Abstract
The chlamydial developmental cycle is characterized by an intracellular replicative form, termed the reticulate body, and an extracellular form called the elementary body. Elementary bodies are characterized by a condensed chromatin, which is maintained by a histone H1-like protein, Hc1. Differentiation of elementary bodies to reticulate bodies is accompanied by dispersal of the chromatin as chlamydiae become transcriptionally active, although the mechanisms of Hc1 release from DNA have remained unknown. Dissociation of the nucleoid requires chlamydial transcription and translation with negligible loss of Hc1. A genetic screen was therefore designed to identify chlamydial genes rescuing Escherichia coli from the lethal effects of Hc1 overexpression. CT804, a gene homologous to ispE, which encodes an intermediate enzyme of the non-mevalonate methylerythritol phosphate (MEP) pathway of isoprenoid biosynthesis, was selected. E. coli coexpressing CT804 and Hc1 grew normally, although they expressed Hc1 to a level equivalent to that which condensed the chromatin of parent Hc1-expressing controls. Inhibition of the MEP pathway with fosmidomycin abolished IspE rescue of Hc1-expressing E. coli. Deproteinated extract from IspE-expressing bacteria caused dispersal of purified chlamydial nucleoids, suggesting that chlamydial histone-DNA interactions are disrupted by a small metabolite within the MEP pathway rather than by direct action of IspE. By partial reconstruction of the MEP pathway, we determined that 2-C-methylerythritol 2,4-cyclodiphosphate dissociated Hc1 from chlamydial chromatin. These results suggest that chlamydial histone-DNA interactions are disrupted upon germination by a small metabolite in the MEP pathway of isoprenoid biosynthesis.
Chlamydia trachomatis is the leading cause of preventable blindness worldwide and a major cause of sexually transmitted disease in developed countries (1). Chlamydiae are bacterial obligate intracellular pathogens with a biphasic developmental cycle that alternates between infectious extracellular forms, termed elementary bodies (EBs), and intracellular replicative forms known as reticulate bodies (RBs) (2). The metabolically inert EBs are characterized by a condensed nucleoid structure. Within a few hours after infection the chromatin becomes dispersed as transcription is initiated and differentiation to the larger, metabolically active RBs begins (3). RBs replicate by binary fission until 18 to 48 h after infection, at which time they begin to differentiate back to EBs, a process typified by recompaction of the nucleoid.
The DNA of EBs is held in a condensed state by the action of two histone H1 homologs, Hc1 and Hc2 (4-8). Hc1 is conserved among all chlamydiae, whereas Hc2 displays a variable molecular weight depending on the C. trachomatis serovar and is absent from some chlamydial species and strains (5). Both hctA and hctB, encoding Hc1 and Hc2, respectively, are transcribed late in the developmental cycle concomitant with RB differentiation back to EBs and nucleoid condensation (3, 9). Expression of either of the histone homologs in Escherichia coli results in the formation of a highly condensed nucleoid (8, 10). Hc1 expression in particular results in a nucleoid ultrastructurally similar to the condensed nucleoid of EBs. Expression levels of Hc1 that result in nucleoid condensation are accompanied by a global downshift in transcription and translation, a phenomenon that also reflects events during differentiation of RBs to EBs (11, 12). Although Hc1 interactions with DNA have been studied extensively both in vivo and in vitro (10-15), the mechanisms of release of chlamydial chromatin from the constraints imposed by Hc1 association are unknown and have not been studied in chlamydiae.
Expression of Hc1 in E. coli is effectively lethal (11, 12), presumably because E. coli lacks the means to release the histone-DNA complex. A heterologous genetic screen was therefore designed to select for chlamydial genes that might release E. coli from growth restriction imposed by histone expression. The results suggest that an intermediate enzyme of the non-mevalonate methylerythritol 4-phosphate (MEP) pathway of isoprenoid biosynthesis is critical to Hc1-DNA disassociation and that a small molecule, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate (MEC), produced by this pathway is responsible for chlamydial nucleoid decondensation.
Materials and Methods
Vector Construction. The two compatible vectors pLac and pTet, designed for coexpression in E. coli, were constructed by using cassettes from the modular vectors pZE12-luc, pZE12-luc, andpZA31-luc, described and kindly provided by Hermann Bujard (16). pLac was designed to contain a ColE1 origin of replication (ori), a PLlacO-1 promoter, and an ampicillin (Amp)-resistance (Ampr) gene. To construct this vector, pZE12-luc was digested with SacI/KpnI and the fragment containing Ampr/PLlacO-1/RBSII was isolated. This fragment was ligated to a second fragment containing the multiple cloning site (MCS) and ColE1 ori that had been isolated from a KpnI/SacI digestion of pZE21-MCS-1. pTet was designed to contain a p15A ori, PLtetO-1 promoter, and the chloramphenicol (Cm)-resistance gene. To construct this vector, the ColE1 ori of pPROTet.E (Clontech) was replaced with the p15A ori isolated from pZA31-luc.
C. trachomatis L2 Genomic Library Construction. A partial Sau3AI digest of C. trachomatis L2 genomic DNA was size fractionated on a 0.8% agarose gel. Fragments of 1-5 kb were electroeluted and ligated into pLac that had been digested with BamHI and dephosphorylated with calf intestinal alkaline phosphatase. Aliquots of the ligation reaction mixture were electroporated into electrocompetent spectinomycin (Spec)-resistant DH5αPRO cells (Clontech), plated on LB plates containing Amp (100 μg/ml) and Spec (50 μg/ml), and incubated for 16 h at 37°C. The genomic library consisted of ≈40,000 clones, providing ≈40× coverage of the 1.043-megabase 875-gene genome (17). Colonies were scraped directly into LB broth and the DNA was isolated by using a Qiagen midi kit.
Genetic Screen. hctA, the gene encoding Hc1 (4), was subcloned into the pTet vector to yield pTetHctA. This construct, expressed in E. coli DH5αPRO, was able to induce the same condensed nucleoid and growth defect as previously described by Barry et al. (10) (data not shown). To identify genomic clones that would rescue the aforementioned phenotype, pTetHctA and pLacCt were cotransformed into DH5αPRO cells, which were plated on LB plates containing Cm (34 μg/ml), Amp, and Spec and incubated for 16 h at 37°C. The resulting colonies were scraped directly into LB broth containing Cm, Amp, and Spec and diluted to an OD550 of 0.2. The L2 genomic library was induced for 2 h at 30°C with 1 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG), at which point Hc1 expression was induced with 100 ng/ml anhydrotetracycline. The culture was grown for an additional 5 h at 30°C before being plated on LB plates containing Cm, Amp, and Spec and incubated for 16 h at 30°C. Eighty surviving colonies were rescreened for their ability to survive Hc1 induction, of which 22 remained positive. Each C. trachomatis genomic clone (Amp-resistant) was purified away from pTetHctA (Cm-resistant) by the following method. Plasmid from each clone was purified by using a Qiagen mini prep isolation kit, and the DNA was used to transform DH5αPRO. Transformants were plated on LB agar plates containing Amp and Spec and incubated for 16 h at 30°C. The resultant colonies were streaked in duplicate onto LB agar plates containing either Amp or Cm. pLac plasmid clones were isolated from colonies that grew on Amp but not Cm plates and examined for duplications by enzymatic digestions. Of the 22 positive clones tested, 11 were duplicates. The 11 unique clones were confirmed for their ability to rescue E. coli chromatin from Hc1 condensation before being sequenced. All sequencing services were performed by the Iowa State University DNA Sequencing and Synthesis Facility (Ames, IA).
C. trachomatis Nucleoid Preparation. C. trachomatis was propagated in HeLa 229 cells and the EBs were purified by Renografin (Bracco Diagnostics, Princeton) density gradient centrifugation (18). C. trachomatis nucleoids were isolated from EBs by extraction with 2% Zwittergent 3-14 (Calbiochem) for 1 h at room temperature with rotation. This procedure yielded structures consisting of the nucleoid surrounded by the freely permeable disulfide-linked outer membrane complex (19). Nucleoid complexes were pelleted at 15,000 × g for 5 min and washed at least three times with PBSE (50 mM sodium phosphate, pH 7.5/150 mM NaCl/1.5 mM EDTA)
Deproteinated E. coli Extract. Fifty milliliters of culture containing the appropriate strains were induced at 30°C for 5 h with 1 mM IPTG before processing. Cells were pelleted, washed twice in TMN (20 mM Tris·HCl, pH 6.8/10 mM MgCl2/50 mM NaCl) and resuspended in 200 μl of TMN. Cells were disrupted by ultrasonication and the cell debris was removed by ultracentrifugation (Optima Max Ultracentrifuge, Beckman Coulter) at 35,000 rpm for 15 min. The supernatant was passed through a Centricon YM-3 centrifugal filter (3,000 molecular weight cutoff, Millipore), and the deproteinated flow-through was collected for further experiments.
Enzyme Reconstitution. Partial reconstitution of the MEP pathway was performed essentially as described (20-22). Briefly, partially purified recombinant chlamydial IspD, IspE, and IspF, dialyzed in a 10,000 molecular weight cut-off dialysis cassette (Pierce) against a 20 mM Tris·HCl, pH 6.8/50 mM NaCl solution to remove any contaminating small molecules, were sequentially added to reaction mixtures containing 5 mM CTP, 5 mM ATP, 1 mM DTT, 10 mM MgCl2, and 100 mM Tris·HCl at pH 6.8, in the presence or absence of 12 μM MEP. Reactions were incubated in a 37°C water bath for 12 h.
DNase I Assays. Either deproteinated extract from recombinant E. coli or reaction products from enzyme reconstitutions were added to purified chlamydial nucleoids. Nucleoid reactions were incubated in a 37°C water bath for 0.5 h before being split into two aliquots, one of which was treated with 150 units of DNase I for 15 min at room temperature. Reactions were stopped with 1 mM EDTA and incubation at 65°C for 10 min; the mixture was then treated with Proteinase K for 1 h at 37°C. After extraction with phenol/chloroform, the DNA was precipitated and the pellet was dissolved in TE (10 mM Tris·HCl/1 mM EDTA) containing RNase A and resolved on a 3% agarose gel.
Transmission Electron Microscopy. Samples were fixed with 2.5% glutaraldehyde/4% paraformaldehyde in 0.1 M sodium cacodylate/0.05 M sucrose buffer. The samples were postfixed with 0.5% osmium tetroxide/0.8% potassium ferricyanide followed by 1% tannic acid and stained overnight at 4°C en bloc in 1% uranyl acetate. Samples were dehydrated in a graded ethanol series and embedded in Spurrr's resin. Thin sections were cut with an RMC MT-7000 ultramicrotome (Ventana, Tucson, AZ), stained with 1% uranyl acetate and Reynold's lead citrate, and observed at 60 kV on a Philips CM-10 electron microscope (FEI, Hillsboro, OR). Images were acquired with an AMT digital camera system (Advanced Microscopy Techniques, Chazy, NY) and processed by using Adobe Photoshop v.7.0 (Adobe Systems, San Jose, CA).
MS Analysis. DNase I assays were performed on C. trachomatis nucleoids in extracts from E. coli expressing pLac or CT804 as described, except that samples for MS analysis were collected directly after DNase I digestion. The histone samples were diluted 1:10 in a matrix solution containing saturated sinapinic acid in acetonitrile/water/trifluoroacetic acid (30:70:0.1, vol/vol) and a 1-μl aliquot was spotted and air dried on a sample plate for matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) MS analysis. Mass spectra were obtained by using a Voyager-DE STR MALDI-TOF instrument (Applied Biosystems). The ion spectra were collected with the following instrument settings: mode of operation, linear; extraction mode, delayed; extraction delay time, 450 nsec; polarity, positive; accelerating voltage, 25,000 V; grid voltage, 91%; acquisition mass range, 5,000-20,000 Da, laser repetition rate, 20 Hz. The spectra were calibrated by using internal standards of E. coli thioredoxin (average molecular mass 11,674.5 Da) and horse apomyoglobin (average molecular mass 16,952.6 Da).
Results
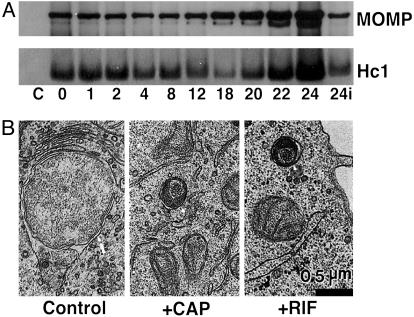
Fate of Hc1 During Early Differentiation. Decondensation of the condensed nucleoid in EBs occurs rapidly after infection, and chlamydiae are transcriptionally active by 2 h after infection (3). To determine the fate of histone present upon infection, the Hc1 content of C. trachomatis L2 was analyzed during the initial 24 h of development by immunoblotting and densitometry. As shown in Fig. 1A, Hc1 levels remain essentially constant through the first 8 h of development (96% of 0-time control), although the nucleoid has clearly dispersed and the chlamydiae are transcriptionally active. This finding suggests that although Hc1 remains present in early chlamydial development forms, it is no longer functional in maintaining the condensed state of the nucleoid. To further explore requirements for nucleoid decondensation, cultures were infected in the presence of Cm (50 μg/ml), an inhibitor of prokaryotic translation, or rifampicin (150 μg/ml), an inhibitor of prokaryotic transcription, and observed 8 h after infection by transmission electron microscopy. By 8 h after infection, EBs in untreated cultures had differentiated into RBs, whereas treatment with either Cm or rifampicin abolished nucleoid decondensation and the ability to progress through the developmental cycle (Fig. 1B). These data demonstrate that de novo chlamydial transcription and translation are required for decondensation of the EB nucleoid upon germination.
Fig. 1.
Properties of chlamydial nucleoid decondensation. (A) Western blot analysis of C. trachomatis L2 Hc1 and major outer membrane protein (MOMP) over the initial 24 h of the developmental cycle. HeLa cells in 24-well plates were infected at a multiplicity of infection of ≈100 and solubilized at the times indicated, and equivalent amounts of each culture were loaded for SDS/PAGE and immunoblotting with an anti-Hc1 antibody. Control (C) indicates uninfected cells, and subsequent times are given as hours after infection; 24i indicates infection carried out in the continuous presence of chloramphenicol. (B) Ultrastructural analysis of C. trachomatis L2 development in the absence (control) or presence of Cm (CAP) or rifampicin (RIF) at 8 h after infection. The arrow in the control panel indicates an RB with a characteristic dispersed nucleoid. The arrowheads in the CAP and RIF panels identify EBs with condensed nucleoids and unable to differentiate and progress through the developmental cycle. (Bar equals 0.5 μm.)
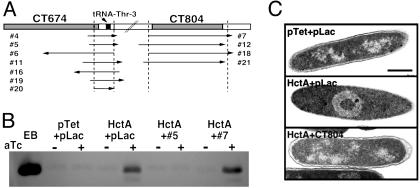
Selection of Chlamydial Genes Required for Nucleoid Decondensation. Because chlamydiae lack a tractable genetic system, we designed a genetic screen in E. coli to identify the molecule or molecules involved in nucleoid decondensation. Expression of Hc1 in E. coli results in condensation of the chromatin to form a nucleoid structure morphologically indistinguishable from that of EBs (10, 12). Concurrent with this phenomenon is a global down-regulation of transcription and translation as the chromatin is occluded in the nucleoid complex (10), an effect believed to reflect events in the terminal differentiation of RBs to EBs. The requirement for chlamydial protein synthesis to initiate nucleoid decondensation coupled with the lethal effect of Hc1 expression on E. coli suggested that chlamydial gene products might overcome the growth inhibition in E. coli. Thus, we constructed a two-plasmid system to coexpress in E. coli both Hc1 under the control of the Tet promoter and a C. trachomatis library under the control of the Lac promoter, and we selected for clones that could rescue the lethal phenotype induced upon Hc1 expression (see Materials and Methods and Fig. 6, which is published as supporting information on the PNAS web site). Eleven clones were selected on the basis of this screening, all of which fell into two distinct loci (Fig. 2A). Clones from the first locus contained inserts in either orientation with the minimal chromosomal overlap encompassing a tRNA but no other recognized ORF. Four independent clones mapping to the second locus were in an expressing orientation relative to the vector promoter and the minimum contiguous sequence overlapped a single ORF, CT804, in the C. trachomatis serovar D genome (23). The effect of expression of representative clones from each locus on Hc1 protein levels were examined by immunoblotting (Fig. 2B). Regardless of orientation, expression of the first locus reduced Hc1 levels below the limits of detection, presumably by inhibiting transcription or translation of hctA. In contrast, in E. coli coexpressing Hc1 and CT804, Hc1 was present at levels equivalent to those sufficient to condense the nucleoid in the parent Hc1-expressing strain. Examination of the nucleoid structure by transmission electron microscopy confirmed that E. coli coexpressing Hc1 and the parent pLac vector displayed the characteristically condensed nucleoid observed previously (10). However, E. coli coexpressing Hc1 and CT804 had a chromatin structure identical to the vector controls (Fig. 2C). Histone was therefore present but not functioning to condense the chromatin in E. coli coexpressing CT804 and Hc1. Because this situation reflects that of germinating EBs, CT804 was selected for further study.
Fig. 2.
Selection of C. trachomatis genetic loci that rescue E. coli from Hc1-induced growth inhibition. (A) Schematic showing overlapping clones and their orientations relative to the corresponding annotated ORFs (www.stdgen.lanl.gov). Vertical lines indicate the minimal contiguous sequence. (B) Western blot analysis of Hc1 content in E. coli rescues. Cultures expressing empty vectors (pLac + pTet), Hc1 alone (HctA + pLac), and Hc1 and a representative rescuing clone from each locus (HctA + #5 and HctA + #7) were grown in the presence of IPTG to induce expression of the rescuing construct and split, and anhydrotetracycline (aTc) was added to one half to induce Hc1. After 5 h of induction, cultures were normalized for OD550 before loading and EBs were used as a control for Hc1 position. (C) Ultrastructural analysis of E. coli induced to express empty vectors (pTet + pLac), Hc1 (HctA + pLac), or Hc1 and CT804 (HctA + CT804). (Bar equals 0.5 μm.)
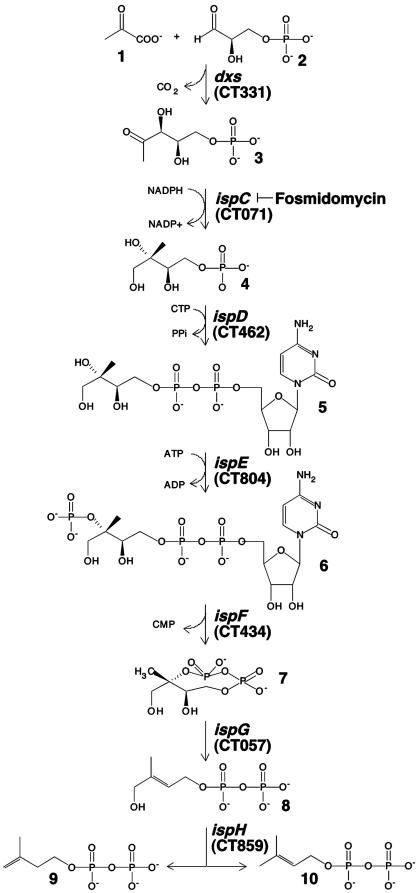
Requirement for Metabolites of the MEP Pathway. CT804 encodes ispE (ychB) or 4-diphosphocytidyl-2-C- methyl-d-erythritol kinase, an intermediate enzyme in the non-mevalonate MEP isoprenoid biosynthetic pathway. The MEP pathway consists of seven enzymes involved in the metabolism of pyruvate and glyceraldehyde 3-phosphate to isopentenyl diphosphate and dimethylallyl diphosphate, the universal five-carbon precursors for isoprenoids (Fig. 3). This pathway, which has only recently been described (24-27), has no known orthologs in mammals but is present in higher plants and is the sole source of isoprenoids in many eubacteria, including numerous microbial pathogens.
Fig. 3.
The MEP pathway. 1, pyruvate; 2, glyceraldehyde 3-phosphate; 3, deoxyxylulose 5-phosphate; 4, 2-C-methyl-d-erythritol 4-phosphate; 5, 4-diphosphocytidyl-2-C-methyl-d-erythritol; 6, 4-diphosphocytidyl-2-C-methyl-d-erythritol 2-phosphate; 7, 2-C-methylerythritol 2,4-cyclodiphosphate; 8, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate; 9, isopentenyl diphosphate; 10, dimethylallyl diphosphate. The genes encoding enzymes of the pathway are indicated in italics and the C. trachomatis ORFs predicted to encode the orthologous enzymes are in parentheses. The enzymatic step inhibited by fosmidomycin is indicated.
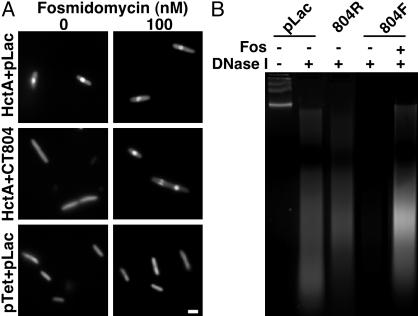
IspE catalyzes the ATP-dependent phosphorylation of 4-diphosphocytidyl-2-C-methyl-d-erythritol (5, Fig. 3) to 4-diphosphocytidyl-2-C-methyl-d-erythritol 2-phosphate (6, Fig. 3) (22). Thus, we considered whether a small metabolite produced from the activity of CT804, rather than a direct action of the enzyme on histone, might control Hc1-DNA interactions. To investigate this possibility, E. coli expressing Hc1 only (HctA + pLac), Hc1 and CT804 (HctA + CT804), and empty vectors (pTet + pLac), were grown in the presence of fosmidomycin, an antibiotic that specifically inhibits IspC, an enzyme involved in an early step of the MEP pathway (Fig. 3). We reasoned that if a small molecule produced downstream of IspC was involved, rescue by CT804 overexpression would be abolished by fosmidomycin, whereas a protein-protein interaction would not. In the absence of fosmidomycin, the nucleoid of E. coli expressing Hc1 was highly condensed compared with both vector only and CT804-expressing cultures, as monitored by acridine orange staining (Fig. 4A). Expression of CT804 in the presence of fosmidomycin abolished the ability of CT804 products to protect the chromatin from condensation by Hc1. Fosmidomycin had no effect on E. coli chromatin structure or Hc1-induced nucleoid condensation. Depletion of precursor substrates within the MEP pathway thus ablates CT804 rescue of Hc1-expressing E. coli, suggesting a role for intermediates in this pathway in histone dissociation.
Fig. 4.
(A) Acridine orange staining of E. coli induced to express Hc1 (HctA + pLac), Hc1 and CT804 (HctA + CT804), or parent vectors only (pTet + pLac), in the absence or presence of 100 nM fosmidomycin. Condensed DNA appears as a central brightly fluorescent sphere (10). (Bar equals 1 μm.) (B) DNase I digestion of C. trachomatis L2 EB nucleoids incubated with deproteinated extracts of E. coli cultures expressing the parent plasmid, pLac; CT804 in a nonexpressing reverse orientation (804R), CT804 in an expressing orientation (804F), or CT804 in an expressing orientation in the presence of fosmidomycin (Fos). Untreated chlamydial chromosomal DNA from EB nucleoids runs as a high molecular weight band or bands. Chlamydial chromatin associated with Hc1 is partially protected from DNase I digestion such that it is nicked and appears as a ladder or smear on the gel. When Hc1 is dissociated, the DNA is exposed and completely digested.
A Small Metabolite Is Required for in Vitro Nucleoid Decondensation. To verify the requirement for metabolites of the MEP pathway, we applied deproteinated extracts from E. coli expressing CT804 to authentic chlamydial nucleoids and assessed protection of the chromatin from DNase I digestion. Deproteinated extracts from E. coli expressing CT804, but not from vector controls or those with CT804 in a nonexpressing orientation, released Hc1 from the DNA, leading to complete digestion by DNase I (Fig. 4B). In addition, deproteinated extracts of E. coli expressing CT804 in the presence of fosmidomycin did not cause dissociation of Hc1 from chlamydial chromatin as indicated by loss of DNase I sensitivity. Decondensation of the nucleoid thus appears to be mediated by a small metabolite within the MEP pathway. Furthermore, the fact that a small-molecule fraction of CT804-expressing E. coli confers DNase I sensitivity to nucleoids suggests that the molecule is able to disrupt preformed DNA-Hc1 complexes rather than simply prevent Hc1 from binding DNA.
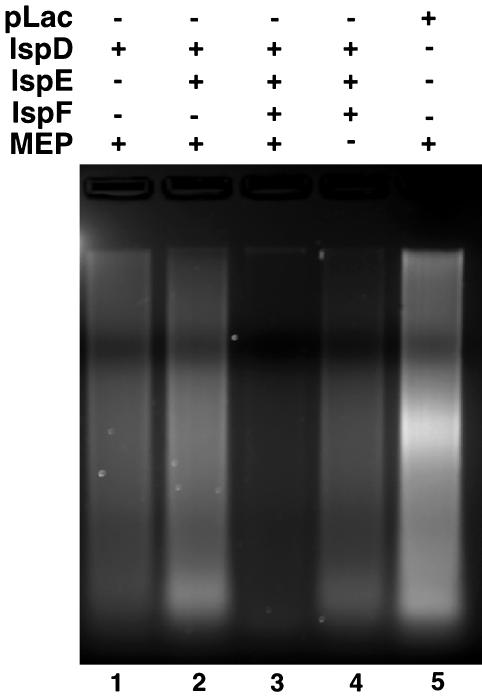
Partial Reconstitution of the MEP Pathway. Orthologs of the entire MEP pathway are present in the genomes of Chlamydia spp. (23, 24) and, consistent with a function early in the chlamydial developmental cycle, each of these genes is transcribed by 2 h after infection (see Fig. 7, which is published as supporting information on the PNAS web site). E. coli also encodes a complete MEP pathway (22, 24). Because both C. trachomatis and E. coli possess this pathway, it is possible that molecules downstream of the CT804 product are produced in increased quantities as a result of CT804 overexpression. Indeed, the subsequent product, MEC, is accumulated to high concentrations in several bacteria under conditions of stress (28). To determine which molecule is actually responsible for nucleoid decondensation, we partially reconstructed the MEP pathway by sequentially adding the expressed and partially purified orthologous chlamydial proteins, IspD (CT462), IspE (CT804), and IspF (CT434) to commercially available MEP. Only when all three enzymes were combined in the presence of MEP was Hc1 dissociated from the chromatin (Fig. 5). Neither MEP, isopentenyl diphosphate, nor dimethylallyl diphosphate alone was able to confer DNase I sensitivity (Fig. 5, lane 5 and data not shown). The actual metabolite dissociating Hc1 thus appears to be MEC, although we have not yet isolated the compound from germinating EBs to confirm the structure chemically.
Fig. 5.
DNase I assay of EB nucleoids incubated with partially reconstructed MEP pathway. Partially purified recombinant chlamydial IspD, IspE, and IspF were dialyzed to deplete small molecules and sequentially combined in the presence or absence of MEP as substrate. DNase I assays were performed on purified EB nucleoids incubated with the enzymatic reaction mixtures containing IspD (lane 1), IspD plus IspE (lane 2), IspD plus IspE plus IspF (lane 3), IspD plus IspE plus IspF minus MEP (lane 4), or pLac (i.e., no exogenously expressed protein) (lane 5).
Discussion
The chromosome of chlamydial EBs exists in a highly compacted state that presumably protects the chromatin during the extracellular phase of the developmental cycle. The chlamydial chromatin is condensed primarily through the actions of a histone H1-like protein, Hc1. The release of the chlamydial chromosome from the constraints imposed by Hc1 has been enigmatic. A single report using partially purified proteins in an in vitro assay suggested proteolysis of Hc1 as a possible mechanism of Hc1 dispersal (29); however, our analysis of Hc1 during the intracellular germination of EBs revealed no change in quantity or properties as the nucleoid was decondensed. Indeed, Hc1 was present in nascent RBs although it no longer functioned in condensation of the chromatin. A requirement for chlamydial transcription and translation to initiate this process suggested the possibility of a heterologous genetic screen to identify the chlamydial genes involved. This genetic screen identified chlamydial ispE as the gene that rescued E. coli from the lethal effects of Hc1 expression and did so with the phenotype observed in EB differentiation. This finding, in addition to a requirement for precursors of that enzymatic pathway and the demonstration that deproteinated extracts of chlamydial ispE-expressing E. coli release Hc1 from purified EB nucleoids, suggests that an intermediary metabolite of the non-mevalonate MEP pathway of isoprenoid biosynthesis is used by chlamydiae to release their chromatin from the constraints of histone-mediated compaction as EBs germinate to initiate intracellular development.
Although C. trachomatis encodes a complete MEP pathway, ispE was the only component of that pathway selected in the rescue of Hc1-expressing E. coli. As the chlamydial library provides 40-fold coverage of the genome, the fact that ispE was the sole member of the pathway identified suggests that it plays an important regulatory role within the MEP pathway in chlamydiae. Overexpression of E. coli IspE in the same vector system did not protect E. coli from the lethal effects of Hc1 expression (data not shown), suggesting that chlamydial IspE displays unique properties and is a central regulator of the MEP pathway in chlamydiae. A detailed comparison of the kinetic properties of chlamydial and E. coli IspE will be required to determine how chlamydial IspE effects such a key role in the pathway. This is of particular interest because it appears not to be the immediate product of IspE enzymatic activity but a downstream metabolite, MEC, that actually disrupts Hc1-DNA interaction. The precise mechanism of Hc1 dissociation from DNA is unclear, but matrix-assisted laser desorption ionization-time-of-flight MS of the released histone revealed no change in mass indicative of a covalent modification, suggesting that the small molecule may act as a competitive inhibitor or an allosteric effector on Hc1 (see Fig. 8, which is published as supporting information on the PNAS web site).
It is somewhat paradoxical that decondensation of the chlamydial nucleoid, a step absolutely required for progress through the developmental cycle, is itself dependant on de novo transcription and translation. Despite the constrained state of EB chromatin, CT804 must be available, or made available, for transcription upon EB internalization. Interestingly, structural predictions of the chlamydial chromosome (www.cbs.dtu.dk) indicate that the highest probability of A-form DNA coincides with CT804. A-form DNA is thought to be more readily available for initiation of transcription (30); however, another possibility is that A-form DNA may simply have a lower affinity for Hc1.
An additional concern is the fate of the MEC during terminal differentiation of RBs to EBs, as its presence would presumably prohibit Hc1-mediated compaction of the chromatin that characterizes this process. Because of the asynchrony of the late chlamydial developmental cycle as RBs are making the transition to EBs, depletion of MEC from intermediate forms committed to late differentiation would, of necessity, be rapid and likely specific to only those chlamydiae in the process of transition to EBs. Because isoprenoids are a ubiquitous class of compounds that serve a role in many essential cellular processes in both prokaryotes and eukaryotes (27), a likely means of its selective removal would be depletion of MEC through utilization in some physiological process that accompanies late differentiation.
Interactions of histones and DNA-binding proteins with DNA to establish chromatin structure are regulated by many mechanisms, including covalent modification by phosphorylation, methylation, or acetylation (31), proteolysis (32), or topology of the DNA itself (33-35). Disruption of chlamydial Hc1-DNA interactions by a small metabolite is an unusual means of releasing histone from chromatin. Release of chlamydial chromatin from the constraints imposed by Hc1 is an essential early event in the chlamydial developmental cycle. That chlamydiae use an intermediate in a metabolic pathway not present in mammalian cells suggests a potential target for development of chemotherapeutic agents that could inhibit this critical event.
Supplementary Material
Acknowledgments
We thank J. Sager for technical assistance and Drs. H. Caldwell, R. Heinzen, R. Carabeo, D. Clifton, K. Fields, and S. Grieshaber for review of the manuscript.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Amp, ampicillin; Cm, chloramphenicol; EB, elementary body; IPTG, isopropyl 1-thio-β-d-galactopyranoside; MEC, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate; MEP, methylerythritol 4-phosphate; RB, reticulate body; Spec, spectinomycin.
References
- 1.Schachter, J. (1988) Curr. Top. Microbiol. Immunol. 138, 109-139. [PubMed] [Google Scholar]
- 2.Moulder, J. W. (1991) Microbiol. Rev. 55, 143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw, E. I., Dooley, C. A., Fischer, E. R., Scidmore, M. A., Fields, K. A. & Hackstadt, T. (2000) Mol. Microbiol. 37, 913-925. [DOI] [PubMed] [Google Scholar]
- 4.Hackstadt, T., Baehr, W. & Ying, Y. (1991) Proc. Natl. Acad. Sci. USA 88, 3937-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hackstadt, T., Brickman, T. J., Barry, C. E. & Sager, J. (1993) Gene 132, 137-141. [DOI] [PubMed] [Google Scholar]
- 6.Tao, S., Kaul, R. & Wenman, W. M. (1991) J. Bacteriol. 173, 2818-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perara, E., Ganem, D. & Engel, J. N. (1992) Proc. Natl. Acad. Sci. USA 89, 2125-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brickman, T. J., Barry, C. E. & Hackstadt, T. (1993) J. Bacteriol. 175, 4274-4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belland, R. J., Zhong, G., Crane, D. D., Hogan, D., Sturdevant, D., Sharma, J., Beatty, W. L. & Caldwell, H. D. (2003) Proc. Natl. Acad. Sci. USA 100, 8478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barry, C. E., Hayes, S. F. & Hackstadt, T. (1992) Science 256, 377-379. [DOI] [PubMed] [Google Scholar]
- 11.Pedersen, L. B., Birkelund, S. & Christiansen, G. (1994) Mol. Microbiol. 11, 1085-1098. [DOI] [PubMed] [Google Scholar]
- 12.Barry, C. E., III, Brickman, T. J. & Hackstadt, T. (1993) Mol. Microbiol. 9, 273-283. [DOI] [PubMed] [Google Scholar]
- 13.Kaul, R., Allen, M., Bradbury, E. M. & Wenman, W. M. (1996) Nucleic Acids Res. 24, 2981-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christiansen, G., Pedersen, L. B., Koehler, J. E., Lundemose, A. G. & Birkelund, S. (1993) J. Bacteriol. 175, 1785-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remacha, M., Kaul, R., Sherburne, R. & Wenman, W. M. (1996) Biochem. J. 315, 481-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutz, R. & Bujard, H. (1997) Nucleic Acids Res. 25, 1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholson, T. L., Olinger, L., Chong, K., Schoolnik, G. & Stephens, R. S. (2003) J. Bacteriol. 185, 3179-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caldwell, H. D., Kromhout, J. & Schachter, J. (1981) Infect. Immun. 31, 1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hackstadt, T. (1991) J. Bacteriol. 173, 7046-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohdich, F., Wungsintaweekul, J., Fellermeier, M., Sagner, S., Herz, S., Kis, K., Eisenreich, W., Bacher, A. & Zenk, M. H. (1999) Proc. Natl. Acad. Sci. USA 96, 11758-11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herz, S., Wungsintaweekul, J., Schuhr, C. A., Hecht, S., Luttgen, H., Sagner, S., Fellermeier, M., Eisenreich, W., Zenk, M. H., Bacher, A. & Rohdich, F. (2000) Proc. Natl. Acad. Sci. USA 97, 2486-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luttgen, H., Rohdich, F., Herz, S., Wungsintaweekul, J., Hecht, S., Schuhr, C. A., Fellermeier, M., Sagner, S., Zenk, M. H., Bacher, A. & Eisenreich, W. (2000) Proc. Natl. Acad. Sci. USA 97, 1062-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens, R. S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., Mitchell, W., Olinger, L., Tatusov, R. L., Zhao, Q., et al. (1998) Science 282, 754-759. [DOI] [PubMed] [Google Scholar]
- 24.Rohdich, F., Kis, K., Bacher, A. & Eisenreich, W. (2001) Curr. Opin. Chem. Biol. 5, 535-540. [DOI] [PubMed] [Google Scholar]
- 25.Lichtenthaler, H. K. (2000) Biochem. Soc. Trans. 28, 785-789. [PubMed] [Google Scholar]
- 26.Rodriguez-Concepcion, M. & Boronat, A. (2002) Plant Physiol. 130, 1079-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lange, B. M., Rujan, T., Martin, W. & Croteau, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13172-13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostrovsky, D., Diomina, G., Lysak, E., Matveeva, E., Ogrel, O. & Trutko, S. (1998) Arch. Microbiol. 171, 69-72. [DOI] [PubMed] [Google Scholar]
- 29.Kaul, R., Hoang, A., Yau, P., Bradbury, E. M. & Wenman, W. M. (1997) J. Bacteriol. 179, 5928-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eisenstein, M. & Shakked, Z. (1995) J. Mol. Biol. 248, 662-678. [DOI] [PubMed] [Google Scholar]
- 31.Grant, P. A. (2001) Genome Biol. 2, reviews0003.1-reviews0003.6. [DOI] [PMC free article] [PubMed]
- 32.Setlow, P. (1988) Annu. Rev. Microbiol. 42, 319-338. [DOI] [PubMed] [Google Scholar]
- 33.Rice, P. A. (1997) Curr. Opin. Struct. Biol. 7, 86-93. [DOI] [PubMed] [Google Scholar]
- 34.Higgins, C. F., Hinton, J. C., Hulton, C. S., Owen-Hughes, T., Pavitt, G. D. & Seirafi, A. (1990) Mol. Microbiol. 4, 2007-2012. [DOI] [PubMed] [Google Scholar]
- 35.Hulton, C. S., Seirafi, A., Hinton, J. C., Sidebotham, J. M., Waddell, L., Pavitt, G. D., Owen-Hughes, T., Spassky, A., Buc, H. & Higgins, C. F. (1990) Cell 63, 631-642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.