Abstract
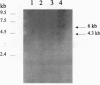
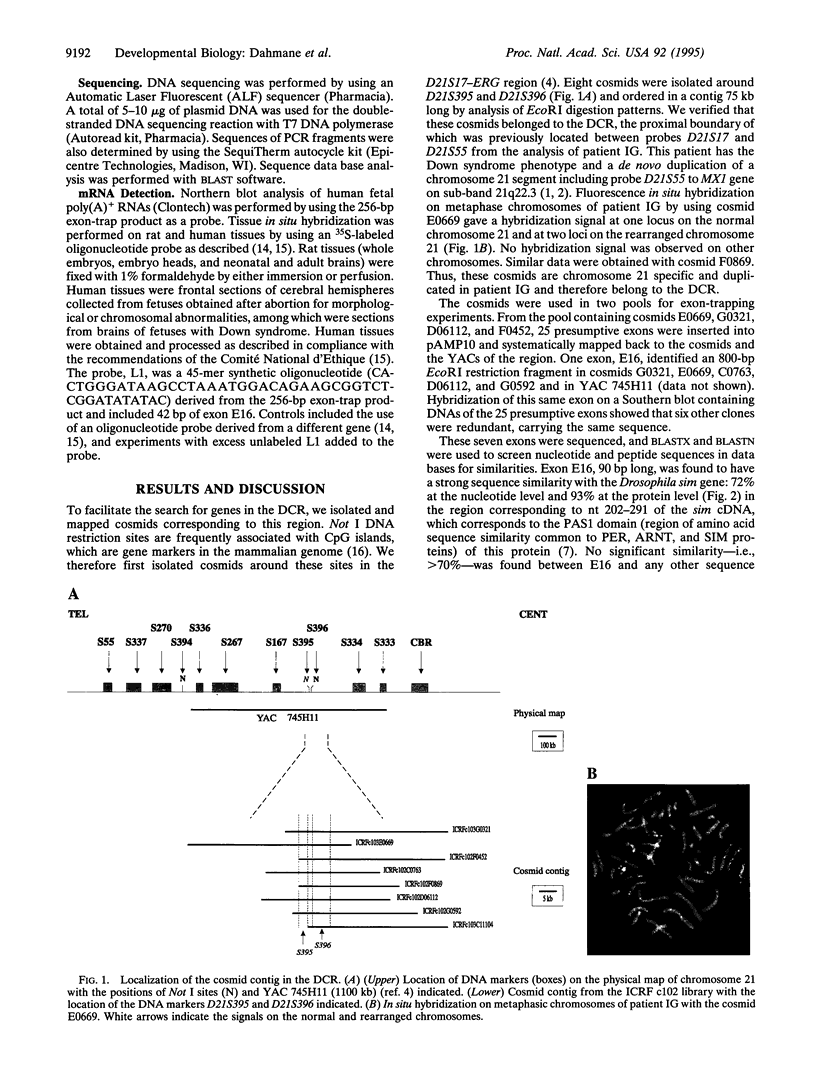
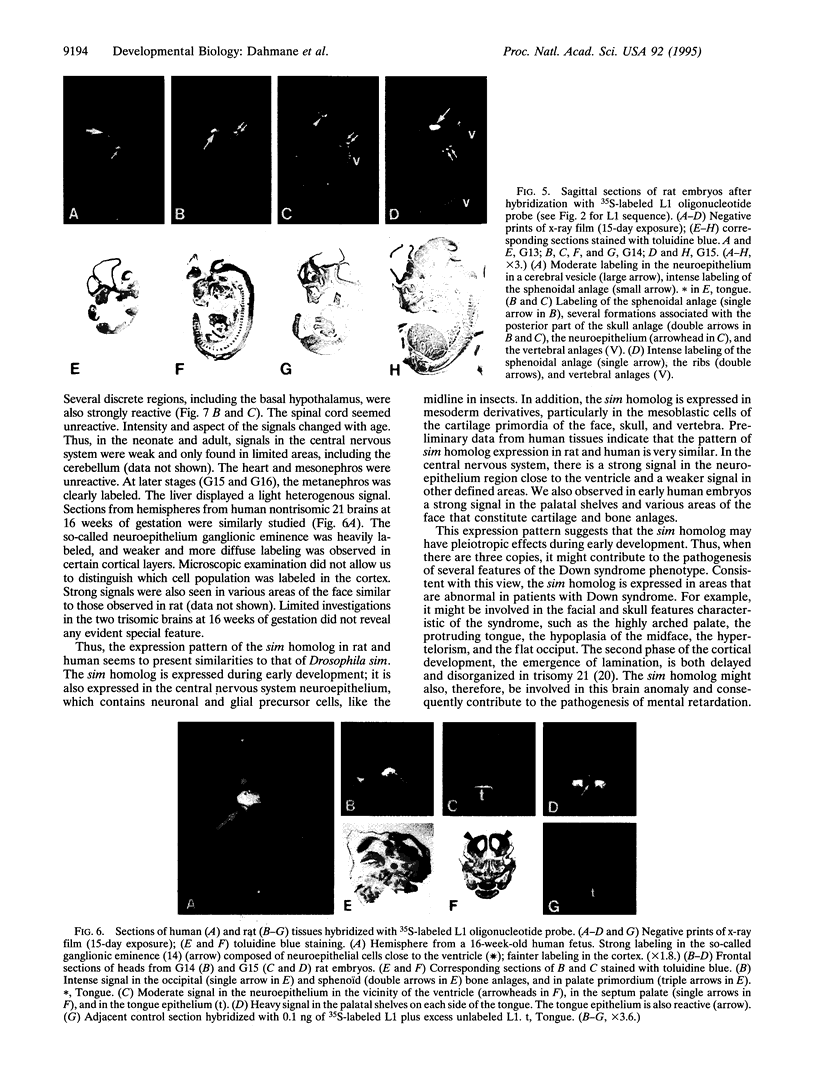
Many features of Down syndrome might result from the overdosage of only a few genes located in a critical region of chromosome 21. To search for these genes, cosmids mapping in this region were isolated and used for trapping exons. One of the trapped exons obtained has a sequence very similar to part of the Drosophila single-minded (sim) gene, a master regulator of the early development of the fly central nervous system midline. Mapping data indicated that this exonic sequence is only present in the Down syndrome-critical region in the human genome. Hybridization of this exonic sequence with human fetal kidney poly(A)+ RNA revealed two transcripts of 6 and 4.3 kb. In situ hybridization of a probe derived from this exon with human and rat fetuses showed that the corresponding gene is expressed during early fetal life in the central nervous system and in other tissues, including the facial, skull, palate, and vertebra primordia. The expression pattern of this gene suggests that it might be involved in the pathogenesis of some of the morphological features and brain anomalies observed in Down syndrome.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aronson D. C., Jansweijer M. C., Hoovers J. M., Barth P. G. A male infant with holoprosencephaly, associated with ring chromosome 21. Clin Genet. 1987 Jan;31(1):48–52. doi: 10.1111/j.1399-0004.1987.tb02766.x. [DOI] [PubMed] [Google Scholar]
- Bloch B., Normand E., Kovesdi I., Böhlen P. Expression of the HBNF (heparin-binding neurite-promoting factor) gene in the brain of fetal, neonatal and adult rat: an in situ hybridization study. Brain Res Dev Brain Res. 1992 Dec 18;70(2):267–278. doi: 10.1016/0165-3806(92)90206-c. [DOI] [PubMed] [Google Scholar]
- Chen H., Chrast R., Rossier C., Gos A., Antonarakis S. E., Kudoh J., Yamaki A., Shindoh N., Maeda H., Minoshima S. Single-minded and Down syndrome? Nat Genet. 1995 May;10(1):9–10. doi: 10.1038/ng0595-9. [DOI] [PubMed] [Google Scholar]
- Church D. M., Stotler C. J., Rutter J. L., Murrell J. R., Trofatter J. A., Buckler A. J. Isolation of genes from complex sources of mammalian genomic DNA using exon amplification. Nat Genet. 1994 Jan;6(1):98–105. doi: 10.1038/ng0194-98. [DOI] [PubMed] [Google Scholar]
- Crews S. T., Thomas J. B., Goodman C. S. The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the per gene product. Cell. 1988 Jan 15;52(1):143–151. doi: 10.1016/0092-8674(88)90538-7. [DOI] [PubMed] [Google Scholar]
- Crété N., Gosset P., Théophile D., Duterque-Coquillaud M., Blouin J. L., Vayssettes C., Sinet P. M., Créau-Goldberg N. Mapping the Down syndrome chromosome region. Establishment of a YAC contig spanning 1.2 megabases. Eur J Hum Genet. 1993;1(1):51–63. doi: 10.1159/000472387. [DOI] [PubMed] [Google Scholar]
- Delabar J. M., Theophile D., Rahmani Z., Chettouh Z., Blouin J. L., Prieur M., Noel B., Sinet P. M. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur J Hum Genet. 1993;1(2):114–124. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- Dufresne-Zacharia M. C., Dahmane N., Theophile D., Orti R., Chettouh Z., Sinet P. M., Delabar J. M. 3.6-Mb genomic and YAC physical map of the Down syndrome chromosome region on chromosome 21. Genomics. 1994 Feb;19(3):462–469. doi: 10.1006/geno.1994.1095. [DOI] [PubMed] [Google Scholar]
- Estabrooks L. L., Rao K. W., Donahue R. P., Aylsworth A. S. Holoprosencephaly in an infant with a minute deletion of chromosome 21(q22.3). Am J Med Genet. 1990 Jul;36(3):306–309. doi: 10.1002/ajmg.1320360312. [DOI] [PubMed] [Google Scholar]
- Golden J. A., Hyman B. T. Development of the superior temporal neocortex is anomalous in trisomy 21. J Neuropathol Exp Neurol. 1994 Sep;53(5):513–520. doi: 10.1097/00005072-199409000-00011. [DOI] [PubMed] [Google Scholar]
- Gosset P., Crété N., Ait Ghezala G., Théophile D., Van Broeckhoven C., Vayssettes C., Sinet P. M., Créau N. A high-resolution map of 1.6 Mb in the Down syndrome region: a new map between D21S55 and ETS2. Mamm Genome. 1995 Feb;6(2):127–130. doi: 10.1007/BF00303257. [DOI] [PubMed] [Google Scholar]
- Ichikawa H., Hosoda F., Arai Y., Shimizu K., Ohira M., Ohki M. A NotI restriction map of the entire long arm of human chromosome 21. Nat Genet. 1993 Aug;4(4):361–366. doi: 10.1038/ng0893-361. [DOI] [PubMed] [Google Scholar]
- Itoh S., Kamataki T. Human Ah receptor cDNA: analysis for highly conserved sequences. Nucleic Acids Res. 1993 Jul 25;21(15):3578–3578. doi: 10.1093/nar/21.15.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. O., Crews S. T. Genetic analysis of the Drosophila single-minded gene reveals a central nervous system influence on muscle development. Mech Dev. 1994 Nov;48(2):81–91. doi: 10.1016/0925-4773(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Lindsay S., Bird A. P. Use of restriction enzymes to detect potential gene sequences in mammalian DNA. 1987 May 28-Jun 3Nature. 327(6120):336–338. doi: 10.1038/327336a0. [DOI] [PubMed] [Google Scholar]
- Muralidhar M. G., Callahan C. A., Thomas J. B. Single-minded regulation of genes in the embryonic midline of the Drosophila central nervous system. Mech Dev. 1993 May;41(2-3):129–138. doi: 10.1016/0925-4773(93)90043-w. [DOI] [PubMed] [Google Scholar]
- Münke M. Clinical, cytogenetic, and molecular approaches to the genetic heterogeneity of holoprosencephaly. Am J Med Genet. 1989 Oct;34(2):237–245. doi: 10.1002/ajmg.1320340222. [DOI] [PubMed] [Google Scholar]
- Nambu J. R., Lewis J. O., Crews S. T. The development and function of the Drosophila CNS midline cells. Comp Biochem Physiol Comp Physiol. 1993 Mar;104(3):399–409. doi: 10.1016/0300-9629(93)90439-b. [DOI] [PubMed] [Google Scholar]
- Nambu J. R., Lewis J. O., Wharton K. A., Jr, Crews S. T. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991 Dec 20;67(6):1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Nizetić D., Zehetner G., Monaco A. P., Gellen L., Young B. D., Lehrach H. Construction, arraying, and high-density screening of large insert libraries of human chromosomes X and 21: their potential use as reference libraries. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3233–3237. doi: 10.1073/pnas.88.8.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani Z., Blouin J. L., Creau-Goldberg N., Watkins P. C., Mattei J. F., Poissonnier M., Prieur M., Chettouh Z., Nicole A., Aurias A. Critical role of the D21S55 region on chromosome 21 in the pathogenesis of Down syndrome. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5958–5962. doi: 10.1073/pnas.86.15.5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisz-Porszasz S., Probst M. R., Fukunaga B. N., Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor nuclear translocator protein (ARNT). Mol Cell Biol. 1994 Sep;14(9):6075–6086. doi: 10.1128/mcb.14.9.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. B., Crews S. T., Goodman C. S. Molecular genetics of the single-minded locus: a gene involved in the development of the Drosophila nervous system. Cell. 1988 Jan 15;52(1):133–141. doi: 10.1016/0092-8674(88)90537-5. [DOI] [PubMed] [Google Scholar]