Abstract
p-Aminobenzoic acid (pABA) plays important roles in a wide variety of metabolic processes. Herein we report the synthesis, theoretical calculations, crystallographic investigation, and in vitro determination of the biological activity and phytotoxicity of the pABA salt, 2-hydroxyethylammonium p-aminobenzoate (HEA-pABA). The ability of neutral and anionic forms of pABA to interact with TIR1 pocket was investigated by calculation of molecular electrostatic potential maps on the accessible surface area, docking experiments, Molecular Dynamics and Quantum Mechanics/Molecular Mechanics calculations. The docking study of the folate precursor pABA, its anionic form and natural auxin (indole-3-acetic acid, IAA) with the auxin receptor TIR1 revealed a similar binding mode in the active site. The phytotoxic evaluation of HEA-pABA, pABA and 2-hydroxyethylamine (HEA) was performed on the model plant Arabidopsis thaliana ecotype Col 0 at five different concentrations. HEA-pABA and pABA acted as potential auxin-like regulators of root development in Arabidopsis thaliana (0.1 and 0.2 mM) and displayed an agravitropic root response at high concentration (2 mM). This study suggests that HEA-pABA and pABA might be considered as potential new regulators of plant growth.
Introduction
The interest in carboxylate compounds in general and benzoic acid derivatives in particular continues to grow because of their chemical and biological properties. Benzoic acids and their derivatives are important structural elements for many natural products, being involved in various physiological processes in plants. Among these compounds, p-aminobenzoic acid (pABA) has displayed biological, medicinal and industrial interest. pABA is a compound of high biological significance, it is present in plant and animal tissues and is sometimes referred to as bacterial vitamin H1, Bx, or B10 [1]. pABA is a well-known precursor of folic acid [2], and has recently been identified as a coenzyme Q precursor [3], [4], which opens new possibilities for its application in medicine. It is a building block used in design of drugs and frequently found as a structure moiety in drugs [5]. pABA exhibits a wide range of therapeutic uses as antioxidant [6], [7], antibacterial [8], [9], antimutagenic [10], anticoagulant [11], [12], fibrinolytic and immunomodulating agent [13], protective drug against UV-irradiation [14]–[16] and in diagnostic tests for the state of the gastrointestinal tract [17], [18].
Recent studies in agriculture report the role of pABA in plant thermotolerance [19] and its action as chemical inducer of systemic acquired resistance against plant pathogens [20]. Considered to be in the B-complex vitamin family and characterized by safety and cost effectiveness, pABA is frequently added along with plant hormones in nutriment media as a stimulator of seed germination [21]. To the best of our knowledge, the role of folate precursor pABA as a plant growth modulator has not been investigated so far.
Besides the plethora of properties noted above, pABA is a versatile reagent for structure extension through linear hydrogen bonding associations, through both the carboxylic and amine functional groups [22]. Keeping in view the structural and biological diversity of pABA and in connection with our interest in supramolecular chemistry of carboxylate salts [23], [24], here we report the synthesis, theoretical calculations, crystallographic investigation, and in vitro phytotoxic activity of 2-hydroxyethylammonium p-aminobenzoate (HEA-pABA). As a base, we chose the natural amino alcohol 2-hydroxyethylamine (HEA), a suitable model of alkanolamine. This compound is cheap, commercially available, environmentally tolerable and has hydrogen-bond donor sites. On the other hand, HEA is an essential component of cell membranes and an important metabolite in plants for the synthesis of choline and membrane lipids, such as phosphatidylethanolamine and phosphatidylcholine [25].
The preparation of multicomponent organic crystals (solvates, hydrates, co-crystals, and salts) from such multifunctional molecules as benzoic acids and alkanolamine display interesting networks and a wide range of different biological properties [23], [24], [26]–[28]. The possible binding modes of pABA, its anionic form and the natural auxin IAA bound to the auxin receptor TIR1 have been evaluated by docking experiments and refined by Molecular Dynamics (MD) and Quantum Mechanics/Molecular Mechanics (QM/MM) calculations. The present work continues our recent investigations of new structural aspects of alkanolamine salts [24], reports the full details of the X-ray crystallographic structure of HEA-pABA and introduces this compound and corresponding acid as new potential regulators of plant growth.
Materials and Methods
General
pABA and HEA used for the synthesis of HEA-pABA were of analytical grade and purchased from Fluka AG (Buchs SG). HEA was freshly distilled before any use. FT-IR spectra in the range 4000−400 cm−1 were recorded with a JASCO − FT/IR-4200 spectrometer, using the KBr pellet technique with a resolution of 4.0 cm−1 and a scanning speed of 16 mm s−1. The optical properties of HEA-pABA were examined by using a UV−Vis spectrophotometer at room temperature. UV−visible spectra were recorded in the 190−800 nm range with PERKIN-ELMER LAMBDA 12 UV−Vis spectrometer. Melting point was found for finely purifying compound (by repeated recrystallization) using a Boetius instrument.
Synthesis and characterization of HEA-pABA
HEA-pABA was prepared by the reaction of HEA and pABA in a 1∶1 molar ratio. Under vigorous stirring, freshly distilled HEA was added drop wise to the solution of the acid in acetone, at reflux for 2 h to complete the reaction. The formed salt was precipitated in a white crystalline state and, after cooling at room temperature, collected by filtration, washed with cold acetone, and dried under vacuum for 3 h. This reaction had a 90% yield. The salt was recrystallized to obtain suitable crystals for X-ray analysis. Crystals were grown by slow evaporation of ethanol solution at room temperature. The purity of synthesized salts was 99.3% (±0.8%), established by spectrophotometric method. This consisted in determining the specific extinction (absorbance at λmax of a solution 1 gl−1, in a cell of 1 cm width) of the free acid (εa) and the corresponding salt (εs), for the UV maxima:
where Ms, and Ma are the molecular weights for the salt and free acid, respectively. The elemental analysis was in agreement with the expected stoichiometry. The FT-IR spectrum (KBr pellets prepared by grinding a 5−10 mg sample with 100 mg KBr) was consistent with salt formation.
C9H14N2O3, (198.22), m.p. 147–150°C; λmax = 265.19 nm; FT-IR spectra (KBr pelet, cm−1): 3392, 3227, 2132, 1591, 1540, 1384, 1361, 1073, 1016; calcd. (%): C 54.53, H 7.12, N 14.13; found (%): C 54.51, H 7.03, N 14.10.
X-ray measurement and refinement
X-ray data for HEA-pABA were collected at room temperature on a Siemens P3/PC diffractometer (CuKα-radiation). The crystallographic data and the experimental details are summarized in Table 1. Structure solution and refinement were carried out using the SHELX-97 program [29]. Non-hydrogen atoms were refined anisotropically. The hydrogen atoms were placed in calculated positions with their isotropic displacement parameters riding on those of parent atoms and the H-atoms of NH2 group of HEA-pABA were found from differential Fourier maps. The geometric parameters for H–bonds are listed in Table 2. The geometric parameters were calculated and the figures were drawn with the use of the PLATON program [30]. Bond lengths (Å) and angles (°) for HEA-pABA are listed in Table 3. The hydrogen atoms that are not involved in the hydrogen bonding were omitted from the representation of the crystal packing. Crystallographic data for HEA-pABA were deposited in the Cambridge Crystallographic Data Center (CCDC 936586).
Table 1. Crystal Data and Structure Refinement for HEA-pABA.
| Compound | HEA-pABA |
| Empirical formula | C9H14N2O3 |
| Formula weight | 198.22 |
| Wavelength | 1.54184 |
| Crystal system | Orthorhombic |
| Space group | Pbca |
| a, Å | 11.898(2) |
| b, Å | 8.3140(17) |
| c, Å | 20.790(4) |
| α, deg | 90 |
| β, deg | 90 |
| γ, deg | 90 |
| V, Å3 | 2056.5(7) |
| Z | 8 |
| Density (calc.), mg/m3 | 1.280 |
| Absorption coefficient, mm-1 | 0.808 |
| F(000) | 848 |
| Crystal size, mm3 | 0.14×0.10×0.08 |
| θ range for data collection, deg | 4.25–70.06 |
| Index ranges | −14≤h≤14, −10≤k≤1, −25≤l≤24 |
| Reflections collected/unique | 1952/1952, R(int) = 0.0000 |
| Completeness (%) | 99.9 (θ = 70.06) |
| Data/parameters | 1952/135 |
| GOF on F2 | 0.998 |
| Final R indices (I>2σ (I)) | R1 = 0.0467, wR2 = 0.0930 |
| R indices (all data) | R1 = 0.1101, wR2 = 0.1105 |
| Largest diff. peak and hole, e.Å−3 | 0.196/−0.193 |
Table 2. Hydrogen-bonding geometry (Å) for compound HEA-pABA.
| No. of HB | D−H…A | d(D…H), Å | d(H…A), Å | d(D A), Å | ∠(DHA), deg. | Symmetry transformation for H-acceptor |
| 1 | N(1)–H…O(3) | 0.89 | 1.91 | 2.801(3) | 175 | −x+2, −y, −z+2 |
| 2 | N(1)–H…N(2) | 0.89 | 2.22 | 3.051(4) | 155 | x, −y+1/2, z+1/2 |
| 3 | N(1)–H…O(2) | 0.89 | 1.87 | 2.752(3) | 171 | x, y, z |
| 4 | O(1)–H…O(3) | 0.82 | 1.90 | 2.715(3) | 176 | x, y, z |
| 5 | N(2)–H…O(1) | 0.91(3) | 2.29(3) | 3.087(3) | 146 | −x+2, y−1/2, −z+1/2 |
| 6 | N(2)–H…O(3) | 0.82(3) | 2.20(3) | 3.025(3) | 174 | x+1/2, y, −z+1/2 |
| 7 | C(2)–H…O(2) | 0.97 | 2.41 | 3.247(3) | 145 | x−1/2, −y+1/2, −z+1 |
Table 3. Bond lengths (Å) and angles (°) for HEA-pABA.
| N(1)-C(1) | 1.481(3) |
| O(1)-C(2) | 1.409(3) |
| C(1)-C(2) | 1.495(4) |
| N(2)-C(6) | 1.404(3) |
| O(2)-C(9) | 1.257(3) |
| O(3)-C(9) | 1.281(3) |
| C(3)-C(4) | 1.384(3) |
| C(3)-C(8) | 1.391(3) |
| C(3)-C(9) | 1.492(3) |
| C(4)-C(5) | 1.377(3) |
| C(5)-C(6) | 1.387(4) |
| C(6)-C(7) | 1.389(4) |
| C(7)-C(8) | 1.379(3) |
| N(1)-C(1)-C(2) | 112.1(3) |
| O(1)-C(2)-C(1) | 113.9(3) |
| C(4)-C(3)-C(8) | 117.8(2) |
| C(4)-C(3)-C(9) | 121.8(2) |
| C(8)-C(3)-C(9) | 120.3(3) |
| C(5)-C(4)-C(3) | 121.4(3) |
| C(4)-C(5)-C(6) | 120.7(3) |
| C(5)-C(6)-C(7) | 118.3(2) |
| C(5)-C(6)-N(2) | 121.5(3) |
| C(7)-C(6)-N(2) | 120.1(3) |
| C(8)-C(7)-C(6) | 120.7(3) |
| C(7)-C(8)-C(3) | 121.1(3) |
| O(2)-C(9)-O(3) | 122.9(3) |
| O(2)-C(9)-C(3) | 118.6(3) |
| O(3)-C(9)-C(3) | 118.5(3) |
Docking and QM/MM calculations
The natural auxin (indole-3-acetic acid, IAA) and pABA, the last one both in the neutral and anionic form, were docked in the auxin binding site as identified in the crystal structure of the TIR1 protein-auxin complex retrieved from protein data bank (PDB id 2P1P) [31] using AutoDock 4.2 software [32]. This software considers only the polar hydrogen atoms during the docking process. A Lamarkian genetic algorithm [33] was employed for the docking simulation, performing 100 independent runs per molecule. In each run, a population of 50 individuals evolved along 27000 generations and a maximum number of 25 million energy evaluations were performed. The best fit (lowest docked energy) solutions of the 100 independent runs were stored for subsequent analysis.
Docking results were refined by performing molecular dynamics simulations using the AMBER 11 program suite [34], the AMBER99SB force field [35] for the protein and the GAFF force field for ligands [36]. Each of the three protein-ligand complexes was first submitted to a geometry optimization consisting of 1000 steps of steepest descent, followed by 9000 steps of conjugate gradient, energy minimization. The systems were then equilibrated for 100 ps in the NVT ensemble and for additional 100 ps in the NPT ensemble. Ten ns long molecular dynamics simulations were then run in the NPT ensemble at 300 K and 1 atm. Water solvent was modelled with the TIP4P water model [37]. A cutoff of 12 Å was used for non-bonded VdW interactions, while electrostatic interactions were treated with the Particle Mesh Ewald approach [38].
QM/MM calculations were performed on the final structures obtained from MD simulations, using the two-layer ONIOM model [39]–[41] as implemented in Gaussian09 [42]. Owing to the high complexity of the examined system, QM/MM calculations were performed in vacuo, including only the ligands and the residues that showed the most favorable interactions with the ligands during the MD simulations (namely Arg403, Ser438 and Leu439) in the QM layer. The protein was included in the MM layer and its geometry was kept fixed. The QM part was treated at Density Functional Theory (DFT) level with the B3LYP functional [43], [44] and the 6–31+G* basis set. Protein atom charges were assigned using the Amber Force Field [34]. The mutual electrostatic interaction between the ligand and the protein was treated using the electrostatic embedding approach [41]. In this model, the point charges of the MM layer are included in the Hamiltonian of the QM layer, introducing a polarization of the wave function.
Biological activity
Plant material and seed germination
Arabidopsis thaliana, ecotype Columbia 0 (Col 0) seeds were surface sterilized by treatment with a 5% (w/v) calcium hypochlorite solution and 0.02% (v/v) Triton X-100 in 80% ethanol (EtOH), for 10–12 minutes at 25–28°C, by continuous shaking. They were then rinsed twice with 80% EtOH, washed with 100% EtOH and finally with sterile distilled water [45]. Thirty sterile seeds were sown on every square Petri plate (12×12 cm) contained: 80 ml of sterile solid MS agar medium (modified from [46]), HEA-pABA, pABA or HEA at five concentrations (0.02, 0.1, 0.2, 1 and 2 mM) and IAA at five concentrations (0.01, 0,1, 1, 10 and 100 nM). The salt and IAA were dissolved in 5 mM MES (2-[N′-morpholino] ethanesulfonic acid) buffer, while pABA, which is slightly soluble in water, was dissolved in MES buffer – EtOH solution, with a final EtOH concentration of 0.2% (v/v). Our previous research on Arabidopsis thaliana (Col 0) [28] and Cucumis sativus L. [27] seeds show that 0.2% (v/v) ethanol concentration in culture media does not alter germination rate of seeds, but inhibit slowly primary root length and lateral root formation, as it is confirmed by some studies reported in the literature [47], [48]. For clarity, control corresponding to pABA was noted CONTROL 0.2% EtOH and for HEA-pABA and HEA, CONTROL. HEA-pABA, pABA or HEA solved in the solvents mentioned above were sterilized by filtration (Millipore) and added to molten control agar (∼50°C). Five repetitions for each compound concentration were performed. The plates were stratified horizontally in darkness at 4°C for 2 days to break dormancy and synchronize germination, and then grown in a climate chamber at 22°C in a vertical position under a photoperiod of 16 h light (220 µmol m−2 s−1) and 8 h darkness.
Data analysis
After 5 and 10 days, respectively, images of the Petri dishes were generated by scanning the individual plate on an EPSON Perfection 4870 Photo scanner at 600 DPI. Obtained images were digitized using the ImageJ software package, and primary root length was measured for each seedling. After 10 days, plants were stained with 0.1% (w/v) safranin for 1–2 h, then rinsed with distilled water and 100, 80, 50, 30, 15% EtOH, respectively, and mounted in 50% (v/v) glycerol. All lateral roots emerging from the primary one were observed using a Nikon YS 100 microscope 10 x objective, and were taken into account for lateral root number data.
Statistical analysis
For all experiments, the overall data were statistically analysed by SYSTAT 10.0 program. All values are expressed as a mean ± S.E. One-way ANOVA analyses, followed by t-test using the Bonferroni correction, were used to compare the data obtained for primary root length, lateral root number at 5 and 10 days for treatments (HEA-pABA, pABA and HEA) at five different concentrations and controls. Different letters are used to indicate means that differ significantly (P<0.05).
Results and Discussion
Synthesis and characterization of HEA-pABA
HEA-pABA is a stable solid salt in accordance with the appropriate “rule of three”. The value of ΔpKa (the difference between the pKa value for HEA and pABA) was estimated to be >3, that determines the extent of proton transfer. UV spectroscopic data obtained in 0.1 M NaOH showed similar λmax values for both salt and acid, that confirms the existence of the same anion in both compounds (see Figure S1). The cut off wavelength for HEA-pABA was found to be 265.19 nm, corresponding to the gap energy of 4.68 eV, which is typical of insulating materials. There is no absorption in the entire visible region. This is the most favourable characteristic for a non-linear optical material.
FT-IR analysis indicates salt formation by the presence of bands arising from the asymmetric and symmetric vibrations of the COO− group occurring at 1591 cm−1 and 1384 cm−1, which do not exist in the spectra of free acid and by the absence of νC = O and νC-OH bands at 1673 cm−1 and 1291 cm−1, characteristic for –COOH group [49], [50]. At the same time, the band of the deformation vibrations of the N–H bonds is observed at 1540 cm−1. A supplementary proof of the salt formation was the appearance of C−O vibrations between 1100 and 1000 cm−1, belonging to −CH2OH from HEA and a broad band near 2100 cm−1, which often appears in amine salt [50] (see Figure S2). The -NH3 + vibrations appears at 3200–2800 cm−1 and δNH3 + at 2400–2800 cm−1. The changes of wavenumbers of the bands of carboxylate group in the case of salt were discussed comparing to the acid. From the spectroscopic data and the elemental analysis it is concluded that pABA reacted with HEA with a 1∶1 ratio.
Crystal structure analysis of HEA-pABA
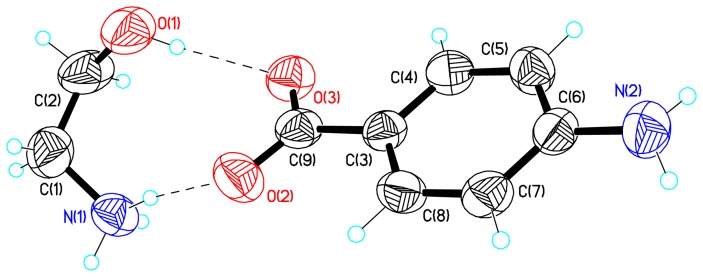
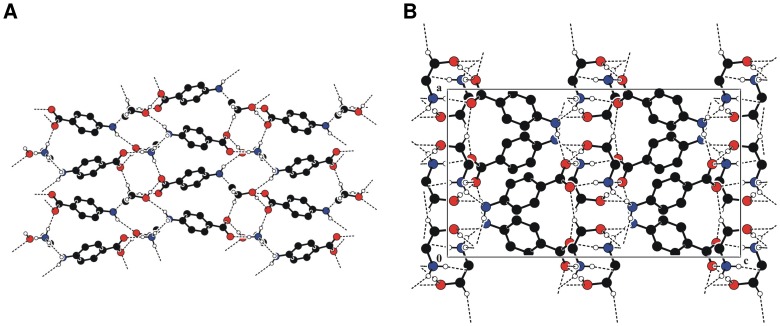
The salt unit of HEA-pABA serves as building block for the supramolecular architecture and in the crystal it is self-assembled via ionic N-H…O and normal O-H…O hydrogen bonds (HB) (Figure 1, Table 2). The anion in HEA-pABA forms a practically planar system because the dihedral angle between the least-square plane of the phenyl ring A(C3C4C5C6C7C8) and the least-square plane of the COO− group is equal to 5.8°. The cation adopts the –Syn-Clinal conformation, [30] the N1C1C2O1 torsion angle in HEA-pABA is equal to −68.8°. In the studied compound, the nitrogen atom in amino group has practically a pyramidal configuration because its valence angles are equal to 111.4, 114.9 and 117.2°. It means that the non-bonding electron pair of N2 could be considered as a fourth substituent on a sp3 hybridized nitrogen atom. Thus, the pyramidal geometry of the amino group allows forming three hydrogen bonds in the crystal. Indeed, the building blocks are assembled into 2-D layers parallel to (100) plane (Figure 2A, Table 2) via N1…O3 (1) HB and two hydrogen bonds with participation of amino group N1…N2 (2) and N2…O1 (5). These layers are further related along a axis by a glide plane perpendicular to [001] with the glide component [0.5 0 0] and a twofold screw axis with direction [100] and screw component [0.5 0 0] due to N2…O3 (6) and C2…O2 (7) HB respectively (Figure 2B, Table 2). According to the criterion proposed by Spek [30], to establish the presence of π–π stacking interactions between phenyl rings J1 and J2 (Cg(J1)···Cg(J2)<6.0 Å, β<60.0°, where Cg are the respective centroids of the rings and β is the angle between the Cg(J1)Cg(J2) vector and the normal to the J1 ring), there is a stacking interaction between the phenyl rings A and A (−x, 1/2+y, 1/2−z) within the layers of HEA-pABA. The distance between their centroids is in fact equal to 5.062 Å and β = 16.7°. There is an effect of hydrogen bonding on the geometry of carboxylate groups. While the carboxylate group in HEA-pABA participates as hydrogen-bonding acceptor, the C-O bond lengths vary significantly with the number and type of hydrogen-bonding donors linked to the oxygen atom. In the absence of hydrogen bonding and other electronic perturbations, the C-O bond lengths should be equal because of electron delocalization [23], [24]. Formation of single or multiple hydrogen bonds at one oxygen atom should cause the associated C-O bond to lengthen. In the studied compound, the difference in C9-O2 (1.257(3) Å) and C9-O3 (1.281(3) Å) bond lengths in carboxylate group is equal to 0.024 Å (Table 3). The transfer of proton in HEA-pABA has led to a significant polarization of the anion in comparison with pABA because the calculated dipole moments using HF method [51] for these systems are equal to 13.952 and 3.13 Debye, respectively, and dipole vectors in both molecules lie very close to C3–C9 bond.
Figure 1. ORTEP drawing for HEA-pABA.
The atomic labeling and charge-assisted hydrogen bonds are represented by dashed lines. Thermal ellipsoids are shown with the 50% probability level.
Figure 2. Crystal packing of HEA-pABA.
A. Crystal packing of HEA-pABA representing the 2-D layers parallel to (100) plane. B. Fragment of molecular packing in the crystal of HEA-pABA.
Molecular properties of HEA-pABA and pABA compared to classical auxin molecules
Beside the importance of salt unit of HEA-pABA as building block for supramolecular architecture, we found a new role of this compound and its precursor (pABA) on plant development as potential regulators of plant growth with an auxin-like effect. Auxins are defined by a set of physiological actions: cell division, elongation, formation of lateral and adventitious roots and gravitropism [52]–[54]. A large number of compounds with auxin and anti-auxin activity were identified so far as a result of structure-activity relationship investigations. More than 200 compounds with auxinic action and with different chemical structures have two common features critical for auxin activity: a planar aromatic ring structure and a carboxyl group-containing side chain [55]. The activity of both natural and synthetic auxins depends strongly on side groups binding to the ring structure. Through such substitutions, it is suggested that a positive charged area at specific distance from the carboxylic group is generated. The distance between the negative charge on the carboxyl group and the partial positive charge on the ring has been claimed to be critically important for detectable activity. According to some authors, the optimal distance (OD) appears to be about 5.0–5.5 Å [56], [57]. The ODs in HEA-pABA and pABA were compared with those of classical auxin molecules: indole-3-acetic acid (IAA), 2,4-dichlorophenoxyacetic acid (2,4-D) and 1-naphthaleneacetic acid (1-NAA), where IAA is the most important member of the auxin family and the natural one, the others being synthetic auxin analogs. We performed a search through the Cambridge Structural Database (version 5.34) [58], [59] and found that the crystal structures of these compounds indeed have an OD ranging from 4 to 6 Å. We observed 11, 4, 7 hits for IAA, 1-NAA and 2,4-D where the ODs range from 4.3 to 5.5, 4.3 to 4.6 and 5.1 to 5.3 Å, respectively. The presence of several conformers for IAA, 1-NAA and 2,4-D occurs due to rotation of molecules' moieties on bonds C1(O)-C2 and C2-C3 (see Figure S3). However, only for the conformers of 2,4-D family the ODs satisfy to specific distance between the negative charge on the carboxyl group and the partial positive charge on the ring. Therefore, this result brought us to look for the low energy conformer for IAA and 1-NAA, and then to check the above mentioned criteria for OD values. This was achieved by using the module Conformational Search implemented in HyperChem 6.03 [60]. The method involves the use of the directed scheme as introduced in the Monte Carlo multiple minimum approach [61]. This method seeks to uniformly sample low-energy regions by cycling through all previously accepted conformations (from lowest energy to highest, in order) when selecting each initial structure. In this process, the initial structures from series conformers of IAA and 1-NAA were modified by variation of the torsion angles along the C1–C2 and C2–C3 bonds followed by energy minimization of these angles using the MM+ force field. Then the point charges of atoms were calculated using HF method [51] for found low calculated energy conformers of IAA, 1-NAA and crystal structures of HEA-pABA, pABA [62] and 2,4-D [63]. The results of ODs calculation are summarized in Figure S3. For the studied compounds, the distances between the negative charges on the carboxyl groups and the partial positive charges on the rings ranged from 5.026 to 5.158 Å and the maximal absolute values of point charges which are used for estimation of ODs are observed in HEA-pABA.
Docking and QM/MM calculations
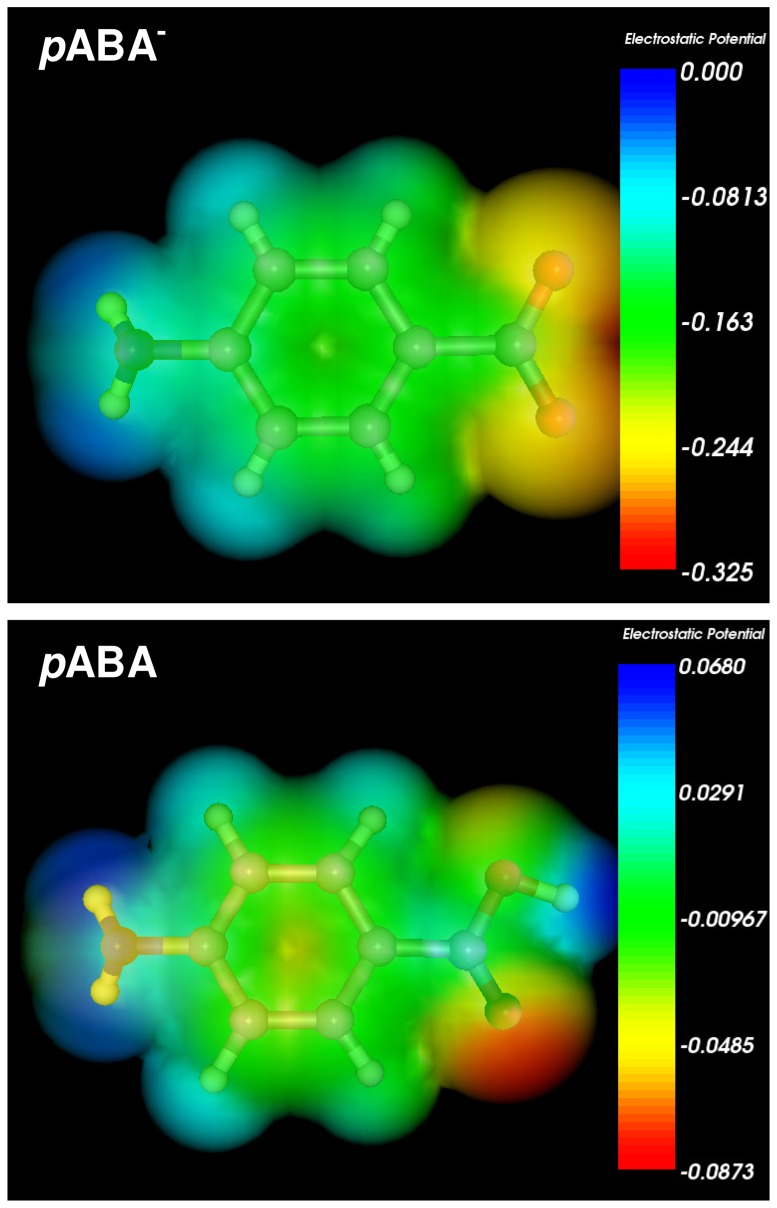
Auxin's receptor, TIR1, belongs to a large family of F-box proteins, which function as part of a protein destruction machinery in the ubiquitin–proteasome system. At the bottom of the TIR1 surface pocket, auxin helps nucleate a hydrophobic core together with TIR1 and its substrate polypeptide [31]. It is known that auxin molecules IAA, 2,4-D, and 1-NAA bind to the TIR1 pocket in a similar manner. Their common carboxyl group interacts with the same positive charged residue at the bottom of the TIR1 pocket [55]. Therefore, in order to estimate the ability of the anionic form of pABA (pABA−) and pABA to interact with positive charged residue at the bottom of the TIR1 pocket, we performed the calculation of molecular electrostatic potential map on accessible surface area (Figure 3). In pABA− the region of maximal attractive potential is located in vicinity of both oxygen atoms, while in pABA it is concentrated in vicinity of carbonyl oxygen. Thus, both oxygen atoms in pABA− are able to bind with positive charged residues at the bottom of the TIR1 pocket. In both molecules the attractive potential, which is located in vicinity of the rings, enhances their association with TIR1 pocket.
Figure 3. Mapping of the molecular electrostatic potential onto p-aminobenzoate anion (pABA−) and pABA accessible surface area.
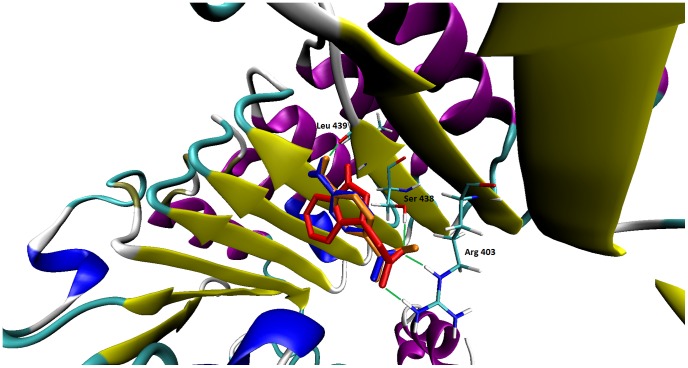
The possible binding modes of pABA to the TIR1 protein were then evaluated by performing docking experiments using the AutoDock 4.2 program. Both the neutral and the anionic form of pABA were docked in the auxin binding site of TIR1 protein as well as auxin itself (IAA), which was redocked to evaluate the performance of the program on this system. The best fit docking poses obtained from the simulation (see Materials and Methods) were then submitted to cluster analysis on a geometrical basis using a 2 Å rmsd threshold. This clustering procedure revealed that in each case only one cluster is present, suggesting a very good convergence of the algorithm. In Figure 4, the docked structure of both neutral and anionic forms of pABA and of auxin IAA are shown. Protonation does not seem to affect binding geometry of pABA. It makes hydrogen bond with Arg403, Ser438 (using the carboxyl function) and with Leu439 backbone oxygen using the amino function. These interactions are the same polar interactions exhibited by IAA, even if in auxin the hydrogen bond of the indolic nitrogen is slightly shorter than the corresponding hydrogen bond in pABA.
Figure 4. Binding modes of IAA (red), pABA (orange) and pABA− (blue).
Hydrogen bonds formed by pABA are drawn as green lines. Only polar hydrogen atoms are shown.
Binding energy calculated by AutoDock for the best fit conformations are as follows (in kcal/mol): IAA, −7.91; pABA−, −6.19; pABA, −5.97. Again, no significant difference exists between the neutral and the anionic form of pABA, while IAA exhibits a slightly stronger binding to the substrate, probably due to the larger size of the hydrophobic moiety. However, it is to be evidenced the quite small difference in binding energy provided by the three compounds, indicating a similar mode of binding of the proposed ligand with respect to auxin. Molecular dynamics simulations, on the other hand, showed that, while auxin and anionic form of pABA conserve their position and orientation in the binding site compared to the docked structures, the neutral pABA is expelled from the binding site during the simulation. Accordingly, the corresponding Root Mean Square Deviations (RMSD) of the three ligands during simulation with respect to the MD starting structure show different trends (see Figure S4). While auxin IAA and anionic form of pABA display small and well equilibrated RMSD values, for neutral pABA the RMSD increase progressively up to more than 8 Å after 10000 ps, indicating a loss of the starting structure as a consequence of its expulsion from the binding site. The MD results therefore suggest that the salt bridge interaction between Arg403 and the charged carboxyl group on auxin or pABA− is crucial for the interaction of ligands with TIR1 protein.
Geometry optimizations at the hybrid QM/MM level were performed on the final MD structures of auxin and pABA− complex. The QM/MM optimized structure of the TIR1-auxin complex revealed no substantial modification with respect to the docked structures. The HB interactions between Arg403 and Ser438 and auxin carboxyl group are preserved, as well as the HB between the indolic nitrogen and the backbone carbonyl of Leu439 (see Figure S4). In the QM/MM optimised structure of the TIR1-pABA− complex, interaction of the carboxyl group with Arg403 and Ser438 is preserved, while no HB is formed between the amino group of pABA and Leu439 (see Figure S5). The binding energies evaluated at the QM/MM level are −117.0 kcal/mol and −85.6 kcal/mol for IAA and pABA−, respectively, in agreement with the observed differences in the network of interactions between the protein and the two ligands.
It is worth noting that the larger magnitude of binding energies with respect to the values obtained by docking calculations should be ascribed to the fact that QM/MM calculations have been performed in vacuo, owing to the high complexity of the investigated systems. Within such approximation, the electrostatic interaction necessarily results to be overestimated with respect to the more realistic values obtained by docking experiments, where the solvent effect is implicitly taken into account by the score function used. In spite of such approximation, QM/MM calculations appear to confirm the similar interaction of anionic form of pABA with respect to auxin IAA. It is to be noted that very similar results have been recently obtained on the binding mechanisms of neutral and charged phenolic compounds with the human transthyretin, hTTR, using a similar approach combining molecular docking with QM/MM optimizations [64]. In agreement with their conclusions, ionization effects should not be neglected when modeling the binding modes of ligands with proteins.
HEA-pABA and pABA activity on root growth
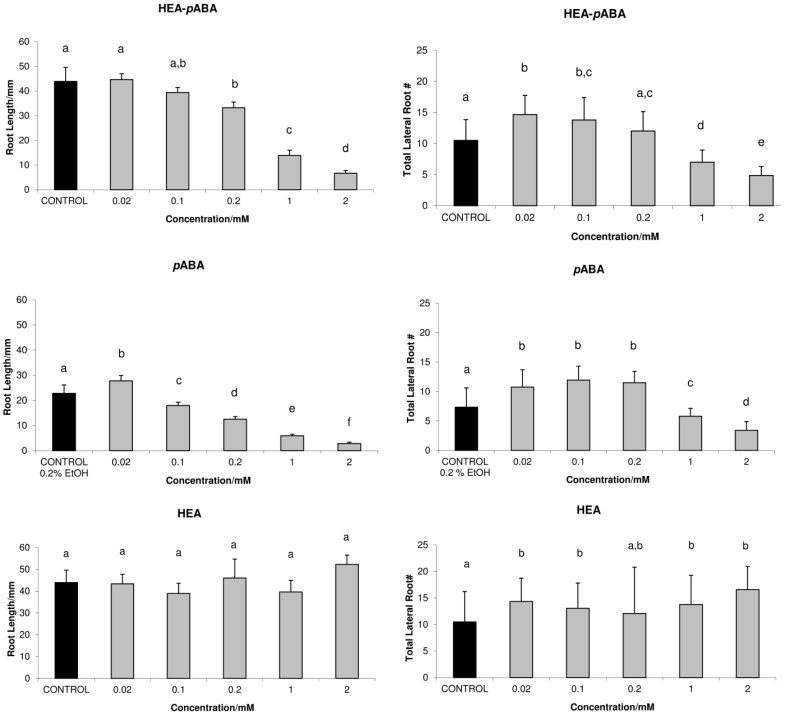
Phytotoxicity tests, especially the seed germination and root elongation tests, have more advantages over those toxicity tests using animals and algae, such as sensitivity, simplicity and low cost [65]–[67]. Phytotoxicity of HEA-pABA, pABA and HEA were determined at five different concentrations. Seeds of the model plant Arabidopsis thaliana have been used for germination, early seedling growth and gravity response tests. Figure 5 shows wild-type (WT) A. thaliana root growth on a medium supplemented with 0.02, 0.1, 0.2, 1 and 2 mM of HEA-pABA, pABA and HEA as compared to controls after 5 and 10 days (see also Figure S6). HEA-pABA and pABA showed a clear difference in primary root growth, and lateral root development after prolonged growth (10 days) (Figure 5). Moreover, the root growth difference was already apparent after 5 days, indicating that the action of tested compounds occurs throughout the period of elongation growth (see Figure S6). After 10 days, HEA treated seedlings exhibited a comparable effect to control for primary root length and an increased number of lateral roots at all concentrations tested. At the lowest concentration (0.02 mM), pABA treatment showed an increased number of lateral roots and primary root length, a common anti-auxin feature (Figure 5). At this concentration, HEA-pABA displays a comparable effect to control, regarding the primary root length and an increased number of lateral roots. At 0.1 and 0.2 mM, both HEA-pABA and pABA promote lateral roots, while they inhibit primary root length (Figure 5). The phenotypes like short root length, increased lateral root number are phenotypes similar to those observed in A. thaliana treated with auxins [68]–[70]. At high concentration (2 mM), primary root length is inhibited but also displayed a different growth behavior with respect to control seedlings. Preliminary tests aimed to compare HEA-pABA, pABA and IAA effects indicate that the latter exerts a 1000-fold higher root length inhibition (data not shown). The agravitropic effect of root growth is specific for pABA and HEA-pABA, never having been reported for an auxinic compound. Therefore, we argue that HEA-pABA and pABA might potentially act as auxin-like regulators of root development in A. thaliana (0.1 and 0.2 mM) and display an agravitropic root response at high concentration (2 mM) (see Figure S7).
Figure 5. Root length and number of lateral roots after 10 days of treated A. thaliana seedlings.
HEA-pABA, pABA and HEA treatments at different concentrations in comparison with control are shown. Control corresponding to pABA was noted CONTROL 0.2% EtOH and for HEA-pABA and HEA, CONTROL. Values with different letter annotation are significantly different (P<0.05). Data are means ± SE of 5 replicates.
Conclusions
This study presents the synthesis and the unique structural and biological properties of HEA-pABA. In the crystal, the salt unit of HEA-pABA serves as building block of the supramolecular architecture and in the crystal it is self-assembled via ionic N-H…O and normal O-H…O hydrogen bonds. The pyramidal geometry of pABA amino group allows to form three hydrogen bonds in the crystal. The transfer of proton in HEA-pABA has led to significant polarization of anion in comparison with pABA and the absolute values of point charges, which are used for estimation of ODs, are higher in HEA-pABA. For both these compounds, ODs are equal to 5.045 and 5.026 Å respectively. Both oxygen atoms in anionic form of pABA are able to bind with positive charged residues at the bottom of the TIR1 pocket. In pABA− and pABA the molecular electrostatic attractive potential, which is located in vicinity of the rings, enhances their association with TIR1 pocket. Docking experiments on complexes of pABA (in both neutral and anionic form) and IAA with TIR1 provide a similar binding mode between the two ligands. MD and QM/MM calculations confirm such result as far as pABA− is concerned. Phytotoxicity test indicates that compound HEA-pABA and pABA act as auxin-like regulators of root development in A. thaliana (0.1 and 0.2 mM) and display an agravitropic root response at higher concentration (2 mM). This study suggests for these compounds a potential plant growth regulatory activity of the auxin type. Between the two ionic species, anionic from acid and cationic from HEA, the anionic species presented an increased phytotoxic effect. However, up to now, the plant growth regulatory properties of HEA-pABA and pABA have not been found in literature, which is needed in order to develop its new application fields and carry out relevant theoretical research.
Supporting Information
UV absorption spectra of HEA- p ABA and p ABA in 0.1 M NaOH solution.
(PDF)
FT-IR spectra of HEA- p ABA (black) and p ABA (red).
(PDF)
Optimal distance calculation for HEA- p ABA, p ABA and classical auxin molecules (IAA, 1-NAA, 2,4-D).
(PDF)
(A) Binding mode of IAA with TIR1. Hydrogen bonds are drawn as green lines. (B). Root Mean Square Deviation of the ligand with respect to the MD starting structure.
(PDF)
Binding mode of p ABA− with TIR1. Hydrogen bonds are drawn as green lines.
(PDF)
Root length (after 5 and 10 days) of treated A. thaliana seedlings. HEA-pABA, pABA and HEA treatments at different concentrations in comparison with control. Values with different letter annotation are significantly different (P<0.05). Data are means ± SE of 5 replicates.
(PDF)
Effects of HEA- p ABA, p ABA and HEA on root development of A. thaliana (Columbia 0) seedlings. The 10 day old A. thaliana seedlings were germinated on vertical agar plates. Scale bars, 0.5 cm.
(PDF)
Funding Statement
This work was partially supported by the Bilateral Project Academy of Sciences of Moldova - National Research Council of Italy 2013–2014 and Project 2.2 of the Institute of Chemistry Timisoara of Romanian Academy. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chang TY, Hu ML (1996) Concentrations and lipid peroxidation in tissues and toxicity of para-aminobenzoic acid fed to rats in drinking water. J Nutr Biochem 7: 408–413. [Google Scholar]
- 2. Basset GJC, Quinlivan EP, Ravanel S, Rebeille F, Nichols BP, et al. (2004) Folate synthesis in plants: The p-aminobenzoate branch is initiated by a bifunctional PabA-PabB protein that is targeted to plastids. Proc Natl Acad Sci USA 101: 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pierrel F, Hamelin O, Douki T, Kieffer-Jaquinod S, Muhlenhoff U, et al. (2010) Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast Coenzyme Q biosynthesis. Chem Biol 17: 449–459. [DOI] [PubMed] [Google Scholar]
- 4. Marbois B, Xie LX, Choi S, Hirano K, Hyman K, et al. (2010) para-Aminobenzoic acid is a precursor in Coenzyme Q6 biosynthesis in Saccharomyces cerevisiae . J Biol Chem 285: 27827–27838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kluczyk A, Popek T, Kiyota T, de Macedo P, Stefanowicz P, et al. (2002) Drug evolution: p-Aminobenzoic acid as a building block. Curr Med Chem 9: 1871–1892. [DOI] [PubMed] [Google Scholar]
- 6. Galbinur T, Obolensky A, Berenshtein E, Vinokur V, Chowers I, et al. (2009) Effect of para-aminobenzoic acid on the course of retinal degeneration in the rd10 mouse. J Ocul Pharmacol Ther 25: 475–482. [DOI] [PubMed] [Google Scholar]
- 7. Akberova SI (2002) New biological properties of p-aminobenzoic acid. Biol Bull (Izv Akad Nauk Ser Biol) 29: 390–393. [PubMed] [Google Scholar]
- 8. Richards RME, Xing DKL (1995) The effect of p-aminobenzoic acid on the uptake of thymidine and uracil by Escherichia-coli. Int J Pharm 116: 217–221. [Google Scholar]
- 9. Richards RMR, Xing DKL, King TP (1995) Activity of p-aminobenzoic acid compared with other organic acids against selected bacteria. J. Appl Bacteriol 78: 209–215. [DOI] [PubMed] [Google Scholar]
- 10. Vasilieva S (2001) Para-aminobenzoic acid inhibits a set of SOS functions in Escherichia coli K12. Mutat Res Genet Toxicol Environ Mutagen 496: 89–95. [DOI] [PubMed] [Google Scholar]
- 11. Stroeva OG, Akberova SI, Drozd NN, Makarov VA, Miftakhova NT, et al. (1999) The antithrombotic activity of para-aminobenzoic acid in experimental thrombosis. Izv Akad Nauk Ser Biol 3: 329–336. [PubMed] [Google Scholar]
- 12. Drozd NN, Makarov VA, Miftakhova NT, Kalugin SA, Stroeva OG, et al. (2000) Antithrombotic activity of para-aminobenzoic acid. Eksp Klin Farmakol 63: 40–44. [PubMed] [Google Scholar]
- 13. Andreenko GV, Karabasova MA, Liutova LA, Podorolskaia LV, Serebriakova TN, et al. (1996) Effect of para-aminobenzoic acid on the fibrinolytic activity of blood. Dokl Akad Nauk 346: 268–270. [PubMed] [Google Scholar]
- 14. Hu Z, Liu J, Shen L, Xu D, Xu Y (2002) Separation of 4-aminobenzoic acid by cocrystallization: Crystal structure of the complex of 4-aminobenzoic acid with (2R,3R)-tartaric acid. J Chem Crystallogr 32: 525–529. [Google Scholar]
- 15. Xavier S, Macdonald S, Roth J, Caunt M, Akalu A, et al. (2006) The vitamin-like dietary supplement para-aminobenzoic acid enhances the antitumor activity of ionizing radiation. Int J Radiat Oncol Biol Phys 65: 517–527. [DOI] [PubMed] [Google Scholar]
- 16. Hanson KM, Gratton E, Bardeen CJ (2006) Sunscreen enhancement of UV-induced reactive oxygen species in the skin. Free Radical Biol Med 41: 1205–1212. [DOI] [PubMed] [Google Scholar]
- 17. Mackie DS, Mackie LE (1999) The PABA story. Australas J Dermatol 40: 51–53. [DOI] [PubMed] [Google Scholar]
- 18. Kostic N, Bozanic M, Vukovic A (1992) Diagnostic value of the NBT-PABA test in the functional evaluation of the exocrine pancreas. Srp Arb Celok Lek 120: 269–272. [PubMed] [Google Scholar]
- 19. Lu Z, Kong X, Lu Z, Xiao M, Chen M, et al. (2014) para-Aminobenzoic acid (PABA) synthase enhances thermotolerance of mushroom Agaricus bisporus . Plos One 9: e91298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Song GC, Choi HK, Ryu CM (2013) The folate precursor para-aminobenzoic acid elicits induced resistance against Cucumber mosaic virus and Xanthomonas axonopodis . Ann Bot 111: 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bekusarova SA, Bome NA, Weisfeld LI, Tzomatova FT, Luschenko GV (2013) On the effect of PABA on germination. In: Zaikov GE, Goloshchapov AN, Lobanov AV, editors. Progress in organic and physical chemistry: Structures and mechanisms. Apple Academic Press. pp. 253–260.
- 22. Etter MC, Frankenbach GM (1989) Hydrogen-bond directed cocrystallization as a tool for designing acentric organic solids. Chem Mater 1: 10–12. [Google Scholar]
- 23. Chumakov YM, Simonov YA, Grozav M, Crisan M, Bocelli G, et al. (2006) Hydrogen-bonding network in the organic salts of 4-nitrobenzoic acid. Central Eur J Chem 4: 458–475. [Google Scholar]
- 24. Crisan M, Bourosh P, Chumakov Y, Petric M, Ilia G (2013) Supramolecular assembly and ab initio quantum chemical calculations of 2-hydroxyethylammonium salts of para-substituted benzoic acids. Cryst Growth Des 13: 143–154. [Google Scholar]
- 25. Gibellini F, Smith TK (2010) The Kennedy pathway—De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62: 414–428. [DOI] [PubMed] [Google Scholar]
- 26. Chicu SA, Grozav M, Kurunczi L, Crisan M (2008) SAR for amine salts of carboxylic acids to Hydractinia echinata . Rev Chim 59: 582–587. [Google Scholar]
- 27. Crisan M, Grozav M, Kurunczi L, Ilia G, Bertea C (2007) Inhibitory effects of some synthetic monoethanolaminesalts of para-substituted benzoic acids and corresponding benzoic acids on cucumber seed germination. J Plant Interact 2: 53–61. [Google Scholar]
- 28. Crisan M, Grozav M, Bertea C (2009) Arabidopsis thaliana seed germination and early seedling growth are inhibited by monoethanolamine salts of para-halogenated benzoic acids. J Plant Interact 4: 271–277. [Google Scholar]
- 29. Sheldrick GM (2008) A short history of SHELX . Acta Cryst A 64: 112–122. [DOI] [PubMed] [Google Scholar]
- 30. Spek AL (2003) Single-crystal structure validation with the program PLATON . J Appl Cryst 36: 7–13. [Google Scholar]
- 31. Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, et al. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. [DOI] [PubMed] [Google Scholar]
- 32. Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, et al. (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, et al. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19: 1639–1662. [Google Scholar]
- 34.Case DA, Darden TA, Cheatham III TE, Simmerling CL, Wang J, et al.. (2010) AMBER 11, University of California, San Francisco.
- 35. Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, et al. (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78: 1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25: 1157–1174. [DOI] [PubMed] [Google Scholar]
- 37. Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79: 926–935. [Google Scholar]
- 38. Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, et al. (1995) A smooth particle mesh Ewald method. J Chem Phys 103: 8577–8593. [Google Scholar]
- 39. Dapprich S, Komaromi I, Byun S, Morokuma K, Frisch MJ (1999) A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J Mol Struct (THEOCHEM) 461: 1–21. [Google Scholar]
- 40. Vreven T, Morokuma K, Frakas O, Schlegel HB, Frisch MJ (2003) Geometry optimization with QM/MM, ONIOM, and other combined methods. I. Microiterations and constraints. J Comput Chem 24: 760–769. [DOI] [PubMed] [Google Scholar]
- 41. Vreven T, Byun KS, Komaromi I, Dapprich S, Montgomery Jr JA, et al. (2006) Combining Quantum Mechanics Methods with Molecular Mechanics Methods in ONIOM. J Chem Theory Comput 2: 815–826. [DOI] [PubMed] [Google Scholar]
- 42.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, et al.. (2009) GAUSSIAN 09 (Revision D.01), Gaussian, Inc., Wallingford, CT.
- 43. Becke AD (1988) Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A 38: 3098–3100. [DOI] [PubMed] [Google Scholar]
- 44. Lee C, Yang W, Parr RG (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B 37: 785–789. [DOI] [PubMed] [Google Scholar]
- 45. Müller H, Eckert H (1989) Simultaneous determination of monoethanolamine and glycine betaine in plants. J Chromatogr 479: 452–458. [Google Scholar]
- 46. Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plantarum 15: 473–497. [Google Scholar]
- 47. Li XQ, Li SH, Chen DF, Meng FR (2004) Induced activity of superoxide dismutase and peroxidase of in vitro plants by low concentrations of ethanol. Plant Cell Tiss Org Cult 79: 83–86. [Google Scholar]
- 48. Hirayama T, Fujishige N, Kunii T, Nishimura N, Iuchi S, et al. (2004) A novel ethanol-hypersensitive mutant of Arabidopsis. Plant Cell Physiol 45: 703–711. [DOI] [PubMed] [Google Scholar]
- 49.Silverstein RM, Webster FX (1998) Spectrometric identification of organic compounds, 6th ed. New York: John Wiley & Sons.
- 50.Pavia DL, Lampman GM, Kriz GS, Vyvyan JR (2009) Introduction to spectroscopy. Brooks/Cole, Cengage Learning. p. 80
- 51.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, et al.. (2003) GAUSSIAN 03 (Revision B.01), Gaussian, Inc., Pittsburgh, PA.
- 52. Ward SP, Leyser O (2004) Shoot branching. Curr Opin Plant Biol 7: 73–78. [DOI] [PubMed] [Google Scholar]
- 53. Masucci JD (1994) Schiefelbein JW (1994) The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol 106: 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen R, Guan C, Boonsirichai K, Masson PH (2002) Complex physiological and molecular processes underlying root gravitropism. Plant Molec Biol 49: 305–317. [PubMed] [Google Scholar]
- 55. Calderon-Villalobos LI, Tan X, Zheng N, Estelle M (2010) Auxin perception-structural insights. Cold Spring Harb Perspect Biol 2: 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Porter WL, Thimann KV (1965) Molecular requirements for auxin action—I. Halogenated indoles and indoleacetic acid. Phytochemistry 4: 229–243. [Google Scholar]
- 57. Farrimond JA, Elliott MC, Clack DW (1978) Charge separation as a component of the structural requirements for hormone activity. Nature 274: 401–402. [Google Scholar]
- 58. Bruno IJ, Cole JC, Edgington PR, Kessler MK, Macrae CF, et al. (2002) New software for searching the Cambridge Structural Database and visualizing crystal structures. Acta Crystallogr B: Struct Sci 58: 389–397. [DOI] [PubMed] [Google Scholar]
- 59. Allen FH (2002) The Cambridge Structural Database: a quarter of a million crystal structures and rising. Acta Crystallogr B: Struct Sci 58: 380–388. [DOI] [PubMed] [Google Scholar]
- 60.Hyperchem 6, Hypercube Inc., Gainesville, FL, USA, 2001.
- 61. Peishoff CE, Dixon JS (1992) Improvements to the distance geometry algorithm for conformational sampling of cyclic structures. J. Comput Chem 13: 565–569. [Google Scholar]
- 62. Gracin S, Fischer A (2005) Redetermination of the β-polymorph of p-aminobenzoic acid. Acta Crystallogr E: Struct Rep Online 61: 1242–1244. [Google Scholar]
- 63.Sieron L, Kobylecka J, Turek A (2011) Crystal packing and supramolecular motifs in four phenoxyalkanoic acid herbicides—low-temperature redeterminations. Org Chem Int, ID 608165, 1–5.
- 64. Yang X, Xie H, Chen J, Li X (2013) Anionic phenolic compounds bind stronger with transthyretin than their neutral forms: Nonnegligible mechanisms in virtual screening of endocrine disrupting chemicals. Chem Res Toxicol 26: 1340–1347. [DOI] [PubMed] [Google Scholar]
- 65. Wang W (1991) Literature review on higher plants for toxicity testing. Water Air Soil Poll 59: 381–400. [Google Scholar]
- 66. Hulzebos EM, Ademn DMM, Van Breemen D, Henzen L, Van Gestel CAM (1991) QSARs in phytotoxicity. Sci Total Environ 109/110: 493–497. [Google Scholar]
- 67. Wang XD, Dong YY, Wang LS, Han SK (2001) Acute toxicity of substituted phenols to Rana Japonica tadpoles and mechanism-based quantitative structure-activity relationship (QSAR) study. Chemosphere 44: 447–455. [DOI] [PubMed] [Google Scholar]
- 68. Woodward AW, Bartel B (2005) Auxin: Regulation, action, and interaction. Ann Bot 95: 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ludwig-Müller J, Vertocnik A, Town CD (2005) Analysis of indole-3-butyric acid induced adventitious root formation on Arabidopsis stem segments. J Exp Bot 56: 2095–2105. [DOI] [PubMed] [Google Scholar]
- 70. Pitts RJ, Cernac A, Estelle M (1998) Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J 16: 553–560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
UV absorption spectra of HEA- p ABA and p ABA in 0.1 M NaOH solution.
(PDF)
FT-IR spectra of HEA- p ABA (black) and p ABA (red).
(PDF)
Optimal distance calculation for HEA- p ABA, p ABA and classical auxin molecules (IAA, 1-NAA, 2,4-D).
(PDF)
(A) Binding mode of IAA with TIR1. Hydrogen bonds are drawn as green lines. (B). Root Mean Square Deviation of the ligand with respect to the MD starting structure.
(PDF)
Binding mode of p ABA− with TIR1. Hydrogen bonds are drawn as green lines.
(PDF)
Root length (after 5 and 10 days) of treated A. thaliana seedlings. HEA-pABA, pABA and HEA treatments at different concentrations in comparison with control. Values with different letter annotation are significantly different (P<0.05). Data are means ± SE of 5 replicates.
(PDF)
Effects of HEA- p ABA, p ABA and HEA on root development of A. thaliana (Columbia 0) seedlings. The 10 day old A. thaliana seedlings were germinated on vertical agar plates. Scale bars, 0.5 cm.
(PDF)