Abstract
The RNA polymerase II (Pol-II) holoenzyme, responsible for messenger RNA production, typically consists of 10–12 subunits. Our laboratory previously demonstrated that maternally deposited, long, noncoding, template RNAs are essential for programmed genome rearrangements in the ciliate Oxytricha trifallax. Here we show that such RNAs are bidirectionally transcribed and transported to the zygotic nucleus. The gene encoding the second-largest subunit of Pol-II, Rpb2, has undergone gene duplication, and the two paralogs, Rpb2-a and -b, display different expression patterns. Immunoprecipitation of double-stranded RNAs identified an association with Rpb2-a. Through immunoprecipitation and mass spectrometry, we show that Rpb2-a in early zygotes appears surprisingly unassociated with other Pol II subunits. A partial loss of function of Rpb2-a leads to an increase in expression of transposons and other germline-limited satellite repeats. We propose that evolutionary divergence of the Rpb2 paralogs has led to acquisition of transcription-independent functions during sexual reproduction that may contribute to the negative regulation of germline gene expression.
Keywords: genome integrity, RNA polymerase, noncoding RNAs, transcription
RNA polymerases play conserved roles in transcription throughout the three domains of life. RNA polymerase II (Pol-II) is a holoenzyme consisting of 10 core subunits and 2 accessory subunits and is responsible for messenger RNA (mRNA) transcription in eukaryotes (Cramer et al. 2000). The accessory subunits, Rpb4 and Rpb7, considered dispensable for transcription, play additional roles in mRNA decay and translation initiation (Lotan et al. 2005; Lotan et al. 2007; Harel-Sharvit et al. 2010). Rpb2, the second-largest subunit of Pol II, interacts with Argonaute 1 (Ago1) and directs RNA interference (RNAi)-mediated heterochromatin silencing at centromeres in Schizosaccharomyces pombe (Kato et al. 2005). Similarly, Rpb7 can promote presmall interfering RNA (pre-siRNA) transcription in S. pombe (Djupedal et al. 2005). Besides these two cases of subfunctionalization of individual subunits, the Pol II holoenzyme is responsible for mRNA transcription. The essential core components of RNA polymerase are well conserved across the tree of life (Zhang et al. 1999; Cramer et al. 2001; Hirata et al. 2008; Korkhin et al. 2009).
Here we describe a duplication of the Rpb2 gene in the ciliate Oxytricha trifallax and its role beyond mRNA transcription in early zygotes. Of the two paralogous copies of Rpb2, one of them, Rpb2-a, has exclusive expression during the sexual phase of the life cycle and displays a dynamic localization pattern, largely independent of the active transcription machinery. Like other ciliates, Oxytricha exhibits nuclear duality within the same cell (Prescott 1994). The gene-rich macronucleus (MAC) is responsible for asexual (vegetative) growth while the germline micronucleus (MIC) is transcriptionally silent during the vegetative stage of the life cycle, but supplies the precursor for the zygotic macronucleus following sexual reproduction. Following meiosis in Oxytricha, a copy of the micronucleus undergoes programmed deletion of ∼95% of the DNA complexity, including transposons and other repetitive elements (Lauth et al. 1976), followed by genome-wide DNA rearrangements that produce a zygotic macronucleus (Prescott 1994) that contains mostly single-gene “nanochromosomes” (Swart et al. 2013) (average 3.2 kb). Previously, Nowacki et al. (2008) reported the expression of maternal, long, noncoding transcripts from the parental macronucleus early in conjugation (Nowacki et al. 2008). These long, noncoding RNAs (lncRNAs) act as a maternal cache that guides DNA rearrangements and can transfer nucleotide substitutions to the genomic DNA in the progeny (Nowacki et al. 2008). Here we show that Rpb2-a, along with the Oxytricha Piwi-1 homolog, Otiwi-1, associates in a putative complex with these lncRNAs and is likely involved in their regulation. We used immunoprecipitation and liquid chromatography–mass spectrometry (LC-MS) to identify and confirm that Rpb2-a is not part of the Pol-II complex in early zygotes. A partial loss of function of Rpb2-a or Otiwi-1, independently, is lethal and leads to an increase in germline gene expression, suggesting a role in maintenance of genome integrity during sexual development in O. trifallax.
Materials and Methods
Cell culture
Two cross-fertile strains of O. trifallax, JRB310 and JRB510, were used in the study. Cells were maintained in Pringsheim buffer [0.11 mM Na2HPO4, 0.08 mM MgSO4, 0.85 mM Ca(NO3)2, 0.35 mM KCl, pH 7.0] with Chlamydomonas reinhardtii and Klebsiella as the food source, as in Fang et al. 2012. To initiate conjugation, cells were starved and mixed in equal numbers after food depletion. Cells started mating 2–3 hr postmixing with 80–90% conjugation efficiency.
Antisense oligonucleotide knockdown
Targeted mRNA (Rpb2-a and Otiwi1) structures were predicted using Mfold (http://mfold.rna.albany.edu/?q=mfold) with default parameters. Unpaired regions illustrated by multiple low-energy folding structures were chosen to design antisense oligonucleotides (Patzel et al. 1999). In knockdown experiments, we mixed six (against Rpb2-a) or eight (against Otiwi1) different oligonucleotides with phosphorothioate linkages in their backbones (IDT DNA Coralville, Iowa) and injected them into parental macronuclei of cells at 2 hr postmixing. Roughly 50 pairs were injected and RNA was harvested from injected cells at 24 hr postmixing for RT–semiquantitative PCR (qPCR) analysis. The experiment was repeated at least three times.
Immunostaining
Cells were centrifuged at 200 × g, followed by fixation using 4% paraformaldehyde at room temperature for 10 min and washing with PBS. Cells were affixed onto polylysine-coated slides, permeabilized with 0.5% Triton-X 100, and immunostained with the following antibodies: Rpb1 (CTD4H8; Millipore, Bedford, MA), J2 (English and Scientific Consulting Bt.), and Rpb2 (Abcam). Anti-mouse and -rabbit Alexa-488 secondary antibodies (Invitrogen, Carlsbad, CA) were used for detection. DAPI (Invitrogen) was used to stain DNA. The images were captured on Nikon (Garden City, NY) A1-RS and Zeiss (Carl Zeiss, Thornwood, NY) LSM 510 confocal microscopes and processed using ImageJ (Schneider et al. 2012) and Adobe Photoshop.
Reverse transcriptase and quantitative PCR
RNA was isolated from cells at indicated developmental time points by using Trizol (Invitrogen) or the mirVana RNA isolation kit (Ambion). Residual DNA was digested from RNA samples, using Turbo-DNA free (Invitrogen). First-strand complementary DNA (cDNA) synthesis was done using Superscript III enzyme (Invitrogen) and random hexamers or oligo(dT) primers, following manufacturer’s instructions. Reverse transcription to identify lncRNAs was performed as described previously (Nowacki et al. 2008). The cDNA was used as the template in the qPCR, using Power Sybr Green master mix (Applied Biosystems, Foster City, CA). The data were analyzed using the ddCt method (Schmittgen and Livak 2008). The experiments were performed in triplicate and the average values were plotted with error bars representing SEM.
Following is the list of primers used in qPCR (5′–3′) (F, forward; rev, reverse):
Pol-α 5′ F: CCAAAACCCCTCAATTCAAA
rev: TCGCAAACTAGACTGTTCACAAA
Pol-α 3′ F: CCTCAGCAGCATATTCAACTTC
rev: CCTGAGTGTAAATTGATTTCTTTGA
Contig 8213 F: TGAAGTCAGGCATCAAACTACG
rev: TACCATGTGCCTTGTTTGTTCT
28S-rDNA F: GGTAAGAACCCTGGCCTTTC
rev: ATCTGATGAGCGTGCAGTTG
17S-rDNA F: GTGCCAATGTCGTCTTGTTG
rev: AGCCTGCCTGGAACACTCTA
RTel-1 F: GACCCTTTAACTAATCAATGGAAGAGA
rev: AAGGGCGTTTGAACACATCTTT
Contig 17756.0 F: CGTAGTAGGGGGTCTCCAAAAA
rev: TGTATCTATGATAGAATGCTGGAGTGAATA
Rpb2-a F: CAGCAGCCAATGAGGGTCAA
rev: TGACAGTCCAGGCGTCATCC
Rpb2-b F: GCATGATATCGCATGGTGCAG
rev: GCTTGACAAATCAGGCCACA
380bp F: TGAGATGACCTGGAATTAAGCA
rev: TATATCTGGTGTTCCGGTGTCA
Spt5 F: CATCCAAGTGAACCCCGACT
rev: TGTTGTGGTTGCGATCCAGT
RTel-MIC F: ATCCAATTGCTGCCCATAATTT
rev: CCATGAAATCCAAAGACACAGC
Rpb2-a-MIC F: AAAGGAAATTTGGCTTTAGATTTTG
rev: TCGATCTAATCTCTTAATATATACAACTCT.
Primers to amplify the 170-bp satellite repeat (Bracht et al. 2012), TEBP-α (Nowacki et al. 2008), and TBE1 (Nowacki et al. 2009) have been previously described.
Chromatin immunoprecipitation and RNA immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using a published protocol with some modifications (Khurana et al. 2010). Cells from 12 or 24 hr postmixing were fixed in 1% formaldehyde for 10 min. Cross-linked chromatin was sheared using a Covaris Ultrasonicator to generate 200–600 bp DNA size. Anti-Rpb1 antibody (4 μg) was used to precipitate chromatin. Mouse IgG was used as the negative control. After DNA elution and purification, the samples were subjected to qPCR for validation. Illumina libraries were prepared using manufacturer’s instructions. For RNA immunoprecipitation (RNA-IP), the ChIP protocol was used with some modifications. Before IP, DNA was digested using DNase I (Roche). The RNA was eluted from magnetic beads (Dynabeads, Invitrogen) and reverse transcribed.
ChIP and deep sequencing analysis
After quality trimming (using Phred quality threshold 20), 12,303,838 input reads and 19,913,676 Pol-II ChIP reads were mapped to the MAC genome [17,107 full-length telomere-masked MAC chromosomes (Swart et al. 2013)] and the mitochondrial genome (Swart et al. 2012) with BLAT (Kent 2002), using default parameters, and then filtered for paired reads aligned in full length with at least 94% identity. ChIP and deep sequencing (ChIP-seq) reads were normalized to input by comparing number of reads mapping to the Oxytricha mitochondrial genome. The normalization factor is 7.83 for Rpb1 ChIP. For each MAC nanochromosome, input reads were subtracted from normalized Rpb1 ChIP reads. All 17,107 full-length MAC chromosomes are covered by normalized Rpb1 ChIP-seq reads. The correlation between input and Rpb1 ChIP and MAC genomic DNA copy number and mRNA-seq level was assessed for 13,410 full-length MAC chromosomes encoding single genes.
Illumina sequence reads from the MAC genomic library were mapped to 13,410 full-length MAC nanochromosomes encoding single genes to determine the copy number of each nanochromosome. RNA-seq reads for cells collected at 10 hr were mapped to the same set of nanochromosomes, using BLAT to determine gene expression level. Numbers of reads mapped to each nanochromosome were plotted for different data sets. Enrichment at 5′ and 3′ regions was assessed relative to the gene body in Rpb1 ChIP-seq samples. For 13,410 full-length single-gene MAC chromosomes, we calculated coverage of the 100-bp regions 5′ and 3′ to each encoded gene and the body of the gene for Rpb1 ChIP and normalized each coverage value to input. The coverage of 5′ and 3′ regions was then normalized to the gene body and plotted as 1 kb of the contig. A paired t-test was performed to calculate statistical significance between mapping at 5′ or 3′ ends (100 bp) to the middle of the contigs (800 bp). P-values are noted on Figure S2 E-G.
mRNA decay measurement
mRNA half-life was measured after treatment with Actinomycin D to stop active transcription. Cells were mixed in equal numbers and treated with 20 μg/ml Actinomycin D (Sigma, St. Louis) after 8 hr to achieve maximum pairing. RNA was collected at 4-hr intervals for six consecutive time points. After RNA isolation and cDNA synthesis, the amount of Dcl-1, Spt5, and Otiwi-1 transcripts was assayed by qPCR. Ct values were fitted onto the respective standard curves to analyze the amount of transcripts in different time points following Actinomycin addition. To prepare the standard curve, PCR for the aforementioned genes was performed on cDNA prepared from a 12-hr conjugation sample, followed by gel purification. The purified PCR products were serially diluted and used as a template in qPCR.
Co-immunoprecipitation and Western analysis
Mated cells were harvested at 12, 15, or 18 hr; washed twice in PBS; and lysed in buffer containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40, 2 mM EDTA, and 10% glycerol. The lysate was shaken at 4° for 30 min, followed by centrifugation at 16,000 × g for 10 min. The clarified extract was stored at −80° until needed. For immunoprecipitation, 5 μg of J2 or Rpb2 antibody was bound to Dynal magnetic beads (Invitrogen). Mouse or rabbit IgG was used as a negative control. Cell lysates were incubated with antibody-bound beads at 4° for 7–8 hr, followed by three washes in lysis buffer. For Western blotting, the beads were boiled in 2× Laemmli buffer at 95° for 10 min. The sample was run on a 6% SDS–polyacrylamide gel and blotted onto a PVDF membrane, using a semidry Western transfer apparatus (Bio-Rad, Hercules, CA). The membrane was blocked with 5% nonfat dry milk and incubated with Anti-Rpb2 or Anti-PiwiL1 (Abcam) antibody diluted 1:1000 and 1:5000, respectively. For samples submitted to mass spectrometric analysis, the immunoprecipitates were run on precast 4–20% Tris-Glycine gels (Thermo Scientific) and stained with GelCode blue staining reagent (Thermo Scientific).
Mass spectrometry sample preparation
Entire gel lanes were analyzed by division into gel slices, which were excised and diced into 1-mm3 cubes. Gel pieces were destained with ammonium bicarbonate buffer; washed extensively; and subjected to DTT thiol reduction, iodoacetamide alkylation, overnight trypsin digestion, and peptide elution as described in Shevchenko et al. (2006). Peptides were desalted using StageTip microscale reversed-phase chromatography (Rappsilber et al. 2003) and then subjected to reversed-phase nano-LC-MS and MS/MS performed on a nanoflow capillary high pressure HPLC system (Nano Ultra 2D Plus; Eksigent, Dublin, CA) coupled to an LTQ-Orbitrap XL hybrid mass spectrometer (ThermoFisher Scientific, Bremen, Germany) outfitted with a Triversa NanoMate ion source robot (Advion, Ithaca, NY) or an Easy nLC 1000 Ultra capillary UPLC system (Proxeon, Odense, Denmark) coupled to a VelosPro-Orbitrap Elite hybrid mass spectrometer (ThermoFisher Scientific) outfitted with a Flex ion source (Proxeon). Sample capture and washing were accomplished online, using a trapping capillary column (150 × ∼40 μm, packed with 3 µm 100-Å Magic AQ C18 resin; Michrom, Auburn, CA) at a flow rate of 4 µl/min for 9 min.
Separation was achieved using an analytical capillary column [75 µm × ∼20 cm, packed with 1.7 µm 100-Å BEH C18 resin (Waters Associates, Milford, MA)] and a linear gradient from 5 to 35% solutions A and B (solution A, 100% water/0.1% formic acid; solution B, 100% acetonitrile/0.1% formic acid) applied over 180 min at a flow rate of ∼250 nl/min. On the Orbi XL platform, nanospray ionization was carried out using the NanoMate ion source at 1.74 kV, with the LTQ heated capillary set to 200°, while on the Orbi Elite platform, nanospray was achieved using Picospray tips (New Objective, Woburn, MA), using a voltage of 2.1 kV, with the Velos heated capillary set to 250°. Full-scan mass spectra were acquired in the Orbitrap in positive-ion mode over the m/z range of 335–1800 at a resolution of 100,000 (Orbi XL) or 120,000 (Orbi Elite). MS/MS spectra were simultaneously acquired using CID in the LTQ for the 7 (Orbi XL) or 15 (Orbi Elite) most abundant multiply-charged species in the full-scan spectrum having signal intensities >1000 NL (normalized largest peak). All spectra were acquired in profile mode. Dynamic exclusion was set such that MS/MS was acquired only once for each species over a period of 120 sec.
Mass spectrometry data analysis
The resulting LC-MS/MS data were subjected to database searching and analysis, using the framework of ProteomeDiscoverer software (v. 1.4, ThermoFisher). The search was conducted using a workflow employing a Mascot search engine server module (v. 2.4, Matrix Science) to match against a proteomic database consisting of translations of open reading frames predicted to be coded by the Oxytricha MAC genome. Mascot search parameters included an initial mass error of 8 ppm for the precursor and 1.2 Da for the fragment ion species, two or fewer missed trypsin cleavages, and carbamidomethylation of cysteines as a fixed modification, with methionine oxidation and N-terminal protein acetylation as variable modifications. Aggregate search results were subjected to statistical analysis and filtering to a 5% peptide-level false discovery rate (FDR), using the Percolator support vector machine module (Kall et al. 2007), which employs semisupervised machine learning to dynamically discriminate between correct and decoy spectral matches, using multiple-match metrics. Fragmentation spectral assignments were subjected to manual inspection and validation using ProteomeDiscoverer and the original tandem mass spectra acquired in profile mode, using Xcalibur software (ThermoFisher).
Results and Discussion
Pol-II involvement in bidirectional transcription of lncRNAs during Oxytricha development
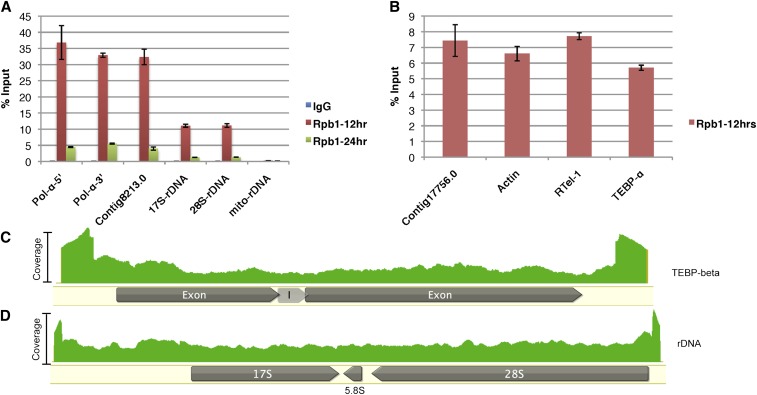
Rpb1, the largest subunit of Pol-II, localizes to both macronuclei and micronuclei during nuclear development when lncRNA production is at its peak (Nowacki et al. 2008) (Supporting Information, Figure S1). This suggested that Pol-II is responsible for lncRNA transcription, which we tested by ChIP-seq, using an antibody that detects total and serine-2 phosphorylated (active) forms of Rpb1. The specificity of this commercially available antibody was tested via immunoprecipitation, followed by mass spectrometry from 24 hr postmixing (Table 1). Mapping of Illumina 100-bp paired-end reads onto the O. trifallax macronuclear genome (Swart et al. 2013) revealed the presence of Pol-II on essentially all full-length nanochromosomes (Figure S2, A and C). In addition, there is poor correlation between Pol-II ChIP-seq and mRNA-seq levels (r = 0.126), but better correlation with chromosome copy number (Swart et al. 2013) (r = 0.671), suggesting Pol-II occupancy on most MAC chromosomes, independent of the encoded mRNA transcript levels (Figure S2, B and D). Noticeable peaks of Pol-II occupancy were present at both the 5′ and 3′ ends of expressed, nonexpressed, or noncoding nanochromosomes (Figure S2, E–G). Taken together, these observations suggest genome-wide nanochromosome transcription at this stage in development, supporting the hypothesis that an RNA cached copy of all maternal nanochromosomes provides a template for the next generation (Nowacki et al. 2008). Conclusions from ChIP-seq data were confirmed with qPCR for three nanochromosomes, including one with no expression at 12 hr (Contig 8213.0; Figure 1A), one that is not normally transcribed by Pol-II (rDNA chromosome; Figure 1A), and one large nanochromosome (DNA Pol-α), all of which displayed relatively equal signal at the 5′ and 3′ ends, a pattern found on all the assembled MAC chromosomes, which suggests bidirectional transcription (Figure 1, C and D; Figure S2). In other eukaryotes, Pol-II can occupy promoters of certain signal-inducible genes without actively transcribing them, a phenomenon known as promoter-proximal pausing (Adelman and Lis 2012). To confirm that the occupancy peaks in the ChIP-seq data represent active Pol-II transcription, instead of promoter pausing, we performed RNA-IP, using the same antibody as above for ChIP, which confirmed active Pol-II transcription (Figure 1B).
Table 1. Peptides from Rpb complex identified via LC-MS after Rpb1-IP from 24-hr cell extracts.
| Identified proteins | Pfam domain (identity) |
|---|---|
| Contig 18775.0.g49.t1_0 | RNA_Pol_Rpb1_1, RNA_Pol_Rpb1_2, RNA_Pol_Rpb1_3, RNA_Pol_Rpb1_4, RNA_Pol_Rpb1_5, RNA_Pol_Rpb1_6, RNA_Pol_Rpb1_7 (Rpb1-a) |
| Contig 19200.0.g86.t1_1 | RNA_pol_Rpb2_5, RNA_pol_Rpb2_4, RNA_pol_Rpb2_7, RNA_pol_Rpb2_6, RNA_pol_Rpb2_1, RNA_pol_Rpb2_3 (Rpb2-a) |
| Contig 486.1.g67.t1_1 | RNA_pol_Rpb5_N, RNA_pol_Rpb5_C (Rpb5) |
| Contig 11798.0.g106.t1_0 | RNA_pol_L_2 (Rpb11) |
| Contig 1141.1.g1.t1_0 | RNA_pol_Rpb2_5, RNA_pol_Rpb2_4, RNA_pol_Rpb2_7, RNA_pol_Rpb2_6, RNA_pol_Rpb2_1, RNA_pol_Rpb2_3 (Rpb2-b) |
| Contig 14143.0.g92.t1_1 | RNA_POL_M_15KD (Rpb9) |
| Contig 21831.0.g92.t1_1 | SHS2_Rpb7-N, S1 (Rpb7) |
| Contig 12705.0.g18.t1_0 | RNA_pol_L_2 (Rpb11) |
| Contig 19419.0.g46.t1_1 | RNA_Pol_Rpb8 (Rpb8) |
Figure 1.
Bidirectional transcription at 12 hr postmating. (A) Chromatin immunoprecipitation (IP) with IgG (negative control) or Rpb1 antibody at 12 and 24 hr postmixing, followed by quantitative PCR (qPCR) for the indicated contigs or gene loci. The graphs are represented as percentage of IP relative to the input. (B) RNA-IP using anti-Rpb1 antibody, followed by RT-qPCR for indicated transcripts. (C and D) Mapping of Rpb1 ChIP-seq reads for (C) Contig 22209.0 (TEBP-β) and (D) Contig 451.1 (ribosomal RNA) at 12 hr. The intensity of the signal (green) represents the number of ChIP-seq reads mapping to that region; I = intron in panel (C).
Because bidirectional transcription should generate complementary transcripts with the potential to form double-stranded RNA (dsRNA), we examined the presence of dsRNA, using a well-characterized antibody that can specifically recognize dsRNA >40 bp (Schonborn et al. 1991; Kaneko et al. 2011). The antibody stains the macronuclear periphery in 15-hr cells and later localizes to the newly formed zygotic macronucleus (Figure S3), consistent with genome-wide transcription of the parental macronuclear genome into lncRNAs, which are later transported as dsRNAs to the zygotic macronucleus to guide genome rearrangements (Nowacki et al. 2008).
Gene duplication of Rpb2, the second-largest subunit of RNA Pol-II
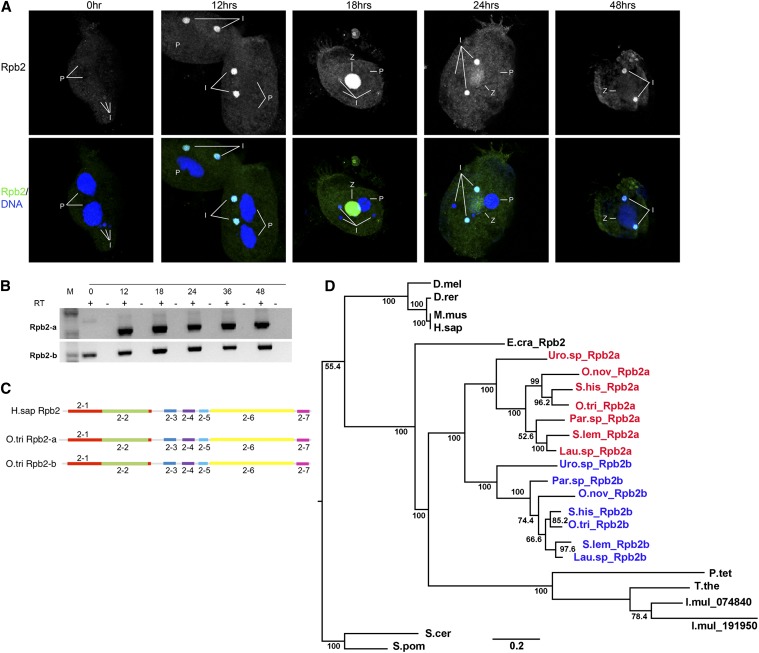
In an effort to identify genes involved in lncRNA production and/or genome rearrangements, we examined a list of the most highly expressed mRNAs at 10 hr after initiating conjugation, relative to vegetative cells (Swart et al. 2013). The third most highly expressed gene in Oxytricha according to this criterion encodes the second-largest subunit of Pol-II, Rpb2 (hereafter named Rpb2-a). In contrast, no other protein from the Pol-II complex was expressed at such high levels at this time point. Considering that the components of a protein complex are typically expressed at similar levels to ensure proper stoichiometry within the complex (Papp et al. 2003), we examined Rpb2 in more detail. Immunofluorescence reveals that Rpb2 localizes in the MIC throughout sexual development and additionally localizes in the zygotic macronucleus at 18 hr (early zygotes) (Figure 2A). This staining pattern differs significantly from the active transcription machinery (Figure S1), suggesting a possible role for this protein in other processes.
Figure 2.
Localization and evolutionary duplication of Rpb2. (A) Cells fixed at indicated times were stained using the Rpb2 antibody (green) and DAPI (blue). Rpb2 localizes to micronuclei (I) at all stages during conjugation and to the zygotic macronucleus (Z) most strikingly at 18 hr. No visible signal is seen in the parental macronuclei (P) or at 0 hr. (B) RT-PCR analysis of Rpb2-a and Rpb2-b during development. (C) Pfam protein domain analysis of human Rpb2 (H.sap) and Oxytricha Rpb2-a and Rpb2-b. The domain annotation is RNA_Pol_Rpb- followed by unique identifiers shown. Protein domains are drawn to scale. (D) Maximum-likelihood phylogenetic analysis of Rpb2 from O. trifallax (O.tri Rpb2-a and Rpb2-b), T. thermophila (T.the), P. tetraurelia (P.tet), Plasmodium falciparum (P.fal), E. crassus (E.cra), D. melanogaster (D.mel), M. musculus (M.mus), H. sapiens (H.sap), S. cerevisiae (S.cer), S. pombe (S.pom), D. rerio (D.rer). Urostyla sp. (Uro.sp_Rpb2a and Rpb2b), O. nova (O.nov_Rpb2-a and -b), S. lemnae (S.lem_Rpb2-a and -b), S. histriomuscorum (S.his_Rpb2-a and -b), Paraurostyla sp. (Par.sp_Rpb2-a and -b), and Laurentialla sp. (Lau.sp_Rpb2-a and -b).
BLASTp against the Oxytricha MAC genome using human Rpb2 (Uniprot ID: P30876) as a query revealed two significant hits (E-value = 0), suggesting a gene duplication in Rpb2 in Oxytricha. The two paralogs display differential expression throughout development (Figure 2B). While the more abundant paralog (Rpb2-a) has conjugation-specific expression, Rpb2-b has constitutive expression throughout development (Figure 2B). To assess the level of divergence between the two paralogs and among homologous proteins from other organisms, we performed a multiple-sequence alignment, using ClustalW (Larkin et al. 2007). The protein domains were analyzed using Pfam (Punta et al. 2012) (Figure 2C and Figure S4). The Pfam analysis indicates that both Rpb2-a and Rpb2-b contain essentially the same domains as metazoa (Figure 2C). The multiple-sequence alignment indicates that Rpb2-a possesses the conserved residues that should bind other proteins in the Pol-II complex, as well as the DNA-binding capabilities of the Zinc-finger binding domain. A notable difference between Rpb2-a and all the other protein sequences in the alignment is the presence of an additional 10-amino-acid sequence at the N-terminal region of all Rpb2-a orthologs. In addition, all studied stichotrich Rpb2-a proteins (see below) are consistently longer than their respective paralog, with the exception of Paraurostyla sp. (Figure S4, File S1). We propose that this could act as a signal peptide to guide protein localization.
To understand the evolutionary timing of gene duplication in Rpb2, we performed a phylogenetic analysis using maximum likelihood (PhyML) (Guindon and Gascuel 2003) (Figure 2D). We chose to include Rpb2 sequences from representative metazoa (Homo sapiens, Mus musculus, Danio rerio, and Drosophila melanogaster) and yeasts (Saccharomyces cerevisiae and S. pombe), providing many outgroups to ciliates. Ciliates include three oligohymenophoreans with completely sequenced somatic genomes (Ichthyophthirius multifiliis, Paramecium tetraurelia, and Tetrahymena thermophila), a spirotrich (Emeus crassus), and seven stichotrichs (Urostyla sp., Paraurostyla sp., Laurentiella sp., O. nova, Sterkiella histriomuscorum, Stylonychia lemnae, and O. trifallax). With the exception of I. multifiliis, the gene duplication appears specific to stichotrich ciliates, a group that shares germline genome scrambling (Chang et al. 2005). The Rpb2 gene duplication in I. multifiliis appears to be an independent duplication event, and its two paralogs could not be classified as Rpb2-a and -b, as defined by the corresponding stichotrich orthologs, based on the sequence alignment. These observations suggest that the Rpb2 duplication likely occurred in the ancestors of stichotrichs. Both Oxytricha paralogs retain amino acid residues essential for transcription, although they have acquired other changes that may permit additional functions.
Rpb2-a is absent from the Pol-II complex in early zygotes
To test the possibility of newly acquired functions of Rpb2-a, we used LC-MS to survey the binding partners that coprecipitated with an Rpb2 antibody at 18 hr (early zygotes). The antibody was tested via Western hybridization and immunoprecipitation, followed by mass spectrometry. A low-coverage LC-MS analysis revealed Rpb2-a as the top-scoring hit, confirming the specificity of the antibody. The same analysis curiously revealed the absence of any Pol-II complex proteins. To increase our confidence in the data, we repeated the analysis with two different replicates of Rpb2 IP, paired with a negative control, at much deeper mass-spectrometry coverage. The analysis was restricted to peptides matching the MAC reference assembly (Swart et al. 2013) with a FDR of 5% (File S2). From this, we generated a list of high-confidence targets based on the peptide spectrum match (PSM) scoring function. These candidates were either enriched at least fivefold in Rpb2 IP vs. IgG control or completely absent from IgG and detected in both the Rpb2 IP replicates (Table 2, Figure 3A). A medium-confidence list was extracted, which included proteins enriched more than twofold in IP vs. negative control.
Table 2. High- and medium-confidence protein hits identified via LC-MS following Rpb2 immunoprecipitation (IP) from 18-hr cell extracts.
| Accession | Pfam domains | Protein identity | Confidence level |
|---|---|---|---|
| Contig 19200.0.g86.t1_1 | RNA_pol_Rpb2_5, RNA_pol_Rpb2_4, RNA_pol_Rpb2_7, RNA_pol_Rpb2_6, RNA_pol_Rpb2_1, RNA_pol_Rpb2_3 | Rpb2-a | High |
| Contig 22488.0.g2.t1_1 | — | — | |
| Contig 13381.0.g48.t1_1 | DEAD, Helicase_C | Ddx5-like | |
| Contig 16219.0.g100.t1_0 | GAS2 | — | |
| Contig 755.0.g44.t1_0 | GAS2 | — | |
| Contig 8173.0.g79.t1_0 | RNA_Pol_L, RNA_Pol_A_bac | Rpb3 | |
| Contig 11442.0.g16.t1_0 | 2x RRM_1 | ||
| Contig 1141.1.g1.t1_0 | RNA_Pol_Rpb2_1, RNA_Pol_Rpb2_2, RNA_Pol_Rpb2_3, RNA_Pol_Rpb_4, RNA_Pol_Rpb2_5, RNA_Pol_Rpb2_6, RNA_Pol-Rpb2_7 | Rpb2-b | |
| Contig 21576.0.g108.t1_0 | Glyco_transf_5, Glyco_transf_1 | Glycosyl transferase | |
| Contig 14597.0.g69.t1_0 | — | — | |
| Contig 10038.0.g32.t1_1 | — | — | |
| Contig 18098.0.g9.t1_1 | RPAP_1N | — | |
| Contig 21174.0.g65.t1_1 | — | — | |
| Contig 14451.0.g111.t1_1 | ECH | Enoyl-CoA hydratase/isomerase | |
| Contig 15609.0.g93.t1_0 | ATP_bind_1 | GPN-loop GTPase like 1 | |
| Contig 21267.0.g52.t1_0 | ATP_bind_1 | GPN-loop GTPase like 2 | |
| Contig 20804.0.g29.t1_1 | — | — | |
| Contig 103.1.g85.t1_1 | UN_NPL4, NPL4 | Nuclear pore-associated protein | |
| Contig 4686.0.g38.t1_0 | RNA_Pol_N | RNA polymerase I/II/III subunit | |
| Contig 15685.0.g23.t1_1 | Ribosomal_L37 | — | |
| Contig 9287.0.g107.t1_0 | KOW | Ribosomal protein L24 | |
| Contig 12508.0.g82.t1_0 | — | Mdp2 | |
| Contig 22520.0.g95.t1_0 | Ribosomal_L17 | rpS11e | |
| Contig 22526.0.g93.t1_0 | Pkinase | ||
| Contig 13396.0.g39.t1_0 | Ribosomal_L4 | Ribosomal protein L4 | Medium |
| Contig 21133.0.g40.t1_1 | — | — | |
| Contig 9049.0.g92.t1_1 | Ribosomal_L27 | Ribosomal protein L27 | |
| Contig 4562.0.g74.t1_1 | Actin | Actin | |
| Contig 850.1.g11.t1_0 | Ribosomal_S3Ae | Ribosomal protein S3a | |
| Contig 1642.0.g21.t1_0 | Ribosomal_L22 | Ribosomal protein L22 | |
| Contig 7058.0.g101.t1_1 | — | — | |
| Contig 12224.0.g3.t1_0 | START | ||
| Contig 11798.0.g106.t1_0 | RNA_Pol_L2 | Rpb11 | |
| Contig 5219.0.1.g44.t1_0 | Ribosomal_S13N, Ribosomal_S15 | Ribosomal protein S13E/S15P | |
| Contig 22158.0.g4.t1_0 | Ribosomal_L14e | Ribosomal L14 | |
| Contig 4552.0.g54.t1_1 | Ribosomal_L6e | Ribosomal L6 | |
| Contig 11869.0.g78.t1_0 | Ribosomal_L11_N, Ribosomal_L11 | Ribosomal protein L11 | |
| Contig 232.1.g122.t1_0 | Img2 | — | |
| Contig 1499.0.g65.t1_0 | — | — | |
| Contig 15153.0.g102.t1_1 | Ribosomal_L20 | Ribosomal L2 protein | |
| Contig 11390.0.g72.t1_0 | Ribosomal_L11N, Ribosomal_L11 | Ribosomal L12e | |
| Contig 11050.0.g68.t1_0 | Ribosomal_S19e | Ribosomal S19 | |
| Contig 9812.0.g20.t1_0 | Ribosomal_L1 | Ribosomal protein | |
| Contig 13946.0.g19.t1_0 | MRP_L27 | MRP-L27 | |
| Contig 20881.0.1.g63.t1_0 | — | — | |
| Contig 13339.0.g92.t1_0 | PDEase_1 | 3′,5′-cyclic nucleotide phosphodiesterase family protein |
Figure 3.
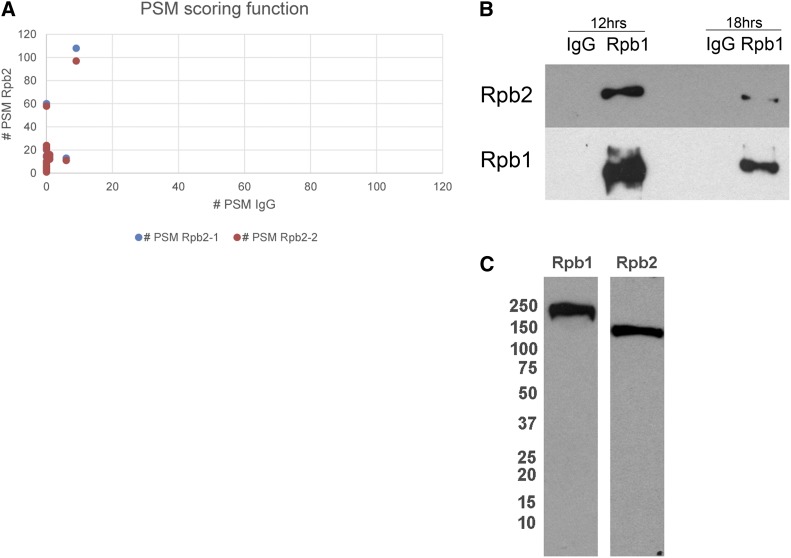
Rpb2-a is not associated with the Rpb1 complex at 18 hr postmixing. (A) Peptide spectrum match (PSM) scoring function for IgG compared to two replicates of Rpb2-IP at 18 hr. (B) Rpb1-IP at 12 and 18 hr postmixing, followed by Western blot analysis of Rpb2 and Rpb1, indicating the abundant presence of Rpb2 in the Pol-II complex at 12 and 18 hr. (C) Western hybridization of cell extracts from 12 hr postmixing, probed with Rpb1 or Rpb2 antibodies and exposed overnight to demonstrate the specificity of the antibody.
Figure 3A compares the PSM score function for the two IP replicates and the corresponding IgG controls, illustrating the concordance between the two data sets. This analysis identified 24 high-confidence and 22 medium-confidence proteins that specifically coprecipitated with an Rpb2 antibody, relative to the IgG control (Table 2, Figure 3A). This analysis also confirmed that either the antibody preferentially binds to Rpb2-a over Rpb2-b or that Rpb2-a is more abundant than Rpb2-b in the cell. Surprisingly, most proteins from the Pol-II complex were absent from the Rpb2 complex at 18 hr, consistent with the lack of colocalization of Rpb1 and Rpb2 by immunostaining. In contrast, Rpb2 immunoprecipitation much later in zygotic development at 48 hr revealed all the components of the Pol-II complex (Table 3), suggesting a role for Rpb2-a outside the Pol-II complex in early zygotes at 18 hr.
Table 3. Peptides from Rpb complex identified via LC-MS after Rpb2-IP from 48-hr cell extracts.
| Identified proteins | Pfam domains (identity) |
|---|---|
| Contig 19200.0.g86.t1_1 | RNA_pol_Rpb2_5, RNA_pol_Rpb2_4, RNA_pol_Rpb2_7, RNA_pol_Rpb2_6, RNA_pol_Rpb2_1, RNA_pol_Rpb2_3 (Rpb2-a) |
| Contig 5780.0.g96.t1_0 | RNA_pol_Rpb1_2, RNA_pol_Rpb1_3, RNA_pol_Rpb1_1, RNA_pol_Rpb1_4, RNA_pol_Rpb1_5 (Rpb1-b) |
| Contig 15041.0.g92.t1_0 | DNA_RNApol_7kD (RPABC4) |
| Contig 11798.0.g106.t1_0 | RNA_pol_L_2 (Rpb11) |
| Contig 1141.1.g1.t1_0 | RNA_pol_Rpb2_5, RNA_pol_Rpb2_4, RNA_pol_Rpb2_7, RNA_pol_Rpb2_6, RNA_pol_Rpb2_1, RNA_pol_Rpb2_3 (Rpb2-b) |
| Contig 13770.0.g40.t1_1 | RNA_pol_N (RPABC5) |
| Contig 12705.0.g18.t1_0 | RNA_pol_L_2 (Rpb11) |
| Contig 486.1.g67.t1_1 | RNA_pol_Rpb5_N, RNA_pol_Rpb5_C (Rpb5) |
| Contig 14143.0.g92.t1_1 | RNA_POL_M_15KD (Rpb9) |
| Contig 21831.0.g92.t1_1 | SHS2_Rpb7-N, S1 (Rpb7) |
| Contig 22041.0.g30.t1_0 | RNA_pol_L, RNA_pol_A_bac (RPAC1) |
| Contig 18775.0.g49.t1_0 | RNA_pol_Rpb1_2, RNA_pol_Rpb1_3, RNA_pol_Rpb1_1, RNA_pol_Rpb1_4, RNA_pol_Rpb1_5 (Rpb1-a) |
Reciprocally, we tested whether Rpb2 would immunoprecipitate with the Rpb1 antibody. Rpb1 IP, followed by Western analysis, revealed the presence of Rpb2 in the Rpb1 complex at 12 hr, but at much-reduced levels from 18-hr cell lysates (Figure 3B). This analysis suggests the potential involvement of Rpb2 in lncRNA production at 12 hr, and its reduced levels at 18 hr suggest a possible function for Rpb2-a outside the Pol-II transcription complex, confirming our LC-MS data and immunofluorescence observations.
Rpb1 IP at 18 hr shows a significantly lower enrichment for Rpb1, compared to that at 12 hr. Combined with the immunostaining (Figure S1), which shows low levels of Rpb1 protein relative to other time points, we conjecture that the Pol-II complex dissociates briefly during early zygotic development. This observation is reminiscent of the maternal-zygotic transition (MZT) in other eukaryotes, which permits the shift of transcriptional burden to the newly formed zygote and involves a brief period of transcriptional quiescence. Immunoprecipitation with the Rpb1 antibody from 24-hr cell extracts revealed all the proteins from the Pol-II complex (Table 1), which is consistent with reassembly of the Pol-II complex at this stage, as also observed by immunostaining. Consistent with this hypothesis, we report the presence of a gene duplication in Rpb1 (Contig 18775.0 and Contig 5780.0), which shows differential expression (Figure S5). Contig 18775.0 (reported in this article; hereby denoted Rpb1-a) is expressed constitutively, with increased expression at 12 hr postmixing, and appears to be involved in lncRNA production at that time and mRNA transcription during vegetative growth. Contig 5780.0 (Rpb1-b) is expressed almost exclusively at 40 and 60 hr during nuclear differentiation, and could have specialized functions in genome rearrangements. The shift in expression of the two paralogs may have either provided an opportunity for Rpb2-a to acquire Pol-II-independent functions in early zygotes or been a consequence of that specialization.
Fang et al. (2012) identified the presence of Rpb2-a in the Otiwi-1 complex by co-immunoprecipitation with a specific Piwi antibody (Fang et al. 2012). Otiwi-1 is expressed in the parental macronuclei at 12 hr and then localizes to the zygotic macronucleus and micronucleus at 18 hr, a pattern similar to that of Rpb2 (Figure S6). Given the similar dynamic localization of dsRNA with respect to the macronuclei, we tested the physical interaction between dsRNA with Rpb2 and Otiwi-1 by co-immunoprecipitation with the dsRNA antibody followed by Western analysis, and this confirmed the association between dsRNA and Otiwi-1 and Rpb2 in 18-hr cell lysates (Figure S3B). LC-MS analysis of Rpb2 IP failed to recover Otiwi-1-specific peptides, which might suggest that the interaction either is transient or occurs via dsRNA. Mass spectrometric analysis of the gel-separated bands from dsRNA-IP further confirmed the presence of Rpb2-a but not the other paralog (Figure S3B; data not shown), suggesting an important interaction among Otiwi-1, Rpb2-a, and dsRNA, possibly in regulating the availability or localization of lncRNAs.
Rpb2-a negatively regulates germline gene expression
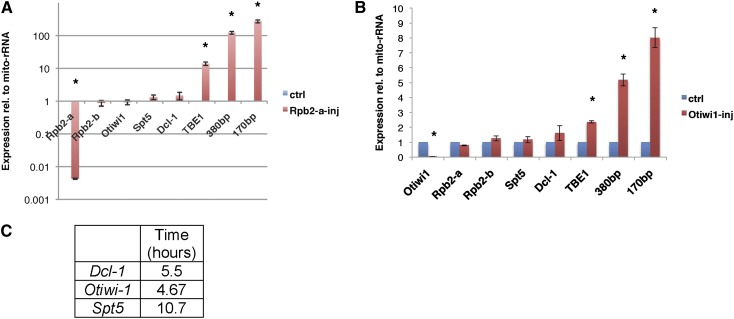
RNA polymerase genes are essential for survival, and inhibition of Pol-II activity affects transcription of all protein-coding genes. Given the differential expression and localization of Rpb2-a, combined with the absence of the Pol-II complex in 18-hr mass-spectrometric analysis of Rpb2-a, we tested whether knockdown of Rpb2-a leads to any changes in gene expression. Rpb2-a knockdown leads to developmental arrest, as paired cells fail to separate even after 24 hr. Eventually, it leads to cell death by 30 hr. Knockdown of Rpb2-a (Figure 4A; 222-fold mRNA reduction, P = 0.0001) has no significant effect on mRNA levels of Rpb2-b (1.19-fold reduction, P = 0.61), indicating specificity of the knockdown. If Rpb2-a were involved in general Pol-II transcription of genes at this time, we would expect a global effect on transcript levels. However, Rpb2-a knockdown (KD) cells show no significant loss of mRNA levels for three unrelated genes, Dcl-1, Otiwi1, and Spt5 (Figure 4A), which suggests that Rpb2-a plays a role independent of the transcription machinery at this time in development. To confirm that the effect is not an artifact of metastable mRNAs, but is transcription independent, we surveyed mRNA stability for the same three genes following conjugation using Actinomycin D treatment. The half-lives of Dcl-1, Otiwi-1, and Spt5 were found to be 5.5, 4.67, and 10.7 hr, respectively (Figure 4C). Because the phosphorothioate antisense oligonucleotides for knockdown were injected 2 hr postmixing and gene expression was assayed 22 hr later, we conclude it is unlikely that Rpb2-a is involved in mRNA transcription during this time in development.
Figure 4.
Rpb2-a or Otiwi-1 knockdown affects germline gene expression. (A and B) Rpb2-a (A) and Otiwi-1 (B) knockdown using phosphorothioate-modified antisense oligonucleotides, followed by mRNA expression analysis via RT-qPCR for the indicated genes at 24 hr postmixing (see text for gene names), relative to mitochondrial rRNA (mito-rRNA) as control uninjected cells. * denotes significant changes relative to control; P-values are noted in the text. Labels 380bp and 170bp refer to MIC-limited satellite repeats (Bracht et al. 2012). The error bars represent standard error of the mean (SEM). (C) mRNA half-life (in hours) for Dcl-1, Otiwi-1, and Spt5 calculated using an Actinomycin D treatment pulse/chase experiment.
In contrast to the lack of effect on somatic gene expression, we observed a dramatic increase in expression of germline-limited sequences in Rpb2-a KD cells. The transcript levels from the MIC-limited transposon TBE-1 (13.8-fold increase, P = 0.01) and 170-bp (272-fold increase, P = 0.01) and 380-bp satellite repeats (122-fold, P = 0.008) were all significantly upregulated relative to wild-type control cells (Figure 4A). Given the previous evidence that the Rpb2-a protein interacts with Otiwi-1, we tested a reciprocal Otiwi-1 knockdown and found that, similar to the Rpb2-a KD, Otiwi-1 KD cells have no general defect in macronuclear gene expression, but have increased gene expression of repetitive elements (Figure 4B).
Piwi proteins and Piwi-interacting RNAs (piRNAs) are part of an ancient pathway for transposon control in diverse eukaryotes (reviewed in Khurana and Theurkauf 2010; Siomi et al. 2011) but have novel roles in protecting DNA against deletion in Oxytricha (Fang et al. 2012). Piwi–piRNA complexes in most studied eukaryotes generally act as sequence-specific nucleases, which target transposons and other repetitive elements, thus maintaining germline genome integrity (Khurana and Theurkauf 2010). An increase in gene expression from transposons and satellite repeats in Otiwi-1 KD cells suggests an additional piRNA-independent role for Otiwi-1 during sexual development in Oxytricha. In support of this argument, Otiwi-1 localizes to the micronucleus in addition to the zygotic macronucleus, which is the likely site for piRNA function at 18 hr (Figure S6 and Fang et al. 2012). Otiwi-1 is essential for accumulation of 27-nt small RNAs, based on a previous knockdown analysis (Fang et al. 2012). Knockdown of Rpb2-a, however, does not show a significant loss of 27-nt small RNAs, suggesting that the protein does not directly affect the Piwi–piRNA pathway (Figure S7).
Gene duplication offers a mechanism to acquire novel gene functions and may facilitate evolution of diverse ciliate lifestyles or genome architectures (Ohno et al. 1968). The Paramecium genome has been shaped by more than one whole-genome duplication (Aury et al. 2006), and the Oxytricha macronuclear genome displays several cases of paralog expansion (Swart et al. 2013), often in genes encoding proteins involved in DNA/RNA metabolism. The recent duplication of Rpb2 most likely permitted its acquisition of novel functions outside the Pol-II complex. Both paralogs have apparently retained their ability to perform their core function in transcription, consistent with the subfunctionalization model of gene duplication, where the duplicated genes perform their original function, while a copy may acquire novel functions. At 12 hr, Rpb2 is found in complex with Pol-II, suggesting that it is potentially involved in production of lncRNAs from the parental macronucleus. In early zygotes, however, Rpb2-a dissociates from the Pol-II complex and instead binds dsRNAs and Otiwi-1 in the zygotic macronucleus. Later in development, we identified both Rpb2-a and Rpb2-b in the Pol-II complex at 48 hr. Thus, the acquisition of transcription-independent functions for Rpb2-a appears specific to early zygotes (18–24 hr).
During conjugation, a duplicate copy of the zygotic micronucleus becomes the new macronucleus, while the others remain transcriptionally inert (Figure S1A). Although germline-encoded TBE transposons are abundantly expressed MIC genes during conjugation (Nowacki et al. 2009 and X. Chen and L. F. Landweber, unpublished results) and are essential to programmed genome rearrangements, their expression appears limited to the zygotic macronucleus (Nowacki et al. 2009). Micronuclear gene expression must be tightly regulated for successful completion of sexual development. Rpb2-a is found in complex with Otiwi-1 and dsRNAs. Given the timing and location of these dsRNAs, it is likely that they could be precursors to piRNAs, which guide retention of the somatic macronuclear-destined sequences. Partial loss of function of Rpb2-a or Otiwi-1 leads to increased expression of germline-limited repetitive elements, suggesting the importance of these noncoding RNAs and this complex in germline gene expression regulation and thus in maintenance of genome integrity.
Supplementary Material
Acknowledgments
We thank Donna Storton, Jessica Buckles, and Wei Wang from the Sequencing Core Facility at Princeton University for Illumina sequencing. We thank Estienne Swart for providing Rpb2 sequences from unpublished E. crassus genome assembly and Seolkyoung Jung and Sean Eddy for sharing unpublished genome sequences from other stichotrichs. We also thank Jingmei Wang for maintenance of cells. The ChIP-seq data are deposited under Gene Expression Omnibus accession no. GSE55703. This study was supported by National Institutes of Health grant GM59708 and National Science Foundation grants 0923810 and 0900544 (to L.F.L.).
Footnotes
Communicating editor: J. Sekelsky
Literature Cited
- Adelman K., Lis J. T., 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13: 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury J. M., Jaillon O., Duret L., Noel B., Jubin C., et al. , 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444: 171–178. [DOI] [PubMed] [Google Scholar]
- Bracht J. R., Perlman D. H., Landweber L. F., 2012. Cytosine methylation and hydroxymethylation mark DNA for elimination in Oxytricha trifallax. Genome Biol. 13: R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W. J., Bryson P. D., Liang H., Shin M. K., Landweber L. F., 2005. The evolutionary origin of a complex scrambled gene. Proc. Natl. Acad. Sci. USA 102: 15149–15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P., Bushnell D. A., Fu J., Gnatt A. L., Maier-Davis B., et al. , 2000. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288: 640–649. [DOI] [PubMed] [Google Scholar]
- Cramer P., Bushnell D. A., Kornberg R. D., 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292: 1863–1876. [DOI] [PubMed] [Google Scholar]
- Djupedal I., Portoso M., Spahr H., Bonilla C., Gustafsson C. M., et al. , 2005. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 19: 2301–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W., Wang X., Bracht J. R., Nowacki M., Landweber L. F., 2012. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell 151: 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O., 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Harel-Sharvit L., Eldad N., Haimovich G., Barkai O., Duek L., et al. , 2010. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 143: 552–563. [DOI] [PubMed] [Google Scholar]
- Hirata A., Klein B. J., Murakami K. S., 2008. The X-ray crystal structure of RNA polymerase from Archaea. Nature 451: 851–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L., Canterbury J. D., Weston J., Noble W. S., MacCoss M. J., 2007. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4: 923–925. [DOI] [PubMed] [Google Scholar]
- Kaneko H., Dridi S., Tarallo V., Gelfand B. D., Fowler B. J., et al. , 2011. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471: 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Goto D. B., Martienssen R. A., Urano T., Furukawa K., et al. , 2005. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309: 467–469. [DOI] [PubMed] [Google Scholar]
- Kent W. J., 2002. BLAT–the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana J. S., Theurkauf W., 2010. piRNAs, transposon silencing, and Drosophila germline development. J. Cell Biol. 191: 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana J. S., Xu J., Weng Z., Theurkauf W. E., 2010. Distinct functions for the Drosophila piRNA pathway in genome maintenance and telomere protection. PLoS Genet. 6: e1001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkhin Y., Unligil U. M., Littlefield O., Nelson P. J., Stuart D. I., et al. , 2009. Evolution of complex RNA polymerases: the complete archaeal RNA polymerase structure. PLoS Biol. 7: e1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., et al. , 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lauth M. R., Spear B. B., Heumann J., Prescott D. M., 1976. DNA of ciliated protozoa: DNA sequence diminution during macronuclear development of Oxytricha. Cell 7: 67–74. [DOI] [PubMed] [Google Scholar]
- Lotan R., Bar-On V. G., Harel-Sharvit L., Duek L., Melamed D., et al. , 2005. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev. 19: 3004–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan R., Goler-Baron V., Duek L., Haimovich G., Choder M., 2007. The Rpb7p subunit of yeast RNA polymerase II plays roles in the two major cytoplasmic mRNA decay mechanisms. J. Cell Biol. 178: 1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M., Vijayan V., Zhou Y., Schotanus K., Doak T. G., et al. , 2008. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature 451: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M., Higgins B. P., Maquilan G. M., Swart E. C., Doak T. G., et al. , 2009. A functional role for transposases in a large eukaryotic genome. Science 324: 935–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., Wolf U., Atkin N. B., 1968. Evolution from fish to mammals by gene duplication. Hereditas 59: 169–187. [DOI] [PubMed] [Google Scholar]
- Papp B., Pal C., Hurst L. D., 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197. [DOI] [PubMed] [Google Scholar]
- Patzel V., Steidl U., Kronenwett R., Haas R., Sczakiel G., 1999. A theoretical approach to select effective antisense oligodeoxyribonucleotides at high statistical probability. Nucleic Acids Res. 27: 4328–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott D. M., 1994. The DNA of ciliated protozoa. Microbiol. Rev. 58: 233–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., et al. , 2012. The Pfam protein families database. Nucleic Acids Res. 40: D290–D301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., Mann M., 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75: 663–670. [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J., 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonborn J., Oberstrass J., Breyel E., Tittgen J., Schumacher J., et al. , 1991. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 19: 2993–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M., 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1: 2856–2860. [DOI] [PubMed] [Google Scholar]
- Siomi M. C., Sato K., Pezic D., Aravin A. A., 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12: 246–258. [DOI] [PubMed] [Google Scholar]
- Swart E. C., Nowacki M., Shum J., Stiles H., Higgins B. P., et al. , 2012. The Oxytricha trifallax mitochondrial genome. Genome Biol. Evol. 4: 136–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart E. C., Bracht J. R., Magrini V., Minx P., Chen X., et al. , 2013. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol. 11: e1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Campbell E. A., Minakhin L., Richter C., Severinov K., et al. , 1999. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell 98: 811–824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.