Abstract
Extensive genetic and genomic studies of the relationship between alcohol drinking preference and withdrawal severity have been performed using animal models. Data from multiple such publications and public data resources have been incorporated in the GeneWeaver database with >60,000 gene sets including 285 alcohol withdrawal and preference-related gene sets. Among these are evidence for positional candidates regulating these behaviors in overlapping quantitative trait loci (QTL) mapped in distinct mouse populations. Combinatorial integration of functional genomics experimental results revealed a single QTL positional candidate gene in one of the loci common to both preference and withdrawal. Functional validation studies in Ap3m2 knockout mice confirmed these relationships. Genetic validation involves confirming the existence of segregating polymorphisms that could account for the phenotypic effect. By exploiting recent advances in mouse genotyping, sequence, epigenetics, and phylogeny resources, we confirmed that Ap3m2 resides in an appropriately segregating genomic region. We have demonstrated genetic and alcohol-induced regulation of Ap3m2 expression. Although sequence analysis revealed no polymorphisms in the Ap3m2-coding region that could account for all phenotypic differences, there are several upstream SNPs that could. We have identified one of these to be an H3K4me3 site that exhibits strain differences in methylation. Thus, by making cross-species functional genomics readily computable we identified a common QTL candidate for two related bio-behavioral processes via functional evidence and demonstrate sufficiency of the genetic locus as a source of variation underlying two traits.
Keywords: data integration, complex traits, behavioral genetics
FINDING the genetic and genomic basis for complex disorders has been a major challenge, particularly for behavioral traits. This is because the disorders are highly heterogeneous in their manifestation and regulation, requiring a match of numerous genes to many aspects of the disorders. A compelling approach to this problem lies in integration of numerous disparate data sets across species, each aimed at characterizing the mechanistic biological underpinnings for the behaviors underlying these disorders as modeled in various species. Alcoholism and alcohol use-related disorders are complex, heritable traits that have been the subject of extensive genomic and genetic investigations (Morozova et al. 2012). These traits are particularly challenging because of their heterogeneity and the presence of multiple overlapping subsystems that subserve them (Gould and Gottesman 2006; Crabbe 2012). Many rodent quantitative trait loci (QTL) have been mapped for alcohol-related traits (Ehlers et al. 2010), gene expression analyses have been performed in a variety of populations, and a long history of alcoholism genetics in humans has led to numerous genome-wide association studies (GWAS) or linkage studies (Enoch 2013). QTL mapping studies historically had very low resolution, and many have been performed using populations for which limited genetic data exist. Publications of gene expression studies typically highlight a few interesting gene centered results, but the bulk of information is rejected due to concern over potential false positives within each study. Integrative strategies have emerged to aggregate this evidence and prioritize recurring relations (Mulligan et al. 2006; Le-Niculescu et al. 2007, 2011; Ponomarev et al. 2012).
Progress toward identification of candidate genes for alcohol-related traits has been limited relative to the number of mapping attempts (Milner and Buck 2010), but there have been compelling successes. Mpdz represents a successfully identified candidate gene for withdrawal seizures (Shirley et al. 2004), and alcohol preference (Milner et al. 2013) with human translational significance (Karpyak et al. 2009) and studies of several alcohol drinking phenotypes in mouse, rat, and human have identified alcohol dehydrogenase and aldehyde dehydrogenase loci as regulators of phenotypic variance (reviewed in Wang et al. 2012). Each of these studies addresses the mediation of a single phenotype, but many alcohol-related phenotypes are highly co-occurring and share biological mediation. We have devised a data integration strategy specifically to identify common mediators of related behavioral phenomena. Here we apply this strategy to examine the relationship between alcohol withdrawal severity and alcohol preference. Studies of these phenomena have been performed across diverse species and experimental protocols, but few common mechanisms are known and the nature of the relationship is an active topic of investigation.
The widespread deployment of genomic technologies has resulted in a wealth of publicly available, whole-genome experimental results, from diverse inquiries to biological and disease-related phenomena. These data sets represent a vast source of information and opportunities for identifying the molecular mechanisms underlying the pathophysiology of disease. In the area of alcoholism alone, there have been at least 142 QTL studies, at least 100 GWAS, and at least 300 gene expression studies performed in numerous species. Each of these technologies is noisy and generates a broad set of candidates. Convergence of evidence is a promising strategy for refinement. However, the data are for the most part poorly integrated, and much currently exists in article supplemental tables or specialty “boutique” databases. Other repositories contain raw data from gene expression and molecular profiling experiments that cannot be efficiently integrated. The diversity and intensiveness of statistical procedures required to re-analyze primary data and the cross-platform and cross-species data integration required for matching identifiers across various experimental platforms place the process out of reach for widespread application. We designed the GeneWeaver software system (Baker et al. 2009, 2012) to address these problems by enabling users to integrate their own data with large numbers of experimental results across multiple species using combinatorial algorithms in a real-time, web-based environment. GeneWeaver contains multiple streams of genomic data, including ontological annotations from Generic Model Organism Databases, QTL positional candidates, transcriptomic results from published experimental studies, user-submitted functional genomics data sets, GWAS candidates, literature annotation, and other strategies. Data are joined across species through the use of Homologene and other identifier integration mappings. Together, these allow seamless aggregation and combinatorial integration of evidence from thousands of empirical results, stored in the form of gene lists that are associated with descriptive text that can be searched to identify gene lists of interest.
To identify gene products that subserve both withdrawal and preference, we made use of data resources and combinatorial data integration tools for integrative functional genomics in the GeneWeaver system. We identified a common candidate gene and assessed whether it satisfied two criteria for causality: first, its functional sufficiency as a cause of phenotypic variation; and second, its genetic sufficiency as a gene or locus that harbors segregating allelic variation consistent with the modifiers of phenotypic variation. We identified such a candidate, confirmed its functional role using an existing gene-targeted knockout in mouse, and detected confirmatory allele distribution patterns across mouse strains that support a causal role for genetic regulation of the gene’s expression in phenotypic variation.
Materials and Methods
Database
GeneWeaver consists of a database of curated functional genomics gene sets. This system has been previously described in detail (Baker et al. 2009, 2012). GeneWeaver tools each operate on bipartite graphs with two sets of vertices, or nodes, representing genes and the gene sets to which they belong (Baker et al. 2009). This data-intensive approach allows for the matching of many genes to many phenotypes through a scalable maximal biclique enumeration algorithm (Zhang et al. 2014). Connections in these bipartite graphs are drawn only from genes to gene sets and therefore represent list membership. The “degree” of a gene vertex in this graph represents the number of gene sets in which it occurs. Key features used in the present study are described below. The GeneWeaver database was queried for “alcohol withdrawal” or for “alcohol preference.” The query results contained overlapping QTL for withdrawal and preference on mouse chromosome 8 among several others. Evaluating the overlapping QTL using the Jaccard similarity tool revealed the extent of this substantial overlap (Jaccard similarity score = 0.5886).
Boolean Gene Set Algebra
The Boolean gene set algebra function enables combination of gene sets by obtaining union, intersection, or high-degree genes—those that are found on many gene sets based on user-defined thresholds. Using this feature, large numbers of gene sets can be collapsed into a limited number of sets representing user-defined conceptual categories. This tool was applied to two data collections: the positional candidates of the overlapping QTL and the numerous preference gene expression sets. The positional candidates for QTL were intersected using Boolean gene set algebra to create a derived gene set consisting of only the candidates in the overlapping region.
A query of the GeneWeaver database for gene sets related to alcohol preference retrieved 86 gene sets from 16 references, each involving different species, strains, and experimental paradigms to assess the association of genes to the concept of alcohol preference. The union of these sets included 8546 genes. GeneWeaver’s Boolean algebra tool was used to derive an aggregate gene set (GS128199) that included genes found in three or more gene expression data sets, narrowing the preference-related candidate set to 1276 unique genes with a high support for preference.
A query of the literature and database revealed that there have been very few alcohol withdrawal genome-wide functional genomic studies; however, there were ample data on the seizure readout. Therefore, the database was queried for withdrawal-related ontology annotations. Associations with two terms in the Mammalian Phenotype Ontology were chosen due to their relatedness to the QTL locus phenotype of withdrawal as measured by handling-induced convulsions (HIC). Gene sets containing murine genes annotated to the Mammalian Phenotype Ontology term MP:0000950 “abnormal seizure responses to pharmacological agent” (Smith and Eppig 2009), as well as the ancestor term MP:0002064 “seizure,” were added to the analysis.
Hierarchical Gene Set Similarity graph
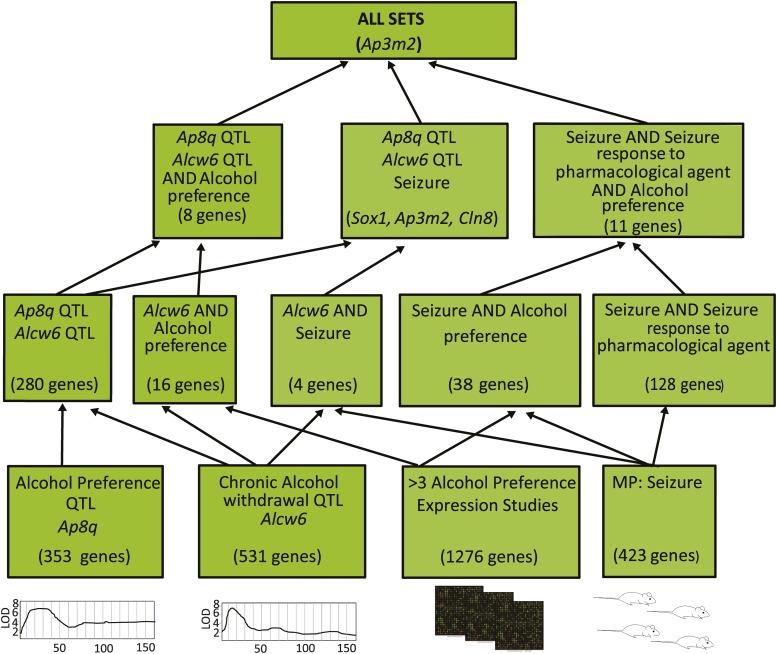
Five Gene sets of interest were then analyzed using GeneWeaver’s Hierarchical Gene Set Similarity (HiSim) graph tool. These include (1) QTL positional candidates for alcohol preference, (2) QTL positional candidates for alcohol withdrawal, (3) aggregate preference-related differentially expressed genes from brain in multiple species from the Boolean analysis above, (4) genes annotated to pharmacological seizure Mammalian Phenotype (MP) Ontology terms, and (5) genes annotated to MP seizure terms. The HiSim tool constructs the hierarchical network of all set–set intersections, and the HiSim Graph tool organizes gene sets based on the genes that they contain, presenting a network graph of hierarchical intersections among a collection of input gene sets and enabling discovery of the relationships among these sets. The network was produced from the overlap of maximal bicliques in discrete bipartite structures, and a directed acyclic graph of intersecting gene sets was produced (Baker et al. 2009, 2012; Zhang et al. 2014). Default options were used, such that bootstrapping of branching was performed, homology was used to integrate gene overlap across species, and the minimum number of intersecting genes required to produce a node was set at 1. This tool presents a graph of hierarchical set intersections in which the terminal nodes each represent individual gene sets and each parent node represents populated gene set intersections, found by enumerating all gene–gene set bicliques found among combinations of these sets. In terms of gene sets, the smallest intersections (fewest gene sets, most genes) are at the lower levels, and the largest intersections (most gene sets, fewest genes) are at the top. A single gene was found in the five-way intersection of the sets, Ap3m2. All members of the combined collection of preference-related gene sets that contained Ap3m2 are listed in Table 1.
Table 1. Description of the genome-wide alcohol preference-related gene sets that contained Ap3m2.
| GeneSet ID | Species | Set type | Reference | Gene set name | No. of genes | Fold change | P-value |
|---|---|---|---|---|---|---|---|
| GS3647 | M. musculus | Differential expression meta-analysis | Mulligan et al. (2006) | Differential expression in three alcohol selected line pairs and six isogenic strains of mice (C57BL/6, BALB/c, DBA/2, LP/NJ, FVB/NJ, and B6xFVB F1) | 3250 | −0.85 | <0.002 |
| GS75568 | Rattus norvegicus | Differential expression | Rodd et al. (2008) | Differentially expressed in the nucleus accumbens of inbred alcohol-preferring rats between ethanol- and saccharin-treated groups | 222 | 1.1 | <0.004 |
| GS137407 | Homo sapiens | Differential expression | Ponomarev et al. (2012). | Basolateral amygdala differential co-expression in alcoholic and non-alcoholic subjects | 3172 | −3.88 | <0.0005 |
| GS137413 | H. sapiens | Differential expression | Ponomarev et al. (2012). | Central nucleus of amygdala differential co-expression in alcoholic and non-alcoholic subjects | 2383 | −2.742 | <0.001 |
Ap3m2 was one of 1276 genes in the node “>3 Alcohol Preference Expression Studies” of the HiSim graph analysis (Figure 1) in GeneWeaver.
Ap3m2-deficient mice
A knockout of Ap3m2 in mice was obtained from RIKEN (Nakatsu et al. 2004; Misawa et al. 2008). Briefly, this strain was created with a targeting vector designed to replace the coding sequence downstream of the ATG in exon 2 with an enhanced green flourescent protein (EGFP) and a floxed neomycin for selection in 129P2/OlaHsd embryonic stem (ES) cells. The selection cassette was removed by crossing to Cre-expressing mice. The mice were then backcrossed to C57BL/6NCrSlc for at least 10 generations, producing B6.129P2-Ap3m2tm1Ohno. However, C57BL/6JJmsSlc may have been used in the backcross when C57BL/6NCrSlc was not available, possibly introducing substrain polymorphisms into the genotype and variations in phenotype compared with a pure inbred strain (Simon et al. 2013). Ap3m2 null embryos were imported to the Jackson Laboratory and implanted in a pseudopregnant mouse. Live litters were introduced into our specific-pathogen free (SPF) facility by hysterectomy derivation. Genotyping was performed using PCR primers specific to the EGFP insert (Supporting Information, Table S1) to detect the null state and primers specific to the targeted genomic region of Ap3m2 that were specific to the wild-type state. Wild-type litter mates were used as controls in all studies. Ap3m2 null mice were used at an age before spontaneous seizures were observed. The Jackson Laboratory follows husbandry practices in accordance with the American Association for the Accreditation of Laboratory Animal Care (AAALAC), and all work was done with the approval of our Institutional Animal Care and Use Committee. Experiments were performed during the light cycle of the mice; all mice were group-housed with pine-shaving bedding (Hancock Lumber) and given environmental enrichment (shepherd shack) and nesting materials. Mice were maintained in conventional SPF barrier facilities and given sterilized, acidified (pH 2.5–3.0) water and 6% fat NIH31 5K52 chow (LabDiet/PMI Nutrition, St. Louis). The mice were identified by ear notching at weaning and moved between cages by forceps.
Chronic alcohol exposure
One-half of a cohort of female mice of each genotype (wild type n = 13 and null n = 17) were exposed to chronic alcohol vapor within their home cages with access to food and water by placing the cages within a vaporization chamber and maintaining their same light–dark cycle. The vaporization chamber, made of clear 0.500-in. thick acrylic (Plas Labs Inc., Lansing, MI), has been previously described and was modified to be 26 in. wide × 24 in. deep and 14 in. high (outside dimensions) (Goldstein 1972; Becker and Hale 1993). The compartment has a stainless steel piano hinged door, two spring tension clamps, and two 1/8-in. National Pipe Thread (NPT) holes on the back wall for vapor introduction. The front door has a gasket that was set with silicone into 0.1875-in. deep grooves on the door. The compartment accommodated two cages (four pens) of mice. Ethanol vaporization was accomplished by forcing a continuous stream of air, using the internal JAX air taps, through a 1-in. spherical diffusing stone in a 1-liter Erlenmeyer flask of 95% ethanol, delivering ∼186.7 µl/min, and mixing with air delivered at ∼10 liters/min. We always maintained the air-flow rate of 10–12 liters/min to meet the animals’ respiratory requirements. The ratios and rate were regulated by flow meters outside the box to create an environment of ∼13.1 mg ethanol/liter of air. The ethanol vapor concentration within the 9–13 mg/liter range in Swiss-Webster mice resulted in a very tolerable blood ethanol concentration of slightly >1 mg/ml that was stable over the 3 days of exposure (Goldstein 1972). Chamber ethanol concentrations were monitored twice daily. Air samples from the inhalation chamber (5 ml) were collected through a septum in the door with a 60-ml syringe and needle, mixed with 55 ml of room air, and forced through a Department of Transportation-certified breathalyzer (Lifeloc FC10, Wheat Ridge, CO). These readings were compared to a standard curve to calculate chamber ethanol concentration (Logan et al. 2010). Pyrazole HCl (Sigma) injections of 68.1 mg/kg were given in experiments to inhibit alcohol dehydrogenase and stabilize blood ethanol concentrations. At the start of the experiment a priming dose of ethanol (1.5 g/kg i.p.) was given to the mice together with the first injection of pyrazole to avoid a 24-hr lag in attaining a stable blood ethanol concentration (BEC). Pyrazole injections were repeated at 24 and 48 hr. BEC was calculated from 30 µl of serum obtained by retro-orbital bleeding and analyzed on a Beckman DXC. The second half of the cohort were control (air-exposed) mice that were given pyrazole injections daily, but a priming dose of saline instead of ethanol.
Handling-induced convulsions
HIC scoring was performed as described (Crabbe et al. 1991) and modified from Goldstein (1972). HIC scores are a measure of alcohol withdrawal. Every hour for the 12 hr after removal from continuous alcohol exposure mice were gently picked up by the tail and, if that failed to elicit a convulsion, the mouse was spun gently through a 180°arc. The HIC scores were independently assigned by an experimenter blind to treatment in our laboratory using the published scales (Crabbe et al. 1991). Briefly, mice scored from a “0” in the absence of convulsion following tail rotation to a “5” with tonic-clonic convulsions with just tail lift and no rotation. The area under the curve was calculated using the trapezoidal rule. Statistical analyses were conducted using JMP 10 (SAS Institute). The best model is
where ε is random error. The β-parameters were estimated by ordinary least squares and the type III sum of squares was considered for ε in the ANOVA model. In all cases, the full model was fit and reduced by dropping nonsignificant interactions followed by main effects. Outliers with an absolute Studentized residual |Z| > 2 were removed.
Two-bottle choice alcohol preference test
The two-bottle free-choice methods were followed as previously described (Bachmanov et al. 2002; Mulligan et al. 2008). Individually housed mice (null n = 20, wild type n = 19) were presented with one tube containing an ethanol solution (The Jackson Lab Store/VWR International, Radnor, PA) made in sterilized, acidified (pH 2.5–3.0) water provided by the research animal facility. Mice always had one additional tube, which contained sterilized, acidified water alone. These sipper tubes were constructed from sterile 50-ml polypropylene centrifuge tubes (#430291 Corning, Corning, NY), fitted with a #6 one-holed rubber stopper (#14-135J Fischer-Scientific) and a 2.5-in. stainless steel sipper (catalog no. SPS-SM tube 2.5 with ball, Sta-Pure Systems, Carnegie, PA). The sippers were placed into the food and drink holder sides of the standard JAX mouse box tops. Food was available ad libitum and spread equally around both drinking tubes to avoid food-associated tube preference. The physical positions of the tubes were switched daily to avoid effects of potential side preference. Sipper tubes were weighed daily during the middle of the light cycle to the nearest 0.01 g to measure the consumption of each solution. Prior to testing, mice were allowed to adapt to single housing with multiple sipper tubes, each containing only sterilized, acidified water for 8 days. The experimental period occurred over 20 days. During the first 4 days the mice received (3%) ethanol (w/v) and during the next 4 days they received (6%) ethanol (w/v) continuing with 9, 12, and 15%. Control bottles of ethanol and water were placed in an empty mouse box. They were also weighed daily and their positions alternated. This value, fluid loss, was subtracted from the daily results to account for any spillage observed in the experimental samples due to handling. Box changes included transfer of some used bedding and nesting materials to the new pen. These were performed by the researcher to decrease stress. Cage changes were done between every second dose, i.e., every 8 days (between 6 and 9% and between 12 and 15%). Leaky bottles affected 33 of 1560 total observations. In these cases, the missing value was replaced with the other observation obtained from the same mouse, dose, and side to allow all cases to be included in the analysis. ANOVA, with dose as a repeated measure, was used to assess genotype effects on alcohol preference. The full model is:
where ε is random error. The β-parameters were estimated by ordinary least squares, and the type III sum of squares was considered ε in the ANOVA model. In all cases, the full model was fit and reduced by dropping nonsignificant interactions followed by main effects.
Sequencing Ap3m2 in QTL cross founders
The alcohol withdrawal QTL (Alcw6) was originally mapped in an F2 cross of the inbred Withdrawal Seizure-Prone (iWSP) and inbred Withdrawal Seizure-Resistant (iWSR) selected lines of mice (Bergeson et al. 2003). Withdrawal Seizure Prone (iWSP-1 and iWSP-2) and Resistant (iWSR-1 and iWSR-2) selected lines (Crabbe et al. 1985) were derived from a heterogeneous stock (HS/Ibg) originally created using eight inbred strains (A, AKR, BALB/c, C3H/2, C57BL, DBA/2, Is/Bi, RIII) (McClearn et al. 1970). After selection for severe or mild alcohol withdrawal HIC for 25 generations, each selected line was subsequently inbred for 38 and 40 generations. The specific mice used for the F2 cross had subsequently been inbred to homozygosity (iWSP-2 and iWSR-1) and retained the large withdrawal severity differences present in the original WSP-2 and WSR-1 selected lines. To determine whether these strains had allelic variation in Ap3m2, the UTR, exons, and intron–exon boundaries were sequenced. Frozen spleens from male and female iWSP-2 mice at S26F57 and iWSR-1 at S26F59 were obtained and later homogenized with Nuclei Lysis Solution (Wizard Genomic DNA Purification Kit (Promega, Madison, WI) using the GentleMacs dissociator (Miltenyi, Bergisch Gladbach, Germany). Genomic DNA was further extracted following the manufacturer’s protocol. Quality control was performed measuring the quantity using a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE). The quality of the DNA was assessed by gel electrophoresis (E-Gel System, Invitrogen, Carlsbad, CA). PCR was performed using genomic DNA as template and various primer pairs throughout Ap3m2 (Table S1) for Sanger sequencing. Primers were designed to amplify exons, UTRs, and intron–exon boundaries. PCR products were purified using Agencout’s AMPure XP magnetic Beads (Beckman Coulter, Brea, CA). The cycle sequencing was performed using Applied Biosystems BigDye Terminator reaction kit Version 3.1 (Life Technologies, Carlsbad, CA). Sequencing reactions were purified using McLab’s BigDye Cleanup sequencing kit (MCLAB, South San Francisco, CA) Purified reactions are run on an Applied Biosystems 3730xl. Sequence data were analyzed using GeneCodes Sequencher 4.10.1 software (Gene Codes, Ann Arbor, MI).
Test for incomplete homozygosity of selected lines
Our sequence analysis of the iWSP-2 vs. iWSR-1 inbreds used iWSP-2 mice at S26G57 and iWSR-1 mice at S26G59 generations, but the F2 cross mice were derived from iWSP-2 at S26G38 and iWSR-1 at S26G40. Original genotype records from the F2 mapping cross were obtained from S. E. Bergeson (Bergeson et al. 2003). Using the microsatellite marker genotype data from the F2 cross, allele frequencies were estimated for a test of segregation distortion. A χ2 test for a lack of fit to expected allele frequencies was calculated using a liberal single locus of α = 0.05.
Expression QTL mapping in BXD recombinant inbred lines
We made use of publicly available gene expression genetic data from the BXD recombinant inbred (RI) strains. These strains are derived from the inbred progeny of an F2 cross of C57BL/6 (B6) and DBA/2 (D2). They were bred in multiple subpopulations first in the late 1970s at The Jackson Laboratory (Taylor et al. 1977). A second (Taylor et al. 1999) and third (Peirce et al. 2004) cohort have since been bred. These strains have been extensively characterized genetically (Williams et al. 2001; Shifman et al. 2006) and on behavioral, anatomical, physiological, and gene expression traits across multiple tissues and disease-related measures. Most of the data is made available for interactive systems genetic analysis in a web-based resource, GeneNetwork (Chesler et al. 2004). Expression QTL mapping for Ap3m2 was performed using GeneNetwork’s Web QTL tool (Wang et al. 2003). Hippocampus messenger RNA (mRNA) expression data from 93 BXD mice of 74 strains were obtained from GeneNetwork [GN206 UMUTAffy Hippocampus Exon (Feb09) RMA] and queried for Ap3m2 (Mulligan et al. 2012). Using RMA normalized hybridization intensity scores for Illumina probe 4465073, an Ap3m2 expression QTL (eQTL) was mapped with 2000 permutations using Interval Mapping and the Haldane mapping function with no control for other QTL.
Alcohol-induced differential expression of Ap3m2 in iWSP-2 and iWSR-1 Mice
Animals were housed within the Portland Veterans Affairs Medical Center (PVAMC) Veterinary Medical Unit, an AAALAC-approved facility. Cages were polycarbonate or polysulfone with corncob bedding (Bed-o-Cob, The Andersons, Inc., Maumee, Ohio). All mice were weaned at 21 ± 1 days of age and separated by sex into groups of two to five, with cages changed once weekly. All animals were fed standard rodent chow (Purina 5001), except during breeding when dietary fat was increased to 9% (Purina 5008, PMI Nutrition International, Brentwood, MO). Water was available ad libitum. The rooms were maintained at 22 ± 1° and on a 12:12 hr light:dark cycle (lights on at 6 am). Animal care and use were approved by the Institutional Animal Care and Use Committee at the PVAMC and were in compliance with National Institutes of Health and U.S. Department of Agriculture guidelines.
qPCR of iWSP-2 and iWSR-1 Lines
Male mice from iWSR-1 (S26F95, n = 29) and iWSP-2 (S26F94, n = 31) at PVAMC were treated with saline, pyrazole, or pyrazole in addition to 72 hr of ethanol vapor chamber exposure. After 72 hr mice were killed, and brains were stored in RNAlater. Brains were then dissected, and the hippocampus was homogenized in TRIzol (Life Technologies, Carlsbad, CA). Total RNA was isolated by a TRIzol Plus kit (Life Technologies) according to the manufacturer’s method including an on-the-column DNase digestion. The quality of the isolated RNA was assessed using an Agilent 2100 Bioanalyzer instrument (Agilent Technologies, Santa Clara, CA) and RNA 6000 Nano LabChip assay. Total RNA (500 ng) was then reverse-transcribed with random decamers and M-MLV reverse transcription using the Message Sensor RT Kit (Life Technologies). A portion of the complementary DNA was then used in a PCR reaction containing Taqman Universal PCR Master Mix (Life Technologies, Applied Biosystems), which includes AmpliTaq Gold DNA Polymerase, AmpErase UNG, dNTPs with dUTP, Passive Reference 1, and other buffer components. The gene-specific Taqman primer and probe sets (Ap3m2, catalog no. Mm00512823_m1) were obtained from Applied Biosystem’s Assay on Demand service and used according to the manufacturer’s protocols. Each sample was assayed in triplicate at a 10-µl volume. Real-Time PCR was performed in a ViiA 7 system (Life Technologies, Applied Biosystems). The standard protocol of 95° for 10 min to activate the DNA polymerase followed by 40 cycles of amplification was used. The threshold cycle (Ct) was determined using the ViiA 7 software. The data were further analyzed using the ΔΔCt method using Gapdh (Mm99999915_g1) and Actb (Mm00607939_s1) as a normalizer.
Reconstruction of WSP and WSR founder haplotypes
We knew the strains with different alleles for the preference cross because it was a simple F2 between inbred strains (B6 × 129). For the withdrawal QTL, we needed to determine which of the eight heterogeneous stock (HS) progenitor strains contributed to the genotype at Alcw6. To determine the founder origin of Alcw6 genetic variation in the region of the Ap3m2 locus, we reconstructed the iWSP-2 and iWSR-1 haplotypes by dense genotyping and mapping to known extant founder-strain haplotypes. DNA from iWSP-2 mice at S26G57 and iWSR-1 mice at S26G59 was obtained from the Portland Alcohol Research Center and genotyped with the 625,000 SNP Mouse Diversity Array (Yang et al. 2009). A restriction digest was carried out with the DNA using Nsp I and Sty I, and the DNA was then ligated to Nsp I and Sty I adaptors (SNP 6 Core Reagent Kit, Affymetrix, Santa Clara, CA). The samples were PCR-amplified followed by an enzymatic fragmentation and biotin labeling. Approximately 4 µg of biotin-labeled and fragmented DNA was then hybridized onto Mouse Diversity Genotyping Arrays (Affymetrix) for 16 hr at 50°. Post-hybridization staining and washing were performed according to the manufacturer’s protocols using the Fludics Station 450 instrument (Affymetrix). The arrays were then scanned with a GeneChipTM Scanner 3000 laser confocal slide scanner.
The resulting CEL files were processed using the MouseDivGeno R package with a 0.1 confidence score threshold. The SNP founder strain calls were retrieved from CGDSNPDB (Hutchins et al. 2010). Where HS founders were not available, the nearest extant relative was chosen (e.g., I/LnJ for Is/Bi, RIIIS/J for RIII). SNPs that were identical across all strains were discarded, and any heterozygous calls were interpreted as noise and ignored. To infer haplotype origins, a simple sliding-window founder-set intersection method was used. First, all SNPs were mapped to their possible founder strains. Then, a sliding window of 1500 SNPs was used to walk down each chromosome and intersect founder sets until a disjoint call was found. This was then repeated in reverse. Finally, for any remaining genotypes with multiple possible founders, the strain that was most prevalent in nearby single haplotypes was selected. The genotype data for these strains are publically available at http://phenome.jax.org/db/q?rtn=projects/projdet&reqprojid=432 and can be used to characterize other iWSP-2 × iWSR-1 QTL.
SNP analysis
Using data obtained from CGDSNPdb (http://cgd.jax.org/cgdsnpdb/), SANGER (http://www.sanger.ac.uk/resources/mouse/genomes/ (accessed on May 20, 2011), and Mouse Phenome Database (MPD; phenome.jax.org), we identified SNPs that had an allele distribution pattern such that I/LnJ (regional haplotype of iWSP-2) and B6 were similar, but differed from an allele common to 129, AKR/J (regional haplotype for iWSR-1), and DBA/2J.
Regulatory sequence identification
The SNPs that segregated were scanned for regulatory features in the Ensembl.org regulatory feature build (release 73), containing the ENCODE data (Stamatoyannopoulos et al. 2012). In addition, the entire region of the overlapping QTL interval containing the 32 SNPs was analyzed using the Transfac Professional Database (http://www.biobase-international.com) (Matys et al. 2006).
Chromatin immunoprecipitation
To prioritize SNPs, we accessed a DNA-chromatin immunoprecipitation (ChIP) data set. ChIP was performed using an antibody against H3K4me3 (Millipore, #07-473) as previously described (Baker et al. 2014). Chromatin was sheared to mononucleosomes using micrococcal nuclease digestion. Briefly, four ChIP reactions from a single preparation of spermatocyte chromatin were pooled after final DNA elution, concentrated using Agencourt AMPure XP beads (Beckman Coulter), and quantitated using the Qubit double-stranded DNA HS assay (Life Technologies) (as described by Baker et al. 2014). Approximately 7–10 ng of ChIP DNA was used for the sequencing library preparation. For all samples, an equal amount of micrococcal nuclease-treated input DNA was sequenced as a control. ChIP was performed on biological replicates for each genotype (C57BL/6J and DBA/2A). Libraries were prepared for sequencing using Bio Scientific’s NEXTflex ChIP-Seq Kit (protocol version V11.11) without size selection. Amplification of the libraries was done with 20 ml of ligation product and 14–18 cycles of PCR.
High-throughput sequencing and data processing
High-throughput sequencing was performed using the Illumina HiSequation 2000 platform. Raw sequences were aligned to the mouse genome National Center for Biotechnology Information (NCBI) Build 37 (mm9) using BWA (v.0.5.10) (Li and Durbin 2009) with default settings. The alignments were subsequently filtered to retain only uniquely mapped reads. Peak calling was performed using MACS (v.1.4.2) (Zhang et al. 2008) with input DNA for control and ChIP samples as treatment, setting the P-value to 0.01, and nodup = ‘‘all.’’ Because nucleosomes are protected from micrococcal nuclease digestion, we chose to keep all mapped reads with the same 5′ starting position in the final coverage profiles and when quantifying ChIP-seq signal. The coverage profiles presented are from MACS bedgraph output after tag shifting normalized between samples as reads per million (RPM). We used NucHunter to predict the position of nucleosomes around our locus from ChIP-seq data (Mammana et al. 2013). To visualize nucleosome positions relative to the ChIP-seq read-depth signal and local SNPs, we rescaled nucleosomes to equal length (150 bp).
Results
Bioinformatic analysis
Of the 60,000 gene sets in the GeneWeaver database (Baker et al. 2012), a query for gene sets containing reference to “alcoholism” in their meta-content revealed 2857 gene sets. A specific query for gene sets annotated with “alcohol preference” revealed 163 gene sets and a separate query for “alcohol withdrawal” revealed 122 gene sets. Prominent among the results were four groups of overlapping mouse QTL for alcohol preference and alcohol withdrawal. For further investigation, we anchored our search on a locus on Mus musculus chromosome 8, corresponding to database records from two independent QTL mapping studies. One record was the set of positional candidates at an alcohol withdrawal QTL, Alcw6 [(chromosome (Chr) 8, 12–58.12 Mb; GeneWeaver GSID:128609), identified by measuring the severity of HICs after chronic alcohol exposure in an F2 intercross population derived from iWSP and iWSR strains of mice (Bergeson et al. 2003). A second record was the set of positional candidates at an overlapping QTL Ap8q (Chr 8, 0–40.29 Mb; GeneWeaver GSID:84201) mapped in a [C57BL/6ByJ (B6) × 129P3/J (129)] F2 cross for alcohol preference in a voluntary consumption two-bottle choice paradigm (Bachmanov et al. 2002). The confidence intervals from these two QTL overlap in a Chr 8 region from 8.12 to 40.29 Mb on M.m. 9 (Build 37). The intersection of positional candidate gene sets for these two QTL contains 280 genes.
To test whether genes in this derived set had functional relevance to alcohol preference and withdrawal, the hierarchical intersections of other preference and withdrawal gene sets were obtained. Our strategy was first to combine preference gene sets and withdrawal gene sets such that we had one composite set for each trait. Each composite set consisted only of highly supported genes. We then used combinatorial analysis tools in GeneWeaver to enumerate the intersections among these sets. The union of all gene sets from 86 alcohol preference-related gene sets contained 8546 genes. This large number of differentially expressed genes undoubtedly contains numerous false positives. Convergent evidence filters implemented in GeneWeaver prioritize genes among these results such that the minimum degree (number of intersecting genes) threshold on connectivity of genes to gene sets was set at 3. The resulting derived set contained 1276 genes and was stored as the “alcohol preference gene set” (GeneWeaver GSID:128199). Gene sets associated with seizure, a phenotypic endpoint of alcohol withdrawal in rodent studies, were retrieved from the database. These included genes annotated to the Mammalian Phenotype Ontology terms “Seizure [MP:0002064]” and “Seizure response to pharmacological agent [MP:0000950]” either directly or through transitive closure of child term annotations. Together, these aggregate gene sets—(1) QTL positional candidates (genes that lie at the intersection the preference QTL Ap8q and the withdrawal QTL Alcw6), (2) preference-related genes, (3) pharmacological seizure-related genes, and (4) seizure-related genes—were analyzed using GeneWeaver’s tools for enumeration of biclique (completely connected subgraph) intersections (Zhang et al. 2014), which computes the hierarchical overlap of gene sets in the query (Figure 1). A single gene, Adapter-related protein complex 3, μ-subunit 2 (Ap3m2), was found to be completely connected to all aggregate gene sets.
Figure 1.
A hierarchical similarity chart of alcohol preference- and withdrawal-related gene sets. At the bottom of the chart are individual gene sets derived from aggregate alcohol preference and withdrawal QTL, differential expression, and mutation characterization. Each successive level from the bottom to the top of the chart represents increasingly higher order intersections among these gene sets, with the most highly connected genes at the top.
Phenotypic validation of Ap3m2 as a preference and withdrawal-related gene
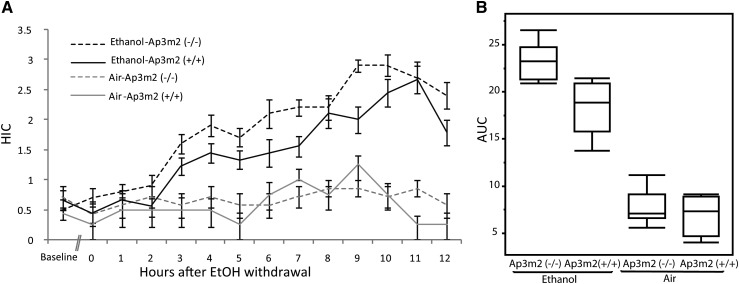
To evaluate the functional role of Ap3m2 perturbation in alcohol preference and withdrawal, B6.129P2-Ap3m2tm1Ohno (Nakatsu et al. 2004; Misawa et al. 2008) female mice of each genotype (wild type n = 13 and null n = 17) were exposed for 72 hr to chronic ethanol vapor or control air (Goldstein 1972; Becker and Hale 1993). There was no baseline difference (baseline in Figure 2A) in seizure severity as measured by HIC between the 19 ethanol-treated mice and the 11 control air mice prior to placement in the chambers F(1, 29) = 0.08, P = 0.7669 (Figure 2A). In a repeated measures Mulitivariate Analysis of Variance (MANOVA) across the 12-hr time span, there was a significant main effect of genotype F(1,27) = 10.8132, P = 0.0027, and a significant time × treatment effect, F(11,17) = 5.1294, P = 0.0014, but no time × treatment × genotype interaction. There was a significant genotype × treatment interaction in the HIC area under the curve (AUC), F(3, 24) = 94.05, P = 0.0325 such that Ap3m2 deletion mutants had a greater effecter of alcohol exposure on HIC AUC (post hoc Tukey–Kramer HSD difference 3.8 ± 0.976, P < 0.0018) than wild-type controls (Figure 2, A and B), demonstrating that the absence of Ap3m2 results in an increased withdrawal severity phenotype. Furthermore, AUC was the trait for which the Alcw6 QTL was mapped. The average blood ethanol concentration at 48 hr was elevated to 1.3 mg/ml and was not statistically different (P = 0.07) between wild-type and Ap3m2tm1Ohno mice, indicating that pharmacokinetics of alcohol are similar in the two groups and that withdrawal was to the same effective amount (Figure S1).
Figure 2.
Increased chronic alcohol-withdrawal effects in Ap3m2 (−/−) knockout mice. B6.129P2-Ap3m2tm1Ohno/EJC [Ap3m2 (−/−)] were exposed to ethanol vapor for 72 hr and then assayed for HIC. (A) HIC curves over time after cessation of ethanol vapor exposure for Ap3m2 (−/−) mice and their wild-type littermates. (B) The area under the curve for withdrawal seizures over time was significantly greater in Ap3m2 (−/−) than in littermate controls with a genotype × treatment (F(3, 24) = 94.05, P = 0.0325).
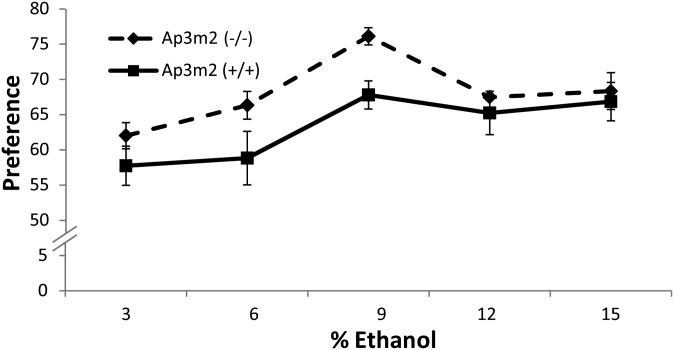
A second cohort of mice (male null n = 14, female null n = 6, male wild type n = 12, female wild type n = 7) were tested using the two-bottle choice paradigm (Bachmanov et al. 1996). Mice were given access to a series of 3, 6, 9, 12, and 15% ethanol solutions vs. water with doses increased every 4 days. The Ap3m2tm1Ohno mice had significantly higher preference for ethanol than the wild-type controls across concentrations (between groups F(1,36) = 4.8646, P = 0.0339) (Figure 3). Post hoc ANOVA at each concentration, with a Bonferroni adjustment for five concentrations, revealed a significant genotype effect at 9% ethanol, F(1,37) = 12.93, and P = 0.0009 even without body weight in the model (Figure 3). No differences were found in total fluid intake over time by genotype F(4,33) = 0.01526, P = 0.9605, but there was an overall genotype effect (F(1,36) = 5.7433, P = 0.0219). For ethanol consumption, there was a significant time × genotype interaction such that Ap3m2−/− mice consumed more ethanol than +/+ mice (F(4,33) = 2.8294, P = 0.0402) (Figure S2). Post hoc ANOVA at each concentration, with a Bonferroni adjustment for five concentrations, revealed no significant genotype effect at each dose.
Figure 3.
Increased alcohol preference in Ap3m2 (−/−) knockout mice. Alcohol preference in B6.129P2-Ap3m2tm1Ohno/EJC [Ap3m2 (−/−)] was evaluated using the two-bottle water vs. ethanol choice test with increasing concentrations of ethanol every 4 days for 20 days. The Ap3m2 (−/−) mice had significantly higher preference for ethanol than the wild-type littermates across concentrations (between groups F(1,36) = 4.8646, P = 0.0339).
Genetic analysis of the Ap3m2 region
Although the functional experiments above indicate a role for Ap3m2 as a compelling functional candidate for alcohol-related phenotypes, they alone do not demonstrate genetic sufficiency. To demonstrate this, one must confirm a functional or regulatory polymorphism at or near Ap3m2 sufficient to enable its detection in QTL mapping studies. Importantly, because the traits reported here were originally mapped in two different populations, it is necessary to show that both QTL could be explained by the same, i.e., the pleiotropic, polymorphisms. In the present study, we hypothesize that an allele segregating among both the iWSP-2 × iWSR-1 and B6 × 129 crosses is responsible for trait variation. Typical of early F2-based studies, these two QTL mapping crosses had very low precision due to the number of markers, the number of mice, and the extent of inherent variation. Several genetic and sequence analyses were performed to investigate whether such a variant exists for Ap3m2.
Sequence analysis of B6J vs. 129S shows one 3′ UTR, three 5′ UTR, four coding synonymous SNP, and 69 intronic SNPs in the Ap3m2 gene (Keane et al. 2011). Sequencing the transcribed region as well as intron–exon boundaries of Ap3m2 in genomic DNA from the iWSP-2 and iWSR-1 strains revealed no SNPs within the Ap3m2-coding region or the immediate 5′or 3′ regulatory region. However, the sequenced samples were obtained from mice that were several generations beyond the F2 cross founders. Therefore, the absence of Ap3m2 polymorphisms could have been the result of a loss of allelic diversity during the additional inbreeding generations that occurred between the mapping samples and the extant sequenced strains. For allele fixation to occur, there would have to be at least one copy of a shared allele in both strains. Although it is possible that both strains were heterozygous for the same variants, this is highly unlikely at S26F38 and S26F40 with eight original founders. Therefore, we expected a 50:50 allele frequency if the locus was unlikely to become fixed and a 75:25 allele frequency if the locus had potential for fixation. A 100:0 allele distribution would be observed if the locus had been fixed in iWSP-2 and iWSR-1 strains, but under this condition the QTL would have been undetectable. Analysis of the original F2 genotype frequencies for chromosome 8 markers revealed no loci out of Hardy–Weinberg equilibrium at 50:50 allele frequencies using a liberal single locus (P < 0.05) (Table S2). This indicates that the Ap3m2 region was most likely displaying allelic variation at the time of mapping and does not account for the absence of polymorphisms in Ap3m2 in the sequenced samples. Therefore, allelic differences in Ap3m2 must be upstream or downstream of the transcript-coding region and likely exerted their allelic effects on expression rather than functional variation.
Mapping expression QTL for Ap3m2 in BXD RI Lines
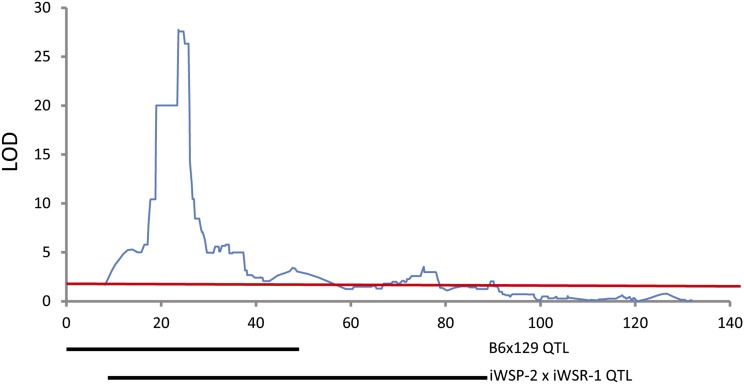
To evaluate whether allelic segregation among inbred strains influenced expression of Ap3m2, we looked for Ap3m2 expression QTL in the BXD RI strains. Alcohol alters the balance between the excitatory pyramidal cell firing and the inhibitory action of the gamma-aminobutyric acid (GABA)-containing interneurons in the hippocampus, affecting the neural circuits underlying preference and withdrawal (Koob and Volkow 2010). We therefore mapped the abundance of Ap3m2 mRNA in the hippocampus using GeneNetwork’s database of BXD recombinant inbred gene expression (Chesler et al. 2005; Mulligan et al. 2012). A significant local (cis-) eQTL for Ap3m2 mapped to proximal chromosome 8, likelihood ratio statistic (LRS) peak = 127.614; P < 0.05 with a 1.5 LOD confidence interval of 22.906–25.498 Mb. (Figure 4 and Figure S3). This finding indicates that a causal polymorphism influencing gene expression must differ between B6 and DBA/2J (D2). The genotype at this locus is correlated with alcohol consumption in two other studies in the BXD RI lines, including sex-specific consumption (GN10138; Fernandez et al. 1999) and withdrawal-induced consumption (GN 12963; Lopez et al. 2011). There is only one directly comparable mapping study of withdrawal seizures in BXD RI, and for this trait only 27 strains were examined (GN 10063-6; Crabbe 1998). No significant or suggestive QTL are found on chromosome 8 in this study. This lack of overlap could be due to the relatively low number of BXD strains that were characterized.
Figure 4.
A cis-eQTL for Ap3m2 expression in the hippocampus of the BXD RI population. The peak eQTL for Ap3m2 maps to chromosome 8 between 22.906 and 25.498 Mb (LOD = 27.74, P < 0.05). Chromosome 8 QTL map is plotted with megabase position (UCSC mm9, NCBI MGSCv37) on the horizontal and with LRS on the vertical [P < 0.05, LOD = 3.79 (red line)].
Confirmation of basal and alcohol-induced expression variation of Ap3m2 iWSP-2 and iWSR-1 lines
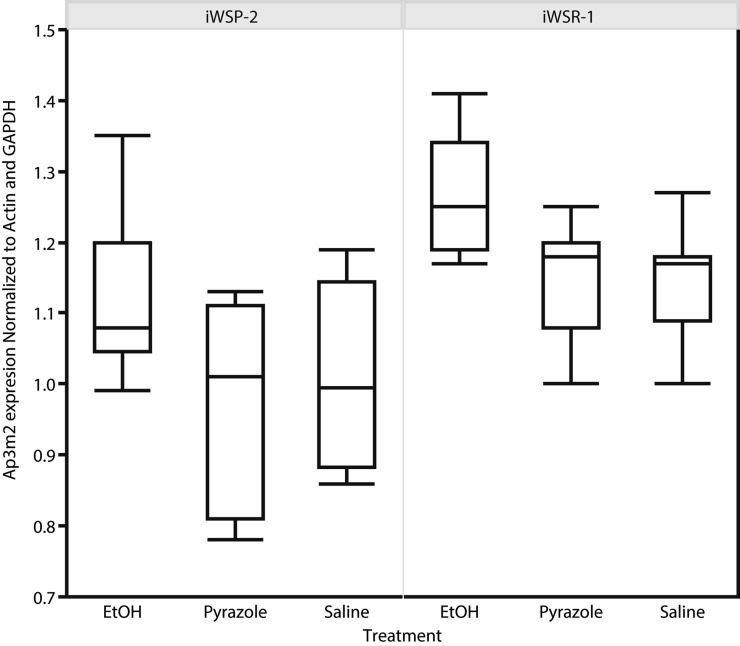
Quantitative PCR was performed to determine Ap3m2 transcript levels in hippocampus from iWSP-2 and iWSR-1 mice after chronic (72 hr) inhalation of ethanol. BECs were in the range of 1.3 mg/ml at 72 hr of chronic exposure. Both genotype and treatment had significant effects on Ap3m2 expression level: full model F(5,59) = 7.9606, P = 0.0002 (Figure 5); Fgenotype (1,56) = 32.9909, P = 0.0001; and Ftreatment (2,56) = 10.5352, P = 0.00001 with no significant genotype × treatment interaction. The genotype effect is such that iWSR1 has greater mean expression than iWSP2 with mean of 1.21 ± 0.02 and 1.06 ± 0.02, respectively. Post-hoc comparisons using Tukey–Kramer’s HSD revealed that there were no significant differences between the pyrazole or saline controls, but that both groups had lower Ap3m2 transcript abundance than the ethanol-treated mice (P < 0.05).
Figure 5.
Genotype and alcohol effects on Ap3m2 expression in iWSP-2 and iWSR-1 mice. ΔΔCt qPCR of iWSP-2 and iWSR-1 mice exposed to 72 hr of chronic ethanol vapor treatment plus pyrazole injection to stabilize blood ethanol concentrations vs. saline-injected and pyrazole-injected air-treated controls. Both genotype and treatment had a significant effect on Ap3m2 expression level: full model F(5,59) = 7.9606, P = 0.0002; Fgenotype (1,56) = 32.9909, P = 0.0001; and Ftreatment(2,56) = 10.5352, P = 0.00001 with no significant genotype × treatment interaction. Post-hoc comparisons using Tukey–Kramer’s HSD revealed that there were no significant differences between the pyrazole or saline controls but that both groups had lower Ap3m2 transcript abundance than the ethanol-treated mice (P < 0.05).
Phylogenetic analysis of the overlapping QTL
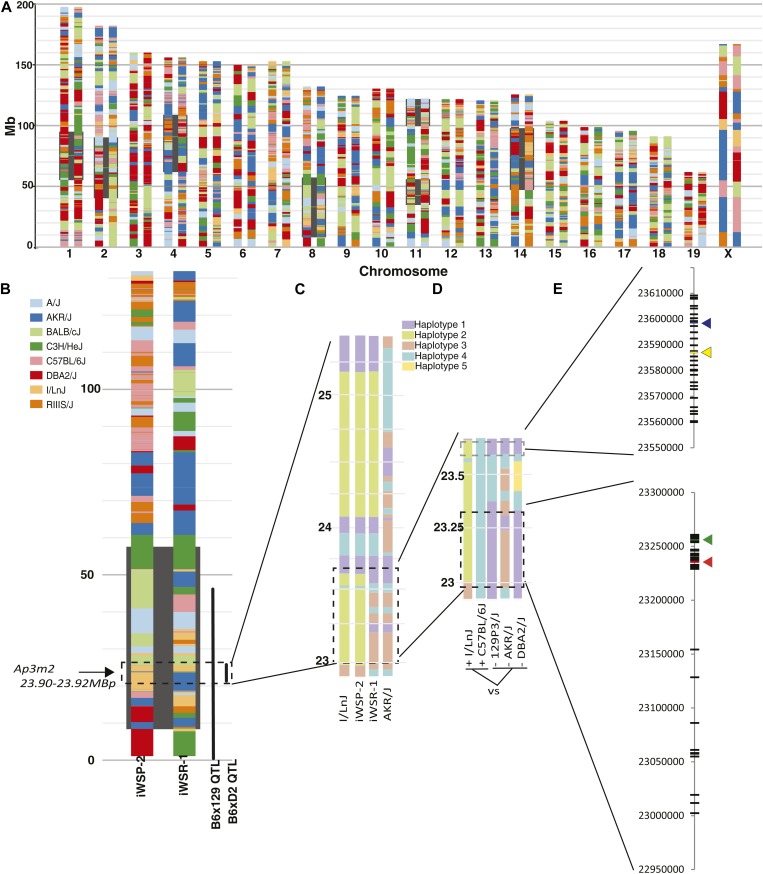
The overlapping loci in this study were originally identified using crosses of mice with different strain backgrounds, enabling the use of shared haplotype and allele distributions to identify likely causative allelic variants using publicly available mouse genome sequence data from multiple strains (Keane et al. 2011). Identifying such SNPs demonstrates the genetic sufficiency of the locus as a plausible source of trait variation, and in this case SNPs should be segregating such that B6 and WSP have one allele and DBA, 129, and WSR have a different allele. A straightforward comparison can be made of B6 haplotype to the 129 haplotype, which confers lower alcohol preference, and of the DBA haplotype, which alters Ap3m2 expression relative to B6. The genetic background of iWSR-1 and iWSP-2 is quite complex and involves eight founder strains (see Materials and Methods). To identify which founder haplotypes were segregating among the iWSP-2 and iWSR-1 and shared with B6/129 and B6/D2, we constructed the haplotypes of the iWSP-2/iWSR-1 strains. Analysis of Mouse Diversity Array genotypes revealed a dense mosaic of different haplotypes from the eight progenitor strains, showing the substantial number of recombination events that occurred within the HS progenitors prior to selection and inbreeding in the QTL region on proximal chromosome 8 (Figure 6, A and B). Thus, even though the F2 cross-derived QTL was large, multiple haplotype fragments of different origins were contained within it. In the iWSR-1 and iWSP-2 strains, the region containing the preference locus was segregating AKR vs. I/LnJ haplotypes, respectively, between 22644764 and 23888988, the region containing Ap3m2 (Figure 6C). This finding further constrains the search for causal variants such that a SNP that segregates in B6, iWSP-2 (represented by surrogate I/LnJ) vs. 129, iWSR-1 (represented by surrogate AKR/J) and D2 must be identified within the region from 22.906 to 23.889 Mb. Analysis of phylogeny in the shared QTL revealed regions for which the AKR haplotype was shared with 129 and D2, while the B6 haplotype was different from these three strains. The I/LnJ haplotype differed from those strains as well (Figure 6D).
Figure 6.
Genetic dissection of shared allelic variants underlying overlapping QTL for alcohol preference and withdrawal. (A) A haplotype reconstruction of the iWSP-2 (left) and iWSR-1 (right) mouse strains based on dense genotyping of the inbred selected lines and existing genotypic data from nearest extant relatives. QTL for alcohol withdrawal seizure previously mapped in crosses of these strains are highlighted in black, including the Alcw6 locus on chromosome 8 (8.12–58.12 Mb) . The contributions of the eight HS progenitor strains are shown as determined by the Mouse Diversity Array using the SNP founder strain calls retrieved from CGDSNPdb (http://cgd.jax.org/cgdsnpdb/; Hutchins et al. 2010). (B) Enlarged reconstruction of chromosome 8 indicating location of QTL overlap of Alcw6, Ap8q (0–40.29 Mb), and BXD Ap3m2 expression QTL (22.906–25.498 Mb). The consensus QTL region on chromosome 8 is segregating I/LnJ and AKR/J haplotypes in the iWSP-2 and iWSR-1 strains. (C) Reconstruction of the iWSP-2 and iWSR-1 strain haplotypes in the region of chromosome 8 containing the consensus QTL (22.906–25.408 Mb-) from the best matching founder surrogate strains I/LnJ and AKR/J. Coloring distinguishes haplotype blocks detected in the Mouse Phylogeny Browser and does not represent genetic origin of each region. (D) Comparison of the overlapping region where iWSP-2 and iWSR-1 haplotypes differ (22.906–23.889 Mb) between reconstructed iWSP-2 and B6 (high alcohol preference, withdrawal, expression) to iWSR-1, 129, and DBA (lower alcohol preference, withdrawal, expression) haplotypes using Mouse Phylogeny Browser. Shared color blocks at a given location represent haplotype similarity across strains. (E) All genetically sufficient SNPs to account for the consensus QTL. SNPs within Ensembl regulatory features are shown with colored arrows. SNPs that contrast the strains used in the Ap8q mapping QTL (129 and B6), Alcw6 iWSP-2/iWSR-1-contributing QTL alleles (AKR/J, I/LnJ), and Ap3m2 BXD eQTL were extracted from Sanger Mouse Genomes (Keane et al. 2011; Yalcin et al. 2011; http://www.sanger.ac.uk/resources/mouse/genomes/) and are plotted in megabase coordinates on chromosome 8. iWSP-2 and iWSR-1 SNP data are publicly available at http://phenome.jax.org/ (accession no. MPD:432).
Regulatory sequence analysis
Inbred strain sequence comparison using data from the Welcome Trust Sanger Institute mouse genomic variation project (Keane et al. 2011; Yalcin et al. 2011) and dense inbred strain SNP genotypes from CGDSNPdb (Hutchins et al. 2010) revealed 33 SNPs (Table S3A) in this region (22.906–25.498 Mb) for which B6 and I were identical and AKR, 129, and DBA differed. Of these SNPs, 11 lie within four different Ensembl regulatory feature types including those identified by the ENCODE project (Table S3A and Figure 6E), and 26 reside within predicted regulatory sequences (Table S3A) according to the predictive software MATCH used in construction of the Transfac database (Matys et al. 2003). Four SNPs are located in regulatory sites reported in both data sources (Table S3B, italicized entries). Together, these regulatory features include transcription factor and polymerase-binding sites, changes in chromatin state by methylated histones, and increased DNaseI sensitivities. Only one of the Ensembl regulatory features (ENSMUSR00000299557) was supported by evidence from a trait-relevant cell type, neural progenitor cells (NPC). The NPC data indicate that there were H3K4me3- and PolII-binding sites within this feature (Table S3B). H3K4me3 (trimethylation of the lysine residue at the fourth position on the N-terminal tail of histone 3) is an important histone mark of active genes, promoting transcription through TFIID recruitment (Lauberth et al. 2013). H3K4me3 has been shown to result in extensive expression changes observed in alcoholic individuals and is therefore considered a component of neuroadaptation in addiction (Zhou et al. 2011). Four different SNPs lie with the regulatory feature ENSMUSR00000299557. Within this H3K4me3 site, two SNPs (rs259599964 and rs52251207) are additionally predicted by TRANSFAC to be transcription factor-binding sites. The sites within ENSMUSR00000299557 satisfy all criteria of the SNP segregation pattern, replicate transcription regulatory indications, and alcohol sensitivity of the regulatory site.
Chromatin immunoprecipitation and DNA sequencing
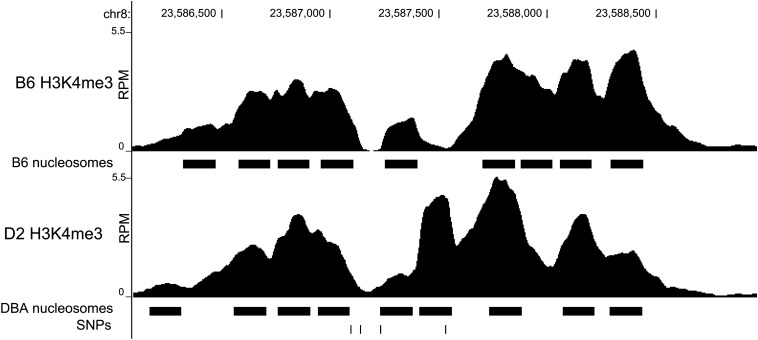
To assess which SNP is mostly likely to be causal within our interval, we sought to investigate the H3K4me3 state of B6 and D2 mice, representative strains of the compared populations. Using existing nucleosome-level H3K4me3 ChIP-seq data, the region containing ENSMUSR00000299557, specifically SNP rs32926479, shows differential abundance and thus differential histone modification in B6 vs. D2 (Baker et al. 2014) (Figure 7).
Figure 7.
Nucleosome resolution H3K4me3 ChIP-seq. The abundance of sequence reads, expressed as RPM, from B6 and D2 covering the region of chromosome 8 containing the ensemble regulatory feature ENSMUSR00000299557. Predicted nucleosome positions are indicated as solid bars below the coverage profiles. The SNP track shows SNPs from left to right: rs32882991, rs52251207, rs259599964, and rs32926479. rs32926479 appears in a region differentially methylated between B6 and D2.
Discussion
Using a two-pronged approach consisting of integrative functional genomics followed by high-precision genetic analysis, we were able to identify a shared candidate gene and regulatory SNP for two related behaviors originally mapped in low-precision mouse crosses (Bachmanov et al. 2002; Bergeson et al. 2003). Ap3m2 is genetically regulated and has a functional role in both alcohol preference and withdrawal. We were able to refine this result using advanced resources for functional genomic data integration with high-precision tools and resources for analysis of mouse genetic diversity. Our GeneWeaver system (Baker et al. 2012) enabled integration of disparate functional genomics experimental results to attack the question of what biological mechanism underlies both alcohol preference and withdrawal. This led us to a wealth of published QTL for these traits, several of which overlapped. High-density genotypes (Yang et al. 2009), phylogeny analysis (Wang et al. 2012), and sequence data (Keane et al. 2011; Yalcin et al. 2011) enabled us to compare allelic variants in closely related inbred mouse strains to narrow the QTL interval to four SNPs in an alcohol-sensitive transcription regulatory region sufficient to explain the pattern of phenotype segregation. We confirmed a functional role for the gene in both alcohol preference and withdrawal using existing knockout mice, established that this gene is differentially expressed in founder strains and in response to alcohol, and identified that the SNPs were in an area of differential histone methylation.
Ap3m2 is a compelling genetic and functional candidate. It is in the intersection of QTL for these behaviors, is found to be differentially expressed in numerous studies of alcohol preference (Mulligan et al. 2006; Rodd et al. 2008; Ponomarev et al. 2012), and has been shown to be related to seizure in gene perturbation studies (Nakatsu et al. 2004; Misawa et al. 2008). The absence of functional polymorphisms in Ap3m2, the discovery of a cis eQTL at Ap3m2 in the BXD population, and the finding of differential expression of Ap3m2 in iWSP-2 and iWSR-1 mice suggest that a common polymorphism affecting Ap3m2 expression is responsible for the heightened alcohol preference and withdrawal. Deletion of this gene in mice results in spontaneous recurrent epileptic seizures, susceptibility to drug-induced seizures, impaired GABA release, fewer synaptic vesicles, enhanced long-term potentiation, and abnormal propagation of neuronal excitability via the temporo-hippocampal pathway (Nakatsu et al. 2004; Misawa et al. 2008).
Knockout mice can carry large introgressions from the ES cell strain of origin, in this case 129/P2OlaHsd-derived cells. The Ap3m2 knockout is no exception and contains a 129 introgression from 0 to D8Mit258 (33.8 Mb). This so called “hitchhiking” DNA can introduce confounding phenotypic effects on behavior, but this is not the case for the present study (Gerlai 1996). The alcohol preference phenotype is increased in the presence of the B6 allele and decreased by the 129 allele based on the analysis of QTL effects at the Ap8q locus. The effects of the Ap3m2-KO genotype are in the opposite direction of the 129 allelic effects in this region, suggesting that the KO phenotype is attributable to the targeted mutation in the Ap3m2 gene rather than to polymorphisms in the donor 129 region. The withdrawal seizure phenotype is also increased in the presence of the B6 allele in this region and decreased by the 129-like allele. Therefore, we would predict that a causative polymorphism in the 129 background would decrease seizure phenotypes, but we observe an increased seizure phenotype in the KO mouse, again suggesting that the difference in seizure phenotypes in the knockout lines are attributable to the targeted mutation and not to the 129 background.
Although Ap3m2 has not been previously associated with alcohol-related behaviors, the known role of Ap3m2 in GABA vesicular transport lends credence to a functional role in alcohol preference and withdrawal. Alcohol withdrawal and preference are known to involve GABAergic transmission, whereby chronic alcohol alters GABA receptor subunit expression and potentiates the action of GABA at the receptors (Buck and Finn 2001; Nakatsu et al. 2004; Finn et al. 2006, 2010; Saba et al. 2011; Sharp et al. 2011; Enoch et al. 2012). In the presence of low levels of alcohol, GABA-A binding is potentiated, as is Cl− influx through the GABA-A ion channel and resulting neuronal inhibition (Clapp et al. 2008). In an Ap3m2 null mouse, impaired GABA release is predicted to result in a lower sensitivity to ethanol’s effects on neuronal inhibition relative to Ap3m2 genotypically wild-type mice. This suggests that knockout mice need to ingest higher concentrations of alcohol to achieve the same effect as a lower concentration in the wild-type controls, a phenomenon that could manifest as an increased preference for ethanol.
Two competing but non-exclusive explanations for the relationship between alcohol preference and withdrawal are (1) that alcohol consumption is motivated by alleviation of withdrawal symptoms; i.e., preference is a manifestation of a serial process emanating from one neurobiological phenomenon that causes increased withdrawal; or (2) that the same biological pathways that predispose to alcohol preference are responsible for susceptibility to withdrawal seizures via gene pleiotropy in a shared or parallel neurobiological process. Identifying the molecular basis of the relationship between alcohol preference and withdrawal may ultimately help to resolve this question. Rodent genetic studies typically indicate a negative correlation between these two processes. However, all such studies (including this report) compared naive mice drinking alcohol and did not study postdependent drinking per se. Here we observe a positive relationship in which loss of Ap3m2 function results in increased withdrawal severity and increased preference in a strain of mouse (B6) with an already high alcohol preference (Mulligan et al. 2008). It is important to note that the allelic effects on behavior in both QTL analyses were in the same direction with respect to B6/I, suggesting that the candidate at the Chr 8 locus should impact both behaviors in the same direction although other segregating loci may dissociate these effects. The negative genetic correlation may be accounted for by the several other overlapping preference and withdrawal loci segregating in inbred mouse populations such as Alcp1 (Melo et al. 1996) and Alcw4 (Crabbe et al. 1994) and Alcp18 (Gill et al. 1998) and Alcw3 (Bergeson et al. 2003). Finding other molecular substrates of the relationships among preference and withdrawal will enable better evaluation of competing mechanistic hypotheses.
The process of going from QTL to gene in conventional two-progenitor mapping crosses has been challenging due to the size of the resulting loci (Nadeau and Frankel 2000; Milner and Buck 2010). Although there have been promising advances in mapping populations, a large number of existing QTL have been reported, and it would be tremendously costly to repeat studies in new high-precision populations. In the present study, we demonstrate efficient identification of candidate genes for large “legacy” QTL due to the availability of extensive genotype, haplotype, phylogeny, and sequence data coupled with a wealth of functional genomics evidence. Numerous integrative strategies for candidate gene identification have been proposed and applied (Belknap and Atkins 2001; Hitzemann et al. 2003, 2004; Aerts et al. 2006; Le-Niculescu et al. 2007, 2011; Ehlers et al. 2010; Guan et al. 2010; Kurian et al. 2011; Leduc et al. 2011; Bhandari et al. 2012; Chesler et al. 2012; Faro et al. 2012). Although these approaches have been effective, their application is limited by the scope and state of existing data resources. Integration of these data in a readily computable form is required for facile identification of highly supported gene–phenotype, gene–gene and phenotype–phenotype relations (Akil et al. 2011). Here we demonstrate the benefit of data integration in the GeneWeaver software system that enabled us to identify a common biological basis for two phenotypic endpoints related to alcoholism. This convergent evidence enabled us to prioritize our search for a biological correlate of these behaviors for which we could then demonstrate functional and genetic validity.
Together, advances made over the past decade in precision characterization of mouse genetic variation along with deep and diverse functional genomics data integration make complex trait genetics tractable in the mouse. Large genetic loci defined in low-precision mapping analyses are readily amenable to deep dissection using this new arsenal of resources. Once a valid candidate is found, advanced genetic resources short-circuit a lengthy and intensive process of genetic refinement through breeding, demonstrating the value of making large-scale genetic and functional genomics data readily computable and interoperable for the resolution of historical genetic mapping results.
Supplementary Material
Acknowledgments
The GeneWeaver.org system was developed collaboratively with Dr. Erich J. Baker and Dr. Michael A. Langston. We thank Jason Schlumbohm at Oregon Health & Science University for producing the iWSP-2/iWSR-1 brain samples and Mount Desert Island High School student Nora Hubbell for Hardy–Weinberg analysis of historic F2 data. In addition, we thank Daniel M. Gatti for sharing R scripts to visualize iWSP-2/iWSR-1 genotypes; Troy Wilcox for helping with the preference assays; Laura C. Anderson for valuable comments on the draft; and Alan Rosenwasser, Ryan Logan, and Walter McCulley for help in establishing the chronic vapor exposure. We acknowledge The Jackson Laboratory Scientific Services supported by NIH grant P30 CA034196 including the importation and reproductive services for the importation and rederivation of the Ap3m2tm1Ohno mice; the gene expression scientific services, specifically of Sonya Kamdar and Janet Pereira for work on the Mouse Diversity Array and/or the qPCR; and Sue Grindle for services in clinical chemistries for the BEC. This project was supported by National Institutes of Health (NIH) grant R01 AA18776 (to E.J.C.). Work on iWSP-2 and iWSR-1 mice was supported by NIH grant P60 AA10760, AA020245 (JCC) and by a grant from the Department of Veterans Affairs (to J.C.C.). National Institute of General Medical Sciences grant F32 GM101736 supported C.L.B.
Footnotes
Communicating editor: D. W. Threadgill
Literature Cited
- Aerts S., Lambrechts D., Maity S., Van Loo P., Coessens B., et al. , 2006. Gene prioritization through genomic data fusion. Nat. Biotechnol. 24: 537–544. [DOI] [PubMed] [Google Scholar]
- Akil H., Martone M. E., Van Essen D. C., 2011. Challenges and opportunities in mining neuroscience data. Science 331: 708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmanov A. A., Tordoff M. G., Beauchamp G. K., 1996. Ethanol consumption and taste preferences in C57BL/6ByJ and 129/J mice. Alcohol. Clin. Exp. Res. 20: 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmanov A. A., Reed D. R., Li X., Li S., Beauchamp G. K., et al. , 2002. Voluntary ethanol consumption by mice: genome-wide analysis of quantitative trait loci and their interactions in a C57BL/6ByJ × 129P3/J F2 intercross. Genome Res. 12: 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. L., Walker M., Kajita S., Petkov P. M., Paigen K., 2014. PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome Res. 24: 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E. J., Jay J. J., Philip V. M., Zhang Y., Li Z., et al. , 2009. Ontological Discovery Environment: a system for integrating gene-phenotype associations. Genomics 94: 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E. J., Jay J. J., Bubier J. A., Langston M. A., Chesler E. J., 2012. GeneWeaver: a web-based system for integrative functional genomics. Nucleic Acids Res. 40: D1067–D1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker H. C., Hale R. L., 1993. Repeated episodes of ethanol withdrawal potentiate the severity of subsequent withdrawal seizures: an animal model of alcohol withdrawal “kindling.” Alcohol. Clin. Exp. Res. 17: 94–98. [DOI] [PubMed] [Google Scholar]
- Belknap J. K., Atkins A. L., 2001. The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mamm. Genome 12: 893–899. [DOI] [PubMed] [Google Scholar]
- Bergeson S. E., Kyle Warren R., Crabbe J. C., Metten P., Gene Erwin V., et al. , 2003. Chromosomal loci influencing chronic alcohol withdrawal severity. Mamm. Genome 14: 454–463. [DOI] [PubMed] [Google Scholar]
- Bhandari P., Hill J. S., Farris S. P., Costin B., Martin I., et al. , 2012. Chloride intracellular channels modulate acute ethanol behaviors in Drosophila, Caenorhabditis elegans and mice. Genes Brain Behav. 11: 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck K. J., Finn D. A., 2001. Genetic factors in addiction: QTL mapping and candidate gene studies implicate GABAergic genes in alcohol and barbiturate withdrawal in mice. Addiction 96: 139–149. [DOI] [PubMed] [Google Scholar]
- Chesler E. J., Lu L., Wang J., Williams R. W., Manly K. F., 2004. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat. Neurosci. 7: 485–486. [DOI] [PubMed] [Google Scholar]
- Chesler E. J., Lu L., Shou S., Qu Y., Gu J., et al. , 2005. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 37: 233–242. [DOI] [PubMed] [Google Scholar]
- Chesler E. J., Plitt A., Fisher D., Hurd B., Lederle L., et al. , 2012. Quantitative trait loci for sensitivity to ethanol intoxication in a C57BL/6J×129S1/SvImJ inbred mouse cross. Mamm. Genome 23: 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp P., Bhave S. V., Hoffman P.L., 2008. How adaptation of the brain to alcohol leads to pependence: a pharmacological perspective. Alcohol Res Health 31: 310–339. [PMC free article] [PubMed] [Google Scholar]
- Crabbe J. C., 1998. Provisional mapping of quantitative trait loci for chronic ethanol withdrawal severity in BXD recombinant inbred mice. J. Pharmacol. Exp. Ther. 286: 263–271. [PubMed] [Google Scholar]
- Crabbe J. C., 2012. Translational behaviour-genetic studies of alcohol: Are we there yet? Genes Brain Behav. 11: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe J. C., Kosobud A., Young E. R., Tam B. R., McSwigan J. D., 1985. Bidirectional selection for susceptibility to ethanol withdrawal seizures in Mus musculus. Behav. Genet. 15: 521–536. [DOI] [PubMed] [Google Scholar]
- Crabbe J. C., Merrill C., Belknap J. K., 1991. Acute dependence on depressant drugs is determined by common genes in mice. J. Pharmacol. Exp. Ther. 257: 663–667. [PubMed] [Google Scholar]
- Crabbe J. C., Belknap J. K., Buck K. J., Metten P., 1994. Use of recombinant inbred strains for studying genetic determinants of responses to alcohol. Alcohol Alcohol. Suppl. 2: 67–71. [PubMed] [Google Scholar]
- Ehlers C. L., Walter N. A., Dick D. M., Buck K. J., Crabbe J. C., 2010. A comparison of selected quantitative trait loci associated with alcohol use phenotypes in humans and mouse models. Addict. Biol. 15: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch M. A., 2013. Genetic influences on the development of alcoholism. Curr. Psychiatry Rep. 15: 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch M. A., Zhou Z., Kimura M., Mash D. C., Yuan Q., et al. , 2012. GABAergic gene expression in postmortem hippocampus from alcoholics and cocaine addicts: corresponding findings in alcohol-naive P and NP rats. PLoS ONE 7: e29369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faro A., Giordano D., Spampinato C., 2012. Combining literature text mining with microarray data: advances for system biology modeling. Brief. Bioinform. 13: 61–82. [DOI] [PubMed] [Google Scholar]
- Fernandez J. R., Vogler G. P., Tarantino L. M., Vignetti S., Plomin R., et al. , 1999. Sex-exclusive quantitative trait loci influences in alcohol-related phenotypes. Am. J. Med. Genet. 88: 647–652. [DOI] [PubMed] [Google Scholar]
- Finn D. A., Douglass A. D., Beadles-Bohling A. S., Tanchuck M. A., Long S. L., et al. , 2006. Selected line difference in sensitivity to a GABAergic neurosteroid during ethanol withdrawal. Genes Brain Behav. 5: 53–63. [DOI] [PubMed] [Google Scholar]
- Finn D. A., Beckley E. H., Kaufman K. R., Ford M. M., 2010. Manipulation of GABAergic steroids: sex differences in the effects on alcohol drinking- and withdrawal-related behaviors. Horm. Behav. 57: 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlai R., 1996. Gene-targeting studies of mammalian behavior: Is it the mutation or the background genotype? Trends Neurosci. 19: 177–181. [DOI] [PubMed] [Google Scholar]
- Gill K., Desaulniers N., Desjardins P., Lake K., 1998. Alcohol preference in AXB/BXA recombinant inbred mice: gender differences and gender-specific quantitative trait loci. Mamm. Genome 9: 929–935. [DOI] [PubMed] [Google Scholar]
- Goldstein D. B., 1972. Relationship of alcohol dose to intensity of withdrawal signs in mice. J. Pharmacol. Exp. Ther. 180: 203–215. [PubMed] [Google Scholar]
- Gould T. D., Gottesman I. I., 2006. Psychiatric endophenotypes and the development of valid animal models. Genes Brain Behav. 5: 113–119. [DOI] [PubMed] [Google Scholar]
- Guan Y., Ackert-Bicknell C. L., Kell B., Troyanskaya O. G., Hibbs M. A., 2010. Functional genomics complements quantitative genetics in identifying disease-gene associations. PLOS Comput. Biol. 6: e1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann, R., B. Malmanger, C. Reed, M. Lawler, B. Hitzemann et al., 2003 A strategy for the integration of QTL, gene expression, and sequence analyses. Mamm. Genome 14: 733–747. [DOI] [PubMed]
- Hitzemann R., Reed C., Malmanger B., Lawler M., Hitzemann B., et al. , 2004. On the integration of alcohol-related quantitative trait loci and gene expression analyses. Alcohol. Clin. Exp. Res. 28: 1437–1448. [DOI] [PubMed] [Google Scholar]
- Hutchins, L. N., Y. Ding, J. P. Szatkiewicz, R. Von Smith, H. Yang et al., 2010 CGDSNPdb: a database resource for error-checked and imputed mouse SNPs. Database (Oxford) DOI: 10.1093/database/baq008. [DOI] [PMC free article] [PubMed]
- Karpyak V. M., Kim J. H., Biernacka J. M., Wieben E. D., Mrazek D. A., et al. , 2009. Sequence variations of the human MPDZ gene and association with alcoholism in subjects with European ancestry. Alcohol. Clin. Exp. Res. 33: 712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob, G. F., and N. D. Volkow, 2010 Neurocircuitry of addiction. Neuropsychopharmacology 35: 217–238. [DOI] [PMC free article] [PubMed]
- Kurian S. M., Le-Niculescu H., Patel S. D., Bertram D., Davis J., et al. , 2011. Identification of blood biomarkers for psychosis using convergent functional genomics. Mol. Psychiatry 16: 37–58. [DOI] [PubMed] [Google Scholar]
- Lauberth S. M., Nakayama T., Wu X., Ferris A. L., Tang Z., et al. , 2013. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152: 1021–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc M. S., Hageman R. S., Verdugo R. A., Tsaih S. W., Walsh K., et al. , 2011. Integration of QTL and bioinformatic tools to identify candidate genes for triglycerides in mice. J. Lipid Res. 52: 1672–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H., McFarland M. J., Mamidipalli S., Ogden C. A., Kuczenski R., et al. , 2007. Convergent Functional Genomics of bipolar disorder: from animal model pharmacogenomics to human genetics and biomarkers. Neurosci. Biobehav. Rev. 31: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H., Case N. J., Hulvershorn L., Patel S. D., Bowker D., et al. , 2011. Convergent functional genomic studies of omega-3 fatty acids in stress reactivity, bipolar disorder and alcoholism. Transl. Psychiatr. 1: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan R. W., Seggio J. A., Robinson S. L., Richard G. R., Rosenwasser A. M., 2010. Circadian wheel-running activity during withdrawal from chronic intermittent ethanol exposure in mice. Alcohol 44: 239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez M. F., Doremus-Fitzwater T. L., Becker H. C., 2011. Chronic social isolation and chronic variable stress during early development induce later elevated ethanol intake in adult C57BL/6J mice. Alcohol 45: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammana A., Vingron M., Chung H. R., 2013. Inferring nucleosome positions with their histone mark annotation from ChIP data. Bioinformatics 29: 2547–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., et al. , 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31: 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., et al. , 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34: D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClearn G. E., Wilson J. R., Meredith J. E., 1970. The Use of Isogenic and Heterogenic Mouse Stocks in Behavioral Research. Appleton-Century-Crofts, New York. [Google Scholar]
- Melo J. A., Shendure J., Pociask K., Silver L. M., 1996. Identification of sex-specific quantitative trait loci controlling alcohol preference in C57BL/ 6 mice. Nat. Genet. 13: 147–153. [DOI] [PubMed] [Google Scholar]
- Milner L. C., Buck K. J., 2010. Identifying quantitative trait loci (QTLs) and genes (QTGs) for alcohol-related phenotypes in mice. Int. Rev. Neurobiol. 91: 173–204. [DOI] [PubMed] [Google Scholar]
- Milner L. C., Shirley R. L., Kozell L. B., Walter N. A., Kruse L. C., et al. , 2013. Novel MPDZ/MUPP1 transgenic and knockdown models confirm Mpdz’s role in ethanol withdrawal and support its role in voluntary ethanol consumption. Addict. Biol. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa H., Fujigaya H., Nishimura T., Moriwaki Y., Okuda T., et al. , 2008. Aberrant trafficking of the high-affinity choline transporter in AP-3-deficient mice. Eur. J. Neurosci. 27: 3109–3117. [DOI] [PubMed] [Google Scholar]
- Morozova T. V., Goldman D., Mackay T. F., Anholt R. R., 2012. The genetic basis of alcoholism: multiple phenotypes, many genes, complex networks. Genome Biol. 13: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M. K., Ponomarev I., Hitzemann R. J., Belknap J. K., Tabakoff B., et al. , 2006. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc. Natl. Acad. Sci. USA 103: 6368–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M. K., Ponomarev I., Boehm S. L., II, Owen J. A., Levin P. S., et al. , 2008. Alcohol trait and transcriptional genomic analysis of C57BL/6 substrains. Genes Brain Behav. 7: 677–689. [DOI] [PubMed] [Google Scholar]
- Mulligan M. K., Wang X., Adler A. L., Mozhui K., Lu L., et al. , 2012. Complex control of GABA(A) receptor subunit mRNA expression: variation, covariation, and genetic regulation. PLoS ONE 7: e34586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J. H., Frankel W. N., 2000. The roads from phenotypic variation to gene discovery: mutagenesis vs. QTLs. Nat. Genet. 25: 381–384. [DOI] [PubMed] [Google Scholar]
- Nakatsu F., Okada M., Mori F., Kumazawa N., Iwasa H., et al. , 2004. Defective function of GABA-containing synaptic vesicles in mice lacking the AP-3B clathrin adaptor. J. Cell Biol. 167: 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce J. L., Lu L., Gu J., Silver L. M., Williams R. W., 2004. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I., Wang S., Zhang L., Harris R. A., Mayfield R. D., 2012. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J. Neurosci. 32: 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd Z. A., Kimpel M. W., Edenberg H. J., Bell R. L., Strother W. N., et al. , 2008. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol. Biochem. Behav. 89: 481–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba L. M., Bennett B., Hoffman P. L., Barcomb K., Ishii T., et al. , 2011. A systems genetic analysis of alcohol drinking by mice, rats and men: influence of brain GABAergic transmission. Neuropharmacology 60: 1269–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp B. M., Chen H., Gong S., Wu X., Liu Z., et al. , 2011. Gene expression in accumbens GABA neurons from inbred rats with different drug-taking behavior. Genes Brain Behav. 10: 778–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S., Bell J. T., Copley R. R., Taylor M. S., Williams R. W., et al. , 2006. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 4: e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley R. L., Walter N. A., Reilly M. T., Fehr C., Buck K. J., 2004. Mpdz is a quantitative trait gene for drug withdrawal seizures. Nat. Neurosci. 7: 699–700. [DOI] [PubMed] [Google Scholar]
- Simon M. M., Greenaway S., White J. K., Fuchs H., Gailus-Durner V., et al. , 2013. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 14: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Eppig J. T., 2009. The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 1: 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos J. A., Snyder M., Hardison R., Ren B., Gingeras T., et al. , 2012. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. 13: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. A., Bedigian H. G., Meier H., 1977. Genetic studies of the Fv-1 locus of mice: linkage with Gpd-1 in recombinant inbred lines. J. Virol. 23: 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. A., Wnek C., Kotlus B. S., Roemer N., MacTaggart T., et al. , 1999. Genotyping new BXD recombinant inbred mouse strains and comparison of BXD and consensus maps. Mamm. Genome 10: 335–348. [DOI] [PubMed] [Google Scholar]
- Wang J., Williams R. W., Manly K. F., 2003. WebQTL: web-based complex trait analysis. Neuroinformatics 1: 299–308. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Kapoor M., Goate A. M., 2012. The genetics of substance dependence. Annu. Rev. Genomics Hum. Genet. 13: 241–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. W., Gu J., Qi S., Lu L., 2001. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol. 2: RESEARCH0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., Wong K., Agam A., Goodson M., Keane T. M., et al. , 2011. Sequence-based characterization of structural variation in the mouse genome. Nature 477: 326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Ding Y., Hutchins L. N., Szatkiewicz J., Bell T. A., et al. , 2009. A customized and versatile high-density genotyping array for the mouse. Nat. Methods 6: 663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Shin H., Song J. S., Lei Y., Liu X. S., 2008. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics 9: 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Phillips C. A., Rogers G. L., Baker E. J., Chesler E. J., et al. , 2014. On finding bicliques in bipartite graphs: a novel algorithm and its application to the integration of diverse biological data types. BMC Bioinformatics 15: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Yuan Q., Mash D. C., Goldman D., 2011. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc. Natl. Acad. Sci. USA 108: 6626–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.