Abstract
We propose a nonlinear regression model for quantitatively analyzing periodic gene expression in studies of experimentally synchronized cells. Our model accounts for the observed attenuation in cycle amplitude by a simple and biologically plausible mechanism. We represent the expression level for each gene as an average across a large number of cells. For a given cell-cycle gene, we model its expression in each cell in the culture as following the same sinusoidal function except that the period, which in any individual cell must be the same for all cell-cycle genes, varies randomly across cells. We model these random periods by using a lognormal distribution. The variability in period causes the measured amplitude of the cyclic expression trajectory to attenuate over time as cells fall increasingly out of synchrony. Gene-specific parameters include initial amplitude and phase angle. Applying the model to data from Whitfield et al. [Whitfield, M. L., Sherlock, G., Saldanha, A. J., Murray, J, I., Ball, C. A., et al. (2002) Mol. Biol. Cell 13, 1977-2000], we fit the trajectories of 18 well characterized phase-marker genes and find that the fit does not suffer when a common lognormal distribution is assumed for all 18 genes compared with a separate distribution for each. We then use the model to identify 337 periodically expressed transcripts, including the 18 phase-marker genes. The model permits estimation of and hypothesis testing about biologically meaningful parameters that characterize cycling genes.
Keywords: bootstrap test, gene expression, microarray, nonlinear regression
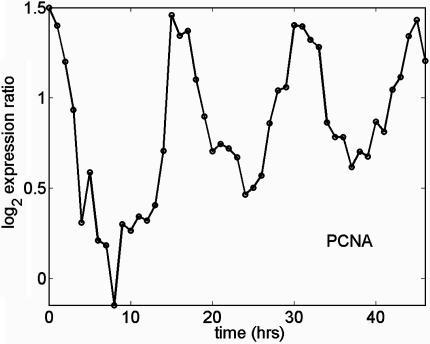
Experimental protocols that arrest cells in vitro at a particular phase of the cell cycle and then release them in a synchronized way allow detailed study of the cycling process. In conjunction with such experiments, cDNA microarray technology allows investigators to assess the temporal expression patterns of thousands of genes simultaneously. Gene-expression studies in yeast (1-3) and in cultured human cells (4, 5) have revealed that expression levels for cell-cycle genes vary periodically and have amplitudes that attenuate through time (Fig. 1). The observed attenuation is generally attributed to cells in the cultures falling increasingly out of synchrony through time.
Fig. 1.
Trajectory of log2-transformed expression ratio for a known cell-cycle gene, PCNA, from synchronized HeLa cultures (data from ref. 5) showing typical attenuation in amplitude.
In cultures of homogeneous cells released from a block on cycling, asynchrony can arise from at least two mechanisms. Cells throughout the culture can differ slightly in the exact timing of their arrest or release, or cells can differ slightly in the duration of their individual cycles. The former mechanism leaves asynchrony constant through time, so only the latter mechanism, where asynchrony increases, produces the characteristic attenuation. Such attenuation may hold biologic interest in itself. The rate of attenuation (i.e., the variation in the duration of the cell cycle across cells) may vary by strain of organism or by cell type (e.g., across tumors with differing metastatic potential). Some influences of cell characteristics on the scheduling of the cell cycle are already known. For instance, cell-size distribution in yeast mutants is related to time spent in the late G1 phase (6).
Few methods proposed for the analysis of cell-cycle expression data address attenuation explicitly. For example, several investigators used a sinusoidal template to identify periodically expressed genes but did not explicitly account for attenuation (2, 5). The basic single-pulse model (SPM) proposed by Zhao et al. (7) addresses asynchrony by assuming that the cell-specific timings with which individual cells in a culture reach a given observation point have a normal distribution whose mean is the observation time itself and whose unknown variance differs across observation times. The magnitudes of the variance parameters measure asynchrony. Stochastic cell-to-cell variation, as mentioned earlier, does not necessarily produce decay in amplitude. Attenuation was built into the SPM separately by an assumed submodel where the log of the variance parameter increases linearly in time; but that submodel's parameters lack direct biologic interpretation.
In this article, we propose an alternative regression model for studying periodically expressed genes. Our model has biologic import: it directly links attenuation in the amplitude of periodic gene expression to stochastic variation across cells in the duration of the cell cycle, while permitting estimation of the phase of the cycle in which the gene is most transcribed. Parameters estimated under our model can be used not only to help identify and characterize periodically expressed genes but also to cluster the identified genes into subgroups based on the estimated phase angle, amplitude, or drift. Our model can facilitate studies of effects of experimental conditions on the variance and median duration of the cell cycle. We describe our model and illustrate its use with a publicly available data set (5). We use the observed expression trajectories of 18 cell-cycle “phase-marker” genes to test a key assumption of our model. We then use estimates based on those genes to help identify additional periodically expressed genes among the remaining transcripts.
Random-Periods Model for Periodically Expressed Genes
To model the expression trajectories of cell-cycle-related genes through time based on cell cultures where synchrony is experimentally induced, we make the following assumptions: (i) measured gene expression is, in effect, the average of mRNA levels across a large number of individual cells in the culture; (ii) in any individual cell, the expression levels (perhaps after logarithmic transformation) of all cell-cycle-related genes have temporal profiles that are well approximated by sinusoidal waves; (iii) the duration of the cell cycle varies stochastically across cells in the culture, following a lognormal distribution with a characteristic median (T) and geometric standard deviation (σ); and (iv) any nonstationary background expression levels are approximately linear in time with gene-specific intercepts and slopes (a and b, respectively). We model the sinusoid using a cosine function with distinct amplitude (K) and phase angle (φ) for each gene.
We model the observed expression of gene g at time t as the sum of an expected response and a random error, that is, Yg(t) = f(t, θg) + εg(t). Here, θg denotes a vector of parameters. We take the εg(t) to have mean zero but make no additional assumptions about their distribution; in particular, we regard them as having possibly different variances across genes or through time and as possibly having serial correlation. In view of the preceding assumptions, we propose the “random-periods” model (RPM) for characterizing the expected periodic expression of a cell-cycle gene:
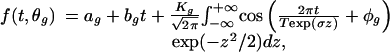 |
1 |
where θg is explicitly (Kg, T, σ, ϕg, ag, bg). The integration in the model computes the expected cosine across the lognormal distribution of periods and thereby accounts for the aggregation of expression levels across a large number of cells. The subscripted parameters in Eq. 1 are gene-specific. The parameter φg corresponds to the phase of the cell cycle where the gene has its peak transcription with φg = 0 corresponding to the point when cells are first released to resume cycling. The parameter Kg is the initial amplitude of the periodic expression pattern. The parameters ag and bg account for any drift in a gene's background expression level.
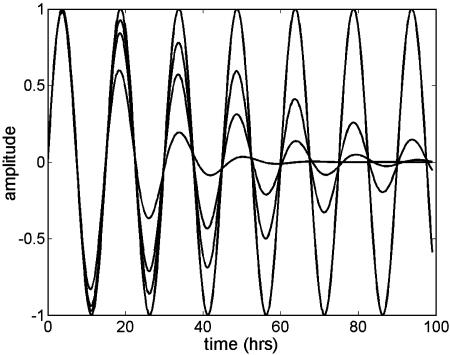
Under our assumptions, the parameters T and σ are specific to the population of cells and the same for all genes, although they can be estimated from data on a single gene or on a set of genes. The parameter σ governs the rate of attenuation in amplitude. If σ is zero, the duration of the cell cycle does not vary across cells, cells remain synchronous through time, and the aggregate expression shows no attenuation in amplitude. If σ is large, cells fall rapidly out of synchrony, and amplitude decays sharply. Increasing σ has two distinct effects on the shape of the aggregate expression trajectory (Fig. 2); the expression level attenuates faster, and the times between successive crossings of 0 increase over time more markedly. The latter feature is more easily seen in the last three cycles of the curve with the largest σ and is less noticeable for curves whose σ values are smaller (<0.075), in particular, within the first three cycles.
Fig. 2.
Trajectories of the RPM for different values of σ For these curves, (K, T, σ, φ, a, b) = (1, 15, σ, π/2, 0, 0) with σ ∈ {0, 0.05, 0.075, 0.13}. Larger values of σ correspond to faster attenuation of peak amplitude.
Statistical Inference: Estimation and Testing
Using numerical quadrature to approximate the needed integral, we estimate the unknown parameters in Eq. 1 by using nonlinear least-squares regression, i.e., we minimize the sum of squared residuals. (For details about nonlinear regression, see refs. 8 and 9.) To help ensure that we reach a global minimum, we repeat the iterative fitting process from multiple distinct starting values (usually 50) and choose the fit with the minimum sum of squared residuals as the best. Our approach to fitting can be applied to one transcript at a time or to several simultaneously and estimates all parameters simultaneously. It ignores, however, possible variance heterogeneity and lack of independence among the εg(t). Calculations were performed with MATLAB software (MathWorks, Natick, MA).
To carry out inference on model parameters, we need to estimate the covariance matrix of their estimates. Suppose that we are fitting Eq. 1 simultaneously to G genes. Each gene g ∈ {1,2,3,..., G} is observed at ng time points, so the total number of observations is 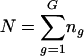 . In general, the overall parameter vector θ has p components indexed by subscript j. For example, P = 2 + 4G if T and σ are estimated jointly for all G genes, but p = 6G if T and σ are estimated separately for each gene. A vector version of the model uses the N observations stacked into a column vector, Y, ordered by genes and by time points within genes, and the corresponding stacked versions of the expected response and the errors: Y = f(t, θ) + ε. Let V be the N × p matrix of partial derivatives of f(t, θ). The (i, j)th element of V is ∂f(t, θi/∂θj, and V̂ is V evaluated at
. In general, the overall parameter vector θ has p components indexed by subscript j. For example, P = 2 + 4G if T and σ are estimated jointly for all G genes, but p = 6G if T and σ are estimated separately for each gene. A vector version of the model uses the N observations stacked into a column vector, Y, ordered by genes and by time points within genes, and the corresponding stacked versions of the expected response and the errors: Y = f(t, θ) + ε. Let V be the N × p matrix of partial derivatives of f(t, θ). The (i, j)th element of V is ∂f(t, θi/∂θj, and V̂ is V evaluated at 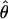 (“hats” denote estimated values). Let V̂g be the ng × p submatrix of V̂ whose rows correspond to the ng time points for gene g. We can express our estimator of Σ, the covariance matrix of
(“hats” denote estimated values). Let V̂g be the ng × p submatrix of V̂ whose rows correspond to the ng time points for gene g. We can express our estimator of Σ, the covariance matrix of 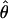 , as:
, as:
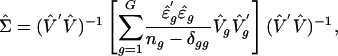 |
2 |
where δgg is the trace of the matrix [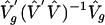 ]. This variance estimator
]. This variance estimator 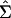 has favorable properties in both heteroscedastic linear models (10, 11) and nonlinear models (12, 13), and it is designed for εg(t) having different variances among genes (but not across time points within genes).
has favorable properties in both heteroscedastic linear models (10, 11) and nonlinear models (12, 13), and it is designed for εg(t) having different variances among genes (but not across time points within genes).
To test hypotheses about the parameters, we use Wald test statistics. Suppose we want to test the null hypothesis h(θ) = 0, where h(·) is vector-valued with q components and is differentiable. The corresponding Wald statistic is 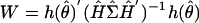 , where Ĥ is the q × p Jacobian of h(·) evaluated at
, where Ĥ is the q × p Jacobian of h(·) evaluated at 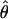 and
and 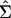 is from Eq. 2. Under technical regularity conditions and for large sample sizes, the Wald statistic would have a χ2 distribution with q degrees of freedom under the null hypothesis. Because the number of time points is not large, and because we expect temporal and gene-to-gene correlations in the εg(t), we opt to evaluate the null distribution of the Wald statistic with a moving-blocks bootstrap procedure (14). Resampling individual values destroys temporal correlation; to retain it, the moving blocks bootstrap resamples fixed-length blocks of consecutive values. Because all genes appear on a single chip at each observation time, we sample observation times and carried along residuals for all G genes at those sampled times when a hypothesis simultaneously involved G genes.
is from Eq. 2. Under technical regularity conditions and for large sample sizes, the Wald statistic would have a χ2 distribution with q degrees of freedom under the null hypothesis. Because the number of time points is not large, and because we expect temporal and gene-to-gene correlations in the εg(t), we opt to evaluate the null distribution of the Wald statistic with a moving-blocks bootstrap procedure (14). Resampling individual values destroys temporal correlation; to retain it, the moving blocks bootstrap resamples fixed-length blocks of consecutive values. Because all genes appear on a single chip at each observation time, we sample observation times and carried along residuals for all G genes at those sampled times when a hypothesis simultaneously involved G genes.
The procedures for generating the bootstrap distribution were, in brief: (i) fit the null model to the original data and compute the residuals; (ii) draw a random sample with replacement from all possible blocks of consecutive residuals with a given length (we sampled six blocks of length 9 and truncated to 47 residuals, the number of time points in the data); (iii) add these sampled residuals to the curve fitted under the null model to obtain a bootstrap data set; (iv) fit the alternative model to the bootstrap data set and compute the Wald statistic; and (v) repeat steps ii through iv a large number of times (our application used 2,000). The resulting collection of bootstrap Wald statistics is used to approximate the null distribution of the test. The bootstrap P value is the proportion of bootstrap Wald statistics that fall above the Wald statistic calculated from the original data.
Identifying Additional Cell-Cycle-Related Transcripts
One approach to identifying additional cell-cycle-related transcripts would fit the RPM to each gene individually and use a testing strategy to see whether the cosine term in the model were necessary for a good fit. Fitting Eq. 1 to the thousands of transcripts typical of cell-cycle expression data would, however, be extremely impractical. Only a small portion of the available transcripts are likely involved in the cell-cycle process (2). Attempts to fit a model designed for cycling trajectories to trajectories without periodicities is time-consuming; convergence of the iterative estimation process is slow, and multiple local minima often present problems. A simple and practical alternative is to adopt template-based correlation methods for selecting genes with periodic expression patterns (3, 5, 15). We propose to use estimates from fitting the RPM to known cell-cycle genes to inform a correlation approach for selecting other cell-cycle-related genes.
We first created a set of model-based templates. A template is a list of fitted values, one at each observation time, generated from Eq. 1, using a prespecified parameter vector. The parameter vectors for the set of templates are chosen so that the set spans trajectories typical of cell-cycle-related genes. We based templates on data from phase-marker genes through parameters estimated by fitting the RPM. We set both a and b to zero, since a has no effect on correlation and since the data for phase-marker genes indicated that b was near zero (Table 1). If a and b are both zero, then K has no effect on the correlation and can be set to 1. Also, under the RPM, T and σ should be constant for all cycle-related genes, but φ can vary from 0 to 2π depending on the cell-cycle phase in which the transcript is expressed. In our application, we chose a set of 24 vectors (K, T, σ, φ, a, b) = (1, T̂, 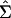 , φ, 0, 0), where T̂ and
, φ, 0, 0), where T̂ and 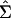 were estimates based on 18 phase-marker genes and φ was one of 24 angles, equally spaced around the circle and starting at 0. For each transcript, we calculate its Pearson correlation with each template in the set and take the maximum correlation over those templates as its score. The higher this score, the more the pattern displayed by the transcript resembles one of the templates. This scoring allows an ordering of any number of transcripts by their similarity to typical cell-cycle genes. Clearly, some transcripts will falsely appear as cycling given that we are examining so many (>44,000 in our application). Accordingly, we based our choice of cut-point for the ordered correlation scores on a permutation procedure (see supporting information, which is published on the PNAS web site) to restrict the expected number of false-positive transcripts, those mistakenly declared as cell-cycle related, to <1%.
were estimates based on 18 phase-marker genes and φ was one of 24 angles, equally spaced around the circle and starting at 0. For each transcript, we calculate its Pearson correlation with each template in the set and take the maximum correlation over those templates as its score. The higher this score, the more the pattern displayed by the transcript resembles one of the templates. This scoring allows an ordering of any number of transcripts by their similarity to typical cell-cycle genes. Clearly, some transcripts will falsely appear as cycling given that we are examining so many (>44,000 in our application). Accordingly, we based our choice of cut-point for the ordered correlation scores on a permutation procedure (see supporting information, which is published on the PNAS web site) to restrict the expected number of false-positive transcripts, those mistakenly declared as cell-cycle related, to <1%.
Table 1. Estimated parameters of the RPM for 18 well characterized phase-marker genes.
| Nominal phase* | Gene symbol | K̂ | T̂, hr | 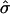 |
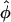 , rad , rad |
â | b̂ | SSE† |
|---|---|---|---|---|---|---|---|---|
| G1/S | CCNE1 | 0.67 | 15.1 | 0.054 | 0.56 | 0.46 | 0.002 | 1.705 |
| CDC6 | 0.69 | 14.7 | 0.056 | 5.96 | 0.46 | 0.000 | 1.774 | |
| PCNA | 0.62 | 15.1 | 0.074 | 5.87 | 0.54 | 0.012 | 1.529 | |
| E2F1 | 0.46 | 14.3 | 0.055 | 5.83 | 0.42 | 0.005 | 1.346 | |
| S | RFC4 | 0.36 | 14.3 | 0.058 | 5.47 | 0.38 | 0.007 | 1.224 |
| RRM2 | 0.69 | 15.3 | 0.075 | 5.36 | 0.76 | −0.008 | 3.281 | |
| G2 | CDC2 | 1.33 | 14.8 | 0.081 | 4.24 | 0.12 | 0.005 | 8.157 |
| TOP2A | 0.81 | 14.6 | 0.080 | 3.74 | 0.14 | 0.008 | 3.345 | |
| CCNA2 | 0.58 | 14.5 | 0.068 | 3.55 | 0.55 | −0.003 | 2.785 | |
| CCNF | 1.00 | 13.9 | 0.083 | 3.25 | 0.44 | 0.000 | 2.946 | |
| G2/M | STK15 | 1.23 | 14.2 | 0.076 | 3.06 | 0.32 | 0.004 | 3.257 |
| CCNB1 | 0.37 | 13.9 | 0.115 | 2.67 | 0.36 | 0.003 | 1.420 | |
| PLK | 1.16 | 14.0 | 0.070 | 2.61 | 0.43 | 0.005 | 1.741 | |
| BUB1 | 0.69 | 13.8 | 0.073 | 2.51 | 0.56 | −0.002 | 1.608 | |
| M/G1 | VEGFC | 0.49 | 14.4 | 0.068 | 2.66 | 0.67 | 0.003 | 1.781 |
| PTTG1 | 0.52 | 14.6 | 0.071 | 2.40 | 0.54 | 0.008 | 1.068 | |
| CDKN3 | 0.51 | 14.0 | 0.096 | 2.25 | 0.30 | 0.007 | 1.842 | |
| RAD21 | 0.36 | 13.2 | 0.084 | 1.81 | 0.29 | 0.009 | 1.745 |
Application of the RPM
To illustrate application of the RPM, we used an experiment where HeLa cells were arrested in S phase by using a double-thymidine block and subsequently released in synchrony (5). Gene expression was assessed with cDNA microarrays by using RNA from asynchronously growing HeLa cells as the reference. We downloaded the “raw” data (nonnormalized mean intensity value on each channel for each spot) for the 46-h experiment (experiment 3) from http://genome-www.stanford.edu/Human-CellCycle/HeLa/data.shtml. These data describe 44,158 transcripts at 47 hourly observation times (approximately three cell cycles). We analyzed base-2 logarithms of the nonnormalized expression ratios.
We illustrate the fit of the RPM using 18 of the phase-marker genes identified in ref. 5 (see supporting information and Table 1). First, we fit Eq. 1 to each gene, estimating T and σ separately for each gene. T̂g ranged from 13.13 to 15.24 h; 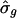 ranged from 0.054 to 0.115; and
ranged from 0.054 to 0.115; and 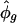 , phase angles estimated in radians, ranged around the circle (Table 1). Reassuringly, the estimates
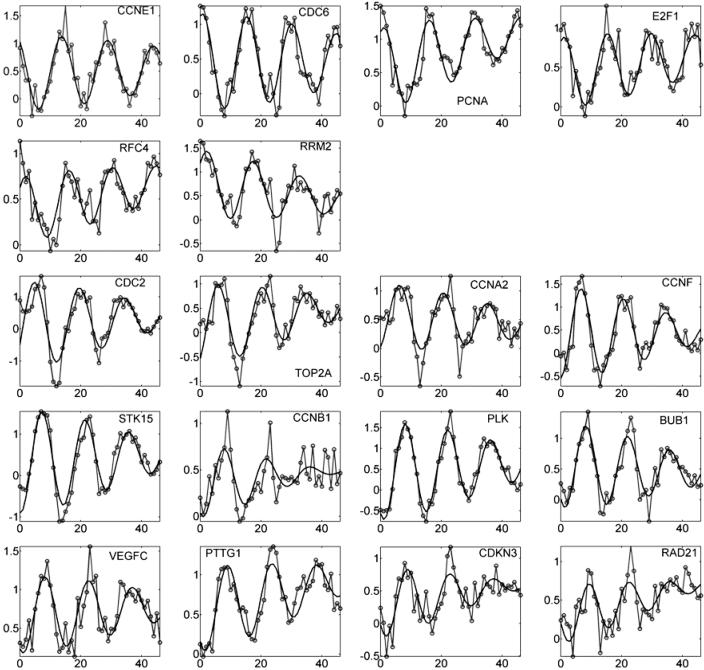
, phase angles estimated in radians, ranged around the circle (Table 1). Reassuringly, the estimates 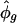 agreed well with the known phases for the 18 genes; the only exception was VEGFC, whose estimated phase of peak transcription came a little early compared with its expected relative position in the cell cycle (5). Our model fit the observed expression trajectories of these genes reasonably well (Fig. 3); the oscillations attenuate with time, with some transcripts showing background drift either upward or downward. Our fitted curves tended to lie below the observations at the first few time points.
agreed well with the known phases for the 18 genes; the only exception was VEGFC, whose estimated phase of peak transcription came a little early compared with its expected relative position in the cell cycle (5). Our model fit the observed expression trajectories of these genes reasonably well (Fig. 3); the oscillations attenuate with time, with some transcripts showing background drift either upward or downward. Our fitted curves tended to lie below the observations at the first few time points.
Fig. 3.
Plots of log2 expression ratio versus time (hr) for 18 well characterized phase-marker genes through about three cell cycles (data from ref. 5). Data (—○—) and fitted trajectory of the RPM (—). Genes in the first row are considered G1/S-phase genes; second row, S phase; third row, G2 phase; fourth row, G2/M phase; fifth row, M/G1 phase.
We tested the null hypothesis H0:σ = 0 to examine whether explicitly modeling attenuation improved fit. In general, it did; the bootstrap P value was <0.05 for 16 of the 18 genes (see supporting information). We also fit the 18 phase-marker genes in a single model with common values for both T and σ. The estimates T̂ and 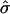 for the 18 marker genes taken together were 14.42 h and 0.073, respectively. The bootstrap P value for comparing the model with the same T and σ for all genes to one with separate values for each gene was 0.12, suggesting that these genes share common values for T and for σ as the biology requires (see supporting information).
for the 18 marker genes taken together were 14.42 h and 0.073, respectively. The bootstrap P value for comparing the model with the same T and σ for all genes to one with separate values for each gene was 0.12, suggesting that these genes share common values for T and for σ as the biology requires (see supporting information).
From the 44,158 transcripts, we identified 337 with a correlation score of 0.6 or greater as cell-cycle-related (see supporting information). Of those 337, we estimated that two would be false positives. One could adjust the cut-point to identify more genes at the cost of more expected false positives. If we lowered the cut-point to 0.5, we identified 675 transcripts overall and expected some 62 of them to be false positives. We considered the latter false-positive rate to be unacceptably high. Comparing our list of 337 transcripts with the list of 1,134 in ref. 5, 219 transcripts were on both lists and 118 were newly identified by our approach.
After selecting the 337 putatively cycling transcripts, we fit the RPM to each one. For most of these, T̂g fell between 13 to 16 h and 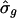 ranged from near 0 to ≈1.1. The scatter plot of
ranged from near 0 to ≈1.1. The scatter plot of 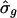 versus
versus 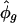 revealed unusually large estimates of σg for some transcripts (see supporting information). The transcripts with
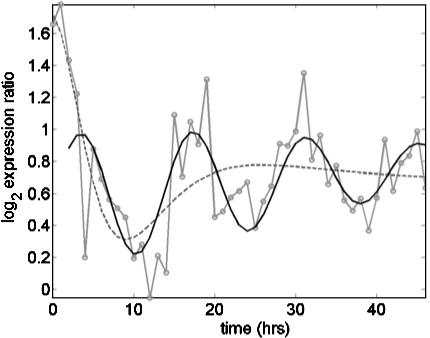
revealed unusually large estimates of σg for some transcripts (see supporting information). The transcripts with 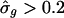 preferentially had phase angles that correspond to late G1 and early S phases, near the point when the cells had been arrested. Further exploration revealed transcripts with aberrant profiles (Fig. 4): an extreme initial mRNA level, with values high enough to distort the model's fit, produced a high estimated
preferentially had phase angles that correspond to late G1 and early S phases, near the point when the cells had been arrested. Further exploration revealed transcripts with aberrant profiles (Fig. 4): an extreme initial mRNA level, with values high enough to distort the model's fit, produced a high estimated 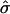 and a correspondingly rapid decay in oscillation and effectively masked the evident cycling. When we removed the first two data points and repeated the fitting, the characteristic cycling pattern was revealed (Fig. 4).
and a correspondingly rapid decay in oscillation and effectively masked the evident cycling. When we removed the first two data points and repeated the fitting, the characteristic cycling pattern was revealed (Fig. 4).
Fig. 4.
Observed log2 expression ratios (—○—) and fitted trajectories based on the RPM with (- - -) and without (—) the first two data points for the transcript with accession no. N95578 (identified as a clone of DKFZp434D0818). The reduced data fit captured the cycling because the influence of extreme values in the first two time points was eliminated.
Because the early time points may be subject to a recovery phenomenon unrelated to steady-state cycling (5), we refit the model for all 337 transcripts after omitting the first two time points to identify transcripts that might be sensitive to such early transient behavior. We regarded the original fit for a transcript as suspect if the estimated value of the parameter vector changed by a sufficiently large amount. For each transcript, we determined the Euclidean distance between the estimated parameter vector based on the original data and the one based on the reduced data, and we calculated the median and interquartile range of this sample of distances. We flagged as exhibiting suspiciously large changes in estimates those transcripts whose distance was more than three interquartile ranges from the median distance. By this criterion, 11 of 337 transcripts were judged subject to distortion. We reported estimates based on the reduced data for these 11 transcripts while retaining the original estimates for the remaining transcripts (see supporting information). This strategy tamed the more extreme estimates of σg.
Of the 337 transcripts, five had 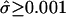 , corresponding to intriguing patterns that exhibited little evident attenuation. When one remembers that we searched for periodicity in >44,000 transcripts, these estimated values were likely due to the random variation that is inevitable with real data. Overall, except for a few extreme values, the estimates T̂g and
, corresponding to intriguing patterns that exhibited little evident attenuation. When one remembers that we searched for periodicity in >44,000 transcripts, these estimated values were likely due to the random variation that is inevitable with real data. Overall, except for a few extreme values, the estimates T̂g and 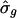 from the set of newly identified transcripts appeared compatible with those from the 18 phase-marker genes.
from the set of newly identified transcripts appeared compatible with those from the 18 phase-marker genes.
Discussion
Regression modeling of gene-expression trajectories can be an important alternative to clustering methods for analyzing the expression patterns of cell-cycle-related genes. By providing estimates of transcript-specific parameters, a regression approach reduces the raw data to a smaller number of biologically interpretable summary parameters that can be used to describe the transcripts or to characterize the effects of experimental interventions.
Recently, several groups have proposed regression models for the analysis of periodically expressed genes. An autoregressive model was able to provide an adequate description of trajectories by using relatively few parameters (16), but its parameters lack natural biologic interpretations. The SPM (7) is based on the simple notion that in a given cell each gene begins full expression abruptly at some point in the cell cycle and is expressed at a constant rate until it reaches the point where it abruptly stops being expressed and the mRNA instantaneously disappears. Under the SPM, stochastic variation among cells in the timing of activation and deactivation smoothes abrupt expression changes and allows a somewhat flexible shape for the observed trajectory of a large number of cells. The SPM accommodates attenuation but without clear biologic mechanism. In addition, the SPM describes the portion of the cycle where the gene is transcribed by two parameters, the activation time and deactivation time, although many investigators use a single “phase angle” to measure the location of peak expression (e.g., refs. 2 and 5). Other methods for describing periodic expression trajectories include singular-value decomposition (17-19), B splines (20), and partial least squares (21). In general, these approaches do not provide the parsimony and biologically interpretable parameters that regression models offer.
An important feature of the RPM is that attenuation arises as a natural consequence of variation in the duration of the cell cycle across cells. The model provides a single parameter, σ, to assess this variation and, hence, to measure attenuation. It allows estimation of a transcript's phase angle, a useful parameter for elucidating its role in the cell cycle. On the other hand, all models involve simplifying assumptions. The cosine functional form provides a rigidly defined shape and does not flexibly adapt to expression trajectories that may vary widely in shape from transcript to transcript while maintaining common periodicity. Nevertheless, as seen in Fig. 3, the cosine curve adequately accommodated differently shaped trajectories while keeping the model parameters as few and as intuitive as possible.
Inference under regression models for cell-cycle expression, whether SPM (7) or RPM, is difficult, however, because the distribution and the correlation structure of the error terms are unknown. In such circumstances, we prefer bootstrap methods for inference to methods that rely on asymptotic distributions, but bootstrapping can be computationally expensive. Better statistical techniques for inference in such models are needed.
One goal in current studies of cell-cycle gene expression is to identify transcripts that are expressed periodically and entrained with the cell cycle. Our approach was to modify widely used correlation-based methods for clustering genes (2, 5, 15). Usually these methods construct templates, the ideal trajectories that represent typical cycling genes, by using averages of a few observed trajectories of known cycling genes with similar phase angles. Our modification replaced the simple averages with predicted trajectories based on the RPM, an approach that smoothes random fluctuations. We used parameter values derived from fitting known cell-cycle genes so that our templates reflected data and incorporated attenuation (see supporting information). We used a range of values only for φ, but larger sets of templates including ranges of values for other parameters might sometimes be warranted (see supporting information). In addition, templates can be formed by the model for possible trajectories for which established cell cycle genes are either unknown or unavailable in the data, features that rule out averaging. We must caution, however, that correlation-based methods can be misleading (22).
In conclusion, we have proposed the RPM for studying periodically expressed transcripts and have demonstrated its use with published data from synchronized HeLa cell cultures. Attenuation in the expression level of cell-cycle genes over time is well characterized by allowing variability across cells in the duration of the cell cycle. Such variability causes the cells to fall increasingly out of synchrony over time, which, in turn, damps the periodic expression. The RPM is parsimonious and can be applied to characterize aggregated levels arising from any studies where initial synchrony among cycling units is experimentally induced. The RPM allows simultaneous estimation of biologically relevant parameters and formal hypothesis testing. Genes can be clustered based on these biologically interpretable parameters, and relationships among cycling genes may be revealed. Several additional applications are envisioned. The approach allows identification of transcripts of periodic expression in the cell cycle. The estimated model parameters could also allow cell cultures, e.g., from normal and tumor tissues, to be characterized and contrasted based on biologically interpretable features of their growth regulation. In studies of genotoxic agents, effects of the agents on the shifts of phase angles of specific checkpoint genes may be studied.
Supplementary Material
Acknowledgments
We thank Barbara Wetmore, Fred Parham, and the two reviewers for their careful reading of and constructive comments on an earlier version of this manuscript.
Abbreviations: RPM, random-periods model; SPM, single-pulse model.
References
- 1.Cho, R. J., Campbell M. J., Winzeler, E. A., Steinmetz, L., Conway, A., Wodicka, L., Wolfsberg, T. G., Gabrielian, A. E., Landsman, D., Lockhart, D. J., et al. (1998) Mol. Cell 2, 65-73. [DOI] [PubMed] [Google Scholar]
- 2.Spellman, P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown, P. O., Bostein, D. & Futcher, B. (1998) Mol. Biol. Cell 9, 3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu, S., DeRisi, J., Eisen, M., Mulholland, J., Botstein, D., Brown, P. O. & Herskowitz, I. (1998) Science 282, 699-705. [DOI] [PubMed] [Google Scholar]
- 4.Cho, R. J., Huang, M., Campbell, M. J., Dong, H., Steinmetz, L., Sapinoso, L., Hampton, G., Elledge, S. J., Davis, R. W. & Lockhart, D. J. (2001) Nat. Genet. 27, 48-54. [DOI] [PubMed] [Google Scholar]
- 5.Whitfield, M. L., Sherlock, G., Saldanha, A. J., Murray, J. I., Ball, C. A., Alexander, K. E., Matese, J. C., Perou, C. M., Hurt, M. M., Brown, P. O., et al. (2002) Mol. Biol. Cell 13, 1977-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jorgensen, P., Nishikawa, J. L., Breitkreutz, B.-J. & Tyers, M. (2002) Science 297, 395-400. [DOI] [PubMed] [Google Scholar]
- 7.Zhao, L. P., Prentice, R. & Breeden, L. (2001) Proc. Natl. Acad. Sci. USA 98, 5631-5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallant, A. R. (1987) Nonlinear Statistical Models (Wiley, New York).
- 9.Seber, G. A. F. & Wild, C. J. (1989) Nonlinear Regression (Wiley, New York).
- 10.Peddada, S. D. & Patwardhan, G. (1992) Biometrika 79, 654-657. [Google Scholar]
- 11.Peddada, S. D. (1993) in Handbook of Statistics, ed. Rao, C. R. (Elsevier North-Holland, New York), Vol. 9, pp. 723-744. [Google Scholar]
- 12.Shao, J. (1990) Stat. Probabil. Lett. 10, 77-85. [Google Scholar]
- 13.Zhang, J., Peddada, S. D. & Rogol, A. (2000) Statistics for 21st Century, eds. Rao, C. R. & Szekeley, G. (Dekker, New York), pp. 459-483.
- 14.Kunsch, H. (1989) Ann. Stat. 17, 1217-1241. [Google Scholar]
- 15.Heyer, L. J., Kruglyak, S. & Yooseph, S. (1999) Genome Res. 9, 1106-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramoni, M. F., Sebastiani, P. & Kohane, I. S. (2002) Proc. Natl. Acad. Sci. USA 99, 9121-9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alter, O., Brown, P. O. & Bostein, D. (2000) Proc. Natl. Acad. Sci. USA 97, 10101-10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holter, N. S., Mitra, M., Maritan A., Cieplak, M., Banavar, J. R. & Fedoroff, N. V. (2000) Proc. Natl. Acad. Sci. USA 97, 8409-8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holter, N. S., Maritan, A., Cieplak, M., Fedoref, N. V. & Banavar, J. R. (2001) Proc. Natl. Acad. Sci. USA 98, 1693-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luan, Y. & Li, H. (2003) Bioinformatics 19, 474-482. [DOI] [PubMed] [Google Scholar]
- 21.Johansson, D., Lindgren, P. & Berglund, A. (2003) Bioinformatics 19, 467-473. [DOI] [PubMed] [Google Scholar]
- 22.Peddada, S. D., Lobenhofer, E. K., Li, L., Afshari, C. A., Weinberg, C. R. & Umbach, D. M. (2003) Bioinformatics 19, 834-841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.