Abstract
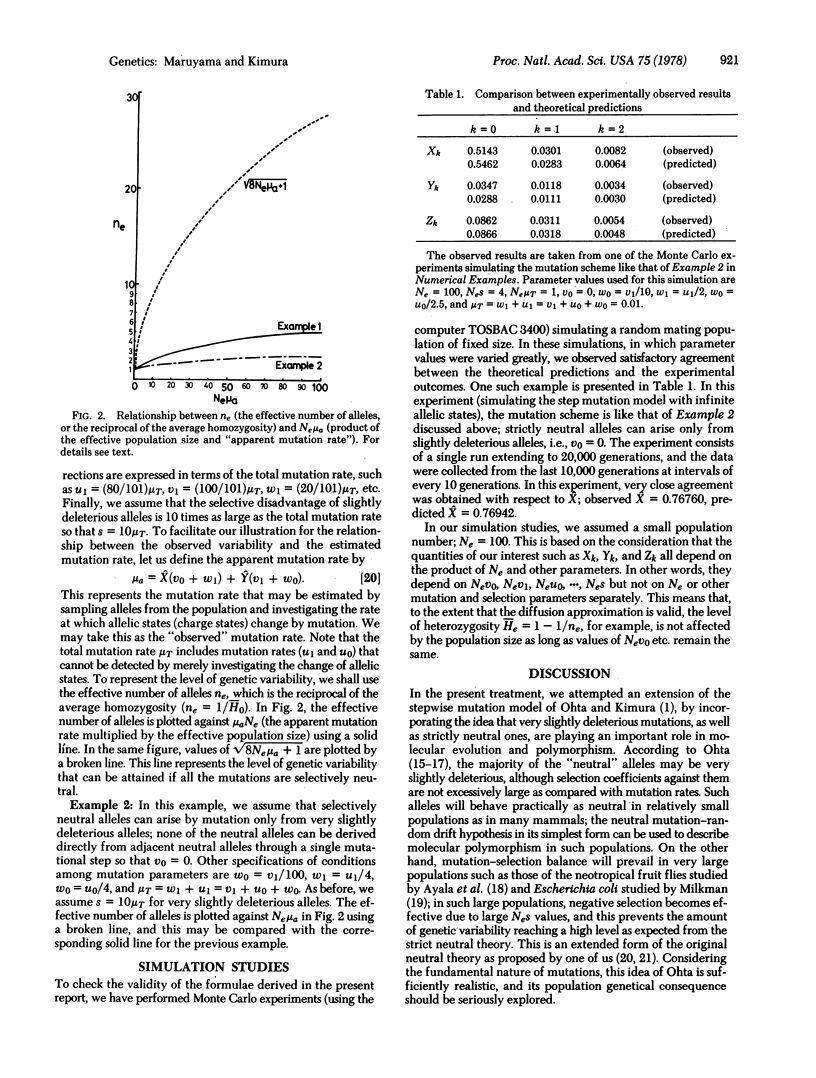
Mathematical treatments are presented that enable us to compute the amount of genetic variability maintained in a finite population, assuming that mutations occur in stepwise fashion and that both selectively neutral and slightly deleterious alleles are involved. Two numerical examples show that, if very slightly deleterious mutations are prevalent, the amount of genetic variability increases much more slowly as the population number increases than is the case when all the mutations are strictly neutral.
Full text
PDF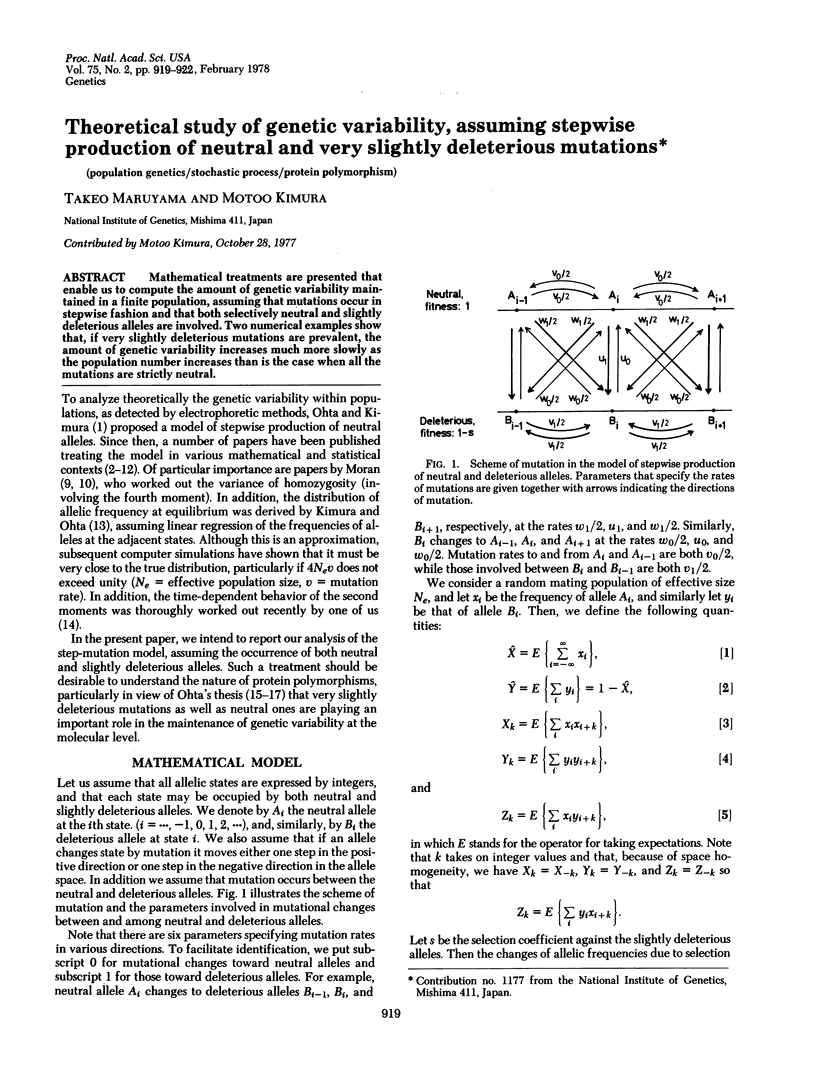


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avery P. J. Extensions to the model of an infinite number of selectively neutral alleles in a finite population. Genet Res. 1975 Apr;25(2):145–153. doi: 10.1017/s0016672300015548. [DOI] [PubMed] [Google Scholar]
- Ayala F. J., Powell J. R., Tracey M. L., Mourão C. A., Pérez-Salas S. Enzyme variability in the Drosophila willistoni group. IV. Genic variation in natural populations of Drosophila willistoni. Genetics. 1972 Jan;70(1):113–139. doi: 10.1093/genetics/70.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. H., Marshall D. R., Albrecht L. Profiles of electrophoretic alleles in natural populations. Genet Res. 1975 Apr;25(2):137–143. doi: 10.1017/s0016672300015536. [DOI] [PubMed] [Google Scholar]
- Chakraborty R., Nei M. Hidden genetic variability within electromorphs in finite populations. Genetics. 1976 Oct;84(2):385–393. doi: 10.1093/genetics/84.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIMURA M., CROW J. F. THE NUMBER OF ALLELES THAT CAN BE MAINTAINED IN A FINITE POPULATION. Genetics. 1964 Apr;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968 Feb 17;217(5129):624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. Genetic variability maintained in a finite population due to mutational production of neutral and nearly neutral isoalleles. Genet Res. 1968 Jun;11(3):247–269. doi: 10.1017/s0016672300011459. [DOI] [PubMed] [Google Scholar]
- Kimura M., Ohta T. Distribution of allelic frequencies in a finite population under stepwise production of neutral alleles. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2761–2764. doi: 10.1073/pnas.72.7.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman J. F. A note on multidimensional models of neutral mutation. Theor Popul Biol. 1977 Jun;11(3):285–290. doi: 10.1016/0040-5809(77)90012-0. [DOI] [PubMed] [Google Scholar]
- Li W. H. A Mixed Model of Mutation for Electrophoretic Identity of Proteins within and between Populations. Genetics. 1976 Jun;83(2):423–432. doi: 10.1093/genetics/83.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H. Electrophoretic identity of proteins in a finite population and genetic distance between taxa. Genet Res. 1976 Oct;28(2):119–127. doi: 10.1017/s0016672300016803. [DOI] [PubMed] [Google Scholar]
- Li W. H. Maintenance of genetic variability under mutation and selection pressures in a finite population. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2509–2513. doi: 10.1073/pnas.74.6.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall D. R., Brown A. H. The charge-state model of protein polymorphism in natural populations. J Mol Evol. 1975 Nov 4;6(3):149–163. doi: 10.1007/BF01732353. [DOI] [PubMed] [Google Scholar]
- Milkman R. Electrophoretic variation in Escherichia coli from natural sources. Science. 1973 Dec 7;182(4116):1024–1026. doi: 10.1126/science.182.4116.1024. [DOI] [PubMed] [Google Scholar]
- Moran P. A. Wandering distributions and the electrophoretic profile. II. Theor Popul Biol. 1976 Oct;10(2):145–149. doi: 10.1016/0040-5809(76)90012-5. [DOI] [PubMed] [Google Scholar]
- Moran P. A. Wandering distributions and the electrophoretic profile. Theor Popul Biol. 1975 Dec;8(3):318–330. doi: 10.1016/0040-5809(75)90049-0. [DOI] [PubMed] [Google Scholar]
- Mukai T., Cockerham C. C. Spontaneous mutation rates at enzyme loci in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2514–2517. doi: 10.1073/pnas.74.6.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Estimation of mutation rate from rare protein variants. Am J Hum Genet. 1977 May;29(3):225–232. [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Kimura M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet Res. 1973 Oct;22(2):201–204. doi: 10.1017/s0016672300012994. [DOI] [PubMed] [Google Scholar]
- Ohta T. Role of very slightly deleterious mutations in molecular evolution and polymorphism. Theor Popul Biol. 1976 Dec;10(3):254–275. doi: 10.1016/0040-5809(76)90019-8. [DOI] [PubMed] [Google Scholar]
- Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973 Nov 9;246(5428):96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- Ota T. Mutational pressure as the main cause of molecular evolution and polymorphism. Nature. 1974 Nov 29;252(5482):351–354. doi: 10.1038/252351a0. [DOI] [PubMed] [Google Scholar]
- Wehrhahn C. F. The evolution of selectively similar electrophoretically detectable alleles in finite natural populations. Genetics. 1975 Jun;80(2):375–394. doi: 10.1093/genetics/80.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B. S. Testing for selective neutrality of electrophoretically detectable protein polymorphisms. Genetics. 1976 Nov;84(3):639–659. doi: 10.1093/genetics/84.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]


