Abstract
Sphingosine kinase inhibitor (SKI) II has been reported as a dual inhibitor of sphingosine kinases (SKs) 1 and 2 and has been extensively used to prove the involvement of SKs and sphingosine-1-phosphate (S1P) in cellular processes. Dihydroceramide desaturase (Des1), the last enzyme in the de novo synthesis of ceramide (Cer), regulates the balance between dihydroceramides (dhCers) and Cers. Both SKs and Des1 have interest as therapeutic targets. Here we show that SKI II is a noncompetitive inhibitor (Ki = 0.3 μM) of Des1 activity with effect also in intact cells without modifying Des1 protein levels. Molecular modeling studies support that the SKI II-induced decrease in Des1 activity could result from inhibition of NADH-cytochrome b5 reductase. SKI II, but not the SK1-specific inhibitor PF-543, provoked a remarkable accumulation of dhCers and their metabolites, while both SKI II and PF-543 reduced S1P to almost undetectable levels. SKI II, but not PF543, reduced cell proliferation with accumulation of cells in the G0/G1 phase. SKI II, but not PF543, induced autophagy. These overall findings should be taken into account when using SKI II as a pharmacological tool, as some of the effects attributed to decreased S1P may actually be caused by augmented dhCers and/or their metabolites.
Keywords: autophagy, cell cycle, enzyme inhibition, molecular docking, mass spectrometry, reduced nicotinamide adenine dinucleotide-cytochrome b5 reductase, sphingolipids, sphingosine kinase inhibitor II
Sphingosine kinases (SKs) are oncogenic lipid kinases that catalyze the formation of the mitogenic second messenger sphingosine-1-phosphate (S1P) at the expense of proapoptotic sphingosine (So) and ceramide (Cer). Thus, as central enzymes in modulating the Cer/S1P balance, SKs are attractive targets for cancer therapy (1). Two SKs exist in humans, SK1 and SK2. SK1 has been extensively studied and there is a large body of evidence that proves its role in promoting cell survival, proliferation, and neoplastic transformation (2–5). SK1 is also elevated in many human cancers, which appears to contribute to carcinogenesis, chemotherapeutic resistance, and poor patient outcome. SK2, however, has not been as well-characterized, and there are contradictions in the key physiological functions that have been proposed for this isoform. Despite this, many studies are now emerging that implicate SK2 in key roles in a variety of diseases, including the development of a range of solid tumors (6).
The potential of SKs as therapeutic targets has boosted the development of small molecule inhibitors (4, 7–14). A detailed characterization of their pharmacology, particularly their selectivity against human SK1 and SK2, has been published (15). The first known SK inhibitors were long chain base analogs such as N,N-dimethyl-D-erythro-sphingosine (DMS) (16, 17) and L-threo-dihydrosphingosine (safingol) (18–21). While DMS inhibits both SK1 and SK2 by competing with the natural substrate So, safingol is a competitive inhibitor of SK1 (18), but unlike DMS, it is a substrate of SK2 (22). A similar behavior is exhibited by FTY720 (23). However, protein kinase C and other kinases are also inhibited by safingol (24), DMS (25, 26), and FTY720 (27), which are not, therefore, considered to be SK-specific inhibitors. A few compounds have been described as SK1-selective inhibitors, including sphingosine kinase inhibitor (SKI) I (28, 29) and SKI 178 (30). Another compound, SKI II, has been widely used as a SK1 inhibitor (31, 32). Nevertheless, SKI II is a dual SK1 and SK2 inhibitor, although it is inactive against other kinases. SKI II is a mixed inhibitor of So and ATP binding to SK1 (33), while the type of inhibition of SK2 has not been reported. Furthermore, SKI II (and also other inhibitors) has also been reported to induce proteasomal degradation of SK (4). The use of SKI II in the context of cancer therapy has been recently reviewed (32).
Dihydroceramide desaturase (Des1) is the last enzyme in the de novo synthesis of Cer. Blockade of Des1 produces an increase in dihydroceramides (dhCers), which have emerged as bioactive lipids and the target of several drugs (34), including fenretinide. Several studies have shown that cells respond to fenretinide treatment with a robust production of dhCers (35–44), and that this increase induces autophagy (45, 46). Furthermore, experimental evidence exists on the connection between resistance to fenretinide and increased S1P production (38, 44) due to an increased SK activity and SK1 mRNA and protein levels (38). In agreement, it has been reported that SK inhibitors abolish fenretinide resistance or synergize with fenretinide to enhance cancer cell death (36, 38, 41). In one of the above studies, the SK inhibitor, SKI II, was used to show the involvement of SK1 in the resistance of A2780 ovary cancer cells to fenretinide (38). The authors showed that treatment of fenretinide-resistant cells with SKI II effectively reduced S1P production and sensitized cells to the cytotoxic effect of fenretinide, and that cells treated with the combination fenretinide/SKI II experienced a dramatic increase in dhCer and sphinganine. Of note, treatment with SKI II alone significantly elevated cellular dhCer levels, but not Cer levels, although this increase was much lower than that observed in fenretinide and fenretinide/SKI II treatments. In another report (15), SKI II was also found to produce an increase in N-hexadecanoyldihydrosphingosine (C16dhCer) in A498 kidney adenocarcinoma cells. Surprisingly, none of the articles addressed and discussed these observations.
In this work, we show that besides inhibiting SK, SKI II reduces the activity of Des1 and increases the levels of dhCers and their metabolic products. This finding should be taken into account when using SKI II to investigate the role of SK in cell biology, as some of the effects attributed to increased S1P may actually be caused by augmented dhCers and/or their metabolites. Conclusions drawn based on the exclusive use of SKI II as a pharmacological tool should be revisited.
EXPERIMENTAL PROCEDURES
Materials
SKI II (Chemical Abstracts Registry number 312636-16-1) and PF-543 (Chemical Abstracts Registry number 1415562-82-1) were from Calbiochem. The compounds N-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]-D-erythro-dihydrosphingosine (dhCerC6NBD) and XM462 (47) were synthesized in our laboratories. Internal standards for lipidomics were from Avanti Polar Lipids. Minimum essential media, fetal bovine serum, nonessential amino acids, penicillin/streptomycin, 3-[4,5-dimethylthiazol-2-yl]-2,5- diphenyltetrazolium bromide (MTT), BSA, NADH, Tween 20, trypsin-EDTA, and the protease inhibitors were from Sigma. Laemmli buffer and acrylamide were from BioRad, SDS from Fluka, and the microBCA protein assay kit from Thermo Scientific. Protease inhibitor cocktail contained 2 μg/ml aprotinin, 5 μg/ml leupeptin, and 1 mM phenylmethylsulphonyl fluoride. Antibodies: anti-SK1 (rabbit) was from Cell Signaling; anti-Des1 (rabbit) and microtubule-associated protein 1-light chain 3 (LC3) II (rabbit) was from Abcam; and β-actin (mouse) was from Sigma. HRP secondary antibodies were from GE Healthcare.
Cell culture
The human gastric cancer cell line, HGC 27, was cultured at 37°C in 5% CO2 in minimum essential medium supplemented with 10% fetal bovine serum, 1% nonessential amino acids, and 100 ng/ml each of penicillin and streptomycin. Cells were routinely grown at a 60% maximum confluence.
Glioblastoma T98G and HeLa cells were maintained at 37°C in 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 100 ng/ml each of penicillin and streptomycin.
Cell viability
Cell viability was examined in triplicate samples by the MTT method. Cells were seeded at a density of 0.1 × 106 cells/ml, 0.1 ml/well (96-well plates). Twenty-four hours after seeding, seven 2:3 serial dilutions of SKI II, from 200 to 18 μM, and vehicle (100% activity) were added and viability was determined 24 h later.
Cell cycle analysis
Cells were seeded at 150,000 cells/ml into 6-well plates (1 ml/well). Cells were allowed to adhere for 24 h, and then they were treated with vehicle (0.1% ethanol) or SKI II (10 μM in 0.1% ethanol in medium). After exposure for 24 h, cell media were discarded and cells were washed with 400 μl PBS-EDTA 1% BSA and harvested with 400 μl trypsin-EDTA 1% BSA (37°C/2 min). Cells were pulled down by centrifugation at 1,300 rpm/3 min; cell pellet was washed once with 400 μl PBS-EDTA 1% BSA and again centrifuged at the same speed/time. Cells were fixed with a −20°C overnight incubation with a 70% ethanol (9.5 ml) in 1× PBS solution (0.5 ml). Fixed cells were pulled down, washed once with PBS-EDTA 1% BSA and stained at 37°C for 2 h with propidium iodide solution (0.1 mg/ml in PBS) and RNase, DNase-free (10 μg/ml). Stained cells were analyzed by using a Guava EasyCyte™ flow cytometer (Merck Millipore, Billerica, MA). Data analysis was performed using the Multicycle AV program (Phoenix Flow Systems, San Diego, CA).
Des1 enzyme assay
To prepare the cell lysate for Des1 activity determination in vitro, a suspension of 106 cells/ml per sample was centrifuged (1,400 rpm/3 min); the pellets were washed twice with PBS and resuspended in 0.1 ml of 0.2 M phosphate buffer (pH 7.4). The ice-cooled suspension was submitted to one round of bath sonication (30 s)/rest on ice (30 s), five rounds of bath sonication (15 s)/rest on ice (15 s), and one final round of bath sonication (30 s)/rest on ice (30 s). A 3.5% (v/v) solution of the required amount of stock substrate solution (0.5, 1.0, 1.5, and 2 mM in ethanol) in a BSA solution [3.3 mg/ml in 0.2 M phosphate buffer (pH 7.4)] was prepared to have the needed substrate concentrations (inhibition experiments, 35 μM; kinetics experiment: 17.5, 35, 52, and 70 μM). To each tube containing lysate from 106 cells was added: 85 μl of BSA-substrate mix (final substrate concentrations: inhibition experiments, 10 μM; kinetics experiment: 5, 10, 15, and 20 μM), 3 μl of SKI II stock solution in ethanol (final concentrations: inhibition experiments, 10 μM; kinetics experiment: 2.5 and 0.6 μM), 3 μl of XM462 stock solution in ethanol (10 μM final concentration), or 3 μl ethanol (vehicle control). Then 30 μl of NADH [20 mg/ml in 0.2 M phosphate buffer (pH 7.4)] and 82 μl of 0.2 M phosphate buffer (pH 7.4) were added to have a final volume of 300 μl. The reaction mixture was incubated at 37°C for 4 h. To stop the reaction, 0.7 ml/sample of methanol was added to each tube, mixed by vortex, and kept at 4°C overnight. The mixture was centrifuged (10,000 rpm/3 min), the clear supernatants were transferred to HPLC vials and 25 μl were injected. Instrumental analysis was carried out by HPLC with a fluorimetric detector as reported (47).
To determine the compound’s activity on Des1 in intact cells, cells were seeded in 24-well plates (106 cell/ml, 0.4 ml/well). Twenty-four hours after seeding, the medium was replaced by fresh complete medium containing substrate and either SKI II or XM462, which was used as a positive control (vehicle in controls) (0.4 ml/well). This solution was prepared as follows: 8 μl each of substrate and test compound solutions (10 mM in ethanol) were taken and diluted with medium to 1 ml and then 50 μl each of substrate and test compound solutions were added to each well (substrate = SKI II = XM462 = 10 μM) prior to addition of 0.3 ml/well of medium. After incubation at 37°C for 4 h, the media were collected, cells were washed with PBS (0.2 ml/well), and the washing solution was mixed with the collected media. Cells were harvested by trypsinization (trypsin/EDTA, 0.2 ml/well), washed with PBS, and the pellet was resuspended in water (0.1 ml) and sonicated (water bath) for 30 s. Methanol (media, 0.4 ml; cell lysate, 0.9 ml) was added to each tube and the mixture was stirred and kept at 4°C overnight. Then the suspension was centrifuged (10,000 rpm for 3 min), the solution was transferred to HPLC vials, and either 25 μl (media) or 0.1 ml (cells) was injected.
Western blotting
For protein analysis, 1–2 × 104 cells were plated in 6-well plates and were allowed to adhere for 24 h. Cells were treated with 10 μM of SKI II, 8 μM of XM462, 1 μM of PF-543, or ethanol as control for 4 or 24 h, collected with trypsin, and then pellets were washed twice with cold PBS. Cell lysis was performed with 30–40 μl of lysis buffer [150 mM NaCl, 1% Igepal-CA630, 50 mM Tris-HCl (pH 8), 2 μg/ml aprotinin, 5 μg/ml leupeptin, and 1 mM PMSF] by three cycles of bath sonication (5 s)/rest on ice (10 s). Then samples were kept on ice for 30 min and centrifuged for 3 min at 10,000 rpm. Supernatants were collected and protein determination was performed using the Micro BCA™ protein assay kit. Supernatants were combined with Laemmli sample buffer and boiled for 5 min. Equal amounts of proteins (Des1 and SK1, 30 μg; LC3, 20 μg) were loaded onto a 12% polyacrylamide gel, separated by electrophoresis at 100 V/90 min, and transferred onto a polyvinylidene fluoride membrane (100 V/1 h). Unspecific binding sites were then blocked with 5% milk in TBS with 0.1% Tween 20 (TBST). Anti-DEGS1 antibody was diluted 1:1,000 in 5% milk in PBS with 0.1% Tween 20, and anti-LC3 and anti-SK antibodies were diluted 1:1,000 in 5% milk in TBST. Membranes were incubated for 1 h at 25°C or overnight at 4°C under gentle agitation. After washing with TBST, membranes were probed with the correspondent secondary antibody for 1 h at 25°C (Des1: 1:2,000 dilution in 5% PBS with 0.1% Tween 20; SK: 1:3,000 dilution in 5% milk in TBST; LC3: 1:1,000 dilution in 3% BSA in TBST). Antibody excess was eliminated by washing with TBST and protein detection was carried out using ECL and membrane scanning with LI-COR C-DiGit® blot scanner. After stripping, membranes were blocked again with 3% BSA in TBST and subsequently incubated with anti-β-actin antibody (1:2,000 in 3% BSA in TBST) for 1 h and the secondary antibody (1:10,000 in 5% milk in TBST) for 1 h. Band intensities were quantified by LI-COR Image Studio Lite software.
Lipid analyses
Cells were seeded at 2,000,000 cells/ml into 6-well plates (1 ml/well) and were allowed to adhere for 24 h. Medium was replaced with fresh medium containing vehicle, SKI II (10 μM), or PF543 (1 μM). After exposure for the specified times, medium was removed, cells were washed with 400 μl PBS and harvested with 400 μl trypsin and 600 μl of medium. Sphingolipid extracts, fortified with internal standards [N-dodecanoylsphingosine, N-dodecanoylglucosylsphingosine, N-dodecanoylsphingosyl phosphorylcholine, C17-sphinganine (0.2 nmol each), and C17-sphinganine-1-phosphate, (0.1 nmol)] were prepared and analyzed as reported by ultraperformance LC (UPLC)-TOFMS (47) or HPLC-MS/MS (48).
Computational docking
Docking simulations were conducted with the package Schrödinger Suite 2013 (49), through its graphical interface Maestro (50). The program Macromodel (51), with its default force field OPLS 2005, a modified version of the OPLS-AA force field (52), and GB/SA water solvation conditions (53) were used for energy minimization. The coordinates of rat NADH-cytochrome b5 reductase (CB5R) (54), which include, in addition to the protein, a molecule of NAD+ and the flavin adenine dinucleotide (FAD) cofactor bound to the NADH and FAD binding sites, were obtained from the Protein Data Bank [(PDB) BC5R code: PDB 1IB0] (55). The structure of the protein was prepared using the Protein Preparation Wizard (56, 57) included in Maestro to remove the solvent molecules, adding hydrogens, setting protonation states (58), and minimizing the energy using the OPLS force field. Ligand structures were built and energy minimized within Maestro. Molecular docking simulations were carried out using the Induced Fit Docking workflow implemented in Maestro, which takes into account ligand and protein flexibility (59–61). Briefly, this protocol consisted of: i) An initial docking step of each ligand with the program Glide (62, 63), using the NAD structure to define the center of the docking box and using a softened potential (van der Waals radii scaling by 0.5) for the ligand and receptor atoms within that box; in this way, up to 20 poses for each ligand were generated. ii) For each protein-ligand complex, a minimization of the ligand plus all protein residues with atoms within 8 Å was carried out using the program Prime (64); the resulting receptor structure from each docked pose then reflected and induced fit to the ligand structure and conformation. iii) Finally, redocking of each ligand against the protein structures derived from the previous step was carried out with Glide using the standard precision settings. Ligand poses were ranked according to their docking scores. Per-residue interaction energies were calculated after docking by selecting the Scoring in Place option within Glide SP.
RESULTS
SKI II inhibits both SK and Des1
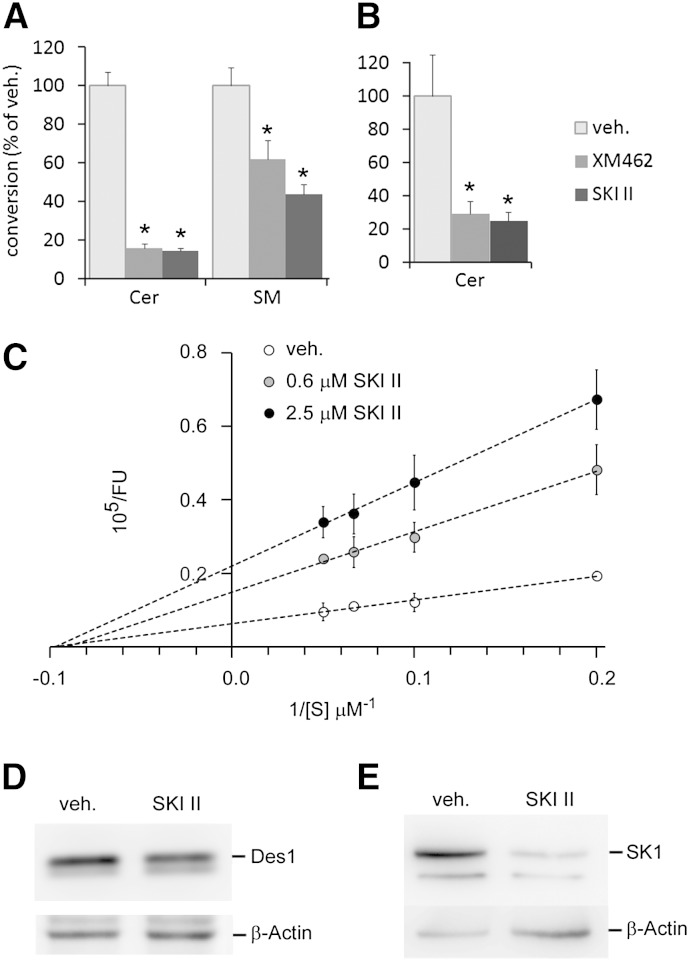
SKI II reduced the viability of HGC 27 cells (MTT) with a LD50 value of 84 μM (supplementary Fig. I). Therefore, the standard concentration used to inhibit SK (10 μM) was not cytotoxic in these cells. Intact HGC 27 cells treated with SKI II (10 μM/4 h) produced significantly lower amounts of N-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]-D-erythro-sphingosine (CerC6NBD) from dhCerC6NBD than controls (Fig. 1A), suggesting a decreased Des1 activity. Because SM is biosynthesized from Cer, levels of 4-nitro-2,1,3-benzoxadiazole (NBD)-labeled SM were also significantly lower in SKI II-treated cells than in controls. Des1 inhibition also occurred with cell lysates incubated for 4 h with equimolar concentrations (10 μM) of substrate and SKI II (Fig. 1B). In both experimental setups, inhibition was similar to that observed with XM462 (10 μM), which was included as positive control (47). To investigate the potency of SKI II as a Des1 inhibitor, cell lysates were incubated with various amounts of compound at different substrate concentrations. As shown in Fig. 1C, a concentration-dependent inhibition of Des1 was observed at all doses. Moreover, while Km did not change, Vmax decreased with increasing SKI II concentrations, which is in accordance with a noncompetitive type of inhibition with a calculated Ki of 0.3 μM. The observed decrease in Des1 activity provoked by SKI II was not paralleled by a reduction in Des1 protein, which showed similar levels to the controls (Fig. 1D). In contrast, SK1 protein levels were substantially reduced in cells exposed to SKI II (10 μM) (Fig. 1E), which agrees with published reports in other cell models (65).
Fig. 1.
Inhibitory activity of Des1 by SKI II. Intact cells (A, D, E) or cell lysates (B, C) were treated with either vehicle (veh., ethanol), SKI II (A, B, D, E; 10 μM), or XM462 (A, B; 8 μM). A–C: The Des1 reaction substrate (dhCerC6NBD, 10 μM) was added together with the test compounds into either the cell culture medium (A) or the reaction mixture (B). After 4 h, cells were collected (A, D, E) or the reaction was stopped by adding methanol (B, C). Samples were processed as described in the Experimental Procedures for analysis by HPLC coupled to a fluorimetric detector (A–C) or Western blot (D, E). Data of (A–C) correspond to the average ± SD of three experiments with triplicates. Asterisks indicate statistical significance (P ≤ 0.05, unpaired two-tail t-test). The Western blot images (D, E) are representative of three to five independent experiments with similar results.
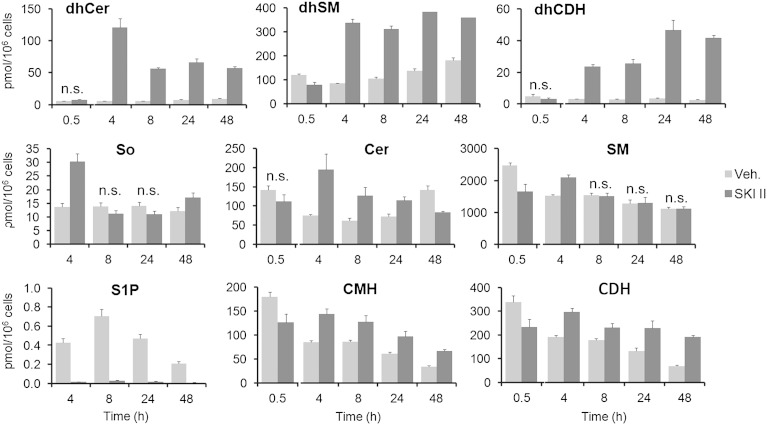
Figure 2 summarizes the results of UPLC/TOFMS analyses of sphingolipids extracted from HGC 27 cells harvested at different time points after incubation with 10 μM SKI II. Levels of dhCer exhibited a 20-fold increase over controls at 4 h after treatment and then decreased slowly to reach a constant level that was maintained up to the latest time point determined (48 h). dhCer metabolites, namely dihydrosphingomyelin (dhSM) and dihydroceramide dihexoside (lactosyldihydroceramide) (dhCDH) (dihydroceramide monohexosides have never been detected in HGC 27 cells under any treatment), also increased over controls at 4, 8, 24, and 48 h. Although similar levels of dhSM were found at all these time points, higher amounts of dhCDH occurred at 24 and 48 h than at 4 and 8 h. As expected from its SK inhibitory activity, S1P was reduced in cells treated with SKI II to almost undetectable levels at all time points sampled. Moreover, inhibition of SK brought about a 2-fold increase of the natural substrate So over controls at 4 h, while Cer and glucosylceramide increased over controls at 4, 8, and 24 h. In contrast, SM was not remarkably affected by SKI II treatment. A different effect on the sphingolipidome was observed after treatment with PF-543, a specific SK1 inhibitor (8). In this case, significant effects were observed at 8 h, but not 24 h, after treatment (supplementary Fig. II). These effects are slight and include a 1.5-fold increase in dhCers and 1.3- and 1.4-fold increases in Cer monohexosides (CMHs) and Cer dihexosides (lactosylceramides) (CDHs), respectively.
Fig. 2.
Effect of SKI II on the sphingolipid content. HGC 27 cells were treated with either ethanol (veh., vehicle) or SKI II (10 μM) for the indicated times and then cells were collected and processed for UPLC/TOFMS analysis as described in the Experimental Procedures. Data are the average ± SD of three experiments with triplicates. In all cases, except for those indicated as not significant (n.s.), the differences between means corresponding to control and treated cells are statistically significant (P ≤ 0.05, unpaired two-tail t-test).
Treatment of two other cell lines, glioblastoma T98G and cervical cancer HeLa cells, with SKI II (24 h) resulted in significantly increased amounts of dhSM and dhCDH over controls (supplementary Fig. III). No increase in dhCer levels was observed, also supporting inhibition of Des1 in these cells with metabolization of dhCer into complex dihydrosphingolipids.
SKI II decreases cell proliferation, provokes accumulation of cells at G1, and induces autophagy
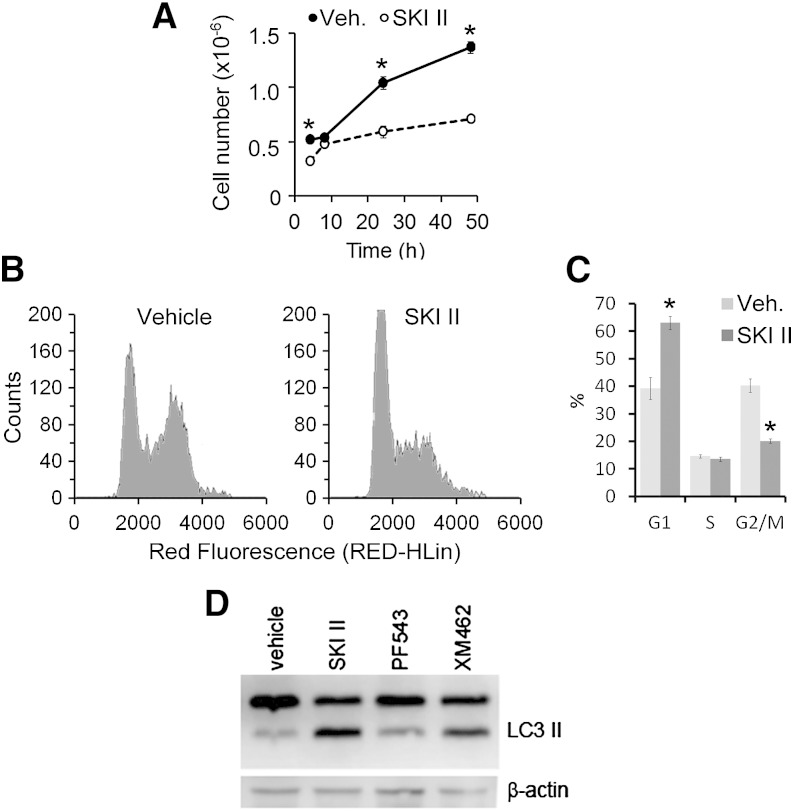
The changes in the sphingolipidome described above were paralleled by a reduction in cell proliferation (Fig. 3A). This effect was not observed when cells were exposed to PF543 (8 h: control, 6.11 ± 0.36 10−5 cells; PF543, 5.04 ± 0.52 10−5 cells; 24 h: control, 0.97 ± 0.78 10−5 cells; PF543, 1.19 ± 0.18 10−5 cells.). Examination of the cell cycle in SKI II-treated cells (10 μM, 24 h, complete medium) showed an increase in the number of cells at the G1 phase (Fig. 3B, C) as compared with vehicle-treated control cells.
Fig. 3.
Effect of SKI II on cell proliferation, cell cycle, and autophagy. A–C: HGC 27 cells were treated with either ethanol (veh., vehicle) or SKI II (10 μM) for 4, 8, 16, 24, and 48 h. Then cells were collected, counted, and processed for cell cycle analysis. A: Cell proliferation as the number of cells counted with a Countess automated cell counter. B: Representative cell cycle histograms obtained from HGC 27 cells after 16 h exposure to vehicle or SKI II. C: quantification of the relative number of cells in each stage of the cell cycle (Multicycle AV software). Data are the average ± SD of three experiments with triplicates. Asterisks denote statistical significance (P ≤ 0.05, unpaired two-tail t-test). D: HGC 27 cells were exposed to SKI-II (40 μM), PF-543 (1 μM), or XM462 (8 μM) for 24 h. Levels of LC3 II were assessed by Western blotting of cell lysates as described in the Experimental Procedures. The images shown are representative of three independent experiments with similar results.
The levels of LC3 II were measured to assess the effect of SKI II on autophagy. As shown in Fig. 3D, levels of LC3 II in cells exposed to 40 μM SKI II for 24 h were higher than in control cells treated with vehicle and similar to those in cells treated with XM462 (10 μM, 24 h), previously shown to induce autophagy in these cells (66).
Molecular docking
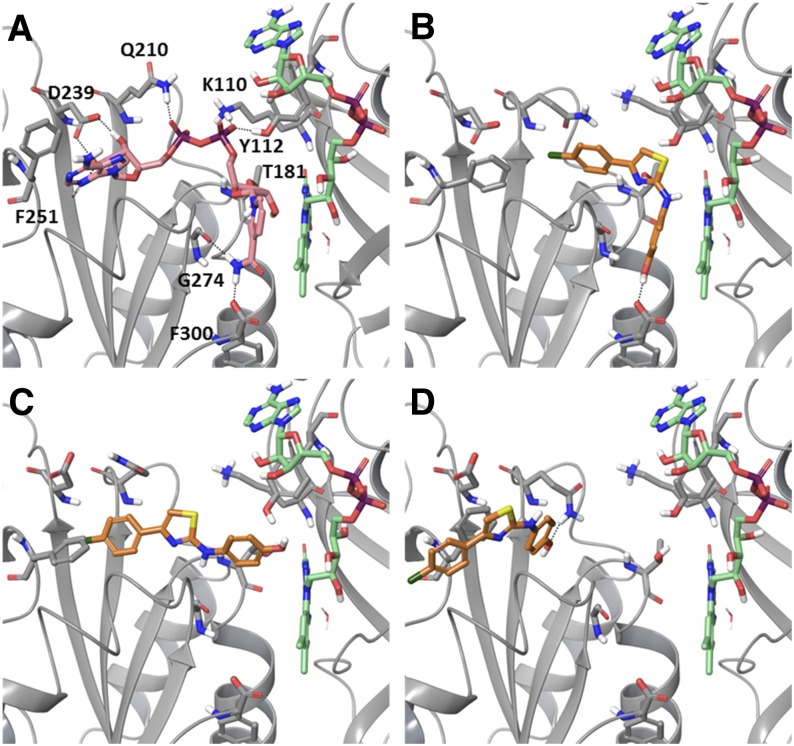
The lack of available structural information on Des1 hampers the application of structure-based methods to obtain information about the interaction between SKI II and this protein. However, it could also be hypothesized that the inhibitory effects of SKI II on Des1 could arise from an effect on an upstream target, such as the CB5R. Indeed, comparison of the structure of SKI II with those of some flavonoids which are known to inhibit CB5R (67, 68) (supplementary Fig. IV), such as luteolin, shows a significant shape and electrostatic character coincidence (supplementary Fig. V and supplementary Table I). It has been proposed that this flavonoid and other related compounds could bind to the NAD binding site on CB5R (69, 70); therefore, we examined the potential binding of SKI II at that site using an in silico docking method. To that end, we used the crystal structure of rat CB5R as docking target (54)2. This structure contains molecules of FAD and NAD+ bound in their respective sites in the protein (Fig. 4A). Both molecules occupy a wide cleft in the protein, with FAD adopting an extended conformation and NAD+ showing an L-shaped conformation, with its ribose-nicotinamide moiety placed in a relatively close disposition with respect to the flavin moiety of FAD. To bind in a productive manner, the sugar and nicotinamide moieties of NADH need to change their conformation to get closer to the flavin (54). This was clearly evidenced in several very recently reported structures of CB5R, which showed a shift on the nicotinamide conformation of reduced NAD to adopt a parallel and closer (∼3 Å) disposition relative to the FAD flavin group (73). Some conformational changes in the protein were also registered between the oxidized and reduced forms, so that “closed” and “open” states for the protein could be distinguished.
Fig. 4.
A: Crystal structure of rat cytochrome b5 reductase [PDB 1IB0 (54)] in complex with FAD (green) and NAD+ (pink). Residues interacting with the NAD+ molecule are labeled. B–D: Docked poses of SKI II (orange) at the NADH binding site on CB5R. Docking scores: −6.97 kcal/mol (B), −7.81 kcal/mol (C), and −7.60 kcal/mol (D).
Taking into account this conformational mobility, in particular at the NADH binding site, a docking protocol which considers some degree of protein flexibility was envisaged as suitable to study the potential binding of SKI II into CB5R. To this end, by applying an induced fit docking protocol that allowed the movement of residues in response to the presence of the ligand, different binding modes of SKI II into the NADH binding site could be determined. Figure 4B–D shows the three best poses which are representative of the docking poses obtained, and suggest that SKI II can adopt a range of binding modes that could be classified as proximal (Fig. 4B), intermediate (Fig. 4C), or distal (Fig. 4D), depending on their disposition relative to the FAD cofactor. Decomposition of the interaction energy of each pose among the residues of the protein (supplementary Table II) shows that the proximal one is mainly stabilized by hydrophobic interactions with the FAD cofactor and residues T181, G274, and P275, as well as polar interactions with F300. Similarly, the intermediate pose is mainly stabilized by hydrophobic interactions with residues G179, Q210, P275, and FAD, and the distal one by interactions with residues Q210, F251, P277, M278, and F281. Residues K110, Q210, D239, and F251 are the ones that experience a larger change in their conformation in response to ligand binding. Although docking scores alone often show little correlation with experimental binding affinities, the similarity between the scores of the three poses shown suggest that there is little preference between one or other binding mode.
Analog docking experiments run with the known CB5R inhibitors, luteolin, quercetin, and (+)-taxifolin, show similar results (supplementary Fig. VI), i.e., the three flavonoids show best docked poses with similar distribution, covering all of the NADH binding site, and with similar docking scores.
DISCUSSION
The validation of SK as a therapeutic target has stimulated research on the discovery of inhibitors for therapeutic purposes. Although not used in therapy, the compound SKI II is a dual SK1/SK2 inhibitor extensively employed as a pharmacological tool. In one such study, SKI II was used to investigate the role of SK on the resistance of an ovary cancer cell line to the antineoplasic drug fenretinide (38). As expected from its reported activity as a Des1 inhibitor, fenretinide treatment provoked an increase in intracellular dhCer levels, which was enhanced by cotreatment with SKI II. Interestingly, treatment with SKI II alone brought about an increase in dhCer that, although much lower than that induced by fenretinide alone or the fenretinide/SKI II combination, appeared to be statistically significant (38). These data prompted us to investigate the possibility of SKI II inhibiting Des1. This hypothesis was confirmed both in intact HGC 27 cells and in cell lysates. In both experimental setups, the amounts of CerC6NBD produced from dhCerC6NBD were lower in treated cells/lysates than in controls treated with vehicle. Western blot analysis showed that SKI II did not modify Des1 protein levels, while in agreement with a previous article (65), a reduction in SK1 protein was evident. Kinetic studies showed that SKI II was a noncompetitive inhibitor with a Ki value in the high nanomolar range. Other Des1 inhibitory compounds produced competitive (GT11, Ki = 6 μM) (74) or mixed-type inhibition (XM462, Ki = 2 μM) (47), while fenretinide functions as a competitive inhibitor (Ki = 8 μM) at short incubation times and as an irreversible inhibitor at long incubation times (40). While GT11 and XM462 are structural analogs of the Des1 substrate, fenretinide and other Des1 inhibitors (34) are not. We hypothesize that, while the structural analogs interact with the terminal desaturase protein, the nonstructural analogs act on other components of the electron transport chain or, alternatively, affect Des1 activity by modifying the redox status of the enzyme environment, which has been reported to affect this enzymatic activity (75). SKI II is a nonstructural analog of Des1 substrate and, as such, it does not probably directly interact with the Des1 protein. SKI II has been reported to induce oxidative stress (76), and so its activity as a Des1 inhibitor may result from its activity at modifying the redox status of cells. However, the occurrence of inhibition in cell lysates is against the involvement of reactive oxygen species. Another possibility is that SKI II inhibits CB5R, the other enzyme present in the electron transport chain associated to Des1 activity (34). Although this possibility has not been experimentally assessed, it is supported by the molecular modeling studies described above, which showed similar results in the docking to CB5R of SKI II and the known CB5R inhibitors luteolin, quercetin, and (+)-taxifolin (Fig. 4 and supplementary Fig. VI). Despite the fact that the precise mechanism by which these flavonoids inhibit the reduction of cytochrome b5 by CB5R is not known, it has been proposed that they could bind to different loci on CB5R which could result in a cooperative behavior (68). Furthermore, molecular dynamics studies show that they could explore more than a single interacting mode at the NADH binding site on CB5R (70). Our results are consistent with these previous reports and suggest that similar interactions could be occurring between SKI II and CB5R. Thus, the three binding modes computationally determined for SKI II could interfere with the binding of different parts of the NADH molecule, namely the nicotinamide (proximal), the pyrophosphate (intermediate), or the adenosine (distal) moieties, competing with its binding or perturbing its function if non-optimal binding is achieved in presence of SKI II.
Results of lipid analysis (Fig. 2) were in accordance with the SK and Des1 inhibitory activity observed for SKI II. Thus, a remarkable accumulation of dhCer was observed in HGC 27 cells at 4 h after treatment and remained significantly higher versus controls up to the last time point investigated (48 h). As expected from its SK inhibitory activity, S1P was reduced in cells treated with SKI II to almost undetectable levels at all time points sampled. Moreover, inhibition of SK brought about a 2-fold increase of the natural substrate So over controls at 4 h. The Cer increases observed at the 4, 8, and 24 h time points are likely the result of So metabolization by Cer synthases. Downstream metabolites of Cer, namely SM, CMHs, and CDHs, were also augmented in the presence of SKI II. While SM increased over controls at only the 4 h time point, CMH and CDH remained higher in SKI II-treated cells than in vehicle-treated cells at all time points. These results suggest that conversion into SM is a first response of cells to the deleterious increase in cytotoxic So, while glycosylation is activated as a sustained response. Similar effects were observed by Gao et al. (15) for SKI II in A498 kidney adenocarcinoma cells. Compared with the control group, the levels of intracellular S1P were decreased by over 90% after treatment with SKI II, while total Cer levels increased and amounts of So were dramatically decreased. These findings agree with metabolization of the accumulated long chain base into Cers. Of note, the levels of C16dhCer were also found to be elevated in SKI II-treated cells, although despite its significant increase over controls, this observation was not addressed by the authors. Other SL inhibitors, such as dimethylsphingosine and the SK2-selective inhibitor ABC294640, also augmented dhCer, although not as much as SKI II. Importantly, high levels of dhCer were also produced upon SK1-selective knockdown, which also resulted in elevated levels of total Cers and decreased amounts of S1P. However, So did not increase, but experienced a small decrease, indicating that it was likely being converted to Cers by Cer synthase. Increased C16dhCer may also result from acylation of sphinganine, although this base and its corresponding phosphate were not analyzed. Levels of dhCer were also shown to increase in A2780 ovarian cancer cells (38) treated with SKI II, although in this case also, no attention was given to this finding. Our findings in T98G and HeLa cells indicating inhibition of Des1 in these cells also, support that inhibition of Des1 by SKI II is not cell line specific.
In accordance with the mitogenic properties of S1P, overexpression of SK promotes growth, while decreasing SK activity by either genetic or pharmacological means reduces cell proliferation (2). In this article, we show that this is also the case in HGC 27 cells. Cell cycle analysis showed a higher population of cells in the G0/G1 phase upon SKI II treatment, which agrees with a previous report in myeloid leukemia K562 cells (77). Similar effects on cell cycle, namely accumulation of the G0/G1 population, were also found with DMS in rat intestinal epithelial cells (78) and by SK1 silencing in MCF-7 breast cancer (79), A498 ovarian cancer (80), and U-1242 MG and U-87 MG glioblastoma cell lines (81). In apparent contrast, Gao et al. (15) reported that SKI II arrested A498 cells in S phase with a concomitant decrease in the G2/M phase, while downregulation of SK1 or SK2 in U87MG cells under hypoxia arrests the cell cycle at G2/M (82). Whether the effect on cell cycle is due to SK or Des1 inhibition cannot be concluded with the available experimental data. However, the latter possibility is supported by two instances of experimental evidence. First, the SK1 specific inhibitor, PF543, did not affect HGC 27 cell proliferation at 8 and 24 h, although it avoided S1P production with no remarkable changes in other sphingolipid species. Second, the effect of dhCers at slowing or arresting the cell cycle with accumulation of cells in the G0/G1 phase is well-documented (35, 83–85). Therefore, we suggest that the effect of SKI II on the cell cycle may be due to the accumulation of dhCer resulting from its Des1 inhibitory activity.
Both Des1 and SK have been implicated in autophagy (86). Several studies have reported on the induction of autophagy by treatment with drugs that increase dhCer levels, such as fenretinide, resveratrol, and γ-tocopherol (34). Moreover, we previously reported on the induction of autophagy with prior increase in dhCer by the Des1 inhibitor, XM462, in HGC 27 cells (66). Autophagy is also induced in the same cell model by SKI II, but not PF543 (this work). These results suggest that autophagy induced by SKI II is actually due to increases in dhCer rather than to decreased production of S1P, which occurs with both SKI II and PF543. Furthermore, increase in LC3 II was also found in A498 cells cultured with SKI II, DMS, or ABC294640, the latter specific for SK2 (87). Interestingly, the three compounds increased C16dhCer, although the highest increments were attained with SKI II. Therefore, the contribution of dhCer to the proautophagic activity of some SK inhibitors cannot be disregarded at this point.
In summary, the dual SK1/SK2 inhibitor SKI II is also an inhibitor of Des1. Treatment of HGC 27 cells with SKI II results in decreased S1P levels and increased amounts of dhCer. Levels of Cer also augment, probably from acylation of So, which does not accumulate despite SK inhibition. The effects of SKI II on the cell cycle (longer duration/arrest of the G0/G1 phase) and autophagy observed here and by others can result from Des1 rather than SK inhibition.
Supplementary Material
Acknowledgments
The authors acknowledge predoctoral contracts from Generalitat de Catalunya (FI to F.C.) and La Marató de TV3 (to M.C. and P.S.), and the excellent technical assistance of Eva Dalmau.
Footnotes
Abbreviations:
- CB5R
- NADH-cytochrome b5 reductase
- C16dhCer
- N-hexadecanoyldihydrosphingosine
- CDH
- ceramide dihexoside (lactosylceramide)
- Cer
- ceramide
- CerC6NBD
- N-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]-D-erythro-sphingosine
- CMH
- ceramide monohexoside (glucosyl and galactosylceramide)
- Des1
- dihydroceramide desaturase
- dhCDH
- dihydroceramide dihexoside (lactosyldihydroceramide)
- dhCer
- dihydroceramide
- dhCerC6NBD
- N-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]-D-erythro-dihydrosphingosine
- dhSM
- dihydrosphingomyelin
- DMS
- N,N-dimethyl-D-erythro-sphingosine
- FAD
- flavin adenine dinucleotide
- LC3
- microtubule-associated protein 1-light chain 3
- MTT
- 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide
- NBD
- 4-nitro-2,1,3-benzoxadiazole
- PDB
- Protein Data Bank
- S1P
- sphingosine-1-phosphate
- SK
- sphingosine kinase
- SKI
- sphingosine kinase inhibitor
- So
- sphingosine
- TBST
- TBS with 0.1% Tween 20
- UPLC
- ultraperformance LC
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (SAF2011-22444), the Generalitat de Catalunya (SGR 2009 1072), and the Fundació la Marató de TV3 (112130 and 112132).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of six figures, two tables, and text.
REFERENCES
- 1.Orr Gandy K. A., Obeid L. M. 2013. Targeting the sphingosine kinase/sphingosine 1-phosphate pathway in disease: review of sphingosine kinase inhibitors. Biochim. Biophys. Acta. 1831: 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heffernan-Stroud L. A., Obeid L. M. 2013. Sphingosine kinase 1 in cancer. Adv. Cancer Res. 117: 201–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alshaker H., Sauer L., Monteil D., Ottaviani S., Srivats S., Böhler T., Pchejetski D. 2013. Therapeutic potential of targeting SK1 in human cancers. Adv. Cancer Res. 117: 143–200. [DOI] [PubMed] [Google Scholar]
- 4.Pyne S., Bittman R., Pyne N. J. 2011. Sphingosine kinase inhibitors and cancer: seeking the golden sword of Hercules. Cancer Res. 71: 6576–6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y., Wang Y., Wan Z., Liu S., Cao Y., Zeng Z. 2014. Sphingosine kinase 1 and cancer: a systematic review and meta-analysis. PLoS ONE. 9: e90362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neubauer H. A., Pitson S. M. 2013. Roles, regulation and inhibitors of sphingosine kinase 2. FEBS J. 280: 5317–5336. [DOI] [PubMed] [Google Scholar]
- 7.Lim K. G., Gray A. I., Pyne S., Pyne N. J. 2012. Resveratrol dimers are novel sphingosine kinase 1 inhibitors and affect sphingosine kinase 1 expression and cancer cell growth and survival. Br. J. Pharmacol. 166: 1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnute M. E., McReynolds M. D., Kasten T., Yates M., Jerome G., Rains J. W., Hall T., Chrencik J., Kraus M., Cronin C. N., et al. 2012. Modulation of cellular S1P levels with a novel, potent and specific inhibitor of sphingosine kinase-1. Biochem. J. 444: 79–88. [DOI] [PubMed] [Google Scholar]
- 9.Gustin D. J., Li Y., Brown M. L., Min X., Schmitt M. J., Wanska M., Wang X., Connors R., Johnstone S., Cardozo M., et al. 2013. Structure guided design of a series of sphingosine kinase (SphK) inhibitors. Bioorg. Med. Chem. Lett. 23: 4608–4616. [DOI] [PubMed] [Google Scholar]
- 10.Baek D. J., MacRitchie N., Pyne N. J., Pyne S., Bittman R. 2013. Synthesis of selective inhibitors of sphingosine kinase 1. Chem. Commun. (Camb.). 49: 2136–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byun H-S., Pyne S., MacRitchie N., Pyne N. J., Bittman R. 2013. Novel sphingosine-containing analogues selectively inhibit sphingosine kinase (SK) isozymes, induce SK1 proteasomal degradation and reduce DNA synthesis in human pulmonary arterial smooth muscle cells. Medchemcomm. 4: 1394–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raje M. R., Knott K., Kharel Y., Bissel P., Lynch K. R., Santos W. L. 2012. Design, synthesis and biological activity of sphingosine kinase 2 selective inhibitors. Bioorg. Med. Chem. 20: 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu K., Guo T. L., Hait N. C., Allegood J., Parikh H. I., Xu W., Kellogg G. E., Grant S., Spiegel S., Zhang S. 2013. Biological characterization of 3-(2-amino-ethyl)-5-[3-(4-butoxyl-phenyl)-propylidene]-thiazolidine-2,4-dione (K145) as a selective sphingosine kinase-2 inhibitor and anticancer agent. PLoS ONE. 8: e56471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z., Min X., Xiao S-H., Johnstone S., Romanow W., Meininger D., Xu H., Liu J., Dai J., An S., et al. 2013. Molecular basis of sphingosine kinase 1 substrate recognition and catalysis. Structure. 21: 798–809. [DOI] [PubMed] [Google Scholar]
- 15.Gao P., Peterson Y. K., Smith R. A., Smith C. D. 2012. Characterization of isoenzyme-selective inhibitors of human sphingosine kinases. PLoS ONE. 7: e44543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yatomi Y., Ruan F., Megidish T., Toyokuni T., Hakomori S., Igarashi Y. 1996. N,N-dimethylsphingosine inhibition of sphingosine kinase and sphingosine 1-phosphate activity in human platelets. Biochemistry. 35: 626–633. [DOI] [PubMed] [Google Scholar]
- 17.Edsall L. C., Van Brocklyn J. R., Cuvillier O., Kleuser B., Spiegel S. 1998. N,N-Dimethylsphingosine is a potent competitive inhibitor of sphingosine kinase but not of protein kinase C: modulation of cellular levels of sphingosine 1-phosphate and ceramide. Biochemistry. 37: 12892–12898. [DOI] [PubMed] [Google Scholar]
- 18.Olivera A., Kohama T., Tu Z., Milstien S., Spiegel S. 1998. Purification and characterization of rat kidney sphingosine kinase. J. Biol. Chem. 273: 12576–12583. [DOI] [PubMed] [Google Scholar]
- 19.Maurer B. J., Melton L., Billups C., Cabot M. C., Reynolds C. P. 2000. Synergistic cytotoxicity in solid tumor cell lines between N-(4-hydroxyphenyl)retinamide and modulators of ceramide metabolism. J. Natl. Cancer Inst. 92: 1897–1909. [DOI] [PubMed] [Google Scholar]
- 20.Kolesnick R. 2002. The therapeutic potential of modulating the ceramide/sphingomyelin pathway. J. Clin. Invest. 110: 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickson M. A., Carvajal R. D., Merrill A. H., Gonen M., Cane L. M., Schwartz G. K. 2011. A phase I clinical trial of safingol in combination with cisplatin in advanced solid tumors. Clin. Cancer Res. 17: 2484–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H., Sugiura M., Nava V. E., Edsall L. C., Kono K., Poulton S., Milstien S., Kohama T., Spiegel S. 2000. Molecular cloning and functional characterization of a novel mammalian sphingosine kinase type 2 isoform. J. Biol. Chem. 275: 19513–19520. [DOI] [PubMed] [Google Scholar]
- 23.Tonelli F., Lim K. G., Loveridge C., Long J., Pitson S. M., Tigyi G., Bittman R., Pyne S., Pyne N. J. 2010. FTY720 and (S)-FTY720 vinylphosphonate inhibit sphingosine kinase 1 and promote its proteasomal degradation in human pulmonary artery smooth muscle, breast cancer and androgen-independent prostate cancer cells. Cell. Signal. 22: 1536–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz G. K., Jiang J., Kelsen D., Albino A. P. 1993. Protein kinase C: a novel target for inhibiting gastric cancer cell invasion. J. Natl. Cancer Inst. 85: 402–407. [DOI] [PubMed] [Google Scholar]
- 25.Igarashi Y., Hakomori S., Toyokuni T., Dean B., Fujita S., Sugimoto M., Ogawa T., el-Ghendy K., Racker E. 1989. Effect of chemically well-defined sphingosine and its N-methyl derivatives on protein kinase C and src kinase activities. Biochemistry. 28: 6796–6800. [DOI] [PubMed] [Google Scholar]
- 26.Igarashi Y., Kitamura K., Toyokuni T., Dean B., Fenderson B., Ogawass T., Hakomori S. 1990. A specific enhancing effect of N,N-dimethylsphingosine on epidermal growth factor receptor autophosphorylation. Demonstration of its endogenous occurrence (and the virtual absence of unsubstituted sphingosine) in human epidermoid carcinoma A431 cells. J. Biol. Chem. 265: 5385–5389. [PubMed] [Google Scholar]
- 27.Sensken S-C., Gräler M. H. 2010. Down-regulation of S1P1 receptor surface expression by protein kinase C inhibition. J. Biol. Chem. 285: 6298–6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paugh S. W., Paugh B. S., Rahmani M., Kapitonov D., Almenara J. A., Kordula T., Milstien S., Adams J. K., Zipkin R. E., Grant S., et al. 2008. A selective sphingosine kinase 1 inhibitor integrates multiple molecular therapeutic targets in human leukemia. Blood. 112: 1382–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapitonov D., Allegood J. C., Mitchell C., Hait N. C., Almenara J. A., Adams J. K., Zipkin R. E., Dent P., Kordula T., Milstien S., et al. 2009. Targeting sphingosine kinase 1 inhibits Akt signaling, induces apoptosis, and suppresses growth of human glioblastoma cells and xenografts. Cancer Res. 69: 6915–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hengst J. A., Wang X., Sk U. H., Sharma A. K., Amin S., Yun J. K. 2010. Development of a sphingosine kinase 1 specific small-molecule inhibitor. Bioorg. Med. Chem. Lett. 20: 7498–7502. [DOI] [PubMed] [Google Scholar]
- 31.French K. J., Schrecengost R. S., Lee B. D., Zhuang Y., Smith S. N., Eberly J. L., Yun J. K., Smith C. D. 2003. Discovery and evaluation of inhibitors of human sphingosine kinase. Cancer Res. 63: 5962–5969. [PubMed] [Google Scholar]
- 32.Truman J-P., García-Barros M., Obeid L. M., Hannun Y. A. Evolving concepts in cancer therapy through targeting sphingolipid metabolism. Biochim. Biophys. Acta. Epub ahead of print. December 30, 2013; 10.1016/j.bbalip.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim K. G., Tonelli F., Berdyshev E., Gorshkova I., Leclercq T., Pitson S. M., Bittman R., Pyne S., Pyne N. J. 2012. Inhibition kinetics and regulation of sphingosine kinase 1 expression in prostate cancer cells: functional differences between sphingosine kinase 1a and 1b. Int. J. Biochem. Cell Biol. 44: 1457–1464. [DOI] [PubMed] [Google Scholar]
- 34.Fabrias G., Muñoz-Olaya J., Cingolani F., Signorelli P., Casas J., Gagliostro V., Ghidoni R. 2012. Dihydroceramide desaturase and dihydrosphingolipids: debutant players in the sphingolipid arena. Prog. Lipid Res. 51: 82–94. [DOI] [PubMed] [Google Scholar]
- 35.Kraveka J. M., Li L., Szulc Z. M., Bielawski J., Ogretmen B., Hannun Y. A., Obeid L. M., Bielawska A. 2007. Involvement of dihydroceramide desaturase in cell cycle progression in human neuroblastoma cells. J. Biol. Chem. 282: 16718–16728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H., Maurer B. J., Liu Y-Y., Wang E., Allegood J. C., Kelly S., Symolon H., Liu Y., Merrill A. H., Gouazé-Andersson V., et al. 2008. N-(4-hydroxyphenyl)retinamide increases dihydroceramide and synergizes with dimethylsphingosine to enhance cancer cell killing. Mol. Cancer Ther. 7: 2967–2976. [DOI] [PubMed] [Google Scholar]
- 37.Valsecchi M., Aureli M., Mauri L., Illuzzi G., Chigorno V., Prinetti A., Sonnino S. 2010. Sphingolipidomics of A2780 human ovarian carcinoma cells treated with synthetic retinoids. J. Lipid Res. 51: 1832–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Illuzzi G., Bernacchioni C., Aureli M., Prioni S., Frera G., Donati C., Valsecchi M., Chigorno V., Bruni P., Sonnino S., et al. 2010. Sphingosine kinase mediates resistance to the synthetic retinoid N-(4-hydroxyphenyl)retinamide in human ovarian cancer cells. J. Biol. Chem. 285: 18594–18602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao Z., Sun W., Xu R., Novgorodov S., Szulc Z. M., Bielawski J., Obeid L. M., Mao C. 2010;. Alkaline ceramidase 2 (ACER2) and its product dihydrosphingosine mediate the cytotoxicity of N-(4-hydroxyphenyl)retinamide in tumor cells. J. Biol. Chem. 285: 29078–29090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rahmaniyan M., Curley R. W., Obeid L. M., Hannun Y. A., Kraveka J. M. 2011. Identification of dihydroceramide desaturase as a direct in vitro target for fenretinide. J. Biol. Chem. 286: 24754–24764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Apraiz A., Idkowiak-Baldys J. K., Boyano M. D., Pérez-Yarza G., Hannun Y. A., Asumendi A. 2011. Evaluation of bioactive sphingolipids in 4-HPR-resistant leukemia cells. BMC Cancer. 11: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Apraiz A., Idkowiak-Baldys J., Nieto-Rementería N., Boyano M. D., Hannun Y. A., Asumendi A. 2012. Dihydroceramide accumulation and reactive oxygen species are distinct and nonessential events in 4-HPR-mediated leukemia cell death. Biochem. Cell Biol. 90: 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bikman B. T., Guan Y., Shui G., Siddique M. M., Holland W. L., Kim J. Y., Fabriàs G., Wenk M. R., Summers S. A. 2012. Fenretinide prevents lipid-induced insulin resistance by blocking ceramide biosynthesis. J. Biol. Chem. 287: 17426–17437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yasuo M., Mizuno S., Allegood J., Kraskauskas D., Bogaard H. J., Spiegel S., Voelkel N. F. 2013. Fenretinide causes emphysema, which is prevented by sphingosine 1-phoshate. PLoS ONE. 8: e53927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng W., Kollmeyer J., Symolon H., Momin A., Munter E., Wang E., Kelly S., Allegood J. C., Liu Y., Peng Q., et al. 2006. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim. Biophys. Acta. 1758: 1864–1884. [DOI] [PubMed] [Google Scholar]
- 46.Holliday M. W., Cox S. B., Kang M. H., Maurer B. J. 2013. C22:0- and C24:0-dihydroceramides confer mixed cytotoxicity in T-cell acute lymphoblastic leukemia cell lines. PLoS ONE. 8: e74768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munoz-Olaya J. M., Matabosch X., Bedia C., Egido-Gabás M., Casas J., Llebaria A., Delgado A., Fabriàs G. 2008. Synthesis and biological activity of a novel inhibitor of dihydroceramide desaturase. ChemMedChem. 3: 946–953. [DOI] [PubMed] [Google Scholar]
- 48.Bedia C., Casas J., Andrieu-Abadie N., Fabriàs G., Levade T. 2011. Acid ceramidase expression modulates the sensitivity of A375 melanoma cells to dacarbazine. J. Biol. Chem. 286: 28200–28209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schrödinger Suite 2013, Update 2. Schrödinger, LLC, New York, NY.
- 50.Maestro, version 9.5. Schrödinger, LLC, New York, NY.
- 51.MacroModel, version 10.1. Schrödinger, LLC, New York, NY.
- 52.Jorgensen W. L., Maxwell D. S., Tirado-Rives J. 1996. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118: 11225–11236. [Google Scholar]
- 53.Still W. C., Tempczyk A., Hawley R. C., Hendrickson T. 1990. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 112: 6127–6129. [Google Scholar]
- 54.Bewley M. C., Marohnic C. C., Barber M. J. 2001. The structure and biochemistry of NADH-dependent cytochrome b5 reductase are now consistent. Biochemistry. 40: 13574–13582. [DOI] [PubMed] [Google Scholar]
- 55.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. 2000. The Protein Data Bank. Nucleic Acids Res. 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schrödinger. Protein Preparation Wizard 2013-2; Epik version 2.4, Schrödinger, LLC, New York, NY. Impact version 5.9, Schrödinger, LLC, New York, NY. Prime version 3.2, Schrödinger, LLC, New York, NY.
- 57.Sastry G. M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. 2013. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 27: 221–234. [DOI] [PubMed] [Google Scholar]
- 58.Olsson M. H. M., Søndergard C. R., Rostkowski M., Jensen J. H. 2011. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7: 525–537. [DOI] [PubMed] [Google Scholar]
- 59.Schrödinger. Induced Fit Docking protocol 2013-2, Glide version 5.9, Prime version 3.2. Schrödinger, LLC, New York, NY. [Google Scholar]
- 60.Sherman W., Day T., Jacobson M. P., Friesner R. A., Farid R. 2006. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 49: 534–553. [DOI] [PubMed] [Google Scholar]
- 61.Sherman W., Beard H. S., Farid R. 2006. Use of an induced fit receptor structure in virtual screening. Chem. Biol. Drug Des. 67: 83–84. [DOI] [PubMed] [Google Scholar]
- 62.Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., et al. 2004. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47: 1739–1749. [DOI] [PubMed] [Google Scholar]
- 63.Halgren T. A., Murphy R. B., Friesner R. A., Beard H. S., Frye L. L., Pollard W. T., Banks J. L. 2004. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47: 1750–1759. [DOI] [PubMed] [Google Scholar]
- 64.Prime, version 3.3. Schrödinger, LLC, New York, NY.
- 65.Loveridge C., Tonelli F., Leclercq T., Lim K. G., Long J. S., Berdyshev E., Tate R. J., Natarajan V., Pitson S. M., Pyne N. J., et al. 2010. The sphingosine kinase 1 inhibitor 2-(p-hydroxyanilino)-4-(p-chlorophenyl)thiazole induces proteasomal degradation of sphingosine kinase 1 in mammalian cells. J. Biol. Chem. 285: 38841–38852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Signorelli P., Munoz-Olaya J. M., Gagliostro V., Casas J., Ghidoni R., Fabriàs G. 2009. Dihydroceramide intracellular increase in response to resveratrol treatment mediates autophagy in gastric cancer cells. Cancer Lett. 282: 238–243. [DOI] [PubMed] [Google Scholar]
- 67.Çelik H., Koşar M. 2012. Inhibitory effects of dietary flavonoids on purified hepatic NADH-cytochrome b5 reductase: structure-activity relationships. Chem. Biol. Interact. 197: 103–109. [DOI] [PubMed] [Google Scholar]
- 68.Çelik H., Koşar M., Arinç E. 2013. In vitro effects of myricetin, morin, apigenin, (+)-taxifolin, (+)-catechin, (-)-epicatechin, naringenin and naringin on cytochrome b5 reduction by purified NADH-cytochrome b5 reductase. Toxicology. 308: 34–40. [DOI] [PubMed] [Google Scholar]
- 69.Havsteen B. H. 2002. The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 96: 67–202. [DOI] [PubMed] [Google Scholar]
- 70.Verma S., Singh A., Mishra A. 2012. Molecular construction of NADH-cytochrome b5 reductase inhibition by flavonoids and chemical basis of difference in inhibition potential: molecular dynamics simulation study. J. Appl. Pharm. Sci. 2: 33–39. [Google Scholar]
- 71.Nishida H., Inaka K., Yamanaka M., Kaida S., Kobayashi K., Miki K. 1995. Crystal structure of NADH-cytochrome b5 reductase from pig liver at 2.4 A resolution. Biochemistry. 34: 2763–2767. [DOI] [PubMed] [Google Scholar]
- 72.Bando S., Takano T., Yubisui T., Shirabe K., Takeshita M., Nakagawa A. 2004. Structure of human erythrocyte NADH-cytochrome b5 reductase. Acta Crystallogr. D Biol. Crystallogr. 60: 1929–1934. [DOI] [PubMed] [Google Scholar]
- 73.Yamada M., Tamada T., Takeda K., Matsumoto F., Ohno H., Kosugi M., Takaba K., Shoyama Y., Kimura S., Kuroki R., et al. 2013. Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer. J. Mol. Biol. 425: 4295–4306. [DOI] [PubMed] [Google Scholar]
- 74.Triola G., Fabriàs G., Casas J., Llebaria A. 2003. Synthesis of cyclopropene analogues of ceramide and their effect on dihydroceramide desaturase. J. Org. Chem. 68: 9924–9932. [DOI] [PubMed] [Google Scholar]
- 75.Idkowiak-Baldys J., Apraiz A., Li L., Rahmaniyan M., Clarke C. J., Kraveka J. M., Asumendi A., Hannun Y. A. 2010. Dihydroceramide desaturase activity is modulated by oxidative stress. Biochem. J. 427: 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tonelli F., Alossaimi M., Williamson L., Tate R. J., Watson D. G., Chan E., Bittman R., Pyne N. J., Pyne S. 2013. The sphingosine kinase inhibitor 2-(p-hyroxyanilino)-4-(p-chlorophenyl)thiazole reduces androgen receptor expression via an oxidative stress-dependent mechanism. Br. J. Pharmacol. 168: 1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ricci C., Onida F., Servida F., Radaelli F., Saporiti G., Todoerti K., Deliliers G. L., Ghidoni R. 2009. In vitro anti-leukaemia activity of sphingosine kinase inhibitor. Br. J. Haematol. 144: 350–357. [DOI] [PubMed] [Google Scholar]
- 78.Kohno M., Momoi M., Oo M. L., Paik J-H., Lee Y-M., Venkataraman K., Ai Y., Ristimaki A. P., Fyrst H., Sano H., et al. 2006. Intracellular role for sphingosine kinase 1 in intestinal adenoma cell proliferation. Mol. Cell. Biol. 26: 7211–7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taha T. A., Kitatani K., El-Alwani M., Bielawski J., Hannun Y. A., Obeid L. M. 2006. Loss of sphingosine kinase-1 activates the intrinsic pathway of programmed cell death: modulation of sphingolipid levels and the induction of apoptosis. FASEB J. 20: 482–484. [DOI] [PubMed] [Google Scholar]
- 80.Gao P., Smith C. D. 2011. Ablation of sphingosine kinase-2 inhibits tumor cell proliferation and migration. Mol. Cancer Res. 9: 1509–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Van Brocklyn J. R., Jackson C. A., Pearl D. K., Kotur M. S., Snyder P. J., Prior T. W. 2005. Sphingosine kinase-1 expression correlates with poor survival of patients with glioblastoma multiforme: roles of sphingosine kinase isoforms in growth of glioblastoma cell lines. J. Neuropathol. Exp. Neurol. 64: 695–705. [DOI] [PubMed] [Google Scholar]
- 82.Zhang H., Li W., Sun S., Yu S., Zhang M., Zou F. 2012. Inhibition of sphingosine kinase 1 suppresses proliferation of glioma cells under hypoxia by attenuating activity of extracellular signal-regulated kinase. Cell Prolif. 45: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gagliostro V., Casas J., Caretti A., Abad J. L., Tagliavacca L., Ghidoni R., Fabrias G., Signorelli P. 2012. Dihydroceramide delays cell cycle G1/S transition via activation of ER stress and induction of autophagy. Int. J. Biochem. Cell Biol. 44: 2135–2143. [DOI] [PubMed] [Google Scholar]
- 84.Spassieva S. D., Rahmaniyan M., Bielawski J., Clarke C. J., Kraveka J. M., Obeid L. M. 2012. Cell density-dependent reduction of dihydroceramide desaturase activity in neuroblastoma cells. J. Lipid Res. 53: 918–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou W., Ye X-L., Sun Z-J., Ji X-D., Chen H-X., Xie D. 2009. Overexpression of degenerative spermatocyte homolog 1 up-regulates the expression of cyclin D1 and enhances metastatic efficiency in esophageal carcinoma Eca109 cells. Mol. Carcinog. 48: 886–894. [DOI] [PubMed] [Google Scholar]
- 86.Bedia C., Levade T., Codogno P. 2011. Regulation of autophagy by sphingolipids. Anticancer. Agents Med. Chem. 11: 844–853. [DOI] [PubMed] [Google Scholar]
- 87.Beljanski V., Knaak C., Smith C. D. 2010. A novel sphingosine kinase inhibitor induces autophagy in tumor cells. J. Pharmacol. Exp. Ther. 333: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.