The MADS box transcription factor AGL16 represses the expression of FT and thus delays flowering time in long-day conditions, while miR824, a direct negative regulator of AGL16 mRNA abundance, accelerates flowering. The magnitude of AGL16 action depends on the joint allelic dosage at both AGL16 and FLC, which may result from a physical interaction between AGL16, SVP, and FLC.
Abstract
The timing of flowering is pivotal for maximizing reproductive success under fluctuating environmental conditions. Flowering time is tightly controlled by complex genetic networks that integrate endogenous and exogenous cues, such as light, temperature, photoperiod, and hormones. Here, we show that AGAMOUS-LIKE16 (AGL16) and its negative regulator microRNA824 (miR824) control flowering time in Arabidopsis thaliana. Knockout of AGL16 effectively accelerates flowering in nonvernalized Col-FRI, in which the floral inhibitor FLOWERING LOCUS C (FLC) is strongly expressed, but shows no effect if plants are vernalized or grown in short days. Alteration of AGL16 expression levels by manipulating miR824 abundance influences the timing of flowering quantitatively, depending on the expression level and number of functional FLC alleles. The effect of AGL16 is fully dependent on the presence of FLOWERING LOCUS T (FT). Further experiments show that AGL16 can interact directly with SHORT VEGETATIVE PHASE and indirectly with FLC, two proteins that form a complex to repress expression of FT. Our data reveal that miR824 and AGL16 modulate the extent of flowering time repression in a long-day photoperiod.
INTRODUCTION
An appropriate timing of flowering is essential for plants to maximize reproductive success and adapt to changing environmental conditions. Molecular genetic pathways controlling the switch from vegetative to reproductive growth have been well characterized, especially in the model plant Arabidopsis thaliana. Various endogenous and exogenous cues, including age, circadian clock, temperature, photoperiod, and hormones, have been reported to be involved in the control of flowering time (Simpson and Dean, 2002; Ausín et al., 2005; Bäurle and Dean, 2006; Dennis and Peacock, 2007; Andrés and Coupland, 2012).
Photoperiod changes are perceived in leaves and transduced to regulate the accumulation of CONSTANS, which encodes a transcription factor that activates the transcription of FLOWERING LOCUS T (FT) in the vascular tissues of the leaves (Samach et al., 2000; An et al., 2004; Kobayashi and Weigel, 2007). The FT protein moves to the shoot apical meristem, where it activates the expression of several floral integrators like APETALA1, SUPPRESSOR OF OVEREXPRESSION OF CO1, FRUITFULL, and LEAFY together with its interaction partners FD and 14-3-3 proteins (reviewed in Andrés and Coupland, 2012). FT expression is negatively regulated by several floral repressors, such as the MADS box transcription factors FLOWERING LOCUS C (FLC), SHORT VEGETATIVE PHASE (SVP), and two distinct groups of APETALA2-like factors, exemplified by TEMPRANILLO1 and SCHLAFMUETZE (SMZ) and their respective closest homologs (Aukerman and Sakai, 2003; Searle et al., 2006; Jung et al., 2007; Castillejo and Pelaz, 2008; Li et al., 2008; Mathieu et al., 2009). FLC, SVP, and other FLC-clade members form repressor complexes governing the integration of flowering signals (Li et al., 2008; Gu et al., 2013). FLC integrates cues from the autonomous and vernalization pathways, both of which cause stable transcriptional repression of FLC (Lee and Amasino, 1995; Michaels and Amasino, 2001; Amasino, 2005; Dennis and Peacock, 2007). High-level expression of FLC depends on the presence of FRIGIDA (FRI) (Johanson et al., 2000; Michaels and Amasino, 2001). Although FRI is inactive in the Columbia-0 (Col-0) accession, ∼60% of individuals in the worldwide Arabidopsis population carry a functional allele delaying flowering time (Johanson et al., 2000; Stinchcombe et al., 2004; Shindo et al., 2005; Korves et al., 2007; Brachi et al., 2010).
MicroRNAs (miRNAs) are 20- to 24-nucleotide regulatory RNA molecules that play critical roles in many aspects of plant development and adaptation to environmental changes (Chen, 2009; Voinnet, 2009; Rubio-Somoza and Weigel, 2011). In Arabidopsis, three miRNAs, miR172, miR156, and miR159, have been shown to play important roles in floral transition (Park et al., 2002; Aukerman and Sakai, 2003; Schmid et al., 2003; Achard et al., 2004; Chen, 2004; Schwab et al., 2005; Wang et al., 2009; Wu et al., 2009; Kim et al., 2012; Yu et al., 2012). miR172, miR156, and miR159 as well as their respective targets are found in many plant families and thus are believed to be ancient modules that regulate developmental timing in plants (Lauter et al., 2005; Chuck et al., 2007; Zhu et al., 2009; Nair et al., 2010; Cho et al., 2012). However, novel regulatory modules seem to evolve frequently in plant lineages, and many miRNA/target gene interactions are family or species specific (Cuperus et al., 2011).
miR824 is a Brassicaceae-specific miRNA (Rajagopalan et al., 2006; Fahlgren et al., 2007; Kutter et al., 2007). miR824 is proposed to have evolved through partial duplication of its target gene, AGAMOUS-LIKE16 (AGL16), which encodes a MADS box transcription factor (Fahlgren et al., 2007; de Meaux et al., 2008). The miR824 precursor displays two alleles and was hypothesized to evolve under balancing selection in Arabidopsis (de Meaux et al., 2008). miR824 is expressed in many tissues including rosette and cauline leaves, shoots, inflorescences, and roots (Kutter et al., 2007). The expression of AGL16, the only confirmed target for miR824, is also detected in these tissues as well as in guard cells, trichomes, and developing siliques (Alvarez-Buylla et al., 2000). In agreement with its expression in guard cells, the miR824/AGL16 module was shown to regulate the development of higher-order stomata complexes by promoting additional divisions of satellite meristemoid cells (Kutter et al., 2007).
However, AGL16 is also expressed in the shoot apex and the root and at a level that is sensitive to light change and environmental stress (https://www.genevestigator.com/gv/plant.jsp). In addition, genome-wide profiling of FLC and SVP binding sites revealed that AGL16 is one of the strongest FLC targets (Deng et al., 2011) and a weak target for SVP (Gregis et al., 2013). These data prompted us to hypothesize that AGL16 might be involved in the control of flowering time. Here, we show that the miR824/AGL16 module participates in the timing of floral transition in Arabidopsis by interacting closely with the FRI/FLC-SVP pathways.
RESULTS
Loss of Function of AGL16 Accelerates Flowering of Nonvernalized Col-FRI in Long-Day Conditions Only
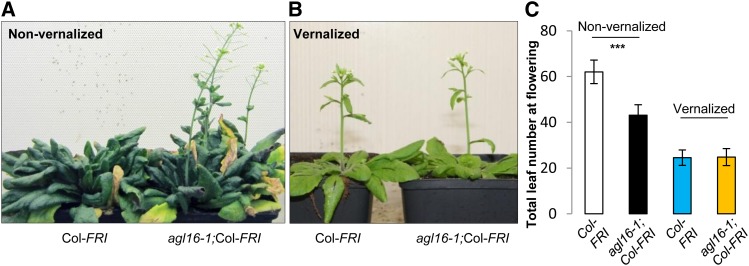
Since AGL16 was reported as one of the major targets of FLC, it was necessary to evaluate its role on flowering time in a background expressing high levels of FLC. We crossed the loss-of-function mutant agl16-1 in the Col-0 background, with Col-FRI, which expresses FLC at high levels due to the introgression of a functional FRI from the accession Sf-2 into Col-0 (Michaels and Amasino, 1999). Under long-day (LD) conditions without vernalization, wild-type Col-FRI flowered at around 62 total leaves, whereas the AGL16 loss-of-function mutant (agl16-1;Col-FRI) flowered much earlier at around 43 leaves (Student’s t test, P < 1E-16; Figures 1A and 1C). This experiment was replicated in three further trials. Although the total number of leaves at flowering fluctuated across experiments, AGL16 loss-of-function mutants always had ∼30% (25 to 35%) fewer leaves upon flowering than the wild type (Table 1; Supplemental Data Set 1). To test the implication of AGL16 function in vernalization, agl16-1;Col-FRI was vernalized together with Col-FRI for 4 weeks. Both lines exhibited a typical vernalization response, indicating that AGL16 is not essential for this response (Figures 1B and 1C).
Figure 1.
Loss of Function of AGL16 Partially Suppressed the Delayed Flowering Conferred by FRI Introgression into the Col-0 Background under LD Conditions but Did Not Influence the Vernalization Response.
(A) and (B) Comparisons between wild-type Col-FRI (left) and the loss-of-function mutant agl16-1;Col-FRI (right) upon flowering under LD conditions without vernalization treatment (A) or with 4 weeks of vernalization at 4°C (B).
(C) The flowering time behavior of loss of function of AGL16 in the Col-FRI background (agl16-1;Col-FRI) grown under LD conditions. Total leaf number including rosette and cauline leaves at flowering was monitored. Note that under nonvernalized LD conditions, agl16-1;Col-FRI showed strongly accelerated flowering compared with Col-FRI (***P < 0.001, two-tailed Student’s t test; black and white bars, respectively), while under vernalized LD conditions, agl16-1;Col-FRI flowered at the same time as Col-FRI (P = 0.9; yellow and blue bars, respectively). Error bars indicate sd (see Supplemental Data Set 1 for number of plants tested).
Table 1. Summary of the Flowering Behavior of Mutants under LD Conditions.
| Line Comparisons | No. of Independent Trials | Cumulated No. of Individuals (Mutant vs. Wild Type) | Differences in Mean Leaf No. | Percentage Difference in Mean Leaf No. |
|---|---|---|---|---|
| agl16-1;Col-FRI vs. Col-FRI | 4 | 73 vs. 94 | 8.5–19 | 65.2–75.3% |
| agl16-1 vs. Col-0 | 10 | 306 vs. 295 | 0.7–2.2 | 82.5–90.5% |
| m3 vs. Col-0 | 4 | 127 vs. 120 | 0.9–2.3 | 83.5–92.2% |
| MIM824s vs. Col-0 (vector) | 3 | 266 vs. 67 | 1.4–3.2 | 111–126% |
See Supplemental Data Set 1 for detailed information. Note that P < 0.01 (two-tailed Student's t test) for all comparisons except for the comparison agl16-1 versus Col-0, where one trial (trial 4 in Supplemental Data Set 1) showed P < 0.05. Differences in mean leaf number reflect the raw observed difference. The total leaf number at flowering varies with the genetic background in which miR824/AGL16 mutations were assessed. The percentage difference in mean leaf number presents the phenotype of the mutant relative to the wild type.
To test if the effect of AGL16 was dependent on a functional copy of FLC, we generated an F2 population segregating for AGL16/agl16-1, FRI/fri, and FLC/flc-3 by crossing the agl16-1 mutant with flc-3, which carries a nonfunctional FLC allele in the Col-FRI background (Michaels and Amasino, 1999). Under inductive LD conditions (16 h light/8 h dark) all plants homozygous for the flc-3 allele flowered after forming ∼10 leaves independent on the presence or absence of either a functional FRI or AGL16 (Figure 2A; Supplemental Data Set 1). A small, but reproducible acceleration of flowering was detected in the absence of FRI for both agl16-1 and flc-3 homozygous single mutants in comparison to Col-0 (see below).
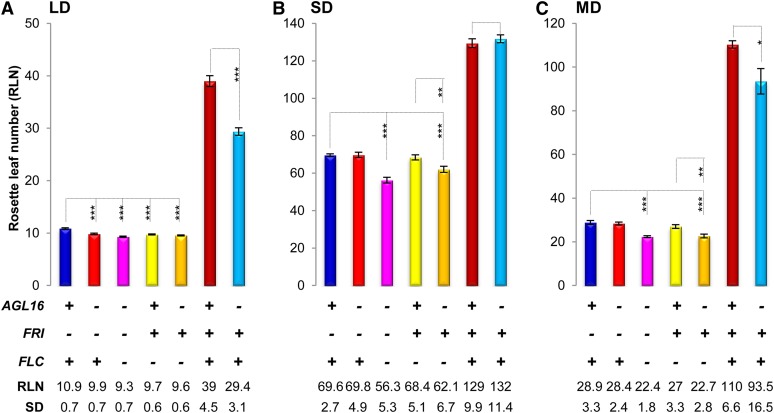
Figure 2.
Effects of the Loss of Function of AGL16 on Flowering Time Depend on the Photoperiod.
RLNs at flowering are shown for each line grown under LD (A), SD (B), and equal light-night (MD; [C]) conditions. Below the graph, the genotypes of each bar are marked for the three genes: AGL16, FRI, and FLC, with a plus sign indicating wild type or functional allele and a minus sign indicating loss of function. The mean values of RLN for each line together with the sd are shown. Statistical analyses were performed with two-tailed Student’s t test with Bonferroni correction (shown here) and confirmed with a Wilcoxon test. ***P < 0.001, **P < 0.01, and *P < 0.05. Note that in the flc-3 background, the absence of AGL16 can accelerate the flowering time under both MD (C) and SD (B) conditions.
The partial suppression of late flowering in Col-FRI by agl16-1 could only be observed when flowering was accelerated by an inductive photoperiod (Figure 2; Supplemental Data Set 1). Under short-day (SD) conditions, irrespective of the presence of FLC, agl16-1 plants flowered at the same time as the corresponding AGL16 siblings (both at around 130 leaves; Figure 2B). Under 12- to 12-h mid-day light cycles conditions, agl16-1;Col-FRI flowered earlier than Col-FRI, but the effect was reduced compared with LD (Figure 2C). In the absence of FRI, the presence or absence of AGL16 did not alter flowering time in SD or mid-day conditions, whereas a small acceleration of flowering was observed in both conditions in the flc-3 background when a functional FRI was present (Figures 2B and 2C). This might suggest that FRI can affect flowering to a limited extent via AGL16 in a FLC-independent manner under noninductive photoperiod conditions.
Taken together, these experiments show that the magnitude of the effect caused by the loss of AGL16 on flowering is not a function of the total time to flowering, but is primarily observed in the presence of strong FLC activity and a positive photoperiod stimulus. Vernalization is epistatic to a loss of AGL16 function, indicating that this gene does not participate in this response.
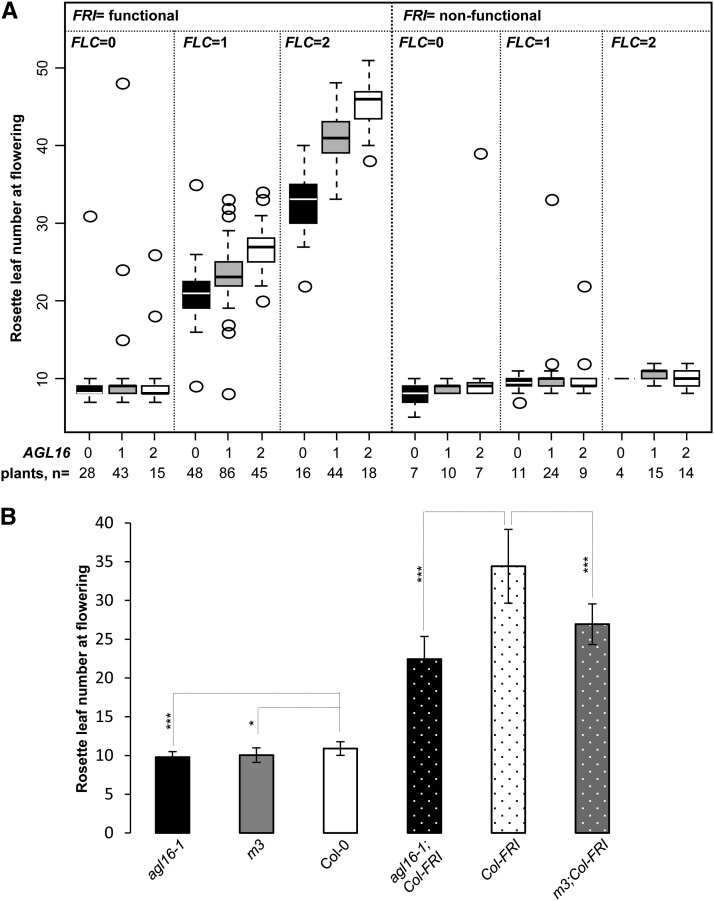
Flowering Time Is Sensitive to the Allelic Dosage of AGL16 and FLC
To further evaluate how sensitive the flowering response was to the dosage of FLC and AGL16, we scored flowering time in a segregating F2 population of the cross between agl16-1 (in Col-0) and flc-3 (in Col-FRI). We grew 451 F2 individuals under LD conditions and scored the flowering phenotype by counting the rosette leaf number at flowering (Figure 3A; Supplemental Data Set 2). All three genes showed a Mendelian inheritance (108:223:116, 132:212:102, and 113:224:113 for wild type:heterozygote:mutant for AGL16, FRI, and FLC, respectively; Fisher’s exact test, P = 1.0 for all three comparisons). As expected, the functionality of FRI (P = 0.008) strongly affected the flowering time in this population, and agl16-1 caused strong changes in flowering time when FRI was functional (P < 10E-10). The number of functional FLC alleles showed a significant influence on flowering time in general (P < 10E-4), but particularly in the FRI functional background (P < 2E-16). Importantly, AGL16 also displayed an allelic dosage effect and significantly influenced the timing of floral transition in general (P = 0.023) and especially in the functional FRI background (P = 4E-12; Figure 3A). In contrast to the previous experiment (Figure 2A), loss of function of AGL16 did not show a significant effect in homozygous fri individuals.
Figure 3.
Flowering Time Is Sensitive to the Quantitative Balance of FLC and AGL16.
(A) RLNs upon flowering are shown for plants of a segregating F2 population of 451 individuals grown under LD conditions. The RLN for each genotype is shown by box plots. Each box encloses the median 50% of the distribution, with the horizontal line marking the median. The lines extending from each box mark the minimum (5%) and maximum (95%) values of the data set. Circles marked the outliers (outside of the 5 to 95% distribution). FRI genotypes are grouped into functional and nonfunctional, as functional FRI is dominant. The allelic numbers of both FLC and AGL16 are coded as 0, 1, and 2, indicating homozygote for knockout allele, heterozygote, and wild-type homozygote, respectively. Numbers at the bottom give the number of plants featuring each multilocus genotype. The AGL16 genotypes alone (P = 0.023) and in interaction with FRI/FLC allelic combinations (P = 0.035) had a significant influence on the flowering time (see main text for details).
(B) Flowering time of plants overexpressing miR824 in both Col-0 (m3) and Col-FRI (m3;Col-FRI) backgrounds. Mean values of rosette leaf number upon flowering for each line together with the sd around the mean were plotted. Differences between lines were evaluated with two-tailed Student’s t test with Bonferroni correction (Supplemental Table 1). ***P < 0.001; *P < 0.05.
Since the number of triple homozygous individuals is small in our F2 segregating population (Figure 3A), this experiment lacks the statistical power to detect small effects. To thoroughly evaluate whether AGL16 loss of function can change the flowering of plants without a functional FRI, we grew agl16-1 together with Col-0 under LD conditions. Col-0 flowered at ∼10.9 rosette leaves (RLN; Figure 3B, Table 1; Supplemental Data Set 1). However, agl16-1 began to flower about one RLN (10%) earlier than Col-0 (Figure 3B; Supplemental Table 1; two-tailed Student’s t test, P = 1E-4). The acceleration of flowering is significant but mild with regard to the variance displayed by individual plants. To confirm this effect in the Col-0 background, we performed nine further independent trials including wild-type Col-0 and agl16-1 grown under LD conditions. Nine out of 10 trials showed that agl16-1 flowered significantly earlier than Col-0, with the differences ranging from 0.7 to 2.2 RLN, with a mean of 1.5 (Figures 2A and 3B; Supplemental Data Set 1). In this early flowering Col-0 background, this corresponds to ∼10 to 16% decrease in the total number of leaves at the time of flowering (Table 1). This indicates that AGL16 delayed flowering, even if FLC is expressed at the low levels characteristic of the Col-0 background, in which FRI is inactivated.
In the Col-FRI background, the contribution of AGL16 and FLC to late flowering was more pronounced. This observation was further corroborated with lines where AGL16 expression was knocked down via overexpressing its negative regulator miR824 in both Col-0 and Col-FRI backgrounds. We crossed Col-FRI with m3 (Kutter et al., 2007), a miR824-overexpressing line in which AGL16 expression is downregulated to ∼50% of the wild type level (Supplemental Figure 1). We compared homozygous m3;Col-FRI plants to Col-FRI. AGL16 expression was decreased to ∼60% of the Col-FRI level due to the presence of m3 (Supplemental Figure 1). Under LD conditions, homozygous m3;Col-FRI lines flowered at ∼26.9 rosette leaves, which was ∼7.5 leaves earlier (22%) than Col-FRI, but ∼4 leaves later than agl16-1;Col-FRI (Wilcoxon test, P = 7E-5; Figure 3B, Table 1; Supplemental Data Set 1).
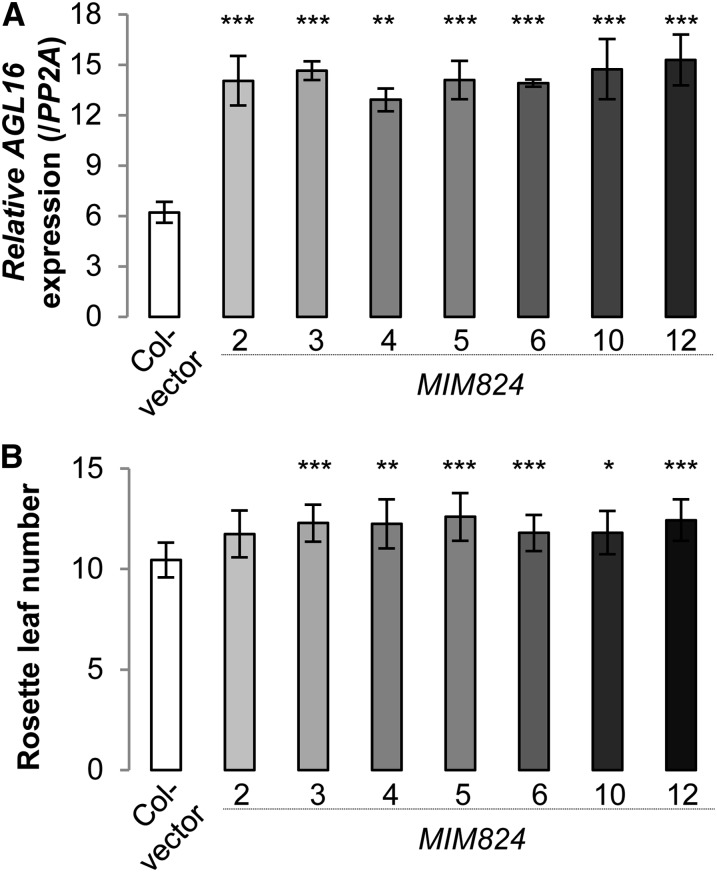
miR824 Mimicry Causes Later Flowering in Col-0
We further made use of miR824 target mimicry lines (MIM824) (Franco-Zorrilla et al., 2007; Todesco et al., 2010), which were created by overexpressing an artificial noncleavable target mimic for miR824, to assess the role of miR824 in regulating flowering. In these lines, a reduced activity of miR824 increases the expression of AGL16 specifically in the cells where AGL16 is naturally expressed. The phenotypic modifications displayed by MIM824 should thus reveal the native function of the miR824/AGL16 module. The expression of the target mimic in seven independent MIM824 lines consistently doubled the level of AGL16 expression (Figure 4A; two-tailed Student’s t test, all P < 0.01). In all seven lines (Figure 4A), the flowering time was consistently delayed under LD conditions (∼1.7 or 11 to 26% leaves more than Col-0 transformed with an empty vector; Wilcoxon test, P = 2E-11; Figure 4B, Table 1; Supplemental Figure 2 and Supplemental Data Set 1; three independent trials), suggesting that enhancing the expression of AGL16 by around 2-fold can delay flowering.
Figure 4.
Moderately Increasing the Expression of AGL16 in MIM824 Lines Can Delay Flowering Time under LD Conditions.
(A) Relative expression level of AGL16 in Col-0 transformed with an empty vector (open bar) and seven independent MIM824 transgenic lines (filled bars in different shades of gray; T3 lines with homozygous single insertions; the same for [B]) in 4-week-old rosette leaves. Expression values are reported as the mean of two biological replicates (each with three technical replicates) examined by real-time quantitative PCR (normalized to PP2A; significant levels tested with Student’s t test after Bonferroni correction. *P < 0.05; **P < 0.01; ***P < 0.001; the same for [B]). AGL16 expression levels in the aerial part of 10-d-old seedlings gave the same pattern (data not shown).
(B) Rosette leaf production upon flowering for the MIM824 lines (filled bars) and Col-0 transformed with empty vector (open bar). Mean leaf numbers to flowering together with sd are given. The flowering time behavior of a second independent experiment is shown in Supplemental Figure 2.
The miR824/AGL16 Module Regulates Flowering Time by Altering FT Expression
Since the miR824/AGL16 module regulates flowering time in a photoperiod-dependent manner, we wondered whether this module could regulate the expression of FT, the key factor controlling flowering time under LD conditions (Kardailsky et al., 1999; Samach et al., 2000). FT is the main target of FLC in leaves and is detected in the phloem companion cells of the leaf vasculature (Takada and Goto, 2003; Adrian et al., 2010). We first evaluated whether miR824 and FT expression overlap using promoter-miR824:GUS lines in the Col-0 background. A 4.7-kb fragment comprising 3.093-kb sequences upstream and 1.614 kb downstream of the transcription start site (TSS) was taken as the promoter of miR824 and fused to a reporter gene encoding β-glucuronidase (GUS) (Figure 5A). In T1 transgenic plants, the strongest GUS activity was detected in vascular tissues of rosette leaves (Figure 5B) as well as in guard cells (Supplemental Figure 3); therefore, the expression of miR824 and FT can colocalize. We further evaluated the role of FT in the process of miR824/AGL16- mediated flowering time regulation. We performed real-time quantitative PCR to quantify the expression of FT in agl16-1 mutants in both the Col-0 and Col-FRI backgrounds.
Figure 5.
The miR824/AGL16 Module Represses the Expression of FT.
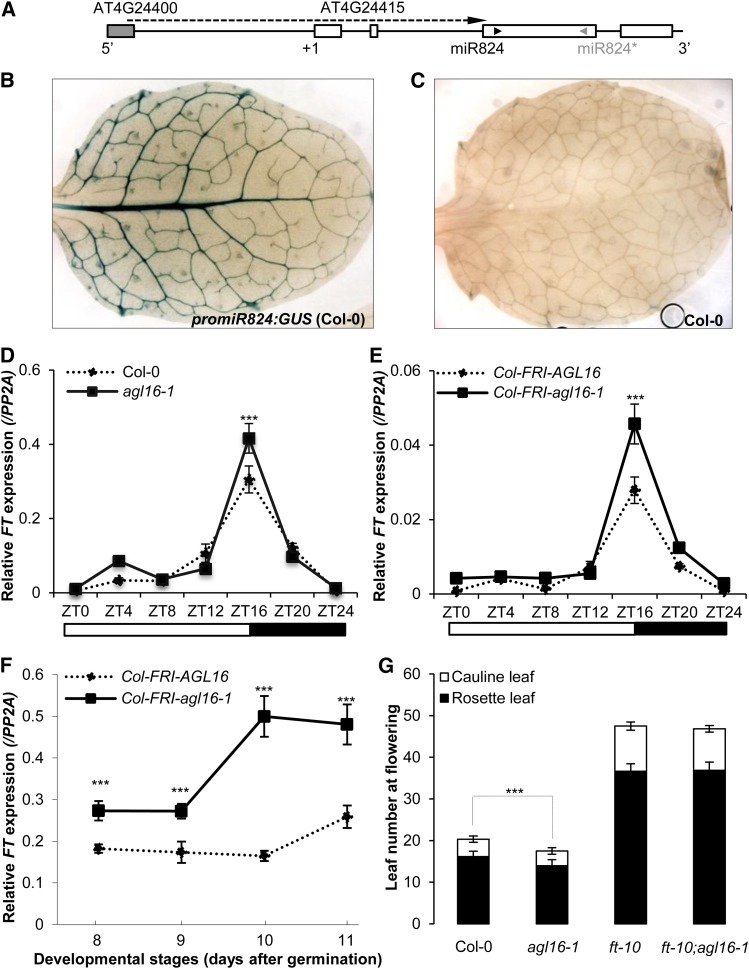
(A) Schematic representation of the miR824 locus. Exons (open bars), introns (horizontal lines), transcription starting site (+1), and positions of miR824 (black arrowhead) and miR824* (gray arrowhead) are indicated. The gray filled bar indicates the last exon of AT4G24400. The long arrow in the dashed line marks the region used as promoter of miR824. The scheme is not drawn to scale.
(B) GUS staining of the first rosette leaf of 2-week-old Col-0 plants transformed with promoter-miR824:GUS. Note the blue stain of the vasculature.
(C) GUS staining of the first rosette leaf of 2-week-old Col-0 plants (negative control).
(D) to (F) Real-time PCR following reverse transcription was used to quantify the level of FT expression, normalized to the expression level of the control gene PP2A, and shown on the y axis. All experiments were performed under LD conditions. Bars indicated sd of three technical replicates of the reactions. Experiments were replicated at least twice and all showed similar patterns (Supplemental Figure 5). Significant levels (Student’s t test with Bonferroni correction): *P < 0.05; **P < 0.01; ***P < 0.001.
(D) and (E) The relative expression of FT was quantified following different zeitgeber time points (from ZT0 to ZT24) with 9-d-old seedlings grown on agar plates. From ZT0 to ZT16 is daytime (open bars), while from ZT16 till ZT24 is nighttime (closed bars) as shown beneath the x axis. Note that at ZT16, the FT mRNA levels in both mutants (agl16-1 [D]; and agl16-1;Col-FRI [E]; solid lines) are higher than their corresponding wild types (dotted lines, P < 0.001).
(F) Time-course expression of FT in Col-FRI background plants grown in soil. Aerial tissues of seedlings of distinct ages (x axis shows days after germination) were harvested at ZT16.
(G) Flowering behavior of single and double mutants between agl16-1 and ft-10 under LD conditions. Mean of rosette (filled bars) and cauline (open bars) leaf number was shown together with sd. Note that agl16-1 flowered significantly earlier than Col-0, ft-10, and agl16-1;ft-10 (Student’s t test with Bonferroni correction, P < 0.001), but agl16-1 ft-10 double mutants flowered at the same time as ft-10 single mutants (P = 0.61).
[See online article for color version of this figure.]
In a 24-h time-course experiment performed with Col-0 plants (9 d after germination), FT expression started to increase strongly between zeitgeber time 12 (ZT12) and 20 (ZT20) (Figure 5D), as expected (Kobayashi et al., 1999). In agl16-1 plants (Supplemental Figure 4), FT mRNA abundance showed a similar diurnal pattern but accumulated to higher levels at ZT16 (Student’s t test, P < 0.001 for two independent trials; Figure 5D; Supplemental Figure 5A). In the Col-FRI background, the diurnal expression of FT was similar to that in Col-0, although the expression levels were more than 10 times reduced (Figure 5E; Supplemental Figure 5B). As in the Col-0 background, the agl16-1 mutation caused a higher expression of FT at ZT16 in Col-FRI (Student’s t test, P < 0.001 for two independent trials; Figure 5E; Supplemental Figures 4 and 5B). We also monitored the FT mRNA level in the Col-FRI background following development from 8 to 12 d after germination at ZT16. For all stages checked, FT levels were consistently higher in the agl16-1 background compared with Col-FRI and the values increased during development (Figure 5F; Supplemental Figure 5C).
Although the loss of function of AGL16 correlated well with elevated expression of FT in both Col-0 and Col-FRI backgrounds, it was unclear if the observed mild differences were sufficient to explain the altered flowering time in these backgrounds. Therefore, we examined whether the early flowering caused by a loss of AGL16 function requires a functional FT by crossing agl16-1 with ft-10 loss-of-function alleles (Yoo et al., 2005). The agl16-1 ft-10 double mutants flowered as late as the ft-10 single mutants (Student’s t test with Bonferroni correction, P = 0.62) and were significantly later than both Col-0 and agl16-1 (both P < 0.001) with agl16-1 significantly earlier than Col-0 (P < 0.001; Figure 5G). These results reveal that FT is required for the early flowering of agl16-1.
AGL16 Is a Potential Partner of Flowering Repressor Complexes Targeting FT
FLC has been shown to directly target the promoter of FT, thereby repressing the response of FT to photoperiod (Searle et al., 2006). As a consequence, a loss of function of FLC massively increases the expression of its direct target FT (Searle et al., 2006). FLC has also been shown to bind to the promoter of AGL16 (Deng et al., 2011), but its loss of function only slightly changes AGL16 expression (Figure 6A; Supplemental Figure 6A; Deng et al., 2011). AGL16 could be required for full functionality of FRI/FLC by increasing the expression of FLC in leaves. However, comparing FLC expression levels between agl16-1 Col-FRI and Col-FRI over a developmental window from 8 to 11 d after germination did not reveal massive differences in FLC expression (Figure 6B) with similar pattern observed in the Col-0 background (Supplemental Figure 6B). These results suggest that the genetic interaction between AGL16 and FLC does not act primarily at the transcription level.
Figure 6.
AGL16 Can Participate in the Repressor Protein Complexes Formed by FLC/SVP.
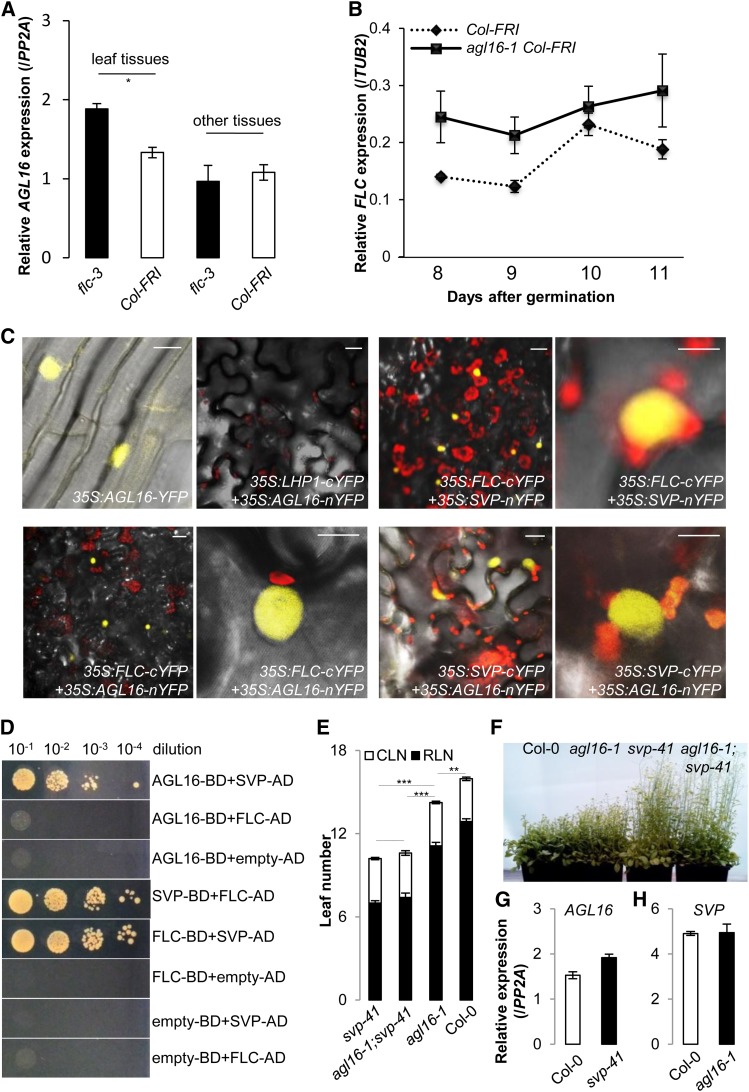
(A) Loss of function of FLC (flc-3) moderately altered the expression of AGL16 in the Col-FRI background (P < 0.05). Relative accumulation of AGL16 in the flc-3 mutant was monitored by real-time RT-PCR (normalized to PP2A) in different tissues of the aerial parts of 12-d-old seedlings grown under LD conditions. Error bars indicate se of three biological replicates. Note that AGL16 expression in the flc-3 mutant was slightly enhanced compared with Col-FRI (P = 0.02, indicated by one asterisk; two-tailed Student‘s t test). The same pattern was observed in a second independent trial (Supplemental Figure 6A).
(B) Loss of function of AGL16 (agl16-1 Col-FRI) mildly altered the expression of FLC in the Col-FRI background. Relative accumulation of FLC in the agl16-1 Col-FRI mutant was monitored by real-time RT-PCR (normalized to TUB2) along developmental stages from 8 to 11 d after germination on soil under LD conditions. Error bars indicate sd of three technical replicates. The experiment was also performed in the Col-0 background and gave the same pattern (Supplemental Figure 6B).
(C) BiFC assay shows that AGL16 can form a heterodimer with both FLC and SVP in N. benthamiana leaf epidermis. From left to right, top panel, (1) AGL16 is localized in the nucleus, (2) no signal, showing that AGL16 and LHP1 do not interact, (3) positive signal, showing that FLC and SVP interact, and (4) enlarged image of (3). From left to right, bottom panel, (1) positive signal showing that FLC and AGL16 interact, (2) enlargement of (1), (3) positive signal showing that SVP and AGL16 interact, and (4) enlargement of (3). Bars = 10 µm.
(D) Yeast two-hybrid assay confirms the direct interaction between AGL16 and SVP but not between AGL16 and FLC. Each protein was fused to either the activation domain (AD) as prey or DNA binding domain (BD) as bait. Serial dilutions (from 10−1 × to 10−4 ×) of J69-4A cells containing different construct combinations indicated on the right were grown on selective medium. The FLC/SVP combination and the protein/empty vector combinations provide positive and negative controls, respectively.
(E) and (F) Genetic analysis reveals that SVP is epistatic to AGL16. The rosette (filled bars) and cauline (open bars) leaves upon flowering (LD growing) are shown for svp-41, agl16-1 svp-41, agl16-1, and Col-0 wild type. Error bars indicate sd of means. Significance level was tested with the Wilcoxon test (***P < 0.001; **P < 0.01).
(G) and (H) Expression of either AGL16 (G) or SVP (H) is not significantly changed in the loss-of-function mutant svp-41 (G) or agl16-1 (H) compared with Col-0 wild type (normalized to PP2A). Error bars indicate sd of means for three biological replicates.
We therefore investigated whether the interaction between AGL16 and FLC may take place at the posttranscriptional level. To this end, we examined whether AGL16 could physically interact with FLC with the bimolecular fluorescence complementation (BiFC) technique (Hu and Kerppola, 2003). We fused the C-terminal half of yellow fluorescent protein (cYFP) with the C terminus of AGL16 (AGL16-cYFP) and the N-terminal half of YFP (nYFP) with the C terminus of FLC (nYFP-FLC). AGL16 was located in the nucleus (Figure 6C). As a negative control, the C terminus of LHP1 (a component of the polycomb complex known to be localized in the nucleus) (Zemach et al., 2006) was fused to nYFP (nYFP-LHP1). As a positive control (Fujiwara et al., 2008; Li et al., 2008), the C termini of SVP and FLC proteins were joined together with nYFP (SVP-nYFP) and cYFP (FLC-cYFP), respectively. Each construct was coinfiltrated into epidermal cells of Nicotiana benthamiana leaves for transient expression (Voinnet et al., 2003). The fluorescent signal of YFP, which depends on the molecular interaction of the fusion proteins, was observed in the nucleus of the cells coinfiltrated with 35S:AGL16-nYFP/35S:FLC-cYFP and 35S:FLC-cYFP/35S:SVP-nYFP but not with 35S:AGL16-nYFP/35S:LHP1-cYFP (Figure 6C). The results demonstrate that AGL16 can interact with FLC in the nucleus, thus supporting the idea that AGL16 might control flowering time as a partner of FLC in a repressive MADS factor complex. As FLC and SVP are known to form a repressive protein complex delaying flowering (Li et al., 2008), we also tested whether AGL16 could interact with SVP. Indeed, a positive YFP signal was detected when 35S:AGL16-nYFP/35S:SVP-cYFP were coinfiltrated (Figure 6C).
To further corroborate these findings, we performed a pairwise yeast two-hybrid assay to test whether AGL16 could directly interact with SVP and FLC (Figure 6D). Interestingly, we only detected a direct interaction between AGL16 and SVP, but not between AGL16 and FLC. This suggests that the interaction between AGL16 and FLC might need another partner. Furthermore, the double mutant agl16-1 svp-41 flowered at the same time as the single mutant svp-41 (Figures 6E and 6F) (Hartmann et al., 2000), and no significant change was observed for the expression of AGL16 in the svp-41 mutant (Figure 6G) or SVP in the agl16-1 mutant (Figure 6H). Therefore, our data show that AGL16 can physically interact with SVP, while SVP is epistatic to AGL16 in regulating flowering time. AGL16 is thus a potential partner of the repressor complexes formed by SVP/FLC to regulate the expression of FT to control flowering.
Natural Variation at AGL16 Associates with Flowering Time
To further investigate the biological relevance of AGL16 in regulating flowering time, we examined the sequence polymorphisms at AGL16 detected in the 250K single nucleotide polymorphism (SNP) array (Atwell et al., 2010). There are two SNPs in the AGL16 locus on chromosome 3: an A/G and A/G at positions 21,179,222 (2nd intron) and 21,180,413 (6th intron), respectively (Supplemental Data Set 3). These two SNPs allowed grouping accessions into four haplotypes: C24 (A,A), Col (A,G), SF-2 (G,A), and UK-4 (G,G).
Because AGL16 is not known to impact flowering time, it was not considered as an a priori candidate gene for flowering time. We reexamined existing data on flowering time variation. The first whole-genome association genetics study in Arabidopsis was based on a relatively limited sample of accessions (Atwell et al., 2010). Nevertheless, this study detected a significant association of the AGL16 locus with flowering time measured in the lab (rank number 63 in all SNPs) and field (rank number 21 in all SNPs). In the field experiment reported by Brachi et al. (2010), AGL16 was also listed as associating significantly with flowering time. We reexamined this association and introduced FRI and FLC alleles as cofactors. In this analysis, we found a significant interaction of AGL16, FRI, and FLC haplotypes on flowering time (P = 0.0014, GLM regression). Despite the small sample size of each haplotype combination, this finding suggests that alleles at AGL16, FRI, and FLC interact to control variation in flowering time in natural populations (Supplemental Figure 7 and Supplemental Table 3 and 4).
DISCUSSION
In this study, we report that a miRNA-regulated MADS box transcription factor represses flowering in Arabidopsis. With a combination of genetic and molecular approaches, we show that the miR824/AGL16 module is most active in a genetic background with high FLC levels and under a LD photoperiod.
We tested the effect of loss of function of AGL16 in two genetic backgrounds, Col-0 and Col-FRI. Col-0, with a nonfunctional fri allele, is only one of the representative genetic backgrounds to study flowering time because this trait is highly variable in Arabidopsis (Brachi et al., 2010; Fournier-Level et al., 2013). In this background, agl16-1 mutant plants flowered with 10% fewer leaves than the wild type, an effect that is difficult to detect when plants flower early. The Col-FRI background is not a native genotype but a combination of natural Arabidopsis alleles. The close relatives of Arabidopsis harbor a functional FRI copy (Kuittinen et al., 2008), and worldwide, some 60% of Arabidopsis genotypes feature a functional FRI allele (Johanson et al., 2000; Stinchcombe et al., 2004; Shindo et al., 2005; Korves et al., 2007; Brachi et al., 2010). Since the linkage disequilibrium between loci is rare in Arabidopsis (Kim et al., 2007), many natural genotypes may exhibit a combination of alleles similar to Col-FRI. This genetic background is thus also relevant to dissect the genetics of flowering time (Michaels and Amasino, 1999). In this background, agl16-1 mutant plants flower with 25 to 35% fewer leaves, and lines overexpressing miR824 flowered with ∼15% fewer leaves than the wild type. The miR824/AGL16 module therefore plays a significant role in the biology of flowering time in Arabidopsis. The magnitude of this role depends on the genetic background and the environmental conditions.
AGL16 belongs to the MADS box family of transcription factors, which are often implicated in controlling structural development and/or transitional timing (Theissen et al., 2000; Becker and Theissen, 2003). Under LD conditions, loss of function of AGL16 in Col-FRI backgrounds counteracted the delayed flowering caused by FRI introgression and the associated strong expression of the floral repressor FLC (Michaels and Amasino, 1999). In contrast to FLC, which delays flowering in both LD and SD conditions, the effect of AGL16 was more specific to the LD photoperiod. This might be due to the repressive action of AGL16 on FT, the major regulator of flowering under LD conditions (Kardailsky et al., 1999; Samach et al., 2000). Interestingly, AGL16 expression seemed to be decreased in plants grown in 24-h light and increased in dark-grown plants (https://www.genevestigator.com/gv/plant.jsp).
Our data show that AGL16 and FLC act additively, in a manner directly proportional to the allelic dosage at the two loci (Figure 3). Our study of protein interactions in yeast and tobacco reveals AGL16 may participate in the repressive complexes formed by SVP and/or FLC (Figure 6), thereby providing a molecular model for the genetic interaction between AGL16 and FLC. AGL16 can directly interact with SVP and potentially form heterodimers with FLC in tobacco leaves, but since the interaction is not detected in yeast, the two proteins might not be in direct contact. SVP could be one partner mediating the interaction between AGL16 and FLC as SVP can directly interact with FLC (Li et al., 2008). FLM could also mediate this interaction as FLM can interact physically with both FLC and SVP (Gu et al., 2013; Lee et al., 2013; Posé et al., 2013). FLC might indeed participate in a large protein complex with a molecular weight exceeding several hundreds of kilodaltons (Helliwell et al., 2006), much larger than if it contained only one FLC protein forming a dimer with SVP (Li et al., 2008) or some other members of the FLC clade (Gu et al., 2013). Therefore, we propose that the miR824/AGL16 module may act as a new trans-factor of FT to regulate the timing of floral transition via interaction with the protein complexes formed by AGL16 and other MADS box proteins.
This model will have to be validated in future studies. Indeed, it is also possible that the activity of AGL16 is regulated at the transcriptional level. AGL16 is one of the major targets bound by FLC (Deng et al., 2011) and a weak target for SVP (Gregis et al., 2013). In loss-of-function mutants for both FLC (flc-3) and SVP (svp-41), the expression of AGL16 was increased (Figure 6; Deng et al., 2011; Gregis et al., 2013). However, this increase was modest, probably due to the negative regulation of AGL16 mRNA abundance by miR824. Multiple negative feedbacks might therefore participate in the regulation of AGL16 activity.
The miR824/AGL16 module was initially reported to promote the development of higher-order stomata complexes, by increasing the number of additional divisions in meristemoid cell lineages (Kutter et al., 2007). The function of AGL16 that we describe here might in fact be the most important for plant fitness. Indeed, the effect of AGL16 is particularly pronounced in a background with high FLC activity, and flowering time has been shown to have a strong impact on the number of branches and plant lifetime fruit production (Fournier-Level et al., 2013). Instead, the effect of agl16-1 on meristemoid division and differentiation did not seem to translate into a macroscopic phenotype such as altered stomatal density. When AGL16 was strongly overexpressed, stomatal density was increased, but these plants also displayed severe growth defects, with leaves that were not fully expanded (Kutter et al., 2007). Nevertheless, it remains possible that AGL16 is involved in additional functions, especially since it is expressed in the root as well (Gan et al., 2010).
Like FLC (Sheldon et al., 2000) and SVP (Hartmann et al., 2000), which control flowering in a quantitative manner, AGL16 controls flowering depending on allelic dosage (Figure 3). The repressive action of AGL16 on flowering time can therefore be modulated by mutations altering the level of AGL16 mRNA, the amount of AGL16 protein, or perhaps the abundance of the putative AGL16-SVP/FLC repressor complexes. FLC, FRI, FLM, and SVP were all reported to control natural variation of flowering time (Johanson et al., 2000; Michaels et al., 2003; Werner et al., 2005; Mendez-Vigo et al., 2013). We reexamined the significant allelic association of AGL16 genotypes with flowering time variation reported in the field (Brachi et al., 2010). Closer inspection suggests that this effect requires the presence of a functional FRI allele and depends on allelic variation at the FLC locus. Therefore, allelic combinations of SVP, FLC, and miR824/AGL16 variants could contribute to natural variation of flowering time in Arabidopsis. AGL16 shows no amino acid polymorphism but two SNPs located in introns. These two SNPs do not have obvious consequences on mRNA maturation because they are not in the splicing elements, but an effect on regulation cannot be excluded. Future GWAS studies including larger sets of genotypes will be useful to validate this effect and investigate the role of AGL16 regulation in adaptive evolution.
METHODS
Plant Materials, Growth Conditions, and Flowering Time Scoring
Arabidopsis thaliana plants including wild-type Col-0, agl16-1, and m3 have been described (Kutter et al., 2007). The knockout mutant of FT (ft-10) was reported by Yoo et al. (2005). The flc-3 mutant in the Col-0 background with the Sf-2 FRI introgression was reported by Michaels and Amasino (1999). To test the effect of modifying AGL16 activity in the Col-FRI background, Col-FRI flc-3 was crossed to Col-0 agl16-1 and Col-0 m3, respectively. Homozygote double mutants at FRI, FLC, or AGL16 were screened by PCR using gene-specific primers (Supplemental Table 2). The F2 population used in Figure 2 was produced by crossing Col-0 agl16-1 with Col-FRI flc-3. The seeds from self-pollinated F1 plants were sown to soil. After all the individuals had bolted, a young leaf was harvested from each plant for rapid genomic DNA isolation using a solution containing 50 mM Tris-HCl (pH 7.2), 0.3 M NaCl, and 10% (w/v) sucrose as extraction buffer. The extract was not purified and directly used for genotyping (see Supplemental Table 2 for primer sequences).
The T1 lines of the miR824 target mimicry lines were described by Todesco et al. (2010). The progeny of seven lines with 3:1 segregation were treated with BASTA reagents to screen for the homozygote single insertion of the mimic. T3 seeds were used for scoring flowering phenotypes using the Col-0 line transformed with an empty binary vector as control (Todesco et al., 2010). To test the genetic relationship between AGL16 and FT, agl16-1 was crossed to the ft-10 mutant and the homozygote double mutant was obtained by screening the segregating F2 population using agl16-1– and ft-10–specific primers (Supplemental Table 2). For investigating the relationship between AGL16 and SVP, agl16-1 was crossed to the svp-41 mutant (Hartmann et al., 2000) with the resulting homozygote double mutant screened by gene-specific primers (Supplemental Table 2).
For promoter-miR824:GUS transgenic lines, the 4.7-kb fragment comprising the 3.093-kb sequences upstream of the TSS (+1) and 1.614-kb segment downstream of TSS was cloned into the Gateway pDONR207 vector (Invitrogen) and then fused with the GUS reporter gene in pGREEN-GW:GUS via Gateway technology (Invitrogen). In this vector, the selection marker BAR is driven by a NOS promoter (Adrian et al., 2010). After sequencing the inserted fragment to confirm the absence of mutations caused by PCR, independent transgenic lines were generated in Col-0 via floral dipping (Clough and Bent, 1998). The first rosette leaf of at least five independent 2-week-old T1 plants was used for GUS staining.
Arabidopsis seeds were stratified in distilled water at 4°C for 72 h and sown in soil and grown under LD (16-h light at 21°C and 8-h night at 18°C) or SD (8-h light at 21°C and 16-h night at 18°C) conditions either in glass houses or growth chambers. Pots and trays were randomized every 2 d to minimize the positional effect on flowering time. For time-course expression of FT, mutant seeds and their corresponding wild-type lines were sterilized with bleach and stratified at 4°C for 72 h. These seeds were then sown on Murashige and Skoog medium plates containing 1% sucrose and grown under LD conditions in growth chambers. For the assays of FT expression across developmental stages and tissues, seeds were sown in soil and grown under LD conditions.
Flowering time was scored for each plant by the RLN or total leaf number (including rosette and cauline leaves), when the first flower was visible. In a few trials, the number of days until the appearance of the first visible flower (days to flowering) was scored in addition to the RLN. Vernalization treatment was applied to LD-grown 10-d-old seedlings in a growth chamber with the temperature set at 4°C under LD conditions. All the experiments were repeated in at least two, and up to 10, independent trials, in glass houses and/or growth chambers. See Supplemental Data Set 1 for detailed information.
BiFC Assay
To reveal the cellular localization of AGL16, full-length cDNA without the stop codon of AGL16 was cloned via Gateway technology (Invitrogen) into pEarlyGate101 containing a 35S promoter and YFP-HA tags after the Gateway cassette (Earley et al., 2006). The resulting construct was used to transform the Col-0 agl16-1 mutant. The cellular localization of AGL16 was examined in T1 plants under a LSM 700 confocal laser scanning microscope (Carl Zeiss). For the BiFC assay, PCR fragments amplified with the specific primers for AGL16, FLC, and LHP1 (see Supplemental Table 2 for primer information) were subcloned into the pDONR221 entry vector (Invitrogen). LHP1 was used as a negative control. The resulting plasmids were inserted by Gateway cloning (LR reaction) into the split YFP vectors RfA-sYFPn-pBatTL-B and RfA-sYFPc-pBatTL-B. Agrobacterium tumefaciens transformant strains carrying plasmids for BiFC and p19 were grown overnight at 28°C in 10 mL YEP medium plus selective antibiotics, collected by centrifugation, and resuspended in infiltration medium (10 mM MgCl2, 150 µg/mL acetosyringone, and 10 mM MES-NaOH, pH 5.6) (Voinnet et al., 2003). Cells were kept at 28°C in the infiltration solution for 3 h in darkness and infiltrated into the abaxial surface of 3-week-old Nicotiana benthamiana plants. The fluorescence signal of YFP was observed and recorded using an LSM 700 confocal laser scanning microscope.
Yeast Two-Hybrid Assay
To test the ability of AGL16 to interact with FLC and SVP, full-length cDNAs excluding the stop codons for these genes were amplified from an Arabidopsis (Col-0) cDNA pool (see Supplemental Table 2 for primer sequences) and cloned into the pDONR201 or pDONR207 vectors to generate pENTRY plasmids. The bait constructs pDEST32-AGL16 and pDEST32-SVP were generated via the LR reaction between the entry plasmids and pDEST32 (Invitrogen), respectively. Similarly, the prey constructs pDEST22-FLC and pDEST22-SVP were produced by the LR reaction between entry plasmids and pDEST22 (Invitrogen), respectively. Bait plasmids and prey plasmids or the blank pDEST22 or pDEST32 were cotransformed into yeast strain J69-4A (James et al., 1996), respectively. Medium lacking SD-Leu-Trp-His was used for selection.
Comparison of Phenotypes
Statistical difference in flowering time in various lines was performed using both Student’s t test (two tailed with two-sample assuming unequal variance) with Bonferroni correction in Microsoft Excel and Wilcoxon rank sum test in R.
Genetic interactions between genotypes in the F2 population were analyzed with the general linear model (glm function) in R according to the following model: flowering time = genotype AGL16 × genotype FRI × genotype FLC + tray number + error, in which AGL16 and FLC genotypes were classified into 0 (knockout homozygote), 1 (heterozygote), or 2 (wild-type homozygote); FRI genotypes were classified into functional or nonfunctional as the functional FRI allele was dominant. Although Poisson distributions were well suited for count data, our data poorly fit to a Poisson distribution and showed clear signs of overdispersion. Therefore, the dispersion parameter of a quasi-Poisson distribution was fitted to the data in the glm function in R, following recommendation by Crawley (2005). Best-fit models were detected by removing nonsignificant interaction items stepwise from the most complex interactions to simpler ones. ANOVA analysis with the Chisq method was used to test whether reducing an interaction item would improve the fit of the model to the data. To test whether the segregation of AGL16, FRI, and FLC follows a Mendelian inheritance, a Fisher’s exact test was performed in R.
GUS Staining
For GUS staining, seedlings were incubated for 30 min in 90% (v/v) acetone on ice, rinsed with 50 mM sodium phosphate buffer, pH 7.0, and incubated overnight at 37°C in staining solution (0.5 mg/mL X-Gluc [5-bromo-4-chloro-3-indolyl-β-d-glucuronide], 50 mM sodium phosphate buffer, pH 7.0, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, and 0.1% [v/v] Triton X-100). After staining, samples were washed with 50 mM sodium phosphate buffer, pH 7.0, and cleared in 70% (v/v) ethanol. The GUS histochemical staining was visualized under a light stereomicroscope (MZ 16 FA; Leica). The first rosette leaves of at least five 2-week-old T1 transgenic plants were stained. A typical GUS pattern is shown in Figures 5B and 5C.
RNA Isolation and Real-Time Quantitative RT-PCR Assays
Total RNA was extracted with TRIzol reagent (Invitrogen). For time-course monitoring of gene expression, both whole seedlings and the aerial parts of the 9-d-old seedlings were used. Tissues were first harvested just before dawn and then collected every 4 h for 24 h. To minimize the duration of sampling at each time point, only one pool of 10 to 15 seedlings was constituted in each of two independent trials. For monitoring tissue-specific expression, the leaf was separated with a forceps from other parts (including the petioles, emerging young rosette leaves, and meristems as well as the hypocotyl without the root) of the 10-/12-d-old seedlings and collected separately. For each sampling, three pools (biological replicates) were made with the tissues of 10 to 15 seedlings in each of at least two independent trials, in LD conditions in glass houses or growth chambers. A similar protocol was followed for monitoring gene expression in leaves during development.
Around 1 µg of total RNA was used for cDNA synthesis after DNase-I treatment (Fermentas) using an oligo-d(T) primer and Superscript III reverse transcriptase (Invitrogen) following previously described protocols (Hu and Saedler, 2007). Quantitative real-time PCR was performed with iQ SYBR Green Supermix (Bio-Rad) on either a Mastercycler Realplex2 (Eppendorf) or a CFX384 Touch real-time PCR detection system (Bio-Rad). The relative expression of FT and AGL16 was normalized to the expression of PP2A and/or Tubulin2. The mRNA levels relative to PP2A mRNA levels are given in Figures 4A, 6G, and 6H. Primers used for each gene are listed in Supplemental Table 2.
Statistical Analysis of Natural Variation at the AGL16 Locus
The 250K SNP array contained two SNPs mapping in AGL16 at positions 2,779,222 (in the 2nd intron; A/G) and 2,780,413 (in the 6th intron; A/G), which defined four haplotypes. Sequencing a fragment containing the 2,780,413 SNPs in a collection of 48 accessions confirmed this SNP. To test whether natural variation at the AGL16 locus is associated with flowering time variation, we explored the flowering time data (scored as accumulated photothermal units at the time of flowering) of 171 accessions collected in a common garden experiment (Brachi et al., 2010). We included the genotypes of FRI and FLC in the analysis. Population structure was also quantified via principal component analysis with the first two principal coordinates (PC1 and PC2; which accounted for more than 50% of the total variation) into the following GLM regression model: flowering time = (AGL16 haplotype) × (FRI functionality) × (FLC haplo-groups) + PC1 + PC2. Main and interaction effects of each genotype on flowering time variation were analyzed as described above for the F2 population. A nonparametric Wilcoxon rank sum test was used to identify significant pairwise differences. The results of these analyses are shown in Supplemental Tables 3 and 4.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: miR824 (At4g24415), AGL16 (At3g57230), FLC (At5g10140), SVP (At2g22540), FT (At1g65480), FRI (At4g00650), PP2A (AT1G13320), LHP1 (AT5G17690), and TUB2 (AT5G62690).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. AGL16 Expression Was Repressed in m3.
Supplemental Figure 2. miR824 Target Mimicries Flowered Later under Long-Day Conditions (Supplementary to Figure 4).
Supplemental Figure 3. Promoter-miR824:GUS Staining Reveals the Expression of miR824 in Guard Cells.
Supplemental Figure 4. The Expression of AGL16 in the Wild Type and Mutants in Both Col-0 and Col-FRI Backgrounds.
Supplemental Figure 5. The Relative FT Expression in a Second Independent Trial Confirming the Pattern Shown in Figure 5.
Supplemental Figure 6. The Relative Expression in an Independent Trial Confirming the Expression Pattern for AGL16 Observed in Figure 6A in the flc-3 Mutant and for FLC in the Col-0 Background (Supplemental for Figure 6B).
Supplemental Figure 7. Polymorphisms at AGL16 Are Associated with Flowering Time Variation in Common Garden Conditions.
Supplemental Table 1. Statistical Significance for Pairwise Mean Flowering Time Differences among Lines Presented in Figure 3B.
Supplemental Table 2. Primers Used in This Study.
Supplemental Table 3. Statistical Test of AGL16 Allele Association with Flowering Time in Field Conditions (Data from Brachi et al. [2011]).
Supplemental Table 4. Wilcoxon Rank Sum Test of Pairwise Mean Flowering Comparison for Various Allelic Comparisons.
Supplemental Data Set 1. Summary of All Flowering Assays.
Supplemental Data Set 2. Statistical Analysis of the F2 Segregating Population Presented in Figure 3A.
Supplemental Data Set 3. AGL16 Genotypes for 171 Accessions Used for Reanalyzing the Brachi Flowering Time Data.
Supplementary Material
Acknowledgments
We thank Frederick Meins Jr. and Renhou Wang for providing seeds of some mutants and for reading and commenting on this article. We appreciate help from Maarten Koornneef and Gregor Schmitz. We are grateful for the technical help provided by Tingting Ning, Li Lei, and Ute Tartler. The work is supported by Germany DFG SFB-680, SPP1530, and the Max-Planck-Society.
AUTHOR CONTRIBUTIONS
J.-Y.H. and J.d.M. designed research. J.-Y.H., Y.Z., F.H., X.D., and L.-Y.L. performed the research. All authors analyzed data. J.-Y.H., F.T., G.C., and J.d.M. wrote the article.
Glossary
- Col-0
Columbia-0
- miRNA
microRNA
- LD
long-day
- SD
short-day
- RLN
rosette leaf number
- TSS
transcription start site
- BiFC
bimolecular fluorescence complementation
- SNP
single nucleotide polymorphism
References
- Achard P., Herr A., Baulcombe D.C., Harberd N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365 [DOI] [PubMed] [Google Scholar]
- Adrian J., Farrona S., Reimer J.J., Albani M.C., Coupland G., Turck F. (2010). cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla E.R., Liljegren S.J., Pelaz S., Gold S.E., Burgeff C., Ditta G.S., Vergara-Silva F., Yanofsky M.F. (2000). MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 24: 457–466 [DOI] [PubMed] [Google Scholar]
- Amasino R.M. (2005). Vernalization and flowering time. Curr. Opin. Biotechnol. 16: 154–158 [DOI] [PubMed] [Google Scholar]
- An H., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Andrés F., Coupland G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Atwell S., et al. (2010). Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausín I., Alonso-Blanco C., Martínez-Zapater J.M. (2005). Environmental regulation of flowering. Int. J. Dev. Biol. 49: 689–705 [DOI] [PubMed] [Google Scholar]
- Bäurle I., Dean C. (2006). The timing of developmental transitions in plants. Cell 125: 655–664 [DOI] [PubMed] [Google Scholar]
- Becker A., Theissen G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Brachi B., Faure N., Horton M., Flahauw E., Vazquez A., Nordborg M., Bergelson J., Cuguen J., Roux F. (2010). Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 6: e1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C., Pelaz S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.H., Coruh C., Axtell M.J. (2012). miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell 24: 4837–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Cigan A.M., Saeteurn K., Hake S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39: 544–549 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crawley, J.M. (2005). Statistics: An Introduction Using R. (Chichester, UK: Wiley). [Google Scholar]
- Cuperus J.T., Fahlgren N., Carrington J.C. (2011). Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meaux J., Hu J.Y., Tartler U., Goebel U. (2008). Structurally different alleles of the ath-MIR824 microRNA precursor are maintained at high frequency in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 8994–8999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Ying H., Helliwell C.A., Taylor J.M., Peacock W.J., Dennis E.S. (2011). FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 6680–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E.S., Peacock W.J. (2007). Epigenetic regulation of flowering. Curr. Opin. Plant Biol. 10: 520–527 [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Carrington J.C. (2007). High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier-Level A., et al. (2013). Paths to selection on life history loci in different natural environments across the native range of Arabidopsis thaliana. Mol. Ecol. 22: 3552–3566 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Fujiwara S., Oda A., Yoshida R., Niinuma K., Miyata K., Tomozoe Y., Tajima T., Nakagawa M., Hayashi K., Coupland G., Mizoguchi T. (2008). Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell 20: 2960–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y., Zhou Z., An L., Bao S., Liu Q., Srinivasan M., Goddard P. (2010). The effects of fluctuations in the nutrient supply on the expression of ANR1 and 11 other MADS box genes in shoots and roots of Arabidopsis thaliana. Botany 88: 1023–1031 [Google Scholar]
- Gregis V., et al. (2013). Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol. 14: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Le C., Wang Y., Li Z., Jiang D., Wang Y., He Y. (2013). Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 4: 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U., Höhmann S., Nettesheim K., Wisman E., Saedler H., Huijser P. (2000). Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 21: 351–360 [DOI] [PubMed] [Google Scholar]
- Helliwell C.A., Wood C.C., Robertson M., James Peacock W., Dennis E.S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46: 183–192 [DOI] [PubMed] [Google Scholar]
- Hu C.D., Kerppola T.K. (2003). Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21: 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.-Y., Saedler H. (2007). Evolution of the inflated calyx syndrome in Solanaceae. Mol. Biol. Evol. 24: 2443–2453 [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., West J., Lister C., Michaels S., Amasino R., Dean C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Jung J.H., Seo Y.H., Seo P.J., Reyes J.L., Yun J., Chua N.H., Park C.M. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. (1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kim J.J., Lee J.H., Kim W., Jung H.S., Huijser P., Ahn J.H. (2012). The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 159: 461–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Plagnol V., Hu T.T., Toomajian C., Clark R.M., Ossowski S., Ecker J.R., Weigel D., Nordborg M. (2007). Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 39: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Weigel D. (2007). Move on up, it’s time for change—mobile signals controlling photoperiod-dependent flowering. Genes Dev. 21: 2371–2384 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Korves T.M., Schmid K.J., Caicedo A.L., Mays C., Stinchcombe J.R., Purugganan M.D., Schmitt J. (2007). Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. Am. Nat. 169: E141–E157 [DOI] [PubMed] [Google Scholar]
- Kuittinen H., Niittyvuopio A., Rinne P., Savolainen O. (2008). Natural variation in Arabidopsis lyrata vernalization requirement conferred by a FRIGIDA indel polymorphism. Mol. Biol. Evol. 25: 319–329 [DOI] [PubMed] [Google Scholar]
- Kutter C., Schöb H., Stadler M., Meins F., Jr and Si-Ammour A. (2007). MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 19: 2417–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter N., Kampani A., Carlson S., Goebel M., Moose S.P. (2005). microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 102: 9412–9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Amasino R.M. (1995). Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 108: 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Ryu H.-S., Chung K.S., Posé D., Kim S., Schmid M., Ahn J.H. (2013). Regulation of temperature-responsive flowering by MADS-box transcription factor repressor complexes. Science 342: 628–632. [DOI] [PubMed] [Google Scholar]
- Li D., Liu C., Shen L., Wu Y., Chen H., Robertson M., Helliwell C.A., Ito T., Meyerowitz E., Yu H. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Yant L.J., Mürdter F., Küttner F., Schmid M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Vigo, B., Martinez-Zapater, J.M., and Alonso-Blanco, C. (2013). The flowering repressor SVP underlies a novel Arabidopsis thaliana QTL interacting with the genetic background. PLoS Genet. 9: e1003289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., He Y., Scortecci K.C., Amasino R.M. (2003). Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S.K., et al. (2010). Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc. Natl. Acad. Sci. USA 107: 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posé D., Verhage L., Ott F., Yant L., Mathieu J., Angenent G.C., Immink R.G.H., Schmid M. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503: 414–417 [DOI] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I., Weigel D. (2011). MicroRNA networks and developmental plasticity in plants. Trends Plant Sci. 16: 258–264 [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schmid M., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012 [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., Amasino R.A., Coupland G. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C.C., Rouse D.T., Finnegan E.J., Peacock W.J., Dennis E.S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA 97: 3753–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C., Aranzana M.J., Lister C., Baxter C., Nicholls C., Nordborg M., Dean C. (2005). Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138: 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G.G., Dean C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Stinchcombe J.R., Weinig C., Ungerer M., Olsen K.M., Mays C., Halldorsdottir S.S., Purugganan M.D., Schmitt J. (2004). A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc. Natl. Acad. Sci. USA 101: 4712–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S., Goto K. (2003). Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Becker A., Di Rosa A., Kanno A., Kim J.T., Münster T., Winter K.-U., Saedler H. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42: 115–149 [PubMed] [Google Scholar]
- Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009). Fly antiviral RNA silencing and miRNA biogenesis claim ARS2. Cell Host Microbe 6: 99–101 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Werner J.D., Borevitz J.O., Warthmann N., Trainer G.T., Ecker J.R., Chory J., Weigel D. (2005). Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc. Natl. Acad. Sci. USA 102: 2460–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.K., Chung K.S., Kim J., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139: 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Galvão V.C., Zhang Y.-C., Horrer D., Zhang T.-Q., Hao Y.-H., Feng Y.-Q., Wang S., Schmid M., Wang J.-W. (2012). Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24: 3320–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Li Y., Ben-Meir H., Oliva M., Mosquna A., Kiss V., Avivi Y., Ohad N., Grafi G. (2006). Different domains control the localization and mobility of LIKE HETEROCHROMATIN PROTEIN1 in Arabidopsis nuclei. Plant Cell 18: 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q.H., Upadhyaya N.M., Gubler F., Helliwell C.A. (2009). Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol. 9: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.