Abstract
The cytosolic sensor MDA5 is crucial for antiviral innate immune defense against various RNA viruses including measles virus; as such, many viruses have evolved strategies to antagonize the antiviral activity of MDA5. Here, we show that measles virus escapes MDA5 detection by targeting the phosphatases PP1α and PP1γ, which regulate MDA5 activity by removing an inhibitory phosphorylation mark. The V proteins of measles virus and the related paramyxovirus Nipah virus interact with PP1α/γ, preventing PP1-mediated dephosphorylation of MDA5 and thereby its activation. The PP1 interaction with the measles V protein is mediated by a conserved PP1-binding motif in the C-terminal region of the V protein. A recombinant measles virus expressing a mutant V protein deficient in PP1 binding is unable to antagonize MDA5 and is growth-impaired due to its inability to suppress interferon induction. This identifies PP1 antagonism as a mechanism employed by paramyxoviruses for evading innate immune recognition.
INTRODUCTION
Pattern recognition receptors (PRRs) are critical components of the host’s innate immune sensing apparatus for detecting microbial pathogens in both non-immune and immune cells. PRRs recognize conserved pathogen-associated molecular patterns (PAMPs) and then activate signaling cascades leading to the production of proinflammatory cytokines and type-I interferons (IFN-α/β), ultimately resulting in an antiviral state and activation of adaptive immune responses (Creagh and O'Neill, 2006; Takeuchi and Akira, 2010). Retinoic acid-inducible gene-I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), the best characterized members of the RIG-I-like receptor (RLR) family, play an essential role in cytosolic detection of RNA viruses (Kato et al., 2006; Loo and Gale, 2011; Yoneyama et al., 2004).
RIG-I is activated by 5’triphosphate short dsRNA structures present in negative-strand RNA viruses as well as polyuridine/cytosine motifs in the positive-strand RNA of hepatitis C virus (Hornung et al., 2006; Pichlmair et al., 2006; Saito et al., 2008). In contrast, MDA5 recognizes long dsRNA or RNA web structures produced during the replication cycle of picornaviruses (Kato et al., 2006). Recent studies have also provided evidence that MDA5 acts in concert with RIG-I to respond to certain flaviviruses, reoviruses, and paramyxoviruses, such as measles virus (MV) and Sendai virus (SeV) (Gitlin et al., 2010; Ikegame et al., 2010; Loo et al., 2008).
Despite their differences in ligand specificity, RIG-I and MDA5 share a common domain structure consisting of tandem caspase activation and recruitment domains (CARDs) at the N-terminus that are necessary and sufficient for signal transduction, as well as a helicase/ATPase domain and a C-terminal domain (CTD), both of which are important for RNA recognition (Cui et al., 2008; Yoneyama et al., 2004). Once activated, RIG-I and MDA5 form a complex with the mitochondrial-localized adaptor molecule MAVS/VISA/IPS-1/Cardif, resulting in downstream signaling to orchestrate activation of the transcription factors NF-κB, AP1 and IRF3/7, leading to IFN-α/β gene expression (Loo and Gale, 2011).
Recent studies demonstrated that host cells are equipped with an elegant system for regulating RLR-induced signaling to avoid aberrant or premature immune activation (Eisenacher and Krug, 2012; Loo and Gale, 2011). Posttranslational modifications of the N-terminal CARDs as well as conformational changes induced by the CTD have been shown to play an important role in regulating RLR signaling activities (Gack et al., 2007; Saito et al., 2007). Recently, we demonstrated that RIG-I and MDA5 signaling activities are tightly controlled by an intricate balance of phosphorylation and dephosphorylation of their CARDs, and identified the phosphatase PP1 – specifically PP1α and PP1γ isoforms – as key regulators of RIG-I and MDA5 activation (Gack et al., 2010; Maharaj et al., 2012; Nistal-Villán et al., 2010; Wies et al., 2013). In uninfected cells, RIG-I and MDA5 signaling is prevented by constitutive phosphorylation of specific serine/threonine residues located in the CARDs: serine-8 (S8) and threonine-170 (T170) in RIG-I and serine-88 (S88) in MDA5. Upon binding to RNA ligands, RIG-I and MDA5 are dephosphorylated by PP1α/γ, allowing RLR interaction with MAVS and IFN-α/β induction (Wies et al., 2013).
Paramyxoviruses are enveloped, non-segmented, negative-strand RNA viruses comprising various human and animal pathogens including MV, mumps virus, parainfluenza virus 5 (PIV5), and the newly emerging Nipah (NiV) and Hendra viruses. To combat recognition and clearance by the immune system, these viruses have evolved sophisticated mechanisms to antagonize both IFN induction and IFN receptor signal transduction (Bowie and Unterholzner, 2008; Horvath, 2004). This immunosuppression is particularly well-known for MV; in fact, many cases of mortality associated with MV infection are due to its potent inhibition of innate and adaptive immune responses (Moss et al., 2004).
The IFN-antagonistic activity of paramyxoviruses is due to one or more gene products of the paramyxovirus P/C/V gene, which encodes the essential phospho-(P) protein and – through alternative reading frames or RNA editing – the virulence factors C, V, and/or W proteins. Of the three, the V protein is the best characterized IFN antagonist. A major target of paramyxovirus V proteins is the immune sensor MDA5 (Andrejeva et al., 2004; Childs et al., 2007; Parisien et al., 2009; Rodriguez and Horvath, 2013); however, the molecular mechanisms by which the V proteins inhibit MDA5 activity have only begun to be elucidated. It has been shown that the V proteins physically interact with the helicase domain of MDA5, inhibiting its ATPase activity and thereby MDA5 filament formation (Childs et al., 2009). Recently, co-crystal structure analysis of porcine MDA5 and the V protein of PIV5 provided evidence that the V protein inhibits the ATP hydrolysis activity of MDA5 through structural unfolding (Motz et al., 2013). Given the large diversity of IFN-antagonistic strategies employed by members of the paramyxovirus family, the mechanism of MDA5 inhibition by their V proteins is likely more complex than the present data suggests.
Here we identify a mechanism of MDA5 inhibition employed by MV and NiV in which their V proteins target PP1α and PP1γ, two essential activators of MDA5 signaling. PP1α/γ binding of the V protein prevents dephosphorylation of MDA5 and thereby its activation. Generation of a PP1 binding-deficient recombinant (r) MV demonstrates that PP1 antagonism by the V protein is an important mechanism for suppressing MDA5-mediated type-I IFN induction.
RESULTS
The V protein of measles and Nipah virus suppresses MDA5 S88 dephosphorylation
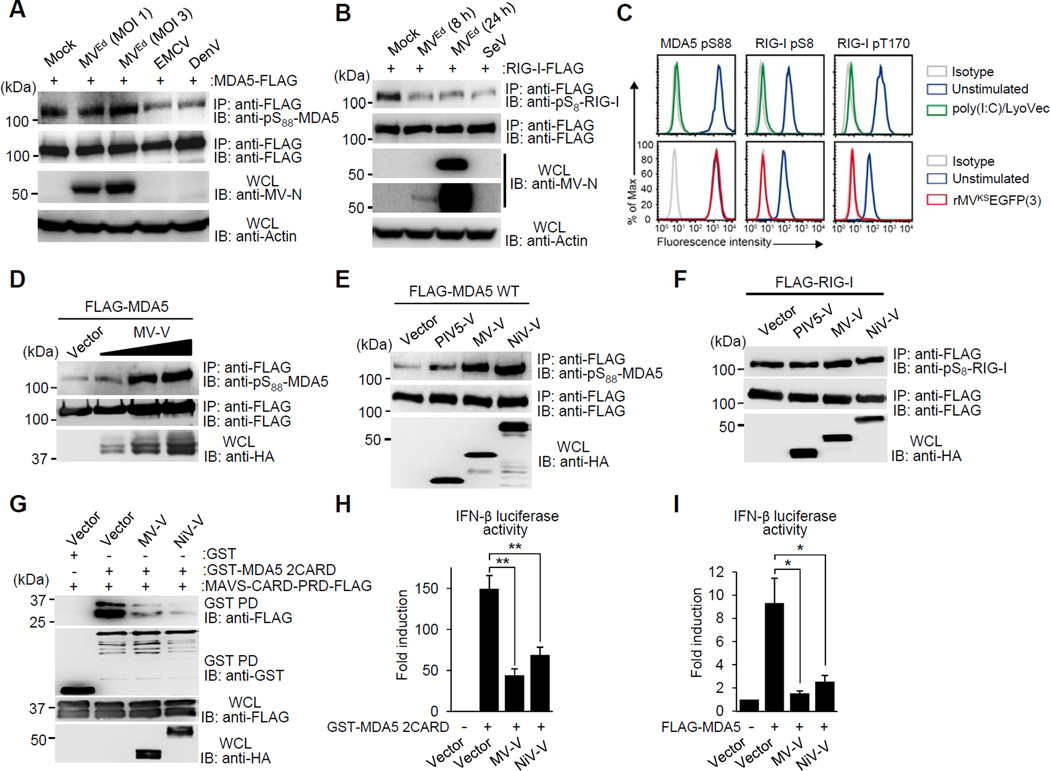
MDA5 signaling activity is tightly regulated by constitutive phosphorylation, keeping it inactive. Upon viral RNA sensing, MDA5 is activated via dephosphorylation by PP1, allowing innate immune signaling (Wies et al., 2013). We hypothesized that viruses may modulate this intricate balance of phosphorylation/dephosphorylation to prevent MDA5 activation, and thus escape innate immune detection. To address this, we tested three RNA viruses, all known to be sensed by MDA5, for their effect on the S88 phosphorylation of FLAG-MDA5 by immunoblot (IB) using a phospho-(p)-S88-MDA5 specific antibody: encephalomyocarditis virus (EMCV), a picornavirus; dengue virus (DenV), a flavivirus; and MV, a paramyxovirus (Figure 1A). Infection with EMCV and DenV induced MDA5 S88 dephosphorylation, indicative of MDA5 activation (Figure 1A). In striking contrast, the Edmonston (Ed) vaccine strain of MV (MVEd) did not induce any change in MDA5 S88 phosphorylation levels, even at a high MOI (Figure 1A), suggesting that MVEd modulates the MDA5 phosphorylation state. However, MVEd triggered RIG-I S8 dephosphorylation as efficiently as SeV, a virus known to induce RIG-I activation (Figure 1B). In line with this, while transfection of polyinosine-polycytidylic acid [poly(I:C)], a potent MDA5 agonist, efficiently triggered S88 dephosphorylation of endogenous MDA5 in primary human dendritic cells (DCs), infection with an enhanced green fluorescent protein (EGFP)-expressing rMV based on the wild-type (WT) Khartoum-Sudan (KS) strain [rMVKSEGFP(3)] did not change the MDA5 S88 phosphorylation levels compared to uninfected cells (Figure 1C). However, rMVKS infection efficiently induced RIG-I S8 and T170 dephosphorylation in DCs at this time point (Figure 1C). Collectively, these results suggest that MV, but not DenV and EMCV, blocks the dephosphorylation of the sensor MDA5.
Figure 1. The paramyxovirus V protein inhibits MDA5 S88 dephosphorylation.
(A) S88 phosphorylation of FLAG-MDA5 in HEK293T infected with MVEd (MOI 1 or 3), or DenV (MOI 5) for 24 h, or with EMCV (MOI 0.5) for 3 h, assessed by IP with anti-FLAG, followed by immunoblot (IB) with anti-pS88-MDA5. Efficient MV infection was determined by IB with anti-MV nucleoprotein (MV-N). (B) S8 phosphorylation of FLAG-RIG-I in HEK293T, either left uninfected or infected with MVEd (MOI 3) for 8 or 24 h, or SeV (50 HAU/ml) for 24 h, assessed by IP with anti-FLAG, followed by IB with anti-pS8-RIG-I. Expression of MV-N was determined as in (A), two different blot exposures are shown. (C) Endogenous MDA5 S88 and RIG-I S8 or T170 phosphorylation in human DCs either unstimulated, treated with poly(I:C)-LyoVec for 3 h (upper panels), or infected with rMVKSEGFP(3) WT for 16 h (lower panels), determined by flow cytometry using phospho-specific pS88-MDA5, pS8-RIG-I and pT170-RIG-I antibodies. Data are representative of three individual donors. (D) S88 phosphorylation of FLAG-MDA5 in HEK293T upon expression of increasing amounts of HA-MV-V protein, assessed as in (A). (E and F) IB analysis of FLAG-MDA5 S88 (E) or FLAG-RIG-I S8 (F) phosphorylation upon expression of HA-tagged V proteins, determined by IP with anti-FLAG and IB using the indicated phospho-antibodies. (G) Interaction of GST-MDA5-2CARD with MAVS-CARD-PRD-FLAG in the presence of HA-MV-V or HA-NiV-V, assessed by GST-pull down (GST-PD). (H and I) IFN-β luciferase activity in HEK293T cells transfected with GST-MDA5 2CARD (H) or FLAG-MDA5 (I) together with vector, MV-V, or NiV-V, normalized to constitutive pGK-β-gal. The results are expressed as means +/− s.d. (n=3). *P < 0.05; **P < 0.01. See also Figure S1.
The V proteins of various paramyxoviruses, including MV, have been shown to interact with and inhibit MDA5 (Andrejeva et al., 2004; Childs et al., 2007); however, the molecular mechanism of this viral antagonism has only begun to be elucidated. We therefore sought to determine whether the MDA5 phosphorylation-modulating activity of MV was caused by the V protein. To address this, we transfected increasing amounts of the MV V protein (MV-V) together with FLAG-MDA5 (Figure 1D). Overexpressed FLAG-MDA5, in contrast to endogenous MDA5, is not fully phosphorylated due to a low-level, constitutive binding to PP1α/γ (Wies et al., 2013), allowing examination of interference with the MDA5 S88 dephosphorylation by PP1. Indeed, MV-V strongly enhanced MDA5 S88 phosphorylation in a dose-dependent manner, indicating interference with MDA5 dephosphorylation. The V protein of NiV also robustly enhanced MDA5 S88 phosphorylation, while that of PIV5 only marginally increased S88 phosphorylation levels (Figures 1E and S1A). Notably, MV-V and NiV-V did not affect RIG-I CARD phosphorylation, indicating that the V protein specifically modulates the phosphorylation state of MDA5, but not RIG-I (Figure 1F and S1B).
Paramyxovirus V proteins have been reported to bind to the helicase domain of MDA5, leading to disruption of the ATP hydrolysis site (Motz et al., 2013). Residue R806 in MDA5 was shown to be critical for V-MDA5 interaction as mutation to leucine (R806L) abrogated or strongly reduced the binding of paramyxovirus V proteins to the helicase (Motz et al., 2013; Rodriguez and Horvath, 2013). To address whether modulation of MDA5 CARD phosphorylation and disruption of ATP hydrolysis are dependent or independent activities of the V protein, we determined the effect of the V protein on S88 phosphorylation of the MDA5 R806L mutant (Figure S1C). As with WT MDA5, ectopic expression of MV-V and NiV-V strongly increased S88 phosphorylation of MDA5 R806L, while PIV5-V had only a marginal effect (Figure S1C). These results indicate that the ability of MV-V and NiV-V to modulate MDA5 phosphorylation is independent of binding to the ATP-hydrolysis domain.
In uninfected cells, MDA5 S88 phosphorylation prevents MDA5 binding to the downstream adaptor MAVS; in contrast, viral RNA binding induces S88 dephosphorylation, allowing MAVS binding and leading to IFN gene expression (Wies et al., 2013). We therefore examined the effect of MV-V and NiV-V, which robustly increased MDA5 S88 phosphorylation, on the CARD-CARD interaction between MDA5 and MAVS (Figure 1G). In the absence of the V protein, GST-MDA5 2CARD efficiently interacted with the CARD-proline-rich domain of MAVS (MAVS-CARD-PRD); however, co-expressed MV-V or NiV-V potently diminished this interaction (Figure 1G). Crucially, the MV-V and NiV-V proteins did not affect the binding of MAVS-CARD-PRD to an MDA5 mutant in which the S88 residue was replaced with alanine (GST-MDA5 2CARD S88A), reinforcing that the inhibition of MDA5-MAVS binding by the V proteins is a direct effect of their ability to enhance MDA5 S88 phosphorylation (Figure S1D). Consistent with their inhibitory effects on MDA5-MAVS binding, MV-V and NiV-V markedly suppressed the IFN-β promoter activation induced by MDA5 2CARD or full-length MDA5 (Figures 1H and 1I). Collectively, these results indicate that the V proteins of MV and NiV inhibit MDA5 S88 dephosphorylation, thereby suppressing MDA5-MAVS binding and downstream signaling.
The V proteins of MV and NiV interact with the phosphatases PP1α/γ
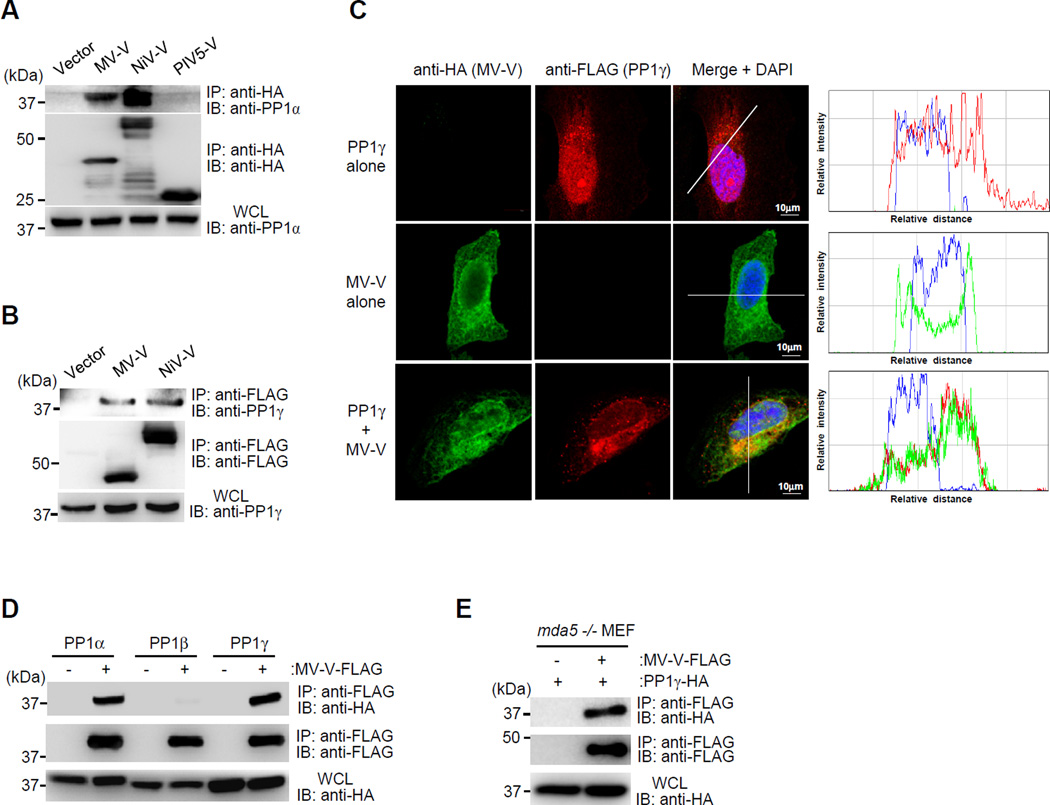
To elucidate the mechanism by which MV-V and NiV-V prevent MDA5 S88 dephosphorylation, we tested their potential interaction with PP1. Co-Immunoprecipitation (Co-IP) showed that MV-V and NiV-V bound efficiently to endogenous PP1α and PP1γ (Figures 2A and 2B). In contrast, PIV5-V did not interact with PP1α/γ (Figures 2A and S2A). Confocal microscopy showed that MV-V preferentially localized to the cytoplasm, with a minor fraction localized to the nucleus, whereas PP1γ was localized both in the nucleus and cytoplasm (Figure 2C). When both proteins were expressed together, however, PP1γ re-localized to the cytoplasm, where it extensively co-localized with MV-V at mitochondria-associated membranes, a subcellular compartment important for MDA5 signaling (Figures 2C and S2B). Furthermore, MV-V bound specifically to PP1α and PP1γ, which dephosphorylate MDA5, but not to PP1β which is not involved in innate immune signaling (Wies et al., 2013) (Figure 2D). To further characterize the PP1-V interaction, we asked whether their binding is mediated by MDA5. To this end, we determined the binding of MV-V to PP1 in mouse embryonic fibroblasts (MEFs) derived from mda5 −/− mice (Figure 2E). MV-V bound efficiently to PP1 in mda5 −/− cells, suggesting that the binding between the V protein and PP1 is not mediated by MDA5 and thus likely to be a direct interaction. These results indicate that the V proteins of MV and NiV, but not that of PIV5, efficiently interact with the phosphatases PP1α and PP1γ.
Figure 2. The V proteins of MV and NiV bind to PP1α/γ.
(A and B) Endogenous PP1α (A) or PP1γ (B) binding to the indicated HA-tagged V proteins in transfected HEK293T cells, determined by Co-IP. (C) Confocal scanning laser images of HA-MV-V (green) and FLAG-PP1γ (red) in transfected HeLa cells. Nuclei were stained with DAPI (blue). Co-localization of MV-V and PP1γ was assessed by histogram profiles of merged images. (D) Co-IP of HA-tagged PP1α, PP1β, or PP1γ and FLAG-MV-V in transfected HEK293T cells. (E) Binding of FLAG-MV-V to HA-PP1γ in transfected mda5 −/− MEFs, assessed by IP with anti-FLAG and IB using anti-HA. See also Figure S2.
The V protein inhibits the PP1-MDA5 interaction and is a substrate for dephosphorylation by PP1
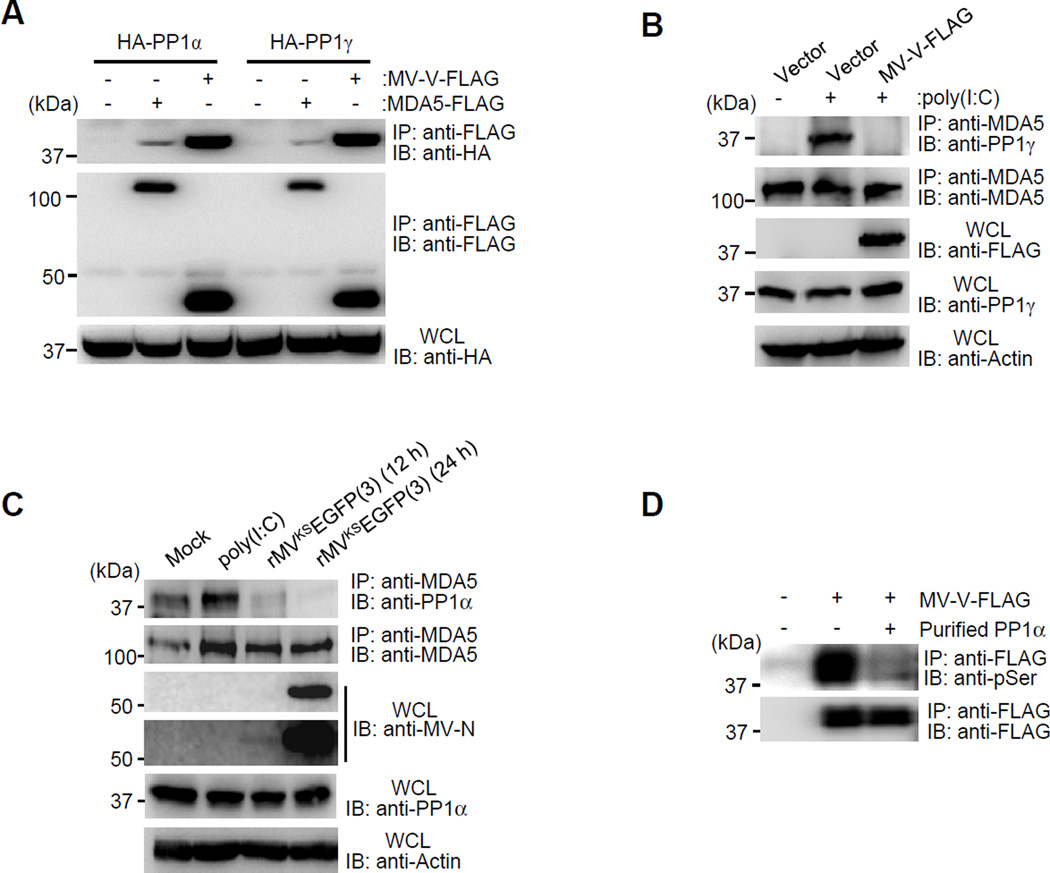
To address the mechanism by which the V protein interaction with PP1α/γ prevents MDA5 dephosphorylation, we asked whether the MV-V protein (i) inhibits PP1’s enzymatic activity, (ii) blocks the interaction of PP1 with MDA5, and (iii) whether it serves as a substrate for PP1-mediated dephosphorylation. As neither measles infection nor MV-V expression suppressed RIG-I dephosphorylation by PP1α/γ (Figures 1B and 1F), it is unlikely that the V protein inhibits the enzymatic activity of PP1. To test whether the V protein competes with MDA5 for PP1 binding, we first compared the PP1α/γ binding of MDA5 and MV-V (Figure 3A). This showed that MV-V had a stronger association with PP1 than did MDA5. Next, we determined the binding of endogenous MDA5 to PP1 induced by poly(I:C) in the presence or absence of MV-V (Figure 3B). Poly(I:C) stimulation efficiently triggered PP1 binding to MDA5 in the absence of MV-V; in contrast, no MDA5-PP1 interaction was observed in poly(I:C)-stimulated cells expressing MV-V (Figure 3B). Finally, we examined endogenous PP1-MDA5 binding in poly(I:C)-stimulated or rMVKS-infected A549 lung epithelial cells stably expressing the entry receptor for WT strains of MV, CD150 (A549-hCD150) (Figure 3C). While poly(I:C) stimulation increased PP1 binding to MDA5, there was a near-complete loss of PP1-MDA5 interaction in rMVKS-infected cells (Figure 3C).
Figure 3. The V protein inhibits PP1 binding to MDA5 and serves as a PP1 substrate.
(A) Binding of FLAG-MDA5 or FLAG-MV-V binding to HA-PP1α and HA-PP1γ in transfected HEK293T cells, determined as described in Fig. 2E. (B) Endogenous MDA5-PP1γ binding in Mock-treated or poly(I:C)-stimulated HEK293T cells, transfected with vector or FLAG-MV-V, assessed by IP with anti-MDA5, followed by IB with anti-PP1γ. (C) Endogenous MDA5-PP1α binding in Mock-treated, poly(I:C)-stimulated, or rMVKSEGFP(3)-infected A549-hCD150 cells, determined by Co-IP. Efficient MV infection was determined by IB with anti-MV-N (two different blot exposures shown). (D) In vitro dephosphorylation of FLAG-MV-V by purified PP1α, assessed by IB analysis using anti-pSer. See also Figure S3.
Paramyxovirus P and V proteins are phosphorylated at multiple Ser/Thr sites (Shiell et al., 2003). The kinases responsible for V protein phosphorylation have been identified (Lu et al., 2008; Ludlow et al., 2008; Pfaller and Conzelmann, 2008), but the phosphatase(s) responsible for its dephosphorylation are unknown. We thus tested whether MV-V, upon binding to PP1, serves as a substrate for dephosphorylation by PP1. We first determined the phosphorylation of FLAG-MV-V after treatment with phosphatase inhibitors specific for PP1 and/or the related phosphatase PP2A (Figure S3A). This showed that specific inhibitors of both PP1/PP2A (Calyculin A and Cantharidic acid), but not the PP2A-specific inhibitor Endothall, or an alkaline phosphatase inhibitor (Bromotetramisole), greatly enhanced the phosphorylation of MV-V, evident from a stronger signal in IB using a pan-phospho-Ser antibody, as well as a shift of the bands of MV-V, representing its phosphorylated forms (Figure S3A). In support of this, while treatment with Cantharidic acid at concentrations inhibiting only PP2A did not affect the phosphorylation state of MV-V, high concentrations of Cantharidic acid also blocking PP1 activity induced a shift in the multiple-band pattern of MV-V, indicating its enhanced phosphorylation (Figure S3B). Finally, purified PP1α efficiently dephosphorylated MV-V in an in vitro dephosphorylation assay, demonstrating direct enzymatic activity of PP1 towards the V protein (Figure 3D). Together, these results indicate that the MV-V protein blocks PP1 binding to MDA5 and serves as a substrate for PP1-mediated dephosphorylation, preventing MDA5 S88 dephosphorylation.
A PP1 binding-deficient V protein is unable to inhibit MDA5 dephosphorylation but retains MDA5 binding and STAT inhibition activities
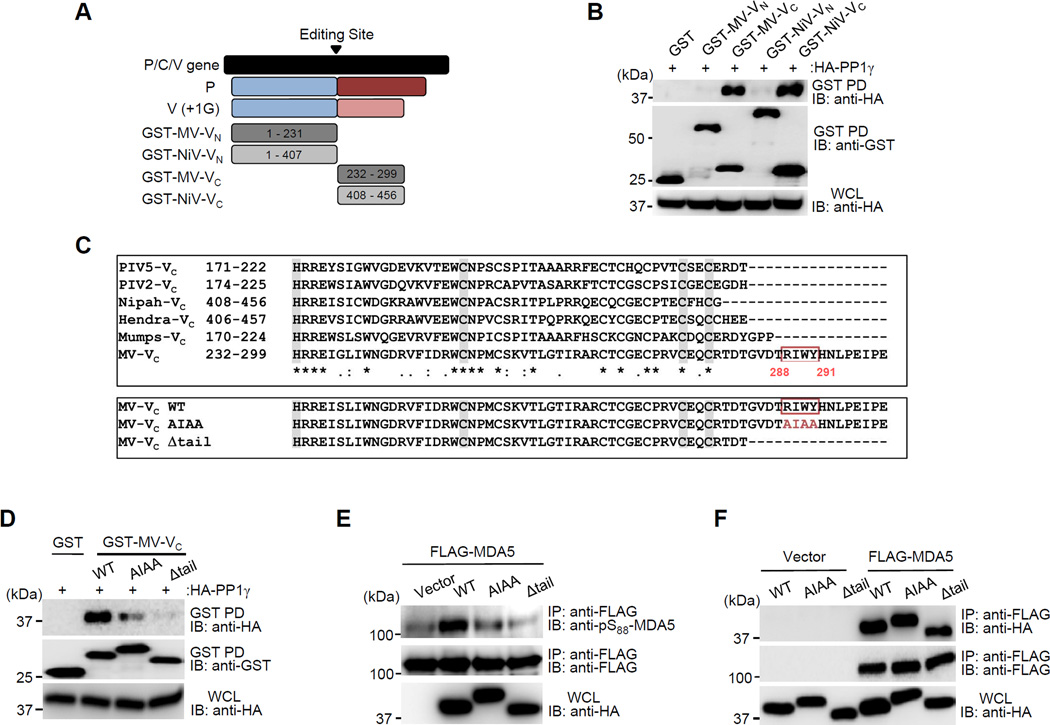
We next sought to identify the site in the V protein that is necessary for PP1 binding. The paramyxovirus V protein is expressed through RNA editing of the P/C/V gene (Cattaneo et al., 1989; Thomas et al., 1988). As such, it shares the N-terminal sequence (VN) with the P and W proteins, but has a unique, cysteine-rich C-terminal domain (VC) which is responsible for many of its specific effector functions (Figure 4A). We thus determined the PP1-binding capacity of the P/V/W-shared VN and the unique VC of the MV-V and NiV-V proteins. Co-IP studies revealed that specifically the MV-VC and NiV-VC bound to PP1, while there was no interaction of PP1 with their VN domains (Figure 4B). Consistent with this binding mode, ectopic expression of MV-VC or NiV-VC fragments was sufficient to enhance MDA5 S88 phosphorylation in a dose-dependent manner (Figures S3C and S3D).
Figure 4. PP1 binding of the measles V protein is required for its inhibitory effect on MDA5 S88 dephosphorylation.
(A) Domain structures of the paramyxovirus P and V proteins, and schematic representation of GST-fused truncation constructs of MV-V and NiV-V. Numbers indicate amino acids. (B) Binding of HA-PP1γ to GST-fused VC or VN of MV or NiV in transfected HEK293T cells, assessed by GST-PD and IB using anti-HA. (C) (Upper panel) Protein sequence alignment of the VC of paramyxoviruses. Alignment was performed using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/). Asterisks (*) indicate a single, fully conserved residue. Colons (:) indicate conservation between groups of strongly similar properties, and periods (.) indicate conservation between groups of weakly similar properties. Conserved residues of the zinc-finger motif responsible for MDA5 interaction are highlighted in grey. (Lower panel) Protein sequences of the MV-Vc WT, AIAA and Δtail mutant. Identified PP1-binding motif in MV-Vc is indicated (red box). Numbers indicate amino acids. (D) Interaction of HA-PP1γ to GST-fused MV-V WT or mutant proteins in transfected HEK293T cells, determined by GST-PD. (E) S88 phosphorylation of FLAG-MDA5 in HEK293T expressing HA MV-V WT or mutant proteins, analyzed by IP. (F) Binding of FLAG-MDA5 to HA-MV-V WT or mutant proteins, determined by Co-IP. See also Figure S3– 5.
A hallmark of PP1-binding proteins is the presence of defined PP1-binding motifs (Roy and Cyert, 2009). Indeed, sequence alignment of the VC of several paramyxoviruses revealed that MV-V harbors a canonical PP1-binding motif, R/K-x(0,1)-V/I-x-F/W/Y (288RIWY291), in the extreme C-terminal portion of VC (from now on referred to as C-terminal ‘tail’) (Figure 4C, upper panel). This PP1-binding motif was conserved among various vaccine and WT strains of MV (Figure S4). In contrast, no conventional PP1-binding motif was identified in the VC of the other paramyxoviruses.
To determine the role of PP1 binding for MDA5 antagonism by the MV-V protein, key residues in the identified PP1-binding motif were mutated to alanine (MV-V AIAA). In addition, a truncation mutant of MV-V was generated in which the C-terminal tail region (aa 284–299) containing the PP1 motif was deleted (MV-VΔtail) (Figure 4C, lower panel). These mutant V proteins were tested for their PP1-binding ability (Figure 4D). While MV-V WT efficiently interacted with PP1, the MV-V AIAA and Δtail mutants showed a reduced and near-complete loss of PP1 binding, respectively (Figure 4D). Consistent with its abolished PP1-binding ability, the Δtail V protein did not inhibit PP1-MDA5 binding, nor did it block MDA5 S88 dephosphorylation; in contrast MV-V WT robustly enhanced MDA5 phosphorylation, indicative of inhibition of MDA5 dephosphorylation, while the AIAA mutant only slightly enhanced MDA5 S88 phosphorylation (Figures 4E and Figure S5A). Together, these results identify a classical PP1-binding motif in the C-terminal tail region of the MV-V protein, which is necessary for PP1 binding and suppression of MDA5 CARD dephosphorylation.
To further characterize the PP1 binding-defective MV-V mutant proteins, we first tested their ability to bind MDA5, an activity mediated by conserved histidine and cysteine residues in the VC domain (Figure 4C, upper panel). In contrast to their defective PP1-binding ability, MV-V AIAA and Δtail mutants interacted with MDA5 as efficiently as MV-V WT (Figure 4F). Furthermore, MV-V is known to prevent the nuclear translocation of Signal transducers and activators of transcription (STAT) proteins to suppress IFN receptor signal transduction (Palosaari et al., 2003). Thus, we also tested the MV-V AIAA and Δtail mutant proteins for their ability to block STAT2 nuclear translocation as compared to MV-V WT. Ectopically expressed MV-V AIAA and Δtail proteins prevented STAT2 nuclear translocation as effectively as MV-V WT (Figure S5B). This demonstrates that a mutant V protein in which the PP1-binding motif is deleted, is unable to bind PP1 and inhibit PP1-mediated MDA5 dephosphorylation, but retains MDA5 binding and STAT inhibition.
PP1 antagonism is required for inhibition of type-I IFN induction and optimal replication of MV in epithelial cells
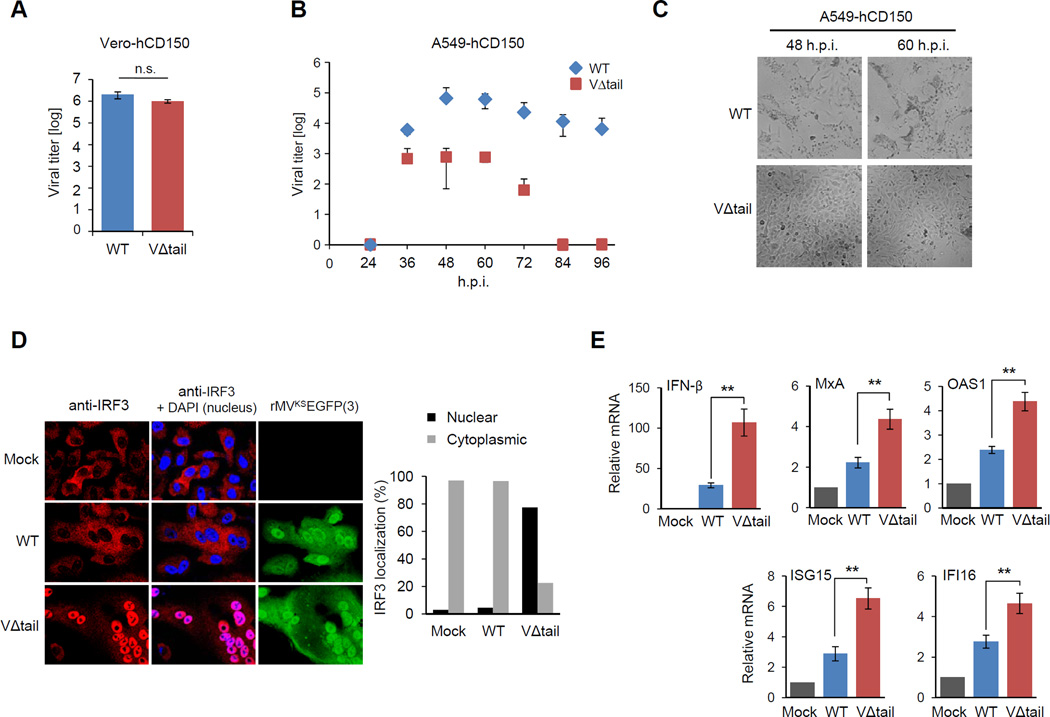
To assess the role of PP1 antagonism by the MV-V protein in innate immune escape of MDA5-mediated IFN induction, we sought to construct an rMV which expresses a mutant V protein deficient in PP1 binding. Based on our biochemical characterization of the MV-V Δtail mutant protein, we argued that an rMV expressing a Δtail V protein will reveal the contribution of PP1 antagonism to IFN suppression and virus replication. We therefore designed a cloning strategy that – by introducing two stop codons in the P/C/V gene – resulted in the deletion of the C-terminal tail region (aa 288–299) of the MV-V protein while leaving P protein expression unaffected (Figure S6A). Using reverse genetics methodology as previously described (Lemon K, 2011), we then generated an EGFP-expressing rMVKS harboring the MV-VΔtail (rMVKSEGFP(3)-VΔtail, referred to as ‘VΔtail virus’). We first examined the replication of the VΔtail virus compared to the parental rMVKSEGFP(3) WT virus (referred to as ‘WT virus’) in Vero cells, which are IFN-defective (Figure 5A). Both viruses replicated with comparable efficiency, indicating that the VΔtail virus does not have a general growth defect (Figure 5A). In striking contrast, the VΔtail virus exhibited profoundly reduced replication capacity compared to WT virus in A549-hCD150 lung epithelial cells, which have an intact IFN system: the titers of the VΔtail virus were reduced by ~2 and ~3 logs compared to WT virus at 60 h.p.i. and 72 h.p.i., respectively. At later time points, the VΔtail virus was not detectable, whereas the WT virus still replicated efficiently (Figure 5B). Depletion of endogenous MDA5 enhanced the replication of the VΔtail virus to titers comparable to those of WT virus, indicating that the reduced replication capacity of the VΔtail virus in A549-hCD150 cells is indeed due to its inability to antagonize MDA5 (Figures S6B and S6C). Consistent with its impaired replication, the VΔtail virus had a greatly reduced ability to induce syncytium formation and cause cell death of A549-hCD150 cells; in contrast, cells infected with the WT virus formed large syncytia and died more rapidly (Figure 5C).
Figure 5. An rMV expressing a PP1-binding deficient mutant V protein shows diminished replication and IFN antagonism in human epithelial cells.
(A) Replication of rMVKSEGFP(3) WT and VΔtail virus in Vero-hCD150 cells infected at an MOI of 0.02. Viral titers were determined at 60 h.p.i. and are expressed as mean 50% tissue culture infectious dose (TCID50/ml) +/− s.d. (n=3). (B) A549-hCD150 cells were infected with WT or VΔtail virus (MOI 0.02). Virus titers in the supernatant were determined by endpoint titration in Vero-hCD150 cells, and expressed as TCID50/ml. (C) Bright-field images of A549-hCD150 cells infected with WT or VΔtail virus at 48 h.p.i. and 60 h.p.i. (D) Confocal microscopy images of endogenous IRF3 (red) in rMVKSEGFP(3) WT or VΔtail virus-infected A549-hCD150 cells (green) at 18 h.p.i. Nuclei were stained with DAPI (blue). Quantification of the percentage of cells with nuclear or cytoplasmic IRF3 (200 cells counted) (right) (E–I) A549-hCD150 cells were infected with WT or VΔtail virus (MOI 0.05). 24 h later, total RNA was extracted and transcript levels of IFNβ and ISGs were determined by quantitative real-time PCR. Transcript levels were normalized to GAPDH and are expressed as fold levels compared to mock-infected cells. Data are expressed as means +/− s.d. (n=3). **P<0.01. See also Figure S6.
We argued that the growth defect of the VΔtail virus in A549-hCD150 cells is due to its diminished ability to suppress the IFN induction pathway activated by MDA5. To address this, we determined the ability of WT and VΔtail virus to induce the nuclear translocation of IRF3, a transcription factor required for type-I IFN gene expression. Infection with the VΔtail virus efficiently triggered nuclear translocation of endogenous IRF3; whereas, cells infected with WT virus exhibited primarily cytoplasmic IRF3 localization, comparable to uninfected cells (Figure 5D). Consistently, infection with the VΔtail virus resulted in significantly higher mRNA levels of IFN-β and the interferon-inducible genes (ISGs), MxA, OAS1, ISG15, and IFI16, compared to infection with WT virus (Figure 5E). Taken together, these results show that an rMV harboring a mutant V protein that is deficient in PP1 binding efficiently induces IRF3-mediated type-I IFN induction in lung epithelial cells, and thus has impaired growth kinetics.
The PP1-binding deficient VΔtail virus is unable to suppress MDA5 S88 dephosphorylation and type-I IFN induction in primary human DCs
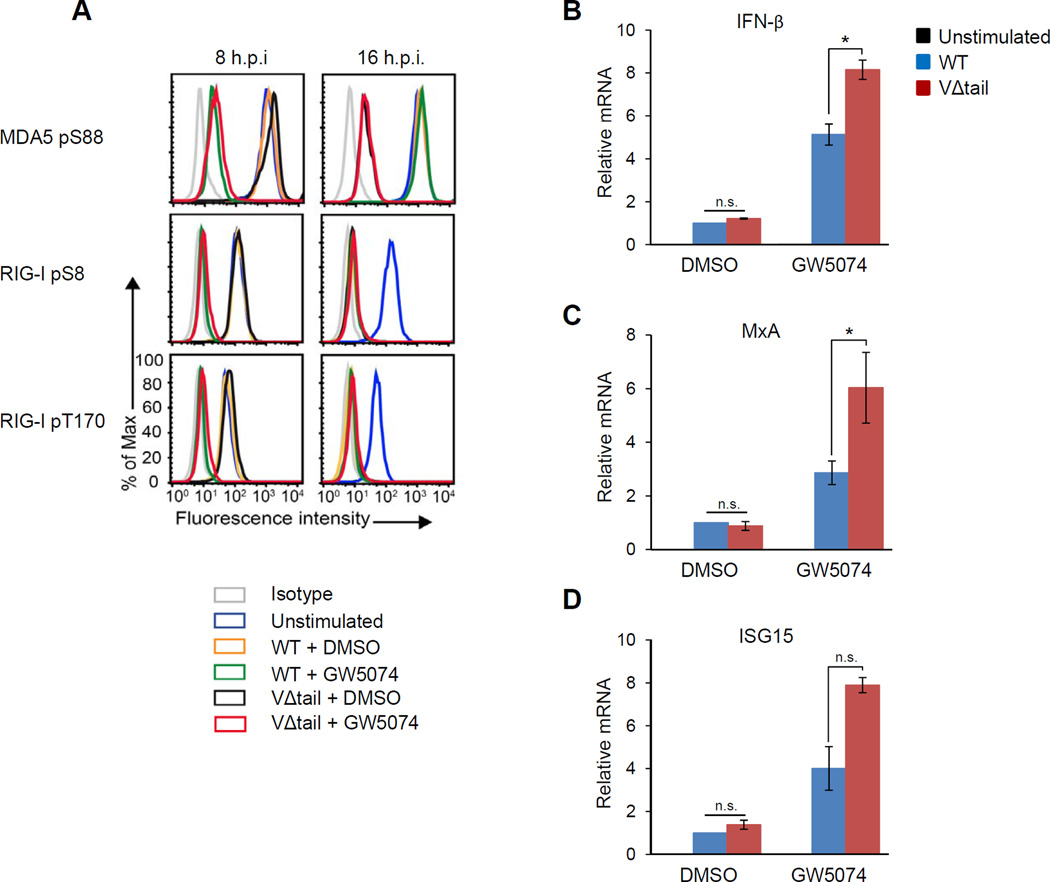
After MV infection of the lung, DCs are among the first cells that become infected (Moss et al., 2004). Thus we investigated whether MV, using its V protein, also targets PP1α/γ in DCs to evade detection by MDA5. We determined the ability of the VΔtail virus and WT virus to modulate MDA5 S88 phosphorylation in primary human DCs at early (8 h.p.i.) and late (16 h.p.i.) time points during infection (Figure 6A, upper panel). In DCs infected with the WT virus, MDA5 remained phosphorylated at both time points, indicating that MV blocks MDA5 phosphorylation early and late during infection. In contrast, in VΔtail virus-infected cells MDA5 was in the dephosphorylated, active state at 16 h.p.i., indicating an inability to block MDA5 dephosphorylation (Figure 6A, upper right panel). Interestingly, at 8 h.p.i. MDA5 was phosphorylated in the VΔtail virus-infected cells. RIG-I was also kept in the S8/T170-phosphorylated, inactive state at 8 h.p.i. in WT and VΔtail virus-infected cells (Figure 6A, middle and lower left panels). In contrast to MDA5 suppression, the inhibition of RIG-I dephosphorylation was transient and not affected by the V protein, as RIG-I was fully dephosphorylated at 16 h.p.i. in both WT and VΔtail virus-infected cells (Figure 6A, middle and lower right panels). These results demonstrate that in DCs, MV uses V-dependent and V-independent mechanisms for preventing RLR activation: while the V-dependent inhibitory mechanism specifically targets MDA5, the V-independent mechanism blocks both RIG-I and MDA5 activation, specifically early during infection (Mesman et al.). This early, V-independent inhibitory mechanism of MV is triggered via DC-specific DC-SIGN signaling and subsequent Raf-1 kinase activation, as treatment with the Raf-1 inhibitor GW5074 induced dephosphorylation of MDA5 and RIG-I at 8 h.p.i., but notably had no effect on both RLRs at 16 h.p.i. (Figure 6A). Our study combined with the results by Mesman et al. (manuscript submitted) thus indicates that early during infection in DCs, MDA5 and RIG-I are inhibited through virus-induced DC-SIGN-Raf-1 signaling; later during infection, however, when viral protein expression occurs, the V protein of MV specifically keeps MDA5, but not RIG-I, in the phosphorylated, repressed state.
Figure 6. The VΔtail rMV is defective in MDA5 suppression and IFN antagonism in primary human DCs.
(A) Primary human DCs were either DMSO-treated or pre-incubated with Raf-1 inhibitor GW5074 for 2 h, and then infected with WT or VΔtail virus (MOI 0.5). Phosphorylation of endogenous MDA5 S88 (upper panel), RIG-I S8 (middle panel), and RIG-I T170 (lower panel) was determined at 8 h.p.i. (left) and 16 h.p.i. (right) by flow cytometry using phospho-specific pS88-MDA5, pS8-RIG-I and pT170-RIG-I antibodies. Data are representative of three independent donors. (B–D) DCs that had been either DMSO-treated or pre-incubated with GW5074 for 2 h, were infected with WT or VΔtail virus (MOI 0.5). IFN-β and ISG mRNA levels were determined by qRT-PCR at 24 h.p.i. Data are pooled from three (B and C) or two (D) independent donors and presented as means +/− s.d. *P<0.05; n.s., not statistically significant.
To determine the contribution of MV-V inhibition of MDA5 to the type-I IFN response in DCs, we compared IFN-β and ISG induction upon infection with WT virus or VΔtail virus (Figures 6B–6D). DCs infected with the VΔtail virus had markedly increased mRNA levels of IFNβ, MxA, and ISG15 compared to cells infected with WT virus, specifically when DC-SIGN signaling was blocked using the Raf-1 inhibitor GW5074, but not upon DMSO treatment (Figures 6B–6D). Collectively, these results demonstrate that in DCs, MV uses V-dependent and V-independent mechanisms to block RLR signaling: in addition to RLR inhibition through DC-SIGN-Raf-1 signaling, the V protein is important for MV to block MDA5 dephosphorylation-dependent activation and thus for optimal IFN antagonism. The V protein specifically targets MDA5, late during infection, while DC-SIGN signaling through Raf-1 blocks both RIG-I and MDA5, early during infection.
DISCUSSION
The Paramyxoviridae family encompasses several clinically relevant human pathogens that pose a serious global health concern. MV, a member of the genus Morbillivirus, continuously causes high morbidity and mortality world-wide, the latter of which is caused by its ability to induce a generalized suppression of the immune system (Moss et al., 2004). It is known that the immunosuppressive effect of MV is partly due to its ability to potently antagonize type-I IFN induction in various cell types including lung epithelial cells and immune cells. Similar to other paramyxoviruses, the IFN-antagonistic activity of MV is largely due to its non-structural V protein, a virulence factor that is not required for virus replication in vitro (Schneider et al., 1997), but enhances pathogenicity in vivo (Devaux et al., 2008; Valsamakis et al., 1998). While it is well known that the V protein antagonizes various host innate immune proteins including MDA5 and STAT1/2, the precise mechanisms and physiological relevance of these virus-host interactions remain largely unknown. In this study, we show that the V protein of MV blocks PP1α/γ-mediated dephosphorylation of MDA5, which keeps MDA5 in the phosphorylated, inactive state. Despite the fact that PP1α/γ are responsible for both MDA5 and RIG-I dephosphorylation, MV-V specifically blocked MDA5, but not RIG-I, dephosphorylation. In addition, the V protein of NiV, of the Henipavirus genus, also robustly modulated the phosphorylation state MDA5 at S88. Maintained phosphorylation of MDA5 by MV-V and NiV-V subsequently blocked its interaction with MAVS, preventing downstream signaling and IFN induction. In contrast, no appreciable enhancement of MDA5 phosphorylation was detected with PIV5-V, of the Rubulavirus genus. Our study thus identifies an important mechanism of MDA5 inhibition that is distinct from that previously described involving a direct interaction with and disruption of the MDA5 ATP hydrolysis domain. Given that paramyxoviruses are equipped with multiple mechanisms to manipulate IFN receptor signaling, it is not surprising that these viruses have also evolved multiple strategies to block MDA5-mediated IFN induction. Our study, combined with previous studies, indicates that whereas the V proteins of some paramyxoviruses, such as PIV5, block MDA5 by disrupting its ATPase activity, the V proteins of other paramyxoviruses, such as MV and NiV, suppress MDA5 activation through modulating the MDA5 phosphorylation state.
Mechanistically, our study revealed a physical interaction of the V protein of MV and NiV with the phosphatases PP1α/γ. In contrast, PIV5-V did not interact with PP1α/γ. Binding of MV-V to PP1 strongly inhibited the PP1-MDA5 interaction. MDA5-PP1 binding was also strongly impaired in MV-infected, but not poly(I:C)-stimulated, cells.
Since PP1 substrates or PP1-interacting proteins usually possess defined consensus motifs that mediate binding (Roy and Cyert, 2009), we searched for known PP1-binding sequences in various paramyxovirus V proteins. A conserved PP1-binding motif 288RIWY291 was identified in the very C-terminal “tail” region of the MV-V protein. The PP1-binding motif in MV-V was required for PP1 interaction and for MV-V’s inhibitory effect on MDA5 dephosphorylation. In contrast, no known PP1-binding motif was found in the V protein of PIV5 or NiV, the latter of which also efficiently interacted with PP1α/γ. It is thus tempting to speculate that the V protein of NiV, which evolved in bats, interacts with PP1α/γ utilizing an as yet unknown PP1-binding motif. Alternatively, it is possible that the binding of NiV-V may be mediated by MDA5 or another cellular protein. Further analyses will be required to define the molecular architecture of the NiV V-PP1 complex and the molecular mechanism of how NiV-V blocks MDA5 activation by PP1. Furthermore, future studies will be directed toward investigating the precise details of how NiV and MV-V block the PP1-mediated activation of specifically MDA5, but not RIG-I.
Several reports indicate that the V protein of paramyxoviruses is a multi-phosphorylated protein (Lu et al., 2008; Ludlow et al., 2008; Pfaller and Conzelmann, 2008; Shiell et al., 2003). Recent studies showed that the V proteins utilize cellular kinases for their own phosphorylation, and that this strategy is employed by some paramyxoviruses to antagonize innate immunity. For example, the V proteins of PIV5 and mumps virus were shown to compete with IRF3 for phosphorylation by TBK1/IKKε, thereby abrogating IRF3-induced gene expression (Lu et al., 2008). Since no phosphatase for V protein dephosphorylation has been identified, we asked whether the MV-V protein serves as a PP1 substrate. Experiments using chemical inhibitors of PP1 and an in vitro dephosphorylation assay indicated that PP1 dephosphorylates MV-V. It is conceivable that the targeting of cellular kinases (TBK1 and IKKs) or phosphatases (PP1α/γ), that play key roles in innate signal transduction pathways, is a common theme of the paramyxovirus V protein for innate immune suppression. However, more detailed studies are needed to determine whether MV-V indeed serves as a ‘decoy’ substrate of PP1α/γ, or whether V dephosphorylation by PP1 is a critical part of MV replication.
The V proteins of paramyxoviruses have been shown to suppress IFN induction and IFN receptor signaling through a number of different mechanisms (Bowie and Unterholzner, 2008; Horvath, 2004); however, the physiological relevance and contribution of specific IFN-antagonistic functions of the V proteins to immune suppression remain largely undetermined. To determine the relevance of PP1 antagonism for innate immune evasion, we generated an rMV in which the PP1-binding motif in the V protein has been deleted. This VΔtail mutant virus was severely growth-impaired compared to the parental virus in human lung epithelial cells which correlated with its ability to robustly trigger IRF3 activation and enhance IFN induction compared to the parental virus, indicating an important role of PP1 antagonism by the MV-V protein for inhibiting type-I IFN induction in epithelial cells. Infection studies in primates or hCD150-transgenic mice are needed to reveal the physiological role of PP1 antagonism in V-mediated innate immune suppression in vivo.
We extended our studies to primary human DCs, which are also key target cells of MV in vivo, to determine the contribution of PP1 antagonism by MV-V in immune cells. In contrast to the parental virus, the VΔtail mutant virus efficiently triggered MDA5 S88 dephosphorylation at 16 h.p.i., demonstrating that the PP1 binding-deficient VΔtail virus loses its ability to suppress MDA5 dephosphorylation and activation. Interestingly, MV also blocked RIG-I dephosphorylation specifically at early time points, but not at later time points during infection of DCs. At this early time point during infection MDA5 dephosphorylation was also blocked in a V protein-independent manner. This early V-independent inhibition of both RIG-I and MDA5 was caused by virus-induced PP1α/γ inhibition through activation of the DC-SIGN-Raf1 signaling cascade (Mesman, et al.). This reveals that in DCs, MV has evolved two mechanisms – V-dependent and V-independent – for blocking the dephosphorylation-dependent activation of RLRs, indicating the importance of modulating this pathway. In the case of MDA5, specifically at later time points when viral gene expression occurs, this involves a direct interaction of the V protein with PP1α/γ, as presented here. In contrast, to block RIG-I and MDA5 dephosphorylation at early time points in infection, MV activates DC-SIGN signaling which triggers Raf-1 kinase activation, ultimately also resulting in PP1α/γ inhibition. Since MV inhibited both RIG-I and MDA5 at early time points but specifically blocks MDA5 activation at later time points, it is tempting to speculate that these receptors sense viral RNAs in DCs at different time points during MV infection: while both RIG-I and MDA5 may contribute to RNA detection at early time points, MDA5 may be the main sensor for detecting viral RNA species generated later during MV infection. A similar model of sequential RLR activation was recently proposed for West Nile virus infection (Errett et al., 2013). However, further studies are needed to determine the temporal contribution of RIG-I and MDA5 to viral RNA sensing during MV infection.
Taken together, these results demonstrate that PP1 antagonism is an important viral strategy for escaping RLR immune signaling and also emphasize the vital role of PP1α and PP1γ in innate immune activation.
EXPERIMENTAL PROCEDURES
GST Pull-Down Assay, Immunoprecipitation (IP), and Immunoblot Analysis
HEK293T and A549-hCD150 cells were lysed in NP-40 buffer [50 mM HEPES, pH 7.4, 150 mM NaCl, 1% (v/v) NP-40, protease inhibitor cocktail (Sigma), and Ser/Thr phosphatase inhibitor cocktail (Sigma)], followed by GST pull-down, IP and western blot analysis as previously described (Gack et al., 2007; Gack et al., 2010). For V-PP1 binding, Co-IPs were rigorously washed with NP40 lysis buffer containing 1 M NaCl.
Antibodies
For immunoblotting the following primary antibodies were used: anti-FLAG (M2; 1:2,000) (Sigma), anti-HA (1:2,000) (Clone HA-7; Sigma), anti-glutathione S-transferase (anti-GST) (1:2,000) (Sigma), anti-measles nucleoprotein [3E1] (1:500) (Abcam), anti-PP1α (1:2,000) (Bethyl laboratories), anti-PP1γ (1:2,000) (Bethyl Laboratories), anti-MDA5 (1:1,000) (Enzo Life Sciences), anti-pan pSer antibody (Abcam), and anti-β-Actin (1:10,000) (Abcam). The phospho-specific pS88-MDA5, pS8-RIG-I, and pT170-RIG-I rabbit polyclonal antibodies have been previously described (Maharaj et al., 2012; Wies et al., 2013).
Luciferase Reporter Assay
HEK293T cells, seeded into 12-well plates, were transfected with 200 ng IFN-β luciferase and 300 ng β-gal-expressing pGK-β-gal as well as 500 ng FLAG-MDA5, 400 ng GST-MDA5 2CARD, and 500 ng V protein. At 48 h post-transfection, whole cell lysates were subjected to a luciferase assay (Promega). Luciferase values were normalized to β-galactosidase to measure transfection efficiency.
In vitro Dephosphorylation Assay
MV-V-FLAG protein was purified from transfected HEK293T cells using anti-FLAG M2 agarose (Sigma). IPs were washed extensively with RIPA buffer, then twice with PBS. The in vitro MV-V protein dephosphorylation reaction was carried out at 30 °C for 1 h in phosphatase buffer (25 mM TRIS-HCl pH 7.5, 150 mM NaCl, 5 mM DTT) using 0.1 units of PP1α protein (Millipore). The reaction was stopped by adding SDS-Laemmli buffer. Samples were subjected to SDS-PAGE, followed by immunoblot analysis.
Microscopy
Bright-field images of infected A549-hCD150 cells were taken on a Nikon Eclipse Ti microscope.
Confocal Microscopy
HeLa cells were transfected with Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. At 36 h post-transfection, cells were stained with 500 µM MitoTracker-Alexa-fluor-633 (Life Technologies) in DMEM for 30 min. Cells were fixed with 2% (w/v) paraformaldehyde for 10 min and permeabilized with 0.1% (v/v) Triton X-100, followed by blocking with 5% (v/v) bovine serum in PBS for 1 h. Cell preparation and confocal microscopy analysis were performed as previously described (Gack et al., 2007). For immunostaining of FLAG-PP1γ and HA-MV-V, rabbit anti-FLAG (Sigma) and mouse anti-HA (clone HA-7, Sigma) antibodies were used, followed by incubation with donkey anti-rabbit Alexa-fluor-594 and donkey anti-mouse Alexa-fluor-488 (Life Technologies), respectively. Nuclei were stained with DAPI. Histogram profiles of confocal images were created using the RGB-Profiler function of ImageJ (Schindelin et al., 2012).
A549-hCD150 cells were infected with rMVKSEGFP(3) WT or VΔtail (MOI 2), or left uninfected. At 18 h post-infection, cells were fixed and permeabilized as described above, followed by blocking with 10% (v/v) goat serum in PBS for 1 h. IRF3 was stained using a polyclonal rabbit anti-IRF3 (Santa Cruz Biotechnology) antibody and a donkey anti-rabbit Alexa-fluor-594 antibody (Abcam). Nuclei were stained with DAPI. All laser scanning images were taken on an Olympus IX8I confocal microscope.
Generation of an rMV expressing EGFP and a truncated V protein
rMVKS is based on a wild-type genotype B3 virus isolated from PBMC collected in 1997 from a severe measles case in Khartoum, Sudan (El Mubarak et al., 2000). The details for generating the rMVKSEGFP(3) VΔtail mutant virus are provided in the SI Materials.
Quantitative Real-time PCR
A549-hCD150 cells were infected with rMVKSEGFP(3) WT or VΔtail virus at an MOI of 0.05. 24 h post-infection, cells were harvested and total RNA was extracted with the E.Z.N.A. HP Total RNA Kit (Omega). RNA was used for qRT-PCR using SuperScript III Platinum One-Step Quantitative RT-PCR System with ROX kit (Invitrogen) with IFNβ, IFIT2, MxA, OAS1, MDA5, and GAPDH gene-specific primers (IDT) on a 7300 Real Time PCR System (Applied Biosystems).
RIG-I and MDA5 Phosphorylation Analysis by Flow Cytometry
For determining RIG-I and MDA5 phosphorylation, DCs were either left unstimulated, or pre-incubated with Raf-1 kinase inhibitor GW507427 (1 µM) (Sigma) for 2 h, and subsequently stimulated with poly(I:C)-LyoVec or infected with rMVKSEGFP(3) WT or VΔtail virus at the indicated MOI. Eight or 16 h after infection, cells were fixed with 4% (w/v) paraformaldehyde and permeabilized in 90% (v/v) methanol. Phosphorylation of endogenous RIG-I and MDA5 was assessed using pS8-RIG-I, pT170-RIG-I, and pS88-MDA5 antibodies. After incubation with PE-conjugated donkey anti-rabbit antibodies (Jackson), fluorescence was measured by flow cytometry.
Statistical Analysis
Statistical analysis was performed by unpaired, two-tailed Student’s t-test.
Supplementary Material
HIGHLIGHTS.
-
□
Measles virus infection inhibits dephosphorylation of MDA5 at S88.
-
□
Measles and Nipah V proteins bind the phosphatases PPlα/γ to block MDA5 dephosphorylation.
-
□
PP1 binding is mediated by a classical PP1-binding motif in the measles V protein.
-
□
A recombinant V mutant measles virus is growth-impaired and strongly induces interferon.
ACKNOWLEDGEMENTS
This work was supported by the Armenise-Harvard Foundation, the William Milton Fund, and the NIH grants R01 AI087846, R21 AI097699, and RR000168 (to M.U.G.), and R01 AI105063 (to W.P.D.). This work was further supported by a fellowship from the German Research Foundation FU 949/1-1 (to F.F.), and by the Dutch Organization for Scientific Research (NWO): ZonMW 9120812 (to A.W.M.), NGI 40-41009-98-8057 (to S.I.G), and NWO 917-46-367, NWO 912-04-025, and VICI 918.10.619 (to T.B.H.G.). We greatly thank C. Basler (Mount Sinai), K.-K. Conzelmann (LMU Munich), L. Gehrke (Harvard), and J. Jung (USC) for providing reagents.
M.E.D. performed and analyzed the majority of the experiments. M.K.W., L.J.R., F.F., S.G. performed experiments. A.W.M. and S.I.G. performed the experiments in DCs. S.I.G. and T.B.H.G. designed the experiments in DCs. W.P.D. and L.J.R designed the experiments to generate the rMVKS. M.U.G. was responsible for the overall conception, design and supervision of the study. M.E.D. and M.U.G. wrote the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andrejeva J, Childs K, Young D, Carlos T, Stock N, Goodbourn S, Randall R. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo R, Kaelin K, Baczko K, Billeter MA. Measles virus editing provides an additional cysteine-rich protein. Cell. 1989;56:759–764. doi: 10.1016/0092-8674(89)90679-x. [DOI] [PubMed] [Google Scholar]
- Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, Randall R, Goodbourn S. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology. 2007;359:190–200. doi: 10.1016/j.virol.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Childs KS, Andrejeva J, Randall RE, Goodbourn S. Mechanism of mda-5 Inhibition by paramyxovirus V proteins. J Virol. 2009;83:1465–1473. doi: 10.1128/JVI.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creagh E, O'Neill L. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends in immunology. 2006;27:352–357. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5'-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Devaux P, Hodge G, McChesney MB, Cattaneo R. Attenuation of V- or C-defective measles viruses: infection control by the inflammatory and interferon responses of rhesus monkeys. J Virol. 2008;82:5359–5367. doi: 10.1128/JVI.00169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenacher K, Krug A. Regulation of RLR-mediated innate immune signaling--it is all about keeping the balance. Eur J Cell Biol. 2012;91:36–47. doi: 10.1016/j.ejcb.2011.01.011. [DOI] [PubMed] [Google Scholar]
- El Mubarak HS, Van De Bildt MWG, Mustafa OA, Vos HW, Mukhtar MM, Groen J, El Hassan AM, Niesters HGM, Ibrahim SA, Zijlstra EE, et al. Serological and Virological Characterization of Clinically Diagnosed Cases of Measles in Suburban Khartoum. Journal of Clinical Microbiology. 2000;38:987–991. doi: 10.1128/jcm.38.3.987-991.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M., Jr The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J Virol. 2013;87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack M, Shin Y, Joo C-H, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Gack MU, Nistal-Villan E, Inn KS, Garcia-Sastre A, Jung JU. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J Virol. 2010;84:3220–3229. doi: 10.1128/JVI.02241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L, Benoit L, Song C, Cella M, Gilfillan S, Holtzman MJ, Colonna M. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 2010;6:e1000734. doi: 10.1371/journal.ppat.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Horvath CM. Silencing STATs: lessons from paramyxovirus interferon evasion. Cytokine Growth Factor Rev. 2004;15:117–127. doi: 10.1016/j.cytogfr.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Ikegame S, Takeda M, Ohno S, Nakatsu Y, Nakanishi Y, Yanagi Y. Both RIG-I and MDA5 RNA helicases contribute to the induction of alpha/beta interferon in measles virus-infected human cells. J Virol. 2010;84:372–379. doi: 10.1128/JVI.01690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Lemon K, dVR, Mesman AW, McQuaid S, van Amerongen G, et al. Early target cell sof measles virus after aerosol infection of non-human primates. PLoS pathogens. 2011;7:e1001263. doi: 10.1371/journal.ppat.1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LL, Puri M, Horvath CM, Sen GC. Select paramyxoviral V proteins inhibit IRF3 activation by acting as alternative substrates for inhibitor of kappaB kinase epsilon (IKKe)/TBK1. J Biol Chem. 2008;283:14269–14276. doi: 10.1074/jbc.M710089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow LE, Lo MK, Rodriguez JJ, Rota PA, Horvath CM. Henipavirus V protein association with Polo-like kinase reveals functional overlap with STAT1 binding and interferon evasion. J Virol. 2008;82:6259–6271. doi: 10.1128/JVI.00409-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharaj N, Wies E, Stoll A, Gack M. Conventional protein kinase C-α (PKC-α) and PKC-β negatively regulate RIG-I antiviral signal transduction. Journal of Virology. 2012;86:1358–1371. doi: 10.1128/JVI.06543-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss WJ, Ota MO, Griffin DE. Measles: immune suppression and immune responses. The international journal of biochemistry & cell biology. 2004;36:1380–1385. doi: 10.1016/j.biocel.2004.01.019. [DOI] [PubMed] [Google Scholar]
- Motz C, Schuhmann K, Kirchhofer A, Moldt M, Witte G, Conzelmann K-K, Hopfner K-P. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science (New York, NY) 2013;339:690–693. doi: 10.1126/science.1230949. [DOI] [PubMed] [Google Scholar]
- Nistal-Villán E, Gack M, Martínez-Delgado G, Maharaj N, Inn K-S, Yang H, Wang R, Aggarwal A, Jung J, García-Sastre A. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. The Journal of biological chemistry. 2010;285:20252–20261. doi: 10.1074/jbc.M109.089912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palosaari H, Parisien JP, Rodriguez JJ, Ulane CM, Horvath CM. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J Virol. 2003;77:7635–7644. doi: 10.1128/JVI.77.13.7635-7644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien JP, Bamming D, Komuro A, Ramachandran A, Rodriguez JJ, Barber G, Wojahn RD, Horvath CM. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J Virol. 2009;83:7252–7260. doi: 10.1128/JVI.00153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller CK, Conzelmann KK. Measles virus V protein is a decoy substrate for IkappaB kinase alpha and prevents Toll-like receptor 7/9-mediated interferon induction. J Virol. 2008;82:12365–12373. doi: 10.1128/JVI.01321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan C, Näslund T, Liljeström P, Weber F, Reis e Sousa C. Science. Vol. 314. New York, NY: 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates; pp. 997–1001. [DOI] [PubMed] [Google Scholar]
- Rodriguez K, Horvath C. Amino acid requirements for MDA5 and LGP2 recognition by paramyxovirus V proteins: a single arginine distinguishes MDA5 from RIG-I. Journal of Virology. 2013;87:2974–2978. doi: 10.1128/JVI.02843-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy J, Cyert MS. Cracking the phosphatase code: docking interactions determine substrate specificity. Sci Signal. 2009;2:re9. doi: 10.1126/scisignal.2100re9. [DOI] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008 doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H, Kaelin K, Billeter MA. Recombinant measles viruses defective for RNA editing and V protein synthesis are viable in cultured cells. Virology. 1997;227:314–322. doi: 10.1006/viro.1996.8339. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012:9. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiell BJ, Gardner DR, Crameri G, Eaton BT, Michalski WP. Sites of phosphorylation of P and V proteins from Hendra and Nipah viruses: newly emerged members of Paramyxoviridae. Virus Res. 2003;92:55–65. doi: 10.1016/s0168-1702(02)00313-1. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Thomas SM, Lamb RA, Paterson RG. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54:891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsamakis A, Schneider H, Auwaerter PG, Kaneshima H, Billeter MA, Griffin DE. Recombinant measles viruses with mutations in the C, V, or F gene have altered growth phenotypes in vivo. J Virol. 1998;72:7754–7761. doi: 10.1128/jvi.72.10.7754-7761.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, Finberg RW, Gack MU. Dephosphorylation of the RNA Sensors RIG-I and MDA5 by the Phosphatase PP1 Is Essential for Innate Immune Signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Mesman AW, Zijlstra-Willems EM, Kaptein TM, MED, Ludlow M, MPD, MUG, SIG, TBHG Measles virus suppresses RIG-I-like receptor activation in dendritic cells via DC-SIGN-mediated inhibition of PP1 phosphatases. doi: 10.1016/j.chom.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.