Abstract
This work probes the mystery of what balance of forces creates the extraordinary mechanical stiffness of DNA to bending and twisting. Here we explore the relationship between base stacking, functional group occupancy of the DNA minor and major grooves, and DNA mechanical properties. We study double-helical DNA molecules substituting either inosine for guanosine or 2,6-diaminopurine for adenine. These DNA variants, respectively, remove or add an amino group from the DNA minor groove, with corresponding changes in hydrogen-bonding and base stacking energy. Using the techniques of ligase-catalyzed cyclization kinetics, atomic force microscopy, and force spectroscopy with optical tweezers, we show that these DNA variants have bending persistence lengths within the range of values reported for sequence-dependent variation of the natural DNA bases. Comparison with seven additional DNA variants that modify the DNA major groove reveals that DNA bending stiffness is not correlated with base stacking energy or groove occupancy. Data from circular dichroism spectroscopy indicate that base analog substitution can alter DNA helical geometry, suggesting a complex relationship among base stacking, groove occupancy, helical structure, and DNA bend stiffness.
Introduction
Double-helical DNA is polymorphic due to local and global differences in groove dimensions, helical diameter, basepair rise, twist, roll, etc. Classical B-form DNA under physiological conditions is characterized by a helical repeat of ∼10.5 bp/turn, a helical rise of ∼3.4 Å, and a helical diameter of ∼24 Å (1–4). B-form DNA describes the low energy global helical conformation under these conditions in the absence of strain (5–8). More than 20 repeated dinucleotide or trinucleotide duplexes (including combinations with inosine or 2-amino adenosine, which is commonly designated by its base diaminopurine) are able to adopt the classical B-form conformation given appropriate conditions of relative humidity, cation type, and retained salt (9), suggesting that B-DNA in solution is dynamic. Canonical values of helical parameters are not constant but depend on environment, including the charge, size, hydration, and concentration of ions. DNA sequence and base composition can also influence the specific values of these parameters in solution (10,11). Recent circular dichroism and x-ray crystallography studies have further added to our understanding of DNA structural polymorphism (12–16). How do the chemical properties of DNA bases relate to DNA structural polymorphism and mechanical properties?
Mechanically, the DNA molecule can be described as a polymer with three independent degrees of freedom: bend, twist, and contraction/extension. Each of these properties is described by an elastic modulus in the framework of the wormlike chain (WLC) polymer model (17–19). Despite several simplifying assumptions (20), the WLC model has proven utility in assays as diverse as ligase-catalyzed cyclization, atomic force microscopy (AFM), force spectroscopy using optical tweezers, transient electric birefringence, fluorescence polarization anisotropy, and small angle x-ray scattering.
Although DNA stiffness is adequately described by the WLC model, its physical basis is not understood. DNA stiffness derives from one or more intrinsic features of DNA. Likely candidates (which may not contribute independently) include
-
1.
Electrostatics (i.e., DNA charge repulsion leading to tension),
-
2.
Basepair stacking energy (i.e., attractive forces leading to compression), and
-
3.
Steric effects altering dimer step motion (e.g., basepair roll) due to sequence-dependent differences in functional group occupancy of the DNA grooves.
Previous studies have revealed that local dimer step conformational flexibility does not determine global mechanical flexibility (21) and that DNA stiffness is not controlled by a mechanism easily interpreted as electrostatic (22). The latter suggested that the invariant residual charge of DNA after Manning’s polyelectrolyte counterion condensation (23) might govern the electrostatic behavior of DNA. In this model, a constant electrostatic stretching contribution to DNA stiffness could arise from repulsive interactions between residual charges, resulting in insensitivity to variations in bare charge density (22). This study seeks to understand the relationship among base stacking, functional group occupancy of the DNA minor and major grooves, and DNA mechanical properties.
We approach the problem by studying double-helical DNA molecules substituting either inosine (I) for guanosine (G) or 2,6-diaminopurine (D) for adenine (A) (Fig. 1). Hypoxanthine is the base found in the nucleoside inosine. Although inosine is considered a guanosine analog (Fig. 1), in some contexts it functions as a universal base that has been used in degenerate polymerase chain reaction (PCR) primers, microarray probes, and triplexes (24). While not truly universal, its incorporation is less destabilizing than mismatches involving the four standard bases. The nucleoside 2-amino adenosine, commonly designated by its base 2,6-diaminopurine, is considered an adenosine analog that alters potential hydrogen bonding in the minor groove (Fig. 1). Investigators have used D-replacement to increase oligonucleotide stability, perhaps interpreting tighter binding to complementary sequences as the result of three hydrogen bonds, although more-favorable stacking effects are presumably the actual explanation.
Figure 1.
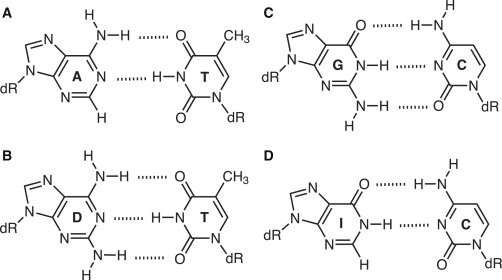
Structures of Watson-Crick basepairs involving natural and modified nucleosides studied initially. Watson-Crick basepairing between A and T (A) D and T (B), G and C (C), and I and C (D), where A, T, G, C, I, and D indicate, respectively, 2′-deoxyadenosine, 2′-deoxythymidine, 2′-deoxyguanosine, 2′-deoxycytidine, 2′-deoxyinosine, and 2-amino-2′-deoxyadenosine (commonly designated by its base 2,6-diaminopurine). Glycosidic bonds to deoxyribose (dR) and hydrogen bonds (dashed lines) are shown. The minor groove appears at the bottom of each basepair.
Molecular mechanics calculations validate that D·T forms a Watson-Crick basepair that is more stable than A·T but less so than G·C (25). Both analogs are found in natural systems: inosine 5′-monophosphate is a branch point in the de novo biosynthesis of purine nucleotides, a role well studied in enterobacteria (26,27), and D completely replaces A in the genome of cyanophage S-2L (28). Both analogs can alter the interaction site preference and affinity for DNA intercalators (29,30) as well as histone octamers (31). DNA conformation and sequence-dependent curvature are also influenced by these base modifications (32,33). These studies highlight the importance of diaminopurine and inosine substitution on DNA groove geometry.
Here we apply three complementary biophysical techniques to measure the mechanical properties of DNA molecules containing these uncharged base analogs and compare the results to AFM measurements for a series of charged and uncharged thymine variants. The fraction of total bases that are modified in the different constructs varies from 21 to 29%. We conclude that nucleoside analogs affect DNA mechanical properties through complex effects that may include their ability to stabilize different double-helical conformations.
Materials and Methods
DNA cyclization kinetics
Sample preparation
pUC19-based plasmids containing intrinsically straight ∼200-bp sequences (34) were flanked by either HindIII (pJ823-pJ833) or NarI (pJ1506 and pJ1741-pJ1750) sites (see Section S1 in the Supporting Material and Peters et al. (22) for details). PCR products (∼400 bp) containing these intrinsically straight sequences were amplified using primers LJM-3222 (5′-G3TA2CGC2AG3T4) and LJM-3223 (5′-TGTGAGT2AGCTCACTCAT2AG2) (Integrated DNA Technologies, Coralville, IA). PCR reactions for natural DNA (100 μL) included 20-ng plasmid template, 0.4-μM forward and reverse primers, 100-μg/mL bovine serum albumin, Taq DNA polymerase buffer (Invitrogen, Carlsbad, CA), 2 mM MgCl2, 0.2 mM each dNTP, and 5 U Taq DNA polymerase (Invitrogen). Cycle conditions were 94°C (3 min), 30 cycles of 94°C (30 s), 60°C (30 s), and 72°C (45 s), followed by 72°C (5 min).
Modified dNTP analogs 2′-deoxyinosine (I) and 2-amino-2′-deoxyadenosine (also called 2,6-diaminopurine; D) were purchased from TriLink BioTechnologies (San Diego, CA). For analog I, PCR reactions (50 μL) included 10-ng purified PCR product from a previous reaction, 0.4-μM forward and reverse primers, 100-μg/mL bovine serum albumin, Taq DNA polymerase buffer (Invitrogen), 1.65 mM MgCl2, 0.2 mM each dNTP (with dGTP completely replaced by dITP), and 5 U Taq DNA polymerase (Invitrogen). Cycle conditions were 94°C (3 min), 30 cycles of 84°C (30 s), 40°C (1 min), and 64°C (5 min), followed by 72°C (10 min) (adapted from Virstedt et al. (35)). For analog D, PCR reactions (100 μL) included 20-ng purified PCR product from a previous reaction, 0.4 μM forward and reverse primers, PrimeSTAR GC buffer (Takara, Clontech Laboratories, Mountain View, CA), 0.2 mM each dNTP (with dATP completely replaced by analog D), 2 M betaine (Sigma-Aldrich), and 5 U PrimeSTAR HS DNA polymerase (Takara). Cycle conditions were 98°C (3 min), 30 cycles of 98°C (15 s), 60°C (5 s), and 72°C (45 s), followed by 72°C (5 min).
PCR products were purified using QIAquick PCR purification kits (Qiagen, Venlo, The Netherlands) and then digested overnight with either HindIII or NarI and phosphatase-treated with Antarctic Phosphatase under conditions recommended by the supplier (New England Biolabs, Ipswich, MA). Reactions were heat-inactivated for 20 min at 65°C followed by radioactive labeling for 2 h at 37°C in T4 polynucleotide kinase buffer (PNK; New England Biolabs) using 600 pmol of (γ-32P)-ATP (PerkinElmer, Waltham, MA) and 40 U T4 PNK (New England Biolabs), with an additional heat inactivation for 20 min at 65°C. Samples were precipitated from ethanol, resuspended in 15 μL of loading buffer, and loaded onto a 5% native polyacrylamide gel (29:1 acrylamide:bisacrylamide; Bio-Rad, Hercules, CA) and visualized by exposure to BioMAX XR film (Kodak, Rochester, NY). The ∼200-bp restriction fragment was cut from the gel, crushed, and eluted overnight at 37°C in 200 mM Tris-HCl (pH 7.5), 300 mM NaCl, 2% SDS (w/v). Eluted DNA was extracted with an equal volume of phenol:chloroform (1:1) and the DNA was precipitated from ethanol and quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, MA) as described in Peters et al. (22).
Cyclization kinetics assay
DNA ligase-catalyzed cyclization reactions (60 μL) were performed at 22°C with 1 nM DNA restriction fragment, T4 DNA ligation buffer (New England Biolabs), and a final concentration of 100 U/mL T4 DNA ligase (New England Biolabs). Aliquots (10 μL) were removed at 5-, 10-, 15-, and 20-min time points (10, 20, 30, and 40 min for I), quenched by addition of EDTA to 20 mM, and then analyzed by electrophoresis through 5% native polyacrylamide gels (29:1 acrylamide:bisacrylamide, Bio-Rad) in 0.5× TBE buffer (50 mM Tris base, 55 mM boric acid, 1 mM EDTA, pH 8.3), followed by drying and storage phosphor imaging. Imaging was performed using a Typhoon FLA 7000 (GE Healthcare) followed by band quantitation, J-factor determination, and WLC analysis using the software R (Ver. 2.14.2, http://www.r-project.org/) as described in Peters et al. (22).
Atomic force microscopy
Sample preparation
DNA fragments 753 basepairs in length were PCR-amplified from pJ1506 with primers LJM-4762 (5′-CG2TGATGACG2TGA4) and LJM-3223 (5′-TGTGAGT2AGCTCACTCAT2AG2) (Integrated DNA Technologies) using conditions described above, purified using QIAquick PCR purification kits (Qiagen), and quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific). Methods for incorporating base substitutions at thymine residues have been described in Peters et al. (22).
AFM imaging
AFM imaging was performed in air at ambient temperature and humidity using a NanoScope IV (Bruker/Veeco/Digital Instruments, Plainview, NY) equipped with a type-E scanner operating in tapping mode. Freshly cleaved mica (grade V1; Ted Pella, Redding, CA) served as a support for sample adsorption. A 10-μL droplet of DNA at a concentration of ∼2 μg/mL in buffer (5 mM Tris-HCl, pH 7.5 supplemented with 10 mM NaCl and 5 mM MgCl2) was deposited for 2–3 min then rinsed carefully with 2–3 mL of Milli-Q water (Millipore, Billerica, MA) and gently dried under a nitrogen flow before imaging. Samples were imaged with silicon cantilevers (FESP; Bruker, Camarillo, CA) at a resonance frequency of 60–80 kHz and a setpoint of 0.6–1.2 V. Images (512 × 512 pixels) were collected with a scan size of 500 nm and scan rate varying between 5 and 15 Hz.
Image processing and data analysis
AFM images were flattened by subtracting from each scan line a least-squares-fitted third-order polynomial using software available with the AFM instrument. No additional background correction was applied to the images. Custom software for all subsequent image analysis was developed using the software R (Ver. 2.14.2). Skeletons of the DNA molecule images were created and digitized using morphological tools (e.g., erosion) in an algorithm previously described in Wang et al. (36). Although the detection and thinning of the molecules was automated, a human supervisor could reject erroneously segmented skeletons or those not meeting the set criteria during interactive steps (see Section S2 in the Supporting Material). After detection of DNA skeletons, trajectories of DNA centerlines were extracted automatically (but with human supervision) using a published routine (37). Statistical descriptors were calculated as a function of separation length along these DNA representations, after which the corresponding predictions from WLC theory were fit to the measured quantities (see Section S2 in the Supporting Material).
Optical tweezers
Sample preparation
DNA fragments 2041 basepairs in length were PCR-amplified from pJ1506 with 5′ modified primers LJM-4762 (5′-/5BiotinTEG/CG2TGATGACG2TGA4) and LJM-4763 (5′-/5DigN/G2ATG2AG2CG2ATA3G) (Integrated DNA Technologies) using conditions described above but increasing the extension time per cycle to 2 min (natural and D) or 10 min (I). The reactions were then twice extracted with an equal volume of phenol:chloroform (1:1) and the DNA was precipitated from ethanol and quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific).
Force extension measurements
The 2041 basepair constructs, terminally labeled with biotin and digoxigenin, were fixed between 2.1-μm diameter and 5.6-μm diameter beads coated with anti-dig antibody and streptavidin, respectively. Force response during cycles of extension and release was recorded in a custom dual-beam optical tweezers described in McCauley et al. (38) and fit using the WLC model
| (1) |
where b is extension, F is force, kB is the Boltzmann constant, T is the absolute temperature, P is the persistence length, and S is the elastic stretch modulus. Absolute lengths (B) of these short DNA constructs cannot be determined with this instrument due to variations in attachment and bead-bead interference at very low extensions. DNA lengths are assumed to be roughly equal across all molecules. Finite length effects in Pfitted were corrected according to
| (2) |
with a = 2.78 and L = 694 nm (39).
Circular dichroism spectroscopy
Circular dichroism (CD) spectroscopy was performed using a J-810 spectropolarimeter (JASCO, Oklahoma City, OK). Briefly, ultraviolet-CD spectra were acquired from 350 to 215 nm, taking measurements every 0.1 nm with a scanning speed of 5 nm/min. DNA fragments 417 basepairs in length were PCR-amplified from pJ1741 using conditions described above. Samples were analyzed in a 0.1-cm cuvette and prepared by diluting 25 μg of DNA into 300 μL of 10 mM phosphate buffer, pH 7.0, containing 1 M NaCl (final DNA concentration of ∼650 μM). Sample temperature was maintained at 20°C throughout. Samples were monitored five times with the average of the five scans reported and buffer contribution subtracted.
Results and Discussion
Preparation and characterization of substituted DNA molecules
The desired substitutions (either D or I) were introduced into an intrinsically straight duplex DNA sequence (see Section S1 in the Supporting Material) by PCR using modified deoxynucleoside triphosphates (22,34). Characterization of polymers with D-substitution is described in Peters et al. (22). Optimal synthesis with I involved Taq DNA polymerase and a lower annealing temperature. Thermal denaturation studies of a 418-basepair duplex revealed that I-substitution decreased DNA melting temperature (Tm) by 14.3°C relative to natural DNA (see Section S1 in the Supporting Material), in agreement with previous studies (28). D-substitution increased melting temperature 5.9°C (see Section S1 in the Supporting Material) in the same sequence context (9,22,35,40–42). Additionally, base-stacking energies (ΔΔG°37) were evaluated from thermodynamic measurements of the melting transition of a duplex formed by the self-complementary DNA oligonucleotide 5′-XCGCGCG (22,43) giving the following stacking stabilization order for the dangling 5′ nucleotide X: D > A > G > I (see also Section S1 in the Supporting Material).
DNA cyclization reveals alterations in both bend and twist stiffness
We determined bend and twist moduli for normal and modified DNA molecules using ligase-catalyzed cyclization experiments. Under appropriate conditions of this kinetic assay, the ratio of the rate of intramolecular DNA cyclization to form monomeric circles (kC1) to the rate of intermolecular dimerization to form linear dimers (kD) gives the cyclization J-factor, equivalent to the intramolecular concentration of one DNA terminus with respect to the other (Table 1) (44). These J-factor data (Fig. 2) were then fit with the WLC model (see equations 1–6 of Peters et al. (22)) to estimate the persistence length, twist persistence length (via torsional rigidity), and helical repeat (Table 2) (22,45). From the spread of the experimental data, estimation of uncertainty was achieved using Monte Carlo simulations (22).
Table 1.
Cyclization J-factor determined from kinetic rates kC1 and kD for the indicated DNA lengths
| DNA variant | DNA length (bp) | J-factor (nM) | kC1 (× 10−3 min−1) | kD (× 10−3 nM−1 min−1) |
|---|---|---|---|---|
| Natural (NarI) | 201 | 5.4 ± 1.3 (5.1 ± 1.2) | 5.0 ± 1.5 | 1.0 ± 0.4 |
| 202 | 3.0 ± 0.1 (2.8 ± 0.4) | 4.7 ± 2.6 | 1.6 ± 0.9 | |
| 203 | 1.6 ± 0.3 (1.4 ± 0.3) | 2.7 ± 0.5 | 1.8 ± 0.8 | |
| 204 | 0.7 ± 0.3 (0.8 ± 0.3) | 1.0 ± 1.0 | 1.3 ± 0.6 | |
| 205 | 0.5 ± 0.1 | 0.4 ± 0.2 | 0.9 ± 0.3 | |
| 206 | 0.7 ± 0.1 | 1.2 ± 0.6 | 1.8 ± 1.0 | |
| 207 | 2.3 ± 0.4 | 2.7 ± 1.1 | 1.2 ± 0.5 | |
| 208 | 6.2 ± 1.4 | 4.2 ± 0.6 | 0.7 ± 0.1 | |
| 209 | 6.9 ± 1.5 | 3.1 ± 1.0 | 0.4 ± 0.1 | |
| 210 | 9.7 ± 2.4 | 5.3 ± 1.5 | 0.6 ± 0.2 | |
| 211 | 8.1 ± 3.3 | 7.4 ± 1.9 | 1.0 ± 0.2 | |
| Natural (HindIII) | 196 | 1.1 ± 0.3 | 9.0 ± 4.5 | 7.6 ± 2.9 |
| 197 | 2.0 ± 0.4 | 19.6 ± 9.2 | 10.0 ± 4.0 | |
| 198 | 3.1 ± 0.4 | 34.1 ± 14.0 | 10.9 ± 3.7 | |
| 199 | 4.1 ± 0.8 | 44.1 ± 14.6 | 10.6 ± 2.2 | |
| 200 | 4.6 ± 0.5 | 47.8 ± 9.1 | 10.3 ± 0.9 | |
| 201 | 4.6 ± 1.0 (5.1 ± 1.2) | 43.6 ± 7.7 | 9.5 ± 0.8 | |
| 202 | 2.6 ± 0.5 (2.8 ± 0.4) | 28.2 ± 2.8 | 11.2 ± 3.1 | |
| 203 | 1.3 ± 0.2 (1.4 ± 0.3) | 13.8 ± 2.9 | 11.2 ± 2.3 | |
| 204 | 0.9 ± 0.2 (0.8 ± 0.3) | 9.9 ± 2.3 | 11.5 ± 2.1 | |
| Diaminopurine | 201 | 3.2 ± 0.1 | 1.1 ± 0.2 | 0.3 ± 0.1 |
| 202 | 2.6 ± 0.9 | 1.3 ± 0.1 | 0.5 ± 0.2 | |
| 203 | 1.6 ± 0.3 | 1.1 ± 0.7 | 0.8 ± 0.6 | |
| 204 | 0.5 ± 0.1 | 0.7 ± 0.1 | 1.5 ± 0.1 | |
| 205 | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.9 ± 0.6 | |
| 206 | 0.4 ± 0.1 | 0.2 ± 0.1 | 0.6 ± 0.3 | |
| 207 | 0.7 ± 0.1 | 0.5 ± 0.1 | 0.7 ± 0.1 | |
| 208 | 1.6 ± 0.2 | 1.5 ± 0.2 | 0.9 ± 0.2 | |
| 209 | 3.5 ± 0.6 | 4.4 ± 0.3 | 1.3 ± 0.1 | |
| 210 | 6.1 ± 1.2 | 6.7 ± 0.8 | 1.1 ± 0.1 | |
| 211 | 6.5 ± 0.8 | 4.1 ± 0.8 | 0.6 ± 0.1 | |
| Inosine | 196 | 5.8 ± 1.6 | 17.9 ± 8.2 | 3.0 ± 0.5 |
| 197 | 6.0 ± 3.2 | 8.1 ± 9.0 | 1.1 ± 0.7 | |
| 198 | 5.4 ± 0.7 | 6.5 ± 0.8 | 1.2 ± 0.2 | |
| 199 | 2.8 ± 0.3 | 3.5 ± 1.7 | 1.3 ± 0.6 | |
| 200 | 1.6 ± 0.2 | 3.7 ± 1.1 | 2.3 ± 0.7 | |
| 201 | 0.8 ± 0.1 | 3.1 ± 0.6 | 3.8 ± 0.7 | |
| 202 | 0.6 ± 0.1 | 2.2 ± 1.4 | 3.6 ± 1.9 | |
| 203 | 1.0 ± 0.2 | 4.8 ± 1.0 | 4.9 ± 0.5 | |
| 204 | 2.3 ± 0.2 | 2.9 ± 1.2 | 1.2 ± 0.4 |
Values are presented as mean ± standard deviation. Pooled J-factor data for natural DNA are indicated in parentheses.
Figure 2.
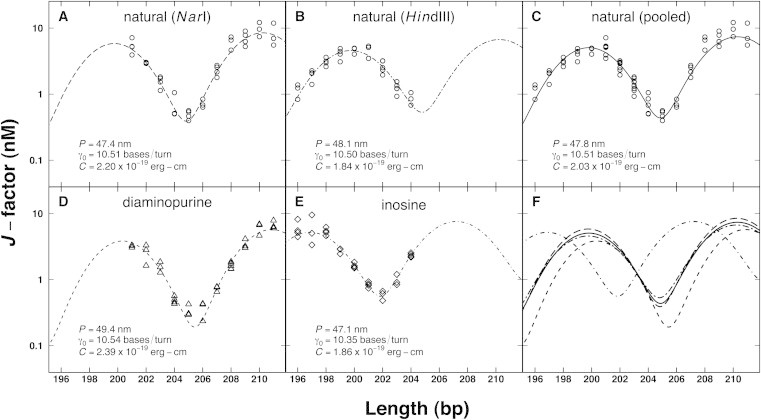
(A–F) J-factor curves from cyclization experiments. Experimental J-factor data (open symbols) for natural DNA (circles), diaminopurine substitution (triangles), and inosine substitution (diamonds) are shown as well as WLC fits (lines) and associated fit parameters: persistence length (P), helical repeat (γ0), and torsional modulus (C). Two different restriction sites, NarI ends (long dash) and HindIII ends (two dash), are shown for natural DNA along with the pooled data set (solid).
Table 2.
Parameters determined from WLC analysis of cyclization data
| DNA variant | P (nm) | γ0 (bases/turn) | C (× 10−19 erg-cm) | Pt (nm) |
|---|---|---|---|---|
| Natural (NarI) | 47.4 ± 0.4 | 10.51 ± 0.01 | 2.20 ± 0.19 | 53.9 ± 4.7 |
| Natural (HindIII) | 48.1 ± 0.2 | 10.50 ± 0.01 | 1.84 ± 0.21 | 45.1 ± 5.2 |
| Natural (pooled) | 47.8 ± 0.3 | 10.51 ± 0.01 | 2.03 ± 0.13 | 49.8 ± 3.2 |
| Diaminopurine | 49.4 ± 0.4 | 10.54 ± 0.01 | 2.39 ± 0.22 | 58.7 ± 5.5 |
| Inosine | 47.1 ± 0.3 | 10.35 ± 0.01 | 1.86 ± 0.13 | 45.6 ± 3.1 |
Persistence length (P), helical repeat (γ0), and torsional rigidity (C) along with the related twist persistence length (Pt) are presented as mean ± standard deviation from Monte Carlo simulations.
The bending persistence length (P) of natural DNA was determined for two sets of molecules. The first set varied in length from 201 to 211 bp and displayed a 5′-CG overhang derived from NarI digestion. The second set (lengths 196–205 bp) displayed a 5′-AGCT overhang derived from HindIII digestion. Different restriction sites were needed to accommodate the different base substitutions: A-to-D substitution necessitated a restriction site devoid of A·T pairs, while the HindIII enzyme tolerated G-to-I substitution within its restriction site. Analyzed individually, P was estimated as 47.4 ± 0.4 nm and 48.1 ± 0.2 nm for natural DNA derived from the NarI and HindIII constructs, respectively (Fig. 2, A and B). When data are pooled, the P estimate is 47.8 ± 0.3 nm (Fig. 2 C), within the accepted range (21,22,34). Substitution with D slightly increased P (49.4 ± 0.4 nm) whereas substitution with I slightly decreased P (47.1 ± 0.3 nm) relative to natural DNA (Fig. 2, D and E). An important conclusion from this study is that in the context of intrinsically straight DNA, alterations of P observed for diaminopurine- and inosine-substituted DNA molecules (Fig. 2 F) are of a similar magnitude to those observed from studies of sequence dependence in natural DNA (21).
The twist persistence length (Pt) values reported in Table 2 were determined from torsional rigidity (C) using C = kBTPt where kB is the Boltzmann constant and T the absolute temperature. Contributions from nicked DNA circles may systematically affect torsional rigidity values determined by cyclization; however, this effect should not alter the rank ordering of apparent twist flexibility. Interestingly, the trend for Pt mirrored that for P in direction; substitution with D increased Pt (58.7 ± 5.5 nm) whereas substitution with I decreased Pt (45.6 ± 3.1 nm) relative to natural DNA (49.8 ± 3.2 nm). However, the magnitude of the changes in Pt is strikingly more pronounced. The observation that DNA-twist persistence length is more sensitive to base modifications has been reported previously for other base substitutions (22). Finally, the helical repeat values (γ0) reported in Table 2 indicate adaptation to D or I substitution by under- or overtwisting relative to natural DNA, again of similar magnitude to those observed from studies of sequence dependence in natural DNA (21).
AFM visualization detects differences in bend flexibility
We confirmed bend flexibility trends using other techniques. Intrinsically straight DNA fragments of length 753 bp and containing the desired base substitutions were prepared for AFM studies. Fig. 3 shows example equilibrium conformations of these DNA fragments when subjected to thermal fluctuations. From a large set of images collected for each DNA substitution, 10 predictions of P from WLC theory (see Section S2 in the Supporting Material) were averaged to estimate the DNA persistence length (Paverage, Table 3). Relative to natural DNA (51.8 ± 3.5 nm), substitution with D increased Paverage (56.1 ± 2.9 nm) whereas substitution with I decreased Paverage (47.6 ± 3.0 nm). Contour length (LC) and helical rise (h) estimates are also reported in Table 3. These data suggest polymorphism among these substituted DNA molecules.
Figure 3.
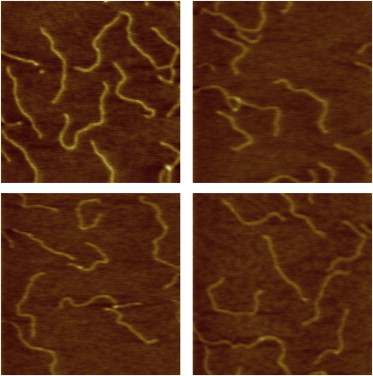
AFM images. 500 × 500 nm (512 × 512 pixel) AFM images of 753-bp substituted double-stranded DNA molecules deposited on mica in 5 mM Tris-HCl, pH 7.5 supplemented with 10 mM NaCl and 5 mM MgCl2. Color scale (from dark to light) is 0–2 nm. Thymine variant 6 (see Fig. 7) is shown as a representative example.
Table 3.
Parameters determined from WLC analysis of AFM data
| DNA variant | N | LC (nm) | h (Å/bp) | Paverage (nm) |
|---|---|---|---|---|
| Natural | 166 | 234 ± 26 | 3.11 ± 0.34 | 51.8 ± 3.5 |
| Diaminopurine | 146 | 229 ± 27 | 3.04 ± 0.36 | 56.1 ± 2.9 |
| Inosine | 172 | 221 ± 30 | 2.93 ± 0.40 | 47.6 ± 3.0 |
| 1 | 126 | 240 ± 30 | 3.18 ± 0.39 | 53.3 ± 2.2 |
| 2 | 113 | 237 ± 30 | 3.14 ± 0.39 | 57.9 ± 4.1 |
| 3 | 187 | 229 ± 34 | 3.04 ± 0.45 | 54.7 ± 2.5 |
| 4 | 113 | 240 ± 39 | 3.19 ± 0.52 | 56.5 ± 3.0 |
| 5 | 188 | 230 ± 35 | 3.05 ± 0.47 | 52.9 ± 4.7 |
| 6 | 131 | 244 ± 28 | 3.25 ± 0.37 | 47.3 ± 2.2 |
| 7 | 101 | 246 ± 39 | 3.27 ± 0.51 | 80.1 ± 3.8 |
N is the number of molecules used in the analysis for each type of DNA, LC is the estimated contour length, and h is the estimated DNA helical rise, each presented as mean ± standard deviation (see Section S2 in the Supporting Material). Persistence length (Paverage) is presented as mean ± standard deviation from 10 distinct estimates of P (see Section S2 in the Supporting Material). Seven thymine variants with functional groups that occupy the major groove (Fig. 7) were also analyzed (numbered 1–7 for simplicity).
Bend flexibilities measured from force-extension curves using optical tweezers
Force-extension curves for natural (circles), D-substitution (triangles), and I-substitution (diamonds) 2041-basepair constructs are shown in Fig. 4, illustrating the reproducibility of the force-extension curves through cycles of extension and release. Fits with the WLC model (Eq. 1) are shown as the associated solid lines (38). To enhance the stability of the fits, the contour length was held fixed at 694 nm so that B = 0.34 nm per basepair for all molecules (46). The adjusted fit parameters were the persistence length (P) and elastic stretch modulus (S), whose values are given in Table 4. The lower basepairing stability of the inosine-substituted molecules skewed initial results for this construct. Extra care was taken for each construct to remove any curves that exhibited hysteresis, which might indicate partial melting, resulting in artificially lower values of fitted persistence lengths.
Figure 4.
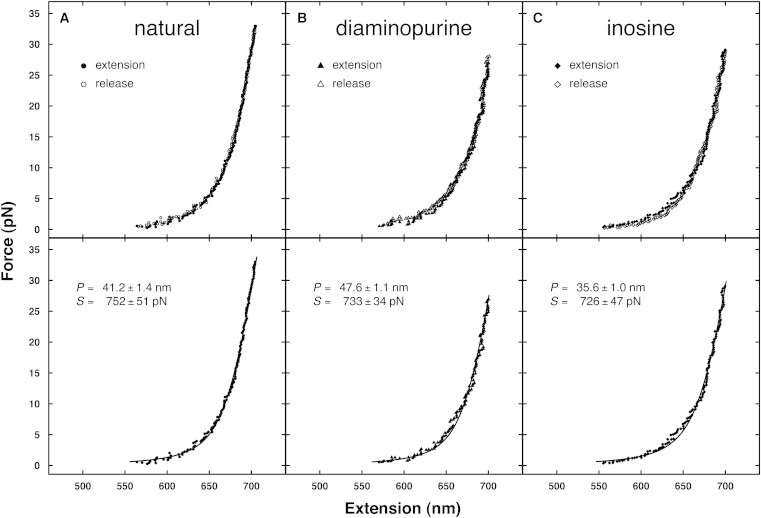
Force-extension curves from optical tweezers experiments. Representative natural (circle), diaminopurine substitution (triangle), and inosine substitution (diamond) extension and release data are shown with WLC fits (solid lines) and the associated fit parameters: persistence length (P) and elastic stretch modulus (S). The contour length per basepair B was fixed at 0.34 nm to enhance the stability of the fits.
Table 4.
Parameters determined from WLC analysis of optical tweezers data
| DNA variant | N | Pfitted (nm) | Pcorrected (nm) | S (pN) |
|---|---|---|---|---|
| Natural | 38 | 40.0 ± 0.7 (4.5) | 47.6 ± 1.1 (6.4) | 719 ± 19 (115) |
| Diaminopurine | 34 | 44.6 ± 0.5 (3.1) | 54.4 ± 0.8 (4.6) | 711 ± 20 (114) |
| Inosine | 34 | 35.0 ± 0.7 (4.2) | 40.9 ± 1.0 (5.7) | 782 ± 22 (128) |
N is the number of fitted curves for each type of DNA, collected across 4–5 distinct molecules. Persistence length (P) and elastic stretch (S) are presented as mean ± SE of the mean (standard deviation). Fitted persistence lengths were corrected for finite length effects (39).
Averaged across several cycles of extension and release (∼35), there are clearly consistent differences between fitted persistence lengths observed for the different constructs: substitution with D increased Pfitted (44.6 ± 0.5 nm) whereas substitution with I decreased Pfitted (35.0 ± 0.7 nm) relative to natural DNA (40.0 ± 0.7 nm). These DNA molecules appear to have reduced persistence lengths in the optical tweezers experiments due to finite length effects that become increasingly noticeable for DNA constructs less than a few thousand basepairs in length when measured by DNA stretching (39). This effect is nearly absent for phage-λ DNA with a length of 48,500 bp, where typical values of Pfitted are ∼50 nm. Using the correction published in Seol et al. (39) (Eq. 2), we find Pcorrected to be 47.6 ± 1.0 nm for natural DNA, 54.4 ± 0.8 nm for D substitution, and 40.9 ± 1.0 for I substitution. Finally, the averaged values of elastic stretch modulus for each construct are very similar, and the distributions overlap well (Table 4).
Comparison of the three methods
Probability histograms of fitted persistence length values from each of the three techniques (Fig. 5) indicate that the probability distributions are approximately normal and well characterized by the mean and standard deviation values reported in Tables 2, 3, and 4. Comparison of the three methods requires consideration of their distinct experimental conditions and the following unique challenges.
-
1.
We note the previously reported finite length effects that are inherent in optical tweezers experiments with short DNA (39). These experiments also rely on measurements from a relatively small number of single molecules. (In contrast, AFM experiments examine hundreds of molecules and cyclization experiments examine billions.)
-
2.
Buffer ionic strength and divalent cation composition must be taken into consideration. In particular, ligase-mediated cyclization methods require low millimolar concentrations of the divalent cation magnesium for ligase catalysis, while AFM requires either divalent cations or polyamines to promote DNA adsorption onto (negatively-charged) mica surfaces via ionic interactions. An abundance of work highlights the ionic strength dependence of DNA persistence length, especially in the presence of divalent ions (47–50). The buffer conditions for the three methods were 50 mM Tris-HCl (pH 7.5) with 10 mM MgCl2, 1 mM ATP, and 10 mM dithiothreitol for cyclization; 5 mM Tris-HCl (pH 7.5) with 10 mM NaCl and 5 mM MgCl2 for AFM; and 10 mM HEPES (pH 7.5) with 100 mM NaCl for optical tweezers. (Because of higher salt (monovalent and particularly divalent magnesium cations), it was anticipated that P would be systematically reduced for cyclization versus AFM experiments.)
-
3.
DNA fragments of different lengths were required for the three techniques. Cyclization is optimal for fragments long enough to detectably cyclize at low ligase concentrations but still short enough to be limited by twist (∼200 bp in this study). AFM requires fragments short enough to avoid excluded volume effects, but still long enough to capture equilibrium conformations on length scales of a few persistence lengths (753 bp in this study). In comparison, a previous study attempted to determine P using molecules that ranged from only half a persistence length to one persistence length (∼150 bp) (35). Optical tweezer experiments are ideal for fragments many thousands of basepairs in length. However, using PCR to prepare samples of substituted DNA molecules on this length scale is impractical. The accessible length of 2041 bp was therefore chosen for this study. Special challenges for optical tweezer analysis of such short DNA lengths have been discussed in Seol et al. (39).
Figure 5.
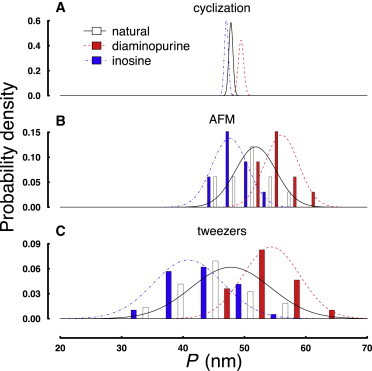
Normalized probability histograms of fitted persistence length values from three methods. Normalized probability histograms of P (along with Gaussian distributions drawn to guide the eye) for natural DNA (solid), diaminopurine substitution (dash), and inosine substitution (dot dash) are shown for the indicated techniques: (A) cyclization, (B) AFM, and (C) force spectroscopy using optical tweezers. For legibility, histograms for cyclization are omitted. Histograms for the AFM data are the result of binning the 10 estimates of P from WLC theory discussed in Section S2 in the Supporting Material. Histograms for the tweezers data come from binning the corrected values from each force-extension curve (∼35 for each construct).
Each of the three methods utilized in this work reports a consistent trend in the direction of flexibility change. Inosine constructs are more flexible than natural constructs, which are more flexible than diaminopurine constructs. Unexpectedly, the magnitude of the observed changes is different for each method. This result may reveal construct-specific differences that are only detected under force, or during the process of deposition onto charged mica or the cation concentration-dependence of persistence length. Further studies beyond the scope of this work are necessary to resolve these possibilities.
Interpretation of nucleoside analog effects on DNA mechanical properties
For natural DNA and the two variants (D or I substitution) studied here, there are strong linear correlations between P and several features of the DNA, including Tm, ΔΔG of base stacking, and van der Waals volume (51) of groove functional groups (solid symbols and dashed lines in Fig. 6). These trends exist for each of the three methods. It is tempting to draw general cause-and-effect conclusions from this limited comparison (35). However, we felt it crucial to test the generality of these results by performing additional analysis of other base-substituted DNA variants. Our goal was to determine which physical and/or thermodynamic feature(s) of base-substituted DNA polymers explain their mechanical properties.
Figure 6.
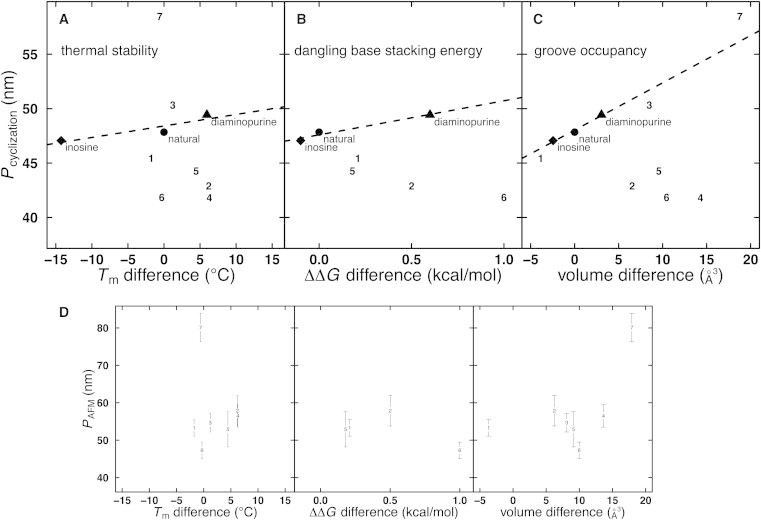
Correlations between physical features and persistence lengths of base-substituted DNA molecules. Linear correlations with P (measured by cyclization) appear for Tm difference from natural DNA (A), base stacking ΔΔG difference (substituted base minus corresponding natural base) (B), and change in van der Waals volume of groove (51) normalized per residue (C) for diaminopurine- and inosine-substituted DNA. Similar correlations exist from analysis by AFM and optical tweezers. Importantly, these correlations vanish when seven thymine variants (data points numbered 1–7, corresponding to Fig. 7) are included in the analysis. Data for these additional variants were taken from Peters et al. (22); base stacking data were not collected for thymine variants 3, 4, and 7. Uncertainty in Pcyclization is smaller than the symbol size. (D) Thymine variants do not display linear trends in PAFM; uncertainty is indicated by error bars.
We used AFM to characterize bending persistence lengths of seven additional DNA variants where all thymine residues have been replaced by different base analogs that modify the C5 position in the major groove (Fig. 7). These variants have been previously characterized using cyclization kinetics experiments (see Peters et al. (22) for details). Table 3 summarizes WLC analysis of AFM data for these substituted DNA molecules (see Section S2 in the Supporting Material). Relative to natural DNA (51.8 ± 3.5 nm), some variants were characterized by increased or decreased values of Paverage, ranging from 47.3 ± 2.2 nm for thymine variant 6 to 80.1 ± 3.8 nm for thymine variant 7. These two most extreme molecules also showed the greatest range in previous characterization by cyclization kinetics experiments (P of 41.8 ± 0.1 nm and 58.5 ± 0.2 nm, respectively) (22).
Figure 7.
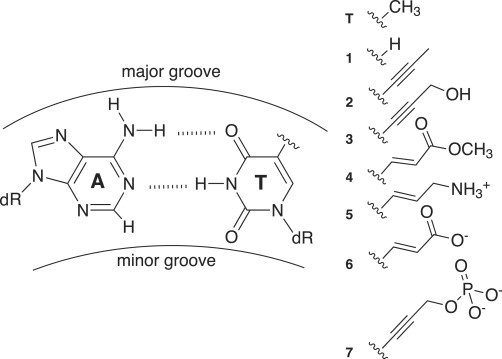
Structure of Watson-Crick basepair involving thymine variants. The glycosidic bond to deoxyribose (dR), hydrogen bonds (dashed lines), and the site of thymine modification (C5 position) are shown. Structures of the functional groups for the seven variants are shown at right.
Importantly, the analysis of physical, thermodynamic, and mechanical properties for a larger number of DNA analogs (characterized by either cyclization kinetics or AFM experiments) provided striking counter examples to the initial correlations implied from the study of only inosine and diaminopurine substitution. For example, the two most extreme molecules with respect to effects on P (thymine variants 6 and 7) have indistinguishable melting temperatures, and, while both add volume in the DNA major groove, they exhibit opposite effects on bending stiffness (Fig. 6). The previous strong correlations observed for thermal stability, dangling base stacking energy, and groove occupancy based on D or I substitution (dashed lines in Fig. 6) are lost when these additional variants are included (data values indicated by analog numbers in Fig. 6). This analysis suggests that no linear correlation exists between bending stiffness (or torsional rigidity (22)) and polymer bare charge, stacking energy measured by melting temperature, stacking energy measured in dangling nucleoside experiments, or functional group volume in the major or minor grooves.
These results argue against any simple cause-and effect relationship between charge or base stacking in bend stiffness (22), challenging contemporary interpretations (35,52). What other possibilities exist? A systematic study modifying each of the four standard DNA bases found that base substitutions promote significant DNA polymorphism and that high-density incorporation of modifications into double-stranded DNA causes conformational transitions from a right-handed B-form DNA to a left-handed form (15). We hypothesize that the main effect of neutral and charged base modifications on DNA mechanical properties is indirect, operating through the ability of these analog substitutions to drive transitions between polymorphic helical conformations different from canonical B-form DNA. We propose that these alternate helical conformations have distinct mechanical properties (especially twist flexibilities).
To test this hypothesis, we performed CD spectroscopy (Fig. 8). Natural DNA exhibits the CD signature of canonical B-DNA, which is characterized by a negative peak in the wavelength range of 245–250 nm and an approximately equal positive peak between 275 and 280 nm so that the CD spectrum is balanced above roughly 220 nm with the two peaks centered at ∼260 nm. In contrast, the CD spectrum of inosine-substituted DNA exhibits a much shallower negative peak shifted to 244 nm and a positive peak shifted to 273 nm that is half as large (Fig. 8). Studies of other inosine-substituted sequences displayed similar (positive and negative) peaks with decreased magnitudes and shifts to shorter wavelengths (35). Finally, diaminopurine-substituted DNA exhibits a positive peak shift to 292 nm and a deep negative peak at 248 nm with a shoulder and shallow crossover, indicative of partial A-type character (22). This analysis revealed significant DNA polymorphism for these substituted DNA molecules, consistent with previous observations (12,15,22,35).
Figure 8.
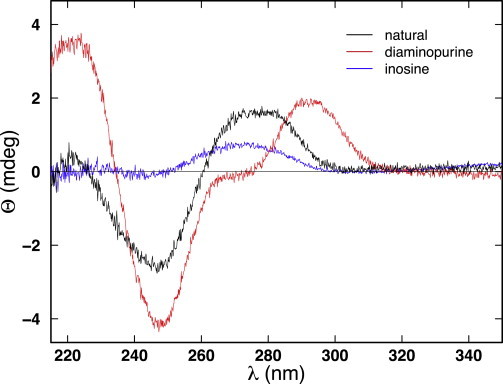
Circular dichroism spectroscopy. Circular dichroism spectra showing ellipticity (Θ) as a function of wavelength (λ) for inosine-substituted DNA. Spectra previously reported for natural and diaminopurine-substituted DNA (22) are shown for comparison.
Although the CD data reported here demonstrate that introduction of modified bases drives DNA between structurally polymorphic forms, it remains unclear what feature(s) of these alternate helical conformations are responsible for their distinct DNA mechanical properties. Addressing this issue may require a systematic, high-resolution structural study of helix geometry for a series of base analogs known to affect DNA mechanical properties.
Acknowledgments
The authors acknowledge Marina Ramirez-Alvarado for technical assistance and for access to instrumentation.
This work was supported by the Mayo Graduate School, the Mayo Foundation, National Institutes of Health grants No. GM075965 to L.J.M. and No. GM72462 to M.C.W., and National Science Foundation grant No. MCB-1243883 to M.C.W.
Supporting Material
Supporting Citations
Refs. (53–58) appear in the Supporting Material.
References
- 1.Watson J.D., Crick F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Dickerson R.E., Drew H.R., Pjura P.E. Helix geometry and hydration in A-DNA, B-DNA, and Z-DNA. Cold Spring Harb. Symp. Quant. Biol. 1983;47:13–24. doi: 10.1101/sqb.1983.047.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Wang J.C. Helical repeat of DNA in solution. Proc. Natl. Acad. Sci. USA. 1979;76:200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnott S., Hukins D.W. The dimensions and shapes of the furanose rings in nucleic acids. Biochem. J. 1972;130:453–465. doi: 10.1042/bj1300453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jayaram B., Sprous D., Beveridge D.L. Free energy analysis of the conformational preferences of A and B forms of DNA in solution. J. Am. Chem. Soc. 1998;120:10629–10633. [Google Scholar]
- 6.Hartmann B., Lavery R. DNA structural forms. Q. Rev. Biophys. 1996;29:309–368. doi: 10.1017/s0033583500005874. [DOI] [PubMed] [Google Scholar]
- 7.Šponer J., Leszczynski J., Hobza P. Electronic properties, hydrogen bonding, stacking, and cation binding of DNA and RNA bases. Biopolymers. 2001-2002;61:3–31. doi: 10.1002/1097-0282(2001)61:1<3::AID-BIP10048>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 8.Arora N., Jayaram B. Energetics of base pairs in B-DNA in solution: an appraisal of potential functions and dielectric treatments. J. Phys. Chem. B. 1998;102:6139–6144. [Google Scholar]
- 9.Leslie A.G., Arnott S., Ratliff R.L. Polymorphism of DNA double helices. J. Mol. Biol. 1980;143:49–72. doi: 10.1016/0022-2836(80)90124-2. [DOI] [PubMed] [Google Scholar]
- 10.Bram S. Secondary structure of DNA depends on base composition. Nat. New Biol. 1971;232:174–176. doi: 10.1038/newbio232174a0. [DOI] [PubMed] [Google Scholar]
- 11.Bram S., Tougard P. Polymorphism of natural DNA. Nat. New Biol. 1972;239:128–131. doi: 10.1038/newbio239128a0. [DOI] [PubMed] [Google Scholar]
- 12.Maehigashi T., Hsiao C., Williams L.D. B-DNA structure is intrinsically polymorphic: even at the level of base pair positions. Nucleic Acids Res. 2012;40:3714–3722. doi: 10.1093/nar/gkr1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowal E.A., Ganguly M., Stone M.P. Altering the electrostatic potential in the major groove: thermodynamic and structural characterization of 7-deaza-2′-deoxyadenosine:dT base pairing in DNA. J. Phys. Chem. B. 2011;115:13925–13934. doi: 10.1021/jp207104w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kypr J., Kejnovská I., Vorlícková M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jäger S., Rasched G., Famulok M. A versatile toolbox for variable DNA functionalization at high density. J. Am. Chem. Soc. 2005;127:15071–15082. doi: 10.1021/ja051725b. [DOI] [PubMed] [Google Scholar]
- 16.Sines C.C., McFail-Isom L., Williams L.D. Cations mediate B-DNA conformational heterogeneity. J. Am. Chem. Soc. 2000;122:11048–11056. [Google Scholar]
- 17.Kratky O., Porod G. X-ray investigations of dissolved chain molecules [Rontgenuntersuchung geloster fadenmolekule] Recl. Trav. Chim. Pays Bas. 1949;68:1106–1122. [Google Scholar]
- 18.Rippe K., von Hippel P.H., Langowski J. Action at a distance: DNA-looping and initiation of transcription. Trends Biochem. Sci. 1995;20:500–506. doi: 10.1016/s0968-0004(00)89117-3. [DOI] [PubMed] [Google Scholar]
- 19.Shimada J., Yamakawa H. Ring-closure probabilities for twisted wormlike chains. Application to DNA. Macromolecules. 1984;17:689–698. [Google Scholar]
- 20.Vologodskii A., Frank-Kamenetskii M.D. Strong bending of the DNA double helix. Nucleic Acids Res. 2013;41:6785–6792. doi: 10.1093/nar/gkt396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geggier S., Vologodskii A. Sequence dependence of DNA bending rigidity. Proc. Natl. Acad. Sci. USA. 2010;107:15421–15426. doi: 10.1073/pnas.1004809107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters J.P., Yelgaonkar S.P., James Maher L., 3rd Mechanical properties of DNA-like polymers. Nucleic Acids Res. 2013;41:10593–10604. doi: 10.1093/nar/gkt808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manning G.S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q. Rev. Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 24.Loakes D. The applications of universal DNA base analogues. Nucleic Acids Res. 2001;29:2437–2447. doi: 10.1093/nar/29.12.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao S.N., Chandra Singh U., Kollman P.A. Molecular dynamics simulations of DNA double helices: studies of sequence dependence and the role of mismatch pairs in the DNA helix. Isr. J. Chem. 1986;27:189–197. [Google Scholar]
- 26.Meng L.M., Nygaard P. Identification of hypoxanthine and guanine as the co-repressors for the purine regulon genes of Escherichia coli. Mol. Microbiol. 1990;4:2187–2192. doi: 10.1111/j.1365-2958.1990.tb00580.x. [DOI] [PubMed] [Google Scholar]
- 27.Cho B.-K., Federowicz S.A., Palsson B.Ø. The PurR regulon in Escherichia coli K-12 MG1655. Nucleic Acids Res. 2011;39:6456–6464. doi: 10.1093/nar/gkr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirnos M.D., Khudyakov I.Y., Vanyushin B.F. 2-aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature. 1977;270:369–370. doi: 10.1038/270369a0. [DOI] [PubMed] [Google Scholar]
- 29.Tseng Y.D., Ge H., Henderson R.M. Atomic force microscopy study of the structural effects induced by echinomycin binding to DNA. J. Mol. Biol. 2005;345:745–758. doi: 10.1016/j.jmb.2004.10.059. [DOI] [PubMed] [Google Scholar]
- 30.Marco E., Negri A., Gago F. Role of stacking interactions in the binding sequence preferences of DNA bis-intercalators: insight from thermodynamic integration free energy simulations. Nucleic Acids Res. 2005;33:6214–6224. doi: 10.1093/nar/gki916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buttinelli M., Minnock A., Travers A. The exocyclic groups of DNA modulate the affinity and positioning of the histone octamer. Proc. Natl. Acad. Sci. USA. 1998;95:8544–8549. doi: 10.1073/pnas.95.15.8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Møllegaard N.E., Bailly C., Nielsen P.E. Effects of diaminopurine and inosine substitutions on A-tract induced DNA curvature. Importance of the 3′-A-tract junction. Nucleic Acids Res. 1997;25:3497–3502. doi: 10.1093/nar/25.17.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lankaš F., Cheatham T.E., 3rd, Šponer J. Critical effect of the N2 amino group on structure, dynamics, and elasticity of DNA polypurine tracts. Biophys. J. 2002;82:2592–2609. doi: 10.1016/s0006-3495(02)75601-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vologodskaia M., Vologodskii A. Contribution of the intrinsic curvature to measured DNA persistence length. J. Mol. Biol. 2002;317:205–213. doi: 10.1006/jmbi.2001.5366. [DOI] [PubMed] [Google Scholar]
- 35.Virstedt J., Berge T., Travers A.A. The influence of DNA stiffness upon nucleosome formation. J. Struct. Biol. 2004;148:66–85. doi: 10.1016/j.jsb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Wang H., Dodd I.B., Finzi L. Single molecule analysis of DNA wrapping and looping by a circular 14-mer wheel of the bacteriophage 186 CI repressor. Nucleic Acids Res. 2013;41:5746–5756. doi: 10.1093/nar/gkt298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiggins P.A., van der Heijden T., Nelson P.C. High flexibility of DNA on short length scales probed by atomic force microscopy. Nat. Nanotechnol. 2006;1:137–141. doi: 10.1038/nnano.2006.63. [DOI] [PubMed] [Google Scholar]
- 38.McCauley M.J., Rueter E.M., Williams M.C. Single-molecule kinetics reveal microscopic mechanism by which high-mobility group B proteins alter DNA flexibility. Nucleic Acids Res. 2013;41:167–181. doi: 10.1093/nar/gks1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seol Y., Li J., Betterton M.D. Elasticity of short DNA molecules: theory and experiment for contour lengths of 0.6–7 microm. Biophys. J. 2007;93:4360–4373. doi: 10.1529/biophysj.107.112995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khudyakov I.Y., Kirnos M.D., Vanyushin B.F. Cyanophage S-2L contains DNA with 2,6-diaminopurine substituted for adenine. Virology. 1978;88:8–18. doi: 10.1016/0042-6822(78)90104-6. [DOI] [PubMed] [Google Scholar]
- 41.Turner D.H. Thermodynamics of base pairing. Curr. Opin. Struct. Biol. 1996;6:299–304. doi: 10.1016/s0959-440x(96)80047-9. [DOI] [PubMed] [Google Scholar]
- 42.Krepl M., Otyepka M., Šponer J. Effect of guanine to inosine substitution on stability of canonical DNA and RNA duplexes: molecular dynamics thermodynamics integration study. J. Phys. Chem. B. 2013;117:1872–1879. doi: 10.1021/jp311180u. [DOI] [PubMed] [Google Scholar]
- 43.Guckian K.M., Schweitzer B.A., Kool E.T. Factors contributing to aromatic stacking in water: evaluation in the context of DNA. J. Am. Chem. Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters J.P., 3rd, Maher L.J., III DNA curvature and flexibility in vitro and in vivo. Q. Rev. Biophys. 2010;43:23–63. doi: 10.1017/S0033583510000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters J.P., Becker N.A., Maher L.J., III Quantitative methods for measuring DNA flexibility in vitro and in vivo. Methods Enzymol. 2011;488:287–335. doi: 10.1016/B978-0-12-381268-1.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wenner J.R., Williams M.C., Bloomfield V.A. Salt dependence of the elasticity and overstretching transition of single DNA molecules. Biophys. J. 2002;82:3160–3169. doi: 10.1016/S0006-3495(02)75658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang M.D., Yin H., Block S.M. Stretching DNA with optical tweezers. Biophys. J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y., Weers B., Stellwagen N.C. DNA persistence length revisited. Biopolymers. 2001-2002;61:261–275. doi: 10.1002/bip.10151. [DOI] [PubMed] [Google Scholar]
- 49.Podestà A., Indrieri M., Dunlap D. Positively charged surfaces increase the flexibility of DNA. Biophys. J. 2005;89:2558–2563. doi: 10.1529/biophysj.105.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savelyev A. Do monovalent mobile ions affect DNA’s flexibility at high salt content? Phys. Chem. Chem. Phys. 2012;14:2250–2254. doi: 10.1039/c2cp23499h. [DOI] [PubMed] [Google Scholar]
- 51.Zhao Y.H., Abraham M.H., Zissimos A.M. Fast calculation of van der Waals volume as a sum of atomic and bond contributions and its application to drug compounds. J. Org. Chem. 2003;68:7368–7373. doi: 10.1021/jo034808o. [DOI] [PubMed] [Google Scholar]
- 52.Manning G.S. The persistence length of DNA is reached from the persistence length of its null isomer through an internal electrostatic stretching force. Biophys. J. 2006;91:3607–3616. doi: 10.1529/biophysj.106.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivetti C., Guthold M., Bustamante C. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 54.Neuman K.C., Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rivetti C., Codeluppi S. Accurate length determination of DNA molecules visualized by atomic force microscopy: evidence for a partial B- to A-form transition on mica. Ultramicroscopy. 2001;87:55–66. doi: 10.1016/s0304-3991(00)00064-4. [DOI] [PubMed] [Google Scholar]
- 56.Faas F.G., Rieger B., Cherny D.I. DNA deformations near charged surfaces: electron and atomic force microscopy views. Biophys. J. 2009;97:1148–1157. doi: 10.1016/j.bpj.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moukhtar J., Faivre-Moskalenko C., Arneodo A. Effect of genomic long-range correlations on DNA persistence length: from theory to single molecule experiments. J. Phys. Chem. B. 2010;114:5125–5143. doi: 10.1021/jp911031y. [DOI] [PubMed] [Google Scholar]
- 58.Abels J.A., Moreno-Herrero F., Dekker N.H. Single-molecule measurements of the persistence length of double-stranded RNA. Biophys. J. 2005;88:2737–2744. doi: 10.1529/biophysj.104.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


