Abstract
Members of the RecQ family of helicases are known for their roles in DNA repair, replication, and recombination. Mutations in the human RecQ helicases, WRN and BLM, cause Werner and Bloom syndromes, which are diseases characterized by genome instability and an increased risk of cancer. While WRN contains both a helicase and an exonuclease domain, the Drosophila melanogaster homolog, WRNexo, contains only the exonuclease domain. Therefore the Drosophila model system provides a unique opportunity to study the exonuclease functions of WRN separate from the helicase. We created a null allele of WRNexo via imprecise P-element excision. The null WRNexo mutants are not sensitive to double-strand break-inducing reagents, suggesting that the exonuclease does not play a key role in homologous recombination-mediated repair of DSBs. However, WRNexo mutant embryos have a reduced hatching frequency and larvae are sensitive to the replication fork-stalling reagent, hydroxyurea (HU), suggesting that WRNexo is important in responding to replication stress. The role of WRNexo in the HU-induced stress response is independent of Rad51. Interestingly, the hatching defect and HU sensitivity of WRNexo mutants do not occur in flies containing an exonuclease-dead copy of WRNexo, suggesting that the role of WRNexo in replication is independent of exonuclease activity. Additionally, WRNexo and Blm mutants exhibit similar sensitivity to HU and synthetic lethality in combination with mutations in structure-selective endonucleases. We propose that WRNexo and BLM interact to promote fork reversal following replication fork stalling and in their absence regressed forks are restarted through a Rad51-mediated process.
Keywords: replication restart, Werner syndrome, double-strand break, DNA replication
MEMBERS of the RecQ family of helicases are known as the “guardians of the genome” due to their roles in DNA replication, repair, and maintenance of genomic integrity. There are five RecQ proteins in humans: RECQ1, RECQ4, RECQ5, BLM, and WRN. Mutations in RECQ4, BLM, or WRN cause Rothmund–Thomson syndrome, Bloom syndrome, and Werner syndrome (WS), respectively. These autosomal diseases are characterized by high cancer incidence, accelerated aging, and developmental defects (Chu and Hickson 2009). Most reported mutations in WS patients result in truncation of the 1432-amino acid WRN protein and loss of the nuclear localization signal (Chun et al. 2011). In culture, WS cells exhibit signs of genomic instability, including early senescence, a high incidence of chromosomal translocations, and prolonged S phase (Sidorova 2008).
Like other RecQ family members, WRN exhibits ATP-dependent 3′–5′ DNA helicase activity (Gray et al. 1997). WRN also contains a RecQ C-terminal (RQC) domain and a helicase and ribonuclease D C-terminal (HRDC) domain, which are largely responsible for DNA and protein binding (Opresko et al. 2002; von Kobbe et al. 2002, 2003). A unique feature of WRN that distinguishes it from other RecQ helicases is its 3′–5′ exonuclease activity (Kamath-Loeb et al. 1998). The WRN exonuclease preferentially digests partial double-strand DNA containing a 5′ overhang, although it will also digest blunt-end DNA containing a fork, a Holliday junction, or a D loop (Kamath-Loeb et al. 1998; Shen and Loeb 2000; Orren et al. 2002). The exonuclease domain also contains DNA-binding sites for replication intermediates, such as forks and 5′ overhangs (Xue et al. 2002; von Kobbe et al. 2003). Additionally, proteins that can modulate WRN activity have been shown to bind the exonuclease domain, including Ku80 and BLM (von Kobbe et al. 2002). Interestingly, DNA binding and protein binding are not dependent upon the exonuclease or helicase activity of WRN (Compton et al. 2008; Kamath-Loeb et al. 2012).
WRN has been shown to play an important role in recovery from replication fork stalling. For example, WRN-depleted cells exhibit a greater number of phosphorylated histone 2AX foci following treatment with hydroxyurea (HU), a reagent that causes replication fork stalling (Opresko et al. 2007; Franchitto et al. 2008; Mao et al. 2010; Murfuni et al. 2012). Similarly, WS cells exhibit spontaneous Rad51 foci, indicating the presence of double-strand breaks (DSBs) and their subsequent repair via homologous recombination (Sakamoto et al. 2001; Rodriguez-Lopez et al. 2007). Together, these results suggest that WRN may prevent accumulation of DSBs caused by unsuccessful recovery from stalled replication forks.
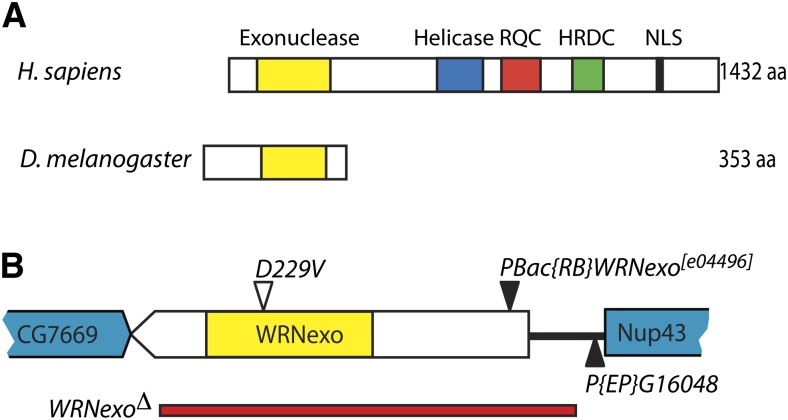
In Drosophila melanogaster, the WRNexo gene encodes a protein with 35% identity and 59% similarity to the exonuclease domain of human WRN (Saunders et al. 2008). However, WRNexo does not contain a helicase domain (Figure 1A). Purified WRNexo exhibits 3′–5′ exonuclease activity on single-strand DNA, double-strand DNA with 5′ overhangs, and substrates representing replication bubbles. However, WRNexo does not digest substrates containing blunt ends or abasic sites (Boubriak et al. 2009; Mason et al. 2013). Activities of WRNexo have been investigated in vivo through use of hypomorphic mutants (Rodriguez-Lopez et al. 2007; Saunders et al. 2008; Boubriak et al. 2009). One such mutant, WRNexoe04496, causes a severe reduction in WRNexo expression, resulting from the presence of a piggyBac {RB} transposable element in the 5′-UTR of WRNexo. WRNexoe04496 flies exhibit high sensitivity to the topoisomerase I inhibitor, camptothecin, as well as hyperrecombination. Female WRNexoe04496 mutants are sterile, but exhibit no other physiological abnormalities (Saunders et al. 2008). A second mutant, WRNexoD229V, contains a point mutation that ablates exonuclease activity at physiological temperatures (Boubriak et al. 2009; Mason et al. 2013). Like WRNexoe04496, WRNexoD229V mutants display hyperrecombination. However, it is important to note that the phenotype of a true null WRNexo allele has yet to be described.
Figure 1.
Generation of a WRNexo null mutant. (A) Conserved regions of Werner protein in humans and Drosophila. (B) A 2.5-kb deletion (red bar) that removes most of the WRNexo coding sequence was generated through imprecise excision of P{EP}G16048. Also shown is the location of PBac{RB}WRNexo[e04496] (Saunders et al. 2008; Boubriak et al. 2009) and the D229V exonuclease dead allele (Boubriak et al. 2009; Mason et al. 2013).
Although much work has been done to delineate the involvement of WRN in responding to replication stress, most hypotheses involve WRN’s helicase activity while the role of the exonuclease remains poorly characterized. In this article, we generate a WRNexo null mutant and show that it has defects in recovering from endogenous and exogenous replication stress. Additionally, we explore a role for WRNexo independent of its exonuclease activity and investigate interactions between WRNexo and the DNA repair proteins BLM and Rad51.
Materials and Methods
Fly stocks
A deletion in WRNexo was created by imprecise P-element excision (Adams et al. 2003). For the screen, we used w1118; P{EP}G16048, which contains a P element located 441 bp upstream of the WRNexo transcription start site (Bellen et al. 2004). The extent of the deletion was determined by Sanger sequencing of a PCR product obtained using the primers WRN -1240F: 5′-GGCAGTCACTTCCTGCT-3′ and 2001R: 5′-GACAACGATCTGCTCAAGCG-3′. The resulting deletion mutant, WRNexoΔ, was male sterile, likely due to a second site mutation generated during P-element mobilization. The WRNexoΔ stock was backcrossed once to w1118 to remove the sterility phenotype.
Other mutants used in this study include Brca2KO, which completely deletes Brca2 (Klovstad et al. 2008); BlmN1, which removes a 2480-bp segment including part of the helicase domain (McVey et al. 2007); and Rad51057, which contains an A205V point mutation in the Rad51 gene (Staeva-Vieira et al. 2003). WRNexoD229V was generated through EMS mutagenesis (Koundakjian et al. 2004). GenZ4325, mus312D1, and mus81NheI were used for analysis of structure-selective endonuclease mutants. Df(3R)Exel6178, which deletes 45 genes between cytological units 90F4–91A5, was used to create WRNexoΔ compound heterozygotes. All double mutants were created by standard meiotic recombination and verified by PCR.
Mutagen sensitivity assays
Sets of five to eight heterozygous virgin females and two to three (heterozygous or homozygous) males were paired in vials containing standard cornmeal agar medium. Females were allowed to lay eggs for 3 days at 25° before transfer into a second vial to lay for an additional 3 days. The first set of vials was treated with 250 μl camptothecin [dissolved in dimethyl sulfoxide (DMSO)], bleomycin, or hydroxyurea (dissolved in ddH2O) 1 day after the transfer of parents. The second set of vials served as the controls and was treated with either 250 μl water (for bleomycin and hydroxyurea experiments) or a matching concentration of DMSO (for camptothecin experiments). Upon eclosion, adults were counted. Relative survival was calculated as (the percentage of viable homozygotes (relative to the total number of viable flies) in mutagen-treated vials)/(the percentage of viable homozygotes in control vials) for each trial. Statistical significance was analyzed using unpaired t-tests.
Hatching frequency assay
WRNexoΔ, WRNexoΔ/WRNexoD229V, and w1118 flies were allowed to lay on grape juice agar plus yeast paste for a period of 8–16 hr at 25°. Each independent experiment consisted of three to four embryo collection periods for a total of 700–4000 embryos per experiment. After 72 hr, embryos were counted and hatching frequency was determined. Statistical significance was analyzed using unpaired t-tests.
Embryo staining
WRNexoΔ and w1118 flies were allowed to lay on grape juice agar plus yeast paste for a period of 3–4 hr at 25°. Embryos were then collected, devitillinized, fixed, and stained with a monoclonal antibody specific to γ-H2Av (Lake et al. 2013) at a dilution of 1:3000. Embryos were then exposed to Texas Red-conjugated rabbit anti-mouse IgG (Abcam) and DAPI at a dilution of 1:1000. Embryos were imaged using a Zeiss (Thornwood, NY) Axio Imager M1 microscope with 3D imaging capability and Slide Book software. Embryos used for analysis of nuclear distribution were within the syncytial division period (mitotic cycles 1–13) whereas embryos used for analysis of γ-H2Av were postcellularization (after cycle 14). Image J was used to calculate the total area of each embryo in pixels. The area of the embryo containing γ-H2Av staining was then calculated to determine the percentage of each embryo in which cells expressed γ-H2Av. Statistical analysis was performed using GraphPad Prism, by combining WRNexoΔ and w1118 images from three separate embryo collection periods, harvested on three consecutive days. Statistical significance was determined using the Mann–Whitney U-test.
Life stage-specific synthetic lethality
Gen1Z4325 WRNexoΔ/TM3 P{w[+mC]=ActGFPJMR2, Ser[1] and mus312D1 WRNexoΔ/TM3 P{w[+mC]=ActGFPJMR2, Ser[1] heterozygotes were each paired in vials and females were allowed to lay eggs for 2–3 days. The resulting progeny were counted daily from the onset of pupariation to eclosion. Heterozygotes and homozygotes were scored by presence or absence of GFP, respectively. Synthetic lethality was determined at the life stages at which no homozygotes were observed.
mus81NheI; WRNexoΔ/TM3 P{w[+mC]=ActGFPJMR2, Ser[1] heterozygotes were paired in bottles and females were allowed to lay eggs for 3–4 days. The resulting adult progeny were scored daily following eclosion. The percentage of adult homozygotes (survival ratio) was calculated and was compared to the expected survival ratio of 33% homozygotes, using the chi-square test. mus81NheI; WRNexoD229V/TM3 P{w[+mC]=ActGFPJMR2, Ser[1] were grown and counted in a similar manner and their survival ratios were compared to mus81; WRNexoΔ survival ratios, using a chi-square test.
Results
Characterization of WRNexo null mutants
Previous studies of Drosophila WRNexo were carried out with hypomorphic alleles. Therefore, we used imprecise P-element excision of a fly stock containing the P{EP}G16048 transposable element to generate a WRNexo null mutant, WRNexoΔ. WRNexoΔ deletes 426 bp upstream of the 5′-UTR to 17 bp upstream of the 3′-UTR. (Figure 1B). Using reverse transcriptase PCR, we showed that this deletion does not affect expression of the upstream gene, Nup43 (data not shown). WRNexoΔ mutants are homozygous viable, are fertile, do not have any observable morphological defects, and eclose at Mendelian ratios (data not shown).
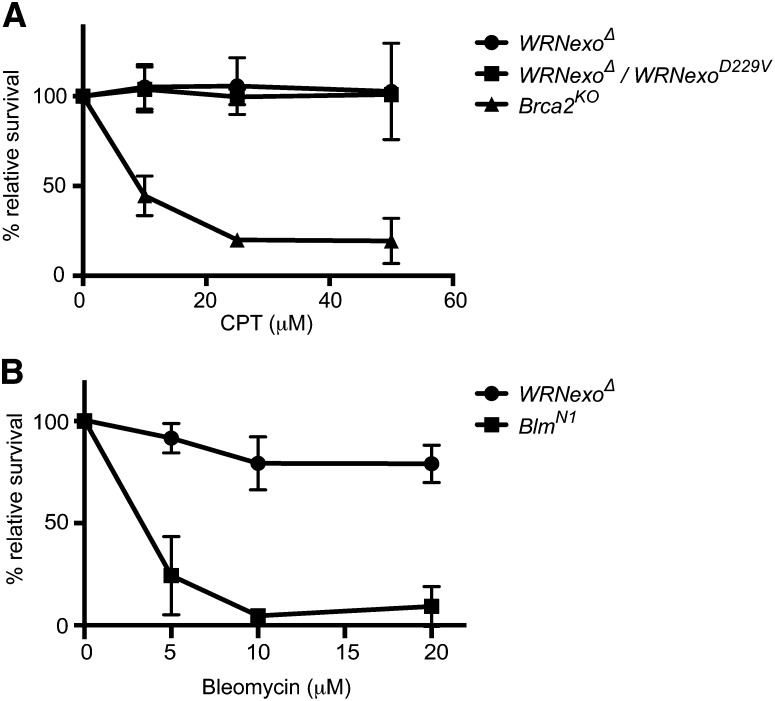
A common phenotype of WS cells is sensitivity to the topoisomerase I inhibitor, camptothecin (CPT), which is due to an inability of these cells to repair DSBs at affected replication forks (Pichierri et al. 2001; Poot et al. 2001). Similarly, the WRNexo hypomorphic fly strain, WRNexoe04496, is sensitive to CPT (Saunders et al. 2008). To assess CPT sensitivity of WRNexoΔ mutants, we treated WRNexoΔ larvae with either CPT or DMSO as a vehicle control. We then calculated relative survival by counting the adult homozygotes eclosed. WRNexoΔ flies were not sensitive to CPT at doses up to 50 μM (Figure 2A). This result is in contrast to CPT sensitivity observed in flies lacking the homologous recombination gene, Brca2 (Thomas et al. 2013). We obtained similar results with flies containing a point mutation in WRNexo that ablates exonuclease activity, WRNexoD229V (Boubriak et al. 2009; Mason et al. 2013). Likewise, when flies were treated with topotecan, a structural analog of CPT, no sensitivity was observed (data not shown). These data suggest that WRNexo does not play an important role in the resolution of DSBs caused by topoisomerase I inhibition.
Figure 2.
WRNexoΔ mutants are not sensitive to double-strand break-inducing agents. (A) Both WRNexoΔ and compound heterozygous WRNexoΔ/WRNexoD229V mutant larvae were exposed to increasing doses of camptothecin (CPT) and adult survival was determined. n = 3 trials for each dose. Brca2KO data were originally reported in Thomas et al. (2013). (B) WRNexoΔ and BlmN1 mutant larvae were exposed to increasing doses of bleomycin and adult survival was calculated. n = 3.
WS cells have also been shown to exhibit a slight sensitivity to ionizing radiation (Bohr et al. 2001; Poot et al. 2001; Yannone et al. 2001), demonstrating involvement of WRN in DSB repair outside of DNA replication. To investigate the possibility that WRNexo is involved in non-replication-based DSB repair, larvae were treated with the radiomimetic agent, bleomycin. Similar to the results with topoisomerase I inhibitors, WRNexoΔ flies were not sensitive to bleomycin at doses up to 25 μM (Figure 2B). In contrast, flies with a deletion of Blm, a RecQ helicase that is important for homologous recombination, were extremely sensitive to bleomycin. Together, these results suggest that WRNexo is not required in homologous recombination-mediated repair of DSBs.
WRNexo is important during early development
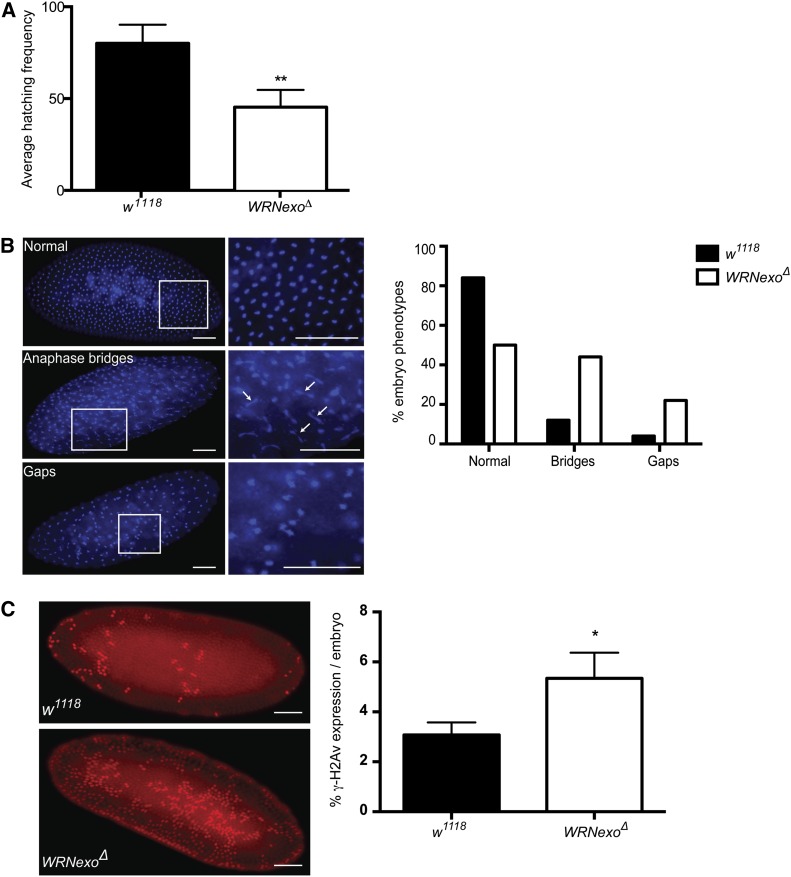
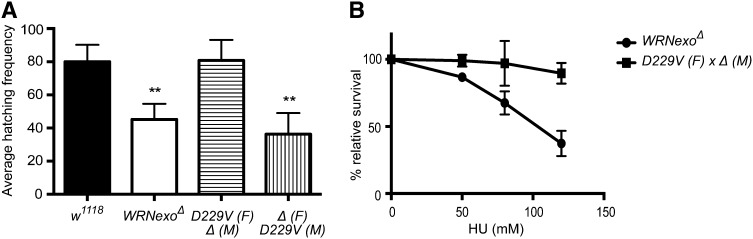
WRNexoΔ mutants exhibit a defect in hatching frequency in which an average of 45% of eggs laid hatch within a 72-hr period compared to 80% of eggs that hatch for w1118 controls (Figure 3A). We hypothesized that this phenotype arose from defects in syncytial nuclear division, a process in which nuclei divide 13 times prior to cellularization in the first 2 hr of embryogenesis (Foe 1993). To test this hypothesis, we stained embryos with the fluorescent DNA marker, DAPI, to visualize syncytial nuclei. Embryos undergoing normal syncytial division exhibit an even spatial pattern of nuclei. In contrast, we observed a range of phenotypes in syncytial WRNexoΔ embryos, including a greater incidence of anaphase bridges, which may indicate incomplete replication or chromosome separation at the time of nuclear division (Figure 3B). We also observed cytoplasmic gaps between nuclei in syncytial WRNexoΔ embryos. This phenotype may be due to the embryo’s response to the presence of DNA damage, in which nuclei containing incompletely replicated DNA fall into the embryo interior (Foe 1993). Together, these phenotypes are consistent with defects in DNA replication and/or proper chromosomal segregation in the absence of WRNexo.
Figure 3.
WRNexo prevents DNA replication defects and the accumulation of double-strand breaks during early embryonic development. (A) Hatching frequencies were determined for eggs laid by w1118 and WRNexoΔ females. n = 3; at least 700 embryos were counted for each independent experiment. **P < 0.01. (B) DAPI staining of WRNexoΔ embryos revealed an increased frequency of nuclear division defects, including the presence of anaphase bridges (arrows) and gaps between nuclei. n = 97 (w1118) and 32 (WRNexoΔ). Bar, 100 μm. (C) Embryos were fixed and stained with an antibody specific for γ-H2Av to determine incidence of double-strand breaks. Image J was used to quantify γ-H2Av staining as a ratio of embryo area and significance was determined by a Mann–Whitney U-test. n = 35 (w1118) and 27 (WRNexoΔ). *P < 0.05. Bar, 100 μm.
Many studies have demonstrated an abundance of DSBs in the absence of WRN either during normal cell growth or following treatment with a replication fork-stalling reagent (Christmann et al. 2008; Franchitto et al. 2008; Liu et al. 2009; Mao et al. 2010; Murfuni et al. 2012). To investigate whether the embryonic nuclear defects we observed were due to an accumulation of DSBs, we stained WRNexoΔ embryos for phosphorylated histone 2Av (γ-H2Av). H2Av is homologous to mammalian H2AX (Madigan et al. 2002) and its phosphorylation is considered a marker for the presence of DSBs and replication stress such as stalled forks (de Feraudy et al. 2010). We observed a greater number of γ-H2Av positive nuclei in WRNexoΔ embryos compared to the w1118 controls (Figure 3C), indicating that WRNexo may be important for the prevention or repair of DSBs during embryogenesis.
WRNexo is important for the stabilization of stalled replication forks
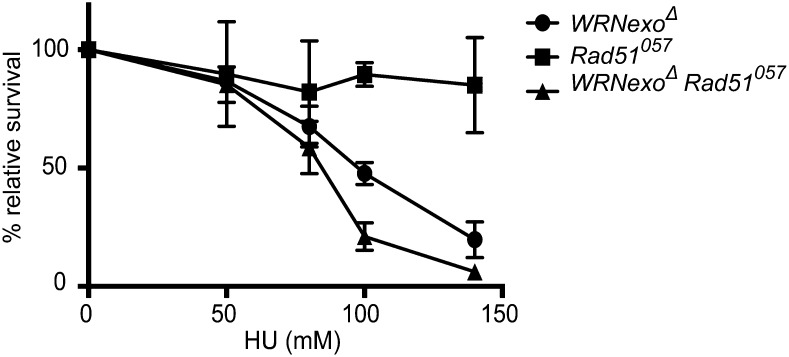
To further investigate a potential role for WRNexo in a replication stress response, we treated WRNexoΔ larvae with increasing concentrations of the fork-stalling reagent, HU. HU induces replication arrest by inhibiting ribonucleotide reductase, leading to localized depletion of dNTPs. WRNexoΔ homozygotes exhibited dose-dependent sensitivity to HU, with only 20% relative survival at 140 mM (Figure 4). Similar HU sensitivity was observed when the WRNexoΔ mutation was combined with a deficiency chromosome, Df(3R)Exel6178, which lacks the WRNexo gene (data not shown). These results demonstrate that the HU sensitivity was caused specifically by loss of WRNexo.
Figure 4.
WRNexo functions in a Rad51-independent pathway in replication. WRNexoΔ, Rad51057, and WRNexoΔ Rad51057 mutant larvae were exposed to hydroxyurea (HU) and adult survival was determined. n = 3–7.
Since stalled replication forks often generate DSBs due to fork collapse, we hypothesized that HU sensitivity in WRNexoΔ mutants could occur because (1) WRNexo is required for repair of DSBs or (2) WRNexo prevents DSBs from occurring through the stabilization or restart of stalled replication forks. To distinguish between these possibilities, we measured HU sensitivity in flies that lack Rad51 and are therefore unable to repair DSBs by homologous recombination (HR) (Staeva-Vieira et al. 2003). Interestingly, Rad51057 mutants were not sensitive to HU (Figure 4), suggesting that when WRNexo is present, treatment with HU does not result in the formation of significant numbers of DSBs that require Rad51-mediated HR repair. WRNexoΔ Rad51057 double mutants were significantly more sensitive to HU than WRNexoΔ single mutants. Thus, HU-induced fork stalling in the absence of WRNexo likely results in the formation of DSBs, at least some of which require Rad51-mediated HR for their repair.
Embryonic defects and HU sensitivity of WRNexo mutants are exonuclease independent
To determine whether the phenotypes observed in WRNexoΔ mutants are due to loss of exonuclease activity, we repeated our experiments with WRNexoD229V flies. The D229V mutant protein has been well characterized in vitro and exhibits no exonuclease activity on WRNexo DNA substrates at physiological conditions (Boubriak et al. 2009; Mason et al. 2013). To control for the effects of potential second-site mutations on the D229V chromosome, WRNexoD229V/WRNexoΔ compound heterozygotes were used in these experiments.
Unlike WRNexoΔ females, WRNexoD229V/WRNexoΔ females did not show a decrease in hatching frequency (Figure 5A). However, embryos laid by WRNexoΔ females that were mated with WRNexoD229V/WRNexoΔ males exhibited a reduced hatching frequency similar to that of WRNexoΔ. The normal hatching frequency of embryos laid by WRNexoD229V females is likely explained by maternal loading of D229V transcript and/or protein into the eggs of WRNexoD229V females since in Drosophila, zygotic transcription does not begin until mitotic cycle 13 or ∼2 hr into embryogenesis (Foe 1993). Together, these data suggest that the presence of WRNexo protein, but not its exonuclease activity, contributes to normal development during the first 2 hr of embryogenesis.
Figure 5.
WRNexo exonuclease activity is not required for normal embryogenesis and hydroxyurea resistance. The D229V point mutation in WRNexo ablates exonuclease activity at physiological conditions (Boubriak et al. 2009; Mason et al. 2013). (A) Hatching frequencies were measured for embryos obtained from crosses between w1118, WRNexoΔ, and WRNexoΔ/WRNexoD229V flies as well as crosses between WRNexoΔ females and WRNexoΔ/WRNexoD229V males. n = 3; at least 700 embryos were counted for each independent experiment. (B) WRNexoΔ/WRNexoD229V compound heterozygous larvae were treated with HU and adult survival was calculated. n = 3. **P < 0.01.
Given this unexpected result, we were interested in investigating whether exonuclease-dead WRNexo protein was sufficient to rescue other WRNexoΔ phenotypic defects. Thus, we assessed the sensitivity of WRNexoD229V mutants to HU. WRNexoD229V/WRNexoΔ virgin females were crossed to WRNexoΔ males and the resulting larvae were treated with HU. Surprisingly, WRNexoD229V mutants were not sensitive to HU (Figure 5B). This result suggests that exonuclease activity is not important for the role of WRNexo in stabilizing or restarting stalled replication forks.
WRNexo may interact with the BLM helicase
In light of our finding that WRNexo exonuclease activity is not important for either normal embryonic development or HU resistance, we speculated that WRNexo may instead recruit another RecQ helicase, such as BLM, to mediate these processes. In humans, both BLM and WRN are important for the resolution of Holliday junctions (Machwe et al. 2011) and have been found to colocalize and physically interact with each other in cell culture (von Kobbe et al. 2002). In Drosophila, BLM is encoded by the mus309 gene and is highly homologous to human BLM. Like WRNexoΔ mutants, Drosophila Blm mutants have nuclear defects during embryogenesis, suggesting that BLM is involved in similar developmental processes (McVey et al. 2007). However, HU sensitivity of Blm mutants has not been reported.
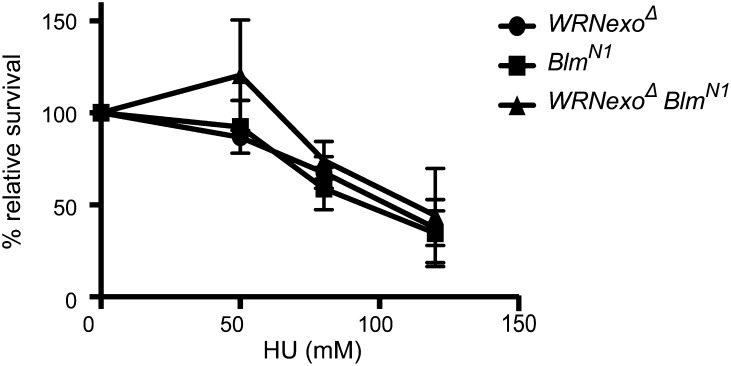
We assessed HU sensitivity in BlmN1 and WRNexoΔ BlmN1 mutants to discern whether BLM shares a role with WRNexo in recovery from fork stalling. BlmN1 and WRNexoΔ BlmN1 mutants exhibited similar sensitivity to WRNexoΔ mutants (Figure 6), suggesting that BLM and WRNexo work in the same pathway following HU-induced replication fork stalling.
Figure 6.
WRNexo and Blm have an epistatic relationship in response to hydroxyurea-induced replication stress. WRNexoΔ, BlmN1, and WRNexoΔ BlmN1 mutant larvae were exposed to hydroxyurea (HU) and adult survival was calculated. n = 3.
Stalled replication forks may form Holliday junctions and intermediates, such as four-way junctions that are cleaved by structure-selective endonucleases (SSEs) such as MUS81, MUS312 (SLX4), and GEN. It is thought that in the absence of BLM, cleavage of these structures by SSEs promotes mitotic crossovers (Andersen et al. 2011). In Drosophila, BLM and SSEs comprise two alternative mechanisms for an essential cellular function, as flies that lack both BLM and a single SSE exhibit developmental stage-specific synthetic lethality. mus81; Blm mutants arrest as pharate adults, while Blm mus312 mutants die as pupae and Blm Gen mutants do not progress past the first-instar larval stage (Andersen et al. 2011).
To determine whether WRNexo is required in the absence of SSEs, we created double mutants and monitored their developmental progression. We observed that WRNexoΔ SSE double mutants also display synthetic lethality, but die at later developmental stages than Blm SSE mutants (Supporting Information, Figure S1). mus81NheI; WRNexoΔ survived to the adult stage; however, homozygotes eclosed at frequencies lower than predicted by Mendelian ratios and demonstrated poor survival (Figure S2). mus312D1 WRNexoΔ mutants survived until the pharate adult stage, while GenZ4325 WRNexoΔ mutants arrested as pupae. Our results are consistent with the observation that Blm Gen mutants have the most deleterious phenotype of all of the Blm SSE mutant combinations (Andersen et al. 2011). Because loss of either BLM or WRNexo results in synthetic lethality in the absence of SSEs, it is likely that these two proteins share a common role in stabilizing or resolving replication intermediates that arise during development.
We were interested to see whether the exonuclease activity of WRNexo is important to prevent the lethality observed in WRNexoΔ SSE double mutants. Thus, we created a mus81NheI; WRNexoD229V mutant, which, like mus81NheI; WRNexoΔ, survived to adulthood. However, in contrast to mus81NheI; WRNexoΔ mutants, mus81NheI; WRNexoD229V homozygotes are healthy and eclose at significantly higher ratios (χ2 = 251, P < 0.001, Figure S2). This result suggests that in the absence of SSEs, the presence of WRNexo, but not its exonuclease activity, is required to produce phenotypically normal adults.
Discussion
The WRN protein is critically important for the maintenance of genome stability, due to its multiple roles in DNA replication and repair and the prevention of aberrant recombination. However, most published WRN studies have focused on potential roles of its helicase domain. We took advantage of Drosophila’s highly conserved exonuclease domain, allowing us to study the role of its exonuclease activity independently from that of the helicase domain. Here, we have demonstrated that Drosophila WRNexo is important for recovery following both endogenous and exogenous replication stress. Importantly, its role is independent of Rad51-mediated homologous recombination repair. Our results also show that the critical role of WRNexo during replication stress does not depend on exonuclease activity, suggesting that it acts as part of a larger protein complex to respond to stalled or collapsed replication forks. Because WRNexoΔ and Blm mutants have similar phenotypes, we speculate that the two RecQ orthologs may constitute or be critical members of this complex.
An important role for WRNexo during early embryogenesis
We have identified a requirement for WRNexo during early embryogenesis as shown by the presence of anaphase bridges and gaps in nuclear distribution in WRNexoΔ embryos. Drosophila embryos go through 13 syncytial nuclear divisions prior to cellularization of the blastoderm, which takes place in the first 2 hr after fertilization (Foe 1993). This rapid replication may result in fork arrests, which, if not restarted, could contribute to improper chromosomal segregation and/or improper nuclear division. These defects can manifest as anaphase bridges and uneven nuclear distribution. It is possible that WRNexoΔ embryos are unable to rapidly process stalled replication forks, resulting in slowed replication that does not allow for proper nuclear division and embryonic development. Human WS cells exhibit a prolonged S phase, indicating slower replication or inhibition of the S-phase checkpoints (Cheng et al. 2007). More specifically, it has been proposed that WRN is required for promoting DNA elongation following replication fork restart, resulting in shorter nascent DNA tracts in cells lacking functional WRN (Rodriguez-Lopez et al. 2002; Sidorova et al. 2008).
Even if WRNexoΔ embryos do successfully complete the syncytial divisions, accumulation of DNA damage may hinder further embryonic development. We observed a greater percentage of γ-H2Av positive nuclei in WRNexoΔ embryos, which can be interpreted as a higher incidence of DSBs. However, it is important to note that the presence of γ-H2Av may not exclusively indicate DSBs, but may also be a signal for replication stress and stalled replication forks (de Feraudy et al. 2010). Our finding is consistent with studies in which elevated levels of endogenous DSBs were observed in WS and WRN-depleted cells (Pichierri et al. 2001; von Kobbe et al. 2004; Szekely et al. 2005; Opresko et al. 2007; Franchitto et al. 2008; Mao et al. 2010).
WRNexo demonstrates a Rad51-independent role in promoting recovery of stalled replication forks
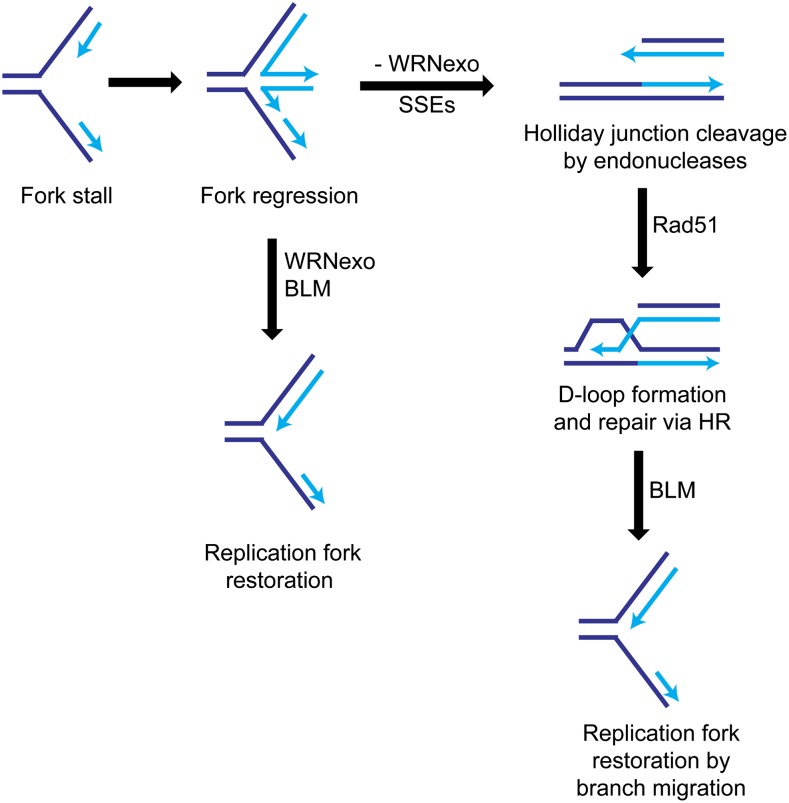
In addition to phenotypic defects caused by endogenous replication stress, WRNexoΔ mutants are sensitive to the fork-stalling reagent, HU. Following HU treatment, the stalled replication fork can either collapse, forming a DSB, or undergo regression, forming an intermediate four-way junction or “chicken foot” structure. Fork restart can occur through reversal of the regressed fork or by cleavage of the Holliday junction by endonucleases, followed by HR-mediated repair (Osborn et al. 2002). To delineate how WRNexo may contribute to stalled fork recovery, we tested HU sensitivity of flies in both WRNexoΔ and Rad51057 mutant backgrounds. WRNexoΔ single mutants were sensitive to HU, whereas Rad51057 larvae lacking Rad51 were resistant to HU. Since Rad51, and therefore HR repair, is not required for HU resistance, it is likely that when WRNexo is present, our treatment protocol does not induce DSBs. Meanwhile, we observed high HU sensitivity in WRNexoΔ Rad51057 double mutants, suggesting that WRNexo and Rad51 operate in separate pathways in response to HU-induced replication fork stalling. We propose that in the presence of WRNexo, regressed replication forks undergo reversal and subsequent recovery and restart (Figure 7). When WRNexo is absent, this reversal process is impaired and the regressed forks can be cleaved by endonucleases. The resultant DSBs can be repaired by HR in a WRNexo-independent manner. This model is supported by evidence that WRN prevents the occurrence of DSBs and subsequent recruitment of Rad51 in human cells (Franchitto et al. 2008; Pichierri et al. 2011).
Figure 7.
A model for the role of WRNexo in recovery from replication fork stalling. Stalled replication forks can undergo regression, forming an intermediate “chicken foot” structure. Fork restart can occur through WRNexo-mediated reversal of the regressed fork, possibly through recruitment of BLM helicase. In the absence of WRNexo, the four-way junctions are cleaved by endonucleases and repaired by HR in a Rad51-dependent manner.
Our finding that WRNexo operates in a Rad51-independent manner in response to HU-induced replication stress contrasts with several lines of evidence supporting a role for WRN in HR. Colocalization of WRN and Rad51 has been observed in human cell culture, although no direct interaction between the proteins has been observed (Sakamoto et al. 2001). Likewise, Sidorova et al. (2013) observed an epistatic relationship between WRN- and Rad51-depleted cells in response to HU treatment, suggesting that these proteins collaborate at stalled forks. We hypothesize that in humans, the role of WRN in HR is helicase mediated, which further supports the use of Drosophila as a model to delineate exonuclease-specific functions of WRN.
Our data demonstrating insensitivity of WRNexoΔ to the topoisomerase I inhibitors camptothecin and topotecan, as well as the radiomimetic agent, bleomycin, further support our hypothesis that WRNexo is not involved in HR repair of DSBs. Both camptothecin and topotecan cause replication-dependent DNA breaks that are usually repaired by HR. Camptothecin sensitivity is a hallmark phenotype of WS cells, likely due to lack of WRN helicase activity. Since WRNexo lacks a helicase domain, a different helicase may be involved in responding to camptothecin-induced damage in Drosophila. Although the WRNexoe04496 hypomorphic mutant is sensitive to camptothecin (Saunders et al. 2008), other observed phenotypic differences between WRNexoe04496 and WRNexoΔ, such as female sterility, lead us to postulate that WRNexoe04496 may contain one or more second-site mutations that could be responsible for these phenotypes.
WRNexo’s role in recovering from replication stress is exonuclease independent
Human WRN exonuclease acts at stalled replication forks, specifically by degrading the leading strand of four-way junctions produced by regression of stalled forks (Machwe et al. 2011). Therefore, we had originally assumed that the defects observed in our WRNexoΔ mutants were due to lack of exonuclease activity. Surprisingly, we found that eggs laid by WRNexoD229V females had normal hatching frequencies and WRNexoD229V mutant larvae were not sensitive to HU.
The biochemical properties of the D229V mutation have been characterized extensively in vitro (Boubriak et al. 2009; Mason et al. 2013). The aspartate at amino acid position 229 is not located within the putative active site of WRNexo. Instead, the D229V mutation is thought to alter the surface structure of the protein, compromising the ability of WRNexo to bind DNA and guide it to the active site (Mason et al. 2013). Under physiological conditions, WRNexo containing the D229V mutation exhibits no exonuclease activity on its preferred substrates: single-strand DNA and double-strand DNA containing a 5′ overhang (Boubriak et al. 2009). Furthermore, the D229V mutation is nonprocessive, limiting digestion to a single nucleotide (Mason et al. 2013). Because D229V ablates exonuclease activity at physiological conditions, it is unlikely that WRNexoD229V mutants possess exonuclease activity that would result in normal phenotypes. In support of this, WRNexoD229V flies exhibit elevated mitotic recombination, suggesting that WRNexo exonuclease activity is required to prevent aberrant HR and excessive recombination (Boubriak et al. 2009).
Since the exonuclease activity of WRNexo is not required for a proper response to endogenous and exogenous replication stress, we hypothesize that WRNexo may instead act as a scaffold for other DNA repair proteins. Human WRN has been shown to physically bind to several proteins within the exonuclease domain, including Ku80 (Li and Comai 2000) and BLM (von Kobbe et al. 2002). Furthermore, it has been suggested that WRN recruits DNA-processing proteins to DNA damage sites due to its ability to bind both proteins and replication intermediates (Kamath-Loeb et al. 2012). Therefore, there is a strong possibility that WRNexo binds similar repair proteins in Drosophila.
WRNexo may interact with BLM at stalled replication forks
We showed that WRNexoΔ, BlmN1, and WRNexoΔ BlmN1 double mutants exhibit similar sensitivity to HU. This epistatic relationship suggests that WRNexo and BLM interact following replication stress and may promote reversal of regressed replication forks (Figure 7). Mao et al. (2010) also discovered an epistatic relationship between WRN and BLM in which codepletion of these proteins suppressed proliferation in cell culture to the same degree as BLM-depleted cells. Similarly, WRN and BLM are both required for fork progression following HU treatment as shown by cell cycle delay when both proteins were depleted (Sidorova et al. 2013). This result demonstrates the ability of WRN and BLM to partially substitute for each other in responding to stalled replication forks, likely due to their shared helicase function. Since WRNexo does not contain a helicase, our results suggest a novel interaction between WRNexo and BLM in recovery of stalled replication forks in Drosophila.
We have also shown that mutants in both WRNexo and the structure-selective endonuclease genes mus312 and Gen are synthetically lethal at different developmental stages. Synthetic lethality was also observed in flies mutant in Blm and mus81, mus312, or Gen, but at earlier developmental time points than observed in WRNexoΔ mutants (Andersen et al. 2011). These results suggest that WRNexo and BLM may have a shared role in development. We hypothesize that WRNexo and BLM are important for an efficient response to replication-related problems that arise during various stages in development. In the absence of WRNexo and BLM, stalled replication forks cannot be restarted and instead, replication intermediates are processed by SSEs (Figure 7). If SSEs are also unavailable, improper chromosome segregation and cell death occur.
Although WRNexo and Blm mutants exhibit similar phenotypes in response to HU treatment and loss of SSEs, it is unlikely that deletion of WRNexo results in destabilization of BLM and a reduction in its activity. We have observed strong sensitivity of BlmN1 mutants to the DSB-inducing reagent, bleomycin, a phenotype not shared by WRNexoΔ mutants (Figure 2B). This result demonstrates that BLM is involved in repair pathways independent of WRNexo and suggests that BLM protein is stably expressed in WRNexoΔ mutants.
We propose that in Drosophila, BLM may serve as a “partner helicase” with WRNexo to carry out functions similar to those of WRN in human cells. This hypothesis is supported by evidence that WRN physically interacts with BLM in human cells and, more importantly, binds BLM within its exonuclease domain (von Kobbe et al. 2002). We showed that the exonuclease activity of WRNexo is not important in recovery from replication stress, using WRNexoD229V mutants. However, because the D229V mutation has been postulated to affect DNA binding (Mason et al. 2013), it is possible that any residual exonuclease activity in this mutant may be enhanced through an interaction with BLM. This seems unlikely, given that in humans, WRN and BLM have different substrate preferences (von Kobbe et al. 2003; Kamath-Loeb et al. 2012), and the exonuclease activity of WRN is inhibited when bound to BLM (von Kobbe et al. 2003). Therefore, our data are most consistent with a scenario in which WRNexo recruits BLM to stalled replication forks where BLM can act to unwind replication intermediates to promote fork progression (Figure 7). WRNexo and BLM may also work together to prevent DSBs from occurring through alternate processing of replication intermediates. This alternate processing can result in unscheduled recombination events and elevated mitotic recombination, which has been described in both Blm and WRNexo mutants (McVey et al. 2007; Saunders et al. 2008).
In summary, our findings support a novel, exonuclease-independent role for WRNexo in recovering from both endogenous and exogenous replication stress in Drosophila. To date, many investigations have attributed WRN’s involvement in replication processes to its helicase activity. Therefore, our findings suggest that further investigation of exonuclease-specific functions of WRN is warranted.
Acknowledgments
We thank Jeff Sekelsky for his generous donation of fly stocks and the γ-H2Av antibody and Adam Thomas for use of his data describing CPT sensitivity in Brca2KO mutants. We also thank the Bloomington Stock Center for mutant fly stocks. This work was funded by the National Institute of General Medical Sciences Institutional Research and Academic Career Development Award (IRACDA) program (K12GM074869) and by National Science Foundation (NSF) grant MCB0643253. Additionally, our undergraduate researchers were supported by the NSF Research Experience for Undergraduates program (DBI1263030).
Footnotes
Communicating editor: S. E. Bickel
Literature Cited
- Adams M. D., McVey M., Sekelsky J. J., 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267 [DOI] [PubMed] [Google Scholar]
- Andersen S. L., Kuo H. K., Savukoski D., Brodsky M. H., Sekelsky J., 2011. Three structure-selective endonucleases are essential in the absence of BLM helicase in Drosophila. PLoS Genet. 7: e1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., et al. , 2004. The BDGP Gene Disruption Project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr V. A., Souza Pinto N., Nyaga S. G., Dianov G., Kraemer K., et al. , 2001. DNA repair and mutagenesis in Werner syndrome. Environ. Mol. Mutagen. 38: 227–234 [DOI] [PubMed] [Google Scholar]
- Boubriak I., Mason P., Clancy D., Dockray J., Saunders R., et al. , 2009. DmWRNexo is a 3′-5′ exonuclease: phenotypic and biochemical characterization of mutants of the Drosophila orthologue of human WRN exonuclease. Biogerontology 10: 267–277 [DOI] [PubMed] [Google Scholar]
- Cheng W.-H., Muftuoglu M., Bohr V. A., 2007. Werner syndrome protein: functions in the response to DNA damage and replication stress in S-phase. Exp. Gerontol. 42: 871–878 [DOI] [PubMed] [Google Scholar]
- Christmann M., Tomicic M. T., Gestrich C., Roos W. P., Bohr V. A., et al. , 2008. WRN protects against topo I but not topo II inhibitors by preventing DNA break formation. DNA Repair 7: 1999–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W. K., Hickson I. D., 2009. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer 9: 644–654 [DOI] [PubMed] [Google Scholar]
- Chun S. G., Shaeffer D. S., Bryant-Greenwood P. K., 2011. The Werner’s Syndrome RecQ helicase/exonuclease at the nexus of cancer and aging. Hawaii Med. J. 70: 52–55 [PMC free article] [PubMed] [Google Scholar]
- Compton S. A., Tolun G., Kamath-Loeb A. S., Loeb L. A., Griffith J. D., 2008. The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J. Biol. Chem. 283: 24478–24483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Feraudy S., Revet I., Bezrookove V., Feeney L., Cleaver J. E., 2010. A minority of foci or pan-nuclear apoptotic staining of γH2AX in the S phase after UV damage contain DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 107: 6870–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe, V. E., G. M. Odell, and B. A. Edgar, 1993 Mitosis and Morphogenesis in the Drosophila Embryo: Point and Counterpoint Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- Franchitto A., Pirzio L. M., Prosperi E., Sapora O., Bignami M., et al. , 2008. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J. Cell Biol. 183: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. D., Shen J. C., Kamath-Loeb A. S., Blank A., Sopher B. L., et al. , 1997. The Werner syndrome protein is a DNA helicase. Nat. Genet. 17: 100–103 [DOI] [PubMed] [Google Scholar]
- Kamath-Loeb A., Loeb L. A., Fry M., 2012. The Werner syndrome protein is distinguished from the Bloom syndrome protein by its capacity to tightly bind diverse DNA structures. PLoS ONE 7: e30189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb A. S., Shen J. C., Loeb L. A., Fry M., 1998. Werner syndrome protein. II. Characterization of the integral 3′ → 5′ DNA exonuclease. J. Biol. Chem. 273: 34145–34150 [DOI] [PubMed] [Google Scholar]
- Klovstad M., Abdu U., Schüpbach T., 2008. Drosophila brca2 is required for mitotic and meiotic DNA repair and efficient activation of the meiotic recombination checkpoint. PLoS Genet. 4: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koundakjian E. J., Cowan D. M., Hardy R. W., Becker A. H., 2004. The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics 167: 203–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake C. M., Holsclaw J. K., Bellendir S. P., Sekelsky J., Hawley R. S., 2013. The development of a monoclonal antibody recognizing the Drosophila melanogaster phosphorylated histone H2A variant (gamma-H2AV). G3 3: 1539–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Comai L., 2000. Functional interaction between Ku and the Werner syndrome protein in DNA end processing. J. Biol. Chem. 275: 28349–28352 [DOI] [PubMed] [Google Scholar]
- Liu F. J., Barchowsky A., Opresko P. L., 2009. The Werner syndrome protein functions in repair of Cr(VI)-induced replication-associated DNA damage. Toxicol. Sci. 110: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A., Karale R., Xu X., Liu Y., Orren D. K., 2011. The Werner and Bloom syndrome proteins help resolve replication blockage by converting (regressed) Holliday junctions to functional replication forks. Biochemistry 50: 6774–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan J. P., Chotkowski H. L., Glaser R. L., 2002. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 30: 3698–3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao F. J., Sidorova J. M., Lauper J. M., Emond M. J., Monnat R. J., 2010. The human WRN and BLM RecQ helicases differentially regulate cell proliferation and survival after chemotherapeutic DNA damage. Cancer Res. 70: 6548–6555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P. A., Boubriak I., Robbins T., Lasala R., Saunders R., et al. , 2013. The Drosophila orthologue of progeroid human WRN exonuclease, DmWRNexo, cleaves replication substrates but is inhibited by uracil or abasic sites: analysis of DmWRNexo activity in vitro. Age 35: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Andersen S. L., Broze Y., Sekelsky J., 2007. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 176: 1979–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfuni I., De Santis A., Federico M., Bignami M., Pichierri P., et al. , 2012. Perturbed replication induced genome-wide or at common fragile sites is differently managed in the absence of WRN. Carcinogenesis 33: 1655–1663. [DOI] [PubMed] [Google Scholar]
- Opresko P. L., von Kobbe C., Laine J. P., Harrigan J., Hickson I. D., et al. , 2002. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 277: 41110–41119 [DOI] [PubMed] [Google Scholar]
- Opresko P. L., Calvo J. P., von Kobbe C., 2007. Role for the Werner syndrome protein in the promotion of tumor cell growth. Mech. Ageing Dev. 128: 423–436 [DOI] [PubMed] [Google Scholar]
- Orren D. K., Theodore S., Machwe A., 2002. The Werner syndrome helicase/exonuclease (WRN) disrupts and degrades D-Loops in vitro. Biochemistry 41: 13483–13488 [DOI] [PubMed] [Google Scholar]
- Osborn A. J., Elledge S. J., Zou L., 2002. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 12: 509–516 [DOI] [PubMed] [Google Scholar]
- Pichierri P., Franchitto A., Mosesso P., Palitti F., 2001. Werner’s syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol. Biol. Cell 12: 2412–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P., Ammazzalorso F., Bignami M., Franchitto A., 2011. The Werner syndrome protein: linking the replication checkpoint response to genome stability. Aging 3: 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M., Yom J. S., Whang S. H., Kato J. T., Gollahon K. A., et al. , 2001. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 15: 1224–1226 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez A. M., Jackson D. A., Iborra F., Cox L. S., 2002. Asymmetry of DNA replication fork progression in Werner’s syndrome. Aging Cell 1: 30–39 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez A. M., Whitby M. C., Borer C. M., Bachler M. A., Cox L. S., 2007. Correction of proliferation and drug sensitivity defects in the progeroid Werner’s Syndrome by Holliday junction resolution. Rejuvenation Res. 10: 27–40 [DOI] [PubMed] [Google Scholar]
- Sakamoto S., Nishikawa K., Heo S. J., Goto M., Furuichi Y., et al. , 2001. Werner helicase relocates into nuclear foci in response to DNA damaging agents and co-localizes with RPA and Rad51. Genes Cells 6: 421–430 [DOI] [PubMed] [Google Scholar]
- Saunders R. D. C., Boubriak I., Clancy D. J., Cox L. S., 2008. Identification and characterization of a Drosophila ortholog of WRN exonuclease that is required to maintain genome integrity. Aging Cell 7: 418–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J. C., Loeb L. A., 2000. Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res. 28: 3260–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova J. M., 2008. Roles of the Werner syndrome RecQ helicase in DNA replication. DNA Repair 7: 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova J. M., Li N., Folch A., Monnat R. J., 2008. The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle 7: 796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova J. M., Kehrli K., Mao F., Monnat R., Jr, 2013. Distinct functions of human RECQ helicases WRN and BLM in replication fork recovery and progression after hydroxyurea-induced stalling. DNA Repair 12: 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staeva-Vieira E., Yoo S., Lehmann R., 2003. An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 22: 5863–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely A. M., Bleichert F., Nümann A., Van Komen S., Manasanch E., et al. , 2005. Werner protein protects nonproliferating cells from oxidative DNA damage. Mol. Cell. Biol. 25: 10492–10506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A. M., Hui C., A. South, and M. McVey, 2013. Common variants of Drosophila melanogaster Cyp6d2 cause camptothecin sensitivity and synergize with loss of Brca2. G3 3: 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kobbe C., Karmakar P., Dawut L., Opresko P., Zeng X., et al. , 2002. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J. Biol. Chem. 277: 22035–22044 [DOI] [PubMed] [Google Scholar]
- von Kobbe C., Thoma N. H., Czyzewski B. K., Pavletich N. P., Bohr V. A., 2003. Werner syndrome protein contains three structure-specific DNA binding domains. J. Biol. Chem. 278: 52997–53006 [DOI] [PubMed] [Google Scholar]
- von Kobbe C., May A., Grandori C., Bohr V. A., 2004. Werner syndrome cells escape hydrogen peroxide-induced cell proliferation arrest. FASEB J. 18: 1970–1972. [DOI] [PubMed] [Google Scholar]
- Xue Y., Ratcliff G. C., Wang H., Davis-Searles P. R., Gray M. D., et al. , 2002. A minimal exonuclease domain of WRN forms a hexamer on DNA and possesses both 3′-5′ exonuclease and 5′-protruding strand endonuclease activities. Biochemistry 41: 2901–2912 [DOI] [PubMed] [Google Scholar]
- Yannone S. M., Roy S., Chan D. W., Murphy M. B., Huang S., et al. , 2001. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J. Biol. Chem. 276: 38242–38248 [DOI] [PubMed] [Google Scholar]