Abstract
Photosynthesis converts solar energy to chemical energy using chlorophylls (Chls). In a late stage of biosynthesis of Chls, dark-operative protochlorophyllide (Pchlide) oxidoreductase (DPOR), a nitrogenase-like enzyme, reduces the C17 = C18 double bond of Pchlide and drastically changes the spectral properties suitable for photosynthesis forming the parental chlorin ring for Chl a. We previously proposed that the spatial arrangement of the proton donors determines the stereospecificity of the Pchlide reduction based on the recently resolved structure of the DPOR catalytic component, NB-protein. However, it was not clear how the two-electron and two-proton transfer events are coordinated in the reaction. In this study, we demonstrate that DPOR initiates a single electron transfer reaction from a [4Fe-4S]-cluster (NB-cluster) to Pchlide, generating Pchlide anion radicals followed by a single proton transfer, and then, further electron/proton transfer steps transform the anion radicals into chlorophyllide (Chlide). Thus, DPOR is a unique iron-sulphur enzyme to form substrate radicals followed by sequential proton- and electron-transfer steps with the protein folding very similar to that of nitrogenase. This novel radical-mediated reaction supports the biosynthesis of Chl in a wide variety of photosynthetic organisms.
In Chl biosynthesis, Pchlide reduction is the final reaction forming the conjugated π-system, the chlorin structure of Chl a, by the stereo-specific reduction of the C17 = C18 double bond (Fig. 1a). There are two evolutionary unrelated Pchlide reductases, one is the light-dependent Pchlide oxidoreductase (LPOR)1, which requires light for catalysis, and the other is DPOR2, which operates in a light-independent manner.
Figure 1.
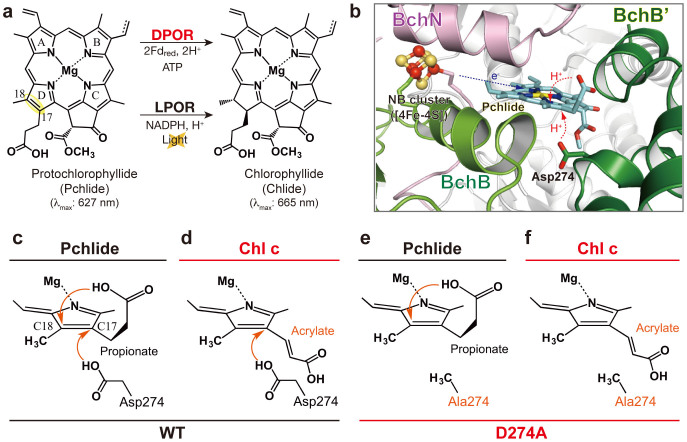
(a) Pchlide reduction. The C17 = C18 double bond of Pchlide D-ring (yellow) is trans-reduced by DPOR (upper) and LPOR (lower) to form Chlide. (b) Close-up view of the catalytic site of NB-protein. The direct electron transfer from the NB-cluster to Pchlide is shown by a dashed blue arrow. The proton transfers from BchB'-Asp274 and the propionate to Pchlide are shown by dashed red arrows. (c–f) Schematic representation of the combination of WT-D274A NB-protein, and Pchlide-Chl c; the native combination (c), WT NB-protein and Chl c (d), D274A NB-protein and Pchlide (e) and D274A NB-protein and Chl c (f).
LPOR is composed of a single polypeptide that belongs to short-chain dehydrogenase/reductase (SDR) family. LPOR is the responsible enzyme for light-dependent greening of angiosperms because LPOR operates as the sole Pchlide reductase in angiosperms1. Since the LPOR reaction is triggered by light, this enzyme provides a unique opportunity to study the reactions by ultrafast timescale. Although crystal structure of LPOR has not yet been revealed, several spectroscopic reaction intermediates were detected by triggering the reaction with a short laser pulse allowing to elucidate the catalytic cycle of LPOR3,4. These kinetic analyses suggested that a hydride transfer from NADPH to C18 is induced by light activation of Pchlide followed by several light-independent reactions including a proton transfer from a Tyr residue to C17, release of Chlide and exchange of cofactors5.
In oxygenic photosynthetic organisms except for angiosperms, DPOR serves as the determinant enzyme mediating the greening in the dark. Anoxygenic photosynthetic bacteria employ DPOR as the sole Pchlide reductase in bacteriochlorophyll biosynthesis. DPOR consists of two separable components, L-protein (a BchL homodimer) and NB-protein (a BchN-BchB heterotetramer), which are structurally related to Fe protein (a NifH homodimer) and MoFe protein (a NifD-NifK heterotetramer) of nitrogenase, respectively. Similar to nitrogenase, L-protein is the ATP-dependent reductase component6 for the catalytic component, NB-protein, where Pchlide reduction takes place7. A reaction cycle of L-protein was proposed based on analysis with a site-directed L-protein variant and an ATPase inhibitor8. Furthermore, using a new specific inhibitor nicotinamide (NA), a reaction cycle of NB-protein was also proposed to consist of 10 steps9 as follows: 1) the oxidized NB-protein binds to Pchlide; 2) the reduced L-protein (that is reduced by ferredoxin10) binds to the NB-protein–Pchlide complex; 3) an electron is transferred to the NB-cluster (that is coupled with ATP hydrolysis by the L-protein); 4) the oxidized L-protein is released from the NB-protein; 5) Pchlide is reduced by the single electron from the NB-cluster; 6) the second reduced L-protein binds to the NB-protein; 7) the NB-cluster is reduced again by the second L-protein; 8) Pchlide reduction is completed by the second electron transfer from the NB-cluster; 9) the oxidized L-protein is released; and 10) Chlide is released.
Crystal structures of individual DPOR components and a hexameric L-protein–NB-protein complex were recently revealed11,12,13,14. Then, DPOR became the first enzyme whose reaction mechanism was proposed on the basis of crystal structures among enzymes of the Mg branch. In our previous study12, we proposed a molecular basis for the stereospecificity of the C17 = C18 reduction based on the crystal structure of the NB-protein isolated from the purple bacterium Rhodobacter capsulatus (Fig. 1b). The model predicted that Asp274 in BchB' and a propionate group at C17 of the substrate donate protons to C17 and C18, respectively (Fig. 1b, c). In this reaction, two electron transfer events from the NB-cluster to Pchlide (steps 5 and 8 mentioned above) are accompanied with two proton transfer events. Considering that the NB-cluster, a [4Fe-4S] cluster, delivers directly one electron at a time to Pchlide, the reaction intermediates must include Pchlide radicals. However, such reaction intermediates including Pchlide radicals have never been detected so far and it remained unclear how these electron and proton transfer events are coordinated.
Radical enzymes catalyse a variety of reactions that are chemically difficult to achieve. Most radical enzymes, including ribonucleotide reductases15, vitamin B12-dependent enzymes16 and radical-SAM enzymes17, generate cofactor radicals. For example, vitamin B12-dependent and radical SAM enzymes generate a 5′-deoxyadenosyl radical, and ribonucleotide reductases generate a thiyl radical to induce the abstraction of a hydrogen atom from the 3′ carbon of ribonucleotide phosphate. In contrast, enzymes generating substrate radicals by direct electron transfer are limited. Recently, 2-hydroxyisocaproyl-CoA dehydratase (2HCD) from a strict anaerobe Clostridium difficile has been demonstrated that a substrate radical (ketyl radical) is generated by electron transfer from a [4Fe-4S] cluster to induce a dehydration reaction18. 2HCD represents a novel class of radical enzymes.
Here we show that DPOR is another unique radical enzyme generating substrate (Pchlide) radicals by a [4Fe-4S] cluster in NB-protein and propose a reaction scheme, in which Pchlide radicals are converted to the product Chlide by a second reduction with the [4Fe-4S] cluster. This radical mediated Pchlide reduction mechanism by DPOR appears to be distributed among not only anoxygenic photosynthetic bacteria but also oxygenic phototrophs including cyanobacteria, green algae and lower land plants.
Results
In the DPOR assay system in this study, a stable substrate-bound catalytic component, Pchlide-bound NB-protein, was prepared and pre-incubated with L-protein, dithionite and an ATP-regeneration system7. The reactions were initiated by the addition of ATP and monitored by absorption and electron paramagnetic resonance (EPR) spectroscopy. Absorption spectra were measured at 5 °C to facilitate the detection of intermediates by slowing down the whole reaction process. If Pchlide radicals are generated during the reaction, they can be detected directly by monitoring absorbance changes or by EPR after rapid freezing of samples. Using the wild-type (WT) Pchlide-bound NB-protein, only the absorption spectrum indicating Chlide formation was detected 30 s after the reaction without any intermediate spectra showing catalytic intermediates (Fig. S1 and inset). The prediction of proton donors from the crystal structure of NB-protein allows us to use a substrate analog Chl c and a site-directed variant to trap reaction intermediates by blocking the proton donor processes. We examined combinations of Chl c that has C17-acrylate instead of C17-propionate and a NB-protein variant D274A that has a site-directed BchB variant with Ala274 instead of Asp274 (Figs. 1d–f).
It is expected that the proton transfer is completely blocked in a combination of D274A and Chl c (Figs. 1f and 2a). Upon the addition of ATP, the absorption peaks were decreased slowly with a time constant (tc) of 1087 s (Fig. 2a). In addition, the reaction mixture showed a clear EPR signal with g = 2.005 and 1.4 mT peak-to-peak width (Fig. 2a, inset), reminiscent of the signals of porphyrin and chlorin radicals19,20. The signal suggests the formation of a Chl c anion radical, which probably has a smaller extinction coefficient in the visible range than a Chl a anion radical21. The g value higher than that of other porphyrin radicals may reflect geometric distortion due to the unique protein environment of the binding cavity, covered by the BchB C-terminal helix that is partly unwound by the binding of the substrate12. The broad EPR signal suggests a magnetic interaction with the spin on NB-cluster. Actually, fast spin relaxation of the Chl c radical was estimated based on the high values of microwave power at 7.6 mW required to half-saturate the signal intensity (P1/2) at 20 K and of inhomogeneity parameter (b) of 1.46 (data not shown). In the class Ib ribonucleotide reductase of Escherichia coli, which contains a Tyr radical located at a short distance ~8 Å from the metal binding site22, the high P1/2 value of the Tyr radical at 1.6 mW at 3.6 K was decreased to 0.03 mW when the ferromagnetic-coupling with a MnIIIMnIII centre was relieved by the exchange of the centre to FeIIIFeIII one with S = 023. On the other hand, a Tyr radical in photosystem II exhibited a rather low P1/2 value of 60 μW at 20 K24 because its distance from the non-haem iron with S = 2 is rather long at ~37 Å25. Therefore, the high values of P1/2 and b above 1 indicate that the radical spin in the substrate binding site interacts with the NB-cluster with S = 3/226 at a short distance of ~10 Å12.
Figure 2.
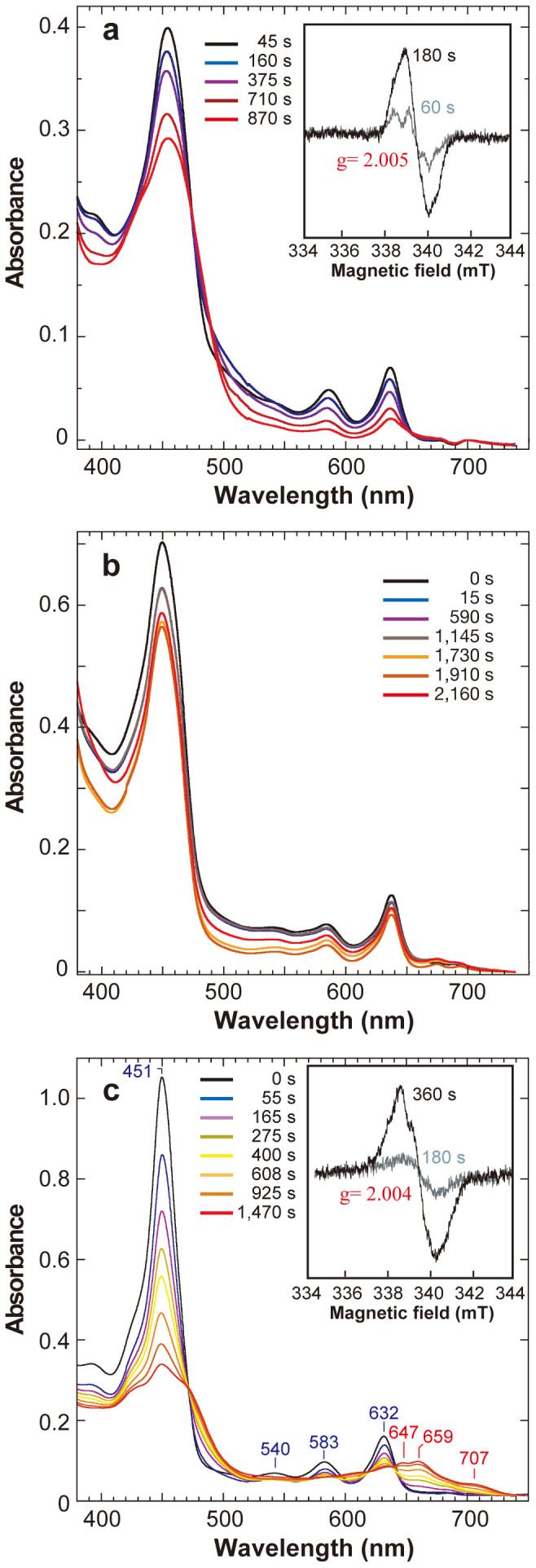
(a) Absorption spectra of the combination of D274A NB-protein and Chl c. Inset: EPR spectra of this combination. (b) Absorption spectra of the combination of WT NB-protein and Chl c. (c) Absorption spectra of the combination of D274A NB-protein and Pchlide. Inset: EPR spectra of this combination. All the absorption spectra were measured at 5 °C at varied times as shown in Figure after initiation of the reaction by adding ATP. EPR spectra were measured at 20 K after rapid freeing of the samples at times indicated in Figures as described in Methods. Note that the time courses are for the reaction at 5 °C and are expected to be significantly slower than those at physiological temperatures.
No modification of chemical structure of Chl c seems to have occurred during this spectral change because given that Chl c was recovered with a high yield of 84% by acetone extraction after the reaction (Table S1). We accordingly suggest that the slow absorption decrease represents the formation of a Chl c anion radical by the one-electron reduction process, and that the electron transfer from the NB-cluster to Chl c occurs even though both proton transfer paths are blocked. The result also implies that the first step of the Pchlide reduction in the NB-protein includes the single electron transfer step from the NB-cluster to Pchlide.
In the WT-Chl c combination (Fig. 1d), the spectral change after the initiation of the reaction was less evident (Fig. 2b). It was smaller than that detected in the reaction of D274A-Chl c. A very weak EPR signal with g = 2.006 was detected (data not shown) in accord with the smaller spectral change. The result suggested that the Chl c anion radical is also generated in this reaction. The lower amount of the Chl c anion radical may come from a lower level of bound Chl c to WT protein (approximately 25%) than that to D274A NB-protein. This result suggests that the Chl c anion radical is formed even when the putative proton donor for C18 is deleted.
The third combination, D274A-Pchlide (Fig. 1e), produced a marked spectral change immediately after the reaction start (Fig. 2c). In a first phase (<400 s), four absorption peaks at 451, 540, 583 and 632 nm, which represent the bound Pchlide, slowly decreased. The reaction mixture after 360 s showed a clear EPR signal with g = 2.004 and a broad 1.7-mT bandwidth (Fig. 2c, inset). The saturation power P1/2 was also high at 8.9 mW and the factor b was estimated to be 1.30, as was the case with Chl c. The broader bandwidth than that of a putative Chl c anion radical may be attributed to larger hyperfine coupling, suggesting stronger or more complex hydrogen bonding to its binding cavity. The appearance of the EPR signal appeared to coincide with the bleaching of the Pchlide peaks, suggesting the formation of Pchlide radicals. In the absorption spectra, new peaks at 647, 659 and 707 nm appeared in a late phase (>400 s), suggesting that further reactions proceeded.
A global fitting analysis suggested that this spectral change could be decomposed into four independent components (Fig. 3a). The first component corresponds to the initial state (State 0; Fig. 2c, 0 s) of the D274A-NB-protein–Pchlide bound form (Fig. 3a, gray). Upon the initiation of the reaction, a second component (State 1) with three absorption peaks at 427, 450 and 660 nm grows with a tc of 300 s (Fig. 3a, blue). Later, a third component (State 2) with peak at 449, 469 and 709 nm appears with a tc of 3138 s (Fig. 3a, green), followed by a fourth component (State 3) with peak at 425, 474, 608 and 659 nm and a tc of 2913 s (Fig. 3c, red). Although the shape of the decomposed spectra (Fig. 3a) still appears complex, suggesting that multiple intermediates are included in addition to the probable major intermediates, the simulated spectral changes fitted the observed ones very well (Figs. 2c and 3b).
Figure 3.
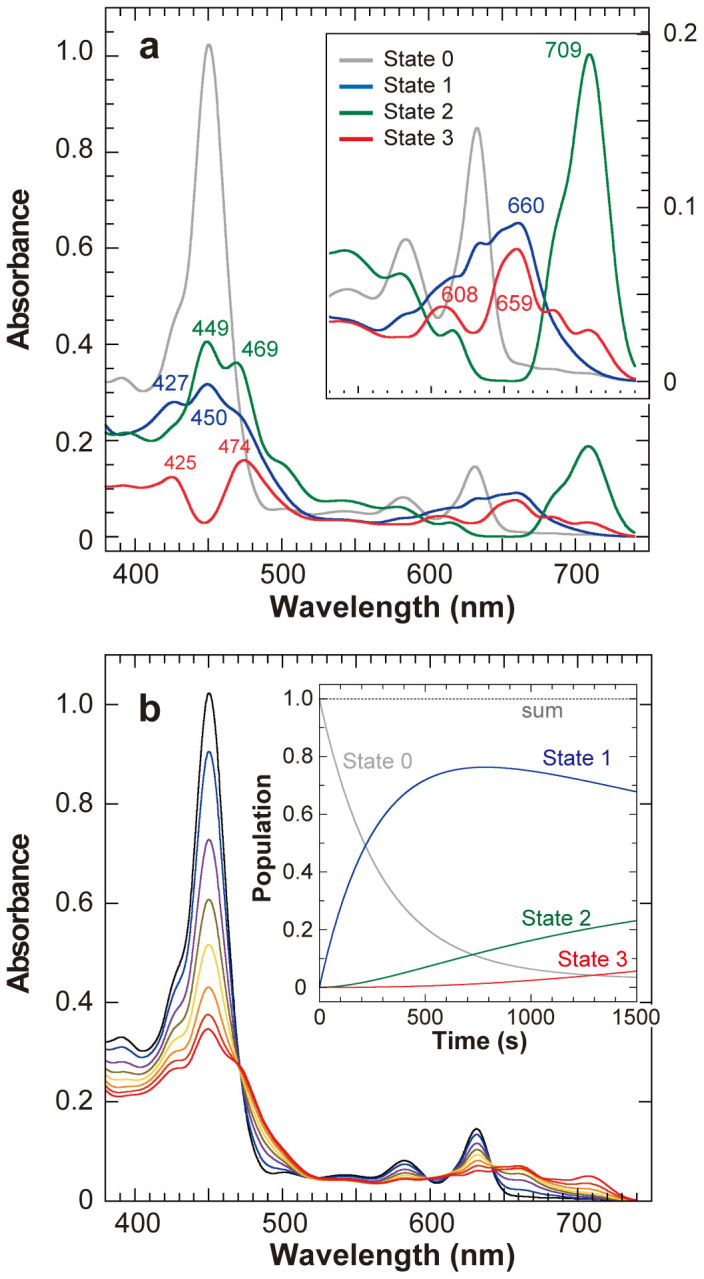
(a) Four absorption spectra (States 0 to 3) decomposed from those of Fig. 2c. State 0 is identical to that at 0 s in Fig. 2c. All spectra represent the states when the respective spectral component occupies 100%. Inset: A close up of the spectra of from 530 to 750 nm. (b) Simulated spectra by the four decomposed spectra. Inset: Time course of the population of the four spectral components.
To determine how the reactions proceeded in these combinations the pigments were extracted with acetone after the reactions (Table S1). In comparison with the high recovery of Chl c (69% and 84%), that of Pchlide was markedly low (32%) in D274A-Pchlide. Furthermore, a small amount of Chlide (2%) was detected in D274A-Pchlide. This indicated that a small portion of Pchlide proceeded further beyond the first radical formation step, even though the proton path to C17 was blocked. In contrast, no products were detected in the other two combinations (WT/Chl c and D274A/Chl c), suggesting that only the first electron transfer occurs and further reactions do not proceed once the proton path to C18 is blocked. Thus, the proton transfer event for C18 from the propionate may precede that for C17 from Asp274.
NA is a specific inhibitor of the electron transfer from L-protein to the NB-protein9. The Pchlide radicals detected in D274A-Pchlide appear to be in a steady state with respect to quenching and generation. Thus, upon NA addition, the electron transfer from the NB-cluster is blocked and unstable Pchlide radical species disappear in a short time. NA was added to the D274A-Pchlide reaction at the beginning of the late phase (420 s). The intensity of the EPR signal at g = 2.004 decreased to approximately 20% of the control (Fig. 4, inset). This decrease of the EPR signal was accompanied by a significant change in the absorption spectra (Fig. 4). A characteristic shoulder at 705 nm disappeared, together with a significant decrease of peaks in the blue region, whereas the increase in the peaks near 650 nm was relatively slight. Pchlide radicals showing the g = 2.004 signal may consist of at least two distinct molecular species, an NA-sensitive and an NA-insensitive species, under steady-state conditions. The concomitant decrease in the peak at 705 nm and the EPR signal (g = 2.004) suggested that the NA-sensitive species represents State 2 with a 709-nm peak.
Figure 4. Absorption spectra of the combination of D274A and Pchlide in the presence (red) and absence (blue) of NA.
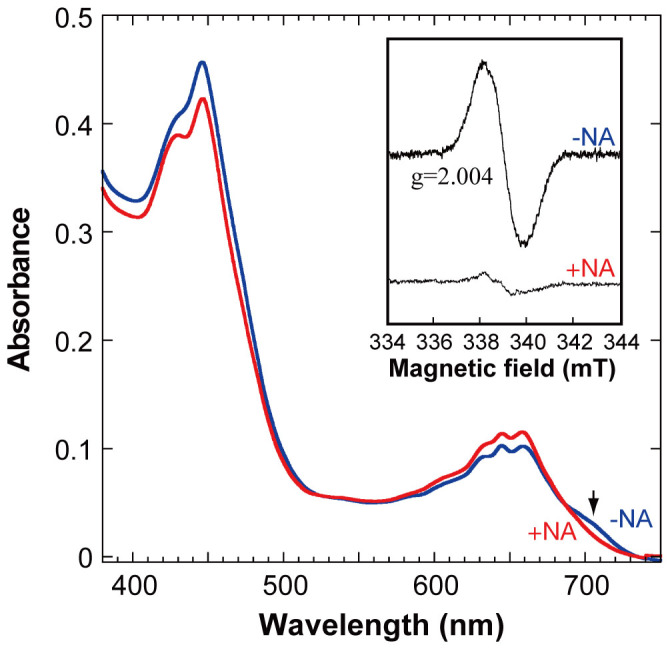
An arrow indicates the decreased peak at 705 nm upon the NA addition. Inset: EPR spectra of the combination of D274A and Pchlide in the presence (lower trace) and absence (upper trace) of NA.
To serve as the first proton donor for C18, the C17-propionate must be protonated before the proton transfer. Considering that the probable pKa of C17-propionic acid of Pchlide is similar to those of the free acids (propionic acid, pKa = 4.87; butyric acid, pKa = 4.82), the C17-propionate is deprotonated under the reaction conditions (pH 7.4). BchB-His378 located 6.5 Å from the propionate, is the best candidate for the proton donor (Fig. S2a). The C17-propionate may be protonated by His378 via a water molecule located 2.6 Å from the propionate. This probable contribution was supported by partial activity of a site-directed variant BchB-H378A (Fig. S2b), which is consistent with that of the equivalent site-directed variant H394A of Prochlorococcus marinus (35% activity)14. The distorted conformation of C17-propionate of Pchlide may contribute to the proton relay from His378 to the propionate via water molecules in the NB-protein (Fig. S2a).
Discussion
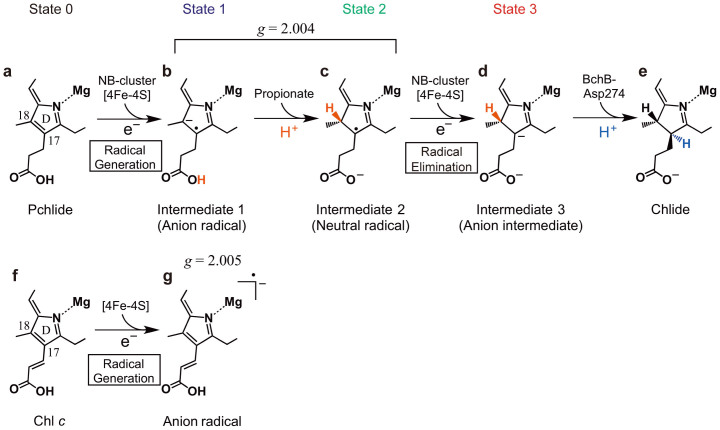
We propose a reaction scheme in Figure 5 based on the results in the present study. In the model, the C17-propionate is already protonated (Fig. 5a, State 0) by proton transfer from His378 (Fig. S2a). The first step is the reduction of Pchlide by a single electron transfer from the NB-cluster, to form a Pchlide anion radical (Intermediate 1, Fig. 5b), which is based on the observation of Pchlide/Chl c radical formation in all three combinations. Following this, proton transfer from the C17-propionate (shown in red) of Pchlide forms a second radical, a Pchlide neutral radical (Intermediate 2, Fig. 5c), which is based on the observation that no chemically altered products were detected in the combinations with Chl c and that a small amount of Chlide was detected in D274A/Pchlide. Both radicals are EPR active, and only a signal with the g value of 2.004 was detected (Fig. 2c, inset). Although we did not identify either of the two intermediates to represent the EPR signal, we propose that States 1 and 2 constitute the radical species. The coincident decrease of the EPR signal and the 705-nm shoulder of absorption spectra upon the NA addition (Fig. 4, inset) suggests that State 2 (with a 709-nm peak) is the NA-sensitive species. A second single electron transfer from the NB-cluster eliminates the radicals to form a non-radical anion (Intermediate 3, Fig. 5d) as State 3. The reaction is completed by the second proton transfer from Asp274 to form Chlide (Fig. 5e).
Figure 5.
Proposed reaction steps in the Pchlide reduction (a–e). First step is the electron transfer from the NB-cluster to Pchlide (a) to form a Pchlide anion radical (b) followed by the proton transfer from the propionate to form a neutral radical (c). The second electron transfer eliminates the neutral radical by reduction to form an anion intermediate (d). The Pchlide reduction is completed by the second proton transfer (e). When Chl c is used instead of Pchlide (f), a Chl c anion radical is generated (g).
Crystallographic analysis of the complex of L-protein and NB-protein from P. marinus suggested that a water molecule just above C18 is the direct proton donor for C18 rather than the C17-propionate in the Prochlorococcus NB-protein14. However, contribution of the water molecule has not yet been experimentally proven to be critical for Pchlide reduction in the Prochlorococcus DPOR. Results we obtained so far in R. capsulatus DPOR support the reaction mechanism of the C17-propionate as the proton donor to C18.
The catalytic mechanism of the other Pchlide reductase, LPOR5, is completely different from that of DPOR. In a photoactive ternary complex of LPOR–NADPH–Pchlide, light absorption by the bound-Pchlide molecule creates a charge-transfer intermediate that triggers a hydride transfer from the NADPH-nicotinamide ring to the C17 carbon. Subsequently, a conserved Tyr residue donates a proton to the C18 carbon27. Although a first photochemical intermediate A696 was once assumed to be a Pchlide radical in initial works19, A696 was later regarded as a charge transfer complex generated by hydride transfer from NADPH by a detailed spectroscopic analysis20. Thus, the LPOR reaction does not appear to involve radical intermediates20. The catalytic steps of LPOR were dissected into one light-driven step and four light-independent steps by a ultrafast pump-probe absorption difference spectroscopy4. Further analyses suggest that a first photon absorption activates the LPOR–NADPH–Pchlide complex and a second photon absorption then induces catalysis4. Photosynthetic organisms, thus, have found two distinct solutions to catalyse the Pchlide reduction using totally different protein architectures, nitrogenase and SDR, during evolution2.
Nitrogenase shares a common architecture with DPOR12. The overall reaction involves transfers of eight electrons and eight protons to produce ammonia and hydrogen, consisting of at least 10 reaction intermediates28. Efforts to trap reaction intermediates lead to characterization of an important intermediate state E4 that carries four electrons stored as two [Fe–H–Fe] bridging hydrides and two protons bound to the sulfides on the FeMo-cofactor28,29. The currently proposed reaction scheme that predicts the E4 state suggests that the coupling between electron and proton transfers is too tight to be discriminated. In contrast, we succeeded to identify the sequential electron and proton transfer steps on the NB-protein in the DPOR reaction, by blocking of the proton transfer step using D274A and Chl c. It is of interest that the two types of reaction mechanisms in DPOR and nitrogenase seem to have diverged from the common structural architectures during evolution.
In terms of reaction mechanism, DPOR appears to be similar to some radical enzymes rather than nitrogenase. Crystal structure of 2HCD30 suggested that one of the two [4Fe-4S] clusters in the heterodimeric structure transfers one electron to the substrate to generate a ketyl radical followed by dehydration of 2-hydroxy group with the β-proton. Then, the radical is eliminated by electron transfer back to the [4Fe-4S] cluster (oxidation). In DPOR, the Pchlide radical is generated by the electron transfer from the NB-cluster similar to 2HCD (Figs. 5a–b). However, the Pchlide radical is eliminated by a second electron transfer (reduction), followed by the proton transfer to produce the final product, Chlide (Figs. 5c–e). Benzoyl-CoA reductase (BCR) may adopt the reaction mechanism more similar to that of DPOR. BCR catalyses two-electron reduction of benzoyl-CoA with two-proton transfer to form stereo-selectively trans-dienoyl-CoA product31, and this enzyme carries two [4Fe-4S] clusters that may act as the radical generator to catalyse the reduction32. Comparison of the reaction mechanism based on crystal structure of BCR would be very interesting.
Phycocyanobilin:ferredoxin oxidoreductase PcyA, which catalyses two vinyl reductions of biliverdin IXα in bilin biosynthesis, is the other example of an enzyme that generates substrate radicals during catalysis33,34. Substrate bilin radicals are generated by electron transfer from a small FeS protein, ferredoxin. The reaction involves sequential electron-coupled proton transfers. PcyA does not carry any metal ions or cofactors, but ferredoxin plays a role as the electron donor. DPOR is distinct from PcyA in that the direct electron donor, the NB-cluster, is embedded in the NB-protein.
Radical-mediated Pchlide reduction by DPOR functions very efficiently under anaerobic conditions. However, such a reaction is potentially dangerous under oxygen-rich conditions because the substrate radical would be a source of reactive oxygen species that would cause severe damage to cells35. In fact, another nitrogenase-like enzyme Chlide oxidoreductase36, a close relative of DPOR, becomes a superoxide-generator in the photosynthetic bacterium R. sphaeroides and Synechocystis sp. PCC 680337,38. Nevertheless DPOR is distributed widely among oxygenic photosynthetic organisms including cyanobacteria, green algae, bryophytes, pteridophytes and gymnosperms (Figs. S3 and S4). As observed in green dark-grown seedlings of some gymnosperms39,40,41, the Pchlide-radical mediated reaction plays a critical role in the greening of many photosynthetic organisms inhabiting light-limited environments.
Methods
Protein purification
All manipulations were performed in an anaerobic chamber7. WT, D274A and H378A NB-protein variants were overexpressed in E. coli harbouring pHANB142, pJnD274A (this plasmid was used for the preparation of D274A variant in the previous report)12 and pJnH378A (see below), respectively. NB-protein was purified from crude extracts on a Strep-Tactin (IBA) affinity chromatography column equilibrated with 100 mM Tris-HCl (pH 8.0) containing 150 mM NaCl and eluted with 100 mM Tris-HCl (pH 8.0) containing 150 mM NaCl and 2.5 mM desthiobiotin. Pchlide- or Chl c-bound forms of WT and D274A were prepared as described previously7. The molar ratio of Pchlide in the obtained WT or D274A variant NB-protein is 1.1–1.2 and that of Chl c in WT or D274A variant NB-protein is 0.3 or 0.9–1.0, respectively.
Site-directed mutagenesis
Nucleotide substitutions causing amino acid change were introduced into bchB region of pHANB142 using two primers, BchBHis378Ala-f1 (5′-ggtgatAtcggcgcccgtcGCGgtgcaggacttccccgcccg-3′) and BchBHis378Ala-r1 (5′-cgggcggggaagtcctgcacCGCgacgggcgccgaTatcacc-3′). The changed nucleotides are shown in upper case and the codons corresponding to the amino acid residues to be changed are capitalised and underlined. To easily confirm the introduction of the mutations, another silent mutation besides the targeted codon was introduced to create a new restriction site (EcoRV). PCR reactions were performed with KOD-plus polymerase (Toyobo, Osaka, Japan). The nucleotide sequences of mutagenised bchB were confirmed by DNA sequencing.
Absorption spectral measurements
To monitor the change in absorption spectra of the DPOR reaction mixture, the assay pre-mixture was prepared in a cuvette with an airtight screw cap in an anaerobic chamber. After incubation of Pchlide- or Chl c-bound NB-protein (5 μM) with L-protein (10 μM), creatine phosphate (20 mM), creatine phosphokinase (21 units) and sodium dithionite (2.7 mM) in 100 mM HEPES-KOH (pH 8.0) for 10 min at 5°C, a pre-chilled mixture of ATP and MgCl2 (final concentration 9 mM and 5 mM, respectively) was added using a syringe and absorption spectra were periodically recorded with a Jasco V550 spectrophotometer (Jasco, Hachioji, Japan) having a temperature control module (model ETC-477, set at 5°C; Jasco). To determine the amount of recovered of pigments, the assay mixture was mixed with acetone (final concentration 80%) and absorption spectra were recorded with a Jasco V550 spectrophotometer.
Preparation of EPR samples
For EPR spectroscopy, the DPOR assay pre-mixtures were prepared in quartz EPR tubes (outer diameter 5 mm) in the anaerobic chamber. Pchlide- or Chl c-bound NB-protein (10 μM) was incubated for 10 min at 3°C with L-protein (20 μM), creatine phosphate (20 mM), creatine phosphokinase (21 units) and sodium dithionite (2.7 mM) in 100 mM HEPES-KOH (pH 8.0). After capping, a pre-chilled mixture of ATP and MgCl2 (final concentration 9 mM and 5 mM, respectively; prepared and packed in an anaerobic chamber) was added to the tube using a syringe outside the anaerobic chamber. After mixing ATP and MgCl2, the reaction mixtures were incubated for 6 min at 3°C and frozen rapidly in liquid nitrogen to quench the reaction and EPR spectra were recorded. EPR measurements were performed using a Bruker ESP-300E X-band spectrometer (Bruker Biospin, Germany) equipped with a liquid-helium flow cryostat and a temperature control system (CF935, Oxford Instruments, UK). Experimental conditions were as follows: microwave frequency, 9.507–9.563 GHz; modulation frequency, 100 kHz; temperature, 20 K; modulation amplitude, 0.4 mT; and time constant, 20 ms.
NA inhibition
After the addition of ATP and MgCl2, the reaction mixtures of D274A/Pchlide were incubated for 7 min at 5°C and NA was subsequently added (70 mM). After additional incubation for 3 min, absorption spectra were recorded. For EPR spectroscopy, the reaction mixtures were frozen rapidly in liquid nitrogen and EPR spectra were recorded. For the control (–NA), only the buffer (100 mM HEPES-KOH; pH 8.0) was added and spectrum were recorded.
Global fitting procedure
The absorption spectra at each time (Fig. 2c) were decomposed into 20 Gaussian bands. The band positions were determined from the second-derivative spectra and the band widths by a global multi-Gaussian fitting analysis. The amplitudes of each band were plotted against time. The time courses were analysed with a global fitting using a rate equation between 4 states, in which the spectra for States 0, 2 and 3 were assumed to be the 0-s spectrum (Fig. 2c), the differential spectra between the presence and absence of NA (Fig. 4) and between the 0-s and 1470-s spectra (Fig. 2c), respectively. The time constants for forward and backward reactions were restricted to 10–5000 s and 10–10000 s, respectively. Absorption spectral changes (Fig. 3b) were reconstructed with the spectra and population changes of each state.
Author Contributions
J.N. purified NB-proteins and performed site-directed mutagenesis, enzymatic assay and absorption spectroscopy; T.K., S.I. and J.N. performed EPR spectroscopy; T.M. and H.T. prepared Chl c; Y.F., J.N. and S.I. contributed to design of the experiments and writing the manuscript. All authors discussed the results and commented on the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Wolfgang Buckel, Kazuyuki Tatsumi, Tatsuo Omata and Kazuki Terauchi for the valuable discussion. This study was supported by Grants-in-Aid for Scientific research Nos 20200063, 23370020, 23000007 (Y.F.) and 22245030 (H.T.) from the Japan Society for the Promotion of Science (JSPS), by the Precursory Research for Embryonic Science and Technology (PRESTO), and by the Advanced Low Carbon Technology Research and Development Program (ALCA) of the Japan Science and Technology Agency (JST). T.K. is also grateful for a Fellowship from the JSPS for Japanese Junior Scientists (24008402).
References
- Masuda T. & Takamiya K. Novel insights into the enzymology, regulation and physiological functions of light-dependent protochlorophyllide oxidoreductase in Angiosperms. Photosynth. Res. 81, 1–29 (2004). [DOI] [PubMed] [Google Scholar]
- Reinbothe C. et al. Chlorophyll biosynthesis: spotlight on protochlorophyllide reduction. Trends Plant Sci. 15, 614–622 (2010). [DOI] [PubMed] [Google Scholar]
- Heyes D., Ruban A., Wilks H. & Hunter C. Enzymology below 200 K: the kinetics and thermodynamics of the photochemistry catalyzed by protochlorophyllide oxidoreductase. Proc. Natl. Acad. Sci. USA 99, 11145–11150 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sytina O. et al. Conformational changes in an ultrafast light-driven enzyme determine catalytic activity. Nature 456, 1001–1004 (2008). [DOI] [PubMed] [Google Scholar]
- Heyes D. & Hunter C. Making light work of enzyme catalysis: protochlorophyllide oxidoreductase. Trends Biochem. Sci. 30, 642–649 (2005). [DOI] [PubMed] [Google Scholar]
- Nomata J., Kitashima M., Inoue K. & Fujita Y. Nitrogenase Fe protein-like Fe-S cluster is conserved in L-protein (BchL) of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. FEBS Lett 580, 6151–6154 (2006). [DOI] [PubMed] [Google Scholar]
- Nomata J., Ogawa T., Kitashima M., Inoue K. & Fujita Y. NB-protein (BchN-BchB) of dark-operative protochlorophyllide reductase is the catalytic component containing oxygen-tolerant Fe-S clusters. FEBS Lett. 582, 1346–1350 (2008). [DOI] [PubMed] [Google Scholar]
- Bröcker M. et al. Biosynthesis of (bacterio)chlorophylls: ATP-dependent transient subunit interaction and electron transfer of dark-operative protochlorophyllide oxidoreductase. J. Biol. Chem. 285, 8268–8277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomata J., Kondo T., Itoh S. & Fujita Y. Nicotinamide is a specific inhibitor of dark-operative protochlorophyllide oxidoreductase, a nitrogenase-like enzyme, from Rhodobacter capsulatus. FEBS Lett. 587, 3142–3147 (2013). [DOI] [PubMed] [Google Scholar]
- Nomata J., Swem L., Bauer C. & Fujita Y. Overexpression and characterization of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. Biochim. Biophys. Acta 1708, 229–237 (2005). [DOI] [PubMed] [Google Scholar]
- Sarma R. et al. Crystal structure of the L protein of Rhodobacter sphaeroides light-independent protochlorophyllide reductase with MgADP bound: a homologue of the nitrogenase Fe protein. Biochemistry 47, 13004–13015 (2008). [DOI] [PubMed] [Google Scholar]
- Muraki N. et al. X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 465, 110–114 (2010). [DOI] [PubMed] [Google Scholar]
- Bröcker M. J. et al. Crystal structure of the nitrogenase-like dark operative protochlorophyllide oxidoreductase catalytic complex (ChlN/ChlB)2. J. Biol. Chem. 285, 27336–27345 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J. et al. Structure of ADP-aluminium fluoride-stabilized protochlorophyllide oxidoreductase complex. Proc. Natl. Acad. Sci. USA. 110, 2094–2098 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund P. & Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 (2006). [DOI] [PubMed] [Google Scholar]
- Dowling D. P., Croft A. K. & Drennan C. L. Radical use of Rossmann and TIM barrel architectures for controlling coenzyme B12 chemistry. Annu. Rev. Biophys. 41, 403–427 (2012). [DOI] [PubMed] [Google Scholar]
- Shisler K. A. & Broderick J. B. Emerging themes in radical SAM chemistry. Curr. Opin. Struct. Biol. 22, 701–710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Darley D. J., Buckel W. & Pierik A. J. An allylic ketyl radical intermediate in clostridial amino-acid fermentation. Nature 452, 239–242 (2008). [DOI] [PubMed] [Google Scholar]
- Lebedev N. & Timko M. Protochlorophyllide oxidoreductase B-catalyzed protochlorophyllide photoreduction in vitro: insight into the mechanism of chlorophyll formation in light-adapted plants. Proc. Natl. Acad. Sci. USA 96, 9954–9959 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyes D. et al. The first catalytic step of the light-driven enzyme protochlorophyllide oxidoreductase proceeds via a charge transfer complex. J. Biol. Chem. 281, 26847–26853 (2006). [DOI] [PubMed] [Google Scholar]
- Fujita I., Davis M. S. & Fajer J. Anion radicals of pheophytin and chlorophyll a: their role in the primary charge separations of plant photosynthesis. J. Am. Chem. Soc. 100, 6280–6282 (1978). [Google Scholar]
- Cox N. et al. A tyrosyl-dimanganese coupled spin system is the native metalloradical cofactor of the R2F subunit of the ribonucleotide reductase of Corynebacterium ammoniagenes. J. Am. Chem. Soc. 132, 11197–11213 (2010). [DOI] [PubMed] [Google Scholar]
- Cotruvo J. A. & Stubbe J. An active dimanganese(III)-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Biochemistry 49, 1297–1309 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y., Nishimura M. & Itoh S. Charge distribution on the membrane surface of the donor side of photosystem II. Effect of the free radical relaxing agent dysprosium on the power saturation of EPR signal IIs in PS-II particles. Plant Cell Physiol. 28, 1493–1499 (1987). [Google Scholar]
- Hirsh D. J., Beck W. F., Innes J. B. & Brudvig G. W. Using saturation-recovery EPR to measure distances in proteins: applications to photosystem II. Biochemistry 31, 532–541 (1992). [DOI] [PubMed] [Google Scholar]
- Kondo T., Nomata J., Fujita Y. & Itoh S. EPR study of 1Asp-3Cys ligated 4Fe-4S iron-sulfur cluster in NB-protein (BchN-BchB) of a dark-operative protochlorophyllide reductase complex. FEBS Lett. 585, 214–218 (2011). [DOI] [PubMed] [Google Scholar]
- Wilks H. & Timko M. A light-dependent complementation system for analysis of NADPH:protochlorophyllide oxidoreductase: identification and mutagenesis of two conserved residues that are essential for enzyme activity. Proc. Natl. Acad. Sci. USA 92, 724–728 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B. M., Lukoyanov D., Dean D. R. & Seefeldt L. C. Nitrogenase: a draft mechanism. Acc. Chem. Res. 46, 587–595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukoyanov D., Barney B. M., Dean D. R., Seefeldt L. C. & Hoffman B. M. Connecting nitrogenase intermediates with the kinetic scheme for N2 reduction by a relaxation protocol and identification of the N2 binding state. Proc. Natl. Acad. Sci. USA 104, 1451–1455 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer S. H., Buckel W. & Dobbek H. Structural basis for reductive radical formation and electron recycling in (R)-2-hydroxyisocaproyl-CoA dehydratase. J. Am. Chem. Soc. 133, 4342–4347 (2011). [DOI] [PubMed] [Google Scholar]
- Thiele B., Rieder O., Golding B. T., Müller M. & Boll M. Mechanism of enzymatic Birch reduction: stereochemical course and exchange reactions of benzoyl-CoA reductase. J. Am. Chem. Soc. 130, 14050–14051 (2008). [DOI] [PubMed] [Google Scholar]
- Unciuleac M. & Boll M. Mechanism of ATP-driven electron transfer catalyzed by the benzene ring-reducing enzyme benzoyl-CoA reductase. Proc. Natl. Acad. Sci. USA 98, 13619–13624 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S. L., Rockwell N. C., Lagarias J. C. & Fisher A. J. Insight into the radical mechanism of phycocyanobilin-ferredoxin oxidoreductase (PcyA) revealed by X-ray crystallography and biochemical measurements. Biochemistry 46, 1484–1494 (2007). [DOI] [PubMed] [Google Scholar]
- Stoll S. et al. Structure of the biliverdin radical intermediate in phycocyanobilin:ferredoxin oxidoreductase identified by high-field EPR and DFT. J. Am. Chem. Soc. 131, 1986–1995 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59, 1073–1082 (2006). [DOI] [PubMed] [Google Scholar]
- Nomata J., Mizoguchi T., Tamiaki H. & Fujita Y. A second nitrogenase-like enzyme for bacteriochlorophyll biosynthesis - Reconstitution of chlorophyllide a reductase with purified X-protein (BchX) and YZ-protein (BchY-BchZ) from Rhodobacter capsulatus. J. Biol. Chem. 281, 15021–15028 (2006). [DOI] [PubMed] [Google Scholar]
- Kim E. J., Kim J. S., Lee I. H., Rhee H. J. & Lee J. K. Superoxide generation by chlorophyllide a reductase of Rhodobacter sphaeroides. J. Biol. Chem. 283, 3718–3730 (2008). [DOI] [PubMed] [Google Scholar]
- Kim E. J., Kim J. S., Rhee H. J. & Lee J. K. Growth arrest of Synechocystis sp. PCC6803 by superoxide generated from heterologously expressed Rhodobacter sphaeroides chlorophyllide a reductase. FEBS Lett. 583, 219–223 (2009). [DOI] [PubMed] [Google Scholar]
- Kusumi J., Sato A., Tachida H. & Investigators S. T.-N. Y. Proceedings of the SMBE Tri-National Young Investigators' Workshop 2005. Relaxation of functional constraint on light-independent protochlorophyllide oxidoreductase in Thuja. Mol. Biol. Evol. 23, 941–948 (2006). [DOI] [PubMed] [Google Scholar]
- Demko V. et al. A novel insight into the regulation of light-independent chlorophyll biosynthesis in Larix decidua and Picea abies seedlings. Planta 230, 165–176 (2009). [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Kurumiya S., Ohashi R. & Fujita Y. Functional evaluation of a nitrogenase-like protochlorophyllide reductase encoded by the chloroplast DNA of Physcomitrella patens in the cyanobacterium Leptolyngbya boryana. Plant Cell Physiol. 52, 1983–1993 (2011). [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Nomata J. & Fujita Y. Functional expression of nitrogenase-like protochlorophyllide reductase from Rhodobacter capsulatus in Escherichia coli. Photochem. Photobiol. Sci. 7, 1238–1242 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information