Abstract
Background & Aims
Mitochondrial dysfunction has been implicated in various functional disorders that are co-morbid to Irritable Bowel Syndrome (IBS) such as migraine, depression and chronic fatigue syndrome. The aim of the current case-control pilot study was to determine if functional symptoms in IBS show a maternal inheritance bias, and if the degree of this maternal inheritance is related to mitochondrial DNA (mtDNA) polymorphisms.
Methods
Pedigrees were obtained from N=308 adult IBS patients, N=102 healthy controls, and N=36 controls with Inflammatory Bowel Disease (IBD), all from Caucasian heritage, to determine probable maternal inheritance. Two mtDNA polymorphisms (16519T and 3010A), which have previously been implicated in other functional disorders, were assayed in mtDNA haplogroup H IBS subjects and compared to genetic data from N=344 published haplogroup H controls.
Results
Probable Maternal Inheritance was found in 17.5% IBS, 2% healthy controls and 0% IBD controls (p < 0.0001). No difference was found between IBS and control for 3010A, and a trend was found for 16519T (p=.05). IBS with maternal inheritance were significantly more likely to have the 16519T than controls (OR=5.8; 95%CI=1.5–23.1) or IBS without maternal inheritance (OR=5.2; 95%CI=1.2–22.6).
Conclusions
This small pilot study shows that a significant minority (1/6) of IBS patients have pedigrees suggestive of maternal inheritance. The mtDNA polymorphism 16519T, which has been previously implicated in other functional disorders, is also associated with IBS patients who display maternal inheritance. These findings suggest that mtDNA-related mitochondrial dysfunction may constitute a sub-group within IBS. Future replication studies in larger samples are needed.
Keywords: Functional gastrointestinal disorders, genetics, cellular energy metabolism, comorbidity
Introduction
Irritable Bowel Syndrome (IBS) is a very common condition afflicting 10% of the population1, and many patients struggle with co-morbid conditions such as fibromyalgia, chronic fatigue syndrome, temporomandibular joint disorder, anxiety disorder, and depression2. These conditions are all of “functional" origin, diagnosed based on a constellation of clinical findings but lack clear biological markers. Functional disorders may run in families, and respond to similar treatments (e.g., tricyclic antidepressants), suggesting a shared genetic component and pathophysiology3–5. Many genes have been implied in each of these disorders separately, but so far none of these can explain the overlap.
Defects in cellular energy metabolism have been observed in the brain or muscle of subjects with migraine, cyclic vomiting syndrome, depression, and chronic fatigue syndrome6–9. The vast majority of energy (ATP) is produced by sub-cellular organelles called mitochondria. The mitochondria contain their own DNA (mtDNA) consisting of only 37 genes, all involved in ATP synthesis. Mutations in these genes can lead to disease caused by low cellular energy production10. mtDNA is inherited from the mother only, which opens the possibility of examining maternal inheritance patterns as a way to examine if disease-associated mtDNA polymorphisms may be present. In fact, IBS has been shown to be aggregated more strongly in mothers than in fathers, which suggests a maternal inheritance pattern could be present3.
Of particular note is that mtDNA disorders do not ‘breed true’, but are associated with extraordinary phenotypic variation among affected individuals11. This means that many different types of functional symptoms are common in these families, with each individual having a unique set of such conditions. When multiple functional disorders are ascertained for, preferential inheritance through the maternal line was reported in 20-60% of patients with migraine12, cyclic vomiting syndrome13, depression14, 15 and complex regional pain syndrome16.
Recently, two common mtDNA sequence variants (polymorphisms 3010A and 16519T) have been associated with cyclic vomiting syndrome and migraine12. Camilleri and colleagues17, found no association with IBS. However, they included all IBS patients rather than focusing on the subgroup with likely maternal inheritance patterns, possibly underestimating the effects of mtDNA in IBS.
In the current pilot study, we aimed to explore if functional symptoms are preferentially inherited through the matrilineal line among IBS patients. We also examined if 3010A and 16519T, may distinguish IBS patients with maternal inheritance patterns from IBS patients without maternal inheritance patterns and controls. A last exploratory aim was to identify other possible mtDNA sequence variants associated with the putative maternally-inherited subset of IBS.
Methods
Subjects
IBS patients had a prior physician diagnosis of IBS, met the Rome III criteria for IBS and had no other diagnoses that could cause bowel symptom (e.g., Crohn’s disease). Healthy control subjects were free of any bowel disorders as well as manifestations suggestive of possible mitochondrial dysfunction. Inflammatory Bowel Disease (IBD) control subjects had a physician diagnosis of Ulcerative Colitis or Crohn’s disease as verified by one of the investigators. IBS patients and healthy controls were recruited from 2009–2011 among patients, faculty, staff and students at the University of North Carolina. IBD controls were recruited among patients at the Gastroenterology clinics in the Medical College of Wisconsin. The study was approved in 2008 by the Institutional Review Boards of the University of North Carolina, Children’s Hospital Los Angeles and the Medical College of Wisconsin.
Measures
Quantitative Pedigree Analysis
Detailed pedigrees were collected over the telephone or in person per established protocol12, 16. In short, health information from all first and second-degree relatives was obtained from the subject using a semi-structured interview (for a copy of the interview see Higashimoto et al16). The average number of functional conditions recorded among the matrilineal and non-matrilineal relatives were calculated and compared to each other (“maternal inheritance ratio”). Pedigrees were labeled as “probable maternal inheritance” if the average number of conditions per matrilineal relative was at least 1.75 and functional conditions was at least 3-fold more common in matrilineal versus non-matrilineal relatives (Maternal Inheritance Ratio ≥ 3.0). Pedigrees that did not qualify for maternal inheritance were labeled as “probable non-maternal inheritance”.
Molecular Analyses
Saliva kits (Oragene; DNA Genotek Inc., Ottawa, ON, Canada) for collecting mtDNA were sent to IBS subjects and returned by mail. DNA was isolated from the saliva kits. In order to increase statistical power and assign appropriate controls, we limited all analyses to haplogroup H. The mtDNA haplogroups denote the major groupings of mitochondrial lineage sharing a similar maternal ancestor. Haplogroup H is the most common among Americans (about 30%)18, show minor interfamilial sequence variation, and previous investigations of mtDNA in functional symptoms have largely been limited to haplogroup H12, 17.
Haplogroup H was defined in the conventional manner as the presence of a C at position 7028 by polymerase chain reaction-restriction fragment length polymorphism (RFLP)—7028: AluI F TTTCGGTCACCCTGAAGTTTA, and R AGCGAAGGCTTCTCAAATCAT. Subjects with 7028C (haplogroup H) were tested for the 16519C>T and 3010G>A polymorphisms by PCR/restriction fragment length polymorphism (16519: HaeIII forward GGATGACCCCCCTCAGATA, reverse CTTATTTAAGGGGAACGTG; 3010: BccI forward CATGCTAAGACTTCACCA, reverse TCGTTGAACAAACGAACC).
In 10 haplogroup H IBS patients with the highest degree of maternal inheritance complete cyclosequencing of the mtDNA was performed by a commercial laboratory (Eton, San Diego, CA). The entire mtDNA was amplified using 30 overlapping primer sets as described previously19. Individual sequences were assembled, aligned and compared on Sequencher® software (Gene Codes Corp., Ann Arbor, MI, USA) vs. our reference sequence (revised Cambridge Reference Sequence- MITOMAP10). Complete mtDNA sequences were assigned to haplogroups and clades according to Phylotree.org Build 1419 (http://www.phylotree.org). Sequences were assigned to the closest matching halogroup/clade.
Mitochondrial DNA rearrangements were investigated in the aforementioned 10 haplogroup H IBS patients with the highest degree of maternal inheritance using Long-Range PCR20. Near complete mtDNA was amplified using two primers LR321 TGGCCACAGCACTTAAACACATCTC and LR16215 TGCTGTACTTGCTTGTAAGCATGGG. PCR amplifications were the resolved on a 1.0% agarose gel.
Statistical analyses
Data analyses were performed with SPSS Statistics 19 (IBM). Percentage of pedigrees with a maternal inheritance patterns was compared between IBS patients and controls with Chi2-test. Odds ratios were calculated to compare presence of 3010A and 16519T in haplogroup H IBS patients compared to genetic data from N= 344 published controls12. For the genetic analyses, missing data was not included in the analyses.
Sequence variants in the 10 haplogroup H IBS subjects with the highest degrees of probable maternal inheritance were compared to the most common haplogroup H nucleotide sequence generated from 344 complete mtDNA published sequences. These sequences were ascertained from individuals as part of a population or control study from Europe or North America. To reduce potential bias, no samples were included that were associated with any known illness or symptoms, from self-selected groups (commercial heritage testing), and from islands with small founding and/or geographically isolated populations (Iceland and Sardinia).
Results
Pedigrees were obtained from N=102 healthy controls N=36 IBD patients and N=308 adult IBS patients (for demographic information see Table 1). No differences were found in age and gender between these groups. Probable maternal inheritance was found in 17.5% of IBS patients versus in 2% of healthy controls (p < 0.0001) and 0% of 36 IBD controls (p < .0001). An example of a pedigree with maternal inheritance is available in supplementary Figure a. In addition, supplementary Figure b shows that the majority of control patients have few maternal symptoms per relative and low maternal inheritance ratios, while a subgroup of IBS patients score high on these.
Table 1.
Sample Demographics
| IBS N=308 |
IBD N=36 |
Healthy Controls N=102 |
|
|---|---|---|---|
| Age | Mean= 40.1 | Mean=47.2 | Mean=38.7 |
| Gender | 88.6% female | 63.4% female | 91.2% female |
| Race | 100% Caucasian | 100% Caucasian | 100% Caucasian |
| IBS subtype | 45.5% IBS-C 16.7% IBS-D |
||
| Number of functional symptoms per maternal relative |
Mean=1.29* | Mean=0.53 | Mean=0.62 |
| Number of functional symptoms per non- maternal relative |
Mean=0.54** | Mean=0.29 | Mean=0.42 |
Significant higher compared to IBD and HC (p < .001) by one-way anova.
Significant higher compared to IBD (p < .01) by one-way anova.
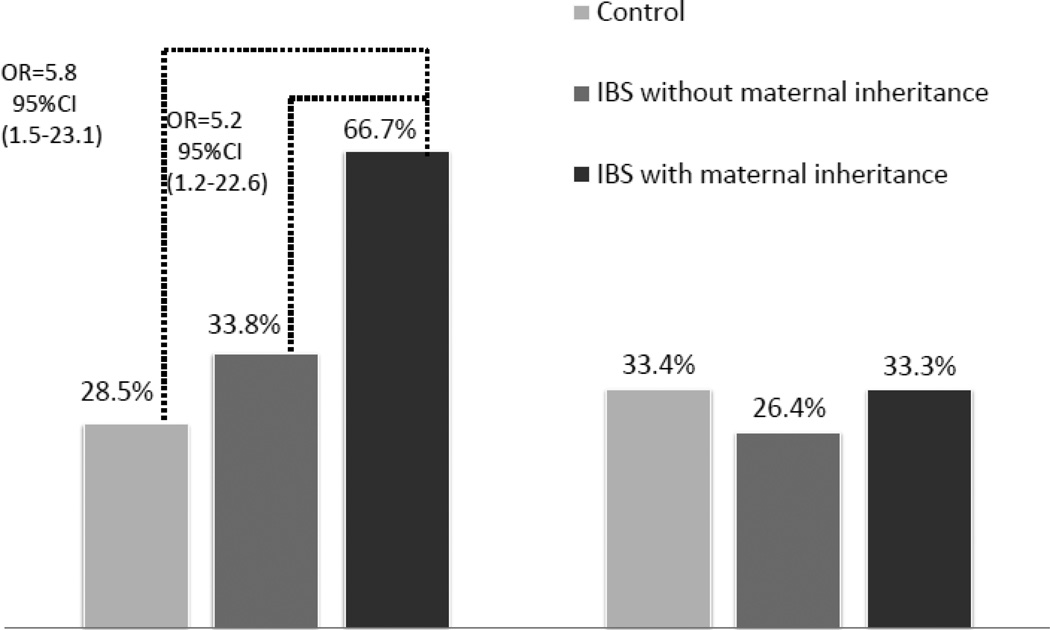
DNA samples were missing or of low quality for 13 IBS patients, leaving mtDNA samples for N=295 IBS patients, and N=86 (29.2%) belonged to haplogroup H. In this group, 3010A was as common as among the 344 population controls (27.4% versus 33.4%; OR=0.74, 95% CI 0.44–1.23), but a trend (p=.05) was found for 16519T (38.4% versus 28.5%; OR=1.6, 95% CI .99–2.63). IBS patients demonstrating probable maternal inheritance were significantly more likely to have the 16519T polymorphism than IBS patients without maternal inheritance or controls (see Figure 1).
Figure 1.
mtDNA polymorphism 16519T is associated with IBS and maternal inheritance.
Full mtDNA sequences of ten IBS patients who had the highest maternal inheritance ratio, identified between 1-11 mtDNA variants per person (see Table 2). Not one variant was shared among all subjects, although 16519T was present in five out of these ten subjects. Of interest, was a high rate of variants in region 110-567: In the IBS patients, 40% had 3 or more variants in this region versus 13.9% in the controls (Chi2=6.64; p < .01).
Table 2.
mtDNA variants in full sequences of 10 IBS patients with high maternal inheritance of functional symptoms.
| Sample | Total Number Of Variants |
Number of Variants in 110–567* |
Variants (Underlined Variants Are HgH Defining sequence variants) |
|---|---|---|---|
| 1 | 12 | 4 |
T239C
302.1 (ins) 302.2 (ins)
302.3 (ins) G366A
G3915A G9380A T11253C T16298C T16362C A16482G C16519T |
| 2 | 12 | 1 |
T152C 302.3 (ins) C3393T C3992T
T4418C
T6776C
10754C 11017G C11563T T12502G T14180C C16519T |
| 3 | 8 | 4 |
302.2 (ins) 302.3 (ins) T309C G316C G366A
G3010A
G6722A C15088T |
| 4 | 6 | 2 | C151T 302.1 (ins) 302.3 (ins) G3010A, T10687T/C C16519T |
| 5 | 9 | 4 |
302.2 (ins) 302.3 (ins) T309C C456T C534A
T4336C
C15833T T16304C C16519T |
| 6 | 10 | 4 |
T146CT195C 302.2 (ins) 302.3 (ins) C456T
G5471A A8343G A14497G T16304C C16519T |
| 7 | 3 | 1 | 302.2 (ins) 302.3 (ins) T14470A |
| 8 | 5 | 1 | C186A 302.3 (ins) G1438A T3396C G4769A |
| 9 | 9 | 2 |
C186A, 302.3 (ins) 567 (ins) G951A G7762A T8715C
C11191T C16173T C16354T |
| 10 | 10 | 1 |
A73G
302.1 (ins), 302.3 (ins) G3010A A8338T G9053A T13830C A16051G A16162G C16465T |
302.3 (ins) occurs in all samples and has therefore been removed from this count of variants.
Discussion
The current pilot study suggests a possible role for mitochondrial DNA in IBS. Our findings indicate that functional symptoms are preferentially maternally inherited in a subset of IBS patients. In addition, the 16519T sequence variant, which has been associated with the functional conditions of migraine and cyclic vomiting syndrome12, may also be associated with the 1/6th of IBS patients who show a maternal inheritance pattern of functional symptoms. The association with all cases of IBS was weak (p=.05), replicating the non-significant results reported earlier17. These findings suggest that a defect in energy metabolism may play a role in IBS among a limited, but substantial, subset of patients. Validation in larger samples are needed.
The location of the 16519T polymorphism may indicate its possible role in IBS and other co-morbid functional disorders. 16519T is located in the 1-kb non-coding mtDNA control region not far from the origin of heavy-strand replication and putative membrane-attachment site10. The control region does not code for proteins or RNA, but replication of mtDNA starts in this region and, therefore, 16519T may be involved in the number or mtDNA genomes available in a cell (cells contain many copies of mtDNA). Future studies are needed to determine the physiological effects of the presence of 16519T.
No reported or obvious disease-associated sequence variants were found on full mtDNA sequencing of the 10 most extreme probable maternally inherited IBS families other than a high prevalence of 16519T as noted above. However, polymorphisms in the 3 side of the control region (#110–567) were far more common among the patient than among the control sequences. A recent study in IBS patients found a similar result in which they reported increased polymorphisms (authors only studied the MT-ATP6-8 regions), but no one polymorphism distinguished IBS patients from controls21. The area identified in the current study, starting at the H-strand replication origin, includes promoters for both strands and conserved sequence blocks. Their locations, plus that of 16519 suggests pathology that predisposes towards functional disease is likely a result of a decreased copy number of mitochondrial-encoded respiratory chain subunits.
If the role of mtDNA in IBS is correct, as suggested by these pilot data, low cellular energy metabolism can contribute towards the development of functional symptoms as varied as gastrointestinal problems, headaches and fatigue. Thus, it may be a mechanism by which comorbid functional symptoms in IBS can be explained. The cause of overlapping co-morbid disorders in IBS is likely complicated and has been associated with increased attention to bodily symptoms, mental health conditions, and wide spread neural hypersensitivity2, 22 some of which are common in patients diagnosed with genetic mitochondrial disorders as well6, 23. Mitochondrial dysfunction warrants investigation as another possible cause for co-morbidity.
The current study has several limitations. First, we studied a relatively small set of haplogroup H patients which yields issues with power, generalizability, and the inability to examine confounders such as age and gender. Second, the control sample for the genetic analyses was drawn from published resources and these samples were not collected for our purpose. Lastly, the quantitative pedigree, to establish phenotype has been used in many other published studies, but almost all of these are among children. Given that both our and pediatric studies have found a positive association with 16519T, suggests that the interview is sensitive in adults as well. More studies are needed to replicate the findings of this small pilot study before any definitive conclusions can be drawn about the role of mitochondrial dysfunction in IBS.
In conclusion, our data suggest a possible role of mitochondrial dysfunction in a subset of IBS patients. Although our study is small and has many limitations, the findings provide rationale for further investigation into mitochondrial dysfunction in IBS. Mitochondrial-targeted therapies have shown some initial promising efficacy in migraine24–27, fibromyalgia28, 29, and cyclic vomiting syndrome30, 31. Thus, investigation of the role of mitochondrial dysfunction in IBS could in the future make an important impact on the care of this disorder.
Supplementary Material
Acknowledgments
Financial support: This study was supported by a research grant from the International Foundation of Functional Gastrointestinal Disorders as well as P01NS045685 and RO1 DK31369. The study sponsors had no role in any part of the study.
Study support
Guarantor of the study: Dr van Tilburg
Abbreviations
- IBD
Inflammatory Bowel Disease
- IBS
Irritable Bowel Syndrome
- mtDNA
mitochondrial DNA
Footnotes
Potential competing interest: None
Conflict of interest
Author contributions:
Dr van Tilburg had primary responsibility of all aspects of the study including concept and design, obtaining funding, recruitment and data collection at UNC, data analysis, and manuscript preparation.
Dr Boles was involved in study concept and design, obtaining funding, data analysis, manuscript preparation and had primary responsibility of analysis and interpretation of the genetic data.
Dr Zaki ran all genetic analyses and was involved in data interpretation and manuscript preparation.
Dr Venkatesan was involved in data collection and critical revision of the manuscript.
References
- 1.Choung RS, Locke GR., 3rd Epidemiology of IBS. Gastroenterol Clin North Am. 2011;40:1–10. doi: 10.1016/j.gtc.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Whitehead WE, Palsson O, Jones KR. Systematic review of the comorbidity of irritable bowel syndrome with other disorders: What are the causes and implications? Gastroenterology. 2002;122:1140–1156. doi: 10.1053/gast.2002.32392. [DOI] [PubMed] [Google Scholar]
- 3.Saito YA, Zimmerman JM, Harmsen WS, et al. Irritable bowel syndrome aggregates strongly in families: a family-based case-control study. Neurogastroenterol. Motil. 2008;20:790–797. doi: 10.1111/j.1365-2982.2007.1077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am. J. Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 5.Russell MB, Iselius L, Olesen J. Migraine without aura and migraine with aura are inherited disorders. Cephalalgia. 1996;16:305–309. doi: 10.1046/j.1468-2982.1996.1605305.x. [DOI] [PubMed] [Google Scholar]
- 6.Gardner A, Boles RG. Mitochondrial energy depletion in depression with somatization. Psychother Psychosom. 2008;77:127–129. doi: 10.1159/000112891. [DOI] [PubMed] [Google Scholar]
- 7.Gardner A, Johansson A, Wibom R, et al. Alterations of mitochondrial function and correlations with personality traits in selected major depressive disorder patients. J Affect Disord. 2003;76:55–68. doi: 10.1016/s0165-0327(02)00067-8. [DOI] [PubMed] [Google Scholar]
- 8.Buchsbaum MS, Wu J, DeLisi LE, et al. Frontal cortex and basal ganglia metabolic rates assessed by positron emission tomography with [18F]2-deoxyglucose in affective illness. J Affect Disord. 1986;10:137–152. doi: 10.1016/0165-0327(86)90036-4. [DOI] [PubMed] [Google Scholar]
- 9.Montagna P. Magnetic resonance spectroscopy in migraine. Cephalalgia. 1995;15:323–327. doi: 10.1046/j.1468-2982.1995.1504323.x. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz-Pesini E, Lott MT, Procaccio V, et al. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smeitink JA. Mitochondrial disorders: clinical presentation and diagnostic dilemmas. J Inherit Metab Dis. 2003;26:199–207. doi: 10.1023/a:1024489218004. [DOI] [PubMed] [Google Scholar]
- 12.Zaki EA, Freilinger T, Klopstock T, et al. Two common mitochondrial DNA polymorphisms are highly associated with migraine headache and cyclic vomiting syndrome. Cephalalgia. 2009;29:719–728. doi: 10.1111/j.1468-2982.2008.01793.x. [DOI] [PubMed] [Google Scholar]
- 13.Boles RG, Adams K, Li BU. Maternal inheritance in cyclic vomiting syndrome. Am J Med Genet A. 2005;133A:71–77. doi: 10.1002/ajmg.a.30524. [DOI] [PubMed] [Google Scholar]
- 14.Burnett BB, Gardner A, Boles RG. Mitochondrial inheritance in depression, dysmotility and migraine? J Affect.Disord. 2005;88:109–116. doi: 10.1016/j.jad.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Bergemann ER, Boles RG. Maternal inheritance in recurrent early-onset depression. Psychiatr Genet. 2010;20:31–34. doi: 10.1097/YPG.0b013e3283351153. [DOI] [PubMed] [Google Scholar]
- 16.Higashimoto T, Baldwin EE, Gold JI, et al. Reflex sympathetic dystrophy: complex regional pain syndrome type I in children with mitochondrial disease and maternal inheritance. Arch Dis Child. 2008;93:390–397. doi: 10.1136/adc.2007.123661. [DOI] [PubMed] [Google Scholar]
- 17.Camilleri M, Carlson P, Zinsmeister AR, et al. Mitochondrial DNA and gastrointestinal motor and sensory functions in health and functional gastrointestinal disorders. Am. J Physiol Gastrointest. Liver Physiol. 2009;296:G510–G516. doi: 10.1152/ajpgi.90650.2008. [DOI] [PubMed] [Google Scholar]
- 18.Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 19.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 20.Cheng S, Fockler C, Barnes WM, et al. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci U S A. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang WF, Li X, Guo MZ, et al. Mitochondrial ATP 6 and 8 polymorphisms in irritable bowel syndrome with diarrhea. World J Gastroenterol. 2013;19:3847–3853. doi: 10.3748/wjg.v19.i24.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitehead WE, Palsson OS, Levy RR, et al. Comorbidity in irritable bowel syndrome. Am. J. Gastroenterol. 2007;102:2767–2776. doi: 10.1111/j.1572-0241.2007.01540.x. [DOI] [PubMed] [Google Scholar]
- 23.Fattal O, Link J, Quinn K, et al. Psychiatric comorbidity in 36 adults with mitochondrial cytopathies. CNS Spectr. 2007;12:429–438. doi: 10.1017/s1092852900015303. [DOI] [PubMed] [Google Scholar]
- 24.Slater SK, Nelson TD, Kabbouche MA, et al. A randomized, double-blinded, placebo-controlled, crossover, add-on study of CoEnzyme Q10 in the prevention of pediatric and adolescent migraine. Cephalalgia. 2011;31:897–905. doi: 10.1177/0333102411406755. [DOI] [PubMed] [Google Scholar]
- 25.Sandor PS, Di Clemente L, Coppola G, et al. Efficacy of coenzyme Q10 in migraine prophylaxis: a randomized controlled trial. Neurology. 2005;64:713–715. doi: 10.1212/01.WNL.0000151975.03598.ED. [DOI] [PubMed] [Google Scholar]
- 26.Hershey AD, Powers SW, Vockell AL, et al. Coenzyme Q10 deficiency and response to supplementation in pediatric and adolescent migraine. Headache. 2007;47:73–80. doi: 10.1111/j.1526-4610.2007.00652.x. [DOI] [PubMed] [Google Scholar]
- 27.Rozen TD, Oshinsky ML, Gebeline CA, et al. Open label trial of coenzyme Q10 as a migraine preventive. Cephalalgia. 2002;22:137–141. doi: 10.1046/j.1468-2982.2002.00335.x. [DOI] [PubMed] [Google Scholar]
- 28.Cordero MD, Cano-Garcia FJ, Alcocer-Gomez E, et al. Oxidative stress correlates with headache symptoms in fibromyalgia: coenzyme Q(1)(0) effect on clinical improvement. PLoS One. 2012;7:e35677. doi: 10.1371/journal.pone.0035677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordero MD, Alcocer-Gomez E, de Miguel M, et al. Coenzyme Q(10): a novel therapeutic approach for Fibromyalgia? case series with 5 patients. Mitochondrion. 2011;11:623–625. doi: 10.1016/j.mito.2011.03.122. [DOI] [PubMed] [Google Scholar]
- 30.Boles RG. High degree of efficacy in the treatment of cyclic vomiting syndrome with combined co-enzyme Q10, L-carnitine and amitriptyline, a case series. BMC Neurol. 2011;11:102. doi: 10.1186/1471-2377-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boles RG, Lovett-Barr MR, Preston A, et al. Treatment of cyclic vomiting syndrome with co-enzyme Q10 and amitriptyline, a retrospective study. BMC Neurol. 2010;10:10. doi: 10.1186/1471-2377-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.