007168). The open reading frame was synthesized (GeneArt, Regensburg, Germany) with an E. coli-optimized codon bias and inserted into pET21a (EMD Biosciences, San Diego, CA) using conventional molecular techniques. The plasmid construct contains the lysin gene between the NdeI and XhoI restriction sites and encodes a C-terminally 6 × His-tagged version of the protein. The truncations of the ΦSH2 gene were created by standard molecular biological methods. Gene fragments were amplified by PCR as previously described (Donovan and Foster-Frey, 2008) using the full length ΦSH2 construct (in pET21a) as template and the primers shown in Table 1, that by design add NdeI and XhoI sites to the 5’ and 3’ ends of each fragment, respectively. PCR products were NdeI and XhoI digested and inserted into similarly digested pET21a. The ΦSH2 B1 and B2 constructs were created by inserting SH3b CBD encoding PCR derived DNA fragments with XhoI restriction sites on both ends into the XhoI site of p ΦSH2_A1 and p ΦSH2_A3, respectively.
Table 1.
Plasmids and Primers
| Plasmid | Protein produced | Forward primer | Reverse primer | Recipient vector |
|---|---|---|---|---|
| pΦSH2 | ΦSH2 complete (1-493) | pET21a | ||
| pΦSH2_A1 | ΦSH2 A (1-166) | ΦSH2 _NdeI_F | ΦSH2 _166_XhoI_R | pET21a |
| pΦSH2_A2 | ΦSH2 A1 (1-210) | ΦSH2 _NdeI_F | ΦSH2 _210_XhoI_R | pET21a |
| pΦSH2_A3 | ΦSH2 A2 (1-245) | ΦSH2 _NdeI_F | ΦSH2 _245_XhoI_R | pET21a |
| pΦSH2_A4 | ΦSH2 A1 (1-287) | ΦSH2 _NdeI_F | ΦSH2 _287_XhoI_R | pET21a |
| pΦSH2_B1 | ΦSH2 B (1-166 + 341-493) | ΦSH2 _341_XhoI_F | ΦSH2 _XhoI_R | pΦSH2 _A1 |
| pΦSH2_B2 | ΦSH2 B (1-246 + 347-493) | ΦSH2 _347_XhoI_F | ΦSH2 _XhoI_R | pΦSH2 _A3 |
| pΦSH2_C1 | ΦSH2 C (1-397) | ΦSH2 _NdeI_F | ΦSH2 _397_XhoI_R | pET21a |
| pΦSH2_C2 | ΦSH2 C (1-429) | ΦSH2 _NdeI_F | ΦSH2 _429_XhoI_R | pET21a |
| pΦSH2_D1 | ΦSH2 D (178-397) | ΦSH2 _178_NdeI_F | ΦSH2 _397_XhoI_R | pET21a |
| pΦSH2_D2 | ΦSH2 D (178-429) | ΦSH2 _178_NdeI_F | ΦSH2 _429_XhoI_R | pET21a |
| pΦSH2_E1 | ΦSH2 E (178-493) | ΦSH2 _178_NdeI_F | ΦSH2 _XhoI_R | pET21a |
| pΦSH2_E2 | ΦSH2 E (148-493) | ΦSH2 _148_NdeI_F | ΦSH2 _XhoI_R | pET21a |
| Primer | Sequence |
|---|---|
| ΦSH2 _NdeI_F | 5′-CGC GCG CAT ATG AAA ACA CAA GCA-3′ |
| ΦSH2 _166_XhoI_R | 5′-CAC CAC CTC GAG AGC TAC TGG TGG AAC-3′ |
| ΦSH2 _210_XhoI_R | 5′-AC ACC TTT CTC GAG GTA ACC TCG T-3′ |
| ΦSH2 _245_XhoI_R | 5′-AAC ATA AGC CTC GAG GAT ACC ACG-3′ |
| ΦSH2 _287_XhoI_R | 5′-TC ACT TGC ACG TAA CTC GAG ATT CAC T-3′ |
| ΦSH2 _341_XhoI_F | 5′-CTC TGA CTC GAG GTG CTT CAT ACT GG-3′ |
| ΦSH2 _XhoI_R | 5′-GTG GTG CTC GAG ACT GAT TAC TCC-3′ |
| ΦSH2 _347_XhoI_F | 5′-GTG CTT CAT ACT CTC GAG GAT CCG TTG-3′ |
| ΦSH2 _397_XhoI_R | 5′-GAC CAC CTC GAG TTT ACG TGT AGC TGG-3′ |
| ΦSH2 _429_XhoI_R | 5′-T ATT TGT GAA CTC GAG AAC GTA ACG-3′ |
| ΦSH2 _178_NdeI_F | 5′-AGT ATC AAC ACA TAT GCA AGC ACC TAA ACA AAA AG-3′ |
| ΦSH2 _148_NdeI_F | 5'-CTAATAAGCATATGAGCCTACGTTGG-3′ |
All constructs contained eight additional amino acids at their C-termini, consisting of Leu-Glu (which is introduced by the XhoI restriction enzyme recognition sequence) and the 6xHis-tag used for nickel affinity chromatography during protein purification. All subcloning was performed in E. coli DH5α, and all constructs were verified by sequencing. E. coli BL21 (DE3) was used for expression of proteins. All E. coli strains were grown in Luria-Bertani (LB) medium, with addition of 150 μg/ml ampicillin for plasmid selection. Staphylococcus aureus strain Newman (NCTC8178) was used for turbidity reduction assays, plate lysis assays, and zymograms, after culturing at 37°C to mid-log phase growth in tryptic soy broth (TSB). Other S. aureus and coagulase negative staphylococcal (CoNS) strains used for plate lysis assays (listed in Table 2) were cultured the same way. Strain 305 (Newbould) was purchased from the American Type Culture Collection (ATCC 29740). The Tanji strains (mastitis isolates) were obtained as a gift from Yasunori Tanji (Tokyo Institute of Technology) (Synnott et al. 2009). The NRS strains (MRSA strains) were obtained from NARSA (Eurofins Medinet, Inc., Chantilly, VA). All CoNS strains are mastitis isolates and gifts from Max Paape (ARS, Beltsville, MD), except for Staphylococcus hyicus, a gift from David Kerr (University of Vermont).
Table 2.
Plate lysis assay results with the ΦSH2 full length lysin and the ΦSH2 A1 construct against multiple Staphylococcus strains.
| Strain | Lytic activity |
|
|---|---|---|
| ΦSH2 full length | ΦSH2 A1 | |
| S. aureus Newman | - | + |
| S. aureus 305 | ++ | ++++ |
| S. aureus Tanji 1 | ++ | +++ |
| S. aureus Tanji 19 | ++ | +++ |
| S. aureus Tanji 20 | ++ | +++ |
| S. aureus Tanji 26 | + | + |
| S. aureus NRS 382 | - | ++ |
| S. aureus NRS 383 | - | - |
| S. aureus NRS 384 | + | ++ |
| S. aureus NRS 385 | - | - |
| S. chromogenes | +++ | ++ |
| S. epidermidis | - | - |
| S. hyicus | ++++++ | ++++ |
| S. simulans | + | +++ |
| S. warneri | +++ | ++ |
| S. xylocus | +++ | ++ |
100 pmoles and 2-fold serial dilutions thereof were spotted on bacterial lawns. The lytic activity against each strain corresponding to the lowest amount of protein producing a lysis zone is indicated as follows. +: 100 pmoles, ++: 50 pmoles; +++: 25 pmoles; ++++: 12.5 pmoles; ++++++: ≤ 3.125 pmoles. ‘-’ means that no lysis was observed at the highest amount tested (100 pmoles).
2.2 Protein expression and purification
E. coli BL21 (DE3) cultures harboring pET21a-derived expression vectors were grown to mid log phase (OD600 nm of 0.4–0.6) under ampicillin selection, chilled on ice for 30 min, induced with 1mM IPTG, and incubated with shaking for 18 h at 19°C. For protein purification under native conditions, cells from 800 mL cultures were harvested by centrifugation, resuspended in 16 mL of Lysis Buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 30% glycerol, pH 8.0), sonicated on ice for 5 min (1 s pulses separated by 1 s rests), and centrifuged at 9000 × g for 30 min. The cleared supernatant containing the soluble form of the target proteins was applied to 1 mL nickel-NTA Superflow resin (QIAGEN, Valencia, CA) and rotated for 1 h at 4 °C. The nickel matrix was packed into empty chromatography columns (QIAGEN) and washed with 20 mL of Lysis buffer, followed by 20 mL of Wash Buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 30% glycerol, pH 8.0). Target proteins were eluted with 2 mL of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 30% glycerol, pH 8.0). The buffer was exchanged to 20 mM NaH2PO4, 150 mM NaCl, 30% glycerol, pH 7.5 using 5 mL ZEBA desalting columns (Thermo Fisher Scientific, Rockford, IL), and protein preparations were 0.22-μm filter sterilized and stored at 4°C until used.
For protein purification under denaturing conditions, pellets from induced cultures were resuspended in 16 mL buffer B (100 mM NaH2PO4, 10 mM Tris, 8 M urea, pH 8.0) per 800 mL culture and incubated at room temperature for 60 min under agitation in order to disrupt the cells and dissolve putative inclusion bodies. Residual insoluble material was eliminated by centrifugation for 25 min at 10000 × g. The supernatant was applied to Ni-NTA resin and packed into a column as described above. The column was washed with 20 mL buffer C (100 mM NaH2PO4, 10 mM Tris, 8 M urea, pH 6.3) and target proteins were eluted with 2 ml of buffer E (100 mM NaH2PO4, 10 mM Tris, 8 M urea, pH 4.5). Eluted fractions were dialyzed against 20 mM NaH2PO4, 150 mM NaCl, 15% glycerol, pH 7.5 using Spectra/Por tubing (MWCO 12000 – 14000; Spectrum Laboratories Inc., Rancho Dominguez, CA), 0.22-μm filter sterilized, and stored at 4 °C until use. Concentrations of all protein preparations were determined spectrophotometrically using a NanoDrop ND-1000 device (NanoDrop Technologies, Wilmington, DE).
2.3 Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and zymogram
All purified proteins were analyzed with 15% SDS-PAGE, using a Kaleidoscope Precision Plus Protein Standard (BioRad, Hercules, CA). For zymograms, a 300 mL culture volume equivalent of live mid-log phase (OD600nm = 0.4 - 0.6) cells of S. aureus strain Newman was embedded in the gel during polymerization. The protein samples were boiled in Laemmli sample buffer (BioRad, Hercules, CA) with β-mercaptoethanol, and electrophoresed with identical buffers and voltage for one hour in both the SDS-PAGE and the zymogram using a Mini-Protean TETRA gel system (BioRad). SDS gels were Coomassie stained and zymograms were washed in excess water and further incubated in deionized water for 60 min, followed by incubation in 10 mM Tris, pH 8.0 with 150 mM or 300 mM NaCl for 80 min. Areas of clearing in the turbid zymogram gel indicate a lytic protein in the gel.
2.4 Plate lysis assay
Purified proteins (100 pmoles in a 10 μl volume and five two-fold serial dilutions thereof) were spotted onto a freshly spread lawn of log phase staphylococcal cells (OD600nm = 0.4 – 0.6, diluted 1/4 before plating) that had air dried for 15 min on gridded TSA (tryptic soy agar) plates. The spotted plates were air dried for 10 min in a laminar flow hood and incubated overnight at 37°C. Cleared spots indicating cell lysis were scored within 24 hours of plating the cells. Staphylococcal strains used for plate lysis are shown in Tab. 2.
2.5 Turbidity reduction assay
The turbidity reduction assay measures a decrease in optical density of a bacterial suspension due to lysis of the target bacteria by a lytic protein. The assays were performed in a 96-well dish format essentially as described previously (Donovan et al., 2006). In order to reduce variability of the assay, frozen stocks of S. aureus Newman substrate cells were prepared in a large batch as described by Becker et al. (Becker et al., 2009b). Immediately prior to the assay, the frozen cells were thawed, washed, and re-suspended in the desired assay buffer such that when 100 μl of cell suspension were added to 100 μl of buffer +/− protein, the final suspension has an OD600nm of ~1.0.
Protein constructs were assayed at a concentration of 1 μM, and the assay was performed at room temperature, measuring the OD600nm at 1 min intervals for 45 min. Buffer without enzyme served as control. The steepest slopes of the resulting lysis curves, which correspond to the lytic activities of the proteins, were determined by a sliding window over each group of three consecutive time points for the entire 45 min period. Specific activities were expressed as ΔOD600nm min−1 μM−1, and the ‘no enzyme’ control value was subtracted from each experimental value.
In order to determine the effect of salt concentration on lytic activity, turbidity reduction assays were performed in 10 mM Tris buffers at pH 7.5 with varying NaCl concentrations between 0 mM and 600 mM. Accordingly, the influence of pH was determined by using a series of different buffers with pH values ranging from 3.5 to 10.0. Citrate buffers were used for pH 3.5, 4.5, 5.5, and 6.0, MOPS buffers for pH 6.5, 7.0, and 7.5, Tris buffers for pH 8.0 and 9.0, and Carbonate/bicarbonate buffer for pH 10.0. The concentration of the buffer substance in all buffers used was 20 mM, and the NaCl concentration was 200 mM. To test the effect of bivalent metal ions, assays were performed in 10 mM Tris, 200 mM NaCl, pH 7.5 supplemented with MgCl2, MnCl2, or CaCl2 at different concentrations (0.1 mM, 1 mM, 10 mM), buffer without metal ions serving as control.
3. Results
3.1 Solubility of ΦSH2 lysin deletion constructs depends on integrity of functional domains
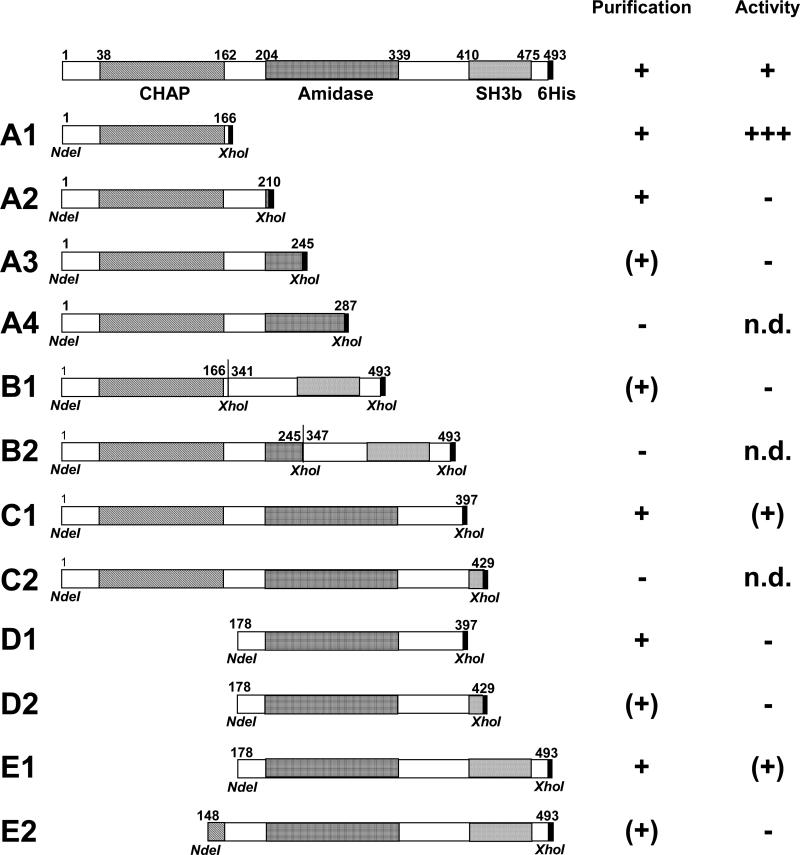
A series of deletion constructs of the ΦSH2 lysin were created in order to determine the contribution of each lytic domain to the lytic activity of the enzyme (Figure 1). These include a series of constructs comprising the complete putative CHAP domain and portions of increasing length of the linker region between the CHAP and the amidase domain, as well as the N-terminal end of the amidase domain (A1 – A4); two versions of the CHAP domain (with and without a portion of the amidase domain) fused to the SH3b cell wall binding domain (B1, B2); and two different length variants each of the CHAP plus the amidase domain (C1, C2), the isolated amidase domain (D1, D2), and the amidase plus the SH3b domain (E1, E2). When purified under native conditions, mostly those constructs comprising only complete functional domains and no partial domains yielded high concentrations (> 0.5 mg/ml) and purities (as shown for the full length protein and the A1, C1, D1, and E1 constructs in Figure 2 A, and indicated in the ‘Purification’ column in Fig. 1). Within the CHAP series (A1 – A4), solubility decreased with increasing length of the constructs and extension across the inter-domain region. For the B1 construct, which is a fusion of the intact ΦSH2 CHAP and SH3b domains (see Fig. 2 A, lane 3), as well as the A3, D2, and E2 constructs, which all contain fractions of functional domains, lower concentrations (< 0.5 mg/ml) were obtained, and preparations were of lower purity. The A4, B2, and C2 constructs did not yield any detectable soluble protein when purified under native conditions, but under denaturing conditions, they were obtained in the elution fraction at concentrations > 1 mg/ml, suggesting that they were present as inclusion bodies in the E. coli cells (data not shown). However, all three constructs completely precipitated during subsequent dialysis, and no soluble protein could be recovered for determination of lytic activity. Mostly those constructs that extended beyond inter-domain regions yielded low concentrations when purified under native conditions and showed very low or no lytic activity against staphylococcal cells in turbidity reduction assays (as indicated in the ‘Activity’ column in Fig. 1; data not shown). Therefore, for each group of constructs, the protein that yielded the highest concentration under native purification conditions and contained only intact functional domains (A1, B1, C1, D1, and E1; subsequently referred to as the ‘selected constructs’) was selected for further characterization in zymograms and turbidity reduction assays in comparison with the full length ΦSH2 lysin, whereas characterization of all other constructs was not further pursued.
Figure 1. Schematic representation of the full length ΦSH2 lysin and deletion constructs.
The full length C-terminally 6 × His-tagged protein (top) and all truncation constructs of the enzyme created in this work are shown. Amino acid positions of functional domains (in the full length protein) and restriction sites used for cloning of fragments (in the deletion constructs) are indicated. The number of amino acids of the full length protein (493 AA) excludes the C-terminal 6 × His-tag. In the ‘Purification’ column, all constructs that could be purified at a concentration of > 0.5 mg/ml and an estimated purity of > 90% under native conditions are rated with ’+’ ; ‘(+)’ marks constructs that yielded lower concentrations and/or purities when purified under native conditions; and ‘–‘ indicates constructs for which no soluble protein could be obtained under native conditions. In the ‘Activity’ column, lytic activity of all constructs as determined by turbidity reduction assays is indicated. The full length enzyme as reference protein is rated with ‘+’; multiple ‘+'s indicate x-fold activity compared to the full length protein; ‘(+)’ indicates approximately half the activity of the full length protein; and constructs marked with ‘−‘ showed very low or no activity. n.d. = not determined.
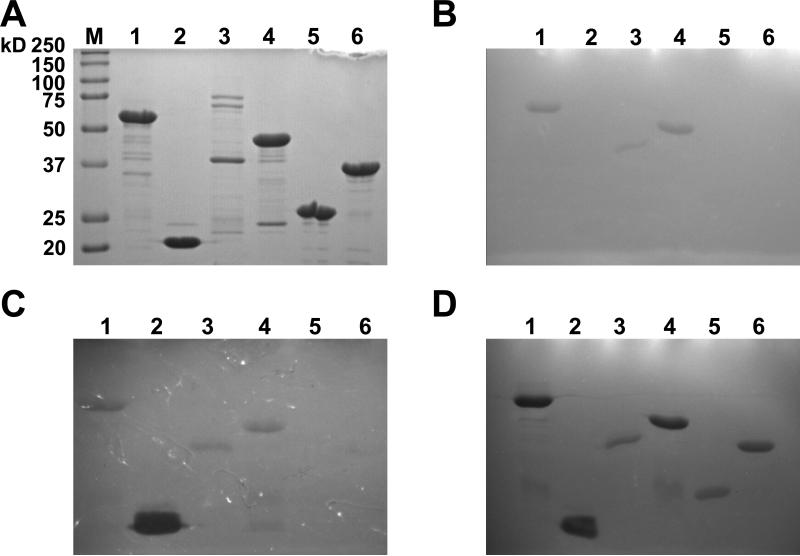
Figure 2. SDS PAGE and zymograms of ΦSH2 full length and deletion constructs expressed in E. coli and purified via Nickel affinity chromatography.
A: SDS PAGE. B: Zymogram with S. aureus Newman cells embedded in the gel after incubation in deionized water for 60 min. C: Zymogram after incubation in deionized water for 60 min and subsequently in 10 mM Tris, 150 mM NaCl, pH 8.0 for 80 min. D: Zymogram shown in B after additional incubation in 10 mM Tris, 300 mM NaCl, pH 8.0 for 80 min. All lanes contain 5 μg of protein. Lane 1: ΦSH2 full length (57.4 kD); lane 2: ΦSH2 A1 (20.4 kD); lane 3: ΦSH2 B1 (38.0 kD); lane 4: ΦSH2 C1 (46.5 kD); lane 5: ΦSH2 D1 (26.1 kD); lane 6: ΦSH2 E1 (37.0 kD); M: Protein standard.
3.2 Lytic activity of the ΦSH2 lysin relies on the CHAP domain
In zymograms impregnated with S. aureus Newman cells, the full length ΦSH2 lysin as well as those constructs containing a CHAP and additionally either an SH3b or an amidase domain (B1 and C1) showed faint zones of lysis after incubation in deionized water for 60 min (Fig. 2 B). When further incubated in buffer containing 150 mM NaCl for up to 80 min, the A1 construct (isolated CHAP domain) developed an intense lytic band, whereas lysis zones of the full length ΦSH2 and the B1 and C1 constructs remained considerably weaker, and the E1 construct (amidase plus SH3b) caused only very faint lysis (Fig. 2 C). When the zymogram was incubated in buffer containing 300 mM NaCl following incubation in water, also the D1 construct (isolated amidase domain) showed a pronounced lytic band, as did all other selected constructs and the full length ΦSH2 lysin (Fig. 2 D).
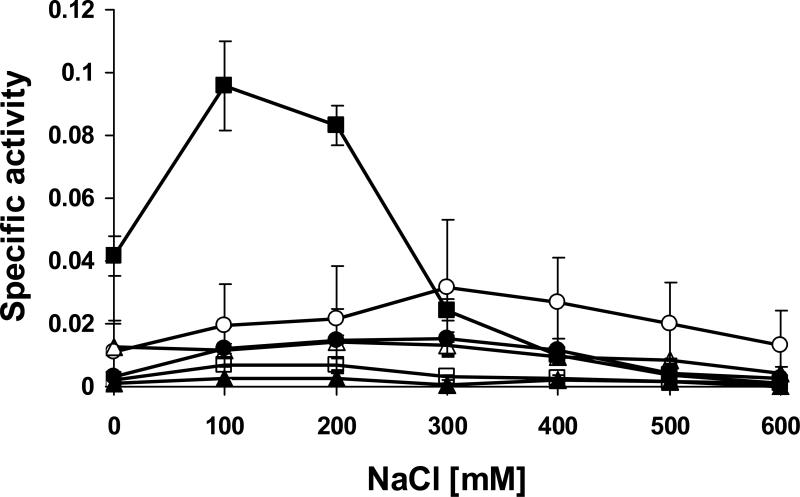
These findings are consistent with the results of turbidity reduction assays performed at different salt concentrations with equimolar concentrations of the selected constructs, using S. aureus Newman as substrate cells (Figure 3). Whereas the full length ΦSH2 lysin showed moderate lytic activity over the whole range of tested NaCl concentrations with optimum activity at 300 mM, the isolated CHAP domain (A1) displayed a more pronounced optimum of activity between 100 mM and 200 mM NaCl. Interestingly, the specific activity of the CHAP domain at its optimum salt concentration was approximately three-fold higher than that of the full length enzyme. This is in accordance with the zymogram, where the A1 construct caused the most prominent lysis zone of all proteins. In contrast, all other deletion constructs exhibited considerably weaker activity than the full length lysin in turbidity reduction assays, suggesting that the enzyme mainly relies on the CHAP domain activity during lysis from without.
Figure 3. Lytic activities of ΦSH2 full length endolysin and deletion constructs against Staphylococcus aureus Newman cells at different salt concentrations.
Turbidity reduction assays were performed with ΦSH2 full length endolysin (open circles), ΦSH2 A1 (black squares), ΦSH2 B1 (black triangles), ΦSH2 C1 (open triangles), ΦSH2 D1 (open squares), and ΦSH2 E1 (black circles) at identical molar concentrations (1 μM). Specific activities are expressed as ΔOD600nm min−1 μM−1. Error bars represent standard deviations from three experiments.
3.3 The SH3b domain is required for full activity of the amidase domain and the full length ΦSH2 lysin
While the CHAP domain alone showed the highest lytic activity of all constructs tested in this study, the addition of an SH3b domain to its C-terminus (in the B1 construct) not only markedly reduced its solubility (see Fig. 1), but also completely abolished its activity (Fig. 3). Also the C1 construct, which combines both enzymatic domains but lacks the SH3b cell wall binding domain, showed drastically reduced activity compared to the CHAP domain alone, and the activity was similar over a wider range of NaCl concentrations, similar to the full length ΦSH2 lysin. The full length lysin, however, which differs from the C1 construct only by the presence of the C-terminal SH3b domain, was about twice as active at its optimum salt concentration. This also holds true for the amidase domain. Whereas the D1 construct (isolated amidase domain) showed only background levels of activity in turbidity reduction assays, the addition of an SH3b domain (E1 construct) increased the activity to approximately the same level as the C1 construct, i.e. about half the activity of the full length ΦSH2 lysin, with a similar salt optimum curve. Overall, these results suggest that the cell wall binding domain is required for full activity of the amidase domain and the complete ΦSH2 lysin, but the function of the CHAP domain in a lysis from without setting is impaired by the addition of a further enzymatic or SH3b domain.
3.4 Ca2+ increases activity of the full length ΦSH2 lysin and its CHAP domain
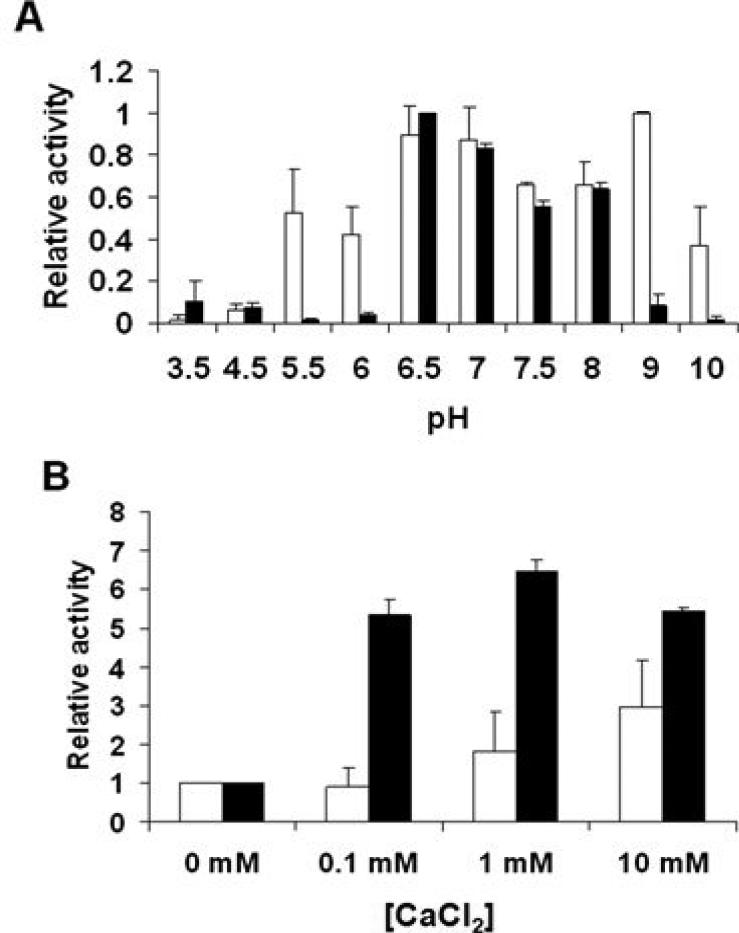
The full length ΦSH2 lysin and the A1 construct were further characterized in turbidity reduction assays, investigating the effect of pH and the presence of various bivalent metal ions on their activity against S. aureus Newman cells. While the complete enzyme showed a higher tolerance to different pH conditions, exhibiting lytic activity in different buffer systems between pH 5.5 and 10, with highest activity between pH 6.5 and 9, the activity range of the CHAP domain was found to be narrower, with activity dropping to background levels below pH 6.5 and above pH 8 (Figure 4 A). Regarding the effect of bivalent metal ions, Mg2+ was found to not significantly alter the lytic activity of the full length enzyme and the CHAP domain at the concentrations tested (0.1 – 10 mM), and Mn2+ had a slightly inhibitory effect (data not shown). In contrast, Ca2+ increased the activity of the full length ΦSH2 lysin three-fold (at a concentration of 10 mM), and that of the A1 construct more than 6-fold (at a concentration of 1 mM) (Fig. 4 B).
Figure 4. Lytic activities of the ΦSH2 full length endolysin and the ΦSH2 A1 construct against Staphylococcus aureus Newman cells at different pH (A) and CaCl2 concentrations (B).
Turbidity assays were performed with identical molar amounts (1 μM) of each protein. Relative activities of ΦSH2 full length (white bars) and ΦSH2 A1 (black bars) are shown. In (A), the highest lytic activity of each protein throughout the whole pH range was defined as 1, and in (B), the activity at 0 mM CaCl2 was defined as 1. Error bars represent standard deviations from three experiments.
3.5 ΦSH2 lysin constructs kill MRSA, mastitis strains, and CoN staphylococci
The ΦSH2 full length enzyme and the A1 construct were further tested in plate lysis assays against a set of different staphylococcal strains, including MRSA strains (NRS 382 – 385), mastitis isolates (305, Tanji 1 – 26), and coagulase negative staphylococci (non S. aureus) (Table 2). The same molar amounts of both proteins (ranging from 100 pmoles to 3.125 pmoles, in a volume of 10 ul) were spotted onto lawns of mid log phase cells of the respective strains, and the plates were analyzed for lysis zones the next day. S. aureus strain Newman, which was used as a reference strain, was not lysed by the highest amount of full length enzyme tested, but 100 pmoles of the A1 construct caused a lysis zone on this strain. For all mastitis and MRSA strains, the CHAP construct showed equal or higher activity (i.e. caused a clearing at a lower amount) than the parental full length ΦSH2 lysin. Whereas all mastitis isolates were lysed by both proteins within the range of amounts tested, the full length enzyme showed activity against only one out of four MRSA strains, and the A1 construct lysed two MRSA strains at 50 pmoles. Regarding the CoN strains, mostly the ΦSH2 full length showed higher activity than the A1 construct, except for S. simulans, which was lysed by 25 pmoles of A1, as opposed to 100 pmoles of the parental protein. S. hyicus was found to be most susceptible to the action of both constructs. The full length lysin caused a lysis zones against this strain at the lowest amount tested, and the A1 construct at 12.5 pmoles. Overall, the parental enzyme displayed lytic activity against 11 out of 16 strains tested in this experimental setup, and the CHAP construct against 13 out of 16.
4. Discussion
When aiming tocreate antimicrobials based on peptidoglycan hydrolases by combining enzymatic and cell wall binding domains from different sources, the identification of potent functional modules is a first important step towards this goal. The Staphylococcus haemolyticus ΦSH2 endolysin characterized in this study exhibits lytic activity ‘from without’ against S. aureus cells in three different in vitro assays and, therefore, is an interesting potential source of such functional domains that could be utilized in modular fusion constructs directed against this important human and animal pathogen.
To more fully define the sequences responsible for the antimicrobial activity of the ΦSH2 lysin, we have created a series of deletion constructs that isolates each domain in a separate construct in an effort to compare their activity alone and when fused pairwise to the other domains in this protein. Our results suggest that the CHAP domain is the most active lytic domain during lysis from without, and that the SH3b domain is important for activity of the full length protein. However, as with any deletion construct, our findings might be complicated by incorrect folding of the truncated proteins. For instance, similar to the low solubility problems encountered with many of the constructs made in this study that extend across inter-domain regions, and which are supposedly due to misfolding of the proteins, also the low solubility and activity of our CHAP-SH3b fusions may be caused by incorrect folding of these specific constructs rather than general incompatibility of the two domains. This is backed by the finding that the CHAP-SH3b construct B1 showed a lytic band in the zymogram, which could be caused by active intermediate products formed during the refolding process within the gel, as opposed to a presumably misfolded and inactive end product which is analyzed in turbidity reduction assays. Except for the isolated CHAP domain construct A1, which showed the most intense lytic band of all tested constructs in a zymogram after incubation in a 150 mM salt buffer, all other proteins displayed their highest activity after incubation at 300 mM NaCl, which could be due to better refolding and/or higher enzymatic activity under these conditions. Some protein preparations yielded minor lytic bands in the zymogram in addition to the dominant bands caused by the ΦSH2 constructs (e.g., lane 4 in Fig. 2 D). This can likely be explained my small amounts of degradation products or co-purified proteins in this highly senstitive assay.
Also turbidity reduction assays with the ΦSH2 lysin and selected deletion constructs revealed that the CHAP domain obviously plays a crucial role in the lytic activity of this endolysin when applied externally. In contrast, the amidase domain showed activity only in presence of the SH3b domain, which was still clearly lower than that of the full length enzyme or the CHAP domain. This is consistent with findings of Becker et al. (Becker et al., 2009a) and Horgan et al. (Horgan et al., 2009) who describe a similar situation in LysK, a staphylococcal phage lysin sharing the same architecture with the ΦSH2 endolysin. However, for LysK, the activity of the CHAP domain was enhanced by addition of an SH3b domain (Becker et al., 2009a), which was not the case for the ΦSH2 lysin. The CHAP domain dominance pattern also holds true for the S. aureus Φ11 phage lysin LytA (Donovan et al., 2006) (also here, the SH3b domain was shown to increase activity of the CHAP and also the amidase domain (Sass and Bierbaum, 2007)), the Staphylococcus warneri M phage ΦWMY lysin (Yokoi et al., 2005), and the Streptococcus agalactiae phage B30 endolysin (Donovan et al. 2006). Low et al. linked the dependence of EAD activity on the presence of the respective CBD to the net charge of the catalytic domain, with positively charged EADs functioning independently of a binding domain (Low et al., 2011). The CHAP domain of the ΦSH2 endolysin features a positive net charge at pH 7.5 (+6; data not shown), and therefore the high activity of the isolated domain is in agreement with this observation.
Turbidity reduction assays with varying NaCl concentrations revealed that the A1 construct exhibits optimum activity at lower ionic strength than the full length ΦSH2 endolysin. A similar effect has been described for the group B streptococcal lytic enzyme PlyGBS, which consists of an N-terminal endopeptidase, a muramidases domain, and a putative C-terminal cell wall binding domain (Cheng and Fischetti, 2007). Here, a deletion construct containing only the endopeptidase domain showed 25-fold higher activity than the full length enzyme and displayed optimum activity shifted towards lower salt concentrations. Also for LysK, the isolated CHAP domain displayed higher activity at low salt concentrations (Fenton et al., 2011), in contrast to the complete endolysin (Becker et al., 2008). This effect can be explained by reduced binding affinity of asingle domain compared to a full length enzyme including a CBD. Also for the staphylococcal phage 187 and Twort endolysins and the Listeria phage endolysin Ply511, higher activity was reported for single domain containing N-terminal portions than for the complete proteins (Gaeng et al., 2000; Loessner et al., 1998; Loessner et al., 1999). However, in these studies lytic activity was not quantified but defined based on the size of lysis zones in overlay assays with S. aureus or M. luteus, so that this effect may be explained by better diffusion of the smaller deletion constructs compared to the full length proteins.
Another important finding with the ΦSH2 lysin is the reduction in lytic activity of the full length enzyme and the amidase domain resulting from a deletion of the SH3b domain. This is consistent with the requirement for a cell wall binding domain for full lytic activity of the bacteriocin Lysostaphin (Baba and Schneewind,1996) and its homologue ALE-1 (Lu et al., 2006), LytA from phage Φ11 (Donovan et al., 2006), the Listeria phage endolysins Ply118 and Ply500 (Loessner et al., 2002), and the LambdaSa2 prophage endolysin where the activity of the isolated endopeptidase domain is reduced in the absence of a CBD (Becker et al., 2009b). Also for the B30 lysin, the glycosidase domain was active only in combination with an SH3b domain (Donovan et al., 2006).
It should be emphasized that all results presented here describe a ‘lysis from without’ situation, whereas in a natural environment, the phage endolysin accesses the peptidoglycan from within the phage's host cell. This being said, the higher activity of the CHAP domain found in this study could be due to better accessibility of the peptidoglycan, which is usually shielded by multiple surface associated molecules from the outside, by the small CHAP domain molecule compared to the larger full length lysin, whereas the latter may work equally well or better from within the cell. Also, a highly diffusible small molecule with high activity may not be beneficial for the phage due to the collateral damage in uninfected cells it would cause after lysis of the host cell (Loessner et al., 2002). Effects of different surface structures such as capsules and teichoic acids, which may show strain-specific variations, on accessibility of the substrate from without may also explain the differences in susceptibility of the various S. aureus strains to the ΦSH2 constructs observed here. Furthermore, there was a trend that the full length ΦSH2 lysin is more effective against the coagulase-negative staphylococci than the CHAP domain construct in plate lysis assays. This could be explained by the fact that the peptidoglycan of coagulase negative species differs from that of S. aureus mainly within the inter-peptide bridge (Schleifer and Kandler, 1972), which may affect the activity of the CHAP domain against these organisms (The CHAP domains of LysK and LytA have been demonstrated to cleave the D-Ala-Gly bond between the stem peptide and the pentaglycine bridge of S. aureus peptidoglycan (Becker et al., 2009a; Navarre et al., 1999)).
It has been reported that different quantitative results can be obtained from multiple lytic activity assays with the same enzymes (Kusuma and Kokai-Kun, 2005). This may explain, for instance, why the A1 construct did not cause a lytic band in the zymogram after incubation in water, whereas it had considerable lytic activity at 0 mM salt in a turbidity reduction assay; why the full length protein did not lyse S. aureus strain Newman in a plate lysis assay at the highest concentration tested, whereas it showed activity in turbidity reduction assays against the same strain; or why the B1 and D1 constructs displayed only background levels of activity in the turbidity reduction assay, whereas they yielded cleared bands in the zymogram. An alternative explanation for the difference in activity of the A1 construct at 0 mM NaCl between the zymogram and the turbidity reduction assay may be that water provides suboptimal re-folding conditions for A1 after SDS PAGE treatment, such that it does not achieve a functional conformation in the zymogram assay.Enhancement of lytic activity by low concentrations of bivalent metal cations as observed for the full length ΦSH2 lysin and the A1 construct has been described previously for a number of different peptidoglycan hydrolases, such as the Φ11 lysin (Donovan et al., 2006) and the B30 endolysin (Pritchard et al., 2004), which both displayed optimum activity in presence of 3 mM and 10 mM Ca2+, respectively. This is an advantageous property of an enzyme when considering an application as antimicrobial against mastitis causing pathogens in a milk environment (the concentration of available Ca2+ in milk is approximately 3 mM).
The idea of using peptidoglycan hydrolases against mastitis causing pathogens is not new. Lysostaphin has been used successfully for preventing Staphylococcus aureus mammary infection in transgenic mice (Kerr et al., 2001) and cattle (Wall et al., 2005). Even though these results are encouraging, it is known that Lysostaphin has a single endopeptidase domain which cleaves within the pentaglycine crossbridge (Browder et al., 1965). As mentioned above, this is the least conserved portion of the peptidoglycan, and mutant strains with alterations in their peptidoglycan crossbridges that render them resistant to Lysostaphin have been described . In contrast, many staphylococcal bacteriophage endolysins including the ΦSH2 lysin feature two different enzymatically active domains, which is anticipated to make resistance formation a rare event, as one bacterial cell would likely require two simultaneous compensatory mutations (Fischetti ,2005). This reduced chance of resistance formation as well as high target cell specificity render native bacteriophage endolysins and especially chimeric constructs consisting of multiple unique and potent enzymatic domains promising alternatives to antibiotics in medical and agricultural applications. To this end, the CHAP domain of the ΦSH2 endolysin could be an interesting addition to the arsenal of enzymatic modules directed against pathogenic staphylococci both on the farm and in human clinical settings.
CHAP domain of ΦSH2 lysin has higher lytic activity than the full length protein
The SH3b domain is required for full activity of the amidase domain
Ca2+ increases activity of the full length ΦSH2 lysin and its CHAP domain
ΦSH2 lysin constructs kill MRSA, mastitis strains, and Coag. Neg. staphylococci
Acknowledgements
This work was supported in part by National Institutes of Health grant 1RO1AI075077-01A1 (DMD); National Research Initiative grant 2007-35204-18395 (DMD) and US State Dept funds supporting a US-Pakistani (DMD) and US Russian collaboration (IA, DMD). The funding sources had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication. Mentioning of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United States Department of Agriculture. The U.S. Department of Agriculture (USDA) prohibits discrimination in all its programs and activities on the basis of race, color, national origin, age, disability, and where applicable, sex, marital status, familial status, parental status, religion, sexual orientation, genetic information, political beliefs, reprisal, or because all or part of an individual's income is derived from any public assistance program. USDA is an equal opportunity provider and employer.
We would like to thank Yasunori Tanji for the gift of strains.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Mentioning of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United States Department of Agriculture.
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- Albrecht T, Raue S, Rosenstein R, Nieselt K, Götz F. Phylogeny of the staphylococcal major autolysin and its use in genus and species typing. J. Bacteriol. 2012;194:2630–2636. doi: 10.1128/JB.06609-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Schneewind O. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 1996;15:4789–4797. [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Rawlings ND. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 2003;28:234–237. doi: 10.1016/S0968-0004(03)00061-6. [DOI] [PubMed] [Google Scholar]
- Becker SC, Dong S, Baker JR, Foster-Frey J, Pritchard DG, Donovan DM. LysK CHAP endopeptidase domain is required for lysis of live staphylococcal cells. FEMS Microbiol. Lett. 2009a;294:52–60. doi: 10.1111/j.1574-6968.2009.01541.x. [DOI] [PubMed] [Google Scholar]
- Becker SC, Foster-Frey J, Stodola AJ, Anacker D, Donovan DM. Differentially conserved staphylococcal SH3b_5 cell wall binding domains confer increased staphylolytic and streptolytic activity to a streptococcal prophage endolysin domain. Gene. 2009b;443:32–41. doi: 10.1016/j.gene.2009.04.023. [DOI] [PubMed] [Google Scholar]
- Becker SC, Foster-Frey J, Donovan DM. The phage K lytic enzyme LysK and lysostaphin act synergistically to kill MRSA. FEMS Microbiol. Lett. 2008;287:185–191. doi: 10.1111/j.1574-6968.2008.01308.x. [DOI] [PubMed] [Google Scholar]
- Billot-Klein D, Gutmann L, Bryant D, Bell D, Van HJ, Grewal J, Shlaes DM. Peptidoglycan synthesis and structure in Staphylococcus haemolyticus expressing increasing levels of resistance to glycopeptide antibiotics. J. Bacteriol. 1996;178:4696–4703. doi: 10.1128/jb.178.15.4696-4703.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borysowski J, Weber-Dabrowska B, Gorski A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp. Biol. Med. (Maywood) 2006;231:366–377. doi: 10.1177/153537020623100402. [DOI] [PubMed] [Google Scholar]
- Browder HP, Zygmunt WA, Young JR, Tavormina PA. Lysostaphin enzymatic mode of action. Biochem. Biophys. Res. Commun. 1965;19:389. doi: 10.1016/0006-291x(65)90473-0. [DOI] [PubMed] [Google Scholar]
- Cattell MB, Dinsmore RP, Belschner AP, Carmen J, Goodell G. Environmental gram-positive mastitis treatment: in vitro sensitivity and bacteriologic cure. J. Dairy Sci. 2001;84:2036–2043. doi: 10.3168/jds.S0022-0302(01)74647-4. [DOI] [PubMed] [Google Scholar]
- Cheng Q, Fischetti VA. Mutagenesis of a bacteriophage lytic enzyme PlyGBS significantly increases its antibacterial activity against group B streptococci. Appl. Microbiol. Biotechnol. 2007;74:1284–1291. doi: 10.1007/s00253-006-0771-1. [DOI] [PubMed] [Google Scholar]
- Croux C, Ronda C, Lopez R, Garcia JL. Interchange of functional domains switches enzyme specificity: construction of a chimeric pneumococcal-clostridial cell wall lytic enzyme. Mol. Microbiol. 1993a;9:1019–1025. doi: 10.1111/j.1365-2958.1993.tb01231.x. [DOI] [PubMed] [Google Scholar]
- Croux C, Ronda C, Lopez R, Garcia JL. Role of the C-terminal domain of the lysozyme of Clostridium acetobutylicum ATCC 824 in a chimeric pneumococcal-clostridial cell wall lytic enzyme. FEBS Lett. 1993b;336:111–114. doi: 10.1016/0014-5793(93)81621-6. [DOI] [PubMed] [Google Scholar]
- De Oliveira AP, Watts JL, Salmon SA, Aarestrup FM. Antimicrobial susceptibility of Staphylococcus aureus isolated from bovine mastitis in Europe and the United States. J. Dairy Sci. 2000;83:855–862. doi: 10.3168/jds.S0022-0302(00)74949-6. [DOI] [PubMed] [Google Scholar]
- Dehart HP, Heath HE, Heath LS, Leblanc PA, Sloan GL. The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl. Environ. Microbiol. 1995;61:1475–1479. doi: 10.1128/aem.61.4.1475-1479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluyker HA, Van Oye SN, Boucher JF. Factors affecting cure and somatic cell count after pirlimycin treatment of subclinical mastitis in lactating cows. J. Dairy Sci. 2005;88:604–614. doi: 10.3168/jds.S0022-0302(05)72724-7. [DOI] [PubMed] [Google Scholar]
- Diaz E, Lopez R, Garcia JL. Chimeric phage-bacterial enzymes: a clue to the modular evolution of genes. Proc. Natl. Acad. Sci. U. S. A. 1990;87:8125–8129. doi: 10.1073/pnas.87.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan DM, Becker SC, Dong S, Baker JR, Foster-Frey J, Pritchard DG. Peptidoglycan hydrolase enzyme fusions for treating multi-drug resistant pathogens. Biotech. International. 2009;21:6–10. [Google Scholar]
- Donovan DM, Dong S, Garrett W, Rousseau GM, Moineau S, Pritchard DG. Peptidoglycan hydrolase fusions maintain their parental specificities. Appl. Environ. Microbiol. 2006;72:2988–2996. doi: 10.1128/AEM.72.4.2988-2996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan DM, Foster-Frey J. LambdaSa2 prophage endolysin requires Cpl-7-binding domains and amidase-5 domain for antimicrobial lysis of streptococci. FEMS Microbiol. Lett. 2008;287:22–33. doi: 10.1111/j.1574-6968.2008.01287.x. [DOI] [PubMed] [Google Scholar]
- Donovan DM, Foster-Frey J, Dong S, Rousseau GM, Moineau S, Pritchard DG. The cell lysis activity of the Streptococcus agalactiae bacteriophage B30 endolysin relies on the cysteine, histidine-dependent amidohydrolase/peptidase domain. Appl. Environ. Microbiol. 2006;72:5108–5112. doi: 10.1128/AEM.03065-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan DM, Lardeo M, Foster-Frey J. Lysis of staphylococcal mastitis pathogens by bacteriophage phi11 endolysin. FEMS Microbiol. Lett. 2006;265:133–139. doi: 10.1111/j.1574-6968.2006.00483.x. [DOI] [PubMed] [Google Scholar]
- Fenton M, Ross RP, McAuliffe O, O'Mahony J, Coffey A. Characterization of the staphylococcal bacteriophage lysin CHAP(K). J. Appl. Microbiol. 2011;111:1025–35. doi: 10.1111/j.1365-2672.2011.05119.x. [DOI] [PubMed] [Google Scholar]
- Ferber D. Antibiotic resistance. Livestock feed ban preserves drugs’ power. Science. 2002;295:27–28. doi: 10.1126/science.295.5552.27a. [DOI] [PubMed] [Google Scholar]
- Ferber D. Antibiotic resistance. WHO advises kicking the livestock antibiotic habit. Science. 2003;301:1027. doi: 10.1126/science.301.5636.1027. [DOI] [PubMed] [Google Scholar]
- Fischetti VA. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 2005;13:491–496. doi: 10.1016/j.tim.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Gaeng S, Scherer S, Neve H, Loessner MJ. Gene cloning and expression and secretion of Listeria monocytogenes bacteriophage-lytic enzymes in Lactococcus lactis. Appl. Environ. Microbiol. 2000;66:2951–2958. doi: 10.1128/aem.66.7.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründling A, Missiakas DM, Schneewind O. Staphylococcus aureus mutants with increased lysostaphin resistance. J. Bacteriol. 2006;188:6286–6297. doi: 10.1128/JB.00457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guariglia-Oropeza V, Helmann JD. Bacillus subtilis sigma(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids. J. Bacteriol. 2011;193:6223–6232. doi: 10.1128/JB.06023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermoso JA, Garcia JL, Garcia P. Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr. Opin. Microbiol. 2007;10:461–472. doi: 10.1016/j.mib.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Horgan M, O'Flynn G, Garry J, Cooney J, Coffey A, Fitzgerald GF, Ross RP, McAuliffe O. Phage lysin LysK can be truncated to its CHAP domain and retain lytic activity against live antibiotic-resistant staphylococci. Appl. Environ. Microbiol. 2009;75:872–874. doi: 10.1128/AEM.01831-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr DE, Plaut K, Bramley AJ, Williamson CM, Lax AJ, Moore K, Wells KD, Wall RJ. Lysostaphin expression in mammary glands confers protection against staphylococcal infection in transgenic mice. Nat. Biotechnol. 2001;19:66–70. doi: 10.1038/83540. [DOI] [PubMed] [Google Scholar]
- Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- Kusuma C, Kokai-Kun J. Comparison of four methods for determining lysostaphin susceptibility of various strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 2005;49:3256–3263. doi: 10.1128/AAC.49.8.3256-3263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 2003;69:6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler JM, Nelson D, Fischetti VA. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science. 2001;294:2170–2172. doi: 10.1126/science.1066869. [DOI] [PubMed] [Google Scholar]
- Loessner MJ. Bacteriophage endolysins--current state of research and applications. Curr. Opin. Microbiol. 2005;8:480–487. doi: 10.1016/j.mib.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Loessner MJ, Gaeng S, Scherer S. Evidence for a holin-like protein gene fully embedded out of frame in the endolysin gene of Staphylococcus aureus bacteriophage 187. J. Bacteriol. 1999;181:4452–4460. doi: 10.1128/jb.181.15.4452-4460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessner MJ, Gaeng S, Wendlinger G, Maier SK, Scherer S. The two-component lysis system of Staphylococcus aureus bacteriophage Twort: a large TTG-start holin and an associated amidase endolysin. FEMS Microbiol. Lett. 1998;162:265–274. doi: 10.1111/j.1574-6968.1998.tb13008.x. [DOI] [PubMed] [Google Scholar]
- Loessner MJ, Kramer K, Ebel F, Scherer S. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 2002;44:335–349. doi: 10.1046/j.1365-2958.2002.02889.x. [DOI] [PubMed] [Google Scholar]
- Low LY, Yang C, Perego M, Osterman A, Liddington R. Role of net charge on catalytic domain and influence of cell wall binding domain on bactericidal activity, specificity, and host range of phage lysins. J. Biol. Chem. 2011;286:34391–34403. doi: 10.1074/jbc.M111.244160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JZ, Fujiwara T, Komatsuzawa H, Sugai M, Sakon J. Cell wall-targeting domain of glycylglycine endopeptidase distinguishes among peptidoglycan cross-bridges. J. Biol. Chem. 2006;281:549–558. doi: 10.1074/jbc.M509691200. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Ton-That H, Faull KF, Schneewind O. Multiple enzymatic activities of the murein hydrolase from staphylococcal phage phi11. Identification of a D-alanyl-glycine endopeptidase activity. J. Biol. Chem. 1999;274:15847–15856. doi: 10.1074/jbc.274.22.15847. [DOI] [PubMed] [Google Scholar]
- O'Flaherty S, Coffey A, Meaney W, Fitzgerald GF, Ross RP. The recombinant phage lysin LysK has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin- resistant Staphylococcus aureus. J. Bacteriol. 2005;187:7161–7164. doi: 10.1128/JB.187.20.7161-7164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard DG, Dong S, Baker JR, Engler JA. The bifunctional peptidoglycan lysin of Streptococcus agalactieae bacteriophage B30. Microbiology. 2004;150:2079–2087. doi: 10.1099/mic.0.27063-0. [DOI] [PubMed] [Google Scholar]
- Rigden DJ, Jedrzejas MJ, Galperin MY. Amidase domains from bacterial and phage autolysins define a family of gamma-D,L-glutamate-specific amidohydrolases. Trends Biochem. Sci. 2003;28:230–234. doi: 10.1016/s0968-0004(03)00062-8. [DOI] [PubMed] [Google Scholar]
- Rohrer S, Ehlert K, Tschierske M, Labischinski H, Berger-Bächi B. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9351–9356. doi: 10.1073/pnas.96.16.9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass P, Bierbaum G. Lytic Activity of Recombinant Bacteriophage {phi}11 and {phi}12 Endolysins on Whole Cells and Biofilms of Staphylococcus aureus. Appl. Environ. Microbiol. 2007;73:347–352. doi: 10.1128/AEM.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler CA, Schuhardt VT. Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc. Natl. Acad. Sci. U. S. A. 1964;51:414–421. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelcher M, Tchang VS, Loessner MJ. Domain shuffling and module engineering of Listeria phage endolysins for enhanced lytic activity and binding affinity. Microb. Biotechnol. 2011;4:651–662. doi: 10.1111/j.1751-7915.2011.00263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuch R, Nelson D, Fischetti VA. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature. 2002;418:884–889. doi: 10.1038/nature01026. [DOI] [PubMed] [Google Scholar]
- Sordillo LM, Streicher KL. Mammary gland immunity and mastitis susceptibility. J. Mammary Gland Biol. Neoplasia. 2002;7:135–146. doi: 10.1023/a:1020347818725. [DOI] [PubMed] [Google Scholar]
- Stranden AM, Ehlert K, Labischinski H, Berger-Bächi B. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 1997;179:9–16. doi: 10.1128/jb.179.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugai M, Fujiwara T, Ohta K, Komatsuzawa H, Ohara M, Suginaka H. epr, which encodes glycylglycine endopeptidase resistance, is homologous to femAB and affects serine content of peptidoglycan cross bridges in Staphylococcus capitis and Staphylococcus aureus. J. Bacteriol. 1997;179:4311–4318. doi: 10.1128/jb.179.13.4311-4318.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synnott AJ, Kuang Y, Kurimoto M, Yamamichi K, Iwano H, Tanji Y. Isolation from sewage influent and characterization of novel Staphylococcus aureus bacteriophages with wide host ranges and potent lytic capabilities. Appl. Environ. Microbiol. 2009;75:4483–4490. doi: 10.1128/AEM.02641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, Kuroda M, Cui L, Takahashi M, Ankai A, Baba S, Fukui S, Lee JC, Hiramatsu K. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 2005;187:7292–7308. doi: 10.1128/JB.187.21.7292-7308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm G, Götz F. Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol. Microbiol. 1997;23:1251–1265. doi: 10.1046/j.1365-2958.1997.2911657.x. [DOI] [PubMed] [Google Scholar]
- Vanderhaeghen W, Hermans K, Haesebrouck F, Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiol. Infect. 2010;138:606–625. doi: 10.1017/S0950268809991567. [DOI] [PubMed] [Google Scholar]
- Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 2008;32:287–306. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- Wall RJ, Powell A, Paape MJ, Kerr DE, Bannerman DD, Pursel VG, Wells KD, Talbot N, Hawk HW. Genetically enhanced cows resist intramammary Staphylococcus aureus infection. Nat. Biotechnol. 2005;23:445–451. doi: 10.1038/nbt1078. [DOI] [PubMed] [Google Scholar]
- Walsh C. Where will new antibiotics come from? Nat. Rev. Microbiol. 2003;1:65–70. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- Wang X, Wilkinson BJ, Jayaswal RK. Sequence analysis of a Staphylococcus aureus gene encoding a peptidoglycan hydrolase activity. Gene. 1991;102:105–109. doi: 10.1016/0378-1119(91)90547-o. [DOI] [PubMed] [Google Scholar]
- Whisstock JC, Lesk AM. SH3 domains in prokaryotes. Trends Biochem. Sci. 1999;24:132–133. doi: 10.1016/s0968-0004(99)01366-3. [DOI] [PubMed] [Google Scholar]
- Wilson DJ, Gonzalez RN, Das HH. Bovine mastitis pathogens in New York and Pennsylvania: prevalence and effects on somatic cell count and milk production. J. Dairy Sci. 1997;80:2592–2598. doi: 10.3168/jds.S0022-0302(97)76215-5. [DOI] [PubMed] [Google Scholar]
- Yokoi KJ, Kawahigashi N, Uchida M, Sugahara K, Shinohara M, Kawasaki K, Nakamura S, Taketo A, Kodaira K. The two-component cell lysis genes holWMY and lysWMY of the Staphylococcus warneri M phage varphiWMY: cloning, sequencing, expression, and mutational analysis in Escherichia coli. Gene. 2005;351:97–108. doi: 10.1016/j.gene.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Young R, Bläsi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol. Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]
- Zoll S, Pätzold B, Schlag M, Götz F, Kalbacher H, Stehle T. Structural basis of cell wall cleavage by a staphylococcal autolysin. PLoS Pathog. 2010;6:e1000807. doi: 10.1371/journal.ppat.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoll S, Schlag M, Shkumatov AV, Rautenberg M, Svergun DI, Götz F, Stehle T. Ligand-binding properties and conformational dynamics of autolysin repeat domains in staphylococcal cell wall recognition. J. Bacteriol. 2012;194:3789–3802. doi: 10.1128/JB.00331-12. [DOI] [PMC free article] [PubMed] [Google Scholar]