Abstract
HIV replication and the cellular miRNA machinery interconnect at several post-transcriptional levels. To understand their regulatory role in the intestine, a major site of HIV/SIV replication, dissemination and CD4+ T cell depletion we profiled miRNA expression in colon following SIV infection (10-Acute-SIV, 5-uninfected). Nine (4-up and 5-down) miRNAs showed statistically significant differential expression. Most notably, miR-190b expression showed high statistical significance (Adjusted p=0.0032), the greatest fold change and was markedly elevated in colon and jejunum throughout SIV infection. Additionally, miR-190b upregulation was detected before peak viral replication and the nadir of CD4+ T cell depletion predominantly in lamina propria leukocytes. Interestingly non-SIV-infected macaques with diarrhea and colitis failed to upregulate miR-190b suggesting that its upregulation was neither inflammation nor immune-activation driven. SIV infection of in vitro cultured CD4+ T cells and primary intestinal macrophages conclusively identified miR-190b upregulation to be driven in response to viral replication. Further miR-190b expression levels in colon and jejunum positively correlated with tissue viral loads. In contrast, mRNA expression of MTMR6, a negative regulator of CD4+ T cell activation/proliferation significantly decreased in SIV-infected macrophages. Luciferase reporter assays confirmed MTMR6 as a direct miR-190b target. This first report describing dysregulated miRNA expression in the intestine identifies a potentially significant role for miR-190b in HIV/SIV pathogenesis. More importantly, miR-190b mediated MTMR6 downregulation suggests an important mechanism that could keep infected cells in an activated state thereby promoting viral replication. In future, the mechanisms driving miR-190b upregulation including other cellular processes it regulates in SIV-infected cells needs determination.
INTRODUCTION
Human/Simian immunodeficiency virus (HIV/SIV) infection of the gastrointestinal (GI) immune system results in massive virus induced depletion of CD4+/CCR5+ memory T cells, uncontrolled inflammation and immune activation that are proximate causes of structural and functional damage to the GI tract (1-6). Most of the current knowledge pertaining to the early molecular immunopathological events in the GI tract have been generated predominantly by transcriptome profiling studies (7-8) including our most recent studies done separately on the lamina propria leukocyte (LPL) (9) and epithelial cellular compartments (10) of the intestine in SIV-infected macaques. These studies have revealed significant alterations in the expression of genes associated with early immune response/inflammation, immune activation, anti-viral signaling and T cell, B cell and macrophage dysfunction in LPLs (7-9). Similarly, genes encoding cell cycle regulators, Wnt-TCF7L2 signaling, Notch signaling, genes associated with the formation of adherens junction, hemidesmosomes, desmosomes, etc. were found to be significantly dysregulated in the intestinal epithelium (10) at 21 and 90 days post SIV infection.
Immune/inflammatory responses against viruses are orchestrated for the most part by signaling pathways activated by interferons and numerous other pro-inflammatory cytokines that stimulate transcription factors including but not limited to, interferon regulatory factors (IRFs), signal transducers and activators of transcription (STATs), NFκB and CAAAT Enhancer binding proteins (C/EBPs) (11-12). In this context, we previously demonstrated that GI disease in HIV/SIV infection is characterized by constitutive activation of proinflammatory transcription factors such as STAT3 and C/EBPβ that can generate a self-perpetuating cycle involving intestinal epithelial damage, inflammation, immune activation and viral replication (13-14). Apart from inducing inflammatory gene expression, proinflammatory transcription factors such as STAT3 (15), C/EBP (16) and NFκB (17) also activate the transcription of another novel class of regulatory non-protein coding RNA called micro ribonucleic acids (miRNAs) and these small RNA molecules have been shown to be potent regulators of early immune and inflammatory responses (18) including HIV pathogenesis (19).
MiRNAs are small (~21-23 nucleotides) regulatory noncoding RNAs that are highly conserved and have been shown to suppress the expression of protein coding genes by targeting mRNAs for translational repression or degradation (20-21). They are initially transcribed by RNA polymerase II as long variable length primary miRNAs which are then processed in the nucleus by the microprocessor complex (Drosha and DGCR8) to precursor miRNAs and subsequently in the cytoplasm by Dicer to generate the mature miRNAs (21). Recent studies have attributed more significant roles for these small RNA molecules in regulating the immune response that includes immune cell development, proliferation, activation and differentiation (18,22-26). Further, several in vitro studies clearly show that HIV and the host miRNA machinery crisscross at several post transcriptional levels (27-31). The restraints exerted by the host miRNA machinery on HIV replication is clearly evident from the finding that viral replication kinetics are enhanced in peripheral blood mononuclear cells following siRNA mediated knockdown of Drosha and Dicer (32). In addition, several recent miRNA profiling studies performed on purified CD4+ T cells, macrophages and peripheral blood mononuclear cells of HIV infected patients provide compelling evidence of their dysregulation in HIV infection (33-36). Recent studies have also described dysregulated miRNA expression in the brain (37) and plasma of SIV-infected rhesus macaques (38) including miRNAs that directly targeted SIV replication in macrophages (39). Accordingly, we hypothesized that SIV replication in the GI tract in association with the ensuing immune response induces the expression of miRNAs that have the potential to regulate viral replication including host genes expressed during the immune response. Given the lack of information on the expression of miRNAs in the GI tract in response to SIV infection, we performed global miRNA expression profiling in the intestine of acutely SIV-infected macaques with a focus on the colon. Expression of nine miRNAs was significantly altered in the colon following acute SIV infection. MiR-190b, in particular, was significantly upregulated in both colon and jejunum through the course of SIV infection. Further, miR-190b upregulation was detected as early as 7 days post SIV infection even before peak viral replication and the nadir of CD4+ T cell depletion in both colon and jejunum. Furthermore, using multiple experimental approaches, we show that miR-190b is upregulated specifically in the intestinal lamina propria in response to SIV infection and targets the expression of MTMR6 (myotubularin related protein 6); a negative regulator of CD4+ T cell activation/proliferation. Thus miR-190b could potentially play a significant role in the pathogenesis of SIV infection in the intestinal mucosa.
MATERIALS AND METHODS
Animal Ethics statement
All experiments using rhesus macaques were approved by the Tulane Institutional Animal Care and Use Committee (Protocol No-3574). The Tulane National Primate Research Center (TNPRC) is an Association for Assessment and Accreditation of Laboratory Animal Care International accredited facility (AAALAC #000594). The NIH Office of Laboratory Animal Welfare assurance number for the TNPRC is A3071-01. All clinical procedures, including administration of anesthesia and analgesics, were carried out under the direction of a laboratory animal veterinarian. Animals were anesthetized with ketamine hydrochloride for blood collection procedures. Intestinal resections were performed by laboratory animal veterinarians. Animals were pre-anesthetized with ketamine hydrochloride, acepromazine, and glycopyrolate, intubated and maintained on a mixture of isoflurane and oxygen. Buprenorphine was given intraoperatively and post-operatively for analgesia. All possible measures are taken to minimize discomfort of all the animals used in this study. Tulane University complies with NIH policy on animal welfare, the Animal Welfare Act, and all other applicable federal, state and local laws.
Animals and Tissue Collection
Colon and jejunum tissues were collected from forty nine Indian-origin rhesus macaques including thirty three animals infected with pathogenic strains of SIV that use CCR5 in vivo and sixteen animals not infected with SIV. In the SIV-infected group all animals were inoculated intravenously with 100TCID50 of SIV with the exception of 5 (CT16, CG32, AT56, H405 and L441) that were inoculated intravaginally. For intravaginal inoculation a viral inoculum dose of 1000TCID50 was used. Animal IDs, duration of infection, plasma and intestinal viral loads in all SIV-infected animals are provided in Table I. Ten acutely SIV-infected animals (Table I) and five uninfected control animals (Table II) were used for the initial TLDA miRNA profiling. An additional twenty-three SIV-infected macaques at various stages of infection (Table I) were used exclusively for characterization of miR-190b expression through the course of SIV infection. Among the sixteen uninfected animals we included eight that were necropsied for chronic diarrhea unresponsive to treatment (Non-SIV infected with diarrhea and colitis) (40-41). These animals were included to assess whether miR-190b upregulation was in response to viral replication or was simply a byproduct of enterocolitis that generally accompanies HIV/SIV infection.
Table I Animal IDs, duration of infection, SIV inoculum and plasma and intestinal viral loads in SIV-infected macaques.
| Animal ID | Duration of infection (days post infection) | Inoculum | Plasma viral loads Copies/mL (106) | Viral load- Colon Copies/mg RNA (106) | Viral load- Jejunum Copies/mg RNA (106) |
|---|---|---|---|---|---|
| IA85* | 7 | SIVmac251 | NA | 0.1 | 0.4 |
| HI52* | 8 | SIVmac251 | 4 | 900 | 200 |
| AV91* | 10 | SIVmac251 | 157 | 50 | 100 |
| M992* | 13 | SIVmac251 | 35 | 200 | 9 |
| HI63* | 13 | SIVmac251 | 24 | 20 | 10 |
| HI58* | 13 | SIVmac251 | 9 | 20 | 50 |
| HC36* | 21 | SIVmac251 | 20 | 1 | 2 |
| HB31* | 21 | SIVmac251 | 10 | 2 | 1 |
| GA19* | 21 | SIVmac251 | 2 | 10 | 0.5 |
| CT16* | 29 | SIVmac239 | NA | 4 | 40 |
| HT44 | 7 | SIVmac251 | NA | 0.4 | 3 |
| T108 | 8 | SIVmac251 | 0.06 | ND | ND |
| HV61 | 14 | SIVmac251 | NA | 0.8 | 3 |
| HV39 | 14 | SIVmac251 | NA | 0.2 | 0.9 |
| HF27 | 21 | SIVmac251 | 9 | 0.06 | 5 |
| HB48 | 21 | SIVmac251 | 40 | 300 | 6 |
| GK31 | 21 | SIVmac251 | 2 | 2 | 0.04 |
| HR57 | 21 | SIVmac251 | 6 | 0.4 | 1 |
| HV95 | 21 | SIVmac251 | 2 | 0.2 | 0.5 |
| CG32 | 29 | SIVmac239 | NA | ND | ND |
| HC36 | 90 | SIVmac251 | 0.1 | 0.2 | ND |
| HB31 | 90 | SIVmac251 | 70 | 7 | 0.97 |
| HF27 | 90 | SIVmac251 | 10 | 40 | 2 |
| HB48 | 90 | SIVmac251 | 20 | 0.1 | 100 |
| GK31 | 90 | SIVmac251 | 30 | 1 | 0.05 |
| GA19 | 90 | SIVmac251 | 100 | 5 | 4 |
| HR57 | 90 | SIVmac251 | 80 | 2 | 0.04 |
| HV95 | 90 | SIVmac251 | 7 | 0.5 | 0.08 |
| FT11 | 145 | SIVmac251 | 500 | 2075 | 1176 |
| HL01 | 180 | SIVmac251 | 7 | 0.8 | 30 |
| L441 | 170 | SIVmac239 | 1.3 | 3 | 21 |
| H405 | 232 | SIVmac239 | 71.4 | 542 | 758 |
| AT56 | 1460 | SIVmac239 | 360 | 10228 | 1482 |
Colon tissue from 10 animals (IA85-CT16) was used for genome wide miRNA expression profiling using the TLDA platform and are shown above the bar in the table.
Colon and jejunum tissues from all 33 animals were used for miR-190b qRT-PCR characterization studies. NA- not available ND- not detectable
Table II.
List of non-SIV-infected macaques with diarrhea and uninfected control macaques used for TLDA and miR-190b qRT-PCR studies.
| Animal ID | Intestinal Histopathology | Bacterial pathogens | |
|---|---|---|---|
| SIV-uninfected with Diarrhea | Colon | Jejunum | |
| DJ15 | 3 and CD | 1 | None |
| DV87 | 1 and CD | NA | None |
| DV98 | 3 and CD | NA | None |
| EB12 | 3 and CD | NA | None |
| J053 | 2 | 1 | None |
| EJ54 | 3 | 1 | None |
| EL71 | 3 and CD, DV | 0 | None |
| EB27 | 3 and CD, DV | 1 | S.flexneri, C.coli |
| Uninfected Controls | |||
| FF15 | 0 | 0 | None |
| FT23 | 0 | 0 | None |
| HT22 | 0 | 0 | None |
| EL66* | 0 | 0 | None |
| EH70* | 0 | 0 | None |
| EH80* | 0 | 0 | None |
| GI92* | 0 | 0 | None |
| FK25* | 0 | 0 | None |
Sections of jejunum and colon were examined and inflammation was scored semiquantitatively on a scale of 0 to 3 as follows: 0, within normal limits; 1, mild; 2, moderate; 3 severe. In addition, the presence of crypt dilatation (CD), villus blunting (VB), diverticulosis (DV), and amyloidosis (AMD) were recorded NA, not applicable.
Denotes uninfected control animals used for TLDA miRNA profiling
Following euthanasia with an intravenous overdose of pentobarbital all animals received a complete necropsy. Tissue samples (colon and jejunum) from all control, SIV-infected and non-SIV-infected macaques with diarrhea and colitis were collected in RNAlater® (Life Technologies, Grand Island, NY) over several years (~6-7 years) and stored at −20° C for total RNA extraction. In addition, serial resection biopsies (~6-8 cm long) of jejunum including colonic wedge biopsies (2 cm) were collected from eight Indian-origin rhesus macaques (HC36, HB31, HF27, HB48, GK31, GA19, HR57 and HV95) at 6 weeks before SIV infection and 21 and 90 days after SIV infection for TLDA and miR-190b RT-PCR confirmation studies. For histopathological evaluation, colon and jejunum tissues were collected immediately after euthanasia and fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned at 6 μM and stained with hematoxylin and eosin for analysis. Sections were examined in a blinded fashion and inflammation was scored semiquantitatively on a scale of 0 to 3 as follows: 0, within normal limits; 1, mild; 2, moderate; 3, severe. In addition, the presence of crypt dilatation, villus blunting, diverticulosis and amyloidosis were recorded (Table II).
Global microRNA profiling using TaqMan Low Density Arrays
Total RNA was extracted from intact colon and jejunum samples using the miRNeasy total RNA isolation kit (Qiagen Inc, CA). RNA integrity was assessed by running an aliquot on a denaturing agarose gel followed by staining with ethidium bromide to visualize intact 28S and 18S rRNA bands. For TLDA miRNA profiling, ~350 ng of total RNA from intact colon tissue was first reverse transcribed following the ABI microRNA TLDA reverse transcription reaction protocol. Briefly, two master mixes were prepared for each RNA sample representing either TLDA panel (panel A and panel B) and consisted of the following reaction components: 0.80 μL MegaPlex RT primers (10×), 0.20 μL dNTPs with dTTP (100 mM), 1.50 μl MultiScribe™ Reverse Transcriptase (50 U/μL), 0.80 μL 10× RT Buffer, 0.90 μl MgCl2 (25 mM), 0.10 μl RNase Inhibitor, 0.20 μl nuclease-free water (20 U/μL). Three microliters of total RNA (350 ng) were loaded into appropriate wells of a 96-well plate containing 4.5 μl of the RT reaction mix and following a brief 5 min incubation on ice was subjected to the following thermal cycling conditions on the ABI 7900 HT Fast PCR system: standard or max ramp speed, 16 °C for 2 min, 42 °C for 1 min, 50 °C for 1 sec (40 cycles) and 85 °C for 5 min (hold).
Approximately 2.5 μl of the resulting cDNA from each sample was mixed with a total of 22.5 μl of pre-amplification reaction mix consisting of 12.5 μl TaqMan® PreAmp Master Mix (2×); 2.5 μl Megaplex™ PreAmp Primers (10×); 7.5 μl nuclease-free water and preamplified on the ABI 7900 HT Fast PCR system according to the TLDA miRNA preamplification protocol outlined by the manufacturer (Life Technologies). The preamplification thermal cycling conditions were as follows: hold 95 °C for 10 min; hold 55 °C for 2 min; hold 72 °C for 2 min; 12 cycles at 95 °C for 15 sec and 60 °C for 4 min.
The preamplified product was first diluted 4-fold with 75 μl of 0.1× TE pH 8.0 mixed, following which 9 μl of the diluted PreAmp product were mixed with 450 μl of 2× TaqMan® Universal PCR Master Mix with no UNG (AmpErase®) and 441 μl of nuclease-free water to bring the final volume to 1 mL. After proper mixing and centrifuging, 100 μls of the PCR reaction mix were loaded into each port of the TaqMan® Array Human MicroRNA A+B Card Set v3.0. The TLDA cards were then centrifuged, sealed and processed on the ABI 7900 HT Sequence Detection System using the 384-well TaqMan Low Density Array default thermal-cycling conditions.
Quantitative Real-Time TaqMan Stem loop microRNA and SYBR Green RT-PCR
Expression of miR-190b was further investigated in both colon and jejunum through the course of SIV infection using the TaqMan microRNA predesigned and preoptimized assays (Life Technologies). Total RNA [500 ng for miR-190b, RNU48 and 100 ng for snoU6 (colon and jejunum), 250 ng for CD4+ T cells and primary intestinal macrophages] was reverse transcribed using the stem loop primers provided in the predesigned kit in a total volume of 15 μL. Similar total RNA concentrations (500-750 ng) have been previously used for miRNA qRT-PCR (42-43). ~4 μL of cDNA was subjected to 40 cycles of PCR in a total volume of 20 μL on the ABI 7900 HT Fast PCR System (Life Technologies) using the following thermal cycling conditions: 95 °C for 10 minutes followed by 40 repetitive cycles of 95 °C for 15 sec and 60 °C for 1 minute. As a normalization control for RNA loading, parallel reactions in duplicate wells to amplify RNU48 or snoU6 or RNU44 in combination with RNU48 (CD4+ T cells) were run in the same or different multi-well plate. While RNU48 worked well for intact intestine we found snoU6 to be better when analyzing miRNA expression in distinct intestinal mucosal compartments such as epithelium and LPLs. RNU44 was used for CD4+ T cells as it has been previously used for normalization of miRNA expression in HIV-infected CD4+ T cells (44). Comparative real-time PCR was performed in duplicate wells including no template controls and relative change in gene expression was calculated using the comparative (ΔΔCT) method.
Expression of MTMR6 in in vitro cultured SIV-infected primary intestinal macrophages was evaluated by Quantitative Real-Time SYBR Green Two-Step RT-PCR assay (Life Technologies). ~1μg of total RNA was first reverse transcribed in a total volume of 50 μL using the SuperScript™ III First-Strand Synthesis System for RT-PCR kit (Life Technologies) following the manufacturer's protocol. Each qRT-PCR reaction (20 μL) contained the following: 2X Power SYBR Green Master Mix without uracil-N-glycosylase (12.5 μL), target forward and reverse primer (200 nM) and cDNA (4 μL). Forward and reverse primer sequence for MTMR6 and GAPDH (Glyceraldehyde 3-phosphate dehydrogenase) is shown in Table III. The PCR amplification was carried out in the ABI 7900 HT Fast PCR System (Life Technologies) using the default thermal cycling conditions for SYBR Green assays. As a normalization control for RNA loading, parallel reactions in the same multiwell plate were performed using GAPDH. Relative changes in gene expression were calculated using the ΔΔCT method. PCR efficiency analysis was performed using serial 10 fold RNA dilutions (500, 50, 5 and 0.5 ng of total RNA) for miR-190b, RNU48, snoU6, or cDNA dilutions (40, 4, 0.4 and 0.04 ng) for MTMR6 and GAPDH. The amplification curves for all assays were linear and based on slope values (−3.09 to −3.24) all assays had 100-105% efficiency.
Table III.
Primer sequences used for real time SYBR Green Two-step qRT-PCR.
| Gene Name | Primer sequence | Product size (bp) | Primer concentration |
|---|---|---|---|
| MTMR6 | For-5'- CAGCAGCCTGGCAGATAATCGTT-3' | 76 | 200 nM |
| Rev -5'- TAAGCTGACCACAGCAGGTTCTGA -3' | |||
| GAPDH | For-5'- CAACAGCCTCAAGATCGTCAGCAA-3' | 100 | 200 nM |
| Rev-5'- GAGTCCTTCCACGATACCAAAGTTGTC-3' |
In situ hybridization and immunofluorescence for cellular localization of SIV and in vitro characterization of primary intestinal macrophages
In situ hybridization for detecting SIV RNA was performed using SIV-digoxigenin-labeled antisense riboprobes (Lofstrand Laboratories, Gaithersburg, MD). Briefly, 7 μM thick formalin-fixed, paraffin-embedded tissue sections were first deparafinized, rehydrated in decreasing concentrations of ethanol and pretreated in a microwave with citrate buffer (antigen unmasking solution; Vector Laboratories, Burlingame, CA) for 20 minutes at high power according to the manufacturer's instructions. Thereafter, sections were thoroughly washed, placed in a humidified chamber, and prehybridized at 45 °C with in situ hybridization buffer containing 50% formamide with denatured herring sperm DNA and yeast tRNA at 10 mg/ml each. SIV-digoxigenin-labeled antisense riboprobes (Lofstrand Laboratories, Gaithersburg, MD) were used at a concentration of 10 ng/slide in hybridization buffer and hybridized overnight at 45 °C. After hybridization slides were washed with 4× SSC (standard saline citrate buffer), 1× SSC, 0.1× SSC, and blocked with Dako protein free blocker (Dako North America, Inc., Carpinteria, CA) for one hour. Fab fragments of an anti-digoxigenin antibody conjugated with alkaline phosphatase (Roche Diagnostics Corporation, Penzberg, Germany) were used to detect digoxigenin-labeled probes. Positive signals were detected using permanent red according to the manufacturer's (Dako) instructions.
The protocol for detection of SIV RNA in SIV-infected in vitro cultured primary intestinal macrophages remained the same as described above. Immunofluorescence for phenotyping SIV positive cells in tissue and in vitro cultured primary intestinal macrophages was done exactly as described earlier (13-14) using unconjugated or Alexa Fluor® 647 conjugated CD68 (KP1) (Dako and Santa Cruz Biotechnology, CA), CD163 (Serotec, Raleigh, NC) and CD3 (Dako) primary and appropriate Alexa fluor conjugated secondary antibodies (Life Technologies).
Cell isolation from intestinal resection segments, peripheral blood and in vitro SIV infection of CD4+ T cells and primary intestinal macrophages
In order to determine the mucosal compartment contributing to miR-190b upregulation we separated the intestinal epithelial cells from the underlying LPLs and fibrovascular stroma as previously described (9-10,45). Finally, the intra epithelial leukocytes (IELs) were separated from the epithelial cells and changes in gene expression were analyzed in the epithelial cells and LPLs separately. In order to successfully separate all 4 tissue compartments and ensure the availability of sufficient starting material we obtained intestinal resection segments (6-8 cm long) from the jejunum at 21 and 90 days after SIV infection. Comparisons in gene expression were made to resection segments collected from the same animal 6 weeks prior to SIV infection. The LPLs consist predominantly of lymphocytes (50-60%) but also contain small numbers of macrophages, neutrophils, plasma and dendritic cells (supplemental figure 1). The intestinal cell isolation protocol established at the TNPRC (45) yields epithelial cells with ~85% purity with minimal contamination with IELs. Similar purity has also been reported for LPLs with minimal contamination with epithelial cells (45).
In order to further determine if miR-190b upregulation was linked to SIV replication we isolated peripheral blood CD4+ T cells and primary intestinal macrophages for in vitro SIV infection. CD4+ T cells from peripheral blood were isolated using the non-human primate specific CD4+ T cell isolation kit (magnetic separation with an LS column) following the manufacturer's recommended protocol (Miltenyi Biotec, Auburn, CA). ~107 cells were first activated with 1ug/ml concanavalin A and cultured for 3 days in 10 ml of RPMI-1640 containing 10% fetal bovine serum, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Following activation, cells were infected with 300 TCID50 of SIVmac239 in the presence of 2 ng/mL of IL-2 or left uninfected as controls. Syncytia formation was detected 48 h post infection at which time cells were pelleted and lysed for total RNA extraction.
For primary intestinal macrophage isolation, LPLs harvested in the previous step were transferred to T75 flasks containing RPMI1640 supplemented with 10% FBS, 100 mM HEPES, L-glutamine, 10,000 units of penicillin/streptomycin. Under these culture conditions, lymphocytes will not attach and become apoptotic by day 3 to 5 in culture. However, inflammatory macrophages or monocytes that have recently extravasated into the tissues and have not fully differentiated into macrophages will attach and proliferate forming a monolayer after ~7-10 days in culture (46-47). In vitro cultured primary jejunal macrophages isolated from SIV-infected (n=5) and uninfected (n=6) macaques were pelleted, activated with 20 ng/mL of recombinant rhesus macaque granulocyte macrophage colony stimulating factor (Cell Sciences, Canton, MA) and infected with 100 TCID50 of SIVmac251 in a total volume of 200 μL for about 2.5 hrs at 37 °C. Cells were then plated in T25 flasks containing RPMI1640 supplemented with 10% FBS, 100 mM HEPES, L-glutamine, 10,000 units of penicillin/streptomycin and cultured for 4 days post SIV infection.
Flow cytometry to quantify intestinal CD4+ T cell dynamics
LPLs were isolated as described above and adjusted to a concentration of 107/ml. For T cell immunophenotyping ~100 μL aliquots were stained with appropriately diluted, directly-conjugated monoclonal antibodies to CD3 (Pacific blue: SP34-2), CD4 (SK3: PerCP-Cy5.5) and CD8 (PE-TR: 3B5) (BD Biosciences, San Jose, CA). Samples were stained for 30 min in the dark at 4 °C, fixed in 2% paraformaldehyde, and stored in the dark at 4 °C overnight for acquisition the next day. Samples were acquired on a LSR II flow cytometry equipment (BD Biosciences) and analyzed with Flow Jo software (Treestar Inc, Ashland, OR). The cells were first gated on singlets followed by lymphocytes, CD3+ T-cells and then on CD3+CD4+ T-cell subsets.
Cloning of 3’-UTR of MTMR6 mRNA and Dual-Glo luciferase reporter gene assay
The 3’ UTR of the rhesus MTMR6 gene contains a single predicted miR-190b binding site (TargetScan 6.2) (48) highly conserved across several species (Table IV). Accordingly, a short 42-44 nucleotide long sequence representing the 3' UTR containing the predicted miR-190b site (5’-TCTGTTTATTAAAGTACATATCT-3’) was synthesized (IDTDNA Technologies Inc., IA) for cloning into the pmirGLO dual luciferase vector (Promega Corp, Madison, WI). A second oligonucleotide with the binding site mutated (5’-TCTGTTTATTAAAGTAGTATAA-3’) was also synthesized to serve as a negative control. Both oligonucleotide sequences were synthesized with a Pme1 site on the 5’ and Xba1 site on the 3’ end for directional cloning. The pmirGLO vector was first cut with Pme1 and Xba1 restriction enzymes, gel purified and ligated with either wild type (WT) sequence containing the miR-190b binding site (pmirGLO-WT-MTMR6) or mutated (MUT) sequence (pmirGLO-MUT-MTMR6). HEK293 cells were plated at a density of 5×104 cells per well of a 96 well plate. At 50% confluence cells were co-transfected with ~100 ng pmirGLO-WT-MTMR6 or pmirGLO-MUT-MTMR6 UTR miRNA luciferase reporter vector and 100 nM of miR-190b mimic using the Dharmafect Duo transfection reagent (ThermoFisher Scientific). In separate wells, cells were also transfected with pmirGLO vector (Promega Corp) as a normalization control. After 48 h, the Dual Glo luciferase assay was performed according to the manufacturer's recommended protocol using the BioTek H4 Synergy plate reader (BioTek, Winooski, VT). The normalized Firefly to Renila ratio was calculated to determine the relative reporter activity. Experiments were performed in 6 replicates and repeated thrice.
Table IV.
Schematic representation of MTMR6 3' UTR depicting predicted binding site for miR-190b. Alignment of MTMR6 mRNA sequence with miR-190b: top strand- MTMR6 mRNA; bottom strand-miR-190b.
Quantitation of Plasma and Mucosal Viral Loads
Total RNA samples from plasma, colon and jejunum tissues of all SIV-infected animals were subjected to quantitative real-time TaqMan two-step RT-PCR analyses to determine viral loads. Briefly, primers and probes specific to the SIV LTR sequence were designed and used in the real-time TaqMan PCR assay. Probes were conjugated with a fluorescent reporter dye (FAM) at the 5′ end and a non-fluorescent quencher dye at the 3′ end. Fluorescence signal was detected with an ABI Prism 7900 HT sequence detector (Life Technologies). Data were captured and analyzed with Sequence Detector Software (Life Technologies). Viral copy number was determined by plotting CT values against a standard curve (y= −3.246x + 39.374) (r2=0.998) generated with in vitro transcribed RNA representing known viral copy numbers.
Data Analysis and availability
TLDA-SDS run files from ten SIV-infected and five uninfected control macaques were loaded onto Applied Biosystems Relative Quantification (RQ) Manager Software v1.2.2 and analyzed using automatic baseline settings and a manual threshold of 0.2. The results from the RQ manager analysis containing five columns (well, sample, detector, task and CT values) were saved as a tab-delimited text file which was later imported and analyzed using the DataAssist™ version 3.01 software (Life Technologies), a data analysis tool designed to compare samples using the ΔΔCT method for relative quantification of gene expression. MiRNA expression data was analyzed using global normalization (49-50) as this method has been reported to be the most sensitive and accurate approach for high throughput miRNA profiling using qRT-PCR compared to endogenous controls. In all experiments, the CT upper limit was set to 33 meaning that all miRNA detectors with a CT value greater than or equal to 33 were excluded. Multiple comparisons correction using Benjamini-Hochberg method for false discovery rate was simultaneously applied to all 768 miRNA target probes (card A and B combined). TLDA data were deposited with GEO (Accession number- GSE56624, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56624).
For miR-190b and MTMR6 qRT-PCR studies, one uninfected control macaque with the highest (for miR-190b) or lowest (for MTMR6) ΔCT value served as the calibrator/reference and assigned a value of 1. All differentially expressed miRNAs or mRNAs in SIV-infected and uninfected macaques with diarrhea including other macaques in the control group are shown as an n-fold difference relative to this macaque. The step wise calculation of fold change using this approach is shown in supplemental table 1. miRNA fold change was also calculated using an average of all control animal ΔCT values providing very similar results (supplemental figure 2A&B). Accordingly, we have used the former approach as it facilitated graphing the control samples so that the variation within the control samples can be displayed (supplemental figure 2A). Intestinal CD4+ T cell data and individual miR-190b qRT-PCR data in colon, jejunum, intestinal epithelium and LPL compartments were analyzed using non-parametric Kruskal-Wallis test and post-hoc analysis was done using Dunn's multiple groups comparison employing the GraphPad Prism 5 software (La Jolla, CA). p values<0.05 were considered as significant. miR-190b and MTMR6 mRNA qRT-PCR data in SIV-infected macrophages was analyzed by non-parametric Wilcoxon's rank sum test for independent samples using RealTime STATMINER™ package, a bioinformatics software developed by Integromics on Spotfire DecisionSite. A Spearman's non-parametric one-tailed correlation analysis was performed to determine the degree of association between tissue viral loads and miR-190b fold expression. Firefly/Renila ratios were statistically analyzed using an unpaired “t” test.
RESULTS
Mucosal and plasma Viral Loads, CD4+ T Cell dynamics and Intestinal histopathology
The viral loads in plasma, colon and jejunum of all SIV-infected macaques are shown in Table I. Intestinal viral loads were substantial and in the colon ranged from 0.06 × 106 to 10,228 × 106 copies/ mg total RNA with a median of 2 × 106 copies/ mg total RNA. In the jejunum, viral loads ranged from 0.04 × 106 to 1482 × 106 copies/ mg total RNA with a median of 3.5 × 106 copies/ mg total RNA. Similar to tissue viral loads, plasma viral loads from 7DPI to terminal disease ranged from 0.006 × 106 to 500 × 106 copies/mL with a median of 10 × 106 copies/mL. Plasma viral loads were not available for 6 animals (IA85, HT44, HV39, HV61, CG32, CT16).
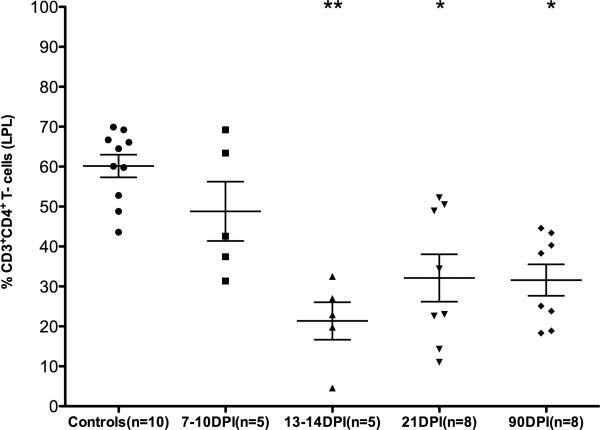
SIV-infected animals at the 13-14DPI, 21DPI and 90DPI time points (Table I) had significant mucosal CD4+ T cell depletion (Fig. 1). Analysis of CD4+ T cell percentages using Kruskal-Wallis (non-parametric method) test revealed statistically significant differences among groups (p=0.0005). Post hoc analysis using Dunn's multiple comparison test identified all post infection time points with the exception of 7-10DPI to be significantly different (p<0.05 to p<0.01) from the uninfected control group (Fig. 1). Data on CD4+ T cell status in the intestine were not available for 7 animals. This included two animals at 29DPI (CG32, CT16) and five macaques that progressed to AIDS (FT11, HL01, AT56, H405, L441) (Table I).
FIGURE 1.
Percentage of CD4+ T cells among lamina propria leukocytes (LPLs) isolated at different time points after SIV infection. The cells were first gated on singlets followed by lymphocytes, CD3 and then on CD3+CD4+ T cell subsets. Data analysis using Kruskal-Wallis test identified significant differences among the different groups (p=0.0004). Post-hoc analysis using Dunn's multiple groups comparison identified 13-14, 21 and 90DPI time points to be significantly different from uninfected controls. Single asterisk (*) = p<0.05 and double asterisk (**) = p<0.01. The error bars represent standard error of mean CD4+ T cell percentage within each group.
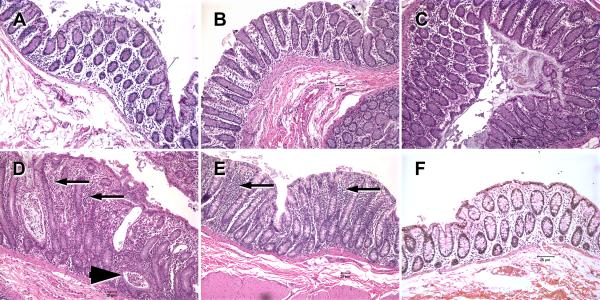
Histologic evaluation of hematoxylin and eosin stained sections of colon and jejunum from all non-SIV-infected animals with diarrhea revealed the presence of moderate to severe colitis including other intestinal lesions such as crypt dilatation/abscess and diverticulosis (Table II and Fig. 2D&E). No bacterial pathogens were detected in 7 out of the 8 non-SIV-infected macaques with diarrhea and colitis. In contrast, the colonic lamina propria of the SIV-infected macaques (7 and 14DPI) showed minimal to no histological signs of inflammation (Fig. 2A-C) and appeared similar to the uninfected control macaque (Fig. 2F).
FIGURE 2.
Hematoxylin and Eosin stained tissue sections of colon from acutely SIV-infected rhesus macaques (A- 7DPI), (B and C- 13DPI), and non-SIV-infected macaques with diarrhea (D&E) and a uninfected normal control macaque (F). In the colon of the non-SIV-infected macaques with diarrhea, moderate to severe colitis is evident from diffuse cellular infiltrates (arrows), crypt dilations and crypt abscesses (arrowhead) and a relative paucity of goblet cells (D&E) compared to acutely SIV-infected (A-C) and normal control (F) macaques. Note that the colons of acutely SIV-infected macaques AV91 (A), HI58 (B) and HI63 (C) show minimal to no histological evidence of inflammation. All figures are 10× magnification.
Acute SIV infection of the intestinal immune system is characterized by marked alterations in miRNA expression
In order to determine if SIV infection of the GI tract/immune system was associated with alterations in miRNA expression we performed global miRNA profiling of colon tissue during acute SIV infection using the human microRNA TLDA cards. As shown in table I, one animal each was at 7, 8 and 10DPI (days post infection), 3 each at 13 and 21DPI, and 1 at 29DPI. After applying multiple comparisons correction (Benjamini-Hochberg adjusted p values for false discovery rate) simultaneously to all miRNA probes (cards A and B combined), 9 miRNAs (4 up and 5 downregulated) were identified as statistically significant (adjusted p<0.05) and differentially expressed following analysis using DataAssist™ software version 3.01 (Table V). Raw CT values shown in table V provide additional information on the cellular abundance (high, medium or low) of each differentially expressed miRNA and the extent of variation across samples.
Among the 4 upregulated miRNAs, the expression of one miRNA, namely, miR-190b exceeded 5-fold (~6.0-fold) and in terms of magnitude showed the highest increase in expression following SIV infection (Table V). The expression levels of the remaining 3 miRNAs (miR-222, miR-22*, miR-223*) ranged from 1.5 to 3.0-fold compared to the uninfected control group. The expression of five miRNAs (hsa-miR-425*, −199a-5p, −221, −324-5p and −361-5p) decreased ~1.5-1.7 fold during acute SIV infection (Table V). These findings suggest that early SIV infection is characterized by significant changes in miRNA expression in the colon and that the profile is slightly dominated by downregulated miRNAs.
MiR-190b upregulation occurs in both colon and jejunum at all stages of SIV infection and its expression follows a trend observed with peripheral blood viral loads during SIV infection
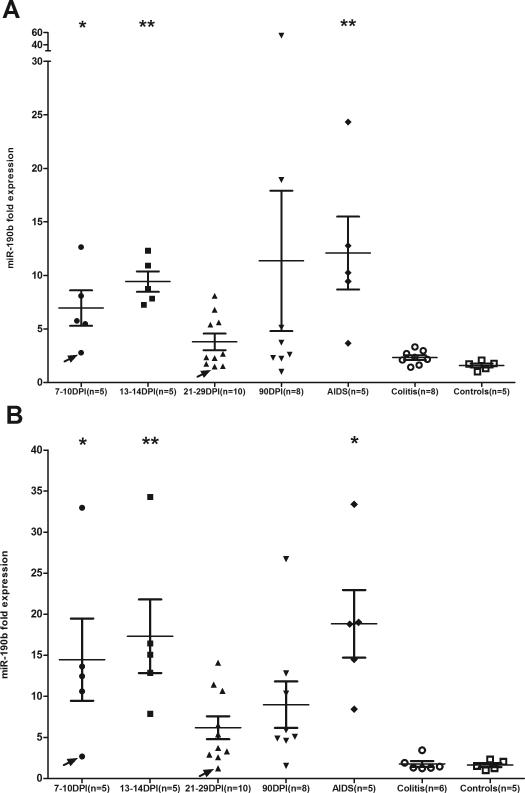
MiR-190b was selected for further characterization as the magnitude of increase was the highest among the four upregulated miRNAs (Table V) and ranked second based on p values (Adjusted p=0.0032) making it an ideal target for further characterization. As shown in figure 3A&B, miR-190b expression significantly increased in the colon and jejunum of SIV-infected compared to uninfected controls and non-SIV-infected macaques with diarrhea and colitis (labeled as Colitis in Fig. 3A&B). With the exception of the 21DPI and 90DPI time points in both colon and jejunum, miR-190b expression at all other time points showed statistical significance (p<0.05) (Fig. 3A&B). However, comparison of both (21 and 90DPI) time points separately with uninfected controls using the non-parametric Wilcoxon's rank sum test or Mann-Whitney U test revealed statistical significance (p<0.05) (data not shown) in colon and jejunum. Further, miR-190b expression in the colon and jejunum of non-SIV-infected macaques with diarrhea and colitis did not differ from normal uninfected controls (Fig. 3A&B). Although not a longitudinal study, some interesting trends in miR-190b expression were observed in the present study. From figure 3A&B, it is clearly evident that enhanced expression of miR-190b occurred as early as 7DPI (p<0.05) and peaked between 13-14DPI (coincident with peak viremia) (p<0.01). The initial elevation in miR-190b at 7DPI occurred even before there was a significant loss of mucosal CD4+ T cells (Fig. 1). This indicates that the increase in miR-190b expression occurs before peak viral loads and the nadir of CD4+ T cell depletion. Subsequent to the peak of viremia between 13-14DPI, average miR-190b expression dropped by ~2-fold in colon and ~3-fold in jejunum at 21-29DPI. Interestingly, this abrupt drop in expression at this time point (21-29DPI) coincides with the nadir of CD4+ T cell depletion which may account for the sudden drop in miR-190b expression. At 90DPI and in animals with terminal disease miR-190b expression showed an upward trend in both colon and jejunum (Fig. 3A&B). Interestingly, miR-190b upregulation was not detected in the colon and jejunum of non-SIV-infected macaques with diarrhea and colitis (Fig. 3A&B). Since intestinal tissues from the non-SIV-uninfected macaques with diarrhea and colitis were collected over a period of 6-7 years there was always the possibility that technical batch effects or RNA degradation could have contributed to the lack of miR-190b upregulation in this group. To specifically address this issue, we collected colon tissues from a second batch of 6 non-SIV-infected macaques necropsied within the past 6 months for chronic diarrhea non-responsive to treatment (colitis). Similar to the findings shown in figure 3A, miR-190b expression did not increase (supplemental figure 2C) and was no different from the uninfected control group. As shown in supplemental table 1, average miR-190b CT values in this group were identical to the uninfected controls (24.2 vs 24.3). Further, the colonic lamina propria of the SIV-infected macaques (7 and 13DPI) did not show any histological signs of inflammation (Fig. 2A-C) and appeared no different from the uninfected control macaque (Fig. 4F). In contrast, both non-SIV-infected macaques with diarrhea (Fig. 2D-E) have moderate to severe colitis (marked inflammatory cell infiltration of the colonic lamina propria along with other lesions such as crypt abscesses) providing strong evidence that inflammation/immune activation is not driving miR-190b upregulation.
FIGURE 3.
Elevated miR-190b expression in the colon (A) and jejunum (B) during SIV infection compared to uninfected normal controls and non-SIV-infected macaques with diarrhea (indicated as Colitis). Note the absence of miR-190b upregulation in non-SIV-infected macaques with diarrhea and colitis suggesting that upregulation of miR-190b is not inflammation driven but occurs in response to viral replication (DPI- days post infection). Arrows in colon (A) and jejunum (B) point to two animals [T108 (7DPI) and CG32 (21-29DPI)] that had no detectable SIV in intestinal tissue and where the levels of miR-190b was no different from controls. Data analysis using non-parametric Kruskal-Wallis test revealed differences among groups in both colon (p=0.0002) and jejunum (p<0.0001). Asterisks (* = p<0.05, ** = p<0.01) indicate groups that showed statistical significance compared to uninfected controls following Dunn's multiple groups comparison test. The error bars represent standard error of mean fold change within each group.
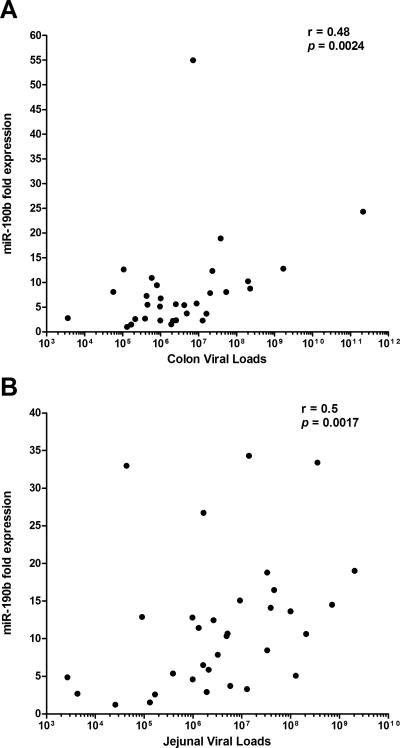
FIGURE 4.
Correlation of miR-190b in colon (A) and jejunum (B) with viral loads. A positive statistical (p<0.05) correlation between miR190b and SIV viral load was found in both colon and jejunum.
The correlation of miR-190b with increased viral replication in the intestine is supported by the finding that miR-190b expression levels in both colon and jejunum of two macaques with undetectable tissue viral load [T108 (7-8DPI), CG32 (21-29DPI), Table I and arrows in Fig. 3A&B] were no different from uninfected controls. Since miR-190b upregulation in intestine was only detected in macaques with detectable tissue viral load, we next examined the statistical correlation between miR-190b expression levels and intestinal tissue viral load. As shown in figure 4A&B, miR-190b expression levels in colon (r=0.48, p=0.0024) and jejunum (r=0.50, p=0.0017) were positively correlated with viral loads. Finally, the absence of miR-190b upregulation in the intestines of non-SIV-infected macaques with diarrhea and colitis suggests that miR-190b is not a general marker of intestinal inflammation or immune activation as these animals had moderate to severe colitis.
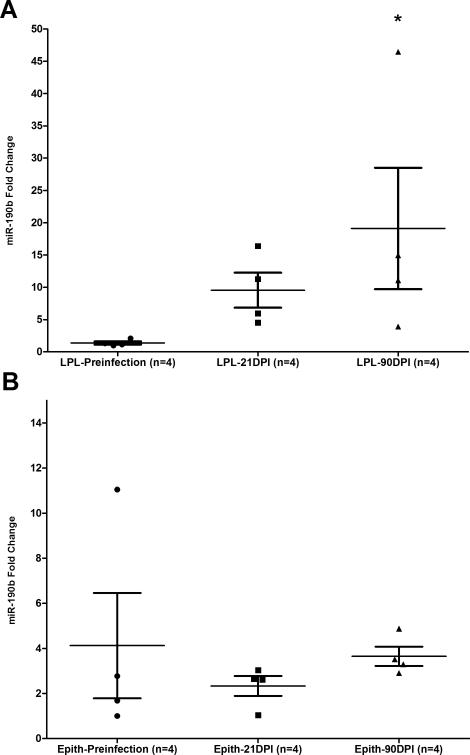
The intestinal lamina propria and not the epithelial compartment contributes to miR-190b upregulation in SIV-infected macaques
The failure to detect miR-190b upregulation in the intestines of non-SIV-infected macaques with diarrhea and colitis (Fig. 3A&B) suggested a strong link between miR-190b upregulation and viral replication. The association with viral replication also suggested that targets of SIV infection such as CD4+ T cells and macrophages may be necessary to drive miR-190b upregulation. If this hypothesis is true then miR-190b upregulation should occur predominantly in the LPL and not the epithelial compartment as both CD4+ T cells and macrophages reside in the LPL compartment. To confirm that the LPL compartment of the intestine was contributing to the upregulation of miR-190b expression in SIV-infected macaques we quantified miR-190b expression in isolated intestinal epithelial cells and LPLs using the individual TaqMan stem loop qRT-PCR assay specific for miR-190b. As shown in figure 5A, miR-190b showed a ~10- and ~20-fold increase in the LPL compartment, at 21 and 90DPI (p<0.05), respectively. In contrast, no significant increase in miR-190b expression was found in the intestinal epithelial compartment at these same time points (Fig. 5B). It is important to note that the presence of a small percentage (10-15%) of IELs in the isolated epithelial cells is not likely to impact miR-190b expression as the vast majority of the IELs are CD8+ and not infected by SIV. Similarly, given the complete absence of miR-190b upregulation in the epithelium (Fig. 5B), the presence of a small percentage of contaminating epithelial cells (10-15%) in LPLs is also not likely to influence miR-190b expression as they are not infected by SIV and express basal levels of miR-190b like other uninfected intestinal cells. miR-190b expression was also quantified in peripheral blood mononuclear cells (PBMCs) isolated from the same 4 animals shown in figure 5A&B at the same time points (Preinfection, 21 and 90DPI) as intestinal resection segments. While miR-190b expression was considerably elevated in the LPL (p<0.05 at 90DPI) (Fig. 5A), parallel statistically significant upregulation was not observed in PBMCs at the same time points as shown in supplemental figure 2D suggesting that PBMCs may not contain a sufficient proportion of SIV-infected cells to result in miR-190b upregulation.
FIGURE 5.
The lamina propria leukocyte (LPL) and not the epithelial compartment of the intestine is the predominant source of miR-190b upregulation in SIV-infected macaques. Statistically significant increase in miR-190b expression was detected in the intestinal lamina propria at 90 days post SIV infection (DPI) (A) but not in the intestinal epithelium (Epith) (B). Data was analyzed using non-parametric Kruskal-Wallis test and post hoc multiple groups comparison was performed using Dunn's test. Single asterisk (*) indicates statistical significance (p<0.05) compared to preinfection samples.
These findings show that under basal conditions, miR-190b is expressed in both the intestinal epithelium and LPLs. However, after SIV infection its expression is significantly increased only in the LPL compartment. This finding along with the correlation between intestinal viral load and miR-190b expression and the absence of miR-190b upregulation in the intestines of non-SIV-infected macaques with diarrhea and colitis (Fig. 3A&B) suggests that miR-190b upregulation is linked to viral replication.
MiR-190b is expressed in CD4+ T cells and macrophages and its expression is significantly increased in in vitro cultured CD4+ T cells and primary intestinal macrophages in response to SIV infection
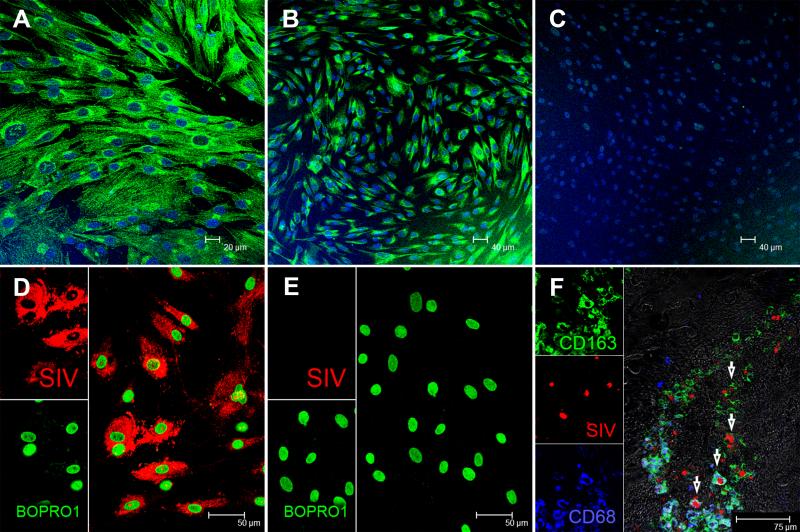
To further explore the linkage between expression of miR-190b and viral replication we examined miR-190b expression in CD4+ T cells and macrophages with and without SIV infection. Accordingly, we isolated peripheral blood CD4+ T cells and primary intestinal macrophages and determined miR-190b expression separately in both cell types using stem loop qRT-PCR assay specific to miR-190b. We preferred this approach to miRNA in situ hybridization as qRT-PCR is highly specific and truly quantitative. As illustrated in figure 6, we were able to obtain high purity macrophage populations that were consistently positive for macrophage specific markers CD68 (Fig. 6A) and CD163 (Fig. 6B) and negative for T lymphocyte markers such as CD3 (Fig. 6C). Further, we successfully infected these cells in vitro with SIVmac251 as demonstrated by the in situ detection of viral RNA 4 days post infection (Fig 6D&E). As shown in supplemental figure 3, miR-190b was strongly expressed by both CD4+ T cells and macrophages. Further as shown in figure 3A&B, miR-190b expression increased as early as 7DPI, peaked at 13-14DPI (peak viral replication) and, particularly, its abrupt drop in expression at 21-29DPI in both colon and jejunum that coincided with the nadir of CD4+ T cell depletion strongly suggests that CD4+ T cells are likely to be a major cellular source of elevated miR-190b expression during acute infection. Nevertheless, as previously published (51), and as illustrated in figure 6F, SIV infection of intestinal macrophages in situ can occur as early as 10 days post infection.
FIGURE 6. Characterization of cultured intestinal macrophages.
In vitro cultured primary intestinal macrophages express classical macrophage markers such as CD68 (A) and CD163 (B) but do not express T cell markers such as CD3 (C). All three panels are double labels with CD68 and CD163 in green and nuclear labeling with Topro3 in blue. Intestinal macrophages were infected with SIVmac251 and in situ hybridization confirmed the presence of viral RNA (D). Uninfected cells are negative for SIV (E). Both panels involve double labels with viral RNA (red) and Bopro1 (green) for nuclear staining. SIV-infected macrophages can be detected in the intestine as early as 10 days post SIV infection (F). Arrows point to SIV-infected (red) macrophages that express CD68 (blue), CD163 (green) or both.
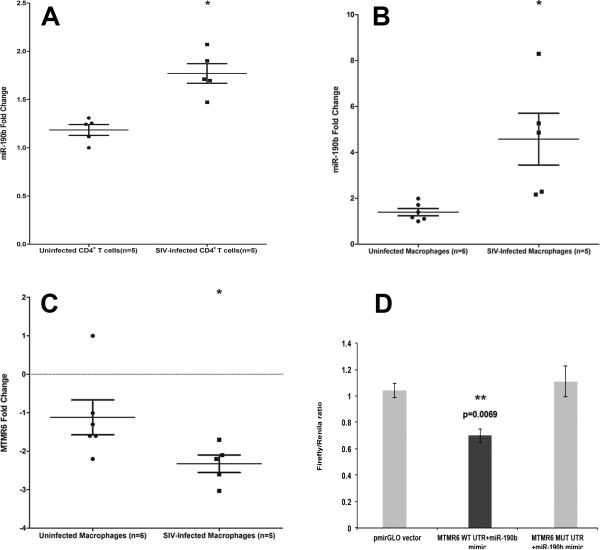
To conclusively verify that miR-190b was upregulated in response to SIV replication we infected peripheral blood CD4+ T cells and primary intestinal macrophages with SIV and found significant upregulation of miR-190b in CD4+ T cells (Fig. 7A) and macrophages (Fig. 7B) by days two and four post infection, respectively. miR-190b expression in CD4+ T cells was normalized to a combination of RNU44 and RNU48 as it yielded better statistical significance (p= 0.0121) compared to RNU44 alone (p= 0.0367). These findings clearly demonstrate that SIV-infected CD4+ T cells and macrophages directly contribute to miR-190b upregulation. In addition, the data also suggests that while SIV-infected CD4+ T cells are likely to be the primary cellular source of miR-190b uregulation during early acute infection (7-10 and 13-14 DPI), SIV-infected macrophages could contribute significantly to miR-190b uregulation at later time points and chronic infection.
FIGURE 7.
miR-190b expression was markedly elevated in in vitro cultured peripheral blood CD4+ T cells (A) (p= 0.0121) and primary jejunal macrophages (B) 2 and 4 days post SIV infection, respectively, suggesting that its upregulation occurs in response to SIV replication. Data was analyzed using non-parametric Wilcoxon's rank sum test for independent samples. Single asterisk (*) indicates statistical significance (p<0.05) compared to uninfected samples. The error bars represent standard error of mean fold change within each group. MTMR6 is a direct target of miR-190b (C-D). mRNA expression of MTMR6 is significantly decreased (p= 0.013) in intestinal macrophages 4 days post SIV infection (C). Data was analyzed using non-parametric Wilcoxon's rank sum test for independent samples. Single asterisk (*) indicates statistical significance (p<0.05) compared to uninfected macrophages. The error bars represent standard error of mean fold change within each group. miR-190b physically associates with the 3’ UTR of MTMR6 mRNA (D). The MTMR6 3′UTR sequences, wild type (WT) or mutated (MUT), were inserted into the multiple cloning sites situated in the 3′ end of firefly luciferase gene in the pmirGLO vector. HEK293 cells were co-transfected with 100 nM miR-190b mimic and 100 ng of luciferase reporter constructs containing WT- or MUT-MTMR6 3′UTR sequences. Firefly and Renila luciferase activities were detected using the Dual-Glo luciferase assay system 48 h after transfection. The ratio of luciferase activities (Firefly/Renilla) was calculated and normalized to the wells transfected with only unmanipulated pmirGLO vector. Double asterisks (**) indicate statistical significance (p<0.01).
MiR-190b can directly regulate the expression of MTMR6 and in doing so potentially play an important role in SIV pathogenesis
To investigate the significance of increased miR-190b expression in response to SIV infection we examined potential targets of miR-190b. TargetScan (version 6.2) (48) lists a total of 186 predicted targets for mml-miR-190b. Among these is MTMR6 which has been shown to negatively regulate CD4+ T cell activation and proliferation by inhibiting Ca++ dependent activated potassium channel KCa3.1, thereby, reducing Ca++ influx (52). Further, KCa3.1 is also required for macrophage activation (53). Consequently, MTMR6 represented a very interesting mR-190b target from the perspective of CD4+ T cell/macrophage activation and HIV replication. Interestingly, MTMR6 mRNA expression decreased significantly (p=0.013) in cultured intestinal macrophages 4 days post SIV infection (Fig. 7C). As shown in figure 7D, co-transfection of pmirGLO-WT-MTMR6 with miR-190b mimic resulted in significant reduction in Firefly/Renila ratios (p=0.0069). In contrast, co-transfection of pmirGLO-MUT-MTMR6 with miR-190b restored the Firefly/Renila ratios to the level observed with unmanipulated pmirGLO vector (Fig. 7D). These results clearly show that miR-190b can physically interact with the 3’ UTR and potentially regulate the expression of MTMR6. The precise effect of this regulation on target cell function including its direct or indirect impact on viral replication in infected cells needs future investigation.
DISCUSSION
HIV/SIV infection is associated with robust viral replication in the intestinal immune system, depletion of mucosal CD4+ T cells and marked immune activation. The dramatic loss of mucosal CD4+ T cells in the intestine has also been associated with intestinal inflammation, damage and microbial translocation. These changes are associated and in some cases preceded by marked changes in gene expression (7-10). Regulation of gene expression associated with the host response to infectious agents is complex and our understanding of this process has for the most part been dominated by the role played by transcription factors, coactivators, corepressors, chromatin modifiers, histone modulators, etc. The discovery of miRNAs has added another layer of complexity to the standard linear concept of DNA being transcribed to mRNA which is then translated to protein. While the regulatory role of miRNAs in immune cell development including several immunopathological and inflammatory conditions (18,22-23) is receiving a lot of attention, their contributions to regulating HIV/SIV replication and host responses, particularly, in the GI tract, an important site of viral replication, CD4+ T cell depletion and viral persistence remain unknown and unexplored. In the present study, using the rhesus macaque model of AIDS we investigated genome wide changes in miRNA expression to better understand their regulatory role in the GI tract following SIV infection. We performed the initial high throughput miRNA profiling on the colon for numerous reasons. First, similar to the jejunum, CD4+ T cell depletion has been well documented to occur in the colon (2,55). Secondly, from our previously published studies (13-14) it is clear that while SIV affects the jejunum, the colon is more severely impacted. Thirdly, the increased concentration of bacteria in the colon (1012 bacterial organisms per mL of contents) (56) makes it an important source of intestinal bacteria/bacterial products that is well documented to translocate into the systemic circulation via a disrupted intestinal epithelial barrier leading to chronic immune activation and AIDS progression (6). The present study for the first time describes dysregulated miRNA expression in the GI tract in response to SIV infection. We specifically, identified miR-190b to be significantly upregulated in both colon and jejunum of acutely SIV-infected macaques. Further, we show that miR-190b upregulation is driven by viral replication and not by the immune/inflammatory processes occurring in response to viral replication. Furthermore, our results provide new insights into the role of miR-190b in regulating host cellular responses, particularly, sustaining the activated state in SIV-infected cells by inhibiting MTMR6 expression.
In recent years, miRNAs have emerged as potent regulators of immune and inflammatory responses (20-22). Consistent with these findings and after applying multiple comparisons correction we identified a total of 9 miRNAs (Table V) to be markedly altered during acute SIV infection. Among these, eNOS suppressing miR-222 has been linked to regulating vascular inflammation (57) and differentiation of dendritic cells (58). Downregulation of miR-199a-5p promoted wound angiogenesis via derepression of the Ets-MMP1 pathway (59) suggesting that its reduced expression in the intestine may be part of the normal host response to repair tissue damage associated with early viral replication. As observed in the present study, miR-221 expression reduced significantly in bronchial epithelial cells in response to respiratory syncytial virus infection (60). Surprisingly, miR-190b exhibited the highest increase in expression following SIV infection and although reported to exhibit altered expression in several cancer studies (61-62) has not been previously linked to an infectious disease. However, miR-190/190a, a closely related miRNA originating from a different chromosome with distinct primary and precursor sequences has been well characterized (63-64). Both miR-190 and miR-190b share identical seed regions and hence have the same predicted targets. Taken together, acute SIV infection of the GI tract results in altered miRNA expression and the markedly elevated miR-190b expression as early as 7 days post SIV infection suggests an important role for this miRNA in SIV pathogenesis.
As mentioned above, the lack of previously published studies linking miR-190b to an infectious disease combined with the fact that it showed the highest fold increase among the 9 differentially expressed miRNAs prompted us to further investigate miR-190b expression at different stages of SIV infection. Surprisingly, miR-190b expression significantly increased in both colon and jejunum as early as 7DPI. Further, its expression peaked at 13-14DPI (time of peak viral replication), dropped abruptly at 21DPI (coinciding with nadir of CD4+ cell depletion) and then gradually increased as disease progressed (90DPI and AIDS). These findings are certainly noteworthy and a longitudinal study is definitely needed in the future to firmly associate alterations in miR-190b expression level with key pathogenic events such as peak viral replication and nadir of CD4+ T cell loss. Subsequently, qRT-PCR quantification of miR-190b expression separately in the intestinal epithelium and LPLs helped us to decisively localize the source of miR-190b upregulation to the immune cells residing in the lamina propria. Since viral replication in the intestinal lamina prorpia elicits an immune/inflammatory response there was definitely the possibility that inflammatory cell infiltration in response to SIV replication may be contributing to miR-190b upregulation. Accordingly, we included a group of macaques with GI disease (non-SIV-infected with diarrhea and colitis) to determine whether inflammatory cell infiltration or SIV replication was driving miR-190b expression as this group of animals has moderate to severe colitis, localized immune activation and consequently massive disruption of the intestinal epithelial barrier. Interestingly, the complete absence of miR-190b upregulation in the colon and jejunum of non-SIV-infected macaques with diarrhea and colitis strongly suggested that SIV replication in the lamina propria target cells likely provides the stimulus to drive miR-190b upregulation. More importantly, the latter finding undoubtedly means that shifts in immune cell composition (infiltration by inflammatory cells) occurring in response to SIV replication does not account for the observed increase in miR-190b expression. Finally, infection of in vitro cultured peripheral blood CD4+ T cells and primary intestinal macrophages with SIV conclusively revealed the SIV-infected cell as the primary cellular source of miR-190b upregulation. Based on these findings we can confidently conclude that miR-190b upregulation occurs primarily in response to SIV replication and is not simply a nonspecific result of immune activation and inflammation.
To gain further insight into the biological significance of miR-190b upregulation we next focused on MTMR6, a predicted mRNA target of miR-190b. We focused on MTMR6 as a direct miR-190b target because of its central importance to CD4+ T cell/macrophage activation/proliferation (52-53). MTMR6 is a phosphatidylinositol-3 phosphatase that has been well demonstrated to negatively regulate Ca++ influx by inhibiting the calcium-dependent activated potassium channel KCa3.1, thereby, preventing CD4+ T cell activation and proliferation (52). In addition to T cells, KCa3.1 channel activity is also critical for macrophage activation (53). KCa3.1 channel activation requires phosphatidylinositol-3 phosphate PI(3)P and MTMR6 downregulates its activity by dephosphorylating PI(3)P (52). This is evident from studies where siRNA mediated silencing of MTMR6 in CD4+ T cells resulted in increased KCa3.1 channel activity, enhanced calcium influx and facilitated T cell activation with 10-fold less antigen concentration compared to untreated cells (53). In the present study, MTMR6 mRNA expression decreased significantly in in vitro cultured primary intestinal macrophages 4 days post SIV infection. Additionally, the luciferase reporter assay clearly demonstrated the existence of a physical interaction between miR-190b and the 3’ UTR of MTMR6 and provided strong evidence for a miR-190b mediated MTMR6 gene silencing in SIV-infected macrophages. The latter possibility is strongly supported by the recent study that demonstrated miRNA mediated destabilization of mRNAs as a major mechanism that accounted for more than 84% of the reduced protein output (65). It has been proposed that MTMR6 functions constitutively to tonically inhibit KCa3.1 channel activity and in doing so sets a threshold stimulus for T cell activation (52). Such a mechanism can be expected to be operational in activated colonic CD4+ T cells and macrophages of non-SIV-infected macaques with diarrhea and colitis maintaining basal miR-190b expression levels. Nevertheless, in SIV-infected intestinal CD4+ T cells and macrophages, miR-190b levels were markedly elevated which in turn provides an additional layer of regulation to augment KCa3.1 activity by reducing MTMR6 levels so that the constitutive Ca++ influx required for increased cytokine production and subsequent immune/inflammatory responses are sustained. Although this represents a protective host response to curtail the spread of the virus, paradoxically; miR-190b mediated silencing of MTMR6 can invariably secure the infected cell in an activated state ultimately promoting viral replication. Collectively, these findings suggest that miR-190b upregulation may be part of the normal cellular stress response to viral replication and its targeting of MTMR6 may be to promote cellular survival, which eventually benefits the virus.
To our knowledge, this is the first report describing genome wide changes in miRNA expression in the GI tract in response to HIV/SIV infection. The fact that miR-190b is markedly elevated predominantly in response to SIV infection suggests important roles for this miRNA in regulating host cell responses to SIV replication. While we have identified and confirmed MTMR6 as a direct target of miR-190b, additional studies employing high throughput approaches are needed to validate and identify the roles of the remaining 185 predicted targets in regulating virus-host cell interactions. It is also intriguing to know whether miR-190b induction is directly triggered by viral proteins or indirectly by host cellular factors responding to viral replication. To gain detailed insight miR-190b knockdown studies to evaluate its role in viral replication and host cell response are also needed. Finally, although miR-190b expression levels increased in response to SIV replication it cannot be concluded that its upregulation is specific to SIV. Future studies are also needed to determine whether miR-190b expression levels increase in response to infection with other viruses.
Supplementary Material
Table V.
Differentially expressed miRNAs in colon during acute SIV infection.
| miRNA ID | Acute SIV (n=10) | Uninfected Controls (n=5) | Fold Change | Adjusted p value | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AV91 | HI52 | HI58 | HI63 | M992 | CT16 | IA85 | HC36 | HB31 | GA19 | EL66 | EH70 | EH80 | GI92 | FK25 | |||
| hsa-miR-425* | 29.3 | 28.8 | 29.3 | 28.7 | 29.5 | 29.1 | 26.8 | 28.0 | 27.7 | 27.9 | 28.2 | 28.6 | 28.4 | 29.4 | 29.0 | −1.7 | 0.0024 |
| hsa-miR-190b | 23.0 | 23.4 | 23.0 | 21.8 | 22.5 | 23.1 | 21.1 | 22.7 | 22.7 | 22.0 | 25.2 | 24.7 | 24.8 | 26.0 | 26.4 | 6.0 | 0.0032 |
| hsa-miR-222 | 15.6 | 15.4 | 15.7 | 15.1 | 15.2 | 15.4 | 14.3 | 14.7 | 14.5 | 14.7 | 15.7 | 15.9 | 15.5 | 16.4 | 16.7 | 1.5 | 0.0114 |
| hsa-miR-199a-5p | 25.3 | 24.5 | 25.6 | 25.4 | 25.8 | 24.4 | 23.8 | 24.7 | 24.7 | 24.4 | 24.3 | 24.3 | 23.8 | 24.9 | 24.6 | -1.7 | 0.0376 |
| hsa-miR-22* | 23.4 | 22.9 | 23.0 | 22.5 | 23.0 | 23.3 | 22.2 | 23.1 | 22.8 | 22.6 | 23.3 | 23.4 | 23.4 | 24.4 | 24.5 | 1.5 | 0.0376 |
| hsa-miR-221 | 21.5 | 20.9 | 21.0 | 21.1 | 22.0 | 21.1 | 20.7 | 20.7 | 21.3 | 21.0 | 20.4 | 20.5 | 20.0 | 21.3 | 21.3 | -1.6 | 0.0376 |
| hsa-miR-223* | 26.7 | 26.1 | 25.4 | 25.6 | 25.0 | 26.6 | 24.7 | 25.0 | 24.4 | 24.5 | 26.5 | 27.2 | 26.7 | 27.0 | 29.0 | 2.8 | 0.0376 |
| hsa-miR-324-5p | 22.8 | 22.5 | 23.0 | 22.5 | 23.3 | 22.3 | 21.5 | 22.6 | 22.7 | 22.7 | 22.0 | 22.2 | 21.6 | 22.7 | 22.7 | −1.6 | 0.0376 |
| hsa-miR-361-5p | 22.0 | 22.2 | 22.5 | 22.0 | 22.4 | 22.0 | 21.6 | 22.2 | 22.1 | 22.1 | 21.5 | 21.6 | 21.1 | 22.6 | 22.3 | −1.5 | 0.0376 |
The table shows raw CT and fold change for all differentially expressed (Adjustedp<0.05) miRNAs after applying multiple comparisons correction (Benjamini-Hochberg adjusted p values for false discovery rate) in the colon of ten acutely SIV-infected macaques.
ACKNOWLEDGMENTS
The authors would like to thank Ronald S Veazey, Maurice Duplantis, Vinay Kumar, Yun Te Lin, Faith R. Schiro, Cecily C. Midkiff, Stephanie Feely and Robin Rodriguez for their technical assistance in the study.
This work was supported by NIH grants: R01DK083929 to MM, AI084793, MH077544 and OD011104 (formerly RR00164)
REFERENCES
- 1.Lackner AA, Mohan M, Veazey RS. The gastrointestinal tract and AIDS pathogenesis. Gastroenterology. 2009;136:1965–1978. doi: 10.1053/j.gastro.2008.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 3.Smit-McBride Z, Mattapallil JJ, McChesney M, Ferrick D, Dandekar S. Gastrointestinal T lymphocytes retain high potential for cytokine responses but have severe CD4(+) T-cell depletion at all stages of simian immunodeficiency virus infection compared to peripheral lymphocytes. J Virol. 1998;72:6646–6656. doi: 10.1128/jvi.72.8.6646-6656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- 5.Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, Boden D, Racz P, Markowitz M. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.George MD, Sankaran S, Reay E, Gelli AC, Dandekar S. High-throughput gene expression profiling indicates dysregulation of intestinal cell cycle mediators and growth factors during primary simian immunodeficiency virus infection. Virology. 2003;312(1):84–94. doi: 10.1016/s0042-6822(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 8.Sankaran S, George MD, Reay E, Guadalupe M, Flamm J, Prindiville T, Dandekar S. Rapid onset of intestinal epithelial barrier dysfunction in primary human immunodeficiency virus infection is driven by an imbalance between immune response and mucosal repair and regeneration. J Virol. 2008;82(1):538–545. doi: 10.1128/JVI.01449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohan M, Kaushal D D, Aye PP, Alvarez X, Veazey RS, Lackner AA. Focused examination of the intestinal lamina propria yields greater molecular insight into mechanisms underlying SIV induced immune dysfunction. PLoS One. 2012;7(4):e34561. doi: 10.1371/journal.pone.0034561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohan M, Kaushal D D, Aye PP, Alvarez X, Veazey RS, Lackner AA. Focused examination of the intestinal epithelium reveals transcriptional signatures consistent with disturbances in enterocyte maturation and differentiation during the course of SIV infection. PLoS One. 2013;8(4):e60122. doi: 10.1371/journal.pone.0060122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang BX, Fish EN. The yin and yang of viruses and interferons. Trends Immunol. 2012;33(4):190–197. doi: 10.1016/j.it.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka N, Hoshino Y, Gold J, Hoshino S, Martiniuk F, Kurata T, Pine R, Levy D, Rom WN, Weiden M. Interleukin-10 induces inhibitory C/EBPbeta through STAT-3 and represses HIV-1 transcription in macrophages. Am J Respir Cell Mol Biol. 2005;33(4):406–411. doi: 10.1165/rcmb.2005-0140OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohan M, Aye PP, Borda JT, Alvarez X, Lackner AA. Gastrointestinal disease in simian immunodeficiency virus-infected rhesus macaques is characterized by proinflammatory dysregulation of the interleukin-6-janus kinase/signal transducer and activator of transcription-3 pathway. Am J Pathol. 2007;171:1952–1965. doi: 10.2353/ajpath.2007.070017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohan M, Aye PP, Borda JT, Alvarez X, Lackner AA. CCAAT/enhancer binding protein beta is a major mediator of inflammation and viral replication in the gastrointestinal tract of SIV-infected rhesus macaques. Am J Pathol. 2008;173(1):106–118. doi: 10.2353/ajpath.2008.080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Löffle D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermüller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, Cvijic H, Ullmann AK, Stadler PF, Horn F. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;15110(4):1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 16.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123(5):819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 17.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157(4):163–179. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swaminathan S, Murray DD, Kelleher AD. The role of microRNAs in HIV-1 pathogenesis and therapy. AIDS. 2012;26(11):1325–1334. doi: 10.1097/QAD.0b013e328352adca. [DOI] [PubMed] [Google Scholar]
- 20.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.El Gazzar M, McCall CE. MicroRNAs regulatory networks in myeloid lineage development and differentiation: regulators of the regulators. Immunol Cell Biol. 2012;90(6):587–593. doi: 10.1038/icb.2011.74. [DOI] [PubMed] [Google Scholar]
- 23.Gracias DT, Katsikis PD. MicroRNAs: key components of immune regulation. Adv Exp Med Biol. 2011;780:15–26. doi: 10.1007/978-1-4419-5632-3_2. [DOI] [PubMed] [Google Scholar]
- 24.Turner ML, Schnorfeil FM, Brocker T. MicroRNAs regulate dendritic cell differentiation and function. J Immunol. 2011;187(8):3911–3917. doi: 10.4049/jimmunol.1101137. Turner ML, Schnorfeil FM, Brocker T. [DOI] [PubMed] [Google Scholar]
- 25.Zhou L, Park JJ, Zheng Q, Dong Z, Mi Q. MicroRNAs are key regulators controlling iNKT and regulatory T-cell development and function. Cell Mol Immunol. 2011;8(5):380–387. doi: 10.1038/cmi.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA PA, Manjunath N. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS One. 2007;2(10):e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houzet L, Jeang KT. MicroRNAs and human retroviruses. Biochim Biophys Acta. 2011;1809:686–693. doi: 10.1016/j.bbagrm.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ouellet DL, Plante I, Barat C, Tremblay MJ, Provost P. Emergence of a complex relationship between HIV-1 and the microRNA pathway. Methods Mol Biol. 2009;487:415–433. doi: 10.1007/978-1-60327-547-7_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sánchez-Del Cojo M, López-Huertas MR, Mateos E, Alcamí J, Coiras M. Mechanisms of RNA interference in the HIV-1-host cell interplay. AIDS Rev. 2011;13:149–160. [PubMed] [Google Scholar]
- 30.Sisk JM, Clements JE, Witwer KW. miRNA profiles of monocyte-lineage cells are consistent with complicated roles in HIV-1 restriction. Viruses. 2012;4(10):1844–1864. doi: 10.3390/v4101844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang HS, Chen XY, Wu TC, Sang WW, Ruan Z. MiR-34a is involved in Tat-induced HIV-1 long terminal repeat (LTR) transactivation through the SIRT1/NFκB pathway. FEBS Lett. 2012;586(23):4203–4207. doi: 10.1016/j.febslet.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 32.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 33.Sun G, Li H, Wu X, Covarrubias M, Scherer L, Meinking K, Luk B, Chomchan P, Alluin J, Gombart AF, Rossi JJ. Interplay between HIV-1 infection and host microRNAs. Nucleic Acids Res. 2012;40(5):2181–2196. doi: 10.1093/nar/gkr961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duskova K, Nagilla P, Le HS, Iyer P, Thalamuthu A, Martinson J, Bar-Joseph Z, Buchanan W, Rinaldo C, Ayyavoo V. MicroRNA regulation and its effects on cellular transcriptome in Human Immunodeficiency Virus-1 (HIV-1) infected individuals with distinct viral load and CD4 cell counts. BMC Infect Dis. 2013;13(1):250. doi: 10.1186/1471-2334-13-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swaminathan S, Hu X, Zheng X, Kriga Y, Shetty J, Zhao Y, Stephens R, Tran B, Baseler MW, Yang J, Lempicki RA, Huang D, Lane HC, Imamichi T. Interleukin-27 treated human macrophages induce the expression of novel microRNAs which may mediate anti-viral properties. Biochem Biophys Res Commun. 2013;434(2):228–234. doi: 10.1016/j.bbrc.2013.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang K, Liu H, Rice AP. miR-132 enhances HIV-1 replication. Virology. 2013;438(1):1–4. doi: 10.1016/j.virol.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yelamanchili SV, Chaudhuri AD, Chen LN, Xiong H, Fox HS. MicroRNA-21 dysregulates the expression of MEF2C in neurons in monkey and human SIV/HIV neurological disease. Cell Death Dis. 2010;1:e77. doi: 10.1038/cddis.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Witwer KW, Sarbanes SL, Liu J, Clements JE. A plasma microRNA signature of acute lentiviral infection: biomarkers of central nervous system disease. AIDS. 2011;25(17):2057–2067. doi: 10.1097/QAD.0b013e32834b95bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sisk JM, Witwer KW, Tarwater PM, Clements JE JE. SIV replication is directly downregulated by four antiviral miRNAs. Retrovirology. 2013;10(1):95. doi: 10.1186/1742-4690-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sestak K, Merritt CK, Borda J, Saylor E, Schwamberger SR, Cogswell F, Didier ES, Didier PJ, Plauche G, Bohm RP, Aye PP, Alexa P, Ward RL, Lackner AA. Infectious agent and immune response characteristics of chronic enterocolitis in captive rhesus macaques. Infect Immun. 2003;71(7):4079–4086. doi: 10.1128/IAI.71.7.4079-4086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramesh G, Alvarez X, Borda JT, Aye PP, Lackner AA, Sestak K. Visualizing cytokine-secreting cells in situ in the rhesus macaque model of chronic gut inflammation. Clin Diagn Lab Immunol. 2005;12(1):192–197. doi: 10.1128/CDLI.12.1.192-197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Behrman S, Acosta-Alvear D, Walter PA. A CHOP-regulated microRNA controls rhodopsin expression. J Cell Biol. 2011;192(6):919–927. doi: 10.1083/jcb.201010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin J, Lwin T, Zhao JJ, Tam W, Choi YS, Moscinski LC, Dalton WS, Sotomayor EM, Wright KL, Tao J. Follicular dendritic cell-induced microRNA-mediated upregulation of PRDM1 and downregulation of BCL-6 in non-Hodgkin's B-cell lymphomas. Leukemia. 2011;25(1):145–152. doi: 10.1038/leu.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swaminathan S, Suzuki K, Seddiki N, Kaplan W, Cowley MJ, Hood CL, Clancy JL, Murray DD, Méndez C, Gelgor L, Anderson B, Roth N, Cooper DA, Kelleher AD. Differential regulation of the Let-7 family of microRNAs in CD4+ T cells alters IL-10 expression. J Immunol. 2012;188(12):6238–6246. doi: 10.4049/jimmunol.1101196. [DOI] [PubMed] [Google Scholar]
- 45.Pan D, Das A, Liu D, Veazey RS, Pahar B. B. Isolation and characterization of intestinal epithelial cells from normal and SIV-infected rhesus macaques. PLoS One. 2012;7(1):e30247. doi: 10.1371/journal.pone.0030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Golder JP, Doe WF. Isolation and preliminary characterization of human intestinal macrophages. Gastroenterology. 1983;84(4):795–802. [PubMed] [Google Scholar]
- 47.Kamada N, Hisamatsu T, Okamoto S, Chinen H, Kobayashi T, Sato T, Sakuraba A, Kitazume MT, Sugita A, Koganei K, Akagawa KS, Hibi T. Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-gamma axis. J Clin Invest. 2008;118(6):2269–2280. doi: 10.1172/JCI34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis BP, Burge CB, Bartel DP. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 49.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D'haene B, Mestdagh P, Hellemans J, Vandesompele J J. miRNA expression profiling: from reference genes to global mean normalization. Methods Mol Biol. 2012;822:261–272. doi: 10.1007/978-1-61779-427-8_18. [DOI] [PubMed] [Google Scholar]
- 51.Heise C, Miller CJ, Lackner AA, Dandekar S. Primary acute simian immunodeficiency virus infection of intestinal lymphoid tissue is associated with gastrointestinal dysfunction. J Infect Dis. 1994;169(5):1116–1120. doi: 10.1093/infdis/169.5.1116. [DOI] [PubMed] [Google Scholar]
- 52.Srivastava S, Ko K, Choudhury P, Li Z, Johnson AK, Nadkarni V, Unutmaz D, Coetzee WA, Skolnik EY. Phosphatidylinositol-3 phosphatase myotubularin-related protein 6 negatively regulates CD4 T cells. Mol Cell Biol. 2006;26(15):5595–5602. doi: 10.1128/MCB.00352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skaper SD. Ion channels on microglia: therapeutic targets for neuroprotection. CNS Neurol Disord Drug Targets. 2011;10(1):44–56. doi: 10.2174/187152711794488638. [DOI] [PubMed] [Google Scholar]
- 54.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2005;3(7):e264. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veazey RS, Tham IC, Mansfield KG, DeMaria M, Forand AE, Shvetz DE, Chalifoux LV, Sehgal PK, Lackner AA. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4+ T cells are rapidly eliminated in early SIV infection in vivo. J Virol. 2000;74:57–64. doi: 10.1128/jvi.74.1.57-64.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8(6):411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 57.Dentelli P, Rosso A, Orso F, Olgasi C, Taverna D, Brizzi MF. microRNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arterioscler Thromb Vasc Biol. 2010;30(8):1562–1568. doi: 10.1161/ATVBAHA.110.206201. [DOI] [PubMed] [Google Scholar]
- 58.Kuipers H, Schnorfeil FM, Brocker T. Differentially expressed microRNAs regulate plasmacytoid vs. conventional dendritic cell development. Mol Immunol. 2010;48(1-3):333–340. doi: 10.1016/j.molimm.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 59.Chan YC, Roy S, Huang Y, Khanna S, Sen CK. The microRNA miR-199a-5p down-regulation switches on wound angiogenesis by derepressing the v-ets erythroblastosis virus E26 oncogene homolog 1-matrix metalloproteinase-1 pathway. J Biol Chem. 2012;287(49):41032–41043. doi: 10.1074/jbc.M112.413294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Othumpangat S, Walton C, Piedimonte G. MicroRNA-221 modulates RSV replication in human bronchial epithelium by targeting NGF expression. PLoS One. 2012;7(1):e30030. doi: 10.1371/journal.pone.0030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patnaik SK, Yendamuri S, Kannisto E, Kucharczuk JC, Singhal S, Vachani A. MicroRNA expression profiles of whole blood in lung adenocarcinoma. PLoS One. 2012;7(9):e46045. doi: 10.1371/journal.pone.0046045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng H, Zeng Y, Zhang X, Chu J, Loh HH, Law PY. mu-Opioid receptor agonists differentially regulate the expression of miR-190 and NeuroD. Mol Pharmacol. 2010;77(1):102–109. doi: 10.1124/mol.109.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beezhold K, Liu J, Kan H, Meighan T, Castranova V, Shi X, Chen F. miR-190-mediated downregulation of PHLPP contributes to arsenic-induced Akt activation and carcinogenesis. Toxicol Sci. 2011;123(2):411–420. doi: 10.1093/toxsci/kfr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Slaby O. MiR-190 leads to aggressive phenotype of neuroblastoma through indirect activation of TrkB pathway. Med Hypotheses. 2013;80(3):325–326. doi: 10.1016/j.mehy.2012.11.033. [DOI] [PubMed] [Google Scholar]
- 65.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.