Abstract
Cardiomyocytes (CMs) differentiated from human embryonic stem cells (hESCs) are a promising and potentially unlimited cell source for myocardial repair and regeneration. Recently, multiple methodologies—primarily based on the optimization of growth factors—have been described for efficient cardiac differentiation of hESCs. However, the role of extracellular matrix (ECM) signaling in CM differentiation has not yet been explored fully. This study examined the role of ECM signaling in the efficient generation of CMs from both H7 and H9 ESCs. The hESCs were differentiated on ECM substrates composed of a range of fibronectin (FN) and laminin (LN) ratios and gelatin and evaluated by the fluorescence activated cell scanning (FACS) analysis on day 14. Of the ECM substrates examined, the 70:30 FN:LN reproducibly generated the greatest numbers of CMs from both hESC lines. Moreover, the LN receptor integrin β4 (ITGB4) and FN receptor integrin β5 (ITGB5) genes, jointly with increased phosphorylated focal adhension kinase and phosphorylated extracellular signal-regulated kinases (p-ERKs), were up-regulated over 13-fold in H7 and H9 cultured on 70:30 FN:LN compared with gelatin. Blocking studies confirmed the role of all these molecules in CM specification, suggesting that the 70:30 FN:LN ECM promotes highly efficient differentiation of CMs through the integrin-mediated MEK/ERK signaling pathway. Lastly, the data suggest that FN:LN-induced signaling utilizes direct cell-to-cell signaling from distinct ITGB4+ and ITGB5+ cells.
Key words: : cardiomyocytes, cell fate, extracellular matrix, fibronectin, human embryonic stem cells, integrin signaling, laminin
Introduction
Cardiovascular disease, which includes heart failure, is the leading cause of death in the United States.1 One common cause of heart failure is the heart's inability to effectively pump blood due to the ischemic, or scarred, tissue from a myocardial infarction. Currently, the most promising cell sources for the restoration of heart function are cells derived from stem cells. Embryonic stem cells (ESCs) and induced pluripotent stem cells have the ability to differentiate into almost all types of cells, including cardiomyocytes (CMs) with cardiac molecular, structural, and functional characteristics.2–4 Moreover, a number of animal studies have shown that hESC-derived CMs can be implanted into the damaged area of the heart to promote the contractile function of the heart.5–9 However, because millions of cells are needed for effective human cell therapies,7,10 efficient induction methods for generating high numbers of functional CMs are critical. Recently, the optimization of Wnt, Activin/Nodal, and bone morphogenetic protein (BMP) soluble signaling levels in cardiac differentiation protocols were identified as exhibiting variable levels of efficiency across the different hESC lines,5,11–15 and a number of highly efficient cell line–specific protocols have been published.13,16
In addition to Wnt, Activin/Nodal, and BMP soluble signaling levels, another important signal mimicking the “niche” of the developing embryo is the extracellular matrix (ECM). The ECM displays an array of macromolecular cues that not only provide the physical support for cell adhesion, but can also direct cell proliferation, migration, and differentiation.17–19 The cellular effects in response to ECM, such as fibronectin, laminins, various collagens, tenascin, vitronectin, and thrombospondin, are mediated by the integrin family of cell surface receptors. Following cell adhesion to ECM, focal adhesion kinase (FAK) becomes phosphorylated at multiple sites, including tyrosines 397, 576, and 577, and the FAK activation phosphorylation loop is significantly elevated in cells expressing activated Src.20 This activation can lead to stimulation of a number of cell-signaling cascades that ultimately activate the Ras/MAPK/ERK pathway leading to cell proliferation, migration, differentiation and/or survival (reviewed by Boudreau and Jones21). Specifically, FAK is known to play a role in BMP4-induced adipogenesis,22 and activation of FAK and ERK1/2 signaling pathway coincides with osteogenic differentiation.23 However, despite the growing interest in this area, no studies to date have linked matrix integrin-mediated FAK and ERK activation in cardiac fate.
Many groups have suggested the role of various ECM molecules in cardiovascular differentiation of hESC. For example, the presence of fibronectin (FN) in three-dimensional (3D) collagen constructs stimulated endothelial cell differentiation and vascularization of differentiating embryoid bodies (EBs),24 while laminin (LN) increased the ability of ESCs to differentiate into beating CM clusters.24 Another group showed that hydrogels, in the absence of supplemental growth factors, containing higher percentages of native cardiac ECM (75%) increased the fraction of cells expressing cardiac marker troponin T compared with lower percentages (25%) of native cardiac ECM.25 Matrigel ECM components have also been implicated in efficient differentiation of CM by promoting the epithelial-to-mesenchymal transition.26
However, the 3D collagen gel,24 cardiac ECM hydrogel,25 cardiogel,27 and Matrigel matrix sandwich26 described to promote in vitro cardiac differentiation are mixed with a variety of biomaterials, many of which (i.e., cardiogel and Matrigel) vary from batch to batch. Therefore, it is not clear which specific molecules are critical in cardiac fate. We set out to explore combinatorial signaling from controlled ratios of the most likely ECM molecules in CM commitment from ESCs. The FN and LN were chosen because they are native cardiac ECM components derived from fibroblasts known to contribute to enhanced numbers of spontaneous beating of CMs from mouse ESCs.27 Moreover, FN is essential for heart and blood vessel morphogenesis28—its absence leads to defects in mesoderm and vascular development29—whereas, LN influences cytoskeletal and myofibrillar organization in neonatal rat CMs30 and increases the ability of mouse ESCs and human adipose-derived stem cells to differentiate into CMs.24,31 Here, using highly optimized stage-specific protocols,16 we demonstrate that a substrate containing a 70:30 FN:LN promotes the generation of the highest numbers of CMs. We also confirm that these ECM-induced CMs involve integrin signaling through activation of integrin β (ITGB)4 receptor for LN, ITGB5 receptor for FN, and subsequent integrin-mediated MEK-ERK signaling pathway.
Materials and Methods
Human ESC culture and differentiation
Native H7 human (h)ESCs (WA07, WISC Bank, Madison, WI) and H9 hESCs (WA09, WISC Bank, Madison, WI), transfected with green fluorescence protein (GFP) expression linked to the α-myosin heavy chain (MHC) promoter (courtesy of Bernstein Lab, UCSF), were maintained in mouse embryonic fibroblast–conditioned medium supplemented with 5 ng/mL basic fibroblast growth factor (bFGF, Sigma) and plated on tissue cultured plates coated with Matrigel (BD) according to previously published protocols.32 Note that although α-MHC GFP H9 hESCs were used in the studies (due to the benefits for downstream cell sorting), it was not used as a marker of cardiac fate. This was due to the fact that α- and β-MHC exhibit oscillatory expression patterns during embryonic development.33 After CM specification, the α-MHC restricts its expression to atrial CMs while β-MHC appears only in ventricular CMs. In vitro differentiation models have shown similar expression patterns as well.33–36 Therefore, cardiac fate was determined by the expression of Nkx2.5 and cardiac troponin, rather than α-MHC.
For CM differentiation, we use our recently published stage-specific optimized protocol for H7 and H9.14 In brief, hESCs were detached using Accutase (Invitrogen) and dissociated into small clumps (three to five cells). The cells were then spun down and resuspended in 2 mL Roswell Park Memorial Institute (RPMI; Invitrogen) medium supplemented with 2% B27 (Invitrogen). To generate EB, a 400-μm Honeycomb Microwell microchip37 was placed into each well of a standard 24-well plate and 6×105 cells were added to the Microwell. Ten micromolar of ROCK inhibitor (Y-27632; EMD) was added during EB formation to enhance cell survival. After 24 h, uniform EBs were formed in Microwell and then transferred to ultra-low attachment plates (Corning) in RPMI/B27 media. EBs were then dissociated into small clusters (three to five cells; treated with Accutase for 1 min) by gently pipetting 1×105 cells into individual wells of a 96-well flat bottom plate (BD) coated with various ratios of FN (BD) and LN (BD) and compared against gelatin control (Table 1) for 5 and 6 days for H7 and H9, respectively. Each 60-mm dish was coated to a final concentration of 5 μg/cm2 of total ECM and incubated for 1 h before use. The growth factors were added with the following sequence: day 0–1, 2 ng/mL BMP4; days 1–4, 5 ng/mL bFGF, 6 ng/mL Activin A, and 30 ng/mL BMP4 for H7, or 5 ng/mL bFGF, 10 ng/mL Activin A, and 60 ng/mL BMP4 for H9; days 4–8, 150 ng/mL Dickkopf-related protein 1 (DKK1) and 10 ng/mL vascular endothelial growth factor (VEGF); days 8–14, 5 ng/mL basic fibroblast growth factor (bFGF) and 10 ng/mL VEGF. All growth factors were purchased from R&D Systems, Inc.
Table 1.
Composition of Six Different Extracellular Matrix Substrates for Testing Cardiomyocyte Differentiation
| Fibronectin | Laminin | Gelatin | |
|---|---|---|---|
| 1 | 100a | — | — |
| 2 | 70 | 30 | — |
| 3 | 50 | 50 | — |
| 4 | 30 | 70 | — |
| 5 | — | 100 | — |
| 6 | — | — | 100 |
Values in the table are percentages.
Quantitative polymerase chain reaction microarray
Total RNA was extracted using TRIzol® (Invitrogen), following the manufacturer's instructions. The concentration of RNA was determined using a Nanodrop spectrophotometer (Thermo Scientific). Two micrograms of RNA was processed with an RT2 First Strand Kit (SA Biosciences) according to the manufacturer's specifications. Quantitative polymerase chain reaction (PCR) analysis of ECM and cell adhesion molecules were assessed using the Human Extracellular Matrix and Adhesion Molecules PCR Array (SA Biosciences), RT2 SYBR® Green qPCR master mix (SA Biosciences), and an 7300 Real-Time PCR System (Applied Biosystems). At least three RNA samples were analyzed per time point.
PCR microarray data analysis
PCR array data were analyzed by the web-based RT2 Profiler PCR Array Data Analysis from SA Biosciences in the ΔΔCt method with five different housekeeping genes. The array data were plotted on volcano plots in which the x-axis indicates the fold change and the y-axis indicates the statistical reliability of the fold change. Genes significantly up-regulated (p<0.1) in both H7 and H9 differentiated cells were identified.
Functional blocking integrin β4/β5 assays
In order to block the ITGB4 and ITBG5 receptors, either 10 μg/mL of mouse monoclonal anti-integrin β4 antibody (Novus Biologicals) or 10 μg/mL mouse monoclonal anti-integrin β5 antibody (Millipore) was added into the differentiation media from day 5 to 14 for H7 or from day 6 to 14 for H9.
MEK inhibitor studies
In order to inhibit the phosphorylation of ERK, 10 μg/mL the MEK inhibitor U0126 (Promega) was added into the differentiation media from day 5 to 14 for H7 or from day 6 to 14 for H9.
Flow cytometry and fluorescent microscopy
EBs and cardiac cells generated from hESC differentiation experiments were dissociated into single cells by incubation with cell dissociation buffer (enzyme-free, phosphate-buffered saline [PBS]-based, Invitrogen). The cells were then fixed with 4% paraformaldehyde (Tousimis Research Corporation) in PBS and permeabilized with 0.7% Triton X100 for 15 min, and immunostained with the following antibodies in 1:300 titration: anti-cardiac Troponin I (cTnI) (US Biological, catalog no. T8665-13F, clone no. 2Q1100, isotype IgG2b), anti-Nkx2.5 (R&D, catalog no. MAB2444, clone no. 259416, isotype IgG1), anti-ITGB4 (Millipore, catalog no. MAB2059, clone no. ASC-8, isotype IgG1), anti-ITGB5 (Novus Biologicals, catalog no. H00003693-M01, clone no. 2C4, isotype IgG1), anti-p-FAK (Santa Cruz Biotech, catalog no. sc-374668 clone no. A12, isotype IgG1), and anti-p-ERK (Santa Cruz Biotech, catalog no. sc-81492, clone no. 12D4, isotype IgG1). Mouse IgG1 and IgG2b (Sigma) were used as isotype controls. A LSRII flow cytometer (BD) was used for data acquisition and data analysis was performed using FlowJo software (Tree Star Inc.). For fluorescent microscopy, cells were stained for cTnI, ITBG4, and ITGB5 and counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Calbiochem) and imaged on our Nikon confocal microscope.
Statistical analysis
Biological samples from three independent differentiation experiments were repeated a minimum of three times (n=3). Statistical significance was measure using one-way ANOVA followed by Tukey's post hoc method between three or more groups (p<0.05). Data are presented as means of the sample set with standard deviation error bars.
Results
Substrate signaling in CM differentiation
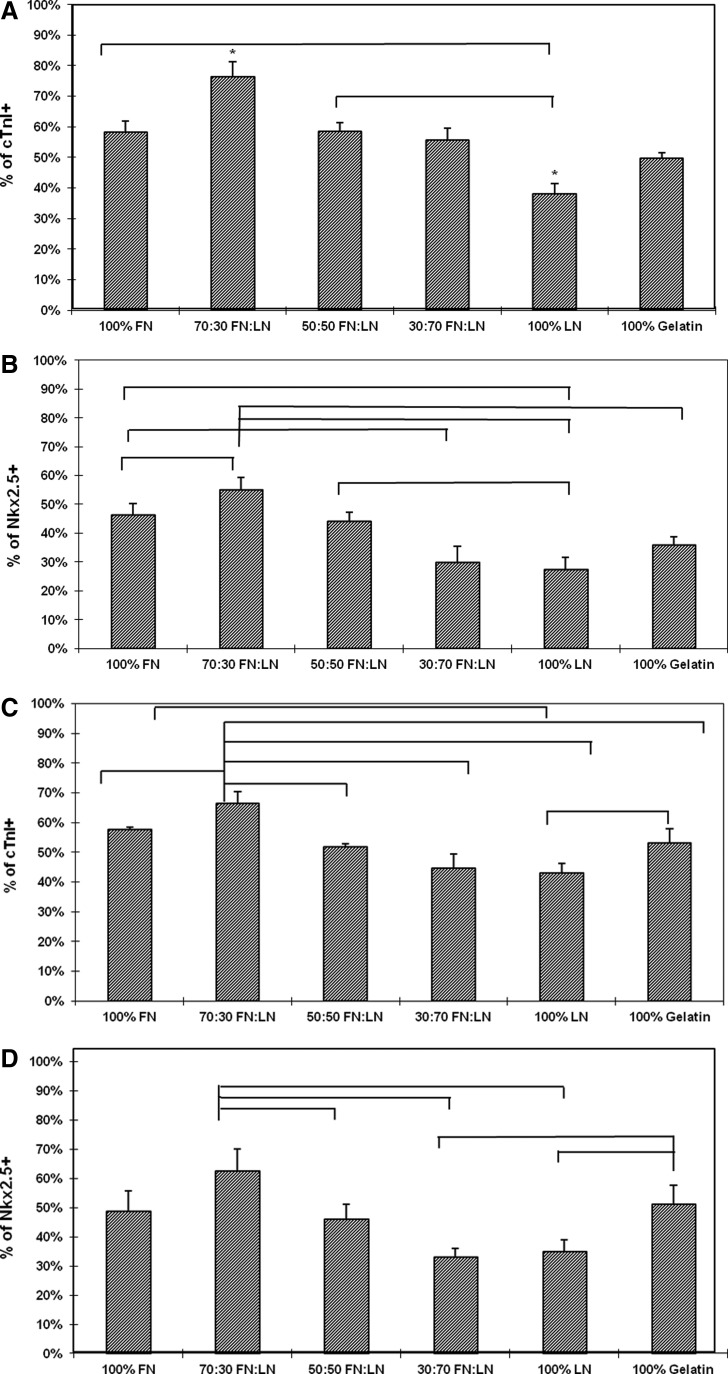
In order to investigate the effect of various proportions of FN and LN ECM proteins on CM differentiation, hESCs were cultured in a series of FN- and/or LN-coated plates and compared with a gelatin control (Table 1). Expression of cardiac markers cTnI and Nkx2.5 was assessed by quantitative flow cytometry analysis (Fig. 1) on day 14 of differentiation. As seen in Figure 1, H7 and H9 cells derived on 70:30 FN:LN substrate gave rise to the greatest percentage of CM compared to other ratios of FN and LN. H7 monolayer on 70:30 FN:LN consisted of 76.5%±4.8% cTnI+ cells and 55.0 %±4.2% Nkx2.5+ cells, whereas the control H7 monolayer on gelatin contained approximately 50% cTnI+ and 35% Nkx2.5+ cells (Fig. 1A, 1B and Supplementary Fig. S1A, S1B). H9 monolayer on 70:30 FN:LN contained over 60% CMs, while H9 on gelatin only contained about 50% CMs (Fig. 1C, 1D and Supplementary Fig. S1C, S1D). Consistent with the literature, the expression of Nkx2.5, although high, remained consistently less compared with the cTnI+.16,38 We expect that this was due to the Nkx2.5 expression peaking earlier on day 10, compared with the more mature cardiac marker, cTnI, expressed in a later stage.39–41 Compared with other ratios of FN and LN, the 70:30 FN:LN was consistently superior in generating CMs, suggesting that FN and LN are both contributing to the cardiogenesis.
FIG. 1.
Percentage of cardiac cells from H7 and H9 human embryonic stem cells (hESCs) on different substrates at day 14. H7 (A, B) differentiated cells cultured on 70% fibronectin (FN) and 30% laminin (LN) contained the highest percentage of (A) cTnI+ and (B) Nkx2.5+ cells H9 (C, D) differentiated cells cultured on 70% FN and 30% LN contained the highest percentage of (C) cTnI+ and (D) Nkx2.5+. Bars indicate statistically significant differences between the indicated groups, and an asterisk (*) indicates statistically significant differences between all groups.
Gene expression of ECM and adhesion molecules
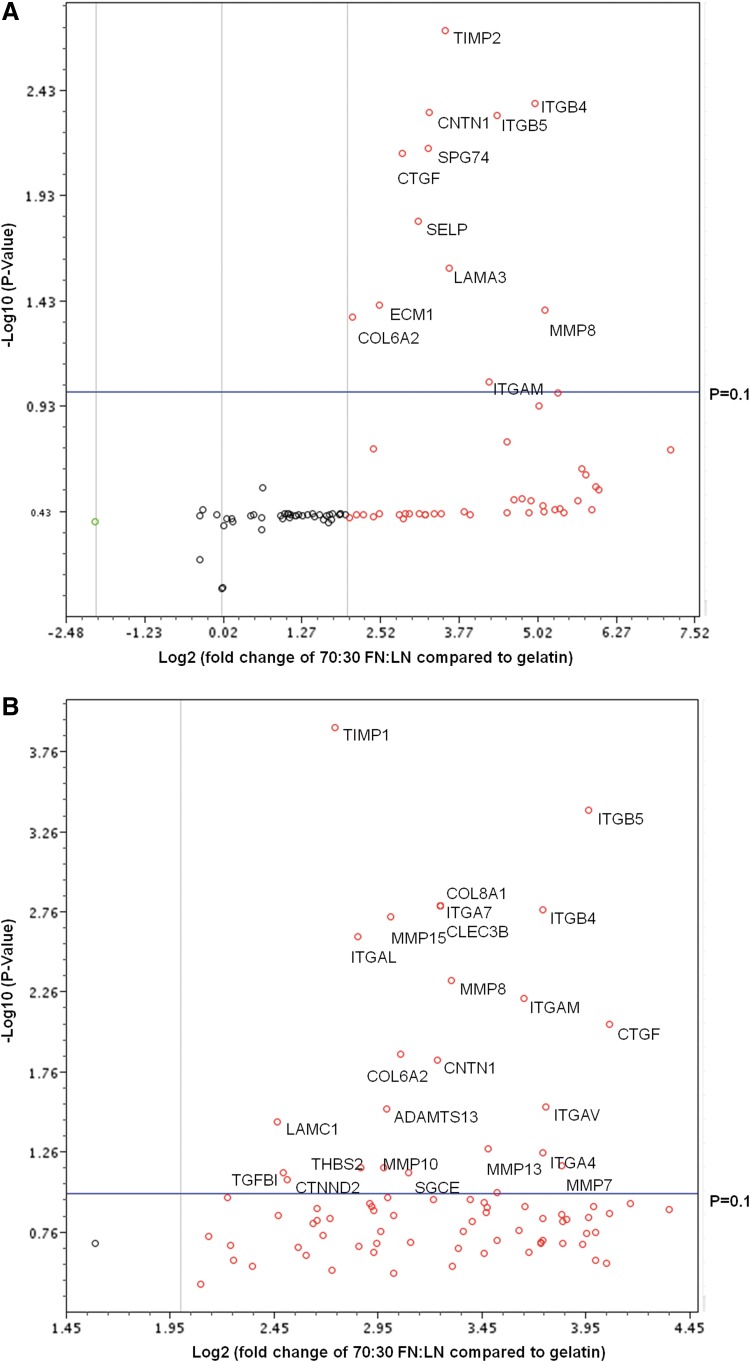
To further understand the molecules involved in FN/LN signaling, we evaluated the gene expression of using a quantitative reverse-transcription PCR micoarray that simultaneously analyzed the gene expression of 84 ECM and adhesion molecules. Here, we compared our optimal substrate 70:30 FN:LN against gelatin (control) based on the fact that some of the most efficient protocols culture cells on gelatin during CM differentiation.13 Based on volcano plots of the array data, 12 genes were significantly up-regulated (p<0.1) in H7, and 25 genes were significantly up-regulated in H9 (Fig. 2, Supplementary Table S1). Based on the categorization of the gene products; 5 out of 12 of the up-regulated genes in H7-derived CMs and 8 out of 25 of the up-regulated genes in H9-derived CMs were transmembrane molecules. The remaining up-regulated genes were adhesion molecules (three and four); ECM proteases (one and five); and cell–cell adhesion molecules (one and four) in H7 and H9, respectively. Based on these data, we deduced that the transmembrane molecules may play the most important roles for directing CM fate.
FIG. 2.
Human extracellular matrix (ECM) and adhesion molecules gene expression of cells cultured on 70:30 FN:LN compared with gelatin. Volcano plots H7 (A) and H9 (B) compared ECM and adhesion molecules gene expression fold change between cells cultured on optimal substrate and cells cultured on gelatin. The x-axis indicates the fold change, while the y-axis indicates the p-value of the fold change. Each dot represents the expression of one gene. Dots above the horizontal line show significant difference (p<0.1). Dots in red represent up-regulated genes, dots in green represent down-regulated genes, while dots in black represent unchanged genes.
Looking more closely at the specific genes up-regulated in both H7 and H9 derived-CM cultured on 70:30 FN:LN compared with 100% gelatin, we find seven genes (CNTN1, COL6A2, CTGF, ITGAM, ITGB4, ITGB5, and MMP8) in common when we set the p=0.1, but only four genes (CNTN1, CTGF, ITGB4, and ITGB5) in common when we set the p=0.01 (Supplementary Table S1). Two of these are integrin transmembrane molecules: ITGB4, a, LN receptor,42 and ITGB5, an FN receptor.43 Because these integrins exhibited much higher fold changes than CNTN1 and CTGF in both H7 and H9 ESCs, the effects of ITGB4 and ITGB5 on CM differentiation were selected for further investigation.
Quantification of ITGB4, ITGB5, p-FAK, and p-ERK positive cells
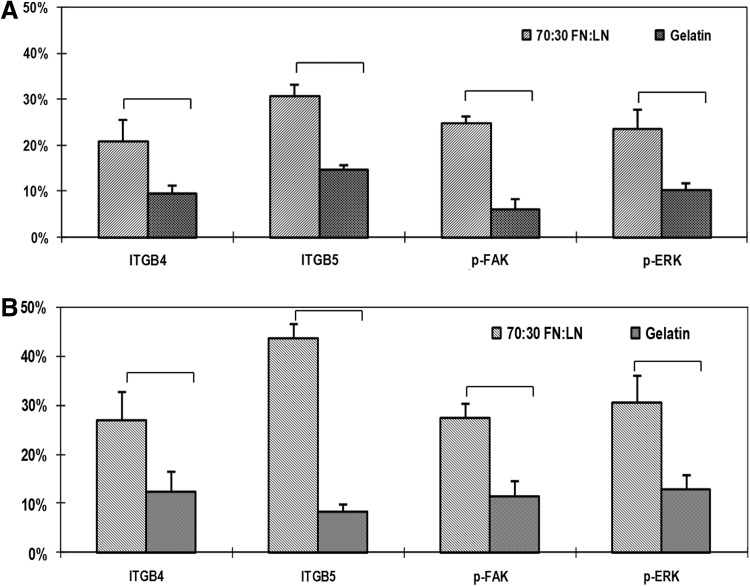
The ITGB4 and ITGB5 protein expression in H7 and H9 differentiation cultures on the optimal substrate and gelatin were first analyzed by flow cytometry analysis. The H7 differentiated on the optimal substrate, 70:30 FN:LN, contained 21% ITGB4 and 31% ITGB5 on day 14; whereas, the same cells on the gelatin contained 6% ITGB4 land 10% ITGB5 (Fig. 3A, Supplementary Fig. S2A). Similarly, H9 ESC differentiating cells on the FN/LN substrate contained higher numbers, 27% of ITGB4+ cells and 44% ITGB5+ cells versus 12% and 13%, respectively, when derived on gelatin (Fig. 3B, Supplementary Fig. S2B). Evidence of downstream integrin signaling was also investigated by analyzing the phosphorylation of both FAK and ERK in cells differentiated on the 70:30 FN:LN compared with gelatin. The significantly increased numbers of p-FAK (25% vs. 9% in H7; 27% vs. 12% in H9) and p-ERK (23% vs. 15% in H7; 31% vs. 8% in H9) were detected in hESCs differentiated on the optimal substrate compared with gelatin, respectively (Fig. 3, Supplementary Fig. S2).
FIG. 3.
Substrate signaling from 70:30 FN:LN compared with gelatin increased the number of p-FAK, p-ERK, ITGB4, and ITGB5 positive cells. Percentages of ITGB 4, ITGB 5, p-FAK, and p-ERK expressing cells from (A) H7and (B) H9 hESCs differentiating on 70:30 FN:LN were higher than those of cells on gelatin at day 14. Bars indicate statistically significant differences between the indicated groups. p-FAK, phosphorylated focal adhesion kinase; p-ERK, phosphorylated extracellular signal-regulated kinase; ITGB, intergrin β.
Blocking ITGB4/ITGB5 signaling reduces CM differentiation
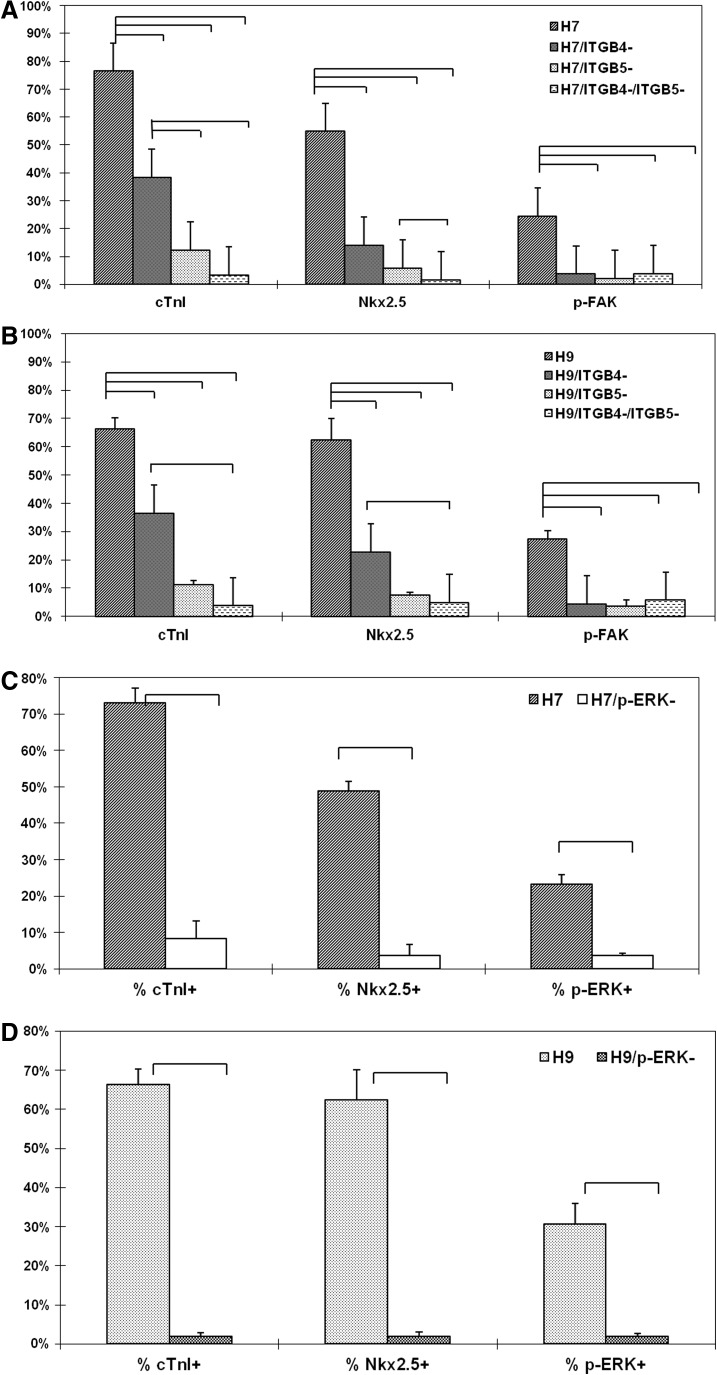
In order to verify the role of ITGB4 and ITGB5 signaling in CM differentiation, cells were treated with ITGB4 and/or ITGB5 blocking antibodies starting from the day the cells were plated as a monolayer on the 70:30 FN:LN (optimized at day 5 and 6 for H7 and H9, respectively16) through day 14. ITGB4 blocking antibody treatment reduced cTnI+ cells in H7 and H9 by 50% and 45%, respectively, and reduced Nkx2.5+ cells in H7 and H9 by 74% and 64%, respectively (Fig. 4, Supplementary Fig. S3). ITGB5 blocking antibody treatment reduced cTnI and Nkx2.5 expression in H7 and H9 by even more, 83%–89% (Fig. 4, Supplementary Fig. S3). Blocking both ITGB4 and ITGB5 simultaneously almost completely eliminated the CM derivation in these culture, reducing cTnI and Nkx2.5 expression in both H7 and H9 by 92%–97% (Fig. 4, Supplementary Fig. S3).
FIG. 4.
Blocking ITGB4 and/or ITGB5 reduced both the percentage of cardiomyocyte (CM) positive cells and p-FAK–expressing cells in differentiating hESC cultures. The reduced numbers of cTnI+ and Nkx2.5+ cells in differentiated hESC cultures (A) H7 and (B) H9 treated with ITGB4 and/or ITGB5 blocking antibodies at day 14 correlate with the reduction in p-FAK. Blocking MEK signaling inhibited cardiomyocyte differentiation from H7 (C) and H9 (D). The cTnI+, Nkx2.5+, and p-ERK+ cells in differentiation cultures were significantly much greater compared with cultures treated with MEK inhibitor U0126 at day 14. Bars indicate statistically significant differences between the indicated groups.
The effects of MEK inhibitor U0126 on CM differentiation
To verify the significance of the MEK signaling pathway, a downstream effector of FAK, in regulating CM differentiation of hESCs, the cells were cultured in the presence of the MEK inhibitor U0126 to inhibit MEK and the subsequent downstream phosphorylation of ERK. U0126 treatment started on the day that the cells were plated as a monolayer on the 70:30 FN:LN (day 5 and 6 for H7 and H9, respectively) through day 14. The flow cytometry data confirm that the U0126 reduced the p-ERK+ cells from 23% to 4% in H7 and from 31% to 2% in H9 (Fig. 4, Supplementary Fig. S4). Moreover, the suppression of MEK/ERK by U0126 treatment also significantly decreased the cTnI+ cells to only 8% and 2% in H7 and H9, respectively (Fig. 4, Supplementary Fig. S4). The Nkx2.5 expression was reduced to 4% and 2% in H7 and H9 treated with U0126, respectively, as well (Fig. 4, Supplementary Fig. S4), indicating that MEK/ERK signaling plays a critical role in integrin signaling during cardiac specification.
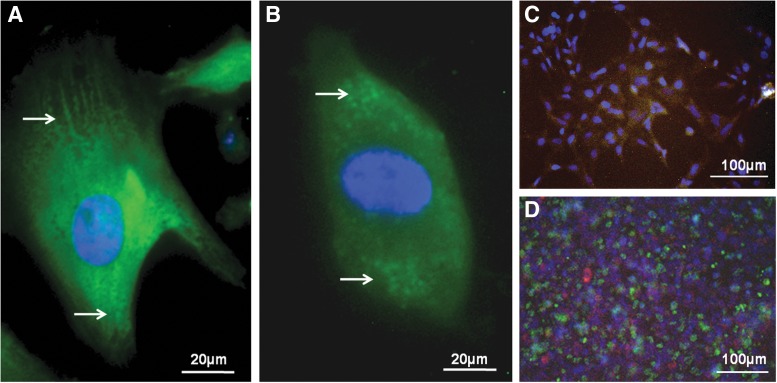
Fluorescent imaging
The differentiated cultures were stained for cTnI and imaged in order to verify the presence of sacromeric structures (Fig. 5A, 5B). The differentiated CM cultures were then co-stained with cTnI tetramethyl rhodamine isothiocyanate (TRITC), ITGB5 fluorescein isothiocyanate (FITC), and DAPI (Fig. 5C). The image shows that cTnI+ cells were also positive for ITGB5 FITC. Interestingly, the co-staining of ITGB5 FITC with ITGB4 TRITC indicates that the cells are not double-positive for both integrins, but that the CM cultures contain cells expressing either ITGB4 or ITGB5 (Fig. 5D). Consistent with FACS measurements (Fig. 3), this image depicts that the cultures contain greater numbers of cells expressing ITGB5 compared with ITGB4, and that the spatial patterning of the ITGB4 and ITGB5 cells is mingled such that each ITGB5 cells is in contact with the ITGB4-expressing cells.
FIG. 5.
Immunofluorescent images of hESC-CM. The (A) H7 and (B) H9 ESC-derived CMs were imaged under confocal microscope at day 14. The cells were immunofluorescently stained with cardiac troponin I (green) and DAPI (blue). The presence of some sacromeric structures are indicated by arrows. (C) The differentiated CM cultures from H9 ESCs were also co-stained with cTnI TRITC (red), ITGB5 FITC (green), and DAPI (blue), as well as (D) co-stained with ITGB5 FITC (green), ITGB4 (red), and DAPI (blue). DAPI, 4′,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanate; TRITC, tetramethyl rhodamine isothiocyanate.
Discussion and Conclusions
Stem cell fate choices, including the regulation of cardiac structure and function of individual cells and tissues,17,44 have been shown to be critically impacted by cell–matrix interactions. Therefore, we hypothesized that the ECM signaling from niche-appropriate molecules FN and LN would play a role in hESC specification toward CMs. Cardiac ECM is defined as a network surrounding and supporting myocardial constitutive cells including CMs, cardiac fibroblasts, endothelial cells (ECs), and vascular smooth muscle cells (SMCs). CM fibroblasts and SMCs produce the FN, while LN is synthesized by SMCs, CMs, and ECs.45 Therefore, the cardiac ECM consists of 3D interstitial collagens—predominantly collagen types I and III—to which other matrix components FN and LN are attached and mediate important functions such as healing and remodeling.46 By investigating the effects of various ratios of FN and LN, we found that hESCs differentiating on 70:30 FN:LN generated the greatest percentage (∼60%–70%) of CMs compared with gelatin (∼50%–60%). In general, collagen I and III expression levels (measure by mRNA) in the heart are comparatively high, while collagen IV, LN, and FN are expressed at intermediate levels.47 However, ECM components also vary by location (epi-, edo-, and myocardium) and over time in the developing heart. Specifically, in the myocardium, FN and collagen I are the highest in early development (E12.5), then lower after birth (P2), while collagen IV is low at E12.5 and increases later in development (P2).47
Because the integrin-mediated activation of MEK/ERK-induced differentiation of a variety of non-CM cell types is also triggered by the binding of FN and LN,48–50 we used a PCR array to highlight the transmembrane molecules playing a dominant role in ECM-induced CM differentiation. Specifically, the expression of FN receptor, ITGB5, and LN receptor, ITGB4, were found to be significantly increased in both H7 and H9 ESCs, and blocking these two molecules confirmed their role in generating CMs. The larger impact of ITGB5 blocking compared with ITGB4 blocking (Fig. 4) is also consistent with the greater amount of the ITGB5 ligand, FN, in the FN/LN combined substrate. Analogous to the importance in optimization of the Activin A/BMP4 signaling levels in CM differentiation13,14; we show here that a precise balance of FN and LN improves the differentiation efficiency of CMs from hESCs.
Although the 70:30 FN:LN directed the highest percentage of CM+ cells for both H7 and H9 hESC lines, the CM derivation of H9 cells on gelatin proved to be as efficient as the 100% FN and 50:50 FN:LN, while the 30:70 FN:LN and 100% LN proved to be the least efficient substrates for CM induction from both hESC lines. Moreover, the 70% FN is also consistent with a larger percentage of cells expressing the corresponding integrin ITGB5 (30% and 45% in H7 and H9, respectively) compared with only 20% and 35% ITGB4-expressing cells in H7 and H9 derivation cultures. Combined, these data indicate that the FN:LN signaling in CM fate is facilitated by the crosstalk between the ITGB5+ and the ITGB4+ cells.
One of the primary mechanisms for ECM signaling is the intracellular signal transduction pathway activated following the binding of ECM molecules to integrin receptors on the cell. The interplay between ECM components and integrin receptors regulate signal transduction in cells by first associating with FAK51 and subsequently initiating several downstream signaling events, including activation of ERK cascades that are known to regulate many developmentally relevant processes,52,53 including cell fate.54,55 The ITGB4, ITGB5, and ERK functional blocking studies, through inactivation of MEK activity, confirm that the 70:30 FN:LN ECM activated integrin signaling pathways result in phosphorylation of FAK and MEK/ERK signaling.
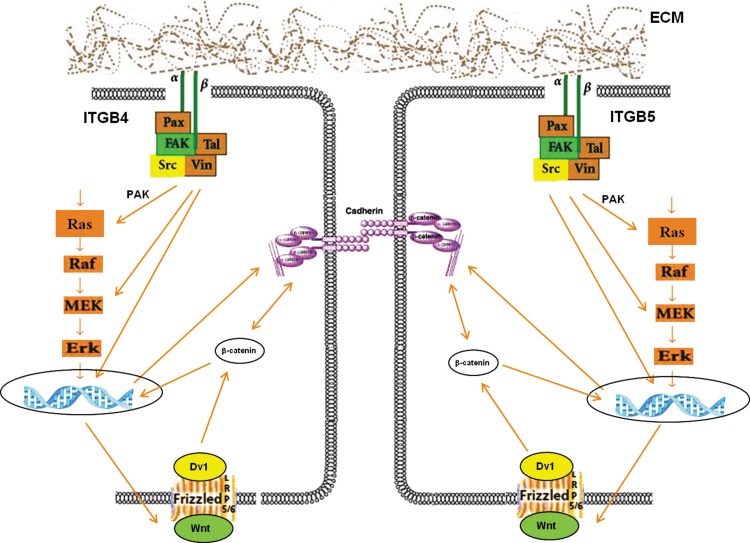
Because integrins and growth factors share common elements in their signaling pathways, there are many opportunities for integrin signals to modulate growth factor signals and for growth factors to modulate integrin signaling.56 Moreover, these activation pathways can be intercepted at a number of locations. For example, Raf activation is thought to be downstream of integrin-regulated p21-activated kinase, whereas MEK activation is downstream of integrin-regulated FAK.57,58 Combining current dogma on integrin signaling with our integrin data, a partial mechanism is proposed (Fig. 6). Integrin signaling on distinct cells begins with the clustering of dimeric (α,β) integrin receptors on distinct cells followed by induced recruitment of focal adhesion proteins (e.g., vinculin, paxillin, talin) and subsequent activation of integrin-associated signaling cascades, which among others, include such protein as FAK and src.59 Additional arrows are included because integrins can also intersect MAP kinase pathways at multiple points, including activation of Raf, activation of MEK, and nuclear translocation of Erk, thus enhancing or mitigating the effects of growth factor signaling, acting as genuine signaling receptors themselves.56
FIG. 6.
Schematic drawing summarizing the mechanisms for ECM signaling in cardiomyocyte differentiation. The FN:LN ECM activates integrin, FAK, and MEK/ERK pathways, and in turn leads to the specification of cardiomyocyte differentiation from hESCs.
However, the spatial organization of the ITGB4+ and ITGB5+ cells indicate a stronger role for direct cell-to-cell communication in ITGB4/5 signaling rather than an indirect communication by release of soluble growth factors. The requirement of cell-to-cell signaling implicates the role of β-catenin signaling associated with cadherin cell adhesion molecules,60 and one of the most powerful pathways directing cardiac fate is the β-catenin/Wnt signaling.61 In general, Wnts are powerful regulators of cell proliferation and differentiation, and their signaling pathway involves proteins that directly participate in both gene transcription and cell adhesion. In fact, the multiple nodes that link cell adhesion to Wnt signaling indicate that these two processes may have coevolved (reviewed by Nelson and Nusse62). Therefore, our proposed mechanism (Fig. 6) highlights the integrin signaling crosstalk cooperating with cadherin-dependent β-catenin signaling to increase β-catenin/Wnt differentiation of cardiac fate. Note that many of the specific components of this mechanism remain unclear, including the specifics of how ITGB4 and ITGB5 signaling are distinct from one another.
Although this manuscript focused on investigating the ITGB4 or ITGB5 signaling, the PCR array results also suggest that adhesion molecules CNTN1 and CTGF are involved in CM differentiation. CNTN1, a ligand for Notch, has been shown to promote the oligodendrocyte maturation.63 The Notch pathway is also a crucial cell-fate regulator for proper heart development,64,65 suggesting the CNTN1 might have a functional role in promoting CM fate through Notch signaling pathway. Additionally, while CTGF has been described as inducing the differentiation of many cells,66–72 a role in CM specification has not yet been suggested.
Supplementary Material
Abbreviations Used
- 3D
three-dimensional
- bFGF
basic fibroblast growth factor
- BMP
bone morphogenetic protein
- CM
cardiomyocyte
- DAPI
4′,6-diamidino-2-phenylindole
- EB
embryoid body
- EC
endothelial cell
- ECM
extracellular matrix
- ERK
extracellular signal-regulated kinase
- ESC
embryonic stem cell
- FACS
fluorescence activated cell scanning
- FAK
focal adhesion kinase
- FITC
fluorescein isothiocyanate
- FN
fibronectin
- GFP
green fluorescence protein
- hESC
human embryonic stem cell
- ITGB
integrin β
- LN
laminin
- MHC
myosin heavy chain
- PBS
phosphate-buffered saline
- SMC
smooth muscle cell
- TRITC
tetramethyl rhodamine isothiocyanate
- VEGF
vascular endothelial growth factor
Acknowledgment
This work was funded by a New Faculty Award from the California Institute of Regenerative Medicine (RN2-00921-1).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Miniño AM, Murphy SL, Xu J, et al. Deaths: final data for 2008. National Vital Statistics Reports. National Center for Health Statistics 59. Available at: www.cdc.gov/nchs/data/nvsr/nvsr59/nvsr59_10.pdf [PubMed]
- 2.Xu C, Police S, Rao N, et al. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res. 2002;91:501–508 [DOI] [PubMed] [Google Scholar]
- 3.Kehat I, Kenyagin-Karsenti D, Snir M, et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mummery C, Ward-van Oostwaard D, Doevendans P, et al. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–2740 [DOI] [PubMed] [Google Scholar]
- 5.Laflamme MA, Chen KY, Naumova AV, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024 [DOI] [PubMed] [Google Scholar]
- 6.van Laake LW, Passier R, Doevendans PA, et al. Human embryonic stem cell-derived cardiomyocytes and cardiac repair in rodents. Circ Res. 2008;102:1008–1010 [DOI] [PubMed] [Google Scholar]
- 7.Shiba Y, Fernandes S, Zhu WZ, et al. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature. 2012;489:322–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caspi O, Huber I, Kehat I, et al. Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. J Am Coll Cardiol. 2007;50:1884–1893 [DOI] [PubMed] [Google Scholar]
- 9.Leor J, Gerecht S, Cohen S, et al. Human embryonic stem cell transplantation to repair the infarcted myocardium. Heart. 2007;93:1278–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bongso A, Fong CY, Gauthaman K.Taking stem cells to the clinic: major challenges. J Cell Biochem. 2008;105:1352–1360 [DOI] [PubMed] [Google Scholar]
- 11.Yang L, Soonpaa MH, Adler ED, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528 [DOI] [PubMed] [Google Scholar]
- 12.Burridge PW, Thompson S, Millrod MA, et al. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6:e18293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kattman SJ, Witty AD, Gagliardi M, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240 [DOI] [PubMed] [Google Scholar]
- 14.Sa S, McCloskey K.Activin A and BMP4 signaling for efficient cardiac differentiation of H7 and H9 human embryonic stem cells. J Stem Cells Regen Med. 2012;8:198–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu C, Police S, Hassanipour M, et al. Efficient generation and cryopreservation of cardiomyocytes derived from human embryonic stem cells. Regen Med. 2011;6:53–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sa S, McCloskey KE.Stage-specific cardiomyocyte differentiation method for H7 and H9 human embryonic stem cells. Stem Cell Rev. 2012;8:1120–1128 [DOI] [PubMed] [Google Scholar]
- 17.Guilak F, Cohen DM, Estes BT, et al. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5:17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozario T, DeSimone DW.The extracellular matrix in development and morphogenesis: a dynamic view. Dev Biol. 2009;341:126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutolf MP, Hubbell JA.Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47–55 [DOI] [PubMed] [Google Scholar]
- 20.Ruest PJ, Roy S, Shi E, et al. Phosphospecific antibodies reveal focal adhesion kinase activation loop phosphorylation in nascent and mature focal adhesions and requirement for the autophosphorylation site. Cell Growth Differ. 2000;11:41–48 [PubMed] [Google Scholar]
- 21.Boudreau NJ, Jones PL.Extracellular matrix and integrin signalling: the shape of things to come. Biochem J. 1999;339(Pt 3):481–488 [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JS, Ha L, Kwon IK, et al. The role of focal adhesion kinase in BMP4 induction of mesenchymal stem cell adipogenesis. Biochem Biophys Res Commun. 2013;435:696–701 [DOI] [PubMed] [Google Scholar]
- 23.Bai B, He J, Li YS, et al. Activation of the ERK1/2 signaling pathway during the osteogenic differentiation of mesenchymal stem cells cultured on substrates modified with various chemical groups. Biomed Res Int. 2013;2013:361906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battista S, Guarnieri D, Borselli C, et al. The effect of matrix composition of 3D constructs on embryonic stem cell differentiation. Biomaterials. 2005;26:6194–6207 [DOI] [PubMed] [Google Scholar]
- 25.Duan Y, Liu Z, O'Neill J, et al. Hybrid gel composed of native heart matrix and collagen induces cardiac differentiation of human embryonic stem cells without supplemental growth factors. J Cardiovasc Transl Res. 2011;4:605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Klos M, Wilson GF, et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ Res. 2012;111:1125–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baharvand H, Azarnia M, Parivar K, et al. The effect of extracellular matrix on embryonic stem cell-derived cardiomyocytes. J Mol Cell Cardiol. 2005;38:495–503 [DOI] [PubMed] [Google Scholar]
- 28.George EL, Baldwin HS, Hynes RO.Fibronectins are essential for heart and blood vessel morphogenesis but are dispensable for initial specification of precursor cells. Blood. 1997;90:3073–3081 [PubMed] [Google Scholar]
- 29.George EL, Georges-Labouesse EN, Patel-King RS, et al. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–1091 [DOI] [PubMed] [Google Scholar]
- 30.Hilenski LL, Ma XH, Vinson N, et al. The role of beta 1 integrin in spreading and myofibrillogenesis in neonatal rat cardiomyocytes in vitro. Cell Motil Cytoskeleton. 1992;21:87–100 [DOI] [PubMed] [Google Scholar]
- 31.van Dijk A, Niessen HW, Zandieh Doulabi B, et al. Differentiation of human adipose-derived stem cells towards cardiomyocytes is facilitated by laminin. Cell Tissue Res. 2008;334:457–467 [DOI] [PubMed] [Google Scholar]
- 32.Xu C, Inokuma MS, Denham J, et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974 [DOI] [PubMed] [Google Scholar]
- 33.Sanchez A, Jones WK, Gulick J, et al. Myosin heavy chain gene expression in mouse embryoid bodies. An in vitro developmental study. J Biol Chem. 1991;266:22419–22426 [PubMed] [Google Scholar]
- 34.Baba S, Heike T, Yoshimoto M, et al. Flk1(+) cardiac stem/progenitor cells derived from embryonic stem cells improve cardiac function in a dilated cardiomyopathy mouse model. Cardiovasc Res. 2007;76:119–131 [DOI] [PubMed] [Google Scholar]
- 35.Kubalak SW, Miller-Hance WC, O'Brien TX, et al. Chamber specification of atrial myosin light chain-2 expression precedes septation during murine cardiogenesis. J Biol Chem. 1994;269:16961–16970 [PubMed] [Google Scholar]
- 36.Wobus AM, Rohwedel J, Maltsev V, et al. Development of cardiomyocytes expressing cardiac-specific genes, action potentials, and ionic channels during embryonic stem cell-derived cardiogenesis. Ann N Y Acad Sci. 1995;752:460–469 [DOI] [PubMed] [Google Scholar]
- 37.Nguyen D, Sa S, Pegan JD, et al. Tunable shrink-induced honeycomb microwell arrays for uniform embryoid bodies. Lab Chip. 2009;9:3338–3344 [DOI] [PubMed] [Google Scholar]
- 38.Hodgkiss-Geere HM, Argyle DJ, Corcoran BM, et al. Characterisation and cardiac directed differentiation of canine adult cardiac stem cells. Vet J. 2012;191:176–182 [DOI] [PubMed] [Google Scholar]
- 39.Elliott DA, Braam SR, Koutsis K, et al. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods. 2011;8:1037–1040 [DOI] [PubMed] [Google Scholar]
- 40.Martin-Puig S, Wang Z, Chien KR.Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell. 2008;2:320–331 [DOI] [PubMed] [Google Scholar]
- 41.Hidaka K, Lee JK, Kim HS, et al. Chamber-specific differentiation of Nkx2.5-positive cardiac precursor cells from murine embryonic stem cells. FASEB J. 2003;17:740–742 [DOI] [PubMed] [Google Scholar]
- 42.Lee EC, Lotz MM, Steele GD Jr, et al. The integrin alpha 6 beta 4 is a laminin receptor. J Cell Biol. 1992;117:671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaudesius G, Miragoli M, Thomas SP, et al. Coupling of cardiac electrical activity over extended distances by fibroblasts of cardiac origin. Circ Res. 2003;93:421–428 [DOI] [PubMed] [Google Scholar]
- 44.Horton RE, Auguste DT.Synergistic effects of hypoxia and extracellular matrix cues in cardiomyogenesis. Biomaterials. 2012;33:6313–6319 [DOI] [PubMed] [Google Scholar]
- 45.Corda S, Samuel JL, Rappaport L.Extracellular matrix and growth factors during heart growth. Heart Fail Rev. 2000;5:119–130 [DOI] [PubMed] [Google Scholar]
- 46.Macfelda K, Kapeller B, Wilbacher I, et al. Behavior of cardiomyocytes and skeletal muscle cells on different extracellular matrix components—relevance for cardiac tissue engineering. Artif Organs. 2007;31:4–12 [DOI] [PubMed] [Google Scholar]
- 47.Hanson KP, Jung JP, Tran QA, et al. Spatial and temporal analysis of extracellular matrix proteins in the developing murine heart: a blueprint for regeneration. Tissue Eng Part A. 2013;19:1132–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen YC, Lee DC, Tsai TY, et al. Induction and regulation of differentiation in neural stem cells on ultra-nanocrystalline diamond films. Biomaterials. 2010;31:5575–5587 [DOI] [PubMed] [Google Scholar]
- 49.Mruthyunjaya S, Manchanda R, Godbole R, et al. Laminin-1 induces neurite outgrowth in human mesenchymal stem cells in serum/differentiation factors-free conditions through activation of FAK-MEK/ERK signaling pathways. Biochem Biophys Res Commun. 2009;391:43–48 [DOI] [PubMed] [Google Scholar]
- 50.Kohan M, Muro AF, White ES, et al. EDA-containing cellular fibronectin induces fibroblast differentiation through binding to alpha4beta7 integrin receptor and MAPK/Erk 1/2-dependent signaling. FASEB J. 2010;24:4503–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlaepfer DD, Hunter T.Signal transduction from the extracellular matrix—a role for the focal adhesion protein-tyrosine kinase FAK. Cell Struct Funct. 1996;21:445–450 [DOI] [PubMed] [Google Scholar]
- 52.Wozniak MA, Modzelewska K, Kwong L, et al. Focal adhesion regulation of cell behavior. Biochim Biophys Acta. 2004;1692:103–119 [DOI] [PubMed] [Google Scholar]
- 53.Robles E, Gomez TM.Focal adhesion kinase signaling at sites of integrin-mediated adhesion controls axon pathfinding. Nat Neurosci. 2006;9:1274–1283 [DOI] [PubMed] [Google Scholar]
- 54.Saleem S, Li J, Yee SP, et al. beta1 integrin/FAK/ERK signalling pathway is essential for human fetal islet cell differentiation and survival. J Pathol. 2009;219:182–192 [DOI] [PubMed] [Google Scholar]
- 55.Salasznyk RM, Klees RF, Williams WA, et al. Focal adhesion kinase signaling pathways regulate the osteogenic differentiation of human mesenchymal stem cells. Exp Cell Res. 2007;313:22–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwartz MA, Ginsberg MH.Networks and crosstalk: integrin signalling spreads. Nat Cell Biol. 2002;4:E65–E68 [DOI] [PubMed] [Google Scholar]
- 57.Juliano RL, Aplin AE, Howe AK, et al. Integrin regulation of receptor tyrosine kinase and G protein-coupled receptor signaling to mitogen-activated protein kinases. Methods Enzymol. 2001;333:151–163 [DOI] [PubMed] [Google Scholar]
- 58.Schwartz MA, Assoian RK.Integrins and cell proliferation: regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J Cell Sci. 2001;114:2553–2560 [DOI] [PubMed] [Google Scholar]
- 59.Ingber DE.Tensegrity II. How structural networks influence cellular information processing networks. J Cell Sci. 2003;116:1397–1408 [DOI] [PubMed] [Google Scholar]
- 60.Gumbiner BM.Regulation of cadherin adhesive activity. J Cell Biol. 2000;148:399–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakamura T, Sano M, Songyang Z, et al. A Wnt- and beta -catenin-dependent pathway for mammalian cardiac myogenesis. Proc Natl Acad Sci U S A. 2003;100:5834–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nelson WJ, Nusse R.Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu QD, Ang BT, Karsak M, et al. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell. 2003;115:163–175 [DOI] [PubMed] [Google Scholar]
- 64.Gude N, Sussman M.Notch signaling and cardiac repair. J Mol Cell Cardiol. 2012;52:1226–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de la Pompa JL, Epstein JA.Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22:244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yosimichi G, Nakanishi T, Nishida T, et al. CTGF/Hcs24 induces chondrocyte differentiation through a p38 mitogen-activated protein kinase (p38MAPK), and proliferation through a p44/42 MAPK/extracellular-signal regulated kinase (ERK). Eur J Biochem. 2001;268:6058–6065 [DOI] [PubMed] [Google Scholar]
- 67.Safadi FF, Xu J, Smock SL, et al. Expression of connective tissue growth factor in bone: its role in osteoblast proliferation and differentiation in vitro and bone formation in vivo. J Cell Physiol. 2003;196:51–62 [DOI] [PubMed] [Google Scholar]
- 68.Croci S, Landuzzi L, Astolfi A, et al. Inhibition of connective tissue growth factor (CTGF/CCN2) expression decreases the survival and myogenic differentiation of human rhabdomyosarcoma cells. Cancer Res. 2004;64:1730–1736 [DOI] [PubMed] [Google Scholar]
- 69.Asano M, Kubota S, Nakanishi T, et al. Effect of connective tissue growth factor (CCN2/CTGF) on proliferation and differentiation of mouse periodontal ligament-derived cells. Cell Commun Signal. 2005;3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang JJ, Ye F, Cheng LJ, et al. Osteogenic differentiation of mesenchymal stem cells promoted by overexpression of connective tissue growth factor. J Zhejiang Univ Sci B. 2009;10:355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morrison BL, Jose CC, Cutler ML.Connective Tissue Growth Factor (CTGF/CCN2) enhances lactogenic differentiation of mammary epithelial cells via integrin-mediated cell adhesion. BMC Cell Biol 2010;11:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee CH, Shah B, Moioli EK, et al. CTGF directs fibroblast differentiation from human mesenchymal stem/stromal cells and defines connective tissue healing in a rodent injury model. J Clin Invest. 2010;120:3340–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.