Abstract
Potassium (K) deficiency is one of the major factors affecting crop growth and productivity. Development of low-K tolerant crops is an effective approach to solve the nutritional deficiency in agricultural production. Tibetan annual wild barley is rich in genetic diversity and can grow normally under poor soils, including low-K supply. However, the molecular mechanism about low K tolerance is still poorly understood. In this study, Illumina RNA-Sequencing was performed using two Tibetan wild barley genotypes differing in low K tolerance (XZ153, tolerant and XZ141, sensitive), to determine the genotypic difference in transcriptome profiling. We identified a total of 692 differentially expressed genes (DEGs) in two genotypes at 6 h and 48 h after low-K treatment, including transcription factors, transporters and kinases, oxidative stress and hormone signaling related genes. Meanwhile, 294 low-K tolerant associated DEGs were assigned to transporter and antioxidant activities, stimulus response, and other gene ontology (GO), which were mainly involved in starch and sucrose metabolism, lipid metabolism and ethylene biosynthesis. Finally, a hypothetical model of low-K tolerance mechanism in XZ153 was presented. It may be concluded that wild barley accession XZ153 has a higher capability of K absorption and use efficiency than XZ141 under low K stress. A rapid response to low K stress in XZ153 is attributed to its more K uptake and accumulation in plants, resulting in higher low K tolerance. The ethylene response pathway may account for the genotypic difference in low-K tolerance.
Introduction
Mineral nutrition is crucial for plant growth and development. However, many plants are often subjected to nutrition stress due to insufficient nutrient supply in soils. Like nitrogen (N), potassium (K) is one of the most abundant elements in plants and performs vital functions in growth, stress adaptation and metabolism, as it is involved in stoma movement, enzyme activation, maintenance of cytosolic pH homeostasis, and stabilization of protein synthesis, etc [1]–[6]. Although K is quite abundant in the lithosphere and soils, being nearly 10 times higher than N and phosphorus (P) in terms of absolute content, most of them (90–98%) exists in the form of unavailability for plants [6], [7]. In other words, available potassium content in soils is commonly very low. In China, most soils show K deficiency for crops, and the case become more severe in recent decades, accompanied by a wide planting of hybrid rice, as it absorbs more K and is more sensitive to low K than inbred rice [8], [9].
On the other hand, plants have developed the strategies of coping with low-K stress. It has been well documented that there is a dramatic difference among plant species and genotypes within a species in the response to low-K stress [6], [7], indicating that K nutrition in plants is a genetically-controlled trait, and can be improved by genetic manipulation. Thus, it is imperative for us to reveal the mechanism or to explore the relevant genes of high K use efficiency. However, narrower genetic diversity in cultivated barley has become a bottleneck for genetic improvement [10]. The Tibetan Plateau is one of the centers of cultivated barley, and well known for its extreme environment [11]. The Tibetan annual wild barley (referred to wild barley thereafter) has been proved to be rich in genetic diversity and high tolerance to abiotic stresses, such as drought and salinity [12]–[14]. In the previous experiments, we found that wild barley grew well in the soils with poor fertility and less fertilizer application. Therefore it is possible that wild barley has the special mechanisms in tolerance to low-K stress.
Transcriptome analysis has been widely used in studies of functional genomics. There are two major approaches in the studies of transcriptomes, i.e. sequencing-based and hybridization-based. With the rapid advancement of High-throughput sequencing or so-called Next Generation Sequencing (NGS), RNA-Seq has recently become an attractive method. Compared with hybridization-based tool, such as microarray, RNA-Seq emerges as higher sensitivity, greater dynamic range of expression and base-pair resolution for transcription profiling [15]–[17]. Furthermore, it also shows clear advantages in revealing novel transcribed regions, single nucleotide polymorphisms (SNPs), the precise location of transcription boundaries and splice isoforms [18], [19]. This technique has been used in many plants to reveal gene annotation and expression under biotic and abiotic stresses [20]–[24].
Previous studies suggested that there is a considerable genetic variation in low-K tolerance among the wild barley accessions [25]. However, a comprehensive transcriptomic analysis of wild barley in response to low-K stress is still not done up to date. Based on the evaluation of low-K tolerance of 99 wild barley genotypes (accessions), we selected 2 wild barley accessions (XZ153, low-K-tolerant and XZ141, low-K-sensitive) as materials in transcriptome analysis using the Illumina RNA-Seq method. The objectives of this study are to determine (1) the possible difference in transcriptome profiles of two wild barley accessions in response to low-K stress; and (2) the signaling pathways and regulatory networks related to low-K tolerance.
Materials and Methods
Plant materials and low-K stress
A hydroponic experiment was conducted in a greenhouse with natural light at Zijingang Campus, Zhejiang University, China. Seeds of two wild barley accessions (XZ153, low-K-tolerant and XZ141, low-K-sensitive) were sterilized with 2% H2O2 for 30 min and rinsed with distilled water for three times, then soaked for 6 h at room temperature. The seeds were germinated on moistened filter papers in the germination boxes, placed into a plant growth chamber (22/18°C, day/night). Ten-days-old seedlings were transplanted into plastic pots (5L) for hydroponic incubation. The full-strength nutrient solution contains: 1 mM Ca(NO3)2.4H2O, 1 mM KCl, 1 mM MgSO4, 0.25 mM NH4H2PO4, 50 µM CaCl2, 20 µM Fe-citrate.nH2O, 12.5 µM H3BO3, 0.5 µM H2MoO4, 0.5 µM CuSO4.5H2O, 2 µM MnCl2.4H2O, 2 µM ZnSO4.7H2O. The pH was adjusted to 6.0±0.1 as required. Plants were supplied with half-strength of the hydroponic solution in the first week and then changed into full strength solution from the next week and renewed every five days. Three-leaf-stage seedlings were subjected to low-K treatment. The potassium concentration was adjusted to 0.01 mM (low-K treatment) and 1 mM as control, respectively.
Biomass and potassium content determination
At 15 d after low-K treatment, the roots of all seedlings were thoroughly rinsed with tap water and dried with tissue papers. Then shoots and roots of seedlings were harvested and separated. All the plant samples were dried at 80°C for 72 h until their weight remained constant for biomass measurement. Dry shoots and roots were ground into powder, and approximately 0.1 g tissue sample was prepared for K content determination using Inductively Coupled Plasma-Optical Emission Spectroscopy (ICP-OES) (Optima 8000DV, PerkinElmer, USA).
Gene expression assay
For time course pre-analysis of the expression of the gene HvHAK1 under low-K stress, seeds of XZ153 were germinated and seedlings were cultivated as described above. All the endosperms were removed away from the seedlings to eliminate any additional supply of nutrition. The plants were incubated with 1/2 strength nutrient solution for 5 d and refreshed with full-strength for another 5 d. Then the two-leaf-stage seedlings were treated under low-K (0.01 mM) and normal K (1 mM) conditions. The roots of XZ153 were sampled with 3 biological replicates at 6 h, 12 h, 24 h, 48 h, 5 d and 7 d after treatment. The root samples were frozen in liquid nitrogen immediately and stored at −80°C for RNA extraction.
RNA-Seq sampling and RNA isolation
For RNA-Seq sampling, seeds of XZ153 and XZ141 were germinated at the same condition and placed into a plant growth chamber. The two-leaf-stage seedlings were exposed to low-K stress (0.01 mM) for 0 h, 6 h and 48 h, respectively. Roots of 10 seedlings were collected and mixed together at each time point to reduce the differences between plant individuals. There were 6 samples [2 genotypes (XZ153, low-K-tolerant and XZ141, low-K-sensitive) ×3 time periods (0 h, 6 h, 48 h)] in total for further RNA-Seq research. RNA isolation was carried out according to the instructions of miRNeasy mini kit (QIAGEN, Germany). RNA abundances and purity was tested for meeting the requirements.
Library construction, sequencing and data processing
mRNA enrichment was obtained from the total RNA by the magnetic beads with poly-T oligonucleotide. Then mRNA was randomly broken into fragments. Double-stranded cDNA was synthesized using reverse transcriptase combined with random primers and with adapters ligated at both ends. With those adapter sequences, DNA fragments were selectively amplified and enriched. Thus, the cDNA libraries were ready for sequencing. Qubit quantitation, insert size tested by Agilent 2100, and the Q-PCR were also conducted for accurate quantification before sequencing.
PCR products were loaded onto Illumina HiSeq2000 platform for 2×100 bp pair-ends sequencing. Then the RNA-Seq reads were generated via the Illumina data processing pipeline (version 1.8). To obtain the clean data, the raw reads were trimmed by removing empty reads, adaptor sequences and low quality bases at the 3 end. Then all the clean reads were considered for further analysis. The barley genome sequence and annotation data was downloaded, and TopHat (http://tophat.cbcb.umd.edu/) was adopted to align RNA-Seq reads to the barley reference genomes using the ultra high-throughput short read aligner, and then analyzes the mapping results to identify splice junctions between exons.
Identification of the differentially expression genes (DEGs) and quantitative RT-PCR analysis
For gene expression analysis, the expression level of each gene was calculated by quantifying the number of reads. Gene expression counts were normalized by a variation of the FPKM (fragments per kilo-base of exon per million fragments mapped reads) method [26]. To identify differentially expression genes (DEGs) between the two different samples, the software Cufflinks was employed to output the T-statistic and the p-values for each gene [27]. We calculated the expression ratio of 6 h/0 h or 48 h/0 h as fold changes, respectively. Differentially expressed genes (DEGs) were required to have a 2-fold change and p≤0.01. In addition, an FPKM value≥2 in at least one of the samples was applied to genes for statistical analysis [28].
The RNA samples for RNA-Seq were also used for real-time PCR assays to confirm the reliability of the RNA-Seq result. 1 µg total RNA was treated with DNase I to eliminate the genomic DNA contamination, used as a template for reverse transcription (Takara, Japan). First strand cDNA was synthesized with oligo dT primer and Random 6 mers in a 20 µl reaction. Real time PCR was performed on a CFX96 system machine (Bio-Rad, USA). The PCR profiles were as follows: Pre-denaturation at 95°C for 30 s, 40 cycles of denaturation at 95°C for 5 s and annealing at 60°C for 30 s, followed by steps for Melt-Curve analysis (60°C–95°C, 0.5°C increment for 5 s per step). The relative expression of the chosen genes was calculated according to the comparative CT method [29]. In order to normalize all the data, the amplification of HvGAPDH sequence was used as endogenous reference. The gene specific primers were designed using primer-blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). All the primers were listed in Table S6.
Gene annotation, GO enrichment and KEGG analysis
The Blast2GO program was used to obtain GO annotation for the DEGs, as well as for KEGG analysis (http://www.blast2go.com/b2ghome) [30]. BLASTx was performed to align against NCBI non-redundant (nr) protein database for homology search. Following the mapping step, the gene ontology (GO) annotation, InterProScan annotation and enzyme code annotation steps were conducted in details with default parameters. The GOs distribution associated with DEGs were then obtained from three levels: molecular functions, biological processes and cellular components. The KEGG maps containing the EC numbers and enzymatic functions in the context of the metabolic pathways, in which they participate as well as annotation results can be available in a variety of formats [31].
Statistical analysis
The significance of difference between the two barley genotypes in physiological traits and gene expression was examined using data processing system (DPS) statistical software package, followed by the Duncan's Multiple Range Test (DMRT) and the difference at P<0.05 and 0.01 is considered as significant and highly significant, respectively.
Results
Effect of K level on biomass, K concentration and accumulation of two wild barley accessions
A total of 99 barley accessions were evaluated in a previous experiment under low-K (0.01 mM) and normal K (1 mM) conditions [25]. XZ153 and XZ141 were identified as low-K tolerant and sensitive, respectively. Although two genotypes grew worse under low K than normal K (control), XZ153 was obviously less affected than XZ141 (Table 1). Hence, relatively dry weight of shoot (low K/control) was 90% for XZ153 and 64% for XZ141 (Table 1).
Table 1. K concentration and accumulation of two wild barley genotypes XZ153 (Low-K-tolerant) and XZ141 (Low-K-sensitive) under low and normal K levels.
| Trait | Genotype | ||||||
| XZ153 | XZ141 | ||||||
| CK | LK | Relative | CK | LK | Relative | ||
| Dry weight (mg plant−1 DW) | Root | 63.17a | 60.33a | 0.96 | 61.33a | 46.17b | 0.75 |
| Shoot | 207.67a | 187.67a | 0.90 | 212.33a | 135.50b | 0.64 | |
| Total | 270.83a | 248.00b | 0.92 | 273.67a | 181.67c | 0.66 | |
| K concentration (mg g−1 DW) | Root | 65.44a | 10.08b | 0.15 | 64.21a | 9.70b | 0.15 |
| Shoot | 82.33a | 31.08b | 0.38 | 81.31a | 26.15c | 0.32 | |
| K accumulation (mg plant−1 DW) | Root | 4.13a | 0.61b | 0.15 | 3.94a | 0.45b | 0.11 |
| Shoot | 17.10a | 5.83b | 0.34 | 17.27a | 3.54c | 0.21 | |
| Total | 21.23a | 6.44b | 0.30 | 21.21a | 3.99c | 0.19 | |
CK: Normal K level (1 mM K); LK: Low K level (0.01 mM K); Relative: LK/CK. For each line, different lowercase letters indicate significant differences (P<0.01) among the treatments and genotypes, n = 3.
Furthermore, the two wild barley accessions differed greatly in K concentration and accumulation (Table 1). There was little difference in both root and shoot concentrations between XZ153 and XZ141 under normal K. However, shoot K concentration of XZ153 was significantly higher than that of XZ141 under low-K (Table 1). K accumulation is a function of plant dry weight and K concentration. As a result, K accumulation of XZ153 was 1.61 times larger than that of XZ141 under low-K (Table 1).
Evaluation of RNA-Seq reads and mapping results
In order to determine suitable time of sampling for RNA-Seq analysis, relative expression of HvHAK1 at 6 h, 12 h, 24 h, 48 h, 5 d and 7 d after low-K treatment was compared (Figure S1). The results showed that the HvHAK1 gene was up-regulated at 6 h after low-K stress, then remained little change at 12 h and 24 h. Obviously, roots has already sensed low-K signal and activated relevant signal transduction at 6 h after treatment, resulting in differential expression of the genes so as to cope with low-K stress. Interestingly, the expression level of the HvHAK1 gene was significantly increased at 48 h in comparison with those at 6 h, 12 h or 24 h, and thereafter, remained little change at 5 d and 7 d (Figure S1). Thus, we took the samples at 6 h and 48 h for RNA-Seq analysis.
To obtain an overview of the transcriptome profiling of the early response to low-K stress in the two wild barley accessions, six sequencing cDNA libraries were constructed, and paired-end reads were sequenced using the Illumina platform. We got raw reads with length ranging from lower than 50 bp to as high as 101 bp. A total of 223 112 382 clean reads were generated by sequencing 6 cDNA libraries. All reads were categorized into three classes, including unmapped, multiple mapped and unique mapped reads. Of the 29–41 million clean reads from each library, 77–81% was mapped to unique locations, whereas 6–10% was mapped to multiple locations in the genome (Table S1). Meanwhile, the number of expressing genes found in each sample ranged from 54 322 to 57 516, thus providing massive data for further expression profiling analysis.
Identification of differentially expressed genes (DEGs) and cluster analysis
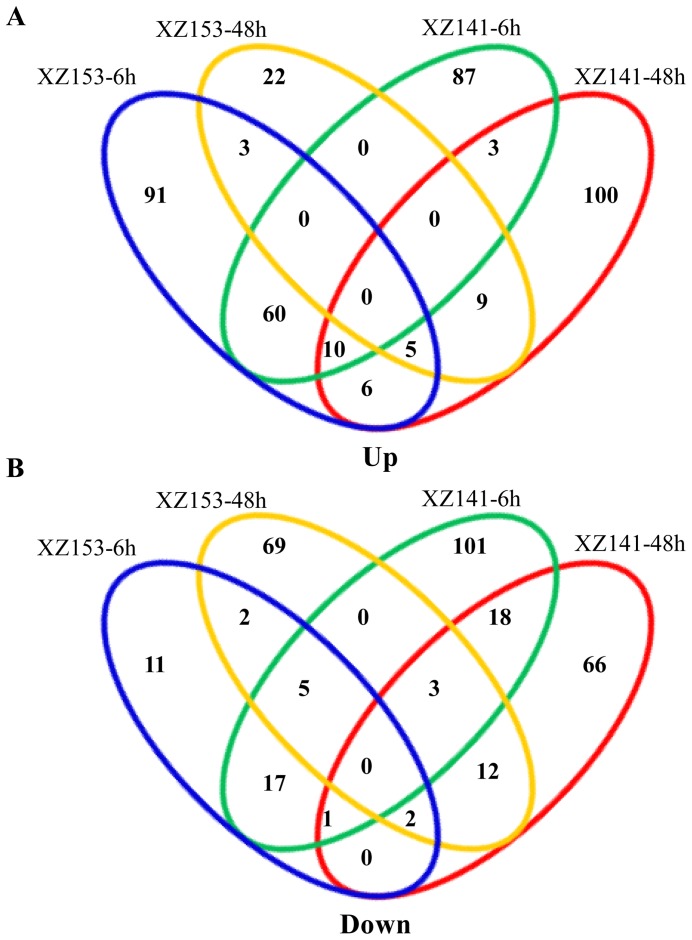
Gene profiles of wild barley roots under both normal and low-K conditions were analyzed. FPKM (Fragments per kilo-base of exon per million fragments mapped reads) method was employed to normalize gene expression counts for the sequence. Previous studies suggested that sequencing with low FPKM may not provide reliable expression data statistically [28]. To minimize false positives, FPKM≥2 was required at least for one of the samples [28]. Additionally, differentially expressed genes were identified according to fold change and P value [32]. Hence, in this study, we set a screening threshold (FPKM≥2 at least in one of the samples, 2-fold change, P≤0.01) of differentially expressed genes. We used the same criteria for both XZ153 and XZ141 to obtain genes that had a significant response to low-K stress. A total of 692 genes showed differential expression at 6 h and 48 h under low-K stress in the two accessions (Table S2, Table S3). XZ141 had more differentially expressed genes than XZ153 (Figure 1). There were only 137 differentially expressed genes, which were commonly found in both XZ153 and XZ141. There were more DEGs at 6 h than at 48 h in the two accessions (Table S4, Table S5). Nearly same amount of DEGs were up-regulated and down-regulated at 6 h and 48 h in XZ141 (Figure 1). However, the gene expression pattern in XZ153 differed from that in XZ141. The number of up-regulated genes in XZ153 was almost four times as large as that of down-regulated ones at 6 h (Figure 1).
Figure 1. A Venn diagram describing overlaps among differentially expressed genes (DEGs) in XZ153 and XZ141.
(A) Up-regulated genes at 6 h and 48 h after low-K treatment. (B) Down-regulated genes at 6 h and 48 h after low-K treatment.
Meanwhile, the 692 DEGs were divided into eight groups, based on their expression pattern by cluster analysis (Genesis 1.7.5) (Figure S2). Clusters 1 and 2 included the genes with continuous positive or negative response along the whole time course; clusters 3 and 4 included the genes with persistent positive or negative response; clusters 5 and 6 included the genes with latent positive or negative response, and cluster 7 and 8 referred to the genes with initial positive or negative response (Figure S2). In view of the differentially expressed gene patterns and DEGs, it can be suggested that XZ153 has a distinct mechanism differing from XZ141 in response to low-K stress. Thus, it is valuable to make further analysis of different responses to low-K stress between the two accessions.
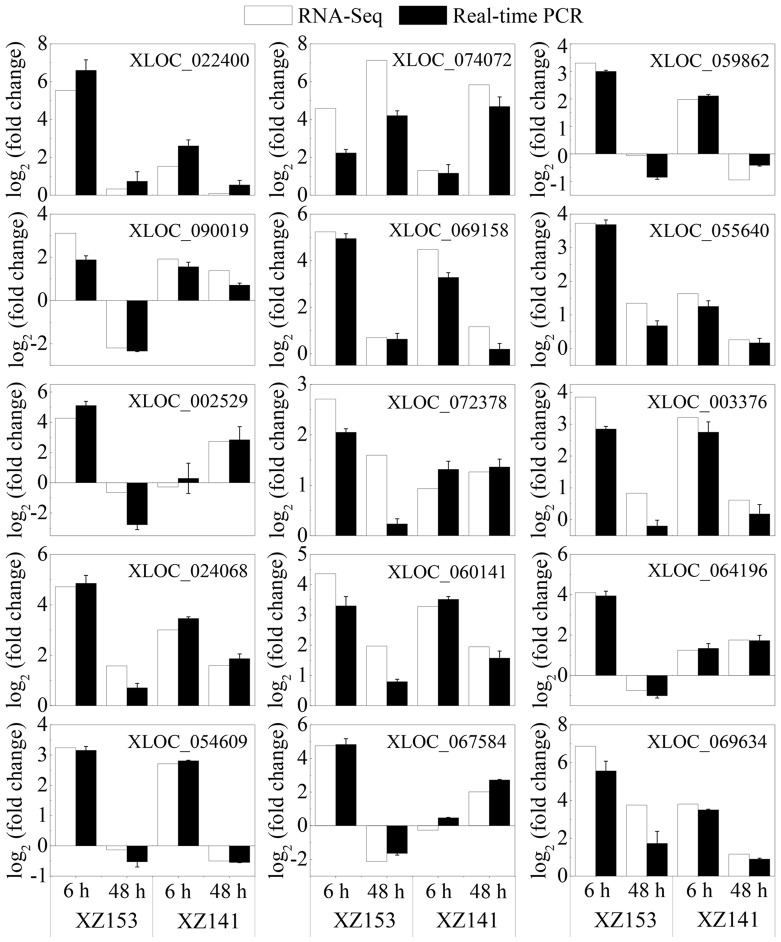
To assess the validity of the RNA-Seq data, 15 DEGs were randomly selected for real-time PCR analysis (Table S6) and they included CBF protein 4 (XLOC_069634), transporter HKT7-like (XLOC_074072), late embryogenesis abundant protein(XLOC_022400), dehydrin 1 (XLOC_090019), kelch repeat-containing F-box family protein (XLOC_069158), chaperone protein DnaJ (XLOC_024068), Ring-h2 finger protein ATL32-like (XLOC_003376), low temperature and salt responsive protein family (XLOC_054609), three pathogenesis-related proteins (XLOC_002529, XLOC_064196, XLOC_067584), response regulator like protein (XLOC_072378), phosphomethylpyrimidine synthase (XLOC_059862) and unknown proteins (XLOC_060141, XLOC_055640) (Table S6). For most of these genes, their expression patterns of the real-time PCR were highly consistent with those shown in the RNA-Seq data (Figure 2).
Figure 2. Quantitative real-time PCR validation of 15 differentially expressed genes.
Gene-specific primers used for real-time PCR are listed in Table S6.
Transcription factors (TFs) in the differentially expressed genes (DEGs)
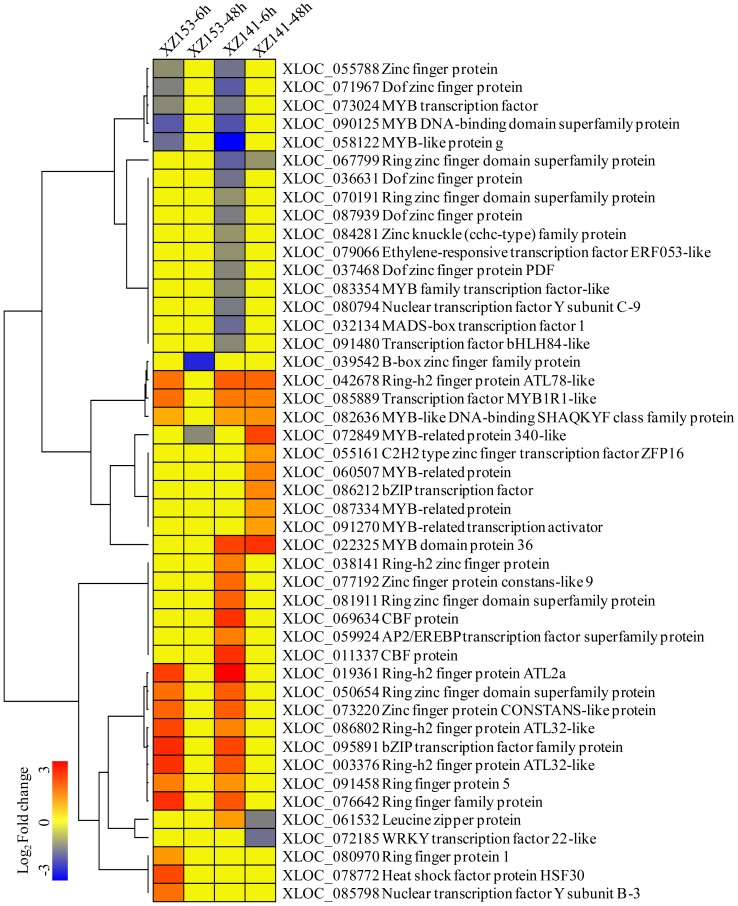
Transcription factors are essential for regulation of gene expression. In this study, 46 DEGs encoding transcription factors were identified, and they belonged to diverse families, such as Zinc finger (22), MYB (11), bZIP (3), CBF (2), NF-Y (2), ERF (1), WRKY (1), bHLH (1), MADS-box (1), AP2/EREBP (1), HSF (1) (Figure 3). Interestingly, we found that the proteins with zinc finger domains were the most enriched among the TFs, accounting for 48% of all DEGs (Figure 3). According to the expression patterns, all the TFs could be clustered into several categories. Overall, most TFs in both two accessions displayed a short-term response, and then returned to the original expression level, although a few of them were only differentially expressed at 48 h (Figure 3). Hence, we were focused on these TFs which were significantly up-regulated in XZ153, but remained little change in XZ141, or remained little change in XZ153 but down-regulated in XZ141.
Figure 3. Average linkage hierarchical cluster analysis of Transcription factors (TFs) identified in differentially expressed genes (DEGs).
The sample and treatments are displayed above each column. Genes are displayed by different colors. Relative levels of expression are showed by a color gradient from low (blue) to high (red).
Transporter and kinase
In the current study, six DEGs encoding K transporters were detected. Surprisingly, expression level of the genes encoding K transporters was not changed obviously until after 48 h stress in XZ141 (Table 2). By contrast, three of them (XLOC_030313, XLOC_086787, XLOC_074072) were up-regulated at 6 h in XZ153, including a HKT7-like gene with higher expression (Table 2). Meanwhile we also found that the expression of the genes encoding nitrate and ammonium transporters changed greatly under low-K stress (Table 2), indicating that K and N uptake is under cross-talking regulation. In addition, the genes associated with ion uptake or translocation were discovered, which is probably involved in ion homeostasis under low-K stress (Table 2). Moreover, some DEGs encoding protein kinases of different groups were also identified (Table 2). Among these genes, there were 4 CBL-interacting protein kinases, 3 leucine-rich repeat receptor-like protein kinases and 2 cysteine-rich receptor-like protein kinases. On the whole, the expression of those groups was mostly down-regulated in XZ141, but up-regulated or unchanged in XZ153.
Table 2. Genes encoding protein transporters and kinases showing genotypic difference expression in response to low K stress.
| Group | Gene id | Log2(Fold change) | Seq description | |||
| XZ153 | XZ141 | |||||
| 6 h | 48 h | 6 h | 48 h | |||
| Potassium | XLOC_030313 | 2.62 | 4.38 | 4.59 | High-affinity potassium transporter | |
| XLOC_086787 | 2.36 | 2.82 | 3.06 | High-affinity potassium transporter | ||
| XLOC_032661 | 2.01 | 2.89 | High-affinity potassium transporter | |||
| XLOC_033262 | 1.89 | Potassium transporter | ||||
| XLOC_035041 | 3.05 | High-affinity potassium transporter | ||||
| XLOC_074072 | 4.59 | 7.13 | 5.84 | Transporter HKT7-like | ||
| Nitrate | XLOC_031163 | 2.14 | Probable nitrate transporter | |||
| XLOC_093217 | 2.04 | 2.22 | Nitrate transporter -like | |||
| XLOC_082123 | −2.61 | −1.89 | −3.79 | Nitrate transporter -like | ||
| XLOC_020339 | 1.94 | High-affinity nitrate transporter -like | ||||
| Ammonium | XLOC_050205 | 2.83 | Ammonium transporter | |||
| XLOC_027452 | 1.86 | Ammonium transporter | ||||
| Yellow-strike | XLOC_082435 | −3.11 | −4.61 | −5.10 | Metal-nicotianamine transporter | |
| XLOC_082568 | −3.19 | −4.05 | −4.89 | Metal-nicotianamine transporter | ||
| XLOC_082319 | −3.19 | −4.62 | −5.00 | Metal-nicotianamine transporter | ||
| XLOC_090161 | −2.26 | Probable metal-nicotianamine transporter | ||||
| Mate | XLOC_071812 | 2.76 | 2.37 | MATE efflux family protein | ||
| XLOC_079551 | 2.34 | MATE efflux family protein | ||||
| XLOC_079205 | −2.23 | MATE efflux family protein, expressed | ||||
| XLOC_035193 | −4.33 | −2.32 | MATE efflux family protein | |||
| CIPK | XLOC_095894 | 4.59 | 4.03 | CBL-interacting protein kinase | ||
| XLOC_050803 | −2.04 | CBL-interacting protein kinase | ||||
| XLOC_083910 | −2.37 | CBL-interacting protein kinase | ||||
| XLOC_065318 | −2.56 | CBL-interacting protein kinase | ||||
| LRR | XLOC_066629 | −3.00 | LRR receptor-like protein kinase | |||
| XLOC_081108 | 3.16 | LRR receptor-like protein kinase | ||||
| XLOC_087240 | 2.80 | LRR receptor-like protein kinase | ||||
| XLOC_084744 | −2.61 | LRR receptor-like protein kinase | ||||
| CRK | XLOC_007825 | −3.52 | −3.99 | Cysteine-rich receptor-like protein kinase | ||
| XLOC_071068 | 5.55 | Cysteine-rich receptor-like protein kinase | ||||
DEGs related to hormone signaling and oxidative stress
Heatmap clustering analysis was performed to study differentially expressed genes involved in hormone signaling in the two barley accessions under low-K stress (Figure S3). A total of 24 hormone-related DEGs were found, including gibberellin (4), jasmonate (5), cytokinin (3), auxin/IAA (8), ethylene (1) and abscisic (3) (Figure S3). Meanwhile, the key enzymes, such as peroxidase (6), cytochrome P450 (8) and glutathione S-transferase (8) were indentified, and they are critical for the regulation of ROS production and reducing cellular damage in the response to low-K stress (Figure S4). At 6 h after low-K treatment, there were few oxidative stress-related genes (Figure S4). However, the changed genes increased dramatically at 48 h after treatment. Moreover, there were more differentially expressed genes in XZ141 than in XZ153 (Figure S4).
Gene ontology (GO) function and KEGG analysis of low-K tolerance related DEGs
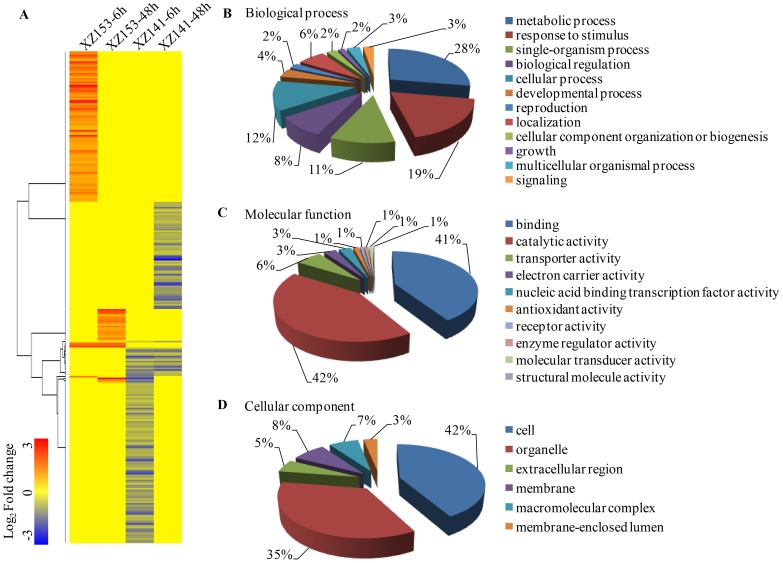
We are focused on those DEGs, whose expression was significantly up-regulated in XZ153 roots, but down-regulated/unchanged in XZ141, or remained little changed in XZ153 but down-regulated in XZ141. In this study, a total of 294 DEGs met the above criteria and were further investigated. Hierarchical clustering analysis of those DEGs was performed and they could be mainly grouped into four classes (Figure 4A). GO functional enrichment analysis revealed that the genes associated with binding (GO: 0005488), catalytic activity (GO: 0003824) and transporter activity (GO: 0005215) were significantly enriched, accounting for as much as 89% of molecular function (Figure 4B). The processes represented by the GO terms ‘metabolic process', ‘response to stimulus’, ‘single-organism process’, ‘biological regulation’ and ‘cellular process’, accounted for the majority of the biological process (Figure 4C). Meanwhile, DEGs related to low-K tolerance also acted as diverse cellular components (Figure 4D).
Figure 4. Hierarchical cluster and gene ontology (GO) categories analysis of low-K tolerance related DEGs.
A total of 294 low-K tolerance related DEGs were performed on (A) Hierarchical cluster analysis. The samples and treatments are displayed above each column. Genes are displayed by different colors and relative levels of expression are showed by a color gradient from low (blue) to high (red). Gene ontology categories from three levels: (B) Molecular function; (C) Biological process; (D) Cellular component.
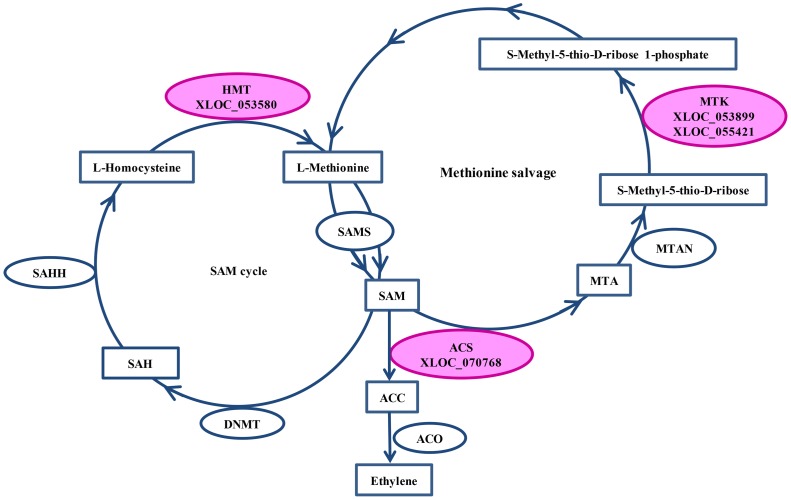
Totally, 294 DEGs encoding various enzymes were further investigated for KEGG pathway enrichment. Forty-seven enzymes were assigned to 32 KEGG pathways, including amino acid, nucleotide, lipid, carbohydrate, energy and other metabolisms (Figure S5). Based on FPKM value, we found 6 enzymes, which were involved in starch and sucrose metabolisms, and altered markedly (Figure S6). In addition, six pathways, classified as lipid metabolism, including alpha-linolenic acid, fatty acid biosynthesis, glycerolipid, glycerophospholipid, linoleic acid and sphingolipid metabolisms, differed in expression patterns between XZ153 and XZ141 (Figure S5). Among them, two DEGs encoding lipase (EC: 3.1.1.3) and 13S-lipoxygenase (EC: 3.2.1.23) showed up-regulation in XZ153, but unchanged in XZ141; whereas three enzymes (EC: 3.2.1.23, EC: 6.3.4.14, EC: 2.3.1.15) were unchanged in XZ153, and down-regulated in XZ141. Meanwhile, the expression of two key enzymes (Homocysteine S-methyltransferase and S-methyl-5-thioribose kinase) participating in S-Adenosyl-L-methionine (SAM) cycle and methionine salvage process, was unchanged in XZ153, but down-regulated in XZ141 (Table S7). Furthermore, one DEGs encoding S-Aminocyclopropane-1-carboxylate synthase (ACS), a key rate-limiting enzyme in ethylene biosynthesis pathway, was also indentified, which showed normal expression in XZ153, and down-regulation in XZ141 (Figure 5, Table S7).
Figure 5. DEGs were mapped to SAM cycle and methionine salvage involving in ethylene biosynthesis process.
The three differentially expressed enzymes were colored in pink. SAM: S-Adenosyl-L-methionine; SAMS: S-Adenosyl-L-methionine synthase; ACC: S-Aminocyclopropane-1-carboxylate; ACS: S-Aminocyclopropane-1-carboxylate synthase; ACO: S-Aminocyclopropane-1-carboxylate oxidase; MTA: S-Methyl-5′-thioadenosine; MTAN: 5′-Methylthioadenosine Nucleosidase; MTK: S-methyl-5-thioribose kinase; SAH: S-Adenosyl-L-homocysteine; SAHH: S-Adenosyl-L-homocysteine hydrolase; DNMT: DNA (cytosine-5-)-methyltransferase; HMT: Homocysteine S-methyltransferase.
Discussion
Potassium is an essential macronutrient for plant growth and development. However, K deficiency in soil is quite common and becomes more severe in crop production [7]. The most effective approach of overcoming K deficiency is to develop the crop cultivars with high tolerance to low-K stress or high K use efficiency. In the present study, we used the RNA-Seq to reveal the transcriptome profiling of two wild barley accessions differing in low K tolerance. Firstly, phenotypic responses of these two genotypes were compared. It was reported by Hermans et al (2006) that biomass was the final result of plant growth and development, so relatively dry weight was often used as indicators of plant tolerance to low nutrition stress [33], [34]. Under low K, root and shoot growth was dramatically inhibited for XZ141, whereas remained less effect for XZ153 (Table 1), proving that XZ153 is more tolerant to low-K stress than XZ141. Meanwhile, the reduction of K concentration and accumulation in the shoot under low K also differed greatly between the two genotypes, XZ153 being less reduced than XZ141, indicating that XZ153 had the higher capability of K absorption and translocation.
The capacity of K absorption and translocation is related to K transporters and channels, which belong to high- and low-affinity uptake system, respectively. A number of K transporters and channels have been functionally characterized in plants [3], [35], [36]. It has been demonstrated that HvHAK1, as well as AtHAK5 and OsHAK1 are all assigned to the KT/KUP/HAK family and could be induced by low external potassium [37]–[41]. The previous studies reported that there were seven HAK-type transporters in the response to low-K stress in rice; including up- and down-regulated expression patterns [42]. However, in this study, all the HAK transporter proteins were up-regulated in the two genotypes. Interestingly, some of them were up-regulated in XZ153 as early as at 6 h after treatment, but not in XZ141 at that time (Table 2). Thus, it may be assumed that a rapid response to low K stress happened in XZ153 is attributed to its more K uptake and accumulation in plants, resulting in higher low K tolerance.
Transcription factors are a kind of proteins that are bound to cis-regulatory elements and can regulate gene expression [43]. The roles of Zinc finger protein and some members of the MYB transcription factor family in abiotic stress tolerance have been well documented [44]–[47]. In the present study, 48 DEGs encoding transcription factors were characterized (Figure 3), with Zinc finger and MYB being the largest components, similar to the results that were obtained in rice plants exposed to K starvation [48]. Obviously, some members of those two families are associated with the responses to low-K stress, at least in rice and barley. Nevertheless, nuclear factor Y (NF-Y), a conserved heterotrimeric and CCAAT-specific domain, composed of NF-YA, NF-YB, NF-YC, was only observed in this study, and was not found in rice and soybean subjected to low-K stress [42], [49]. Although the members of NF-Y are involved in multiple biological functions, its relation with low-K stress tolerance has been not reported up to data [50], [51]. Meanwhile, many TFs, mainly involved in plant growth and development, were repressed in XZ141. It may be assumed that more growth inhibition of XZ141 is described to lower K concentration in plant tissues under low-K stress.
The plant hormone, ethylene is involved in many aspects of the plant life cycle and its production is tightly regulated in response to environmental stimuli from both of biotic and abiotic stresses [52]. Hence, the mechanisms related to biosynthesis of ethylene have been intensively studied. Using chemical and genetic approaches, Jung et al., (2009) found the ethylene-ROS pathway in Arabidopsis that ethylene stimulated the production of ROS and played an important role in low-K stress [53]. As S-Aminocyclopropane-1-carboxylate (ACC) is a direct precursor of ethylene biosynthesis, the step from the (S-Adenosyl-L-methionine synthase) SAM to ACC, catalyzed by ACC synthase (ACS) and usually considered as a rate-limiting step in ethylene synthesis, is very important in this process. In addition to the two enzymes involved in SAM cycle and methionine salvage, a key enzyme ACS was also found in the current study (Figure 5). In particular, the DEGs encoding these three enzymes were inhibited in sensitive genotype XZ141 under low-K stress, but unchanged in the tolerant genotype XZ153 (Table S7). Meanwhile, an ethylene-responsive transcription factor ERF053-like was down-regulated simultaneously in XZ141, while little change in XZ153 (Figure 3). The AP2/ERF transcription factor, RAP2.11 in Arabidopsis, is a factor in regulating transporter through biding to promoter of AtHAK5 and has been also identified as a component of ethylene-ROS pathway under low-K stress [54]. Obviously, the ethylene response pathway may account for the genotypic difference in low-K tolerance, which may be reflected by K uptake capability.
KEGG is a major public pathway-related database, which is valuable for research on the complex biological behaviors of genes. In the present study, 294 DEGs should be specially addressed in view of their expression patterns, i.e. the genes being up-regulated in tolerant genotype XZ153 and remained unchanged or down-regulated in sensitive genotypes XZ141, or unchanged in XZ153, and down-regulated in XZ141 (Figure 4A). KEGG metabolic pathway enrichment of these genes will likely make insight into the mechanism of low-K tolerance. The difference was found in the regulation of some metabolisms between the two genotypes, including carbohydrate, cysteine, methionine and lipid metabolisms (Figure S5). Armengaud et al (2009) reported that the genes involved in carbohydrate metabolism played a vital role in the regulation of potassium nutrition in plants [55]. In this context, these differences in metabolic pathways may lead to different energy distribution and capacity of adaptation to low-K stress in the two genotypes.
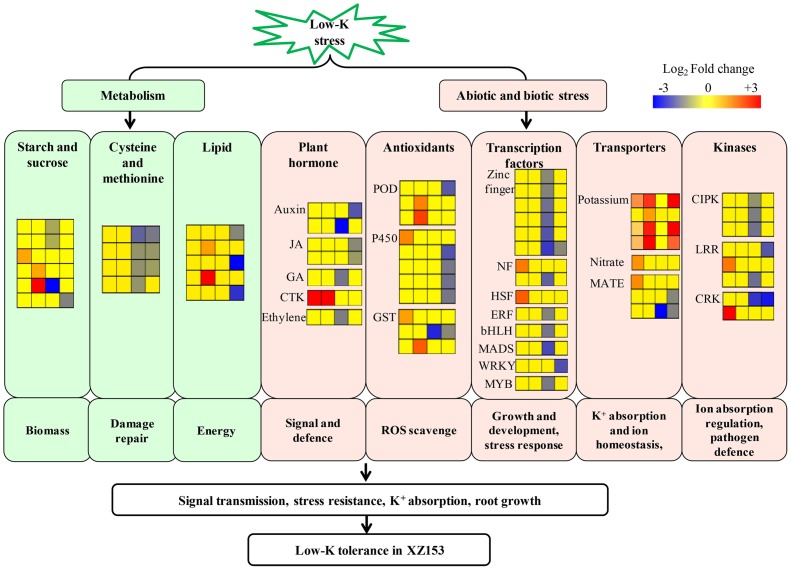
In conclusion, we applied high-throughput Next Generation Sequencing (NGS) technique to investigate gene expression profiling of two Tibetan wild barley genotypes in response to low-K stress. The results demonstrate that the response of XZ153 and XZ141 to low-K stress differed dramatically in the transcriptional level. Despite the complexity of responses to low-K, a hypothetical model could be suggested for low-K tolerance mechanism in XZ153, based on the available results (Figure 6). While the difference in K absorption and accumulation in plant tissues between the two genotypes may explain the different growth inhibition under low K stress. In our understanding, this is a first comprehensive study of low K tolerance in Tibetan wild barley at transcriptome level. In addition, the current results provide some candidate genes, which can be used in barley breeding for improving low-K tolerance.
Figure 6. A hypothetical model of low-K tolerance mechanism underlying in XZ153.
Genes are shown by different colors and relative expression levels are shown by a color gradient from low (blue) to high (red). For each heatmap from left to right: XZ153-6h (first column), XZ153-48h (second column), XZ141-6h (third column), XZ141-48h (fourth column).
Supporting Information
Real-time PCR analysis of the HvHAK1 gene under low K treatment. * represents significant difference according to the Duncan's multiple range, P<0.05, n = 3. Primers of HvHAK1 and GAPDH for RT-PCR are listed in Table S6.
(PDF)
Cluster analysis of the DEGs in the two genotypes. Y-axis represents the gene expression values (FPKM) transformed by logarithms, base 2. The middle white line indicates the gene expression trend of each cluster.
(PDF)
Heat Map analysis of DEGs involved in hormone signaling in XZ153 and XZ141. The sample and treatments are displayed above each column. Genes are displayed by different colors. Relative levels of expression are showed by a color gradient from low (blue) to high (red).
(PDF)
Heat Map analysis of DEGs involved in oxidative stress-related in XZ153 and XZ141. The samples and treatments are displayed above each column. Genes are displayed by different colors. Relative levels of expression are showed by a color gradient from low (blue) to high (red).
(PDF)
KEGG overview of low-K tolerance related DEGs under low K stress. X-axis represents the number of enzymes participating in each pathway; Y- axis depicts the different pathway.
(PDF)
DEGs related to low-K tolerance were mapped to the starch and sucrose metabolism pathway. The column marked in color indicates the differentially expressed enzymes. Four small squares from left to right represent different treatment time of the two varieties (XZ153-6h, XZ153-48h, XZ141-6h, XZ141-48h). Gene expression is displayed by different colors and the relative levels of expression are showed by a color gradient from low (blue) to high (red).
(PDF)
Summary of RNA-seq data and mapping results.
(DOC)
The FPKM value of 692 DEGs.
(XLS)
Gene accession numbers and sequences of 692 DEGs.
(XLS)
DEGs at 6 h and 48 h after low K treatment in XZ153.
(XLS)
DEGs at 6 h and 48 h after low K treatment in XZ141.
(XLS)
The primers used in real-time PCR.
(DOC)
DEGs are involved in Starch and Sucrose metabolism or Cysteine and Methionine metabolism.
(DOC)
Acknowledgments
We thank Prof. Dongfa Sun (Huazhong Agricultural University, China) for providing Tibetan wild barley accessions and Dr. Gulei Jin (Hangzhou Guhe Info-technology Co., Ltd.) for excellent technical assistance and valuable suggestions.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Relevant data are available within the Supporting Information files.
Funding Statement
The authors are grateful for financial support by Natural Science Foundation of China (31330055 and 31171544), “948” Project of Ministry of Agriculture, China (2012-Z25) and China Agriculture Research System (CARS-05). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Amtmann A, Troufflard S, Armengaud P (2008) The effect of potassium nutrition on pest and disease resistance in plants. Physiol Plant 133: 682–691. [DOI] [PubMed] [Google Scholar]
- 2. Britto DT, Kronzucker HJ (2008) Cellular mechanisms of potassium transport in plants. Physiol Plant 133: 637–650. [DOI] [PubMed] [Google Scholar]
- 3. Amtmann A, Armengaud P (2009) Effects of N, P, K and S on metabolism: new knowledge gained from multi-level analysis. Curr Opin Plant Biol 12: 275–283. [DOI] [PubMed] [Google Scholar]
- 4. Maathuis FJM (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12: 250–258. [DOI] [PubMed] [Google Scholar]
- 5. Szczerba MW, Britto DT, Kronzucker HJ (2009) K+ transport in plants: physiology and molecular biology. J Plant Physiol 166: 447–466. [DOI] [PubMed] [Google Scholar]
- 6. Römheld V, Kirkby EA (2010) Research on potassium in agriculture: needs and prospects. Plant Soil 335: 155–180. [Google Scholar]
- 7. Rengel Z, Damon PM (2008) Crops and genotypes differ in efficiency of potassium uptake and use. Physiol Plant 133: 624–636. [DOI] [PubMed] [Google Scholar]
- 8. Fageria NK, Slaton NA, Baligar VC (2003) Nutrient management for improving lowland rice productivity and sustainability. Adv Agron 80: 63–152. [Google Scholar]
- 9. Zhang Y, Zhang CC, Yan P, Chen XP, Yang JC, et al. (2013) Potassium requirement in relation to grain yield and genotypic improvement of irrigated lowland rice in China. J Plant Nutr Soil Sci 176: 400–406. [Google Scholar]
- 10. Ellis RP, Forster BP, Robinson D, Handley LL, Gordon DC, et al. (2000) Wild barley: a source of genes for crop improvement in the 21st century? J Exp Bot 51: 9–17. [PubMed] [Google Scholar]
- 11. Dai F, Nevo E, Wu DZ, Comadran J, Zhou MX, et al. (2012) Tibet is one of the centers of domestication of cultivated barley. Proc Natl Acad Sci USA 109: 16969–16973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao J, Sun HY, Dai HX, Zhang GP, Wu FB (2010) Difference in response to drought stress among Tibet wild barley genotypes. Euphytica 172: 395–403. [Google Scholar]
- 13. Qiu L, Wu DZ, Ali S, Cai SG, Dai F, et al. (2011) Evaluation of salinity tolerance and analysis of allelic function of HvHKT1 and HvHKT2 in Tibetan wild barley. Theor Appl Genet 122: 695–703. [DOI] [PubMed] [Google Scholar]
- 14. Wu DZ, Qiu L, Xu LL, Ye LZ, Chen MX, et al. (2011) Genetic variation of HvCBF genes and their association with salinity tolerance in Tibetan annual wild barley. PLoS One 6: e22938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-Seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ozsolak F, Milos PM (2010) RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cloonan N, Grimmond SM (2008) Transcriptome content and dynamics at single nucleotide resolution. Genome Biol 9: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA -Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 20. Marguerat S, Bähler J (2010) RNA-seq: from technology to biology. Cell Mol Life Sci 67: 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oshlack A, Robinson MD, Young MD (2010) From RNA-seq reads to differential expression results. Genome Biol 11: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mochida K, Shinozaki K (2011) Advances in omics and bioinformatics tools for systems analyses of plant functions. Plant Cell Physiol 52: 2017–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang L, Chen F (2012) Genotypic variation of potassium uptake and use efficiency in cotton Gossypium hirsutum. J Plant Nutr Soil Sci 175: 303–308. [Google Scholar]
- 24. Postnikova OA, Shao J, Nemchinov LG (2013) Analysis of the Alfalfa root transcriptome in response to salinity stress. Plant Cell Physiol 54: 1041–1055. [DOI] [PubMed] [Google Scholar]
- 25. Zhu B, Zeng JB, Wu DZ, Cai SG, Yang LN, et al. (2014) Identification and physiological characterization of low potassium tolerant germplasm in Tibetan Plateau annual wild barley. J Zhejiang Univ (Agric & Life Sci) 40: 165–174. [Google Scholar]
- 26. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, et al. (2012) Differential gene and transcript expression analysis of RNA-Seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zenoni S, Ferrarini A, Giacomelli E, Xumerle L, Fasoli M, et al. (2010) Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol 152: 1787–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 30. Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 31. Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'Rourke JA, Yang SS, Miller SS, Bucciarelli B, Liu JQ, et al. (2013) An RNA-Seq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol 161: 705–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Broadley MR, Bowen HC, Cotterill HL, Hammond JP, Meacham MC, et al. (2004) Phylogenetic variation in the shoot mineral concentration of angiosperms. J Exp Bot 55: 321–336. [DOI] [PubMed] [Google Scholar]
- 34. Hermans C, Hammond JP, White PJ, Verbruggen N (2006) How do plants respond to nutrient shortage by biomass allocation? Trends in Plant Science 11: 610–617. [DOI] [PubMed] [Google Scholar]
- 35. Dreyer I, Uozumi N (2011) Potassium channels in plant cells. FEBS J 278: 4293–4303. [DOI] [PubMed] [Google Scholar]
- 36. Wang Y, Wu WH (2013) Potassium transport and signaling in higher plants. Annu Rev Plant Biol 64: 451–476. [DOI] [PubMed] [Google Scholar]
- 37. Santa-María GE, Rubio F, Dubcovsky J, Rodríguez-Navarro A (1997) The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell 9: 2281–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bañuelos MA, Garciadeblas B, Cubero B, Rodríguez-Navarro A (2002) Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol 130: 784–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gierth M, Maser P, Schroeder JI (2005) The potassium transporter AtHAK5 functions in K+ deprivation induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol 137: 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gierth M, Mäser P (2007) Potassium transporters in plants-involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett 581: 2348–2356. [DOI] [PubMed] [Google Scholar]
- 41. Hong JP, Takeshi Y, Kondou Y, Schachtman DP, Matsui M, et al. (2013) Identification and characterization of transcription factors regulating Arabidopsis HAK5. Plant Cell Physiol 54: 1478–1490. [DOI] [PubMed] [Google Scholar]
- 42. Ma TL, Wu WH, Wang Y (2012) Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biol 12: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee TI, Young RA (2000) Transcription of eukaryotic protein-coding genes. Annu Rev Genet 34: 77–137. [DOI] [PubMed] [Google Scholar]
- 44. Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, et al. (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581. [DOI] [PubMed] [Google Scholar]
- 45. Lin PC, Pomeranz MC, Jikumaru Y, Kang SG, Hah C, et al. (2011) The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA-and GA-mediated growth, stress and gene expression responses. Plant J 65: 253–268. [DOI] [PubMed] [Google Scholar]
- 46. Zhang X, Ju HW, Chung MS, Huang P, Ahn SJ, et al. (2011) The RR-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol 52: 138–148. [DOI] [PubMed] [Google Scholar]
- 47. Jan A, Maruyama K, Todaka D, Kidokoro S, Abo M, et al. (2013) OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol 161: 1202–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shankar A, Singh A, Kanwar P, Srivastava AK, Pandey A, et al. (2013) Gene expression analysis of Rice seedling under potassium deprivation reveals major changes in metabolism and signaling components. PLoS One 8: e70321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang C, Chen H, Hao Q, Sha A, Shan Z, et al. (2012) Transcript profile of the response of two soybean genotypes to potassium deficiency. PLoS One 7(7): e39856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, et al. (2007) Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA 104: 16450–16455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kumimoto RW, Zhang Y, Siefers N, Holt BF (2010) NF-YC3, NF-YC4 and NF-YC9 are required for CONSTAN S-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J 63: 379–391. [DOI] [PubMed] [Google Scholar]
- 52. Merchante C, Alonso JM, Stepanova AN (2013) Ethylene signaling: simple ligand, complex regulation. Curr Opin Plant Biol 16: 554–560. [DOI] [PubMed] [Google Scholar]
- 53. Jung JY, Shin R, Schachtman DP (2009) Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21: 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim MJ, Ruzicka D, Shin R, Schachtman DP (2012) The Arabidopsis AP2/ERF transcription factor RAP2.11 modulates plant response to low-potassium conditions. Mol Plant 5: 1042–1057. [DOI] [PubMed] [Google Scholar]
- 55. Armengaud P, Sulpice R, Miller AJ, Stitt M, Amtmann A, et al. 2009. Multi level analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol 1502: 772–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Real-time PCR analysis of the HvHAK1 gene under low K treatment. * represents significant difference according to the Duncan's multiple range, P<0.05, n = 3. Primers of HvHAK1 and GAPDH for RT-PCR are listed in Table S6.
(PDF)
Cluster analysis of the DEGs in the two genotypes. Y-axis represents the gene expression values (FPKM) transformed by logarithms, base 2. The middle white line indicates the gene expression trend of each cluster.
(PDF)
Heat Map analysis of DEGs involved in hormone signaling in XZ153 and XZ141. The sample and treatments are displayed above each column. Genes are displayed by different colors. Relative levels of expression are showed by a color gradient from low (blue) to high (red).
(PDF)
Heat Map analysis of DEGs involved in oxidative stress-related in XZ153 and XZ141. The samples and treatments are displayed above each column. Genes are displayed by different colors. Relative levels of expression are showed by a color gradient from low (blue) to high (red).
(PDF)
KEGG overview of low-K tolerance related DEGs under low K stress. X-axis represents the number of enzymes participating in each pathway; Y- axis depicts the different pathway.
(PDF)
DEGs related to low-K tolerance were mapped to the starch and sucrose metabolism pathway. The column marked in color indicates the differentially expressed enzymes. Four small squares from left to right represent different treatment time of the two varieties (XZ153-6h, XZ153-48h, XZ141-6h, XZ141-48h). Gene expression is displayed by different colors and the relative levels of expression are showed by a color gradient from low (blue) to high (red).
(PDF)
Summary of RNA-seq data and mapping results.
(DOC)
The FPKM value of 692 DEGs.
(XLS)
Gene accession numbers and sequences of 692 DEGs.
(XLS)
DEGs at 6 h and 48 h after low K treatment in XZ153.
(XLS)
DEGs at 6 h and 48 h after low K treatment in XZ141.
(XLS)
The primers used in real-time PCR.
(DOC)
DEGs are involved in Starch and Sucrose metabolism or Cysteine and Methionine metabolism.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Relevant data are available within the Supporting Information files.