Abstract
Hepatitis C virus (HCV) and Classical swine fever virus (CSFV) mRNAs contain related (HCV-like) internal ribosome entry sites (IRESs) that promote 5’-end independent initiation of translation, requiring only a subset of the eukaryotic initiation factors (eIFs) needed for canonical initiation on cellular mRNAs1. Initiation on HCV-like IRESs relies on their specific interaction with the 40S subunit2–8, which places the initiation codon into the P site, where it directly base-pairs with eIF2-bound Met-tRNAiMet to form a 48S initiation complex. However, all HCV-like IRESs also specifically interact with eIF32,5–7,9–12, but the role of this interaction in IRES-mediated initiation has remained unknown. During canonical initiation, eIF3 binds to the 40S subunit as a component of the 43S pre-initiation complex, and comparison of the ribosomal positions of eIF313 and the HCV IRES8 revealed that they overlap, so that their rearrangement would be required for formation of ribosomal complexes containing both components13. Here, we present a cryo-electron microscopy reconstruction of a 40S ribosomal complex containing eIF3 and the CSFV IRES. Strikingly, although the position and interactions of the CSFV IRES with the 40S subunit in this complex are similar to those of the HCV IRES in the 40S/IRES binary complex8, eIF3 is completely displaced from its ribosomal position in the 43S complex, and instead interacts through its ribosome-binding surface exclusively with the apical region of domain III of the IRES. Our results suggest a role for the specific interaction of HCV-like IRESs with eIF3 in preventing ribosomal association of eIF3, which could serve two purposes: relieving the competition between the IRES and eIF3 for a common binding site on the 40S subunit, and reducing formation of 43S complexes, thereby favoring translation of viral mRNAs.
Canonical translation initiation begins with assembly of a 43S preinitiation complex, comprising a 40S subunit, eIF1, eIF1A, the Met-tRNAiMet/eIF2/GTP ternary complex (eIF2-TC) and the ~800-kDa five-lobed multi-subunit eIF31. The 43S complex attaches to the cap-proximal region of mRNA and then scans to the initiation codon, whereupon it forms a 48S initiation complex with established codon-anticodon base-pairing. Attachment and scanning are mediated by eIF4A, eIF4B and eIF4F, but scanning on structured mRNAs additionally requires DHX2914,15, a DExH-box protein that also binds directly to the 40S subunit13,14. 48S complex formation on the homologous HCV and CSFV IRESs, which comprise two principal domains, II and III (Extended Data Fig. 1a), does not involve scanning and requires only a 40S subunit and the eIF2-TC. The process is based on the specific interaction of the IRES with the 40S subunit, which involves the pseudoknot and subdomains IIId and IIIe3–5,16. Binding to the 40S subunit positions the initiation codon of the IRES in the P site where it directly base-pairs with the anticodon of Met-tRNAiMet as a part of the eIF2-TC, leading to formation of the 48S complex. Subsequent joining of the 60S subunit to this complex is mediated by eIF5 and eIF5B. Although domain II of HCV-like IRESs stimulates eIF5-mediated hydrolysis of eIF2-bound GTP and joining of a 60S subunit17–19, it does not influence the affinity of the IRES to the 40S subunit5, only moderately affects 48S complex formation, and is not essential for initiation on the CSFV IRES3,18,20,21.
An unresolved aspect of initiation on HCV-like IRESs is the role of eIF3, which interacts specifically with the apical region of domain III (helices IIIb and III4)5,9,11 (Extended Data Fig. 1a). Although eIF3 is not essential for 48S complex formation on these IRESs and only slightly stimulates this process in the in vitro reconstituted translation system2,6,7,18, mutations in the apical region of domain III that impair binding of eIF32,9,19 lead to severe translation initiation defects in cell-free extracts22. Importantly, the position of the eIF3 core in 43S complexes13 and of the HCV IRES in 40S/IRES binary complexes8 overlap, with a clash between eIF3’s left arm and the pseudoknot13. The simultaneous presence of eIF3 and the IRES in ribosomal complexes would therefore require their rearrangement.
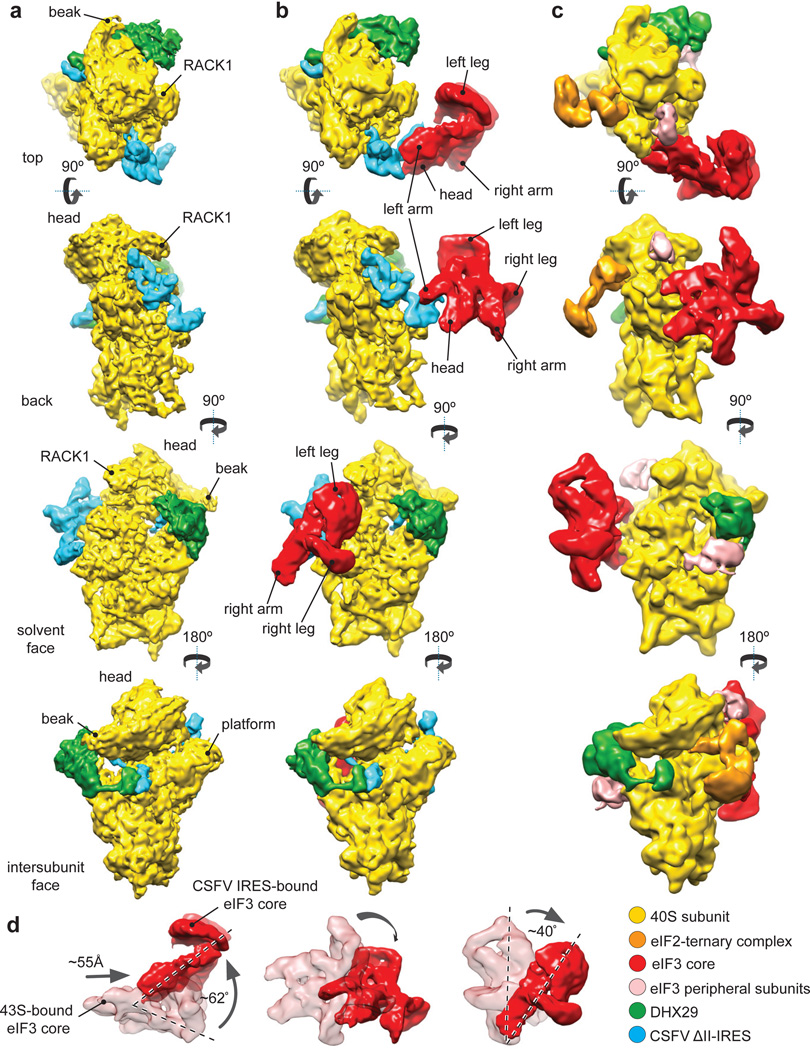
To shed light on eIF3’s role in initiation on HCV-like IRESs and to investigate how the predicted eIF3/IRES clash is resolved, we determined the cryo-electron microscopy (Cryo-EM) structure of the 40S subunit in complex with eIF3 and the CSFV IRES lacking the non-essential domain II (“ΔII-IRES”). The CSFV IRES was chosen since it has higher translational activity than the HCV IRES2, likely because it interacts more strongly with eIF3 and/or the 40S subunit, and would thus yield complexes with higher stability for structural analysis. Domain II was omitted to reduce complexity and to reduce conformational heterogeneity. DHX29 was also included in these complexes because it stabilizes eIF3’s peripheral domains in 43S complexes13 without affecting the interaction of the IRES with the 40S subunit14. The 40S/ΔII-IRES/eIF3/DHX29 complexes were assembled in vitro by incubating individual purified components. Toe-printing analysis of these complexes revealed that they maintained the full complement of interactions of the IRES with the 40S subunit and eIF32,18, and were quantitatively converted into 48S complexes upon addition of eIF2-TC (Extended Data Fig. 2). Although DHX29 does not interfere with 48S complex formation on the ΔII-IRES (Extended Data Fig. 2), only 10% of 43S complexes in cells are bound to DHX2914, and whether it is present in IRES-bound ribosomal initiation complexes in the cytoplasm has not been determined. Processing of ~630,000 particle images (see Methods) yielded several classes containing different combinations of components (Extended Data Figs 3 and 4). The present analysis is focused on the 40S/DHX29/ΔII-IRES complex (class 2, ~72,900 particles) and the first of three 40S/DHX29/ΔII-IRES/eIF3 classes (4–6), which differed slightly in the orientation of eIF3 and eIF3-bound subdomain IIIb of the IRES (class 4, ~26,000 particles). They yielded 8.5Å and 9.3Å reconstructions, respectively, which revealed three well-defined masses of density on the 40S subunit (Fig. 1a and b).
Figure 1. Cryo-EM structures of the CSFV ΔII-IRES•40S•DHX29 complex alone and bound to eIF3 compared to the structure of the DHX29-bound 43S preinitiation complex.
(a) CSFV ΔII-IRES•40S•DHX29 complex (class 2, Extended Data Fig. 3). (b) CSFV ΔII-IRES•40S•DHX29 complex bound to eIF3 (class 4, Extended Data Fig. 3). (c) 43S preinitiation complex13. (, from top to bottom) Complexes are viewed from the top, the back, the solvent and the intersubunit faces, respectively. In panels a–c, the 40S subunit is displayed in yellow, DXH29 in green, the eIF3 structural core in red and the CSFV ΔIIIRES in cyan. (d) Comparison between different positions and orientations of eIF3 in the 43S complex and in the CSFV ΔII-IRES•40S complexes, as indicated.
The shape and location of a mass of density around the tip of h16 and of a smaller mass on the subunit interface near the A site connected to it via a clearly defined linker (green in Fig. 1a and b) matched the density of DHX29 in the 43S complex13 (Fig. 1c). Another density at the back of the platform (cyan in Fig. 1a and b) was assigned to the CSFV ΔII-IRES, since it fitted the shape and location of the related HCV IRES domain III in 40S/IRES and 80S/IRES binary complexes8,23 (Extended Data Fig. 1b). A visible segment of the CSFV coding region emerges from the mRNA entrance and forms a small additional mass near the A site and the small intersubunit domain of DHX29. This mass could be modeled as an ~11-bp stem-loop separated from the P site codon by 5–7 nt and could include a predicted hairpin formed by CSFV nts. 387–406.
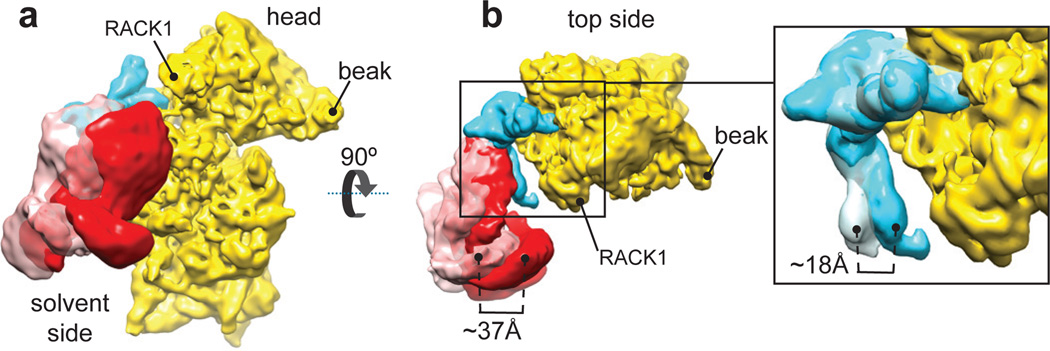
The third mass attached to the ΔII-IRES at the back of the 40S subunit (red in Fig. 1b) was attributed to the core of eIF3, based on its shape12,13,24 and interaction with the apical part of IRES domain III2,5,9,11. Strikingly, the position of the eIF3 core in the IRES-containing complex differs from that in the 43S complex: whereas in the latter, eIF3’s left arm and head interact with ribosomal proteins rpS1e/rpS26e and rpS13e/rpS27e, respectively13 (Fig. 1c), in the former complex, they bind to the apical part of IRES domain III (Fig. 1b), consistent with eIF3’s position on HCV-like IRESs5,9,11. The orientation of the CSFV ΔII-IRES on eIF3 is consistent with the position of scattered density attributed to the HCV IRES in a lower-resolution cryo-EM reconstruction of the eIF3/IIIabc binary complex11. Thus, the IRES effectively usurps ribosomal contacts of eIF3, leading to displacement of eIF3 from the 40S subunit and leaving it interacting exclusively with the IRES. Compared to its position in the 43S complex, eIF3’s core is shifted by ~55Å and rotated by approximately 60° (Fig. 1d). Assignment of eIF3’s left arm and head to eIF3a and eIF3c subunits, respectively24 (Extended Data Fig. 5a and b), makes interaction of the IRES with the left arm and head of eIF3 consistent both with reports of UV cross-linking of the HCV IRES to eIF3a and eIF3c subunits9–11 and with the observed impairment of binding of eIF3 to the HCV IRES by mutations in eIF3a and eIF3c11. Interestingly, eIF3 and the subdomain IIIb bound to it adopt a number of orientations in the 40S/DHX29/ΔII-IRES/eIF3 complex (classes 4–6). Between the two most divergent orientations, shown by classes 4 and 6, the tip of domain IIIb moves by up to 18Å, inducing in turn a movement of the eIF3 core that reaches up to 37Å in the region of the legs (Fig. 2a and b; Supplementary Video 1). Thus, the inherent flexibility of the IIIabc/III4 four-way junction25 allows the eIF3-bound domain IIIb to move while the IRES maintains its contacts with the 40S subunit. In the class 4 map, the high variance and the low resolution in the region of eIF3 legs, when compared to the rest of the eIF3 core (Extended Data Fig. 5c), points to a continuum of orientations of subdomain IIIb and eIF3. The same explanation likely applies to the other two classes, 5 and 6, based on the lower resolution observed in the eIF3 legs. Despite the presence of DHX29, none of the peripheral domains of eIF3 was clearly observed, which is likely a consequence of the displacement of the eIF3 core from its binding site on the 40S subunit.
Figure 2. Different orientations of eIF3 and subdomain IIIb in the CSFV ΔII IRES•40S•DHX29 complex.
(a) Solvent-side view of eIF3 in the two most divergent orientations, as it appears in classes 4 (solid red surface) and 6 (transparent pink surface) of the CSFV ΔII-IRES•40S•DHX29•eIF3 complex, bound to the CSFV ΔII-IRES (cyan) on the 40S subunit (yellow). (b, Left). Top view of eIF3 in the two most divergent orientations, bound to the CSFV ΔII-IRES on the 40S subunit. (b, Right) blowup focused on domain IIIb of the CSFV IRES, showing the extent of its reorientation. The brackets display the magnitude of the movement of eIF3 and of IRES domain IIIb in the two most divergent orientations.
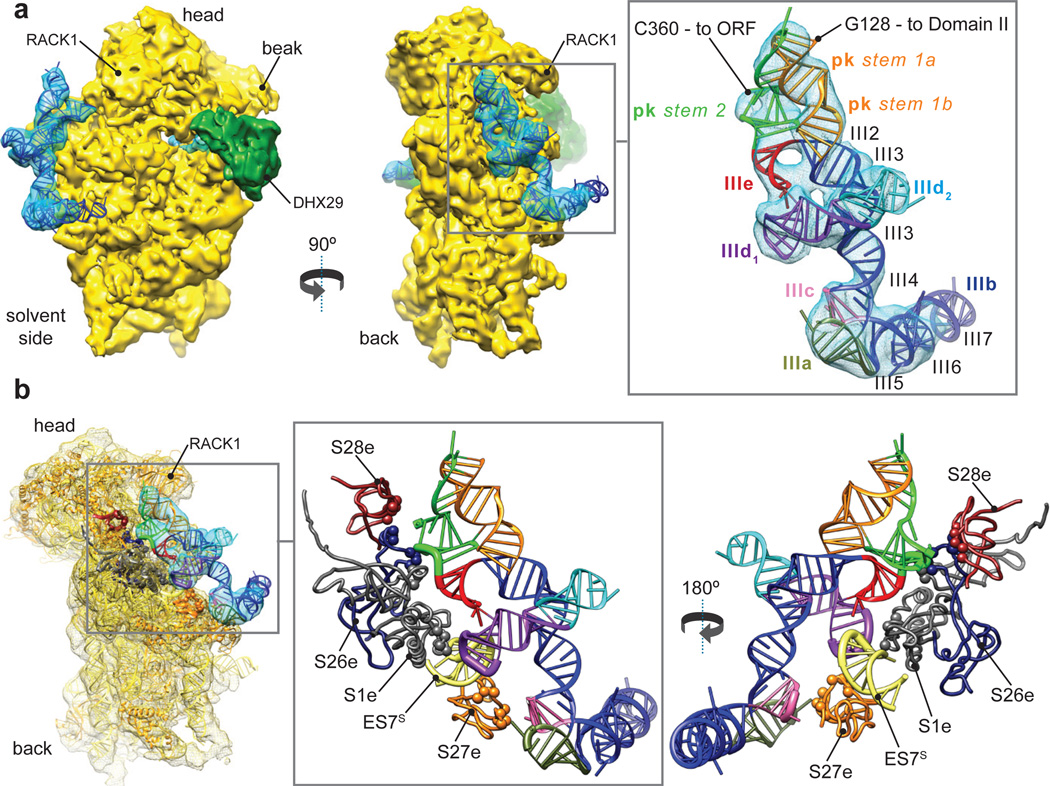
The sub-nanometer resolution of the class 2 map allowed us to attempt pseudo-atomic modeling of the CSFV ΔII-IRES (nts. 129–361), for which no high-resolution structure is available. The model (Fig. 3a) was built and fitted into its density mass segmented from both class 2 and class 4 reconstructions (bound with eIF3 in position 1), yielding a final cross-correlation coefficient with the classes 2 and 4 IRES densities of 0.92 and 0.93, respectively (see Methods). The model is consistent with the results of phylogenetic comparisons, chemical/enzymatic probing and mutational analyses of CSFV and related IRESs3,20,26. The apical region consists of a long cylindrical stem, from which the subdomains IIId1 and IIId2 protrude, and is kinked at the flexible elbow formed by the four-way junction of helix III4 and subdomains IIIa, IIIb and IIIc. Subdomains IIIb and IIId2 extend away from the 40S subunit surface, consistent with their lack of involvement in 40S subunit binding2,3,19. Domain IIIb is not well resolved in the 40S/DHX29/ΔII-IRES complex but is stabilized in the complex containing eIF3. The basal region of domain III is formed by the pseudoknot, subdomain IIIe and helix III1, which together form two sets of coaxially stacked helices, angled at ~40° with respect to each other, that are directly comparable to the ‘main’ and ‘sidecar’ helices of the analogous region in the HCV IRES27 (Extended Data Fig. 6).
Figure 3. Structure and atomic model of the CSFV ΔII-IRES bound to the 40S subunit.
(a) Atomic model of the CSFV ΔII-IRES fitted into its density map (blue mesh), seen from the solvent (Left) and back (Middle) sides. Right panel displays a blowup on the CSFV ΔII-IRES atomic model (ribbon), colored variably to highlight its different subdomains. (b) Ribosomal proteins contacting the CSFV ΔII-IRES when bound to the 40S subunit, seen from the back (Left and Middle) and the front (Right).
The higher resolution of the present maps also allows confident assignment of individual interactions of the IRES with the 40S subunit and eIF3. In addition to the mRNA flanking the initiation codon, five distinct elements of the ΔII-IRES (subdomains IIIa, IIIc, IIId1, IIIe and the pseudoknot) contact the 40S subunit (Fig. 3b, Extended Data Fig. 7a). These elements are highly conserved in HCV-like IRESs in members of several genera of Flaviviridae and Picornaviridae (e.g.28), and each of them coincides precisely with sites in HCV and CSFV IRESs that are protected from enzymatic cleavage or chemical modification by the bound 40S subunit3–5,16. On the 40S subunit, the interactions mostly involve ribosomal proteins (rpS1e, rpS26e, rpS27e and rpS28e) (Fig. 3b, Extended Data Fig. 7a) and, consistent with these contact sites, interactions of corresponding elements of the HCV IRES with rpS1e and rpS27e have been observed29,30. Importantly, rpS1e, rpS26e and rpS27e are also involved in the interaction of the 40S subunit with eIF313, which accounts for the displacement of eIF3 from it by the IRES. However, the apical loop of subdomain IIId1 also contacts the apical loop of ES7 of 18S rRNA (Fig. 3b), likely through base-pairing between the conserved GGG nucleotides on subdomain IIId1 and CCC nucleotides in the apical loop of ES7. This interaction, which induces a small-scale shift in the position of the apex of ES7 towards the head, was previously suggested by a low-resolution cryo-EM study23, and is consistent with the strong ribosomal protection of this region of the IRES in foot-printing studies3–5,16. Interestingly, the ribosomal elements that HCV-like IRESs exploit for binding to the 40S subunit (rpS1e, rpS26e, rpS27e, rpS28e and ES7) are all eukaryote-specific.
The conserved GGG motif in subdomain IIId1 is a major determinant of ribosome binding and initiation activity for all HCV-like IRESs3,5,6. However, whereas the apical UCCC loop of ES7 is highly conserved in vertebrates, the equivalent element in plants has the sequence CUUA, which would not base-pair stably to the GGG motif, likely contributing to the inability of wheat 40S subunits to bind to the HCV IRES2. The observed interactions with the 40S subunit also account for the severe effects of substitutions in the apical loop of subdomain IIId3,5,6,16,19 and of disruption of base-pairing in pseudoknot stem 22,5,20,26.
Binding of the CSFV ΔII-IRES to the eIF3 core is mainly restricted to two regions, helix III4 and domain IIIb (Fig. 4a). Residues G247-C249 and U159-A162 in helix III4, residues G184-U185 and G233-A234 in helix III5, residues C214-U221 in helix III6 and residues C197-G198 in helix III7 are all in close proximity to eIF3’s left arm (eIF3a) and likely interact with it directly. Residues U228-G230 appear to contact eIF3’s head (eIF3c) and are thus also likely involved in binding of the IRES to eIF3. These interactions are consistent with eIF3’s position on HCV and CSFV IRESs determined by foot-printing5,9,11 (Extended Data Fig. 7b to d). Our finding that the interaction of eIF3 with the CSFV IRES primarily involves eIF3’s left arm (eIF3a) is supported by the observations that mutations in eIF3a have a stronger effect than mutations in eIF3c on the binding of eIF3 to the HCV IRES11.
Figure 4. Binding of eIF3 to subdomain IIIb of the CSFV IRES and effects on translation of the eIF3/IRES interaction.
(a) eIF3 binding site on the CSFV IRES. The residues potentially interacting with eIF3 from domain IIIb in the CSFV IRES are highlighted by blue spheres and labeled (bottom). (b) Toeprinting analysis of 48S complexes assembled on wild-type HCV (nt. 1–349)–CAT mRNA (“wt HCV IRES”) and ΔIIIb-HCV IRES or ΔIIIc-HCV IRES derivatives22 lacking either domain IIIb (nt. 172–227) or domain IIIc (nt. 229–238) with 40S subunits, Met-tRNAiMet, eIF2 and eIF3 as indicated. Primer extension was arrested at nts. 342–345 by stably bound 40S subunits2 and at nts. 355–359 by 48S complexes, as indicated. Lanes C, T, A and G show the cDNA sequence corresponding to wt HCV IRES mRNA. The position of the initiation codon AUG373 is indicated on the left. (c) Inhibition of 43S complex formation by subdomain IIIabc of HCV and CSFV IRESs, assayed by sucrose density gradient centrifugation (SDG). The protein composition of ribosomal peak fractions was analyzed by SDS-PAGE and fluorescent SYPRO staining. (d) Inhibition of 48S complex formation on β-globin mRNA by IIIabc subdomains and by complete HCV and CSFV IRESs containing a 4nt. deletion in helix III2, assayed by toe-printing. Lanes C, T, A and G show the cDNA sequence corresponding to β-globin mRNA. The position of the initiation codon is indicated on the left. Each gel reported in the figure is representative of results obtained from three technical replicates.
The fact that in CSFV ΔII-IRES/eIF3-containing ribosomal complexes, the 40S subunit interacts only with the IRES, which in turn also binds to eIF3 through its ribosome-binding surface, suggests that in the case of the HCV-like IRES mutants lacking the eIF3-binding site, eIF3 would more readily compete with them for the conventional site on the 40S subunit, thus reducing 48S complex formation. To test this prediction, we compared 48S complex formation in the presence and in the absence of eIF3 on the wt HCV IRES and those IRES mutants that lacked subdomains IIIb or IIIc and could therefore no longer bind eIF32,22. Consistent with the prediction of our hypothesis, inclusion of eIF3 reduced 48S complex formation on the mutants, whereas in the presence of only 40S subunits and the eIF2-TC, the level of 48S complex formation on all three mRNAs was very similar (Fig. 4b). The small stimulatory effect of eIF3 on 48S complex formation on the wt HCV IRES could be due to stabilization of the IRES structure at the junction of domains IIIa, b and c, which in turn might stabilize interaction between IIIa and rpS27e. This stabilization could play a more significant in the cell by counteracting the dissociative influence of eIF1 on 48S complexes formed on HCV-like IRESs18. However, eIF3 is not essential for subsequent stages in initiation, because 48S complexes formed in its absence on both wt and ΔIIIb mutant HCV IRESs readily underwent subunit joining, forming elongation-competent 80S ribosomes (Extended Data Fig. 8).
Consistent with the IIIabc domain of HCV-like IRESs and the 40S subunit binding to a common site on eIF3, HCV and CSFV IIIabc domains impaired 43S complex formation by reducing ribosomal association of eIF3 by 60–70% (Fig. 4c). Consequently, inclusion of these domains in reaction mixtures strongly inhibited 48S complex formation on β-globin mRNA (Fig. 4d). Inhibition by IIIabc domains was almost as potent as by complete IRESs with a 4nt deletion in helix III2, which could no longer bind 40S subunits but retained eIF3-binding activity2. The CSFV IIIabc domain was a stronger inhibitor, paralleling the CSFV IRES’s higher translational activity in cell-free extracts2, which could therefore be due to its greater ability to compete for eIF3.
In conclusion, our unexpected finding that the CSFV ΔII-IRES displaces the core of eIF3 from its position on the 40S subunit sheds light on the role of eIF3’s interaction with HCV-like IRESs in the mechanism of initiation and provides a plausible explanation for the reason why mutations in the apical region of domain III that impair binding of eIF3 lead to severe translation defects in cell-free extracts22. Thus, by binding to eIF3, HCV-like IRESs would reduce the competition with this factor for binding to the 40S subunit and would also impair formation of 43S complexes, which in turn, might aid the ability of these IRESs to compete with cellular mRNAs.
ONLINE METHODS
Plasmids
The plasmid HCV-MSTN-Stop was made (GenScript Corp.) by inserting a 497 nt. DNA fragment between Xba1 and EcoR1 sites of pUC57 that consisted of a T7 promoter sequence, HCV Type 1b nt. 40–375 (modified to include a UAA stop codon in place of the fifth coding triplet and a BamH1 restriction site 20nt. downstream of it), followed by a segment of the Influenza NS coding sequence2.
The plasmid HCV(ΔIIIb)-MSTN-Stop was constructed similarly, except that the HCV sequence contained a deletion of nt. 172–227 (corresponding to subdomain IIIb) in addition to the UAA stop codon and BamH1 restriction site.
The plasmid HCV(Δ136–139) was also constructed similarly, except that the wt HCV sequence was modified to include a deletion of nt. 136–139, a Sca1 restriction site 2nt. upstream of the HCV initiation codon and HindIII and BamH1 restriction sites 13nt. and 32nt. downstream of it, respectively.
The plasmid HCV(IIIabc) was made (GenScript Corp.) by inserting a 142 nt. DNA fragment between Xba1 and Sal1 sites of pUC57 that consisted of a T7 promoter sequence and HCV Type 1b nt. 143–250, followed by a Nae1 restriction site.
The plasmids pWT-CAT, pΔE-CAT and pΔF-CAT22 consist of nt1–349 of the HCV-H strain, or variants thereof lacking subdomain IIIb (nt. 172–227) or subdomain IIIc (nt. 229–238) respectively, linked to a CAT reporter cistron.
The plasmid CSFV(Δ145–148) was made (GenScript Corp.) by inserting a 556 nt. DNA fragment between Xba1 and HindIII sites of pUC57 that consisted of a T7 promoter sequence, CSFV (Alfort/Tuebingen) nt. 1–442 (modified to delete nt. 145–1482, and to include a Fsp1 restriction site 6nt. upstream of the CSFV initiation codon and a Dra1 restriction site 12nt. downstream of it), followed by a segment of the Influenza NS coding sequence2.
The plasmid CSFV(IIIabc) was made (GenScript Corp.) by inserting a 137 nt. DNA fragment between Xba1 and HindIII sites of pUC57 that consisted of a T7 promoter sequence and CSFV (Alfort/Tuebingen) nt. 152–256, followed by a Sma1 restriction site.
The plasmid pCSFV(128–442)NS’(3) was generated from pCSFV(1–442)NS’(2) and consists of CSFV (Alfort/Tuebingen) nt.128–442 linked to a segment of the Influenza NS coding sequence.
The MVHL-Stop33 modified β-globin transcription vector consists of a DNA fragment corresponding to a T7 promoter sequence, four CAA repeats and the complete human β-globin sequence, modified to convert the 5th coding triplet to a UAA termination codon and the downstream UGUGU sequence to AGUGA, cloned between BglII and XhoI sites of pET28a.
CSFV ΔII-IRES mRNA was transcribed after linearization of pCSFV(128–442)NS’(3) with Pml1, 235 nt. downstream of the CSFV initiation codon. The wt, ΔIIIb (Δ172–227 nt) and ΔIIIc (Δ229–238 nt) HCV IRES mRNAs were transcribed from pWT-CAT, pΔE-CAT and pΔF-CAT plasmids, respectively, that had been linearized with HindIII. HCV-MSTN-Stop and HCV(ΔIIIb)-MSTN-Stop mRNAs were transcribed after linearization of corresponding plasmids with EcoR1. HCV(Δ136–139), CSFV(Δ145–148), HCV IIIabc and CSFV IIIabc mRNAs were transcribed after linearization of corresponding plasmids with Sca1, Fsp1, Sal1 and Hind III, respectively. All mRNAs were transcribed using T7 RNA polymerase.
Purification of ribosomal subunits, initiation and elongation factors, DHX29, and aminoacylation of tRNA
Native 40S and 60S ribosomal subunits, eIF2, eIF3, eIF5B, eEF1H, eEF2 and total aminoacyl-tRNA synthetases were purified from rabbit reticulocyte lysate, and recombinant human DHX29, eIF1, eIF1A, eIF4A, eIF4B and eIF4G736–1115, and E. coli methionyl tRNA synthetase were expressed and purified from E. coli as described previously31–33. Native total rabbit tRNA (Novagen) was aminoacylated with Met, Ser, Thr and Asn using native aminoacyl-tRNA synthetases, whereas in vitro transcribed tRNAiMet(34) was aminoacylated using E. coli methionyl tRNA synthetase as described31.
Assembly of 40S/eIF3/DHX29/CSFV ΔII-IRES complexes for cryoEM analysis
Complexes for cryoEM analysis were assembled by incubating 20 pmol 40S subunits, 30 pmol eIF3, 30 pmol DHX29 and 24 pmol CSFV ΔII-IRES RNA in 50 µl of buffer A (20 mM Tris pH 7.5, 75 mM KCl, 5 mM Mg, 2 mM DTT and 0.25 mM spermidine) for 10 min at 37°C. Before applying onto grids, the reaction mixture was diluted with buffer A to the concentration of 40S subunits of 32 nM.
Toe-printing analysis of 48S initiation and 80S pre-termination complex (pre-TC) formation on wt and mutant CSFV and HCV IRES mRNAs
48S complexes were assembled by incubating 2 pmol mRNA with 2 pmol 40S subunits, 4 pmol eIF2 and 5 pmol Met-tRNAiMet, 3 pmol eIF3 and 2 pmol DHX29, as indicated, in 20 µl buffer A (20 mM Tris pH7.5, 100 mM KCl, 2.5 mM MgCl2, 2 mM DTT, 0.25 mM spermidine) supplemented with 1 mM ATP and 0.4 mM GTP for 10 min at 37°C. For 80S initiation complex formation, reaction mixtures were supplemented with 4 pmol 60S subunits, 4.5 pmol eIF5 and 2 pmol eIF5B, and incubated at 37°C for an additional 10 min to allow formation of 80S initiation complexes. To form pre-TCs, 80S initiation complexes were supplemented with 4 pmol eEF1H, 12 pmol eEF2 and ~10 µg appropriately aminoacylated total native tRNA, and incubated at 37°C for an additional 10 min. Ribosomal complexes were analyzed by toe-printing31 using AMV reverse transcriptase and a [32P]primer. cDNA products were resolved in 6% polyacrylamide sequencing gels.
Toe-printing analysis of 48S complex formation on β-globin mRNA
48S complexes were assembled by incubating 2 pmol of a derivative of β-globin mRNA containing four 5’-terminal CAA repeats (MVHL-STOP mRNA33) with 2 pmol 40S subunits, 4 pmol eIF2, 5 pmol Met-tRNAiMet, 3 pmol eIF3, 5 pmol eIF4A, 2 pmol eIF4B, 3 pmol eIF4G736–1115, 10 pmol eIF1, 10 pmol eIF1A in the presence and in the absence of 15 pmol IIIabc subdomains and by complete HCV and CSFV IRESs containing a 4nt. deletion in helix III2 in 20 µl buffer A supplemented with 1 mM ATP and 0.4 mM GTP for 10 min at 37°C. Ribosomal complexes were analyzed by toe-printing31 using AMV reverse transcriptase and a [32P]primer. cDNA products were resolved in 6% polyacrylamide sequencing gels.
Inhibition of 43S complex formation by subdomain IIIabc of HCV and CSFV IRESs assayed by sucrose density gradient centrifugation (SDG)
43S complexes were assembled by incubating 20 pmol 40S subunits, 50 pmol eIF2, 70 pmol Met-tRNAiMet, 100 pmol eIF1, 100 pmol eIF1A and 30 pmol eIF3 in the presence and in the absence of 150 pmol IIIabc subdomains of HCV and CSFV IRESs in 200 µl buffer A supplemented with 1 mM ATP and 0.4 mM GTP for 10 min at 37°C. The reaction mixtures were then subjected to centrifugation through a 10%–30% sucrose density gradient (SDG) prepared in buffer A in a Beckman SW55 rotor at 53,000 rpm for 1h 15 min. Fractions that corresponded to 43S ribosomal complexes were analyzed by SDS-PAGE with subsequent fluorescent SYPRO (Molecular Probes) staining.
Electron Microscopy
4 µL of each sample was applied to holey carbon grids (carbon-coated Quantifoil 2/4 grid, Quantifoil Micro Tools GmbH) containing an additional continuous thin layer of carbon35. Grids were blotted and vitrified by rapidly plunging into liquid ethane at −180°C with a Vitrobot (FEI)36,37. Data acquisition was done under low-dose conditions (12 e−/Å2) on a FEI Tecnai F20 (FEI, Eindhoven) operating at 120 kV with a Gatan CT3500 side-entry cryo-holder. The dataset was collected automatically using Leginon38 at a calibrated magnification of 51,570× on a 4k × 4k Gatan Utrascan 4000 CCD camera with a physical pixel size of 15 µm, thus making the pixel size 2.245 Å on the object scale.
Image Processing
The data were preprocessed using pySPIDER (Langlois and Frank, unpublished) and Arachnid. Arachnid is a Python-encapsulated version of SPIDER39, replacing SPIDER batch files with Python. It also contains novel procedures such as Autopicker, which was used for the automated particle selection, yielding a total number of particles of ~630,000, picked from ~12,000 micrographs images. Those selected particles were classified with RELION40, ultimately yielding six classes (see Details on the 3D classification and Extended Data Fig. 9); Class 1, 40S●ΔII-IRES, ~56,000 particles. Class 2, 40S●ΔII-IRES●DHX29, ~72,900 particles. Class 3, 40S●DHX29●eIF3, ~18,000 particles. Class 4, 40S●ΔII-IRES●DXH29●eIF3, ~26,000 particles, where eIF3 is in “orientation 1”, Class 5, 40S●ΔII-IRES●DXH29●eIF3, ~18,000 particles, where eIF3 is in “orientation 2”, and Class 6, 40S●ΔII-IRES●DXH29●eIF3, ~17,000 particles, where eIF3 is in “orientation 3”. We focused our analysis on classes 2 and 4, scoring resolutions of 8.5Å and 9.5Å, respectively, estimated with the gold standard Fourier Shell Correlation (FSC) = 0.143 (Extended Data Fig. 4a)40,41. To assess the quality of our reconstruction, we performed a reference-free 2D classification using RELION40 and compared the obtained class-averages with projections generated from our final reconstruction (Extended Data Fig. 4b).
Details on the 3D classification
The unsupervised 3D classification of IRES-bound ribosomal complexes consisted of six rounds of 3D classifications (Extended Data Fig. 9) conducted in a quasi-hierarchical fashion, using Relion40 (version 1.2b7). The classes generated by each round were analyzed and either regrouped and reclassified in a subsequent round or rejected because of their structural inconsistency with the known structures of the different components of the complex. At the end of the different rounds of classifications, particles from similar classes were regrouped and refined together as one class. For each run of classification and refinement the small ribosomal 40S subunit55 (PDBID: 2XZM) was used as an initial reference. The reference was generated by simulating a cryo-EM density map from the atomic coordinates file of the 40S subunit using Chimera UCSF47. The reference map was filtered to 40Å for classification and auto-refinement runs. For all classification runs the regularization factor T in Relion was set to 3.
The first run of classification had the purpose of eliminating those data windows containing obvious contaminants from the rest of the dataset and was set for 10 classes, with the following sampling parameters: Angular sampling interval of 30°, an offset search range of 21 pixels and an offset search step of 3 pixels. The sampling parameters were progressively narrowed in the course of the 36 classification iterations, to 3.7° for the angular sampling interval, 8 pixels for the offset search range and 2 pixels for the offset search step. At the end of the first classification round, three classes (representing 33% of the 630,000 picked windows of particles) were inconsistent with the known structure of the 40S ribosomal subunit and were thus rejected (labeled “rejects”). Out of the first round of classification, particles from 7 classes (representing 67% of the dataset) were pooled together for the second round of classification.
The second round of 3D classification and all of the subsequent rounds of classification started with the following initial sampling parameters: Angular sampling interval of 15°, an offset search range of 14 pixels and an offset search step of 2 pixels. The sampling parameters were progressively narrowed in the course of the 42 classification iterations, to 1.8° for the angular sampling interval, 4 pixels for the offset search range and 1 pixel for the offset search step. All 10 classes of this second classification round were considered to be potentially consistent with the structure of the 40S and thus no rejects were singled out. Based on the visual analysis of these classes, it was possible to regroup structurally similar classes of particles into two groups. Two separate 3D classification rounds were conducted in parallel on the particles of each group, round 3 and round 4.
Round 3 was performed using particles collated from 7 different classes (representing 44% of the full dataset) originating from the previous round, round 2 (Extended Data Fig. 9), and was set to generate 8 classes. This round of classification was conducted for 46 iterations and resulted in 4 classes of particles inconsistent with the structure of the 40S subunit and/or eIF3 and the CSFV IRES (classes 3 to 6 representing 21% of the full dataset, Extended Data Fig. 9).
Round 4 of classification was performed using particles pooled from 2 different classes (representing 23% of the full dataset) originating from the previous round (round 2, Extended Data Fig. 9) and was set to generate 8 classes.
Based on the visual analysis of the classes originated from rounds 3 and 4, it was possible to regroup some of their different classes into two groups and the particles from structurally similar classes forming each group were collated. Two other separate 3D classification rounds were conducted in parallel on the particles of each group, round 5 and round 6.
Round 5 was performed using particles from class 2 originating from classification round 3 (representing 5% of the full dataset, Extended Data Fig. 9) and due to the low number of particles it was set to generate 4 classes only. This round was conducted for 37 iterations.
Round 6 was performed using particles from classes 6 and 7 originating from round 4 and class 1 originating from round 3 (representing 14% of the full dataset) and was set to generate 8 classes. The round yielded 7 classes of rejects (representing 13% of the full dataset). Classes 1, 3, 5 and 6 were rejected because of the inconsistency of the shape of eIF3 with its known structure, which appears to be bound to the CSFV IRES but assumed a scattered and/or deformed aspects, probably due to a very large degree of flexibility. Classes 2, 4 and 7 were rejected because of the low-resolution appearance of the 40S subunit and the inconsistency of the shape of the CSFV IRES with its structure.
Based on the similarities among different classes originating from rounds 3, 4, 5 and 6, particles from certain classes were pooled and auto-refined, using Relion’s Auto-Refine module, into six final classes (Extended Data Fig. 9) as follows: 1- Particles from class 8 of round 3, and from class 8 of round 4 (9% of the full dataset) displaying 40S●ΔII-IRES complexes. 2- Particles from class 7 of round 3, and from classes 3 and 5 of round 4 (12% of the full dataset) displaying 40S●ΔII-IRES●DHX29 complexes. 3- Particles from class 4 of round 4 and from class 8 of round 6 (3% of the full dataset) displaying 40S●DHX29●eIF3 complexes. 4- Particles from class 2 of round 5 and from class 2 of round 4 (4% of the full dataset) displaying 40S●ΔII-IRES●DXH29●eIF3, where eIF3 is in “orientation 1”. 5- Particles from class 3 of round 5 and from class 1 of round 4 (3% of the full dataset) displaying 40S●ΔII-IRES●DXH29●eIF3, where eIF3 is in “orientation 2”. 6- Particles from classes 1 and 4 of round 5 (3% of the full dataset) displaying 40S●ΔII-IRES●DXH29●eIF3, where eIF3 is in “orientation 3”. In all of the auto-refinements, Relion was set with the following initial sampling parameters: Angular sampling interval of 15°, an offset search range of 14 pixels and an offset search step of 2 pixels. The number of iterations for each refinement was determined automatically by Relion based on the improvement of the resolution between consecutive iterations.
3D variance estimation
The particle windows were binned three-fold in order to reduce the memory and CPU requirements for variance estimation, yielding a pixel size of 6.735 Å. The 3D variance map was computed using the bootstrapping method42–45 for the class 4 map (Extended Data Fig. 3) presenting eIF3 in orientation 1, as follows: forty thousand bootstrap reconstructions were generated, each of which was obtained from N = 26,317 particles projections that were randomly sampled with replacement from the total set of the N particles. The bootstrap volumes were filtered to about twice the first zero-crossing of the contrast transfer functions (CTFs) to boost the signal-to-noise ratio of the 3D variance. The structural variance was estimated as the sample variance of the bootstrap volumes minus the variance of the noise, and the difference was then multiplied by N43. In this estimation, the noise variance is assumed to be uniform across the map43.
Density maps segmentation and display
Cryo-EM reconstructions were segmented using the SEGGER module46 implemented in UCSF Chimera47. Segments counting less than 10,000 voxels were discarded. Segments were refined manually using the VOLUME ERASER module implemented in UCSF Chimera. Finally, the obtained segments were smoothed using a Gaussian filter in the VOLUME FILTER module also implemented in Chimera. The final maps were displayed and rendered with Chimera.
CSFV IRES modeling and fitting
The ΔII-IRES RNA was modeled based on the established secondary structure of the CSFV IRES20. The secondary structure was loaded into the S2S nucleic acid alignment and modeling tool48 and the CSFV IRES secondary structure was exported to Assemble, a nucleic acid 2D/3D modeling tool49. As domain II is absent in ΔII-IRES RNA, only domain III was modeled from the IRES sequence (nucleotides 129–361; GenBank J04358) and a three-dimensional model was generated in Assemble and placed into the EM map.
The model was relaxed and fitted into the IRES map using Molecular Dynamics Flexible Fitting (MDFF)50. MDFF is an MD simulation-based fitting procedure, which applies an extra potential to the system, related to the gradient of the cryo-EM density map. The initial system was prepared for MDFF using VMD51 and consisted of the atomic model of the CSFV ΔII-IRES and its corresponding segmented map from class 2 particles, which correspond to the 40S●ΔII-IRES●DHX29 complex. As the model was built into the EM map directly, no rigid-body fitting was required. In order to achieve a better representation of the inter- and intra-molecular interactions, the system was embedded in a solvent box of TIP3P water molecules, with an extra 12 Å padding in each direction, and neutralized by potassium ions, and an excess of ~ 0.2 M KCl was added. The system was minimized for 2000 steps in NAMD52 followed by MDFF in explicit solvent. The run was stopped after 400ps of simulation time, when the cross-correlation coefficient between the model and the map, as well as the RMSD of the model during the trajectories have stabilized. The simulated system was prepared using CHARMM force field parameters (Combined CHARMM All-Hydrogen Topology File for CHARMM22 Proteins and CHARMM27 Lipids53,54. The same protocol was reapplied in order to fit the CSFV ΔII-IRES into its corresponding density segmented from class 4, 40S●ΔII-IRES●DXH29●eIF3, where eIF3 is in orientation 1. This last MDFF endeavors to fit flexible domain IIIb of the CSFV IRES into one specific conformation in interaction with eIF3 in order to identify the IRES residues interacting with the latter (Fig. 4a).
To identify the CSFV IRES binding site on the body of the 40S subunit in terms of ribosomal proteins and rRNA, the crystal structure of Tetrahymena thermophila 40S55 was rigid-body fitted into the class 2, 40S●ΔII-IRES●DHX29, using UCSF Chimera47. The highest CCC guided the optimal fit.
Supplementary Material
AKNOWLEDGEMENTS
We thank Melissa Thomas for assistance in the preparation of figures. This work was supported by HHMI and NIH R01 GM29169 (to J.F.), NIH R01 GM51340 (to C.U.T.H.) and NIH R01 GM59660 (to T.V.P.).
Footnotes
The electron microscopy (EM) map has been deposited in the EMBL-European Bioinformatics Institute EM Data Bank under accession codes EMD-2450 and EMD-2451. Coordinates of EM-based model of the CSFV IRES have been deposited in the RCSB Protein Data Bank under accession number 4c4q. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
Y.H., A.d.G., V.D, T.V.P, C.U.T.H and J.F. interpreted the data and wrote the manuscript. V.D, T.V.P and C.U.T.H prepared the sample. Y.H., A.d.G. and R.A.G. performed the cryo-EM experiments. H.Y.L. performed the 3D variance estimation. Y.H. and R.L. performed the cryo-EM data processing. Y.H. modeled the CSFV IRES. C.U.T.H and J.F. directed research.
REFERENCES
- 1.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nature Rev. Mol. Cell. Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolupaeva VG, Pestova TV, Hellen CU. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA. 2000;6:1791–1807. doi: 10.1017/s1355838200000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Breyne S, Yu Y, Pestova TV, Hellen CU. Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site. RNA. 2009;14:367–380. doi: 10.1261/rna.696508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pisarev AV, et al. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 2004;78:4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spahn CM, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 9.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5' nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buratti E, Tisminetzky S, Zotti M, Baralle FE. Functional analysis of the interaction between HCV 5'UTR and putative subunits of eukaryotic translation initiation factor eIF3. Nucleic Acids Res. 1998;26:3179–3187. doi: 10.1093/nar/26.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun C, Querol-Audí J, Mortimer SA, Arias-Palomo E, Doudna JA, Nogales E, Cate JHD. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. [published online June 13, 2013];Nucl. Acids Res. doi: 10.1093/nar/gkt510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 13.Hashem Y, et al. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013;153:1108–1119. doi: 10.1016/j.cell.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. Translation initiation on mammalian mRNAs with structured 5'UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abaeva IS, Marintchev A, Pisareva VP, Hellen CU, Pestova TV. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J. 2011;30:115–129. doi: 10.1038/emboj.2010.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 17.Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fletcher SP, Jackson RJ. Pestivirus internal ribosome entry site (IRES) structure and function: elements in the 5' untranslated region important for IRES function. J. Virol. 2002;76:5024–5033. doi: 10.1128/JVI.76.10.5024-5033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friis MB, Rasmussen TB, Belsham GJ. Modulation of translation initiation efficiency in classical swine fever virus. J. Virol. 2012;86:8681–8692. doi: 10.1128/JVI.00346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rijnbrand R, et al. Almost the entire 5' non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 23.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H, et al. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. (2005). [DOI] [PubMed] [Google Scholar]
- 24.Querol-Audi J, et al. Architecture of Human Translation Initiation Factor 3. Structure. 2013;21:920–928. doi: 10.1016/j.str.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melcher SE, Wilson TJ, Lilley DM. The dynamic nature of the four-way junction of the hepatitis C virus IRES. RNA. 2003;9:809–820. doi: 10.1261/rna.5130703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rijnbrand R, van der Straaten T, van Rijn PA, Spaan WJ, Bredenbeek PJ. Internal entry of ribosomes is directed by the 5' noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J. Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19:1456–1466. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellen CU, de Breyne S. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 2007;81:5850–5863. doi: 10.1128/JVI.02403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Babaylova E, Graifer D, Malygin A, Stahl J, Shatsky I, Karpova G. Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res. 2009;37:1141–1151. doi: 10.1093/nar/gkn1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malygin AA, Shatsky IN, Karpova GG. Proteins of the human 40S ribosomal subunit involved in hepatitis C IRES binding as revealed from fluorescent labeling. Biochemistry (Mosc) 2013;78:53–59. doi: 10.1134/S0006297913010069. [DOI] [PubMed] [Google Scholar]
- 31.Pisarev AV, Unbehaun A, Hellen CU, Pestova TV. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- 32.Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skabkin MA, et al. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pestova TV, Hellen CU. Preparation and activity of synthetic unmodified mammalian tRNAiMet in initiation of translation in vitro. RNA. 2001;7:1496–1505. doi: 10.1017/s135583820101038x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grassucci RA, Taylor DJ, Frank J. Preparation of macromolecular complexes for cryo-electron microscopy. Nat. Protoc. 2007;2:3239–3246. doi: 10.1038/nprot.2007.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dubochet J, et al. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 37.Wagenknecht T, Frank J, Boublik M, Nurse K, Ofengand J. Direct localization of the tRNA-anticodon interaction site on the Escherichia coli 30 S ribosomal subunit by electron microscopy and computerized image averaging. J. Mol. Biol. 1988;203:753–760. doi: 10.1016/0022-2836(88)90207-0. [DOI] [PubMed] [Google Scholar]
- 38.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–50. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 40.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henderson R, et al. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penczek PA, Yang C, Frank J, Spahn CM. Estimation of variance in single-particle reconstruction using the bootstrap technique. J. Struct. Biol. 2006;154:168–183. doi: 10.1016/j.jsb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 43.Zhang W, Kimmel M, Spahn CM, Penczek PA. Heterogeneity of large macromolecular complexes revealed by 3D cryo-EM variance analysis. Structure. 2008;16:1770–1776. doi: 10.1016/j.str.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, Klaholz BP. Structure of the 30S translation initiation complex. Nature. 2008;455:416–420. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 45.Liao HY, Frank J. Classification by bootstrapping in single particle methods. IEEE Int. Symp. Biom. Imaging. 2010:169–172. doi: 10.1109/ISBI.2010.5490386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 48.Jossinet F, Westhof E. Sequence to Structure (S2S): display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics. 2005;21:3320–3321. doi: 10.1093/bioinformatics/bti504. [DOI] [PubMed] [Google Scholar]
- 49.Jossinet F, Ludwig TE, Westhof E. Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics. 2010;26:2057–2059. doi: 10.1093/bioinformatics/btq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. Model. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 52.Phillips JC, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brooks BR, et al. CHARMM: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J. Comp. Chem. 1983;4:187–217. [Google Scholar]
- 54.MacKerell AD, Jr, et al. HARMM: The Energy Function and Its Parameterization with an Overview of the Program. In: Schleyer PvR, et al., editors. The Encyclopedia of Computational Chemistry. Vol. 1. Chichester: John Wiley & Sons; 1998. pp. 271–277. [Google Scholar]
- 55.Rabl R, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.