An Arabidopsis C2H2-type zinc finger protein increases both biotic and abiotic stress resistances through gene activation, including salicylic acid-related genes.
Abstract
The cysteine2/histidine2-type zinc finger proteins are a large family of transcription regulators, and some of them play essential roles in plant responses to biotic and abiotic stress. In this study, we found that expression of C2H2-type ZINC FINGER OF ARABIDOPSIS THALIANA6 (AtZAT6) was transcriptionally induced by salt, dehydration, cold stress treatments, and pathogen infection, and AtZAT6 was predominantly located in the nucleus. AtZAT6-overexpressing plants exhibited improved resistance to pathogen infection, salt, drought, and freezing stresses, while AtZAT6 knockdown plants showed decreased stress resistance. AtZAT6 positively modulates expression levels of stress-related genes by directly binding to the TACAAT motifs in the promoter region of pathogen-related genes (ENHANCED DISEASE SUSCEPTIBILITY1, PHYTOALEXIN DEFICIENT4, PATHOGENESIS-RELATED GENE1 [PR1], PR2, and PR5) and abiotic stress-responsive genes (C-REPEAT-BINDING FACTOR1 [CBF1], CBF2, and CBF3). Moreover, overexpression of AtZAT6 exhibited pleiotrophic phenotypes with curly leaves and small-sized plant at vegetative stage and reduced size of floral organs and siliques at the reproductive stage. Modulation of AtZAT6 also positively regulates the accumulation of salicylic acid and reactive oxygen species (hydrogen peroxide and superoxide radical). Taken together, our findings indicated that AtZAT6 plays important roles in plant development and positively modulates biotic and abiotic stress resistance by activating the expression levels of salicylic acid-related genes and CBF genes.
In nature, plants live in complex environmental conditions in which various abiotic stresses and multiple microbial pathogens with different infection strategies and lifestyles influence plant growth and development (Bent and Mackey, 2007; Shi et al., 2013a, 2013b). As sessile organisms, plants cannot avoid unfavorable circumstances by adjusting their location. Therefore, they have evolved complex strategies to perceive stress signals and further translate the perception into effective plant responses (Gimenez-Ibanez and Solano, 2013; Shi et al., 2014a, 2014b).
In recent years, much attention has been paid to the roles of the hormones in biotic and abiotic stress responses and especially hormone interactions under stress conditions (Pieterse et al., 2012; Yang et al., 2012a; Liu et al., 2013c). Several plant hormone receptors are located in the nucleus, such as jasmonate and auxin, while the signaling perception of salicylic acid (SA), ethylene, and abscisic acid initiates in the cytoplasm and then translocates to the nucleus. Plant transcription factors (TFs) serve as important mediators during hormone cross talks under biotic and abiotic stress conditions (Kazan and Manners, 2009; Kieffer et al., 2010; Liu et al., 2013c). Currently, various TFs have been shown to be involved in biotic and abiotic stress responses via activating stress-responsive gene expression, such as C-repeat-binding factors (CBFs)/dehydration-responsive element-binding proteins (DREBs), WRKYs, ethylene-responsive element-binding factors, MYBs, MYCs, basic domain-Leu zipper families, and zinc finger proteins (ZFPs; Qiu et al., 2008; Yu et al., 2008; Bi et al., 2010; Li et al., 2010a, 2010b; Seo and Park, 2010; Zhang et al., 2010a, 2010b; Zhu et al., 2010; Cheng et al., 2011).
ZFPs are a large family of transcription regulators in plants for modulation of downstream stress-responsive genes (Ciftci-Yilmaz et al., 2007; Devaiah et al., 2007; Kodaira et al., 2011; Liu et al., 2013d). ZFPs have been classified into at least nine types based on the number and location of characteristic residues (Cys and His), including C2H2, C2HC, C2HC5, C3H, C3HC4, C4, C4HC3, C6, and C8 (Nguyen et al., 2012; Bogamuwa and Jang, 2013; Zhou et al., 2013). In Arabidopsis (Arabidopsis thaliana), the involvement of C2H2-type ZFPs, including Arabidopsis Zinc-Finger Protein1 (AZF1), AZF2, AZF3, Drought induced19 (Di19), Zinc Finger of Arabidopsis6 (ZAT6), ZAT7, ZAT10, and ZAT12, in plant abiotic stress response has been revealed (Liu et al., 2013b, 2013d). Kodaira et al. (2011) found that AtAZF1 and AtZAF2 negatively regulated abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Constitutive expression of AtZAT12 conferred improved resistance to high light and osmotic and oxidative stresses, whereas knockout plants of AtZAT12 exhibited increased sensitivity to osmotic and salt stresses (Rizhsky et al., 2004; Davletova et al., 2005). AtZAT10 played a dual role in response to abiotic stress because both gain- and loss-of-function mutations of AtZAT10 displayed enhanced resistance to drought and osmotic and salt stresses (Mittler et al., 2006). Recently, AtZAT10 was identified as a substrate of mitogen-activated protein kinase (Nguyen et al., 2012). Ciftci-Yilmaz et al. (2007) revealed that AtZAT7 positively mediated salt stress tolerance via modulating expression of several defense-responsive genes and that the ethylene-responsive element-binding factor-associated amphiphilic repression motif is required for abiotic stress response. AtZAT6 is a repressor of primary root growth that modulates phosphate homeostasis through the control of root architecture (Devaiah et al., 2007). Moreover, AtZAT6 was identified as a novel target of MITOGEN-ACTIVATED PROTEIN KINASE6 (MPK6), and phosphorylation of AtZAT6 is essential for its positive regulation of seed germination under salt and osmotic stresses (Liu et al., 2013d). Overexpressing chimeric repressors derived AtZAT6 and also conferred enhanced salt tolerance (Mito et al., 2011). However, the molecular mechanisms of ZAT6 in plant response to abiotic stress remain unclear, and the possible role of ZAT6 in plant-pathogen interaction remains unknown.
In this study, we found that AtZAT6 positively regulated biotic and abiotic stress resistances, and overexpression of AtZAT6 resulted in distinct developmental phenotypes. Additionally, the regulatory mechanism underlying AtZAT6-mediated stress responses was also characterized. The results indicated that AtZAT6 plays important roles in stress responses through activating the expression levels of SA-related genes and CBF genes.
RESULTS
Expression Pattern of AtZAT6 under Stress Treatments
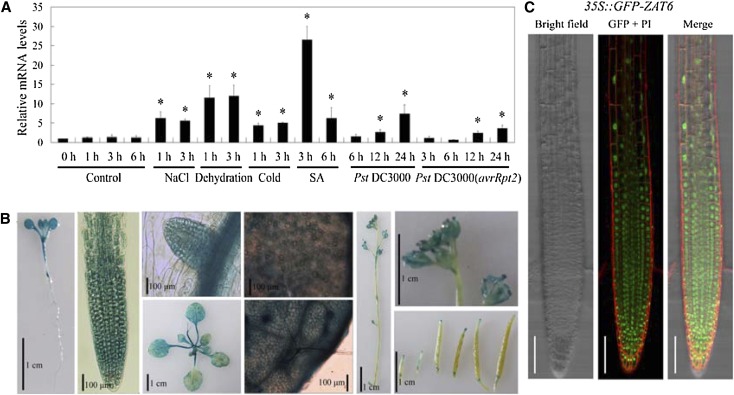
Based on publicly available gene expression data analysis (Winter et al., 2007), we found that AtZAT6 was highly induced by most abiotic and biotic stress treatments (Supplemental Fig. S1). The expression pattern of AtZAT6 was further examined through real-time PCR. Consistent with microarray data, infections with Pseudomonas syringae pv tomato (Pst) DC3000 and Pst DC3000(harboring the avirulence gene of Rpt2 [avrRpt2]) significantly induced the transcript level of AtZAT6 in a time-dependent manner (Fig. 1A). In addition, expression level of AtZAT6 was also transcriptionally induced after abiotic stress treatments, including salt, dehydration, and cold stresses (Fig. 1A). Interestingly, the transcript level of AtZAT6 was largely induced by SA treatment (Fig. 1A). These results indicated that AtZAT6 might play some roles in plant responses to biotic stress and abiotic stress.
Figure 1.
The expression pattern of AtZAT6. A, The expression levels of AtZAT6 after stress treatments. For real-time PCR, 28-d-old Col-0 plants were treated by 300 mm NaCl, dehydration, 4°C, 100 μm SA, Pst DC3000, and Pst DC3000(avrRpt2) for designed times. The expression level of AtZAT6 at 0 h of treatment was normalized as 1.0. The results shown are the means ± sds of at least three independent experiments. Asterisks indicate the significant difference of P < 0.05 compared with the wild type under control condition for 0 h. B, GUS staining of ProZAT6::GUS transgenic plants in different organs. C, Subcellular localization of AtZAT6. GFP signals were detected in 5-d-old seedling roots of 35S::GFP-ZAT6 transgenic plants. The red fluorescence indicates cell wall by propidium iodide staining, and the green fluorescence indicates the localization of AtZAT6. Bars = 100 µm. [See online article for color version of this figure.]
Using ProZAT6::GUS transgenic plants, AtZAT6 was found to be strongly expressed in cotyledons, leaves, and roots, but weakly in flowers, siliques, and stems (Fig. 1B). In the transgenic 35S::GFP-ZAT6 plants, GFP signals were detected in 5-d-old seedling roots. As shown in Figure 1C, 35S::GFP-ZAT6 fluorescence was predominantly localized in the nucleus and partially in the cytosol. This result indicated that AtZAT6 is a nuclear protein, which is consistent with the results of Devaiah et al. (2007). Additionally, yeast (Saccharomyces cerevisiae) transformation assay showed that AtZAT6 had transactivation activity in yeast (Supplemental Fig. S2).
AtZAT6 Regulates Plant Growth and Development
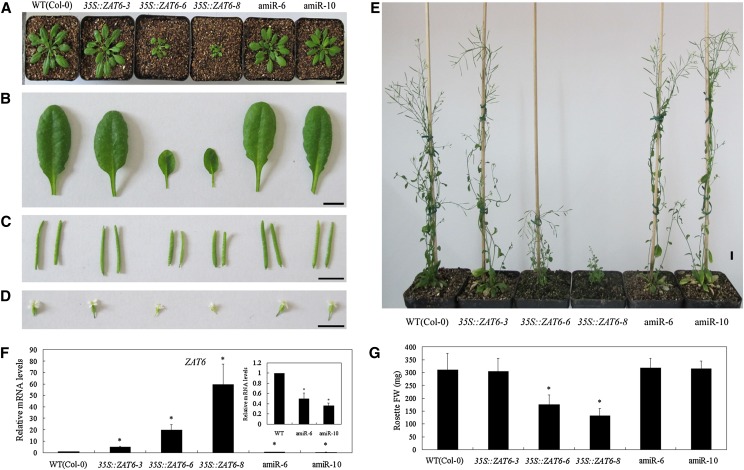
To reveal the in vivo role of AtZAT6, we constructed AtZAT6-overexpressing and knockdown transformations in the background of Columbia (Col-0) Arabidopsis plants. After selection by kanamycin and Basta resistance, the expression levels of AtZAT6 in the T1 generation of 35S::ZAT6 and artificial microRNA (amiR)-ZAT6 transgenic plants were verified by real-time PCR (Supplemental Figs. S3 and S4). The T1 generation of AtZAT6 transgenic plants exhibited pleiotropic developmental phenotypes, including retarded growth with small and curly leaves, and several lines with relatively high transgene expression (lines 5, 10, and 11 with 243.42-, 504.04-, and 337.85-fold relative to the wild type [Col-0], respectively) could not flower and set seeds (Supplemental Fig. S3). Consistently, T2 and T3 generations of AtZAT6 transgenic plants (lines 6 and 8) with relatively low transgene expression showed similar growth to those of the T1 generation, further confirming the pleiotropic developmental phenotypes of AtZAT6-overexpressing plants (Fig. 2A). It has been reported that RNA interference suppression of AtZAT6 appears to be lethal (Devaiah et al., 2007). In our study, we also found that AtZAT6 knockdown lines with relatively lower AtZAT6 transcripts could not flower and set seeds. Therefore, two AtZAT6 knockdown lines (lines 6 and 10) with about 40% expression level of the wild type were used for further study (Fig. 2A).
Figure 2.
Developmental phenotypes of AtZAT6-overexpressing and knockdown plants. A, Twenty-eight-day-old plants in soil. B to D, Size of leaves (B), siliques (C), and floral organs (D) in wild-type (WT) and AtZAT6-overexpressing and knockdown plants. Bars = 1 cm. E, Seventy-day-old wild-type and AtZAT6-overexpressing and knockdown plants. F, AtZAT6 expression levels in wild-type and AtZAT6-overexpressing and knockdown plants. The expression level of AtZAT6 in the wild type was normalized as 1.0 (n = 3). G, Fresh weight per plant of 28-d-old wild-type and AtZAT6-overexpressing and knockdown plants. Means ± sds (n = 15) are shown, and asterisks indicate the significant difference of P < 0.05 compared with the wild type. [See online article for color version of this figure.]
When grown on Murashige and Skoog (MS) medium, AtZAT6-overexpressing plants showed significantly shorter primary roots and displayed abnormal cotyledons (Supplemental Fig. S5, A–C). At the vegetative stage on soil, AtZAT6-overexpressing plants (lines 6 and 8) exhibited retarded growth with bleached leaves, but line 3 with the lowest (6.4-fold) AtZAT6 overexpression level and AtZAT6 knockdown plants showed no significant difference with the wild type (Fig. 2, A, B, F, and G). At the productive stage, AtZAT6-overexpressing plants (lines 6 and 8) exhibited significantly retarded growth, with a greater rosette leaf number, lower plant height, lesser cauline branch number, smaller leaf length and width, and reduced size of siliques and floral organs, including sepals, petals, stamens, and carpels (Fig. 2, B–E; Supplemental Table S1). However, AtZAT6 knockdown plants (lines 6 and 10) displayed no significant difference with wild-type plants for the whole growth stage (Fig. 2, B–E; Supplemental Table S1). All of these results provide genetic evidence that AtZAT6 is involved in plant growth and development.
AtZAT6 Positively Regulates Defense Resistance to Bacterial Pathogen Infection
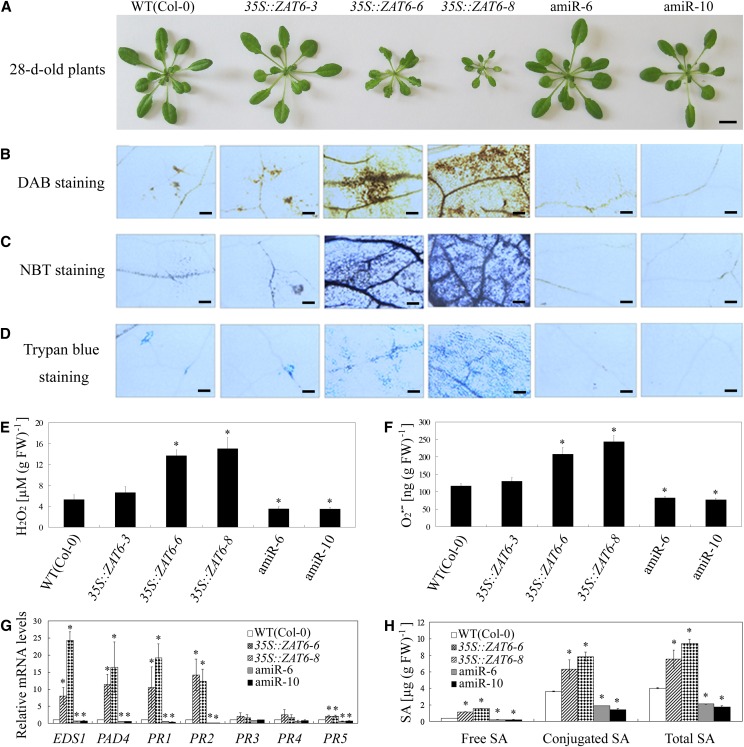
As shown in Figure 3A, AtZAT6-overexpressing plants (lines 6 and 8) exhibited dwarf morphology, whereas overexpressing line 3 and AtZAT6 knockdown plants (lines 6 and 10) exhibited wild-type-like morphology. Diaminobenzidine (DAB) staining and nitro blue tetrazolium (NBT) staining showed that the AtZAT6-overexpressing plants accumulated higher levels of hydrogen peroxide (H2O2) and superoxide radical (O2•–) than those in the wild type, while AtZAT6 knockdown plants accumulated lower H2O2 and O2•– contents (Fig. 3, B and C). Quantification of H2O2 and O2•– contents in leaves of 28-d-old soil-grown AtZAT6-overexpressing and knockdown plants also confirmed the results of DAB and NBT staining (Fig. 3, E and F). Trypan blue staining showed that there was extensive cell death in the AtZAT6-overexpressing plants, with little in the AtZAT6 knockdown plants (Fig. 3D). The glutathione antioxidant pool and redox state were also modulated by AtZAT6 (Supplemental Fig. S6, A–C). AtZAT6-overexpressing plants accumulated lower reduced glutathione (GSH) and higher oxidized glutathione (GSSG), while AtZAT6 knockdown plants displayed increased GSH and decreased GSSG (Supplemental Fig. S6, A–C). In addition, some pathogen-related marker genes, including ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1), PHYTOALEXIN DEFICIENT4 (PAD4), PATHOGENESIS-RELATED GENE1 (PR1), PR2, and PR5, were constitutively expressed in the AtZAT6-overexpressing plants but exhibited lower mRNA levels in the AtZAT6 knockdown plants (Fig. 3G). Moreover, free and conjugated SA contents were also much higher in the overexpressing transgenic plants but were lower in the AtZAT6 knockdown plants than those of wild-type plants (Fig. 3H). These data indicated that cell death and defense responses were constitutively activated in AtZAT6-overexpressing plants but were repressed in AtZAT6 knockdown plants.
Figure 3.
Modulation of AtZAT6 expression affects defense responses. A, Morphology of 28-d-old soil-grown wild-type (WT) and AtZAT6-overexpressing and knockdown plants. Bars = 1 cm. B to D, DAB staining (B), NBT staining (C), and trypan blue staining (D) of 28-d-old soil-grown wild-type and AtZAT6-overexpressing and knockdown plants. Bars = 100 µm. E and F, Quantification of H2O2 content (E) and O2•– content (F). G, Pathogen-related genes’ expression in wild-type and AtZAT6-overexpressing and knockdown plants. H, The accumulation of free SA, conjugated SA, and total SA assayed by HPLC. The results shown are the means ± sds of at least three independent experiments. Asterisks indicate the significant difference of P < 0.05 compared with the wild type. [See online article for color version of this figure.]
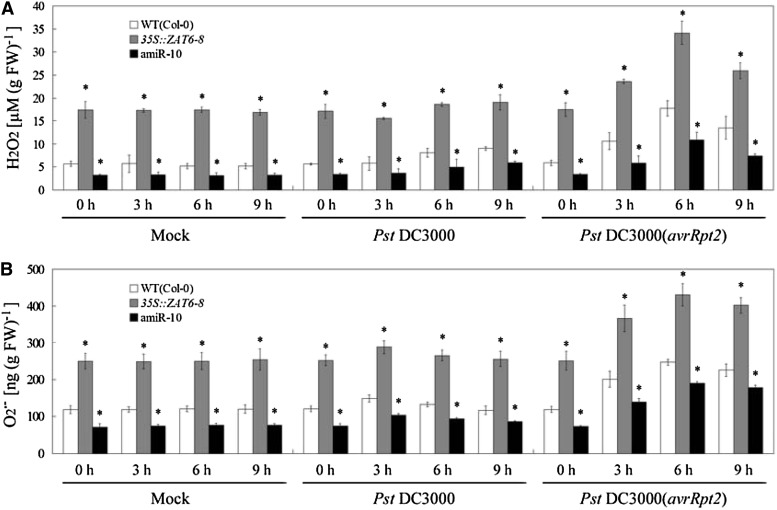
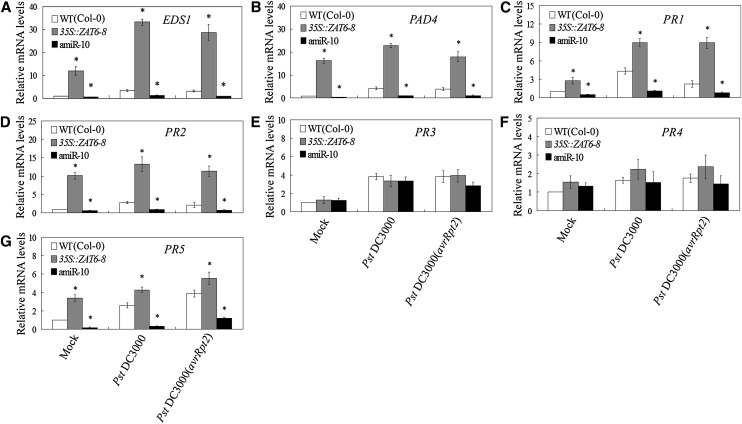
As reviewed by Lamb and Dixon (1997), oxidative burst and associated changes in defense gene expression following bacterial recognition occurs about 3 to 10 h after inoculation. Then, we examined the effects of AtZAT6 expression on oxidative burst and associated changes in defense gene expression at early periods after inoculations by mock, Pst DC3000, and Pst DC3000(avrRpt2). During the period between 0 and 9 h post inoculation (hpi) of Pst DC3000(avrRpt2), oxidative burst, including the accumulations of H2O2 and O2•–, was significantly displayed in wild-type plants (Fig. 4, A and B). Compared with wild-type plants, AtZAT6-overexpressing plants exhibited higher levels of H2O2 and O2•– at 3, 6, and 9 hpi of Pst DC3000(avrRpt2), while AtZAT6 knockdown plants displayed relatively lower levels (Fig. 4, A and B). After inoculation of Pst DC3000, the H2O2 content was only slightly induced at 6 and 9 hpi in wild-type plants, while the O2•– content was not significantly regulated (Fig. 4, A and B). During the period between 0 and 9 hpi of Pst DC3000, AtZAT6-overexpressing plants exhibited higher levels of H2O2 and O2•– than those in wild-type plants, while AtZAT6 knockdown plants displayed relatively lower levels (Fig. 4, A and B). After pathogen infection for 6 h, the expression levels of EDS1, PAD4, PR1, PR2, PR3, PR4, and PR5 were transcriptionally induced compared with those of mock treatment; however, AtZAT6-overexpressing plants exhibited higher transcripts of EDS1, PAD4, PR1, PR2, and PR5 than those in the wild type, while AtZAT6 knockdown plants displayed relatively lower levels (Fig. 5, A–G). These results indicated that AtZAT6 positively modulated reactive oxygen species (ROS) level and SA-related gene expressions under both control (mock) and pathogen-infected conditions.
Figure 4.
Modulation of AtZAT6 expression affects oxidative burst under mock and pathogen-infected conditions. A and B, Quantification of H2O2 content (A) and O2•– content (B) in plants under mock and pathogen-infected conditions. For the assays, 28-d-old plant leaves were infected with mock (10 mm MgCl2), Pst DC3000 plus 10 mm MgCl2, and Pst DC3000(avrRpt2) plus 10 mm MgCl2 for designed times. Means ± sds of three independent experiments are shown in the results, and asterisks indicate the significant difference of P < 0.05 compared with the wild type (WT).
Figure 5.
Modulation of AtZAT6 expression affects defense gene expression under mock and pathogen-infected conditions. A to G, Expression analysis of pathogen-related genes in wild-type (WT) and AtZAT6-overexpressing and knockdown plants. The expression levels of EDS1 (A), PAD4 (B), PR1 (C), PR2 (D), PR3 (E), PR4 (F), and PR5 (G) in plants after mock and pathogen infection for 6 h. The expression level of each gene in wild-type plants by mock infection was normalized as 1.0. For the assays, 28-d-old plant leaves were infected with mock (10 mm MgCl2), Pst DC3000 plus 10 mm MgCl2, and Pst DC3000(avrRpt2) plus 10 mm MgCl2 for 6 h. Means ± sds of three independent experiments are shown in the results, and asterisks indicate the significant difference of P < 0.05 compared with the wild type.
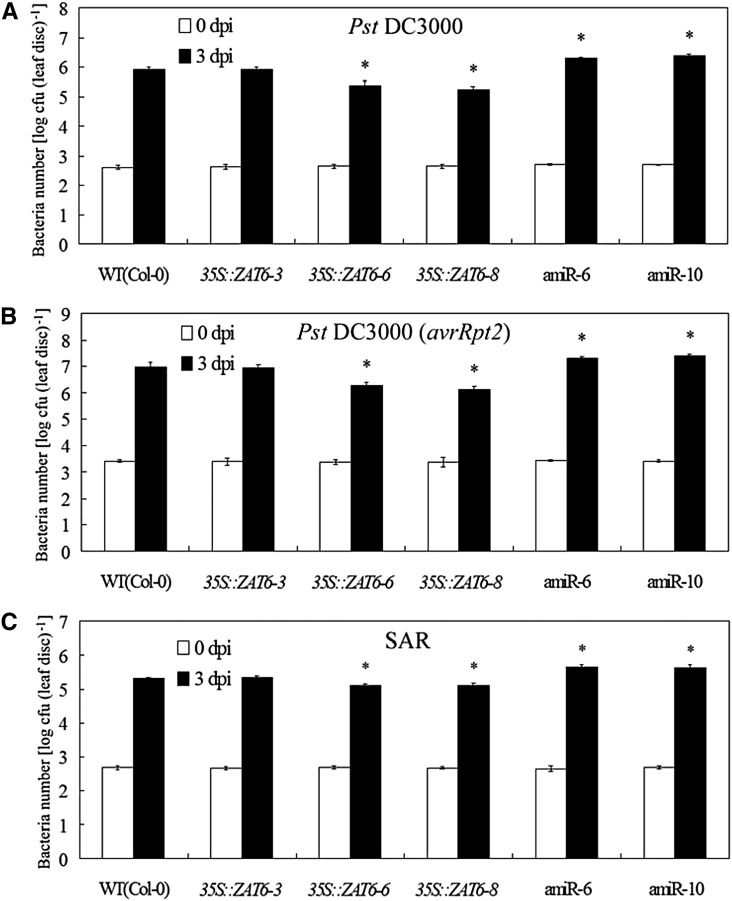
To test whether AtZAT6 regulates disease resistance against pathogenic bacteria, we examined the basal resistance and systemic acquired resistance (SAR) to Pst DC3000 and Pst DC3000(avrRpt2) in the AtZAT6-overexpressing and knockdown plants. Quantification of bacteria number in the pathogen-infected Arabidopsis leaves showed that the AtZAT6-overexpressing plants exhibited significantly less bacterial propagation than the wild type at 3 d post infection (dpi) of these pathogens, while the AtZAT6 knockdown plants exhibited significantly more bacterial propagation than the wild type (Fig. 6, A–C). Therefore, these results indicated that AtZAT6 positively regulated basal resistance and SAR against pathogenic bacteria.
Figure 6.
Modulation of AtZAT6 expression affects disease resistance against bacterial infection. A and B, Growth of Pst DC3000 (A) and Pst DC3000(avrRpt2; B) on plants at 0 and 3 dpi of bacterial infection. C, SAR assay of growth of Pst DC3000 on plants at 0 and 3 dpi of bacterial infection. The results shown are the average means ± sds of four independent experiments, and 20 independent leaf discs were harvested in each independent experiment. Asterisks indicate the significant difference of P < 0.05 compared with the wild type (WT).
AtZAT6 Positively Modulates Abiotic Stress Resistance
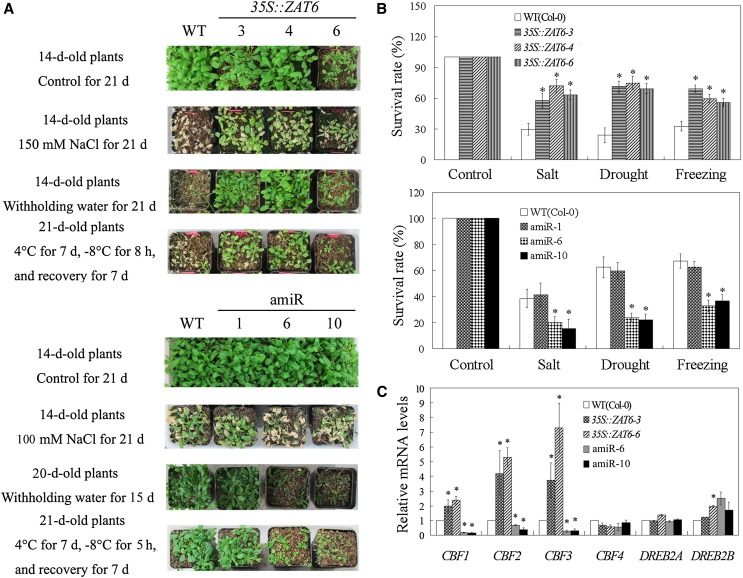
Because the expression of AtZAT6 was transcriptionally induced by multiple abiotic stresses, including salt, dehydration, and cold (Fig. 1A; Supplemental Fig. S1), the involvement of AtZAT6 in abiotic stress resistance was also dissected using AtZAT6-overexpressing and knockdown plants. After salt, drought, and chilling stress treatments, AtZAT6-overexpressing plants showed better growth with higher survival rate than the wild type and conferred improved resistance to abiotic stresses, while AtZAT6 knockdown plants exhibited severely inhibited growth with lower survival rate than the wild type (Fig. 7, A and B). Additionally, the expression levels of CBF1, CBF2, and CBF3 were constitutively expressed in the AtZAT6-overexpressing plants but exhibited lower mRNA levels in the AtZAT6 knockdown plants (Fig. 7C). However, modulation of AtZAT6 had no significant effect on the expression of CBF4, DREB2A, and DREB2B (Fig. 7C). These results indicated that AtZAT6 positively modulated abiotic stress resistance and the expressions of CBF genes (CBF1, CBF2, and CBF3).
Figure 7.
Modulation of AtZAT6 expression affects abiotic stress resistance. A, Phenotype of plants after abiotic stress treatments for designed days. B, Survival rate of the wild type (WT) and different AtZAT6 lines at 7 d after recovery from abiotic stress treatments. C, The expression of CBF genes in wild-type and AtZAT6-overexpressing and knockdown plants. Means ± sds of at least three independent experiments are shown in the results, and asterisks indicate the significant difference of P < 0.05 compared with the wild type. [See online article for color version of this figure.]
AtZAT6 Can Bind to the TACAAT Motif of Several Pathogen-Related Genes and CBF Genes
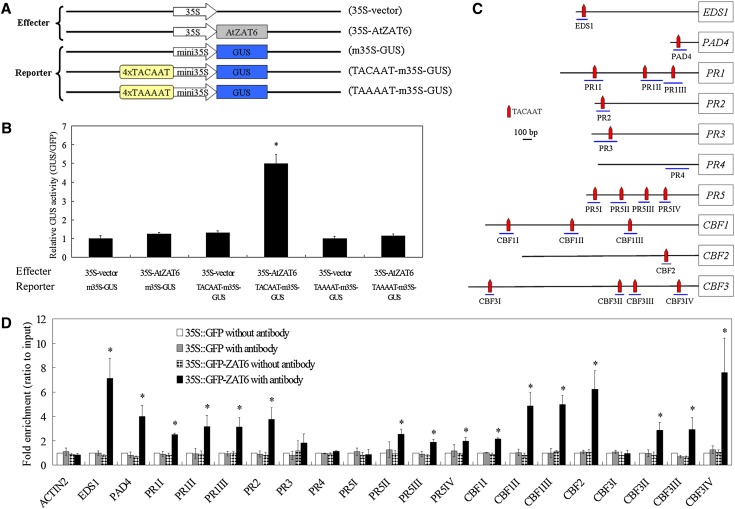
To reveal how ATZAT6 modulated the expressions of the pathogen-related genes and CBF genes, we analyzed the promoter sequences of these genes, and the results showed that TACAAT motifs were enriched (Fig. 8C). Liu et al. (2013b) found that AtDi19 (one C2H2 ZFP) could modulate the expression levels of PR1, PR2, and PR5 through binding to the TACAAT elements within the PR1, PR2, and PR5 promoters; thus, whether AtZAT6 could interact with the TACAAT element was investigated in this study. Then, transient expression assays in tobacco (Nicotiana tabacum) leaves were performed using 35S-vector and 35S-AtZAT6 as the effectors and m35S-GUS, TACAAT-m35S-GUS, and TAAAAT-m35S-GUS as the reporters (Fig. 8A). As shown in Figure 8B, only the leaf pieces cotransformed with 35S-AtZAT6, and TACAAT-m35S-GUS largely activated the expression of GUS, indicating the in vivo interaction of AtZAT6 and the TACAAT motif. To further investigate the interaction of AtZAT6 and the promoters of several pathogen-related genes, chromatin immunoprecipitation (ChIP)-PCR was performed using 35S::GFP-ZAT6-22 transgenic plants, which had the similar phenotype with AtZAT6-overexpressing plants (Supplemental Fig. S7, A–D). The results showed that the AtZAT6 protein strongly interacted with several fragments in the promoters of EDS1, PAD4, PR1, PR2, PR5, CBF1, CBF2, and CBF3 that contained the TACAAT motif but could not interact with the fragments in the promoters of ACTIN2 and PR4 (Fig. 8, C and D), indicating that the TACAAT motif is important for the binding of AtZAT6 to regulate the expressions of pathogen-related genes and CBF genes.
Figure 8.
AtZAT6 binds to the promoters of pathogen-related genes and CBF genes. A, Schematic diagrams of the effectors (35S-vector and 35S-AtZAT6) and reporters (m35S-GUS, TACAAT-m35S-GUS, and TAAAAT-m35S-GUS) used for transient expression analysis. B, Transient expression of representative tobacco leaves infiltrated with different combinations of effector and reporter. 35S::GFP plastid was coinjected as an internal standard in each infection. C, Schematic structure of the TACAAT motifs in the promoter of pathogen-related genes and CBF genes. The relative positions were chosen for ChIP-PCR analysis. D, ChIP enrichment to show the in vivo binding ability of 35S::GFP-ZAT6 to the DNA fragments in the promoters of pathogen-related genes and CBF genes. The ChIP results were normalized to input chromatin, and a fragment in the ACTIN2 promoter was used as the negative control. The data represent the means ± sds of three independent experiments, and asterisks indicate the significant difference of P < 0.05. [See online article for color version of this figure.]
DISCUSSION
Plant hormones play essential roles in integrating developmental and defense cues into complex signaling networks that not only establish plant developmental architecture, but also build up plant defense responses to biotic and abiotic stresses (Kazan and Manners, 2009; Seo and Park, 2010; Liu et al., 2013c). Among these hormones, SA serves as the most important defense hormone associated with microbial pathogenesis (Kieffer et al., 2010; Xu et al., 2011; Zhang et al., 2012).
In this study, a novel function for AtZAT6 in plant development and biotic and abiotic stress responses has been elucidated based on phenotypic, molecular, and physiological characterizations. We found that AtZAT6-overexpressing plants exhibited constitutively activated defense responses, with increased levels of ROS accumulation (H2O2 and O2•–) and extensive cell death in the leaves, while AtZAT6 knockdown plants showed decreased defense responses, with lower ROS accumulation and less cell death (Fig. 3, B–F). In accordance with other studies, Pst DC3000(avrRpt2) triggered ROS production in wild-type plants at the period of 3 to 9 hpi because of the recognition of avrRpt2 by the Resistance to P. syringae2 resistance protein (Kunkel et al., 1993; Lamb and Dixon, 1997). Moreover, AtZAT6-overexpressing plants exhibited even higher levels, while AtZAT6 knockdown plants displayed relatively lower levels of H2O2 and O2•– at these periods after inoculation of Pst DC3000(avrRpt2; Fig. 4, A and B). The rapid induction of ROS by Pst DC3000(avrRpt2) and AtZAT6-mediated ROS accumulation after infection with Pst DC3000(avrRpt2) might be largely contributed to hypersensitive response (Kunkel et al., 1993; Lamb and Dixon, 1997). However, Pst DC3000 only slightly induced the H2O2 content at 6 and 9 hpi but had no significant effect on the O2•– content at the period of 3 to 9 hpi. Additionally, AtZAT6-overexpressing plants exhibited higher levels of H2O2 and O2•– at the period of 3 to 9 hpi of Pst DC3000, while AtZAT6 knockdown plants displayed relatively lower levels than those in wild-type plants (Fig. 4, A and B). Thus, AtZAT6 positively modulates ROS level under both mock and pathogen-infected conditions (Fig. 4, A and B), indicating putative links between AtZAT6-mediated ROS accumulation and disease resistance. Miller et al. (2008) have reviewed that AtZAT proteins (AtZAT7, AtZAT10, and AtZAT12) are key regulators of ROS signaling and could function as integrators of ROS signaling and plant stress responses. Our study indicated that AtZAT6 is another important factor of the ZAT network-involved ROS signaling. As Miller et al. (2008) reviewed, further studies are needed to investigate how ZAT proteins mediate ROS signaling.
Moreover, the dwarf and retarded phenotype of AtZAT6-overexpressing plants is similar to reported dwarf mutants that displayed constitutive activation of ROS and SA-dependent responses, such as activation-tagged mutant of GH3.5 (Zhang et al., 2007), myb96-1d (Seo and Park, 2010), snc2-1D (for suppressor of npr1-1 [knockout mutant of NON-INDUCIBLE IMMUNITY1, NONEXPRESSER OF PR GENES1], constitutive2; Zhang et al., 2010b), snc1 and Toll-like/interleukin-1 receptor1-human influenza virus hemagglutinin (Zhu et al., 2010), loss-of-function of CULLIN1 (Cheng et al., 2011), mos14-1 snc1 npr1 containing the MOS14 [modifier of snc1-1, 14] transgene under its native promoter; Xu et al., 2011), mpk4 (Kong et al., 2012), loss-of-function mutant of BDA1 (for bian da; becoming big in Chinese; Yang et al., 2012b), mkk1/mkk2 for mutant of MAP kinase kinase1 [MKK1]/MKK2; Zhang et al., 2012), and mutant of BAK1-INTERACTING RECEPTOR-LIKE KINASE1 (Liu et al., 2013a). Additionally, the expression of EDS1 and PAD4, which are responsible for SA biosynthesis, were significantly up-regulated after overexpression of AtZAT6 (Fig. 3G). SA accumulation is necessary for the activation of SA downstream gene expression (Zhang et al., 2010b, 2012; Zhu et al., 2010; Kong et al., 2012). Consistently, AtZAT6-overexpressing plants showed increased endogenous SA and up-regulated expression of SA-dependent PR genes, resulting in enhanced immunity resistance against pathogen bacterials, while AtZAT6 knockdown plants exhibited decreased immunity resistance (Fig. 3). Similar to AtDi19 (one C2H2 ZFP; Liu et al., 2013b), AtZAT6 can bind to the TACAAT elements within the PR1, PR2, and PR5 promoters, as well as the TACAAT elements in the EDS1 and PAD4 promoters (Fig. 8). Thus, AtZAT6 positively regulates endogenous SA and the expression of SA-dependent PR genes via directly binding to the TACAAT elements of pathogen-related genes.
In addition to biotic stress, AtZAT6 positively modulated abiotic stress resistance, including salt, drought, and cold stresses. Liu et al. (2013d) found that phosphorylation of AtZAT6 by AtMPK6 was essential for its positive regulation of seed germination under salt and osmotic stresses (Liu et al., 2013d). According to Liu et al. (2013b), the expressions of PR1, PR2, and PR5 were largely induced by drought stress treatment. Overexpression of these PR genes and pretreatment with SA analogs 2,6-dichloroisonicotinic acid conferred improved drought resistance. In Arabidopsis, CBF1, CBF2, and CBF3 (also known as DREB1b, DREB1c, and DREB1a, respectively) play key roles in cold, salt, and drought stress responses via binding to the C-repeat/dehydration-responsive element cis-acting element of several stress-responsive genes (Novillo et al., 2012). Because AtZAT6 positively regulates the expression of SA-dependent PR1/PR2/PR5 and CBF1/CBF2/CBF3 via directly binding to the TACAAT elements in the promoters, the positive regulation of pathogen-related genes and CBF1/CBF2/CBF3 was also contributed to AtZAT6-mediated abiotic stress resistance.
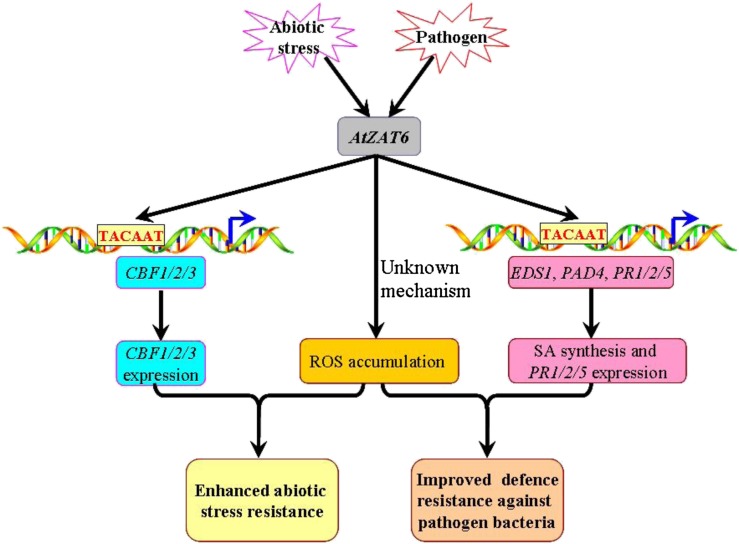
It should be mentioned that ectopic overexpression of tagged TFs obviously does not reflect the natural situation, and it may have some limitations in dissecting the in vivo roles of the TF. However, the ChIP result (the direct binding of AtZAT6 to the TACAAT elements of the promoters of EDS1, PAD4, PR1, PR2, PR5, CBF1, CBF2, and CBF3) exhibited high consistency with changes of gene expression levels in AtZAT6-overexpressing and knockdown plants relative to wild-type plants as well as the phenotypes of stress resistance. Based on the above observations, a working model is proposed in Figure 9 to depict the mechanism of AtZAT6 involved in plant biotic and abiotic stress responses. Biotic and abiotic stresses largely induced the expression of AtZAT6. AtZAT6 could bind to the TACAAT elements of the promoters of EDS1, PAD4, PR1, PR2, PR5, CBF1, CBF2, and CBF3, leading to accumulation of SA and up-regulation of stress-responsive genes. Moreover, increased expression of AtZAT6 also resulted in higher ROS accumulation. All of these changes resulted in improved resistance to biotic and abiotic stress in AtZAT6 overexpressors.
Figure 9.
Model depicting the mechanisms of AtZAT6 involved in plant stress responses. Pathogen and abiotic stress could largely induce the expression of AtZAT6. First, AtZAT6 could bind to the TACAAT elements of the promoters of EDS1, PAD4, PR1/PR2/PR5, and CBF1/CBF2/CBF3, leading to accumulation of SA and up-regulation of these genes’ expression. Second, AtZAT6-overexpressing plants could result in higher ROS concentration. All of these results resulted in improved resistance to biotic and abiotic stress. [See online article for color version of this figure.]
Taken together, this study indicates that AtZAT6 plays important roles in plant development and positively modulates biotic and abiotic stress resistances by activating the expression levels of SA-related genes and CBF genes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) seeds of Col-0 ecotypes were first surface sterilized with 50% (v/v) bleach and 0.1% (v/v) Triton X-100 and then thoroughly washed three times with sterile water. After stratification at 4°C for 3 d in darkness, Arabidopsis seeds were grown in soil or on MS medium containing 1% (w/v) Suc and cultured in a growth chamber. The growth chamber was controlled at an irradiance of 120 µmol quanta m–2 sec–1 at 23°C ± 2°C with 65% relative humidity under 16-h-light and 8-h-dark cycles. Nutrient solution was supplied with water every three days to keep plant growth.
Generation of AtZAT6 Transgenic Lines
For the ProZAT6::GUS transgenic construct, the promoter regions of AtZAT6 were amplified by PCR and inserted into SalI/BamHI sites of pBI101.2 vector with kanamycin resistance. For the overexpressing AtZAT6 transgenic construct, the full-length coding sequence of AtZAT6 was inserted into SmaI/XhoI sites of pBIM vector with kanamycin resistance under the control of the Cauliflower mosaic virus 35S promoter (Yang et al., 2005). For the AtZAT6 and GFP coexpressing construct, the coding regions of AtZAT6 were amplified by PCR and inserted into SmaI/XhoI sites of the 35S::EGFP vector with Basta resistance (Cutler et al., 2000). For the amiR-ZAT6 transgenic construct, amiR-ZAT6 fragments were amplified from the plasmid pRS300 by PCR using specific primers from Web MicroRNA Designer 3 (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi; Schwab et al., 2006), and then the amiR-ZAT6 fragment was inserted into the SmaI site of pBARN vector with Basta resistance (LeClere and Bartel, 2001). The primers for vector constructs are listed in Supplemental Table S2. After introduction into Arabidopsis wild-type (Col-0) plants, homozygous transgenic plants were selected on MS medium containing 1% (w/v) Suc by kanamycin or Basta resistance and were confirmed by PCR analysis.
RNA Isolation and Quantitative Real-Time PCR
RNA isolation and quantitative real-time PCR was performed as described by Shi et al. (2013b). The quantitative real-time PCR was carried out with Sybr-green fluorescence using a CFX96 Real Time System (Bio-Rad). System software by the comparative ΔΔcycle threshold method was used for quantification, and the expression levels of target genes were normalized to the amount of the housekeeping gene ubiquitin10 (UBQ10). At least three independent experiments were performed for real-time PCR, and three technical replicates were used in each independent experiment. The specific primers of analyzed genes for real-time PCR are listed in Supplemental Table S3.
Subcellular Localization Analysis
For subcellular localization analysis, 5-d-old seedling roots of 35S::GFP-ZAT6 transgenic plants were analyzed. GFP signals were detected using the Olympus FluoView 1000 confocal laser scanning microscope, and 20 µg mL–1 propidium iodide was used for cell wall staining in GFP lines.
GUS Staining and Quantification of GUS Activity
GUS staining and quantification of GUS activity was performed using the method described by Jefferson et al. (1987).
Determination of ROS Level
DAB staining and NBT staining were performed as previously described by Shi et al. (2013b). The concentrations of H2O2, O2•–, GSH, and GSSG were also quantified as described by Shi et al. (2013a, 2013b).
Trypan Blue Staining
Trypan blue staining was carried out on 28-d-old plant leaves in lactophenol trypan blue solution as described by Yang et al. (2005). Then, the stained plants were transferred to 70% (v/v) ethanol to remove the chlorophyll and photographed.
Quantification of SA Content
Free and conjugated SA in 28-d-old Arabidopsis leaves were extracted and quantified by HPLC as previously described (Qiu et al., 2007) with minor modification. For HPLC assay, samples were passed over a column with a length of 15 cm and an internal diameter of 4.6 mm filled with 5 µm of reverse phase material (HeSep C18-T; Weltech) at 25°C; the solvent flow was 0.8 mL min–1 (methanol:water with 0.1% [w/v] H3PO4 = 1:1).
Plant Disease Resistance Assay
For disease resistance assay, 28-d-old plant leaves were infected as described by Shi et al. (2012) with the virulent Pseudomonas syringae pv tomato (Pst) DC3000 and the avirulent Pst DC3000(avrRpt2) at optical densities at 600 nm of 0.001 and 0.02, respectively. Inoculation with 10 mm MgCl2 was used as mock treatment. At 0 and 3 dpi, leaf discs within the infiltrated area were taken immediately, and the bacterial growth from infected Arabidopsis leaves and symptoms were monitored as described (Shi et al., 2012). For SAR assay, local leaves were first infected with Pst DC3000(avrRpt2) for 1 d. Then, the distal, uninfected plant leaves were infected with Pst DC3000, and the growth of Pst DC3000 on distal plant leaves was assayed at 0 and 3 dpi of bacterial infection. At least three independent experiments were performed, and 20 independent leaf discs were harvest in each independent experiment.
Plant Abiotic Stress Resistance Assay
Plant abiotic stress resistance was assayed as described by Shi et al. (2013b), and survival rate was determined 7 d after recovery from the abiotic stress treatments.
Transient Expression Assay in Tobacco Leaves
To confirm the interaction between AtZAT6 and the TACAAT element, the transient expression assay in tobacco (Nicotiana tabacum) leaves were performed as described by Huang et al. (2013). The m35S-GUS, TACAAT-m35S-GUS, and TAAAAT-m35S-GUS were constructed by inserting PCR products of mini35S, 4×TACAAT-mini35S, and 4×TAAAAT-mini35S from the minimal-100 Cauliflower mosaic virus 35S-pCAMBIA1391 plastid (Huang et al., 2013) into HindIII and BamHI sites of the pCAMBIA1391Z vector. The corresponding primers for vector construction are listed in Supplemental Table S2. For the transient expression analysis, the pBIM vector and AtZAT6-pBIM construct were used as the effectors, and the m35S-GUS (mini35S-pCAMBIA1391Z), TACAAT-m35S-GUS (4×TACAAT-mini35S-pCAMBIA1391Z), and TAAAAT-m35S-GUS (4×TAAAAT-mini35S-pCAMBIA1391Z) constructs were used as the reporters. The effector and reporter were transformed into Agrobacterium tumefaciens strain GV3101 and coinfected into tobacco leaves as described by Huang et al. (2013), and the 35S::GFP plastid (Cutler et al., 2000) was coinjected as an internal standard in each infection. The GUS activity and GFP fluorescence intensity were quantified using the Infinite M200 Microplate reader (Tecan).
ChIP Assay
ChIP was performed as described by Bowler et al. (2004). Briefly, 2 g of 14-d-old 35S::GFP-ZAT6 transgenic plants were harvested and then immersed in 1% (v/v) formaldehyde for cross linking the DNA with DNA-binding proteins. Next, the chromatin pellets were extracted and sheared by sonication as described by Bowler et al. (2004), and the anti-GFP antibody (AG281; Beyotime) was used to immunoprecipitate the DNA-ZAT6 complexes. The DNA was released with proteinase K, and the enrichment of DNA fragments was determined using quantitative real-time PCR with the specific primers listed in Supplemental Table S3.
Statistical Analysis
All experiments in this study were repeated at least three times, and every experiment was harvested from at least 15 seedlings per genotype. Student’s t test was used to determine the significant difference between wild-type and transgenic lines, and asterisks indicate the significant difference of P < 0.05 compared with the wild type.
Nucleotide sequence data for the genes described in this article are available from The Arabidopsis Information Resource under the following accession numbers: ZAT6 (At5g04340), UBQ10 (At4g05320), ACTIN2 (At3g18780), EDS1 (At3g48090), PAD4 (At3g52430), PR1 (At2g14610), PR2 (At3g57260), PR3 (At3g12500), PR4 (At3g04720), PR5 (At1g75040), CBF1 (At4g25490), CBF2 (At4g25470), CBF3 (At4g25480), CBF4 (At5g51990), DREB2A (At5g05410), and DREB2B (At3g11020).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression level changes of AtZAT6 under abiotic and biotic stress conditions.
Supplemental Figure S2. Transactivation activity of AtZAT6 in yeast.
Supplemental Figure S3. Phenotypes of 21-d-old wild-type and AtZAT6-overexpressing T1 generation plants.
Supplemental Figure S4. The expression level of AtZAT6 in AtZAT6 knockdown plants.
Supplemental Figure S5. Phenotypes of 7-d-old wild-type and AtZAT6-overexpressing plants on MS plate.
Supplemental Figure S6. Modulation of AtZAT6 expression affects glutathione pool and redox state.
Supplemental Figure S7. Phenotype of 35S::GFP-ZAT6 transgenic plants.
Supplemental Table S1. Modulation of morphological phenotypes by AtZAT6 expression.
Supplemental Table S2. Primers used for vector construction.
Supplemental Table S3. Primers used for quantitative real-time PCR.
Supplementary Material
Acknowledgments
We thank Jianmin Zhou for providing bacterial strains of Pst DC3000 and Pst DC3000(avrRpt2), Bonnie Bartel for providing the pBARN plastid, Woo Sik Chung and Dr. Xiaomin Liu for providing seeds of ProZAT6::GUS for comparison, Jihong Liu and Dr. Xiaosan Huang for providing the mini35S-pCAMBIA1391 plastid, Dr. Jun You for providing the pCAMBIA1391Z vector, and Rebecca Grumet for helpful review of the manuscript.
Glossary
- SA
salicylic acid
- TF
transcription factor
- ZFP
zinc finger protein
- Pst
Pseudomonas syringae pv tomato
- Col-0
Columbia
- MS
Murashige and Skoog
- DAB
diaminobenzidine
- NBT
nitro blue tetrazolium
- O2•–
superoxide radical
- H2O2
hydrogen peroxide
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- hpi
hours post inoculation
- ROS
reactive oxygen species
- SAR
systemic acquired resistance
- dpi
days post infection
- ChIP
chromatin immunoprecipitation
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31370302 to Z.C. and 31200194 to H.S.), the Hundred Talents Program, the Knowledge Innovative Key Program of Chinese Academy of Sciences (grant nos. 54Y154761O01076 and Y329631O0263 to Z.C.), the Youth Innovation Promotion Association of Chinese Academy of Sciences (grant no. Y429371O04 to H.S.), and the Outstanding Young Talent Program of Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture (grant nos. Y352811O03 and Y452331O03 to H.S.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Bent AF, Mackey D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Bi D, Cheng YT, Li X, Zhang Y. (2010) Activation of plant immune responses by a gain-of-function mutation in an atypical receptor-like kinase. Plant Physiol 153: 1771–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogamuwa S, Jang JC. (2013) The Arabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light-, abscisic acid- and gibberellic acid-mediated regulation of seed germination. Plant Cell Environ 36: 1507–1519 [DOI] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Cheng YT, Li Y, Huang S, Huang Y, Dong X, Zhang Y, Li X. (2011) Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci USA 108: 14694–14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, Warren D, Cushman J, Connolly EL, Mittler R. (2007) The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem 282: 9260–9268 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Nagarajan VK, Raghothama KG. (2007) Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol 145: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Solano R. (2013) Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Front Plant Sci 4: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XS, Wang W, Zhang Q, Liu JH. (2013) A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol 162: 1178–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. (2009) Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci 14: 373–382 [DOI] [PubMed] [Google Scholar]
- Kieffer M, Neve J, Kepinski S. (2010) Defining auxin response contexts in plant development. Curr Opin Plant Biol 13: 12–20 [DOI] [PubMed] [Google Scholar]
- Kodaira KS, Qin F, Tran LSP, Maruyama K, Kidokoro S, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. (2011) Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol 157: 742–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Qu N, Gao M, Zhang Z, Ding X, Yang F, Li Y, Dong OX, Chen S, Li X, et al. (2012) The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24: 2225–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ. (1993) RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon RA. (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- LeClere S, Bartel B. (2001) A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol Biol 46: 695–703 [DOI] [PubMed] [Google Scholar]
- Li Y, Li S, Bi D, Cheng YT, Li X, Zhang Y. (2010a) SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog 6: e1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tessaro MJ, Li X, Zhang Y. (2010b) Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol 153: 1425–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ding P, Sun T, Nitta Y, Dong O, Huang X, Yang W, Li X, Botella JR, Zhang Y. (2013a) Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol 161: 2146–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WX, Zhang FC, Zhang WZ, Song LF, Wu WH, Chen YF. (2013b) Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol Plant 6: 1487–1502 [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang HQ, Luan S, Li J, He ZH. (2013c) Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc Natl Acad Sci USA 110: 15485–15490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XM, Nguyen XC, Kim KE, Han HJ, Yoo J, Lee K, Kim MC, Yun DJ, Chung WS. (2013d) Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochem Biophys Res Commun 430: 1054–1059 [DOI] [PubMed] [Google Scholar]
- Miller G, Shulaev V, Mittler R. (2008) Reactive oxygen signaling and abiotic stress. Physiol Plant 133: 481–489 [DOI] [PubMed] [Google Scholar]
- Mito T, Seki M, Shinozaki K, Ohme-Takagi M, Matsui K. (2011) Generation of chimeric repressors that confer salt tolerance in Arabidopsis and rice. Plant Biotechnol J 9: 736–746 [DOI] [PubMed] [Google Scholar]
- Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu JK. (2006) Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett 580: 6537–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen XC, Kim SH, Lee K, Kim KE, Liu XM, Han HJ, Hoang MHT, Lee SW, Hong JC, Moon YH, et al. (2012) Identification of a C2H2-type zinc finger transcription factor (ZAT10) from Arabidopsis as a substrate of MAP kinase. Plant Cell Rep 31: 737–745 [DOI] [PubMed] [Google Scholar]
- Novillo F, Medina J, Rodríguez-Franco M, Neuhaus G, Salinas J. (2012) Genetic analysis reveals a complex regulatory network modulating CBF gene expression and Arabidopsis response to abiotic stress. J Exp Bot 63: 293–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S. (2007) OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant Microbe Interact 20: 492–499 [DOI] [PubMed] [Google Scholar]
- Qiu JL, Zhou L, Yun BW, Nielsen HB, Fiil BK, Petersen K, Mackinlay J, Loake GJ, Mundy J, Morris PC. (2008) Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol 148: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R. (2004) The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279: 11736–11743 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Park CM. (2010) MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol 186: 471–483 [DOI] [PubMed] [Google Scholar]
- Shi H, Ye T, Chan Z. (2013a) Comparative proteomic and physiological analyses reveal the protective effect of exogenous polyamines in the bermudagrass (Cynodon dactylon) response to salt and drought stresses. J Proteome Res 12: 4951–4964 [DOI] [PubMed] [Google Scholar]
- Shi H, Ye T, Chen F, Cheng Z, Wang Y, Yang P, Zhang Y, Chan Z. (2013b) Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: effect on arginine metabolism and ROS accumulation. J Exp Bot 64: 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ye T, Zhong B, Liu X, Jin R, Chan Z. (April 16, 2014a) AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytol http//dx..org/10.1111/nph.12812 [DOI] [PubMed] [Google Scholar]
- Shi H, Ye T, Zhu JK, Chan Z. (May 27, 2014b) Constitutive production of nitric oxide leads to enhanced drought stress resistance and extensive transcriptional reprogramming in Arabidopsis. J Exp Bot http//dx..org/10.1093/jxb/eru184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HT, Li RJ, Cai W, Liu W, Wang CL, Lu YT. (2012) Increasing nitric oxide content in Arabidopsis thaliana by expressing rat neuronal nitric oxide synthase resulted in enhanced stress tolerance. Plant Cell Physiol 53: 344–357 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Zhang Z, Jing B, Gannon P, Ding J, Xu F, Li X, Zhang Y. (2011) Transportin-SR is required for proper splicing of resistance genes and plant immunity. PLoS Genet 7: e1002159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. (2012a) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LX, Wang RY, Ren F, Liu J, Cheng J, Lu YT. (2005) AtGLB1 enhances the tolerance of Arabidopsis to hydrogen peroxide stress. Plant Cell Physiol 46: 1309–1316 [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang Y, Ding P, Johnson K, Li X, Zhang Y. (2012b) The ankyrin-repeat transmembrane protein BDA1 functions downstream of the receptor-like protein SNC2 to regulate plant immunity. Plant Physiol 159: 1857–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB. (2008) Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu S, Ding P, Wang D, Cheng YT, He J, Gao M, Xu F, Li Y, Zhu Z, et al. (2010a) Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 107: 18220–18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang Y, Fang B, Gannon P, Ding P, Li X, Zhang Y. (2010b) Arabidopsis snc2-1D activates receptor-like protein-mediated immunity transduced through WRKY70. Plant Cell 22: 3153–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, He Z. (2007) Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol 145: 450–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y. (2012) Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Sun L, Zhao Y, An L, Yan A, Meng X, Gan Y. (2013) Zinc Finger Protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. New Phytol 198: 699–708 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X, Zhang Y. (2010) Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci USA 107: 13960–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.