Abstract
Accurate detection of epimutations in tumor cells is crucial for understanding the molecular pathogenesis of cancer. Alterations in DNA methylation in cancer are functionally important and clinically relevant, but even this well-studied area is continually re-evaluated in light of unanticipated results, including a strong connection between aberrant DNA methylation in adult tumors and polycomb group profiles in embryonic stem cells, cancer-associated genetic mutations in epigenetic regulators such as DNMT3A and TET family genes, and the discovery of abundant 5-hydroxymethylcytosine, a product of TET proteins acting on 5-methylcytosine, in human tissues. The abundance and distribution of covalent histone modifications in primary cancer tissues relative to normal cells is a largely uncharted area, although there is good evidence for a mechanistic role of cancer-specific alterations in epigenetic marks in tumor etiology, drug response and tumor progression. Meanwhile, the discovery of new epigenetic marks continues, and there are many useful methods for epigenome analysis applicable to primary tumor samples, in addition to cancer cell lines. For DNA methylation and hydroxymethylation, next-generation sequencing allows increasingly inexpensive and quantitative whole-genome profiling. Similarly, the refinement and maturation of chromatin immunoprecipitation with next-generation sequencing (ChIP-seq) has made possible genome-wide mapping of histone modifications, open chromatin and transcription factor binding sites. Computational tools have been developed apace with these epigenome methods to better enable the accuracy and interpretation of the data from the profiling methods.
Keywords: cancer epigenetics, DNA Methylation, MeDIP, Microarray, RRBS, Shotgun Bisulfite Sequencing, MRE, ChIP-seq
I. INTRODUCTION
DNA methylation is required for genome function through its roles in maintenance of chromatin structure, chromosome stability and transcription [1–4]. 5-methylcytosine (5MC) is found at a subset of 5’-CpG-3’ dinucleotides and is also sometimes observed at CpNpG, notably in embryonic stem cells[5–7] but also in adult tissues[8]. The modified DNA base 5-hydroxymethylcytosine (5HMC) is also present in mammalian genomes, albeit at a much lower levels compared to 5MC [9,10]. TET proteins catalyze the hydroxylation of 5MC to generate 5HMC, and can act further on 5HMC to yield 5-formylcytosine and carboxylcytosine[10–12].
The N-terminal tails of histone proteins are modified by acetylation, methylation, phosphorylation, ubiquitylation, crotonylation[13] and other covalent modifications. At some histone residues, such as histone H3 lysine 4 (H3K4), methylation can be mono-, di-, or tri-methyl. Furthermore, multiple types of modifications can exist on a single histone molecule. In addition to DNA methylation and histone modifications, there are other interrelated, potentially epigenetic mechanisms including specific deposition of histone variants, noncoding RNAs, chromatin remodeling, and nuclear organization, which are not discussed here. Current epigenomic methods, especially those making use of next-generation sequencing, provide powerful tools to map 5MC, 5HMC and histone modifications at high resolution across the genome. However, there are many considerations for selecting the most suitable method, including ease of use, cost, resolution, specificity, quantitation and availability of computational methods to analyze the data. We describe current epigenomic methods below, focusing primarily on genome-scale mapping methods that use next-generation sequencing-based approaches.
II. METHODS FOR DNA METHYLATION AND HYDROXYMETHYLATION
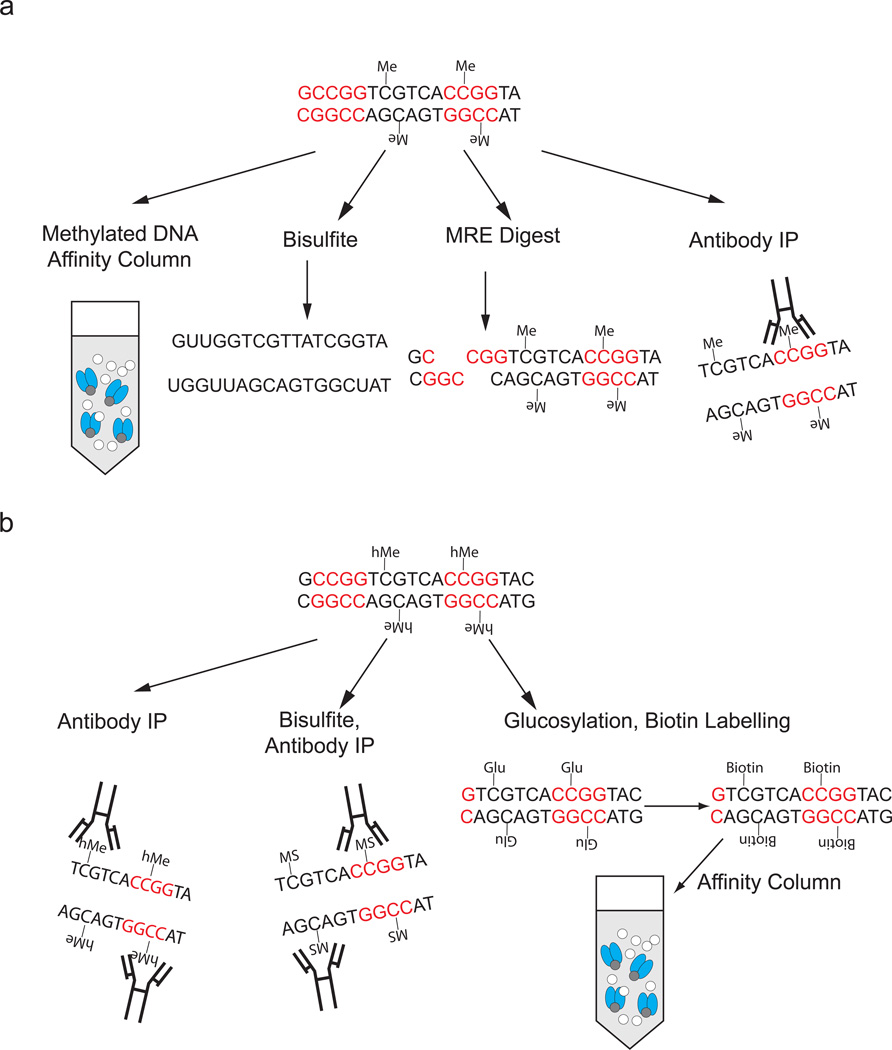
There are three main approaches to detect 5MC and 5HMC. Methyl-sensitive restriction enzymes (MRE) cut DNA based on methylation status of cytosines within their recognition sequences (Figure 1a). A second approach includes differential chemical conversion or enzymatic modification of cytosine according to methylation/hydroxymethylation status, such as sodium bisulfite conversion and 5HMC-specific glucosylation. Third, enrichment methods include methyl DNA immunoprecipitation (MeDIP), hydroxyMeDIP (hMeDIP) and methyl binding domain (MBD) affinity purification that are used to enrich for methylated or hydroxymethylated regions. These approaches can be applied to investigate a single locus, hundreds of thousands of loci, or to all mappable sites genome-wide.
Figure 1.
A summary of methods for direct detection of cytosine methylation and hydroxymethylation. (a) Methylated DNA can be detected with methyl-sensitive restriction enzymes (MRE), the use of antibodies specific for 5-methylcytosine (5MC), by binding to affinity columns that contain methylated DNA binding domains or by the conversion of DNA with sodium bisulfite. It is important to note that some methyl-sensitive restriction enzymes are also sensitive to hydroxymethylation. (b) Several methods have been developed to detect 5 hydroxymethylcytosine (5HMC). These include the addition of a biotin tag to 5HMC through glucosylation and subsequent chemical steps which is followed by an affinity pulldown of the biotin tag, the use of antibodies specific for 5HMC and conversion of 5HMC to 5-cytosine methylenesulfonate (MS) which is then immunoprecipitated with an antibody specific to 5CMS. Me = Methylated Cytosine. hMe = hydroxymethylated cytosine. Glu = glucosylated cytosine.
II.1 Overview of DNA Methylation Reagents
Methyl-sensitive restriction enzymes (MRE) have been used widely for precise, reliable and inexpensive methylation detection. MRE only assay CpGs within their recognition sites but when multiple non-redundant and frequent-cutting MRE are used in parallel this limitation is less problematic. There are approximately 50 unique MRE, though only a few have a methylation-insensitive isoschizomer. MRE can resolve the methylation status regionally or at individual CpGs, depending on the platform used following MRE digestion. Some MRE are inhibited by methylation or hydroxymethylation, for example HpaII[10]. The reliability of MRE enables their straightforward application to next-generation sequencing (MRE-seq) allowing analysis of greater than one million CpGs.
Antibodies against 5-methylcytosine and 5-hydroxymethylcytosine, and columns containing methylated DNA binding proteins (domains of MBD2 or MeCP2 alone, or MBD2b combination with MBD3L) allow enrichment for 5MC/5HMC independent of DNA sequence (Figure 1a,b) [14–17]. Enrichment is greater for regions with higher methylated CpG content relative to fully methylated regions with lower CpG content. These reagents are simple to use and many are commercially available. The lower-limit of resolution is determined initially by the size range of DNA prior to enrichment, generally 100–300bp, and subsequently by the platform used to assess the enrichment, commonly oligonucleotide arrays and next-generation sequencing.
Chemicals including sodium bisulfite and hydrazine react differentially with unmethylated versus methylated cytosine and allow DNA methylation mapping at single base resolution (Figure 1a) [18–20]. Of these, sodium bisulfite is the most commonly used as it results in a positive display of methylation, among other advantages. Sodium bisulfite initiates conversion of cytosine to uracil, which is replaced by thymine during PCR amplification. In contrast, methylated cytosines are non-reactive, and remain as cytosine after bisulfite treatment. Sequencing of individual clones of the PCR product allows assessment of methylation status of contiguous CpGs derived from a single genomic DNA fragment. Bisulfite has many advantages, including single CpG resolution, detection of strand and allele-specific methylation, and detection of non-CpG cytosine methylation. Unlike other methylation-detection reagents, bisulfite provides estimates of absolute rather than relative DNA methylation levels, depending on the platform used. The reduced sequence complexity of the genome following bisulfite treatment complicates its application to oligonucleotide arrays [21], but is not a major issue when a sequencing platform is used. Hydroxymethylated cytosines are resistant to conversion to uracil and are indistinguishable from 5MC in bisulfite sequencing. The reaction of 5HMC with bisulfite yields cytosine methylenesulfonate, which can be specifically detected with an affinity method[22]. Alternatively, the hydroxyl group of 5HMC can be enzymatically glucosylated and biotin labeled to detect 5HMC[23,22].
II.2 Methyl-Sensitive Restriction Enzyme Methods
The HTF (HpaII tiny fragments) enrichment by ligation-mediated PCR, or HELP assay, uses the methyl-sensitive HpaII along with its methylation-insensitive isoschizomer MspI to identify unmethylated CpG sites within the sequence 5’-CCGG-3’ [24]. Genomic DNA digested separately with each enzyme is size-selected to capture small DNA fragments. Custom adaptors complementary to digest ends are ligated and the adaptor-ligated molecules are amplified by PCR. The amplification products can be analyzed using a variety of platforms, including next-generation sequencing on the Illumina platform (HELP-seq) [25]. Methyl-seq is a second Illumina sequencing-based assay that uses HpaII/MspI[26]. Similar to HELP, the protocol involves separate HpaII and MspI digests, adaptor ligation and Illumina sequencing. Approximately 65% of the CpG islands (CGIs) in the human genome are sampled using Methyl-seq. MRE methods are generally biased to CGIs, which constitute 1–2% of the genome and 7% of all CpGs in the genome. Methyl-seq is similarly biased, though non-CGI sites account for ~61% of the regions assayed, including a variety of genomic sequences such as promoters, exons, introns, and intergenic regions.
Ball et al. reported a third variation of MRE-seq, using HpaII/MspI digestion with Illumina sequencing to analyze DNA methylation in the PGP1 EBV-transformed B-lymphocyte cell line [27]. This approach, termed methyl-sensitive cut counting (MSCC), assayed ~ 1.4 million unique HpaII sites. Using MSCC and a complementary method, bisulfite padlock probe sequencing (BSPP) to assay the methylation status of approximately 10,000 CpGs, highly expressed genes were found to be associated with high gene-body methylation and low promoter methylation. MSCC read counts were linearly related to BSPP percent methylation at 381 CpG sites that were assayed with both methods, suggesting that MSCC allows relative quantification of methylation levels.
DNA methylation has also been assessed through traditional Sanger sequencing combined with MRE in digital karyotyping [28,29]. Using a combination of MRE that recognize 6–8bp sites and methylation insensitive restriction enzymes, a library of short sequence tags is generated. The number of tags sequenced reflects the level of methylation at each recognition site, with lower tag counts representing greater methylation levels. In this method, the number of sites analyzed depends on the MRE used – use of AscI for example can generate over 5000 unique tags that correspond to > 4000 genes.
These sequencing-based methods demonstrate the utility of MRE for analysis of DNA methylation. The single CpG resolution and ability to assay a significant portion of the methylome with next-generation sequencing, including most CGIs, makes this a powerful, accurate and straightforward way to assess methylation across the genome. When used alone, the MRE-seq methods enable relative rather than absolute methylation levels to be estimated. An integrative method[30,31] that combines MRE-seq in parallel with MeDIP-seq to increase resolution, CpG coverage, and accuracy in quantitation is discussed below.
II.3 McrBC And CHARM
The methylation-dependent restriction enzyme McrBC recognizes methylated DNA and cuts near its recognition sequence. McrBC recognizes RmC(N)55–103RmC, and cuts once between each pair of half-sites, close to one half-site or the other. The cuts can be distributed over several base pairs and approximately 30 base pairs distant from the methylated base, generating a distribution of DNA ends rather than precisely defined DNA ends. McrBC is useful to size-separate methylated DNA from unmethylated DNA, since the unmethylated DNA remains high-molecular weight after digestion. McrBC was initially applied to microarrays[32].
The “comprehensive high-throughput arrays for relative methylation” (CHARM) method is an array-based technique for methylation profiling using McrBC [33]. To improve specificity and sensitivity, probes were optimized based on location and CpG density on custom arrays. Because neighboring CpG sites tend to have a highly correlated methylation status, neighboring probe signals are averaged to reduce background noise without loss of sensitivity or specificity, though modestly reducing resolution. By comparing CHARM to MeDIP or HpaII on arrays, Irizarry et al. showed that McrBC yields better methylome coverage than HpaII and less bias for CpG density than MeDIP. Using CHARM, aberrant DNA methylation was found in colon cancer at sequences up to 2 kb flanking CGIs, referred to as CGI shores [34]. These data demonstrate the utility of McrBC-based methylation detection, and the new biological insights afforded by the CHARM method.
II.4 Methyl DNA Immunoprecipitation (MeDIP)
In addition to MRE and McrBC, methylation can be assessed by immunoprecipitation of methylated DNA with a monoclonal antibody against 5-methylcytidine (MeDIP) [14]. This antibody does not recognize 5HMC[35], which can be specifically immunoprecipitated with an anti-5HMC antibody[36–39]. A major advantage of MeDIP-based detection is that it is not limited to a specific restriction site and theoretically any fragment with a methylated cytosine is immunoprecipitated. One approach involves the coupling of MeDIP with DNA microarrays to obtain relative methylation levels at the loci represented on the array[14,40–44].
MeDIP combined with next-generation sequencing (MeDIP-seq) can be used to interrogate the majority of mappable CpG and non-CpG cytosines in the genome. In a step forward from array-based methods, MeDIP-seq allows analysis of monoallelic methylation and methylation in a significant number of repeat sequences. Most protocols generate a MeDIP sequencing library by sonicating DNA followed by end-repair, adaptor ligation, immunoprecipitation with the anti-methylcytidine antibody and PCR amplification. The methylation-enriched library is sequenced and the reads are mapped back to a reference genome. A specific genomic region shows higher read density when methylated in one sample compared to when the same region is unmethylated in another sample, although read density between different regions is affected by the density of methylated CpGs, DNA copy number and potentially other factors (discussed in Robinson et al. 2010a and Robinson et al 2010b)[45,46]. These considerations are also important for MBD affinity-based approaches. MeDIP-seq has been applied to a variety of sample types from multiple organisms including human cancer [47,48,30,49–52,31,53].
Several computational methods have been specifically designed for analyzing MeDIP data while addressing local density of methylated CpGs. MEDME (modeling experimental data with MeDIP enrichment) is a combination of analytical and experimental methodologies that improve the interpretation of MeDIP-chip data, and addresses the non-linear relationship between enrichment signal and CpG density that is particular to MeDIP-chip [54]. A second analytical method for MeDIP-chip and also MeDIP–seq data called Bayesian tool for methylation analysis (BATMAN) uses a CpG density-derived coupling factor to quantify methylation levels across a range of CpG densities[47]. MEDIPS is a third approach that, like BATMAN, uses a CpG density coupling factor and in addition provides a framework for evaluating quality control parameters, estimating absolute methylation and comparing samples to detect regions of statistically significant differential methylation[51]. MeDIP-chip and MeDIP–seq are lower resolution compared to bisulfite-based methods. On the other hand, MeDIP-seq provides comprehensive methylome coverage at a fraction of the cost of shotgun bisulfite sequencing. Experimental and computational advances should enable increased resolution and quantitation of methylation levels using MeDIP-seq alone or in combination with MRE-seq.
II.5 Affinity-Based Enrichment Using Methyl Binding Domains
The Methylated CpG Island Recovery Assay (MIRA) is an alternative to MeDIP for selecting/enriching for methylated DNA, particularly at CGIs [15–17]. MIRA involves size fractionation of DNA, either by sonication or with MseI which recognizes 5’-TTAA, a site that is typically found outside of CGIs. After digestion, adaptors are ligated to the DNA followed by selective binding of methylated fragments on a column with full-length MBD2b and MBD3L1 proteins. MBD2b is a methyl-binding protein that exhibits a high affinity for methylated DNA relative to unmethylated DNA [15]. MBD3L1 lacks a methyl-CpG binding domain but can interact with MBD2b and improves enrichment of methylated DNA [15]. The methylated DNA eluted from the column is amplified by PCR, fluorescently labeled and hybridized to a microarray.
There are several similar approaches that combine affinity enrichment with Illumina sequencing. In MethylCap-seq, the methyl binding domain of MeCP2 is used to capture methylated DNA fragments after sonication [55,52]. Binding occurs at low salt concentration and then a step-wise elution of captured DNA is performed by increasing the salt concentration, allowing collection of fractions with differing methylated CpG density, with highly methylated, CpG-dense fragments eluting at the higher salt concentrations. The eluates can be sequenced separately or pooled. The MBD2 methyl binding domain alone can be used for enrichment followed by Illumina sequencing, called MBD-isolated Genome Sequencing (MiGS) [56]. In this protocol, a single elution is performed. MBD2 enrichment with serial elution in increasing salt has been called MBD-seq [31,57] or MBDCap-seq [45].
Several studies have directly compared MeDIP-seq with MBD affinity-based sequencing. Harris et al. found that MeDIP-seq and MBD-seq were 99% concordant using binary methylation calls in 200bp windows or 1000 bp windows [31]. MeDIP-seq enriched more at regions of low methylated CpG density compared to MBD-seq. Also, MeDIP-seq appeared to detect non-CpG methylation (i.e., at CpNpG) but MBD-seq did not, as predicted. Bock et al. compared MeDIP-seq with MethylCap-seq and observed similar levels of accuracy in quantifying methylation when comparing each to Infinium 27K data. In both of these studies, MeDIP-seq and MBD affinity-based sequencing performed well in comparison with bisulfite next-generation sequencing.
II.6 Integrative MeDIP- and MRE-seq
MeDIP-seq and other affinity-based methods provide a positive display of methylated loci, and the absence of signal usually represents unmethylated loci, but also could be a result of regions that are difficult to PCR amplify or sequence, or insufficient sequencing depth. A method that combines MeDIP-seq with MRE-seq leverages their complementarity [30,31,58]. Independent MeDIP-seq and MRE-seq libraries are generated from the same DNA sample and sequenced separately. For MRE-seq, three to five parallel digests are performed using the MRE HpaII, AciI, Hin61, Bsh1236I and HpyCH4IV; the digests are size-selected and combined into a single library. Because the restriction sites from these enzymes are non-overlapping, each additional enzyme greatly increases coverage of unique CpG sites. At a moderate sequencing depth integrated MeDIP- and 3 enzyme MRE-seq together interrogate either uniquely or as multimapping sites ~22 million of the ~29 million CpGs in the haploid human genome[31]. The integrative method is useful for detecting intermediate methylation, including regions of allelic methylation that overlap with monoallelic histone modifications and monoallelic gene expression[31]. This illustrates another significant advantage of sequencing based epigenome analyses – the ability to assign an epigenetic state to a given genetic allele. For extensive DNA methylation profiles of human cells and tissues, see http://vizhub.wustl.edu/.
II.7 Indirect Methylation Detection with Demethylating Agents and Expression Arrays
Genetic or chemical inhibition of DNA methylation followed by expression array analysis can identify genes that may have been silenced by DNA methylation [59–63]. siRNA or shRNA can be used to knock down the DNA methyltransferases, or cell lines can be treated with demethylating agents such as 5-aza-2’deoxycytidine (5-aza) alone, or 5-aza in combination with histone deacetylase inhibitors. 5-aza is a cytidine analog that is incorporated into DNA and covalently binds and inhibits DNA methyltransferase, resulting in passive demethylation. 5-aza treatment results in activation of genes that were silenced by DNA methylation, provided that the appropriate transcription factors are present. However, interpretation of this indirect assessment of methylation is complicated by the fact that genes lacking promoter methylation may also exhibit an increase in expression following 5-aza treatment [64]. Presumably this results from demethylation at other loci within the same gene or in genes upstream that are required for its expression, though direct effects on unmethylated regulatory elements cannot be ruled out. Furthermore, this approach is best applied to cells grown in culture such as cell lines or early passage primary cells [65], as 5-aza requires replication to induce passive demethylation. The application of this approach to primary tumor cells addresses epigenetic silencing that is from long-term culturing rather than cancer or cell type-specificity, though 5-aza may cause growth arrest in non-transformed cells.
II.8 Reduced Representation Bisulfite Sequencing (RRBS)
Bisulfite treatment converts unmethylated cytosines to uracil but methylcytosine and hydroxymethylcytosine are resistant to conversion. When followed by cloning and Sanger sequencing, this approach yields quantitative, allelic, contiguous and base resolution of cytosine methylation. However, the shotgun bisulfite approach has been quite expensive for mammalian methylomes. It is important to note that hydroxymethylcytosine and methylcytosine cannot be distinguished by bisulfite sequencing as both block conversion.
To retain the advantages of methylation detection by bisulfite while reducing the cost of shotgun bisulfite sequencing, Meissner et al. developed a technique that interrogates DNA fragments from a reduced representation of the bisulfite-treated genome [66–68]. The reduction comes from DNA digestion with methylation-insensitive restriction enzyme MspI and fragment size selection. After digestion, the ends of the DNA are filled-in with dGTP and methylated dCTP, followed by the addition of an A overhang to enable adaptor ligation. The adaptors used for this assay are methylated at cytosine residues to prevent conversion during bisulfite treatment. The adaptor-ligated DNA is then size selected on a gel and two fractions are excised – the sizes of which depend on the organism. For mouse DNA, approximately 300,000 MspI fragments that span 40 to 220 bp are analyzed, which corresponds to nearly 1.4 million CpG sites analyzed at the nucleotide level [67]. These fragments are then bisulfite treated, PCR amplified and size selected again to generate a sequencing library. Several factors must be considered with this approach. First, the choice of a restriction enzyme to fractionate the DNA will bias the portion of the genome that is represented. A second consideration is the process of mapping reads of bisulfite converted DNA to the genome. Several mapping algorithms for “bisulfite genomes” have been developed [69,70,67,71]. Compared to other sequencing methods, RRBS provides an efficient way to generate absolute quantification of methylation of more than 1 million CpG sites at single base pair resolution. Methylation at non-CpG cytosines can also be assessed by RRBS[8]. RRBS has been successfully applied to nanogram quantities of genomic DNA[72] and to large numbers of human cell and tissue types (http://vizhub.wustl.edu/).
II.9 Shotgun Bisulfite Sequencing
Shotgun sequencing of bisulfite treated DNA has been successfully applied to several organisms, including humans [69,70,73,7,74–78] and provides comprehensive, single cytosine quantification of methylation level when sequence coverage is sufficiently deep. A single-CpG-resolution shotgun bisulfite experiment on human DNA requires hundreds of millions of sequencing reads, with the exact number varying depending on the desired sequencing depth and on read lengths[78]. Many regions >200bp in the mammalian genome do not contain CpGs and thus a large number of sequence reads may be uninformative, at least for CpG methylation. Prior selection of sequences, for example through sequence capture methodology, or enrichment of methylated DNA or unmethylated DNA followed by shotgun sequencing could increase the efficiency and decrease the cost of this approach. Bisulfite sequencing that first employs selective “reduction” of the genome (e.g. RRBS) is far less expensive. Nevertheless, the cost of sequencing full DNA methylomes has decreased 20-fold since the first human methylome[7]. Shotgun bisulfite methylomes have been generated for a breast cancer cell line and primary human mammary epithelial cells[79] and primary colorectal cancer and adjacent normal colon tissue[80].
RRBS and shotgun bisulfite sequencing require algorithms that are tailored to mapping the sequence reads from bisulfite treated DNA back onto the genome. Several algorithms have been developed for this computationally intensive problem [69,67,70,71,81,82]. The reduction in base complexity from the bisulfite conversion and the fact that a CpG can be methylated or unmethylated are issues that are addressable though complex when aligning bisulfite reads. Due to the bisulfite conversion process, the forward and reverse strands of DNA are no longer complementary and the sequence reads therefore are aligned to four different bisulfite-converted genomes: forward BS, forward BS reverse complement, reverse BS, reverse BS reverse complement). Thus, for this mapping there is increased search space along with a reduction of sequence complexity, requiring significant computation time for the read mapping [31].
II.10 Other Bisulfite Methods
Illumina Infinium methylation assays are mid-range platforms using bisulfite conversion and bead arrays to quantify methylation levels at individual CpGs. The HumanMethylation27 and HumanMethylation450 formats interrogate 27,578 and >450,000 CpGs, respectively. Bead-bound oligonucleotides corresponding to the methylated and unmethylated states of a single CpG site are hybridized to bisulfite-converted DNA and differentially labeled with Cy3 or Cy5. The methylation level is determined by the ratio of Cy3 and Cy5 fluorescence on the bead array. The HumanMethylation27 BeadChip interrogates 12 samples at a time and includes probes from 1,000 cancer-related genes and from putative promoters of 110 miRNA, among others. While there are on average 2 CpG sites assayed per gene for the majority of genes, 150 genes known to exhibit aberrant tumor-specific methylation are assayed at 5–10 CpGs each. The vast majority of 27K probes are located in promoters. The 450K platform expands the genomic regions that are assayed by Infinium. Genes are broadly profiled, with probes in the promoter, 5’UTR, first exon, gene body and 3’ UTR. 99% of CpG islands have probes, and the CpG island shores, 2kb regions flanking CpG islands, and regions flanking shores, called “shelves”. Like the 27K assay, a single 450 BeadChip can assay 12 samples. Both versions require 500 ng of DNA prior to bisulfite conversion. These methods do not assess multiple closely apposed CpGs individually, and such regions are generally avoided in the assay development. This bias is likely to impact biological insights drawn from this data.
Another bisulfite-based method, the Sequenom EpiTyper assay, utilizes MALDI-TOF mass spectrometry to analyze RNA cleavage fragments derived from post-bisulfite PCR products that contain a promoter to drive transcription [83,84]. This unique assay allows high-throughput quantitative methylation analysis at hundreds of loci, usually at single CpG resolution, and is quite useful for candidate loci in hundreds of samples, or as a follow-up to genome-wide profiling.
Bisulfite padlock probes are molecular inversion probes designed to target and capture specific CpG sites from bisulfite-converted DNA[27,85]. The strategy is similar to RRBS in that a subset of CpG sites are analyzed by bisulfite sequencing to reduce the genomic space that must be covered, but with the advantage that particular CpGs can be assayed, instead of only those located within a set of restriction fragments. Tens of thousands of bisulfite padlock probes can be amplified in single reaction and sequenced on the Illumina platform. Deng et al. were able to assay ~66,000 CpG sites, primarily in CpG islands[85]. A prominent advantage of this technology is that it is customizable and can target a specific set of CpG sites of interest to the investigator.
III. DETECTION OF 5-HYDROXYMETHYLCYTOSINE
5-hydroxymethylcytosine (5HMC) is abundant in mammalian genomes. The tissue-specificity, genomic distribution and functional significance of 5HMC are under investigation. Pre-existing 5MC is hydroxylated by the TET family of dioxygenases (TET1, TET2 and TET3) to yield 5HMC[10,86]. TET proteins can further modify 5HMC resulting in formylmethylcytosine, carboxymethylcytosine, and possibly through steps mediated by base excision repair, unmodified cytosine[12,11]. TET1 is an MLL translocation partner in acute myeloid leukemia[87,88] and TET2 mutations occur in myeloid malignancies associated with decreased 5HMC[89], suggesting that dysregulation of 5HMC plays a role in cancer.
Detecting and quantifying 5HMC is challenging because many reagents used for detecting 5MC do not distinguish 5HMC from 5MC. Like 5MC, 5HMC is resistant to C-to-U transition following bisulfite treatment[90], and these bases are indistinguishable by bisulfite cloning and sequencing or other bisulfite-based methods. In addition, 5HMC reacts with bisulfite to yield cytosine 5-methylenesulfonate (CMS) and DNA with dense CMS is inefficiently amplified during PCR due to Taq polymerase stalling at CMS sites[90]. As a result, quantification of hydroxymethylation in regions of dense 5HMC, if they exist in some biological contexts, may be underestimated with bisulfite-based methods. MRE-based methods also do not distinguish 5MC from 5HMC, depending on the enzymes used, such as HpaII, which is inhibited by 5MC or 5HMC in its recognition sequence[10]. Finally, affinity-based 5MC methods (MeDIP-seq, MBD-seq, etc.) are specific to 5MC and do not detect 5HMC directly, but could indirectly enrich for regions with 5HMC when it occurs on the same DNA fragment as 5MC[35].
Global quantification of 5HMC levels (measuring the relative or absolute amount of 5HMC present within a DNA sample) can be assayed by thin layer chromatography (TLC)[10,9] and high-performance liquid chromatography-mass spectrometry (HPLC-MS)[9,91]. Recently, a profusion of 5HMC mapping techniques have also been developed, many of which can be employed for genome-wide analysis.
III.1 5HMC Glucosylation Methods
There are several methods based on in vitro glucosylation of 5HMC in DNA that can be used for global quantification or mapping of 5HMC. These methods use bacteriophage T4 beta-glucosyltransferase (BGT) to catalyze the addition of a glucose moiety to the hydroxyl group of 5HMC. For global quantification, a radiolabeled substrate (uridine 5’-diphosphate-[3H]-glucose) is used in the BGT-catalyzed reaction. The amount of labeled substrate incorporated is compared to standards, allowing absolute quantification[92]. A mapping method called GLIB (glucosylation, periodate oxidation, biotinylation) combines glucosylation by BGT with subsequent chemical reactions, resulting in the addition of two biotin molecules to each 5HMC[22]. The biotin-tagged 5HMC DNA is then pulled down with streptavidin and sequenced on the Helicos single molecule platform. GLIB has high sensitivity, with 90% recovery of DNA fragments containing a single 5HMC molecule. Song et al. present a second mapping method, in which a chemically engineered glucose containing an azide group is transferred to 5HMC by BGT[23]. The azide group is then chemically tagged with biotin and affinity enriched, with global quantification performed using avidin-horseradish peroxidase and genome-wide mapping through Illumina sequencing. Finally, a method has been developed utilizing the restriction endonuclease MspI, which cuts CmCGG and ChmCGG, but not CglucCGG sites. Locus-specific 5HMC can be estimated using MspI digestion on BGT-modified DNA followed by quantitative PCR across the cleavage site[36,93].
III.2 5HMC Affinity Enrichment Methods
There are two enrichment methods for 5HMC based on antibodies that detect 5HMC itself or 5-cytosine methylenesulfonate (CMS), the product of reacting 5HMC with sodium bisulfite. The 5HMC antibody with sequencing approach, hMeDIP-seq[36–39], is similar to MeDIP-seq, and informatic tools originally developed for MeDIP-seq data have been employed in hMeDIP-seq. Monoclonal and polyclonal anti-5HMC antibodies are commercially available, but their 5HMC-density dependence[89,22], along with the relatively low genomic abundance of 5HMC in some tissues, might result in inefficient pulldown of 5HMC-sparse regions. The anti-CMS antibody sequencing approach was developed as a more sensitive, less density-dependent alternative to hMeDIP-seq[22]. CMS pulldown had lower background and decreased density dependence compared to commercial anti-5HMC antibodies. CMS-enriched libraries were sequenced on the Illumina platform. Since Illumina library construction protocols usually require at least one PCR step, the tendency of Taq polymerase to stall at regions of dense CMS could be problematic.
The rapid development of methods for the detection and quantification of 5HMC has paralleled the exciting pace of discovery of the distribution and potential functional roles of this “sixth base”. Computational tools that are specific for hMeDIP-seq and CMS-pulldown have not been reported yet. For hMeDIP-seq, tools developed for MeDIP-seq, such as MEDIPS[51] have been adapted [38]. Stroud et al. used SICER, which was originally developed for analyzing ChIP-seq data for diffusely distributed histone modifications, to define regions of 5HMC enrichment[39]. The next generation of genome-wide mapping methods for 5HMC may involve direct detection of the modified base by single molecule sequencing [94,23].
IV. CHROMATIN IMMUNOPRECIPITATION-SEQUENCING (CHIP-SEQ)
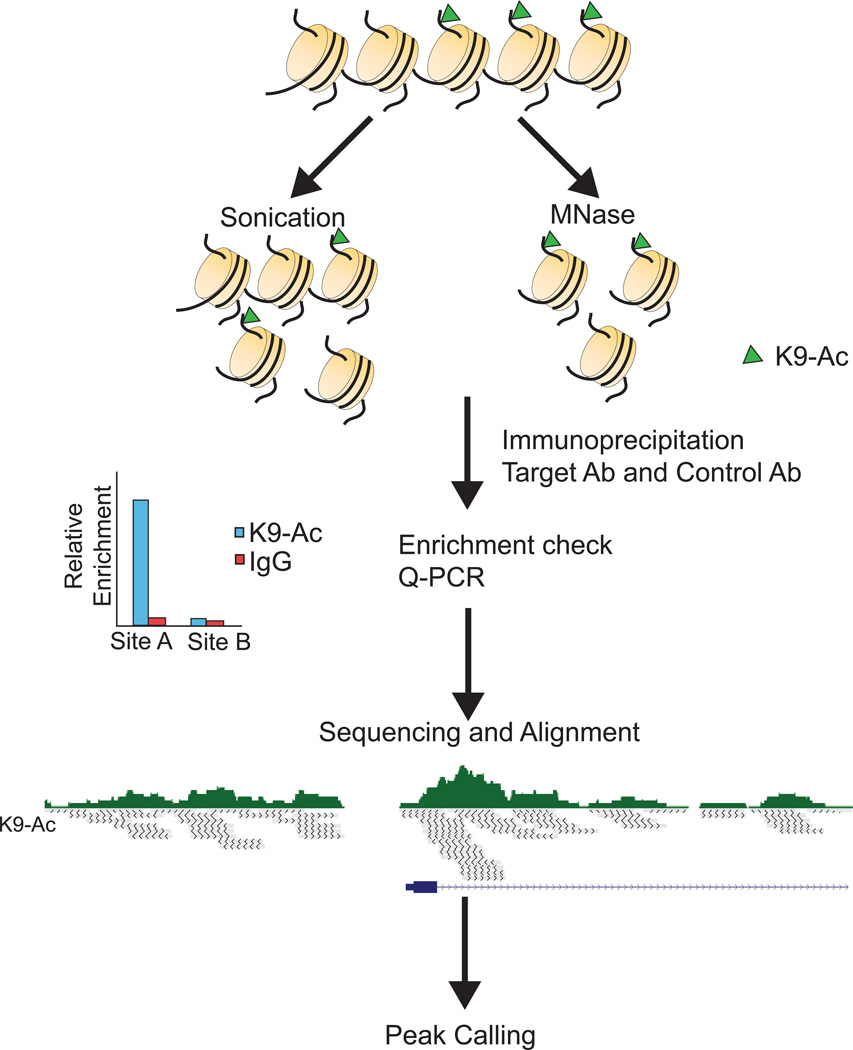
Alterations in histone modification patterns and transcription factor binding impact gene expression and have been implicated in tumorigenesis, cancer cell stemness, metastasis, and drug resistance[95–98]. Chromatin immunoprecipitation coupled with next generation sequencing (ChIP-seq) has become the gold standard to study histone modifications and transcription factor binding genome-wide. It provides higher resolution, improved signal-to-noise ratios, and when using indexed libraries, it is less expensive than coupling ChIP with microarrays (ChIP-chip)[99]. Fresh or fresh frozen tissue or cells are either kept native (N-ChIP)[100] or formaldehyde cross-linked to preserve weaker DNA-protein interactions (X-ChIP)[101], followed by cell lysis (Figure 2). N-ChIP is primarily used for histone modifications, where the DNA histone interactions are inherently strong[99]. Antibody specificity and immunoprecipitation are more efficient with N-ChIP as epitopes can be disrupted by formaldehyde[100], however N-ChIP cannot be applied to proteins with lower DNA binding affinities such as transcription factors. Cross-linking ameliorates this problem, and minimizes stochastic nucleosome movement that can occur during N-ChIP[100], however it also may fix transient non-functional interactions and reacts at lysines which may create biases. Native or cross linked chromatin is then fragmented by sonication or microccocal nuclease (MNase) digestion. Both methods impart bias in downstream sequencing[102]. MNase creates higher resolution, primarily mononucleosome (~146bp) fragments, but is less efficient at cutting between G and C bases, creating greater fragmentation bias[103,104]. In contrast, sonication provides decreased resolution (200–600bp) but is more uniform[99]. Fragmented chromatin is immunoprecipitated with an antibody that specifically recognizes the epitope of interest. The success of ChIP reactions is dependent on antibody quality. Polyclonal antibodies are advantageous for X-ChIP experiments, as they reduce the chance of crosslinking destroying antibody interactions[101], but may have increased cross-reactivity. Relative enrichment of ChIP DNA is assayed via qPCR. Enrichment varies greatly with the protein of interest, antibody quality, and positive and negative control regions of the genome that are used. To minimize the number of reads contributing to background noise, it is common to require greater enrichment in ChIP-seq (5–50 fold) when compared to single locus ChIP-PCR[102]. Purified ChIP DNA sequencing libraries are constructed by end repair, A base addition, adapter ligation, PCR amplification and size selection. Additional bias may occur during library construction and PCR amplification, as both GC-rich and GC-poor regions are underrepresented[102,99]. The total number of sequence reads required depends on the quality of ChIP enrichment, the expected number of peaks and peak size, but sequencing multiple indexed ChIP libraries in a single lane is common practice.
Figure 2.
Overview of chromatin immunoprecipitation-sequencing. DNA is fractionated via sonication (~200–600bp) or with micrococcal nuclease (~146bp). The fractionated DNA is then immunoprecipitated (IP) with a target antibody and an isotype control antibody. The efficiency of the immunoprecipitation is assayed by quantitative PCR, testing regions that are known to be bound (Site A, positive control) or not bound (Site B, negative control). The enriched DNA is then used to generate a DNA sequencing library, which is sequenced and reads are aligned to the appropriate genome. Each read is depicted as a grey line, the read densities are displayed above in green and a gene is shown in blue. Finally, the aligned reads are used to generate peaks that mark regions of statistically significant enrichment of reads for the IP of the histone mark or chromatin protein of interest.
IV.1 ChIP-seq Data Analysis
Transforming the millions of sequencing reads generated by ChIP-seq into biologically interpretable data is a computationally demanding, multi-step process for which a variety of tools have been developed. While many tools address the same problem, each tool is different and can impact the final result. The first and most resource-intensive step is aligning the sequence reads to the genome. Most sequencing platforms come with alignment pipelines, however third party aligners are commonly used, such as MAQ[105], Bowtie[106], BWA[107], SOAP[108,109] and PASH[110]. These packages differ by alignment algorithm, as well as how multi-aligning reads and gapped vs. un-gapped alignments are handled, resulting in differences in sensitivity and specificity. For most cancer samples a gapped aligner is preferred to allow for the variety of genetic aberrations accumulated in the tumor. Aligned reads are then analyzed to find enriched areas or ‘peaks’ in the genome, for which a number of ‘peak calling’ algorithms have been created[111,99]. Though the exact method varies between programs, most shift tags based on chromatin fragment size to accumulate tags near the true binding site and increase peak resolution[111]. Regions of statistical enrichment of IP tags relative to a background control are calculated. The most commonly used control is input DNA isolated from the same chromatin batch as the ChIP[99]. This reduces false positives introduced from fragmentation and mappability biases, and controls for genetic differences such as copy number alterations that affect read density. Finally, peaks are filtered based on uneven distributions of sense and antisense tag accumulation[111]. Most current peak callers identify focal enrichments such as transcription factor binding sites, however some have been developed for broader marks like histone modifications associated with heterochromatin [112–114]. Many groups are actively researching ways to reduce noise and increase true positives.
IV.2 Application of ChIP-seq to Cancer Epigenomes
The network of transcription regulatory factor interactions and their effects on gene expression in cancer are under investigation. ChIP-seq was initially used to profile T-cells, and since then a main focus has been on embryonic stem cells and cell lines[115–117]. Recently, distinct chromatin states or ‘signatures’ comprised of combinatorial histone marks have been linked to specific functional genomic elements by integrating multiple ChIP-seq data across human cell lines[118–120]. The combinatorial histone signatures identified in these studies have not been investigated in the context of tumor progression. Multidimensional epigenomic profiles of tumors also provide a novel means of sub-type classification, identifying prognostic markers, and insight into tumor cell of origin. ChIP-seq will also help the annotation and functional characterization of non-genic susceptibility loci, as has been recently performed in prostate cancer[121] and in GWAS studies[120]. New techniques are being developed to perform ChIP-seq on a small number of cells, creating an opportunity to better analyze intratumoral heterogeneity of epigenomic patterns [122,123]. Finally, chromosome conformation capture (3C) technology[124] and its high-throughput derivatives (4C[125], 5C[126], Hi-C[127], ChIP-Loop[128,129], ChIA-PET[130]) detect distal DNA-DNA interactions (e.g. promoter-enhancer), but can also be used to identify complex genomic rearrangements in cancers[131]. Coupling ChIP with 3C technologies followed by sequencing will likely be a powerful way to study how both epigenetic patterns, and associated structural interactions change during the process of tumorigenesis.
V. FUTURE DIRECTIONS
Recent unanticipated data present new directions for cancer epigenomic studies. First, promoters with polycomb-mediated histone modifications in ES cells are associated with aberrant DNA hypermethylation in adult tumors[132–134]. Second, cancer-associated mutations occur in the DNA methyltransferase DNMT3A[135,136]. Similarly, the occurrence of TET1 translocations[87,88] and TET2 mutations in cancer points to an etiologic role for these epigenetic regulators. Finally, human tissues harbor abundant 5-hydroxymethylcytosine, a product of TET proteins acting on 5-methylcytosine.
The future of cancer epigenomic methods will be shaped by two technological trends. First, the breathtaking pace of advances in next-generation sequencing will continue to improve 5MC/5HMC, histone modification and chromatin conformation mapping. Genome-wide epigenomic experiments will become increasingly inexpensive and accessible, though paralleled with needs for increased computational power and data storage. Second, direct single molecule sequencing that distinguishes between modified bases without bisulfite conversion could revolutionize mapping of 5MC and 5HMC. For example, in single molecule real-time (SMRT) sequencing, fluorescently labeled nucleotides are incorporated by DNA polymerase on complementary DNA strands. Real-time monitoring of the kinetics of this process can identify both unmodified and modified bases, including N6-methyladenine, 5MC and 5HMC[94]. SMRT sequencing has also been combined with selective glucosylation and cleavable biotin labeling of 5HMC to improve detection kinetics[23]. Similarly, the direct detection of modified bases via inexpensively produced nanopores, if they become amenable to high-throughput, could be technologically transformative [137].
Abbreviations
- 5MC
5-methylcytosine
- 5HMC
5hydroxymethylcytosine
- ChIP-seq
chromatin immunoprecipitation-sequencing
- MBD
methyl binding domain
- MeDIP
methyl DNA immunoprecipitation
- MRE
methyl-sensitive restriction enzyme
- RRBS
reduced representation bisulfite sequencing
REFERENCES
- 1.Trasler JM, Trasler DG, Bestor TH, Li E, Ghibu F. DNA methyltransferase in normal and Dnmtn/Dnmtn mouse embryos. Dev Dyn. 1996;206(3):239–247. doi: 10.1002/(SICI)1097-0177(199607)206:3<239::AID-AJA2>3.0.CO;2-J. 10.1002/(SICI)1097-0177(199607)206:3<239::AID-AJA2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 2.Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395(6697):89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 3.Maraschio P, Zuffardi O, Dalla Fior T, Tiepolo L. Immunodeficiency, centromeric heterochromatin instability of chromosomes 1, 9, and 16, and facial anomalies: the ICF syndrome. J Med Genet. 1988;25(3):173–180. doi: 10.1136/jmg.25.3.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu GL, Bestor TH, Bourc'his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402(6758):187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 5.Clark SJ, Harrison J, Frommer M. CpNpG methylation in mammalian cells. Nat Genet. 1995;10(1):20–27. doi: 10.1038/ng0595-20. [DOI] [PubMed] [Google Scholar]
- 6.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97(10):5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ziller MJ, Muller F, Liao J, Zhang Y, Gu H, Bock C, Boyle P, Epstein CB, Bernstein BE, Lengauer T, Gnirke A, Meissner A. Genomic Distribution and Inter-Sample Variation of Non-CpG Methylation across Human Cell Types. PLoS Genet. 2011;7(12):e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z, Zhu Q, Wysocka J, Ye Y, Khochbin S, Ren B, Zhao Y. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008. 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37(8):853–862. doi: 10.1038/ng1598. 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 15.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66(16):7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 16.Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, Kernstine KH, Riggs AD, Pfeifer GP. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A. 2007;104(13):5527–5532. doi: 10.1073/pnas.0701059104. 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci U S A. 2009;106(3):671–678. doi: 10.1073/pnas.0812399106. 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfeifer GP, Riggs AD. Genomic sequencing by ligation-mediated PCR. Mol Biotechnol. 1996;5(3):281–288. doi: 10.1007/BF02900367. [DOI] [PubMed] [Google Scholar]
- 20.Clark SJ, Statham A, Stirzaker C, Molloy PL, Frommer M. DNA methylation: bisulphite modification and analysis. Nat Protoc. 2006;1(5):2353–2364. doi: 10.1038/nprot.2006.324. 10.1038/nprot.2006.324. [DOI] [PubMed] [Google Scholar]
- 21.Gitan RS, Shi H, Chen CM, Yan PS, Huang TH. Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 2002;12(1):158–164. doi: 10.1101/gr.202801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. doi: 10.1038/nature10102. 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29(1):68–72. doi: 10.1038/nbt.1732. 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C, Hatchwell E, Selzer RR, Richmond TA, Green RD, Melnick A, Greally JM. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16(8):1046–1055. doi: 10.1101/gr.5273806. 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oda M, Glass JL, Thompson RF, Mo Y, Olivier EN, Figueroa ME, Selzer RR, Richmond TA, Zhang X, Dannenberg L, Green RD, Melnick A, Hatchwell E, Bouhassira EE, Verma A, Suzuki M, Greally JM. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res. 2009;37(12):3829–3839. doi: 10.1093/nar/gkp260. 10.1093/nar/gkp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, Oyolu CB, Schroth GP, Absher DM, Baker JC, Myers RM. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009;19(6):1044–1056. doi: 10.1101/gr.088773.108. 10.1101/gr.088773.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27(4):361–368. doi: 10.1038/nbt.1533. 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, Polyak K. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37(8):899–905. doi: 10.1038/ng1596. 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 29.Bloushtain-Qimron N, Yao J, Snyder EL, Shipitsin M, Campbell LL, Mani SA, Hu M, Chen H, Ustyansky V, Antosiewicz JE, Argani P, Halushka MK, Thomson JA, Pharoah P, Porgador A, Sukumar S, Parsons R, Richardson AL, Stampfer MR, Gelman RS, Nikolskaya T, Nikolsky Y, Polyak K. Cell type-specific DNA methylation patterns in the human breast. roc Natl Acad Sci U S A. 2008;105(37):14076–14081. doi: 10.1073/pnas.0805206105. 10.1073/pnas.0805206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–257. doi: 10.1038/nature09165. 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, Olshen A, Ballinger T, Zhou X, Forsberg KJ, Gu J, Echipare L, O'Geen H, Lister R, Pelizzola M, Xi Y, Epstein CB, Bernstein BE, Hawkins RD, Ren B, Chung WY, Gu H, Bock C, Gnirke A, Zhang MQ, Haussler D, Ecker JR, Li W, Farnham PJ, Waterland RA, Meissner A, Marra MA, Hirst M, Milosavljevic A, Costello JF. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28(10):1097–1105. doi: 10.1038/nbt.1682. 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabinowicz PD, Schutz K, Dedhia N, Yordan C, Parnell LD, Stein L, McCombie WR, Martienssen RA. Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat Genet. 1999;23(3):305–308. doi: 10.1038/15479. [DOI] [PubMed] [Google Scholar]
- 33.Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18(5):780–790. doi: 10.1101/gr.7301508. 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38(11):e125. doi: 10.1093/nar/gkq223. 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 37.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, Min J, Nicholson T, Chen T, Xu G, Shi Y, Zhang K, Shi YG. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42(4):451–464. doi: 10.1016/j.molcel.2011.04.005. 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–348. doi: 10.1038/nature10066. 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12(6):R54. doi: 10.1186/gb-2011-12-6-r54. 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, Simon I. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38(2):149–153. doi: 10.1038/ng1719. 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 41.Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2(2):160–169. doi: 10.1016/j.stem.2007.12.011. 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD, Fan G. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci U S A. 2008;105(12):4709–4714. doi: 10.1073/pnas.0712018105. 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126(6):1189–1201. doi: 10.1016/j.cell.2006.08.003. 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 44.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39(1):61–69. doi: 10.1038/ng1929. 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 45.Robinson MD, Stirzaker C, Statham AL, Coolen MW, Song JZ, Nair SS, Strbenac D, Speed TP, Clark SJ. Evaluation of affinity-based genome-wide DNA methylation data: effects of CpG density, amplification bias, and copy number variation. Genome Res. 2010;20(12):1719–1729. doi: 10.1101/gr.110601.110. 10.1101/gr.110601.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson MD, Statham AL, Speed TP, Clark SJ. Protocol matters: which methylome are you actually studying? Epigenomics. 2010;2(4):587–598. doi: 10.2217/epi.10.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, Thorne NP, Backdahl L, Herberth M, Howe KL, Jackson DK, Miretti MM, Marioni JC, Birney E, Hubbard TJ, Durbin R, Tavare S, Beck S. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26(7):779–785. doi: 10.1038/nbt1414. 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pomraning KR, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47(3):142–150. doi: 10.1016/j.ymeth.2008.09.022. 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 49.Ruike Y, Imanaka Y, Sato F, Shimizu K, Tsujimoto G. Genome-wide analysis of aberrant methylation in human breast cancer cells using methyl-DNA immunoprecipitation combined with high-throughput sequencing. BMC Genomics. 2010;11:137. doi: 10.1186/1471-2164-11-137. 10.1186/1471-2164-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li N, Ye M, Li Y, Yan Z, Butcher LM, Sun J, Han X, Chen Q, Zhang X, Wang J. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods. 2010;52(3):203–212. doi: 10.1016/j.ymeth.2010.04.009. 10.1016/j.ymeth.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 51.Chavez L, Jozefczuk J, Grimm C, Dietrich J, Timmermann B, Lehrach H, Herwig R, Adjaye J. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 2010;20(10):1441–1450. doi: 10.1101/gr.110114.110. 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28(10):1106–1114. doi: 10.1038/nbt.1681. 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feber A, Wilson GA, Zhang L, Presneau N, Idowu B, Down TA, Rakyan VK, Noon LA, Lloyd AC, Stupka E, Schiza V, Teschendorff AE, Schroth GP, Flanagan A, Beck S. Comparative methylome analysis of benign and malignant peripheral nerve sheath tumors. Genome Res. 2011;21(4):515–524. doi: 10.1101/gr.109678.110. 10.1101/gr.109678.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pelizzola M, Koga Y, Urban AE, Krauthammer M, Weissman S, Halaban R, Molinaro AM. MEDME: an experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res. 2008;18(10):1652–1659. doi: 10.1101/gr.080721.108. 10.1101/gr.080721.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brinkman AB, Simmer F, Ma K, Kaan A, Zhu J, Stunnenberg HG. Whole-genome DNA methylation profiling using MethylCap-seq. Methods. 2010;52(3):232–236. doi: 10.1016/j.ymeth.2010.06.012. 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 56.Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 38(2):391–399. doi: 10.1093/nar/gkp992. 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lan X, Adams C, Landers M, Dudas M, Krissinger D, Marnellos G, Bonneville R, Xu M, Wang J, Huang TH, Meredith G, Jin VX. High resolution detection and analysis of CpG dinucleotides methylation using MBD-Seq technology. PLoS One. 2011;6(7):e22226. doi: 10.1371/journal.pone.0022226. PONE-D-11-02256 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA, Koebbe BC, Nielsen C, Hirst M, Farnham P, Kuhn RM, Zhu J, Smirnov I, Kent WJ, Haussler D, Madden PA, Costello JF, Wang T. The human epigenome browser at washington university. Nat Methods. 2011;8(12):989–990. doi: 10.1038/nmeth.1772. nmeth.1772 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karpf AR, Peterson PW, Rawlins JT, Dalley BK, Yang Q, Albertsen H, Jones DA. Inhibition of DNA methyltransferase stimulates the expression of signal transducer and activator of transcription 1, 2, and 3 genes in colon tumor cells. Proc Natl Acad Sci U S A. 1999;96(24):14007–14012. doi: 10.1073/pnas.96.24.14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, Sato F, Meltzer SJ, Sidransky D. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2(6):485–495. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 61.Foltz G, Yoon JG, Lee H, Ryken TC, Sibenaller Z, Ehrich M, Hood L, Madan A. DNA methyltransferase-mediated transcriptional silencing in malignant glioma: a combined whole-genome microarray and promoter array analysis. Oncogene. 2009 doi: 10.1038/onc.2009.122. 10.1038/onc.2009.122. [DOI] [PubMed] [Google Scholar]
- 62.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21(1):103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 63.Shames DS, Girard L, Gao B, Sato M, Lewis CM, Shivapurkar N, Jiang A, Perou CM, Kim YH, Pollack JR, Fong KM, Lam CL, Wong M, Shyr Y, Nanda R, Olopade OI, Gerald W, Euhus DM, Shay JW, Gazdar AF, Minna JD. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med. 2006;3(12):e486. doi: 10.1371/journal.pmed.0030486. 10.1371/journal.pmed.0030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gius D, Cui H, Bradbury CM, Cook J, Smart DK, Zhao S, Young L, Brandenburg SA, Hu Y, Bisht KS, Ho AS, Mattson D, Sun L, Munson PJ, Chuang EY, Mitchell JB, Feinberg AP. Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell. 2004;6(4):361–371. doi: 10.1016/j.ccr.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 65.Mueller W, Nutt CL, Ehrich M, Riemenschneider MJ, von Deimling A, van den Boom D, Louis DN. Downregulation of RUNX3 and TES by hypermethylation in glioblastoma. Oncogene. 2007;26(4):583–593. doi: 10.1038/sj.onc.1209805. 10.1038/sj.onc.1209805. [DOI] [PubMed] [Google Scholar]
- 66.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33(18):5868–5877. doi: 10.1093/nar/gki901. 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. High-throughput bisulfite sequencing in mammalian genomes. Methods. 2009;48(3):226–232. doi: 10.1016/j.ymeth.2009.05.003. 10.1016/j.ymeth.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6(4):468–481. doi: 10.1038/nprot.2010.190. 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 73.Xiang H, Zhu J, Chen Q, Dai F, Li X, Li M, Zhang H, Zhang G, Li D, Dong Y, Zhao L, Lin Y, Cheng D, Yu J, Sun J, Zhou X, Ma K, He Y, Zhao Y, Guo S, Ye M, Guo G, Li Y, Li R, Zhang X, Ma L, Kristiansen K, Guo Q, Jiang J, Beck S, Xia Q, Wang W, Wang J. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat Biotechnol. 2010;28(5):516–520. doi: 10.1038/nbt.1626. 10.1038/nbt.1626. [DOI] [PubMed] [Google Scholar]
- 74.Schroeder DI, Lott P, Korf I, LaSalle JM. Large-scale methylation domains mark a functional subset of neuronally expressed genes. Genome Res. 2011;21(10):1583–1591. doi: 10.1101/gr.119131.110. 10.1101/gr.119131.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010;107(19):8689–8694. doi: 10.1073/pnas.1002720107. 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–919. doi: 10.1126/science.1186366. 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 77.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20(3):320–331. doi: 10.1101/gr.101907.109. 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, Camargo AA, Stevenson BJ, Ecker JR, Bafna V, Strausberg RL, Simpson AJ, Ren B. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2011 doi: 10.1101/gr.125872.111. 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, Van Den Berg D, Laird PW. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2011;44(1):40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572. doi: 10.1093/bioinformatics/btr167. 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xi Y, Bock C, Muller F, Sun D, Meissner A, Li W. RRBSMAP: A Fast, Accurate and User-friendly Alignment Tool for Reduced Representation Bisulfite Sequencing. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr668. 10.1093/bioinformatics/btr668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102(44):15785–15790. doi: 10.1073/pnas.0507816102. 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raval A, Tanner SM, Byrd JC, Angerman EB, Perko JD, Chen SS, Hackanson B, Grever MR, Lucas DM, Matkovic JJ, Lin TS, Kipps TJ, Murray F, Weisenburger D, Sanger W, Lynch J, Watson P, Jansen M, Yoshinaga Y, Rosenquist R, de Jong PJ, Coggill P, Beck S, Lynch H, de la Chapelle A, Plass C. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129(5):879–890. doi: 10.1016/j.cell.2007.03.043. 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, Daley GQ, Eggan K, Hochedlinger K, Thomson J, Wang W, Gao Y, Zhang K. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27(4):353–360. doi: 10.1038/nbt.1530. 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62(14):4075–4080. [PubMed] [Google Scholar]
- 88.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17(3):637–641. doi: 10.1038/sj.leu.2402834. 2402834 [pii] [DOI] [PubMed] [Google Scholar]
- 89.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 5(1):e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5(12):e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38(19):e181. doi: 10.1093/nar/gkq684. 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kinney SM, Chin HG, Vaisvila R, Bitinaite J, Zheng Y, Esteve PO, Feng S, Stroud H, Jacobsen SE, Pradhan S. Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. J Biol Chem. 2011;286(28):24685–24693. doi: 10.1074/jbc.M110.217083. 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7(6):461–465. doi: 10.1038/nmeth.1459. 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ke X-S, Qu Y, Rostad K, Li W-C, Lin B, Halvorsen OJ, Haukaas SA, Jonassen I, Petersen K, Goldfinger N, Rotter V, Akslen LA, Oyan AM, Kalland K-H. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, PÈrez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37 doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 97.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RGAB, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419 doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 98.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA. A Chromatin-Mediated Reversible Drug-Tolerant State in Cancer Cell Subpopulations. Cell. 2010;141 doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10 doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O'Neill LP, Turner BM, Turner B. ChIP with Native Chromatin: Advantages and Problems Relative to Methods Using Cross-Linked Material. Methods. 2003;31 [PubMed] [Google Scholar]
- 101.Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci. 2000;25 doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 102.Barski A, Zhao K. Genomic location analysis by ChIP-Seq. J Cell Biochem. 2009;107 doi: 10.1002/jcb.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dingwall C, Lomonossoff GP, Laskey RA. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981;9 doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hörz W, Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981;9 doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18 doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10 doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26 doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25 doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 109.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24 doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 110.Coarfa C, Yu F, Miller CA, Chen Z, Harris RA, Milosavljevic A. Pash 3.0: A versatile software package for read mapping and integrative analysis of genomic and epigenomic variation using massively parallel DNA sequencing. BMC Bioinformatics. 2010;11:572. doi: 10.1186/1471-2105-11-572. 10.1186/1471-2105-11-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat Methods. 2009;6 doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu H, Handoko L, Wei X, Ye C, Sheng J, Wei C-L, Lin F, Sung W-K. A signal-noise model for significance analysis of ChIP-seq with negative control. Bioinformatics. 2010;26 doi: 10.1093/bioinformatics/btq128. [DOI] [PubMed] [Google Scholar]
- 113.Xu H, Wei C-L, Lin F, Sung W-K. An HMM approach to genome-wide identification of differential histone modification sites from ChIP-seq data. Bioinformatics. 2008;24 doi: 10.1093/bioinformatics/btn402. [DOI] [PubMed] [Google Scholar]
- 114.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25 doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129 doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 116.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316 doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 117.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448 doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39 doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 119.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459 doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011 doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wasserman NF, Aneas I, Nobrega MA. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 2010;20 doi: 10.1101/gr.105361.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Adli M, Bernstein BE. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat Protoc. 2011;6 doi: 10.1038/nprot.2011.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Goren A, Ozsolak F, Shoresh N, Ku M, Adli M, Hart C, Gymrek M, Zuk O, Regev A, Milos PM, Bernstein BE. Chromatin profiling by directly sequencing small quantities of immunoprecipitated DNA. Nat Methods. 2010;7 doi: 10.1038/nmeth.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295 doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 125.Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38 doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 126.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16 doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326 doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38 doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 129.Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods. 2007;4 doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 130.Fullwood MJ, Wei C-L, Liu ET, Ruan Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19 doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Simonis M, Klous P, Homminga I, Galjaard R-J, Rijkers E-J, Grosveld F, Meijerink JPP, de Laat W. High-resolution identification of balanced and complex chromosomal rearrangements by 4C technology. Nat Methods. 2009;6 doi: 10.1038/nmeth.1391. [DOI] [PubMed] [Google Scholar]
- 132.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, Berman DM, Jenuwein T, Pruitt K, Sharkis SJ, Watkins DN, Herman JG, Baylin SB. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39(2):237–242. doi: 10.1038/ng1972. 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39(2):157–158. doi: 10.1038/ng1941. 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 134.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, Bergman Y, Simon I, Cedar H. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39(2):232–236. doi: 10.1038/ng1950. 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 135.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O'Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL, Ueno T, Soda M, Hamada T, Haruta H, Takada S, Miyazaki Y, Kiyoi H, Ito E, Naoe T, Tomonaga M, Toyota M, Tajima S, Iwama A, Mano H. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29(25):3723–3731. doi: 10.1038/onc.2010.117. 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- 137.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4(4):265–270. doi: 10.1038/nnano.2009.12. 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]