Abstract
Functional nucleic acid (FNA)-based sensing systems have been developed for efficient detection of a wide range of biorelated analytes by employing DNAzymes or aptamers as recognition units. However, their intracellular delivery has always been a concern, mainly in delivery efficiency, kinetics, and the amount of delivered FNAs. Here we report a DNA dendrimer scaffold as an efficient nanocarrier to deliver FNAs and to conduct in situ monitoring of biological molecules in living cells. A histidine-dependent DNAzyme and an anti-ATP aptamer were chosen separately as the model FNAs to make the FNA dendrimer. The FNA-embedded DNA dendrimers maintained the catalytic activity of the DNAzyme or the aptamer recognition function toward ATP in the cellular environment, with no change in sensitivity or specificity. Moreover, these DNA dendrimeric nanocarriers show excellent biocompatibility, high intracellular delivery efficiency, and sufficient stability in a cellular environment. This FNA dendrimeric nanocarrier may find a broad spectrum of applications in biomedical diagnosis and therapy.
Keywords: nanocarrier, DNA dendrimer, functional nucleic acids, DNAzyme, aptamer, intracellular molecular sensing
Monitoring the distribution of intracellular biological molecules is critical for understanding their physiological and pathological functions, validating disease biomarkers and diagnosing disease in its early stages.1 Fluorescent imaging through staining with a molecular probe might be the most attractive molecular imaging technique for the detection of intracellular species by virtue of its high sensitivity, real-time spatial imaging, and detection of targets in living cells or tissues with minimal damaging effects.2−4 Toward this goal, many such small organic molecular probes have been designed and developed for fluorescent sensing of intracellular biomolecules.5−14 However, these organic molecules usually suffer from poor water-solubility and poor biocompatibility, as well as limited membrane-permeability, obstacles which strictly limit their intracellular applications. Moreover, the design of an efficient small molecular probe for biomolecules is very challenging, as no general molecular design strategy is available.
Functional nucleic acids (FNAs) serve as an emerging class of biomolecules that either possess cofactor-dependent catalytic activities toward their substrate (known as DNAzymes)15 or bind to a target molecule (known as aptamers)16−19 with the ability to recognize a broad range of targets from metal ions to small molecules, drugs, proteins, and even whole cells.20,21 In many respects, the binding performance of FNAs to specific targets can rival that of protein antibodies. FNAs are selected in vitro and can be isolated to recognize any target of interest, even molecules that are not suitable for producing antibodies by their poor immunogenicity or high toxicity. In comparison with small molecular probes, FNAs are naturally water-soluble and biocompatible. Moreover, it is relatively simple to synthesize FNAs with a commercial DNA synthesizer and equally easy to modify them with different functional groups during the synthesis process. Importantly, FNAs do not require complicated engineering or advanced design knowledge in order to construct a target-binding site, and the binding affinity toward target analytes can be fine-tuned by varying the selection conditions. All these unique features make FNAs an attractive and general platform for sensing intracellular biomolecules.
However, although a large number of FNA-based sensing systems have been developed for efficient detection of a wide range of biorelated analytes by employing DNAzymes22,23 or aptamers24,25 as recognition units, the strict application of these sensing systems in intracellular environments is compromised for many reasons. First, FNAs are negatively charged hydrophilic biomolecules and, hence, cannot freely penetrate the cell membrane,26 thus requiring additional instruments or carriers for efficient intracellular delivery.27 Second, nucleic acid probes can be unstable, even after successful cellular delivery because of endogenous nuclease digestion,28−30 or they can nonspecifically bind with intracellular nontarget protein,31,32 leading to high false positive signals. Even though inorganic nanocarriers, such as gold nanoparticles (AuNPs) and graphene, have been developed to solve these challenges by providing efficient cellular uptake33 and enhanced enzymatic stability,34−36 FNAs still suffer from non-negligible cytotoxicity at relatively high concentrations as a result of incorporating such inorganic nanomaterials.37 In addition, the preparation of these nanocarriers is tedious and very time-consuming.27 DNA micelles have also been successfully developed as nanocarriers for aptamers39 or DNA probes40 for intracellular molecular imaging or therapy. However, these micelle nanocarriers may dissociate at concentrations lower than the critical micelle concentration (CMC) (i.e., the concentration of surfactant above which micelles form and all additional surfactants added to the system go to micelles), thereby limiting their applications in complex biological samples, especially at low concentration. Therefore, the development of a nanocarrier that is easy to synthesize, highly biocompatible, capable of self-delivery, and sufficiently stable in a cellular environment, even at low concentration, is highly desired.
As a naturally occurring biomacromolecule, DNA exhibits excellent biocompatibility, can afford a large vocabulary of available sequences though programmable design, and can be easily synthesized by automated instruments in large quantities.41 These unique properties, together with the outstanding specificity of A-T and G-C Watson–Crick hydrogen-bonding interactions, make DNA a particularly promising candidate to serve as a building block material for a wide variety of nanostructures with potential applications in biomedicine and biotechnology.42−45 Among them, dendritic DNA nanostructures have attracted increasing interest in the past decade on the basis of their monodispersity, excellent stability, and globular, highly branched, and porous structures.46,47 Herein, by integrating a DNAzyme or an aptamer into the DNA dendrimeric scaffold, we, for the first time, employ DNA dendrimer as an efficient nanocarrier of FNAs for in situ monitoring of biological molecules in living cells. A histidine-dependent DNAzyme48 and an anti-ATP aptamer49,50 were chosen separately as the model FNAs. As shown in Figure 1, the FNA-embedded DNA dendrimers maintain the histidine-dependent catalytic activity of the DNAzyme or the recognition function of the anti-aptamer toward ATP in buffer solution, with no obvious change in sensitivity or specificity observed. Most importantly, the dendritic nanocarriers exhibit excellent biocompatibility and cell membrane permeability, with remarkably enhanced intracellular stability, when compared to free FNAs. All these features favor the use of FNA-embedded DNA dendrimers for intracellular molecular probe applications. The proposed sensing systems were then applied for imaging of histidine or ATP in living cells with satisfactory results, demonstrating that the dendritic DNA nanocarriers are promising for delivery of FNAs for in situ intracellular molecular probing applications.
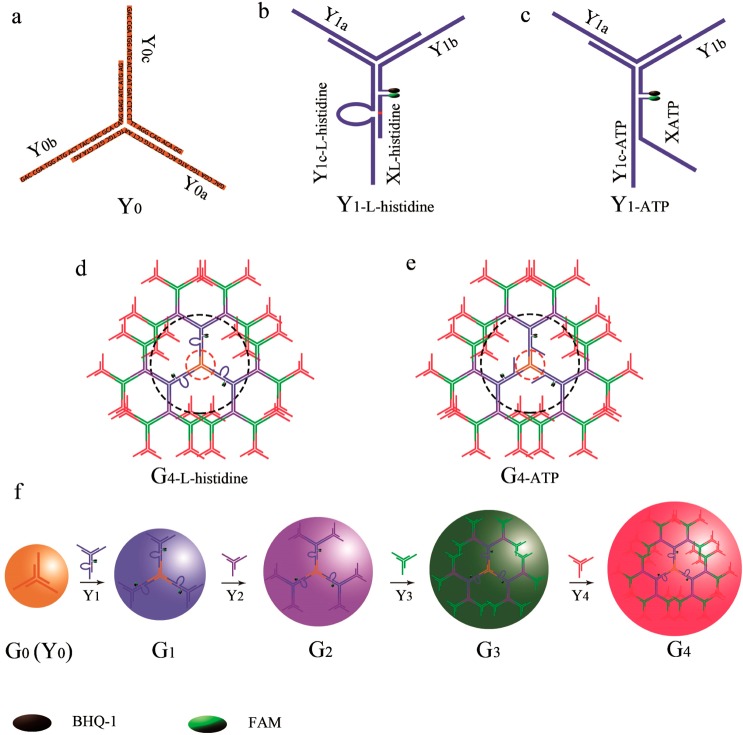
Figure 1.
(a) Y0 was assembled from three different single strands: Y0a, Y0b, and Y0c. The other Y-DNA scaffolds were prepared according to same strategy, except for Y1. (b) Y1-l-histidine was achieved by the assembly of Y1a, Y1b, Y1c-l-histidine, and Xl-histidine. (c) Y1-ATP was achieved by the assembly of Y1a, Y1b, Y1c-ATP, and XATP. (d) G4-l-histidine was assembled from Y0, Y1-l-histidine, Y2, Y3, and Y4. (e) G4-ATP was assembled from Y0, Y1-ATP, Y2, Y3, and Y4. (f) Preparation of the first to fourth generation of DNA dendrimers.
Results and Discussion
Design, Preparation, and Characterization of FNA-Embedded DNA Dendrimers
FNA-embedded DNA dendrimers were prepared from Y-DNA using an enzyme-free and step-by-step assembly strategy. To prepare the Y-DNA, equal moles of three or four oligonucleotides were first mixed together. As shown in Figure 1, the Y-DNA called Y0 was assembled from the hybridization of three different single strands: Y0a, Y0b, and Y0c. Y-DNAs Y2, Y3, and Y4 were prepared according to same procedure. In our design, the DNAzyme or aptamer was incorporated in the second layer (Y1) of the DNA dendrimer, which contains four different single strands and serves as the signal reporter part of the sensing system. Each arm of the Y-DNA contains a 13-base pair duplex and a 13-base pair single-stranded sticky end. The three sticky ends of Y0 were designed to be identical, while the other Y-DNAs (Yn, n > 0) were designed to have three sticky ends; two were identical, and the third end was complementary to the sticky ends of Yn–1. The histidine-dependent DNAzyme was chosen as a model to construct the DNAzyme-based sensing system, in which the second layer of the dendrimer (Y1 named Y1-l-histidine) consisted of Y1a, the Y1b enzyme strand of DNAzyme Y1c-l-histidine, and its substrate strand Xl-histidine. The anti-ATP aptamer was chosen as a model to construct the aptamer-based sensing system, in which the second layer of the dendrimer (Y1 named Y1-ATP) consisted of Y1a, Y1b, Y1c-ATP, and anti-ATP aptamer strand XATP. The as-prepared Y-DNAs were then characterized by agarose gel electrophoresis (Supporting Information Figure S1).
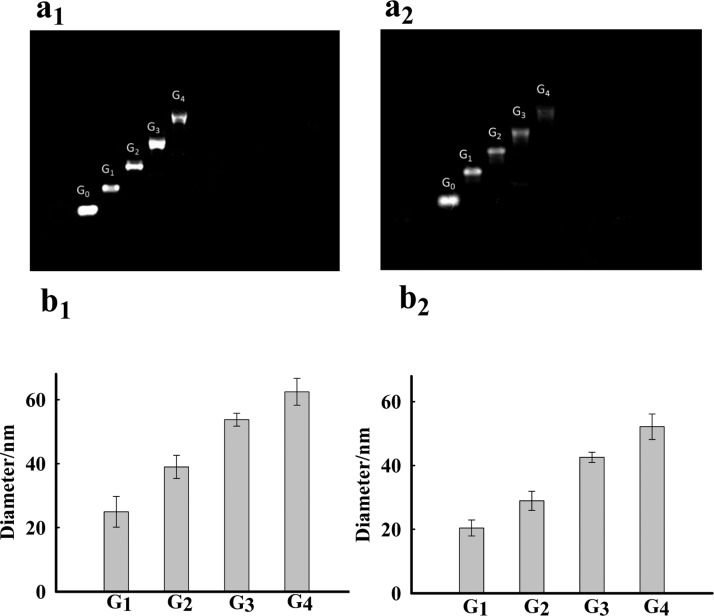
Different generations of DNA dendrimer (Gn) were then prepared from Y-DNAs following the procedure shown in Figure 1. The sequences of Y-DNA were carefully designed to guarantee that the hybridization of sticky ends would only occur between Yn and Yn+1. In this way, hybridization could only occur in one direction: Y0 → Y1 → Y2 → Y3 → Y4. The formation of different generations of DNA dendrimer was confirmed by both agarose gel electrophoresis and dynamic light scattering. As shown in Figure 2a, G4 ran the slowest on the agarose gel, followed sequentially by G3, G2, G1, and G0, indicating that the size of the DNA dendrimer increased with the increase of layers of Y-DNA involved in the assembly process. Moreover, the as-prepared DNA dendrimers without any purification show only a single band, demonstrating that no byproduct is formed in the assembly procedure, confirming the efficiency of our assembly strategy. DLS measurements (Figure 2b) showed that the average diameters for DNAzyme-conjugated DNA dendrimer of G1, G2, G3, and G4 were 24.93, 38.99, 53.75, and 62.44 nm, respectively. The sizes of the ATP aptamer-conjugated DNA dendrimer were also detected, with diameters of 20.44, 28.93, 42.56, and 52.15 nm observed, respectively. These results demonstrated that higher generations of DNA dendrimer are larger in size.
Figure 2.
(a) Agarose gel electrophoresis of G0–G4. G0 is Y-DNA (Y0); G1–G4 are DNA dendrimers: (a1) G-l-histidine, (a2) G-ATP. (b) DLS data of the diameter of G1–G4: (b1) G-l-histidine, (b2) G-ATP.
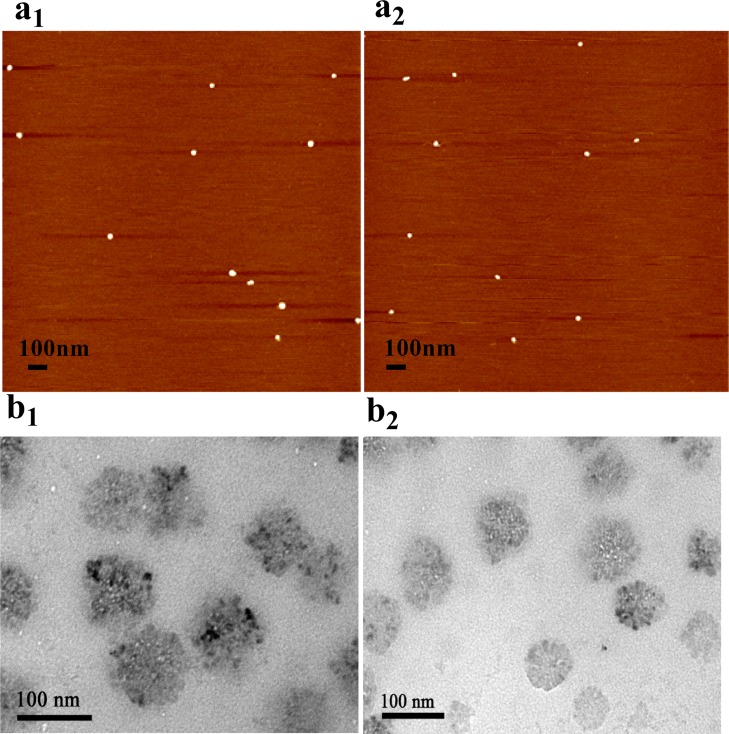
To confirm that the mobility-shifted species were indeed DNA dendrimer, we examined G4 by atomic force microscopy (AFM). The measured diameter of the G4 nanostructure was 50 ± 5 nm (Figure 3a). Although the AFM pictures revealed the dendritic size of G4, they were not suitable for determining the distribution of sizes as a result of the many commonly occurring problems associated with AFM, including sample damage by AFM tips, cleanliness of the substrate, dehydration of DNA, or contaminants during the preparation. To further explore the size distributions, G4 was also visualized by transmission electron microscopy (TEM). Figure 3b shows that the structure of the assembled G4 DNA dendrimers has good monodispersity and that the size (diameter) of G4 was measured at about 60 nm, which agreed with the size measured from AFM.
Figure 3.
(a) AFM images of G4 on mica surface: (a1) G4-l-histidine, (a2) G4-ATP. (b) TEM images of G4: (b1) G4-l-histidine, (b2) G4-ATP. Scale bars correspond to 100 nm.
Sensing Mechanism
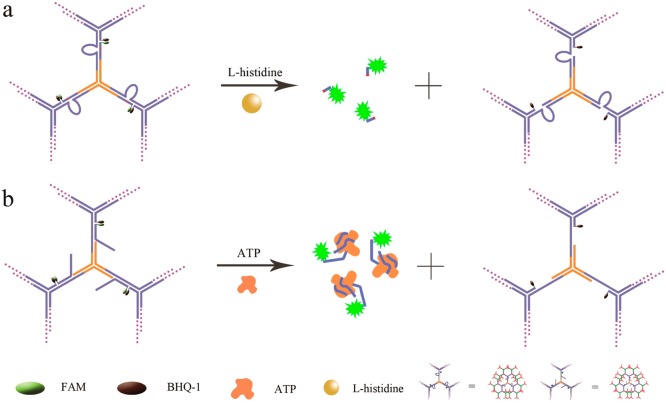
In our design for the DNAzyme-based DNA dendrimer sensing system, the 3′ end of the Y1b strand was functionalized with a quencher (Black Hole Quencher-1, BHQ-1), while the 5′ end of the substrate strand X was functionalized with the fluorophore carboxyfluorescein (FAM). The four oligonucleotides Y1a, Y1b, Y1c-l-histidine, and Xl-histidine will hybridize each other to form the Y-shaped structure Y1-l-histidine, which brings the quencher and fluorophore into close proximity, thereby effectively quenching FAM fluorescence. In the presence of l-histidine, the substrate strand X is cleaved, resulting in a shorter DNA strand which shows a lower melting temperature (12.7 °C) with Y1b than that of the original full-length substrate strand (44.3 °C). At room temperature, a shorter DNA strand containing FAM is then released from the sensing system, and the fluorescent signal of the sensing system is enhanced by the separation of FAM from the BHQ-1 quencher (Figure 4a).
Figure 4.
(a) Schematics of DNAzyme-based and DNA dendrimer sensing system for the detection of l-histidine. (b) Schematics of aptamer-based and DNA dendrimer sensing system for the detection of ATP.
A similar strategy was also employed to design the aptamer-based DNA dendrimer sensing system. The ATP aptamer strand was modified with the fluorophore FAM on its 5′end (denoted XATP). The 3′ end of the Y1b strand was functionalized with a quencher (Black Hole Quencher-1, BHQ-1), and the Y1c-ATP strand was designed to partly hybridize with XATP. In the absence of the target, the four oligonucleotides Y1a, Y1b, Y1c-ATP, and XATP will hybridize each other to form Y1-ATP, bringing the fluorophore and quencher into close proximity, thereby effectively quenching the fluorescence of FAM. However, as a result of the strong affinity of the anti-ATP aptamer toward ATP, the introduction of the target ATP into the DNA dendrimer sensing system will induce the formation of an ATP–aptamer complex, releasing the aptamer from the dendrimer to produce a large increase of fluorescence intensity of the sensing system (Figure 4b).
Optimization of the DNA Dendrimer Sensing System
To achieve the best analytical performance of the DNAzyme-based sensing system, the sequence of substrate strand X of the DNAzyme was first optimized. Two different oligonucleotides, Xl-histidine(6) and Xl-histidine(8), containing a different number of bases (see the Supporting Information, Table S1), were chosen as the substrate strands of DNAzyme. By using the substrate sequence Xl-histidine(8), we observed a 5.0-fold fluorescence enhancement in the presence of 500 μM l-histidine, which was obviously higher than that of Xl-histidine(6), with only a 2.1-fold signal increase observed (see Supporting Information, Figure S2). Therefore, Xl-histidine(8) was chosen as the optimized substrate strand to assemble the DNAzyme-based DNA dendrimer. For the aptamer-based sensing system, the sequence of the Y1c-ATP was also optimized. Four different Y1c-ATP sequences with a different number of bases, including Y1c-ATP9, Y1c-ATP11, Y1c-ATP12, and Y1c-ATP13, were then designed to prepare the DNA dendrimer. A Y1c-ATP with a 12 base pairs stem (Y1c-ATP12) was found to provide satisfactory performance with a 3.4-fold fluorescence enhancement in the presence of 1 mM ATP (Figure S3), and it was therefore chosen for further assembly investigation.
In Vitro Sensing Performance of the FNA-Embedded Dendrimers
The final dendrimeric product G4, containing functional nucleic acids, was then chosen for sensing applications. The analytical performance of these DNA dendrimer sensing systems was first evaluated in a buffer system. The fluorescence signal increase as a function of time for an intermediate, low, and high histidine were monitored (Figure S4). Figure 5a shows the fluorescence emission spectra of the DNAzyme-based DNA dendrimer sensing system upon the addition of l-histidine with different concentrations. A dramatic increase of fluorescence intensity of the FAM was observed when the l-histidine concentration was increased from 5 μM to 10 mM, and the fluorescence response nearly reached a plateau after 30 min incubation of the sensing system with l-histidine at room temperature. Figure 5b depicts the relationship between the fluorescence intensity and the different concentrations of l-histidine, with a dynamic range from 5 μM to 10 mM. A detection limit of 500 nM for l-histidine was estimated based on the 3σ/slope rule. The sensitivity of the dendrimeric sensing system is comparable to that of previously reported monomeric DNAzyme-based electrochemical or fluorescent biosensors for l-histidine, indicating that the DNAzyme embedded in the DNA dendrimeric scaffold maintains its histidine-dependent catalytic activity. For the aptamer-based DNA dendrimer sensing system, a satisfactory sensitivity was also achieved for the detection of ATP, with a detection limit of 20 μM observed (see Supporting Information, Figures S5 and S6). These results indicate that the two DNA dendrimer-based FNA sensing systems worked well in a buffer system and that the FNAs could maintain their functions after they were embedded in the DNA dendrimeric scaffold, favoring their application in extracellular molecular sensing.
Figure 5.
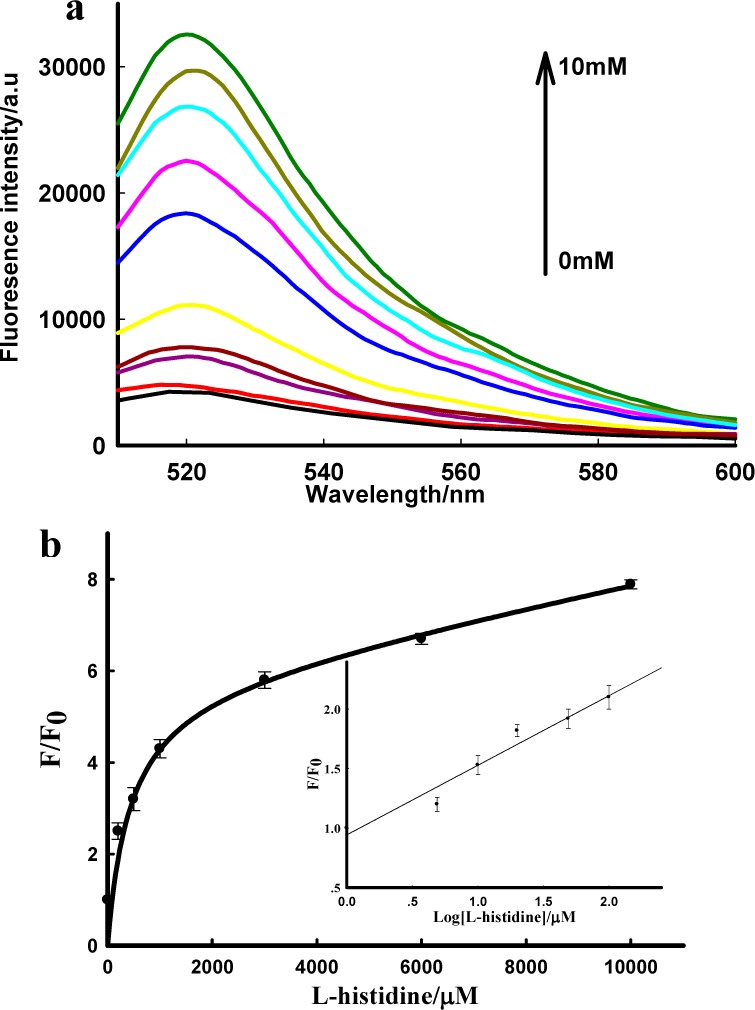
(a) Fluorescence emission spectra in the presence of different concentrations of l-histidine, ranging from 0 to 10 mM; the concentration for G4-l-histidine is 10 nM. (b) Relationship between fluorescence enhancement and the target concentration. Inset shows the responses of the sensing system to l-histidine at low concentration. F0 and F are the fluorescence intensity of the sensing system in the absence and presence of target, respectively.
For a sensing system with potential application in practical biological samples, a highly selective response to the target in the presence of other potential interferences is a necessity. Therefore, the selectivity experiments of the dendrimeric sensing systems were extended to various potentially competing biomolecules. For the DNAzyme-based dendrimeric sensing system G4-l-histidine, significantly larger fluorescence enhancement was observed with 10 mM of l-histidine than with other amino acids when the concentration reached 100 mM (Figure S7a), indicating a high specificity of our proposed DNAzyme-based dendrimeric sensing system to l-histidine. Similar high specificity was observed for the aptamer-based dendrimeric sensing system G4-ATP to ATP. Significant enhancement was shown upon the introduction of 1 mM of ATP (Figure S8), while no obvious fluorescence signal change was observed upon the addition of GTP, UTP, or CTP at the same concentration. These results clearly confirm that the embedding of FNAs into the DNA dendrimeric scaffold does not affect the selectivity of FNAs toward their targets.
Stability of a nanocarrier is critical to its intracellular molecular sensing applications. For example, when it is delivered into a physiological environment, DNA nanocarriers can be subject to (1) digestion by nucleases in the body or (2) dissociation as a result of nonspecific binding of monomeric DNA with intracellular proteins. Therefore, to evaluate the stability of the DNA dendrimeric nanocarriers we designed toward enzymatic digestion, the aptamer-based dendrimeric sensing system G4-ATP and Y-shaped DNA building block Y1-ATP (containing the aptamer sequence and acting as a control) were treated separately and directly with endonuclease DNase I (1 U/mL, a considerably higher concentration than would be found in living cells), and their fluorescence signal change was then separately recorded as a function of time. As shown in Figure S7b, a much slower enhancement of fluorescence signal was observed for G4-ATP compared to that of Y1-ATP. We also estimated the effect of a more than 5 h incubation of the sensing system in cell lysate (Figure S9) and found that no change of fluorescence signal was observed for G4-ATP. These results indicated that the dendrimeric sensing system showed greater stability against enzymatic digestion than that shown by the monomeric DNA-based building block, again favoring the application of this FNA in intracellular applications.
To further assess the stability of the dendrimeric sensing systems toward nonspecific binding dissociation by intracellular proteins, the dendrimeric sensing systems G4-l-histidine and G4-ATP, as well as the Y-shaped DNA building blocks Y1-l-histidine and Y1-ATP, were incubated in buffer, RPMI 1640 medium, and RPMI 1640 medium containing 10% FBS for 4 h at 37 °C, respectively, and the fluorescence spectra were then recorded (Figure S10). No obvious changes of fluorescence signal were observed for G4-l-histidine and G4-ATP in RPMI 1640 medium in the presence and absence of FBS. For G4-l-histidine, a fluorescence increase was observed when changing from the buffer system to the RPMI 1640 medium. This may have been induced by the l-histidine already existing in the RPMI 1640 medium. However, for Y1-l-histidine and Y1-ATP, remarkable fluorescent signal enhancements were observed. These experimental results demonstrate that these two dendrimeric nanocarriers are sufficiently stable to resist nonspecific binding dissociation by intracellular proteins and can therefore reduce the false positive signals induced by nontarget proteins.31
To test the practicality of the sensing systems in complex samples, we further employed the DNAzyme-based dendrimeric sensing system to detect l-histidine in cell lysate, a realistically complex media containing a variety of proteins and other contaminants. In order to avoid the interference of background fluorescence signal of the cellular homogenate, varying amounts of l-histidine were added to the diluted cellular homogenate samples (1.0 × 106 cells/mL). As shown in Figure S11 (see Supporting Information), the l-histidine titration curve in the cellular homogenate was similar to that in the buffer solution, with an observed linear detection range of l-histidine between 5 and 100 μM, thus confirming that the proposed sensing system can be applied to the practical application of l-histidine detection in real samples in the presence of other potentially competing species.
Intracellular Applications of the DNA Dendrimer Sensing Systems
The FNA-embedded DNA dendrimers maintain the recognition functions of FNAs with high selectivity in vitro and show high stability to enzymatic digestion or the nonspecific binding dissociation of intracellular proteins, features which should be favorable for their intracellular sensing applications. Therefore, their ability to permeate the cell membrane and sense intracellular targets was then investigated via confocal fluorescence imaging experiments using MCF-7 (human breast cancer) cells as the model system. MCF-7 cells were first incubated with G4-l-histidine in a DPBS buffer supplemented with 10% (v/v) cell culture medium for 4 h and then with 100 mM of l-histidine and 150 mM of K+ for another 1 h. After rinsing and washing with PBS, cells were then imaged under a confocal laser scanning microscope. A control experiment under the same conditions was also carried out on the DNA dendrimer G4-l-histidine-R, which used an inactive enzyme strand (Y1c-l-histidine-R) to replace the enzyme strand of l-histidine-dependent DNAzyme (Y1c-l-histidine). As shown in Figure 6a, a significant fluorescence signal of FAM could be observed for the MCF-7 cells which were treated with the active G4-l-histidine, while a much weaker fluorescence signal was observed for MCF-7 cells treated with inactive G4-l-histidine-R (Figure 6b). Because K+ serves as an important cofactor in the catalytic process of histidine-dependent DNAzyme,48 for the in vitro experiment, both l-histidine and K+ are necessary for the DNAzyme to carry out its catalytic function. The same conditions would also be necessary for DNAzyme to maintain its catalytic activity in living cells. To verify our hypothesis, confocal fluorescence imaging experiments for G4-l-histidine under different conditions were then carried out, and the results are also shown in Figure 6. The incubation of MCF-7 cells with G4-l-histidine alone (Figure 6c), G4-l-histidine and l-histidine (Figure 6d), or G4-l-histidine and K+ (Figure 6e) afforded much weaker fluorescence signals than that observed in the presence of all three compounds, indicating that both l-histidine and K+ are indeed necessary for DNAzyme to maintain its catalytic activity in living cells. These results indicate that the DNA dendrimeric nanocarrier could successfully deliver the DNAzyme probe into living cells and that the DNAzyme embedded in the dendrimer scaffold could also maintain its l-histidine recognition capability in living cells.
Figure 6.
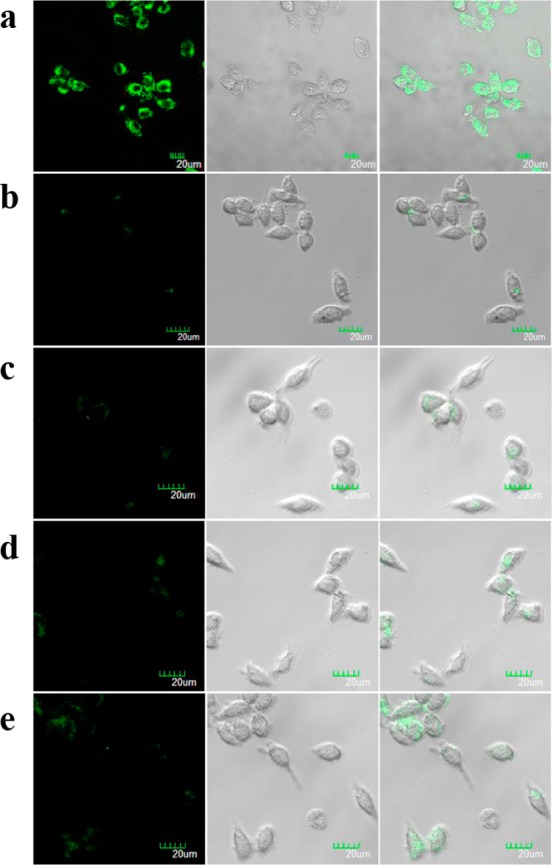
Confocal fluorescence microscopy images of MCF-7 cells treated with (a) 100 mM l-histidine, 150 mM K+, and 60 nM active G4-l-histidine; (b) 100 mM l-histidine, 150 mM K+, and 60 nM in active G4-l-histidine-R; (c) 60 nM active G4-l-histidine; (d) 100 mM l-histidine and 60 nM active G4-l-histidine; (e) 150 mM K+ and 60 nM active G4-l-histidine. The green channel is FAM fluorescence. Differential interference contrast (DIC) microscopy images of cells are shown in the second column. Overlap of fluorescence and DIC images are shown in the third column. Scale bar = 20 μm.
To demonstrate the universality of the DNA dendrimeric nanocarrier for delivery of FNAs in living cells, the ability of aptamer-embedded dendrimer G4-ATP to permeate the cell membrane and sense intracellular ATP was also investigated with confocal fluorescence imaging experiments. DNA dendrimer with the anti-ATP aptamer replaced by a random DNA (G4-ATP-R) was designed as a control probe to evaluate the specificity of the sensing system in living cells. As shown in Figure 7a, MCF-7 cells incubated with 20 nM of G4-ATP for 4 h exhibited an obvious fluorescence signal of FAM, while a much weaker fluorescence signal was observed when the cells were treated with 20 nM of random dendrimeric probe G4-ATP-R (Figure 7d). Moreover, when the concentration of incubated G4-ATP was increased from 20 to 60 nM, the induced fluorescence signal of MCF-7 cells also increased (Figure 7b,c), while changing the concentration of the random probe G4-ATP-R triggered no obvious change of fluorescence signal for MCF-7 cells (Figure 7e,f). These results might be ascribed to the high concentration level of ATP in living cells (typically 1–10 mM),51,52 which cannot trigger the saturated response of the sensing system. The introduction of the control probe can well distinguish the fluorescence change induced by an increase in sensor concentration from that by an increase in ATP concentration. These results indicate that this dendrimeric nanocarrier could also successfully deliver the aptamer probe into living cells and that the aptamer embedded in the dendrimer scaffold could maintain its recognition function in living cells to realize in situ monitoring of ATP. To further confirm the uptake of DNA dendrimers by MCF-7 cells, we carried out Z-scanning (Figure S12), captured by confocal microscopy. In addition, to ensure that the observations made via confocal microscopy apply to the whole cell population, we examined the fluorescence coming from a population of cells and quantified intracellular fluorescence in cells using flow cytometry (Figure S13). Finally, a colocalization assay demonstrated that most of the fluorescence came from the cytoplasm, instead of the endosomes or lysosomes (Figure S14). Moreover, a delivery efficiency of ∼3.0% was estimated for our dendrimeric nanocarrier by lysing the cells, adding ATP to release the dye, and quantifying fluorescence signal.
Figure 7.
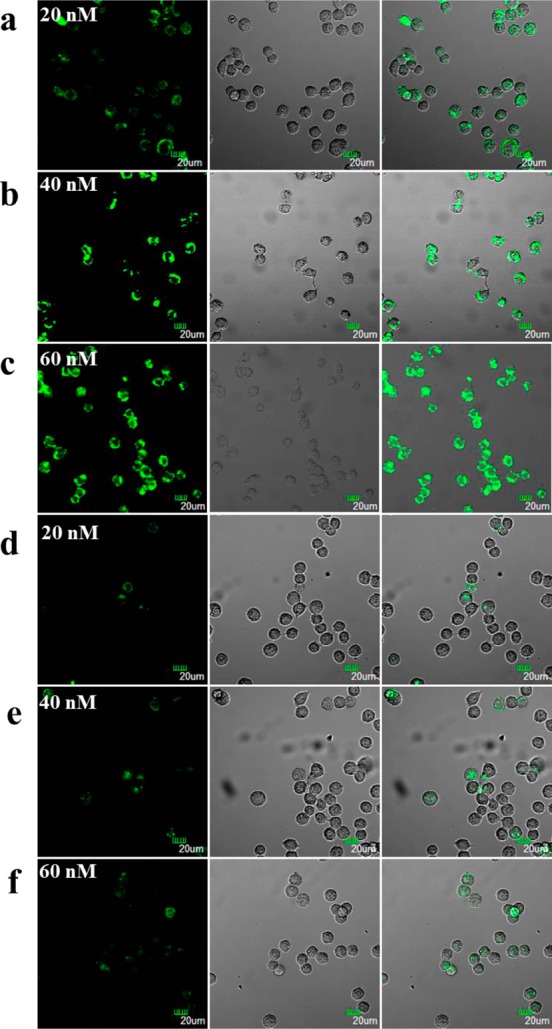
Confocal fluorescence microscopy images of MCF-7 cells treated with (a) 20 nM active G4-ATP; (b) 40 nM active G4-ATP; (c) 60 nM active G4-ATP; (d) 20 nM random G4-ATP-R; (e) 40 nM random G4-ATP-R; (f) 60 nM G4-ATP-R. The green channel is FAM fluorescence. Differential interference contrast microscopy images of cells are shown in the second column. Overlap of fluorescence and DIC images are shown in the third column. Scale bar = 20 μm.
An obvious advantage of our dendrimeric nanocarrier in comparison with previously reported micelle nanocarriers lies in its high stability at very low concentration, which may be ascribed to the strong Watson–Crick hydrogen-bonding interaction between the hybridized oligonucleotides in the dendrimeric structure. In contrast, micelle nanocarriers can dissociate at concentrations lower than the CMC, thereby limiting their application in complex biological samples, such as living cells. In Figure 7a,d, it can be seen that both the aptamer-embedded dendrimer G4-ATP and the random dendrimeric probe G4-ATP-R at a concentration as low as 20 nM can still permeate the cell membrane and work well in living cells. In comparison with inorganic nanocarriers, the unique advantage of our DNA dendrimeric nanocarrier results from its excellent biocompatibility by the naturally occurring feature of DNA. The biocompatibility of our DNA dendrimeric carrier was then verified by MTS assay. Experimental results showed negligible inhibition of proliferation in cancer cells, indicating good biocompatibility of the DNA dendrimers (Figure S15).
Conclusions
In summary, we have developed a DNA dendrimeric nanocarrier for FNAs through DNA molecular assembly for in situ monitoring of biological molecules in living cells. The DNA dendrimer was prepared by an enzyme-free method using step-by-step assembly of Y-shaped DNA structures. As the two main classes of FNAs, both DNAzyme and aptamer were embedded in the DNA dendrimer scaffold to construct sensing systems for intracellular molecular sensing. These sensing systems maintained both the catalytic activity of DNAzyme and the recognition function of aptamer toward in vitro targets. Their further intracellular monitoring of histidine and ATP demonstrated that these FNA-embedded dendrimeric sensing systems could successfully self-deliver into living cells, while maintaining their target recognition capabilities. Instead of incorporating potentially biohazardous inorganic materials for efficient FNA delivery, the proposed nanocarrier employs naturally occurring DNA molecules as building blocks, exhibiting excellent biocompatibility. Moreover, such nanocarriers possess several other advantages, such as easy preparation, enhanced enzymatic stability, high stability under very low concentration (no CMC effect), and good self-delivery capability. These advantages, coupled with the fact that DNAzymes and aptamers for any target can be produced via in vitro selection, make this DNA dendrimeric nanocarrier a promising new platform for efficient intracellular monitoring for targets of interest and, hence, for wide applications in biomedicine.
Experimental Section
Reagents and Instruments
DNA oligonucleotides used in this work were synthesized and purified by Takara Biotechnology Co., Ltd. (Dalian, China), and their sequences are shown in Table S1 (see Supporting Information). Adenosine triphosphate (ATP), uridinetriphosphate (UTP), guanosine triphosphate (GTP), cytidine triphosphate (CTP), and l-histidine were obtained from Sigma-Aldrich. The MCF-7 cell lines were kind gifts from Prof. Jinfeng Zhao (National Hepatobiliary and Enteric Surgery Research Center, Department of Surgery, Xiangya Hosiptal, Central South University). Cell culture media were purchased from Thermo Scientific HyClone (MA, USA). All other chemicals were of analytical grade, were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China), and were used without further purification. All solutions were prepared using ultrapure water, which were prepared through a Millipore Milli-Q water purification system (Billerica, MA, USA), with an electrical resistance >18.3 MΩ.
All fluorescence measurements were carried out on a Fluoromax-4 spectrofluorometer (HORIBA JobinYvon, Edison, NJ) with a 200 μL quartz cuvette. The excitation wavelength was fixed at 494 nm with both excitation and emission bandwidths set at 5.0 nm.
The detection of electrophoresis gels was performed with the Bio-Rad ChemiDoc XRS System. Dynamic light scattering (DLS) was measured on the Malvern Zetasizer Nano ZS90 (Malvern Instruments, Ltd., Worcestershire, UK). Atomic force microscopy of samples was observed on a Multimode 8 (Bruker/USA). Transmission electron microscopy (TEM) images were obtained on an H-7000 NAR transmission electron microscope (Hitachi) with a working voltage of 100 kV. Bioimaging was performed using the FV1000-X81 confocal microscope (Olympus). MTS assay was used with a Synergy 2 Multi-Mode Microplate Reader (Bio-Tek, Winooski, VT).
Preparation of FNA-Embedded DNA Dendrimers
DNA dendrimers were prepared via an enzyme-free self-assembled procedure.53 The Y-shaped junction scaffolds (Y-DNA) were first prepared as building blocks. In the experiment to prepare Y1, four oligonucleotide strands (Y1a, Y1b, Y1c-l-histidine, and Xl-histidine for the DNAzyme-based sensing system, or Y1a, Y1b, Y1c-ATP, and XATP for the aptamer-based sensing system; see Table S1 in Supporting Information) were mixed stoichiometrically in HEPES buffer (25 mM, pH 7.9, containing 500 mM KCl) or Tris-HCl buffer (20 mM, pH 8.3, containing 300 mM NaCl and 5 mM MgCl2), respectively, to give a final concentration of 20 μM for each strand. The mixed solution was annealed by heating at 90 °C for 5 min and then cooling to 4 °C slowly. The annealed sample was then taken to room temperature and incubated for another 30 min to afford Y-shaped DNA building block Y1. The other Y-shaped building blocks, Y0, Y2, Y3, and Y4, were prepared following a similar procedure from three different oligonucleotide strands (see Table S1 in Supporting Information). Y0, Y1, Y2, Y3, and Y4 were then directly used without further purification to assemble the different generations of DNA dendrimers (Gn). In a typical procedure, 1 molar of Gn–1 was mixed with 3 × 2n–1 molar of Yn, resulting in the formation of Gn. Y0 (G0) was first assembled with Y1 at a 1:3 molar stoichiometry through hybridization of the sticky ends, forming the first generation of DNA dendrimer (G1) and leaving six free sticky ends available for further assembly. The mixture was then incubated at room temperature for 1 h, allowing hybridization to be completed. G1 could be further assembled with Y2 at a 1:6 molar stoichiometry to afford the second generation of DNA dendrimer G2. Using this iterative process, different generations of DNA dendrimers could be easily prepared in several hours.
Fluorescence Imaging of Living Cells Using DNA Dendrimers
MCF-7 cell lines were grown in Roswell Park Memorial Institute (RPMI) 1640 medium containing 10% (v/v) fetal bovine serum (FBS), penicillin (100 units mL–1), and streptomycin (100 μg mL–1). MCF-7 cells (2 × 105) were seeded in a 30 mm glass-bottomed dish and incubated for 24 h. After removing cell medium, cells were first incubated with DNA dendrimer in 1 mL of Dulbecco’s PBS (1.47 mM KH2PO4, 8.06 mM Na2HPO4, 137.93 mM NaCl, 2.67 mM KCl) supplemented with 100 μL 1640 medium at 37 °C for 4 h to allow sufficient uptake. For l-histidine detection, MCF-7 cells pretreated with G4-l-histidine were further incubated with 100 mM of l-histidine containing 150 mM K+ for another 1 h. However, for ATP detection, such treatment was unnecessary. Cells were washed three times with PBS, and fluorescence imaging experiments were then performed with a 20× objective. Excitation wavelength and emission filters were as follows: FAM, 488 nm laser line excitation; emission BP, 520 ± 20 nm filter.
Cell Lysate Preparation
Breast cancer MCF-7 cells were plated in a 35 mm cell culture dish and grown to ∼80% confluence before the experiments. Cells were washed twice with 1 mL of PBS, and 200 μL of trypsin was then added to the cell culture dish. After 5 min, MCF-7 cells were centrifuged for 5 min at 25 °C (800 rpm), the supernatant fraction was removed, and the cell precipitate was redispersed in 1 mL of buffer solution. Then, the redispersed cells were subjected to sonication treatment in an ice–water bath using a probe-type sonicator (200 W). The resulting cell lysate was stored at 4 °C.
Agarose Gel Electrophoresis
Without further purification, all Y-DNAs were dissolved in buffer to give a final concentration of 10 μM. For DNA dendrimer, 10 μL of different of DNA dendrimer generations were directly used for electrophoresis experiments. Electrophoresis was carried out in 1× Tris-borate-EDTA (TBE) buffer (90 mM Tris, 90 mM boric acid, and 10 mM EDTA, pH 8.0) at 100 V for 1.5 h. The gels were then stained with ethidium bromide.
Acknowledgments
This work was supported by the National Institutes of Health (GM079359 and CA133086), and by National Key Scientific Program of China (2011CB911000), NSFC (Grants 21325520, J1210040, 20975034, and 21177036), the Foundation for Innovative Research Groups of NSFC (Grant 21221003), the National Key Natural Science Foundation of China (21135001), National Instrumentation Program (2011YQ030124), the Ministry of Education of China (20100161110011), and Hunan Provincial Natural Science Foundation (Grants 12JJ6012 and 11JJ1002).
Supporting Information Available
Additional materials and methods: DNA sequences and figures characterizing DNA dendrimers and their application. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Spiller D. G.; Wood C. D.; Rand D. A.; White M. R. H. Measurement of Single-Cell Dynamics. Nature 2010, 465, 736–745. [DOI] [PubMed] [Google Scholar]
- Yang Y. M.; Zhao Q.; Feng W.; Li F. Y. Luminescent Chemodosimeters for Bioimaging. Chem. Rev. 2013, 113, 192–270. [DOI] [PubMed] [Google Scholar]
- Quang D. T.; Kim J. S. Fluoro- and Chromogenic Chemodosimeters for Heavy Metal Ion Detection in Solution and Biospecimens. Chem. Rev. 2010, 110, 6280–6301. [DOI] [PubMed] [Google Scholar]
- Nolan E. M.; Lippard S. J. Tools and Tactics for the Optical Detection of Mercuric Ion. Chem. Rev. 2008, 108, 3443–3480. [DOI] [PubMed] [Google Scholar]
- Li X. H.; Gao X. H.; Shi W.; Ma H. M. Design Strategies for Water-Soluble Small Molecular Chromogenic and Fluorogenic Probes. Chem. Rev. 2014, 114, 590–659. [DOI] [PubMed] [Google Scholar]
- Ruracka K.; Resch-Gengera U. Rigidization, Preorientation and Electronic Decoupling—The ‘Magic Triangle’ for the Design of Highly Efficient Fluorescent Sensors and Switches. Chem. Soc. Rev. 2002, 31, 116–127. [DOI] [PubMed] [Google Scholar]
- Peng X. J.; Du J. J.; Fan J. L.; Wang J. Y.; Wu Y. K.; Zhao J. Z.; Sun S. G.; Xu T. A Selective Fluorescent Sensor for Imaging Cd2+ in Living Cells. J. Am. Chem. Soc. 2007, 129, 1500–1501. [DOI] [PubMed] [Google Scholar]
- Zhang M.; Yu M. X.; Li F. Y.; Zhu M. W.; Li M. Y.; Gao Y. H.; Li L.; Liu Z. Q.; Zhang J. P.; Zhang D. Q.; et al. A Highly Selective Fluorescence Turn-on Sensor for Cysteine/Homocysteine and Its Application in Bioimaging. J. Am. Chem. Soc. 2007, 129, 10322–10323. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Zhang X. B.; Han Z. X.; Qiao L.; Li C. Y.; Jian L. X.; Shen G. L.; Yu R. Q. Highly Sensitive and Selective Colorimetric and Off–On Fluorescent Chemosensor for Cu2+ in Aqueous Solution and Living Cells. Anal. Chem. 2009, 81, 7022–7030. [DOI] [PubMed] [Google Scholar]
- Jin Z.; Xie D. X.; Zhang X. B.; Gong Y. J.; Tan W. H. Bifunctional Fluoroionphore-Ionic Liquid Hybrid for Toxic Heavy Metal Ions: Improving Its Performance via the Synergistic Extraction Strategy. Anal. Chem. 2012, 84, 4253–4257. [DOI] [PubMed] [Google Scholar]
- Mao G. J.; Wei T. T.; Wang X. X.; Huan S. Y.; Lu D. Q.; Zhang J.; Zhang X. B.; Tan W. H.; Shen G. L.; Yu R. Q. High-Sensitivity Naphthalene-Based Two-Photon Fluorescent Probe Suitable for Direct Bioimaging of H2S in Living Cells. Anal. Chem. 2013, 85, 7875–7881. [DOI] [PubMed] [Google Scholar]
- Fan J. L.; Hu M. M.; Zhan P.; Peng X. J. Energy Transfer Cassettes Based on Organic Fluorophores: Construction and Applications Inratiometric Sensing. Chem. Soc. Rev. 2013, 42, 29–43. [DOI] [PubMed] [Google Scholar]
- Li C. Y.; Zhang X. B.; Qiao L.; Zhao Y.; He C. M.; Huan S. Y.; Lu L. M.; Jian L. X.; Shen G. L.; Yu R. Q. Naphthalimide–Porphyrin Hybrid Based Ratiometric Bioimaging Probe for Hg2+: Well-Resolved Emission Spectra and Unique Specificity. Anal. Chem. 2009, 81, 9993–10001. [DOI] [PubMed] [Google Scholar]
- Gong Y. J.; Zhang X. B.; Zhang C. C.; Luo A. L.; Fu T.; Tan W. H.; Shen G. L.; Yu R. Q. Through Bond Energy Transfer: A Convenient and Universal Strategy toward Efficient Ratiometric Fluorescent Probe for Bioimaging Applications. Anal. Chem. 2012, 84, 10777–10784. [DOI] [PubMed] [Google Scholar]
- Liu J. W.; Cao Z. H.; Lu Y. Functional Nucleic Acid Sensors. Chem. Rev. 2009, 109, 1948–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C.; Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- Ellington A. D.; Szostak J. W. InVitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- Willner I.; Zayats M. Electronic Aptamer-Based Sensors. Angew. Chem., Int. Ed. 2007, 46, 6408–6418. [DOI] [PubMed] [Google Scholar]
- Famulok M.; Mayer G.; Blind M. Nucleic Acid Aptamers: From Selection in Vitro to Applications in Vivo. Acc. Chem. Res. 2000, 33, 591–599. [DOI] [PubMed] [Google Scholar]
- Fang X. H.; Tan W. H. Aptamers Generated from Cell-SELEX for Molecular Medicine: A Chemical Biology Approach. Acc. Chem. Res. 2010, 43, 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R. M.; Zhang X. B.; Chen Z.; Tan W. H. Aptamer-Assembled Nanomaterials for Biosensing and Biomedical Applications. Small 2011, 7, 2428–2436. [DOI] [PubMed] [Google Scholar]
- Yin B. C.; Ye B. C.; Tan W. H.; Wang H.; Xie C. C. An Allosteric Dual-DNAzyme Unimolecular Probe for Colorimetric Detection of Copper(II). J. Am. Chem. Soc. 2009, 131, 14624–14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R. M.; Zhang X. B.; Chen Z.; Meng H. M.; Song Z. L.; Tan W. H.; Shen G. L.; Yu R. Q. Unimolecular Catalytic DNA Biosensor for Amplified Detection of l-Histidine via an Enzymatic Recycling Cleavage Strategy. Anal. Chem. 2011, 83, 7603–7607. [DOI] [PubMed] [Google Scholar]
- Tang Z. W.; Mallikaratchy P.; Yang R. H.; Kim Y.; Zhu Z.; Wang H.; Tan W. H. Aptamer Switch Probe Based on Intramolecular Displacement. J. Am. Chem. Soc. 2008, 130, 11268–11269. [DOI] [PubMed] [Google Scholar]
- Zuo X. L.; Xiao Y.; Plaxco K. W. High Specificity, Electrochemical Sandwich Assays Based on Single Aptamer Sequences and Suitable for the Direct Detection of Small-Molecule Targets in Blood and Other Complex Matrices. J. Am. Chem. Soc. 2009, 131, 6944–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F. S.; Wang Y.; Zhang R.; Li Z. C. Intelligent Nucleic Acid Delivery Systems Based on Stimuli-Responsive Polymers. Soft Matter 2010, 6, 835–848. [Google Scholar]
- Seferos D. S.; Giljohann D. A.; Hill H. D.; Prigodich A. E.; Mirkin C. A. Nano-Flares: Probes for Transfection and mRNA Detection in Living Cells. J. Am. Chem. Soc. 2007, 129, 15477–15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S. M.; Fu P. P.; Shirsat R. N.; Hwang H. M.; Leszczynski J.; Yu H. T. UVA Light-Induced DNA Cleavage by Isomeric Methylbenz[a]anthracenes. Chem. Res. Toxicol. 2002, 15, 400–407. [DOI] [PubMed] [Google Scholar]
- Connelly J. C.; De Leau E. S.; Leach D. R. F. DNA Cleavage and Degradation by the SbcCD Protein Complex from Escherichia coli. Nucleic Acids Res. 1999, 27, 1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins J. B.; Prudent J. R.; Marshall D. J.; Ruppen M.; Thorson J. S. A Continuous Assay for DNA Cleavage: The Application of “Break Lights” to Enediynes, Iron-Dependent Agents, and Nucleases. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 13537–13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X. H.; Chen T.; Xiong X. L.; Mao Y.; Zhu G. Z.; Yasun E.; Li C. M.; Zhu Z.; Tan W. H. Semiquantification of ATP in Live Cells Using Nonspecific Desorption of DNA from Graphene Oxide as the Internal Reference. Anal. Chem. 2012, 84, 8622–8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y.; Zhu Z.; Han D.; Liu H.; Liu J.; Liao J.; Zhang K.; Tan W. H. Insulin-Binding Aptamer-Conjugated Graphene Oxide for Insulin Detection. Analyst 2011, 136, 4138–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P. C.; Giljohann D. A.; Daniel W. L.; Zheng D.; Prigodich A. E.; Mirkin C. A. Scavenger Receptors Mediate Cellular Uptake of Polyvalent Oligonucleotide-Functionalized Gold Nanoparticles. Bioconjugate Chem. 2010, 21, 2250–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seferos D. S.; Prigodich A. E.; Giljohann D. A.; Patel P. C.; Mirkin C. A. Polyvalent DNA Nanoparticle Conjugates Stabilize Nucleic Acids. Nano Lett. 2009, 9, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P. W.; Hwang K.; Lan T.; Lu Y. A DNAzyme-Gold Nanoparticle Probe for Uranyl Ion in Living Cells. J. Am. Chem. Soc. 2013, 135, 5254–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Li Z. H.; Hu D. H.; Lin C. T.; Li J. H.; Lin Y. H. Aptamer/Graphene Oxide Nanocomplex for In Situ Molecular Probing in Living Cells. J. Am. Chem. Soc. 2010, 132, 9274–9276. [DOI] [PubMed] [Google Scholar]
- Jayagopal A.; Halfpenny K. C.; Perez J. W.; Wright D. W. Hairpin DNA-Functionalized Gold Colloids for the Imaging of mRNA in Live Cells. J. Am. Chem. Soc. 2010, 132, 9789–9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.; Wu C. S.; Jimenez E.; Zhu Z.; Dajac J. G.; You M. X.; Han D.; Zhang X. B.; Tan W. H. DNA Micelle Flares for Intracellular mRNA Imaging and Gene Therapy. Angew. Chem., Int. Ed. 2013, 125, 2066–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. C.; Chen T.; Han D.; You M. X.; Peng L.; Cansiz S.; Zhu G. Z.; Li C. M.; Xiong X. L.; Jimenez E.; et al. Engineering of Switchable Aptamer Micelle Flares for Molecular Imaging in Living Cells. ACS Nano 2013, 7, 5724–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruthers M. H. Gene Synthesis Machines: DNA Chemistry and Its Uses. Science 1985, 230, 281–285. [DOI] [PubMed] [Google Scholar]
- Zhu G. Z.; Hu R.; Zhao Z. L.; Chen Z.; Zhang X. B.; Tan W. H. Noncanonical Self-Assembly of Multifunctional DNA Nanoflowers for Biomedical Applications. J. Am. Chem. Soc. 2013, 135, 16438–16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C. Nanomaterials Based on DNA. Annu. Rev. Biochem. 2010, 79, 65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac R.; Sparks A. B.; Callow M. J.; Halpern A. L.; Burns N. L.; Kermani B. G.; Carnevali P.; Nazarenko I.; Nilsen G. B.; Yeung G.; et al. Human Genome Sequencing Using Unchained Base Reads on Self-Assembling DNA Nanoarrays. Science 2010, 327, 78–81. [DOI] [PubMed] [Google Scholar]
- Genereux J. C.; Barton J. K. Mechanisms for DNA Charge Transport. Chem. Rev. 2010, 110, 1642–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astruc D.; Boisselier E.; Ornelas C. Dendrimers and Dendrons: Concepts, Syntheses, Applications. Chem. Rev. 2010, 110, 1857–1959. [DOI] [PubMed] [Google Scholar]
- Mintzer M. A.; Grinstaff M. W. Biomedical Applications of Dendrimers: A Tutorial. Chem. Soc. Rev. 2011, 40, 173–190. [DOI] [PubMed] [Google Scholar]
- Roth A.; Breaker R. R. An Amino Acid as a Cofactor for a Catalytic Polynucleotide. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 6027–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizenga D. E.; Szostak J. W. A DNA Aptamer That Binds Adenosine and ATP. Biochemistry 1995, 34, 656–665. [DOI] [PubMed] [Google Scholar]
- Lin C. H.; Patel D. J. Structural Basis of DNA Folding and Recognition in an AMP-DNA Aptamer Complex: Distinct Architectures but Common Recognition Motifs for DNA and RNA Aptamers Complexed to AMP. Chem. Biol. 1997, 4, 817–832. [DOI] [PubMed] [Google Scholar]
- Hiromi I.; Kim P. H. N.; Hiroko T.; Kenta S.; Ryota I.; Yasuyuki K. Y.; Takeharu N.; Hiroyuki N. Visualization of ATP Levels Inside Single Living Cells with Fluorescence Resonance Energy Transfer-Based Genetically Encoded Indicators. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 15651–15656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beis I.; Newsholme E. A. The Contents of Adenine Nucleotides, Phosphagens and Some Glycolytic Intermediates in Resting Muscles from Vertebrates and Invertebrates. Biochem. J. 1975, 152, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T.; Chen P.; Niu L.; Jin J.; Liang D. H.; Li Z. B.; Yang Z. Q.; Liu D. S. pH-Responsive Size-Tunable Self-Assembled DNA Dendrimers. Angew. Chem., Int. Ed. 2012, 124, 11433–11436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.