Abstract
MYC locus rearrangements – often complex combinations of translocations, insertions, deletions, and inversions - in multiple myeloma (MM) were thought to be a late progression event, which often did not involve immunoglobulin genes. Yet germinal center activation of MYC expression has been reported to cause progression to MM in an MGUS prone mouse strain. Although previously detected in 16% of MM, we find MYC rearrangements in nearly 50% of MM, including smoldering MM, and they are heterogeneous in some cases. Rearrangements reposition MYC near a limited number of genes associated with conventional enhancers, but mostly with super-enhancers (e.g., IGH, IGL, IGK, NSMCE2, TXNDC5, FAM46C, FOXO3, IGJ, PRDM1). MYC rearrangements are associated with a significant increase of MYC expression that is monoallelic, but MM tumors lacking a rearrangement have bi-allelic MYC expression at significantly higher levels than in MGUS. We also show that germinal center activation of MYC does not cause MM in a mouse strain that rarely develops spontaneous MGUS. It appears that increased MYC expression at the MGUS/MM transition usually is bi-allelic, but sometimes can be mono-allelic if there is a MYC rearrangement. Our data suggests that MYC rearrangements, regardless of when they occur during MM pathogenesis, provide one event that contributes to tumor autonomy.
Keywords: Monoclonal Gammopathy of Undetermined Significance; Multiple Myeloma; Genes, MYC; Enhancer and Super-Enhancer Elements, Genetic; Translocation, Genetic; Mice, Transgenic
INTRODUCTION
The dysregulation of MYC by a chromosomal translocation to one of three immunoglobulin (IG) loci (IGH, IGK, IGL) is a nearly invariant and probably primary event in murine plasmacytoma and human Burkitt's lymphoma tumors.1, 2 The translocation is thought to be mediated by an error in one of three B cell specific DNA modification processes (V[D]J recombination, IGH switch recombination, somatic hypermutation),3 which repositions MYC near the intronic and/or 3’ IG enhancers. The non-translocated MYC usually is not expressed or expressed at very low levels, as expected for the corresponding non-transformed cell.4, 5
Multiple myeloma (MM), a post-germinal center tumor with a phenotype that is similar to terminally differentiated long-lived bone marrow plasma cells, usually is preceded by a pre-malignant MGUS (monoclonal gammopathy of undetermined significance) tumor.6, 7 Similar to MGUS, early stages of MM have only a small fraction of cycling cells. However, the fraction of cycling cells increases during progression, and especially as the tumor extends to extramedullary locations, including secondary plasma cell leukemia (PCL). Virtually all MM cell lines (MMCL) are derived from PCL or extramedullary MM tumors.8 Previously, we used metaphase FISH analyses to show that complex rearrangements involving MYC (rarely MYCN or MYCL) were present in about 80% of MMCL and nearly 50% of advanced MM tumors.9-11 Avet-Loiseau et al. reported that interphase FISH identified MYC rearrangements in 16% of newly diagnosed MM tumors and 3% of MGUS tumors.12 Others reported that 8 of 13 patients with PCL had 244K CGH abnormalities that targeted or were close to MYC, with only 4 of these detected by interphase FISH analyses.13 Strikingly, a substantial fraction of MYC rearrangements in MM do not involve IG sequences.10, 11, 13-15 Similarly, MYC rearrangements occurring during progression of B lymphoma sometimes do not involve IG loci.16-18 Together, these results suggest that MYC rearrangements in MM are mostly late progression events. However, gene expression profiling and immunohistochemistry studies show that MYC RNA expression is significantly increased in MM tumors compared to MGUS tumors.19-21 Moreover, activation of a MYC transgene in germinal center B cells results in a nearly universal occurrence of MM in an MGUS prone mouse strain that rarely develops MM.19
In the present study we used a combination of molecular approaches for 238 MM tumors and a panel of 53 MMCL to determine the prevalence of MYC rearrangements and corresponding MYC expression (RNA levels and mono- vs. bi-allelic expression) in tumors and MMCL with differing molecular phenotypes. We also characterized MYC rearrangement breakpoints in an attempt to identify non-IG sequences that are responsible for dysregulating MYC in MM. Finally we have examined the consequences of activating a MYC transgene in the germinal center B cells in a mouse strain that rarely develops MGUS.
MATERIAL AND METHODS
Patient selection
Patients with SMM (9), untreated PCL (3), untreated symptomatic MM (108), and previously treated MM (118) were diagnosed according to established criteria22 and were included in the MMRC Genomics Initiative if more then two million CD138-selected bone marrow cells were available for analysis (~one third of submitted samples).
Comparative Genomic Hybridization
The Agilent 244k CGH raw data was downloaded from the Multiple Myeloma Genomics Portal (http://www.broad.mit.edu/mmgp), and segmented using circular binary segmentation (Bioconductor). The integrated copy number for the region flanking MYC (chr8: 126000000-130000000) was normalized to two copies. In four patients (MMRC0028, MMRC0172, MMRC0440, MMRC0034) a single probe abnormality in the telomeric amplified region was segmented based on amplification of more then one copy (see Figure S3). The hyperdiploid index for each patient was calculated as the average chromosome copy number of chromosomes 3, 5, 7, 9, 11, 15, 19 and 21.
FISH analyses
Metaphase FISH assays, which used MYC, IGH, IGL, IGK, chromosome painting probes, and sometimes other specific probes, were performed on all MMCL (Table S2), as described previously.10 Interphase FISH assays to identify MYC:IGH and MYC:IGL fusions were done on 218 MMRC MM tumors using the cytoplasmic immunoglobulin light chain (cIg-FISH) technique and probes described previously.10, 23 Fosmid G248P85602G8 (chr4: 71533320-71573442) encompassing the IGJ enhancer was used in interphase FISH assays to confirm its insertion adjacent to MYC in MMRC0408.
Gene Expression Profiling
The Affymetrix Hu133Plus2 raw data was downloaded from the Multiple Myeloma Genomics Portal (http://www.broad.mit.edu/mmgp) for the MMRC MM samples and MM cell lines and summarized using MAS5. The Hu133Plus2 MAS5 summarized data for normal plasma cells and MGUS was downloaded from NCBI (GSE5900). Each probe set was normalized to the expression of the 75th percentile on the chip, and then to the median across the samples. The proliferation index24 and UAMS 70 gene prognostic index (GEP70)25 were calculated as described previously.
MYC polymorphisms to assess allele specific expression of MYC RNA
Exonic and intronic polymorphisms were determined in the MMCL and some tumor samples by Sanger sequencing of PCR products generated from different regions of the MYC gene. Expression of MYC polymorphisms that crossed an intron for exonic polymorphisms or total RNA treated with DNase for intronic polymorphisms (Table S2, Table S4). Restriction enzyme polymorphisms of genomic variants rs4645958 and rs2070582 were used to analyze allele specific expression in some MMRC MM tumors (Figure 2C, Table S4). Additional details in Supplemental Methods.
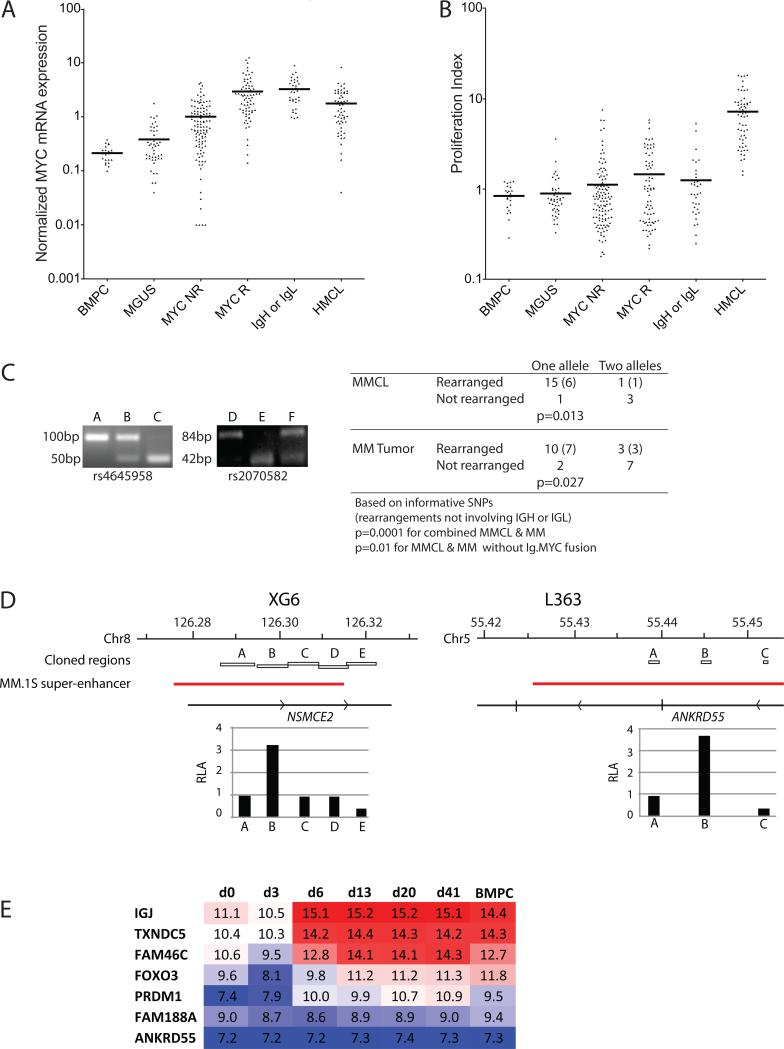
Figure 2. Rearrangements cis-dysregulate MYC expression.
A. Affymetrix Hu133Plus2 arrays were used to estimate the normalized expression of MYC mRNA in CD138-selected plasma cells from normal bone marrow (BMPC), MGUS, MM without MYC rearrangements (MYC NR), MM with MYC rearrangements (MM R), MM with MYC rearrangements involving IgH or IgL (IgH or IgL) and MM cell lines (MMCL).
B. The arrays were also used to calculate the proliferation index in the same cell populations.
C. MM RNA from tumors heterozygous for SNPs rs4645958 or rs2070582 was analyzed by RT-PCR for allele specific expression. For both SNPs the polymorphism alters an Ici1 restriction site allowing the two alleles to be distinguished after restriction digest of the PCR product. Shown is the 4% agarose gel electrophoresis of the PCR digest for three patients with each polymorphism, providing examples of MM tumors that express each allele alone, and both together. Also provided is a table summarizing the allele specific expression of MYC in MM tumors and cell lines.
D. Regions adjacent to MYC breakpoints in XG6 and L363 are shown, with the location of fragments cloned for enhancer assays indicated. The fragments were located in predicted MM.1S super-enhancers35. The relative ability of the various fragments to enhance the transcription of a MYC promoter driven luciferase construct is shown as relative luciferase activity (RLA) normalized to the activity of the MYC promoter without enhancer.
E. Expression of genes adjacent to putative super-enhancers involved in MYC locus rearrangements during in vitro B cell differentiation to long lived plasma cells. The log2-normalized data from Cocco et al is shown for the progressive stages of in vitro plasma cell differentiation23. In addition to IgH, IgK, and IgL (not shown), also IGJ, TXNDC5, FAM46C, FOXO3, PRDM1 (but not FAM188A or ANKRD55) are markedly up regulated with plasma cell differentiation.
Monoclonal gammopathy in Vk*MYC mice
Transgenic Vk*MYC mice in C57Bl/6 were crossed for ten generations into Balb/c. In the Balb/c background 38 Vk*MYC transgenic mice, and 11 wildtype mice were aged and followed for the presence of a monoclonal spike by serum protein electrophoresis every ten weeks as described previously19.
Supplemental methods
Chromosome conformation capture (3C), enhancer assays, somatic cell hybrids, mate pair libraries, MYC rearrangements from whole genome sequences, and other details.
RESULTS
MYC locus rearrangements detected by karyotypic or CGH assays in MM cell lines
Fifty-three independent MMCL were analyzed for MYC locus rearrangements by metaphase FISH assays as described previously.10 Including two MMCL that express MYCL or MYCN, MYC rearrangements involving a MYC family member were detected in 42/53 (79%) MMCL (Table S1, Table S2). Most rearrangements were complex translocations or insertions. The 244K Agilent array CGH platform also detected MYC locus (but not MYCL or MYCN) copy number abnormalities (CNA) identifying unbalanced rearrangements in 42/53 (79%) MMCL, including five for which rearrangements were not detected by the karyotypic assays (Figure 1,Table S1, Table S2). No MYC locus rearrangements were detected by either assay in 6/53 (11%) of MMCL. Twenty-eight of the 47 MMCL with MYC rearrangements repositioned a MYC family member near one of the IG 3’ enhancer sequences that are included in the FISH probes: 21 with IGH, 5 with IGL, and 2 with IGK (Table S2). Rearrangements in the remaining 19 MMCL did not reveal recurrent partner loci.
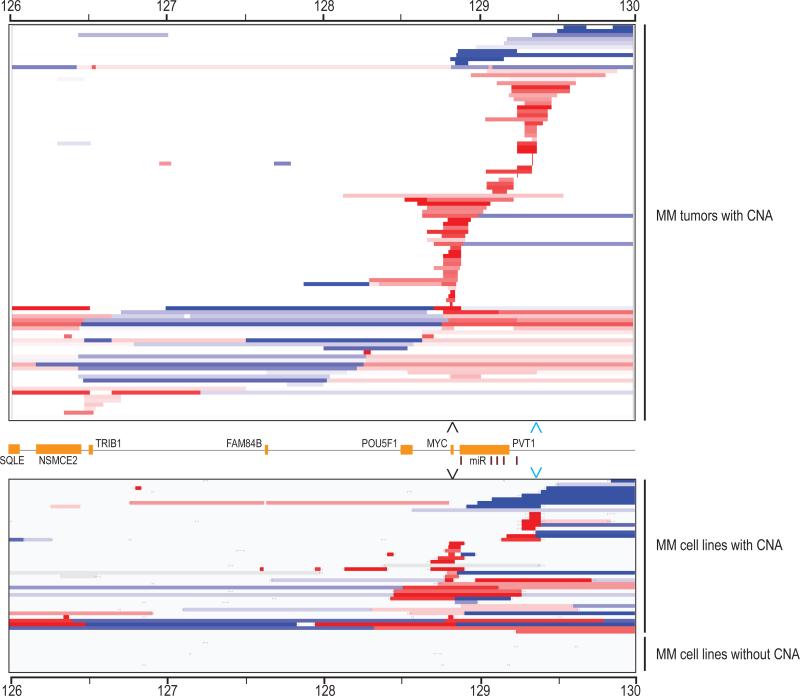
Figure 1. Copy number abnormalities in the MYC locus in MM tumors and cell lines.
Segmented Agilent 244k aCGH of the MYC locus (chr8: 126000000-130000000) in 101 of 238 MM tumors and 42 of 53 MMCL with CNA viewed in IGV is shown. The overall copy number for the region for each sample has been normalized to two copies in order to highlight local changes in copy number. More then a one log gain is deep red, more then one log loss is deep blue, and copy number within 0.2 log of diploid is white. The samples are ordered starting with telomeric deletions, segmental telomeric gains (blue arrowhead), segmental MYC gains (black arrowhead) and centromeric deletions and/or gains.
MYC locus rearrangements detected by interphase FISH and CGH in 238 primary MM tumors
Using publically available 244K CGH data generated by the MMRC from 235 MM and 3 PCL tumor samples, we normalized the copy number of sequences within a 4 Mb region that includes MYC (Figure 1). Recurring patterns of CNA for 101/238 (42%) tumors are similar to what was seen for MMCL. Using MYC as a reference location, we classified these changes into four groups: MYC segmental gain, telomeric segmental gain that was located mostly in a region 350-500 kb telomeric to MYC, centromeric gain or loss, and other telomeric CNA. A summary of these results for 218 MM tumors that also had interphase FISH assays to identify MYC:IGH or MYC:IGL fusions (Table S1) is shown in Table 1. Ectopic MYCN expression (two tumors) or apparent MYC locus rearrangements, detected by CGH and/or FISH assays, were identified in 108/218 (49%) of tumors. There were 33/218 (15%) tumors that had MYC:IGH (11%) or MYC:IGL fusions, with 22 of these tumors also having CNA in the MYC locus. Although only one of the 25 tumors with centromeric CNA had a MYC:IGH or MYC:IGL fusion, approximately 30% of tumors in each of the other three CGH groups had a MYC:IG fusion.
Table 1.
MYC locus rearrangements in 218 MMRC tumors
| Type of MYC rearrangement | # (%) | MYC.Ig fusions | MYC.IGH fusions | MYC.IGL fusions |
|---|---|---|---|---|
| Centromeric gain/loss | 25 (11) | 1 | 1 | 0 |
| MYC segmental gain | 28 (13) | 9 | 8 | 1 |
| Telomeric segmental gain | 30 (14) | 9 | 5 | 4 |
| Telomeric, other | 11 (5) | 3 | 2 | 1 |
| CGH negative, Ig.MYC fusion | 11 (5) | 11 | 9 | 2 |
| All MYC locus rearrangements | 106 (49) | 33 | 25 | 8 |
| N-MYC RNA (no MYC rearrangements)* | 2 (1) | 0 | 0 | 0 |
| No MYC rearrangement or N-MYC RNA | 110 (50) | 0 | - | - |
| All tumors | 218 (100) | 33 | 25 | 8 |
FISH not done for N-MYC.Ig fusions
Consistent with previous FISH studies showing that MYC rearrangements sometimes are not present in all tumor cells10-12, we found that a MYC:IG fusion signal was heterogeneous in 6 (23, 26, 46, 64, 80, 84% of tumor cells) of 33 tumors that had MYC:IG fusion signals (Table S1). Nine of the 218 MM tumors had been classified as smoldering MM (SMM), an asymptomatic MM tumor with a relatively stable tumor burden that can sporadically progress to symptomatic MM at a rate of about 10% per year.26-28 Yet 5 (55%) of these SMM tumors had MYC locus CNA detected by CGH, with one having a MYC:IGH fusion in 100% of tumor cells, and another having a MYC:IGL fusion in 23% of tumor cells.
MYC rearrangements in MM are associated with increased MYC RNA expression
The mean level of MYC RNA is threefold higher in the 106 MM tumors with a MYC rearrangement than in tumors with no detectable rearrangement (P<0.0001)(Figure 2A, Table S3). The mean level of MYC RNA was similar in each of the four groups defined by the different patterns of CNA detected by CGH. In addition, for tumors with MYC rearrangements, the level of MYC expression was not significantly different for tumors with or without MYC:IG fusions. However, the mean level of MYC RNA was significantly higher in MM tumors without a detectable MYC rearrangement compared to MGUS tumors (P<0.0001). Although the mean value of MYC RNA in MGUS tumor cells is significantly higher than in normal bone marrow plasma cells (BMPC)(P=0.03), it is possible that some of the MGUS tumor samples might have been MM or SMM tumors that were incorrectly classified.29 Consistent with the low mitotic index of most MM tumors, a proliferation index based on the expression of 12 genes was only marginally increased in MM vs. MGUS vs. BMPC samples despite the substantial differences in MYC RNA (Figure 2B).24
Although the mean proliferation index is substantially higher in MMCL compared to MM tumors (Figure 2B), MYC RNA is not significantly different in MMCL compared to MM tumors (Figure 2A). Similar to MM tumors, the mean level of MYC RNA was not significantly different for MMCL with MYC:IG rearrangements compared to MMCL with MYC rearrangements not involving an IG locus (Table S3).
Allele specific MYC expression is correlated with rearrangements detected by FISH or CGH
Previously we reported that there was tumor–specific dysregulation of MYCL, MYCN, or only one MYC allele in 13 informative MMCL, all with MYC rearrangements.9, 11 We now have results for nine additional MMCL (Figure 2C, Table S2). Fifteen of 16 MMCL with MYC locus rearrangements selectively expressed one MYC allele, whereas three of four MMCL with no detectable MYC locus rearrangement expressed both MYC alleles (P=0.013). The one MMCL (XG6) with a MYC locus rearrangement that expresses two MYC alleles was unexpected, but is explained by a complex MYC locus rearrangement that appears to have resulted in four copies of MYC, two of which have a somatic mutation (Table S2, Figure S2A, and DISCUSSION).
We identified heterozygous germline polymorphisms in 22 primary MM tumors. Ten of 13 tumors with MYC locus rearrangements selectively expressed one MYC allele, whereas 7 of 9 tumors with no detectable MYC locus rearrangement expressed both MYC alleles (P=0.027) (Figure 2C; Table S4). The most likely explanation for the expression of one MYC allele by the two tumors in the latter group is that our FISH and CGH analyses did not detect all MYC locus rearrangements (see DISCUSSION). However, the expression of both MYC alleles in 3 of 13 MM tumors that had a MYC locus rearrangement was more surprising. Perhaps the MYC locus rearrangement had no effect on MYC expression in some of these tumors. Alternatively, perhaps the rearrangement was present in only a fraction of tumor cells, with either no MYC rearrangement or a different cryptic rearrangement affecting the other MYC allele in other tumor cells, e.g., MMRC0006 had an ~20% segmental copy number gain (e.g., 2.4 vs. 2.0 copies of flanking sequences).
Prevalence of MYC rearrangements is significantly higher in hyperdiploid tumors
We did not detect a significantly different prevalence of ectopic MYCN expression or MYC locus rearrangements in 107 untreated tumors (46%) compared to 111 treated tumors (52%) (Table S5). In addition, heterogeneity of MYC:IG fusions occurred with a similar prevalence in untreated tumors (3/15) and treated tumors (3/18). There were two initial proposals for classifying MM tumors into different molecular groups based on gene expression profiling analyses (Table S6). The TC classification, which identifies IGH Translocation groups and patterns of CYCLIN D expression, mostly reflects early pathogenic events common to MGUS and MM.24, 30 While mostly similar, the seven molecular group (7MG) classification includes a PR group of tumors that generally are more proliferative.31, 32 As summarized in Table S6, there are significant differences in MYC rearrangements for some of the groups: 1) ~22% in the 11+6 TC groups, which have IGH translocations dysregulating CYCLIN D1 or CYCLIN D3, respectively, and in the similar CD1+CD2 7MG (P<0.0001); and 2) ~63% in the D1+D1/D2 TC groups (P=0.003). These results suggested that the differences in the prevalence of MYC locus rearrangements might be related to hyperdiploidy. We developed a hyperdiploid index, which is the average normalized content of 8 odd numbered chromosomes as determined by CGH analyses (Table S1 and Methods). The analysis summarized in Table S7 confirms that there is an increasing prevalence of MYC rearrangements that is significantly correlated with an increasing hyperdiploid index even after removing tumors with IGH translocations dysregulating CYCLIN D1 or CYCLIN D3.
Association of MYC rearrangements and molecular prognostic markers
The PR 7MG group also has an increased prevalence (~70%) of MYC locus rearrangements (P=0.002) (Table S6), indicating that MYC dysregulation might be contributing to the PR phenotype. Gene expression profiling data on a large number of MM tumors has shown that prognostic predictions can be generated from a proliferation index, and even better with the UAMS-70 gene prognostic index (GEP70).25, 33 We determined a proliferation index24 and the GEP70 for the 218 MMRC tumors that had been analyzed by FISH and CGH for MYC locus rearrangements (Table S1). In comparing the prevalence of MYC locus rearrangements in the top quintile vs. the bottom four quintiles (Table S8), we found a significant difference for both the proliferation index (P=0.027) and the GEP70 (P=0.011). Moreover, the GEP70 is significantly correlated with rearrangements in some groups (6+11,P=0.001; CD1+CD2, P=0.005; D1, P=0.01; 4 and MS, P~0.023) but not others (D2, D1/D2, MAF, LB, HY, PR)] (Figure S1, Table S9).
MYC rearrangements in MM often involve enhancers but mostly super-enhancers
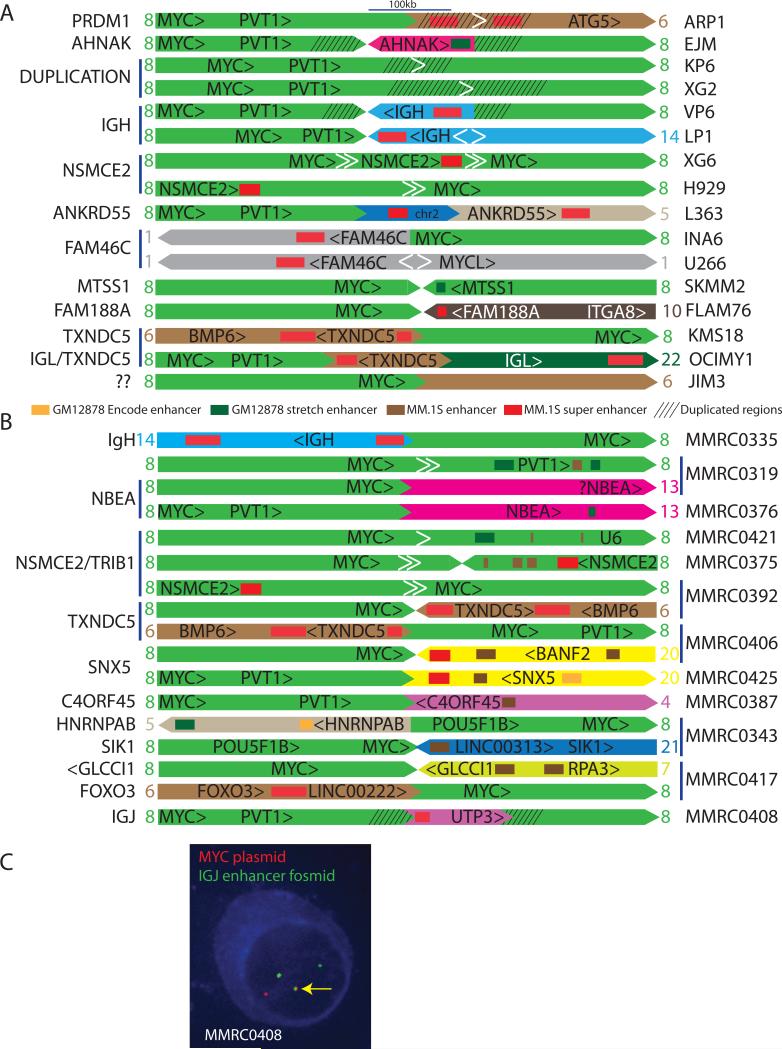
The structures of rearrangements involving MYC (or MYCL) in 17 MMCL (Figure 3A, Table 2, Table S10) were deduced from a combination of FISH, CGH, and mate pair or cloned sequences. We also indicate enhancer elements repositioned near the MYC locus, using published reference data for two cell lines: 1) enhancers and super enhancers (SEs) in the MM.1S MMCL34, 35; and 2) strong enhancers and stretch enhancers identified in the GM12878 lymphoblastoid cell line (LCL), which is phenotypically similar to MMCL, from ENCODE data on the UCSC genome browser (http://genome.ucsc.edu)36 and Parker et al37. Although the enhancers and SEs in these two cell lines might not apply to all MMCL and tumors, we have additional evidence suggesting that they generally are applicable. First, most of the putative SEs associated with MYC locus rearrangements (Tables 2 and 3; DISCUSSION) have been shown to be markedly up-regulated with differentiation of B cells to plasma cells (Figure 2E)23. Second, we have preliminary DNaseI HS.seq and ChIP.seq (H3K4Me3; H3K27Ac) data for 4 MMCL that identify the same enhancers and putative SEs that were identified in the MM.1S MMCL. A description of the structures of rearrangements involving MYC in 12 MMRC tumors, which were deduced mostly from whole genome sequences,38 but also from FISH, CGH, and cloned sequences in some cases, is presented in Table 3 and Figure 3B, with additional details in Table S11.
Figure 3. MYC locus rearrangements in 16 MMCL and 12 MMRC tumors.
A. The anatomy of MYC locus rearrangements on chr8 (light green) shows MM.1S enhancers and super enhancers, and duplicated regions, with the genes associated with the enhancer regions indicated at the left. Enhancer elements in the MYC locus are not shown here but are shown in Figure 4. The orientations of the chromosomes and transcription units are indicated. The white arrowheads indicate intrachromosomal breakpoints, with the pair of white arrows in XG6 and H929 indicating a deletion, the pair of arrows in U266 and LP1 indicating an inversion, and the single white arrow in ARP1, KP6, and XG2 indicating tandem duplication. A more detailed diagram of the XG6 and LP1 rearrangements is shown in Figure S1. Additional information regarding these rearrangements is included in Results and Table S10.
B. The anatomy of MYC locus rearrangements for 12 MMRC tumors is presented as described in Figure 3. Several of the MM tumors have two different rearrangements. Additional information regarding these rearrangements is summarized in Results, Table 2, and Table S11.
C. cIg-FISH was performed in MMRC0408 demonstrating co-localization of probes for MYC (red) and the IgJ enhancer (green) in the nuclei of plasma cells with light chain restriction (blue cytoplasmic staining).
Table 2.
MYC rearrangements and associated enhancers in 17 MM cell lines
| MMCL | Description of MYC locus structural variation |
|---|---|
| LP1 | FISH analyses have shown that variant IGH translocations involving MYC have 3’ IGH SE (E⍰) and VH telomeric sequences located telomeric to MYC.10, 11 The chr8 breakpoint is ~390 kb downstream of MYC and the chr14 breakpoint is located at the telomeric end of the Ea1 SE, but with chr 14 sequences proceeding in a centromeric direction away from the breakpoint as a result of an inversion within the IGH locus that was detected from mate pair sequences (SF1 and Discussion) |
| H929 | a deletion of 8:126344414-128713154 positions MYC ~100 kb from a SE and other enhancer elements in intron 4 of NSCME2 |
| XG6 | has a deletion 8 breakpoint 126324977>|<128737494 but also a second breakpoint downstream of MYC: 128775373>|<126287002. There is a copy number gain by CGH related to these breakpoints, i.e., 126287002-126324977 (~38kb) and 128737494-128775373 (~38kb). These data indicate that MYC is flanked by a SE and other enhancer sequences located in intron 4 of NSMCE2 and vice versa, with several copies of both regions on the rearranged chromosome (see SF1) |
| INA6 | der(1) of a reciprocal t(1;8) translocation has a chr1 breakpoint within or just beyond FAM46C, so that the downstream SE region is near the breakpoint. MYC is predicted to be ~1209 Mb from the chr8 breakpoint, but numerous intrachromosomal rearrangements in the MYC locus suggest that MYC might be much closer to the breakpoint |
| U266 | a 77 Mb inversion on 1p has breakpoints located 7 kb downstream of MYCL and in the 3’ untranslated region of FAM46C; MYCL is repositioned near the SE region downstream of FAM46C |
| KMS18 | an interchromosomal translocation has a chr6 breakpoint ~50kb upstream of TXNDC5 so that SEs associated with TXNDC5 are predicted to be located ~500 kb upstream of MYC |
| OCIMY1 | a complex translocation shows that ~15kb of chr6 sequences that contain a SE region located downstream of TXNDC5 are positioned ~560 kb downstream of MYC; the other end of the chr 6 segment is juxtaposed to chr22, with the breakpoint located ~530 kb centromeric to the 3’ IGL SE |
| ARP1 | this der(8)t(6;8) translocation has a chr 6 breakpoint immediately downstream of PRDM1 so that a pair of flanking SEs are repositioned ~530 kb downstream of MYC. In addition, there is an ~60 kb tandem duplication of the sequences that contain the pair of super enhancers that flank PRDM1 |
| SKMM2 | an ~3.1 Mb inversion on chr8 (125.697-128.848) positions stretch enhancers in intron 3 of MTSS1 ~95kb downstream of MYC |
| FLAM76 | an interchromosomal translocation into FAM188A on chr 10 positions a SE ~25 kb from the breakpoint, which is ~30 kb downstream of MYC |
| L363 | a complex translocation indicates that an ~30 kb segment of chr2, which includes an SE flanking the 3’ end of CTLA4, is positioned ~560kb downstream of MYC; the downstream side of the chr 2 fragment is juxtaposed into the 3’ end of ANKRD55, so that a SE within ANKRD55 is located ~32 kb from the chr2 fragment and ~ 610 kb from MYC |
| VP6 | chr14 sequences including a 3’ IGH SE are inserted downstream of MYC between duplicated flanking sequences (8:129.216-129.305) |
| EJM | a 58 kb fragment from chr11, which contains a stretch enhancer in the ultimate intron of AHNAK, is inserted downstream of MYC between duplicated flanking sequences (8:129.265-129.346) |
| KP6 | an ~90kb tandem duplication ~500 kb downstream of MYC (8:129.239-129.329) |
| XG2 | an ~341 kb tandem duplication ~500 kb downstream of MYC (8:129.072-129.413) |
| JIM3 | an interchromosomal translocation repositions a chr6 “gene dessert” with no apparent enhancers ~45 kb downstream of MYC; mate pair sequences did not detect other rearrangements on the region of chr6 repositioned downstream of MYC |
| XG7 (not in Fig.3) | chr8 and chr6 segments inserted into chr17 with a chr 6 TXNDC5 SE repositioned ~38 kb upstream of MYC; and chr 17 UBRE2G1 enhancers located ~110 kb downstream of MYC |
Table 3.
MYC rearrangements and associated enhancers in 12 MMRC tumors
| MM | Description of MYC locus structural variation |
|---|---|
| MMRC0335 | a reciprocal t(8:14) translocation, with the chr 14 breakpoint occurring within the Eα1 SE and the chr 8 breakpoint ~120 kb upstream of MYC |
| MMRC0408 | a 34kb fragment, which contains a SE element that is located immediately 5’ of IGJ, is inserted into a region ~500 kb downstream of MYC; the insertion is flanked by a duplicated chromosome 8 sequence of ~61 kb |
| MMRC0375 | an inversion has juxtaposed a sequence in intron 4 of NSMCE2 (8:126.359) with a sequence ~280 kb downstream of MYC, but an ~237 kb deletion immediately downstream of MYC suggests that the inversion breakpoint would be located ~40 kb downstream of MYC, i.e., very near a SE and other enhancer elements in intron 4 of NSMCE2 |
| MMRC0421 | this complex rearrangement is manifested by a breakpoint ~69 kb downstream of MYC and ~120 kb downstream of TRIB1 (both strands with the same orientation), suggesting that stretch enhancers ~75 kb downstream of the breakpoint could dysregulate MYC |
| MMRC0392 | there are two MYC rearrangements in this tumor, suggesting the possibility of two clones with different MYC rearrangements: 1) a deletion repositioning intron 4 of NSMCE2 and its SE and enhancer elements within ~20 kb of MYC; and 2) an interchromosomal rearrangement that repositions a SE upstream of TXNDC5 so that it would be ~50kb upstream of MYC |
| MMRC0406 | there were two different interchromosomal breakpoints involving MYC. The first occurs within a chr 6 SE region upstream of TXNDC5 so that it would be located ~32 kb upstream of MYC. The second occurs in a chr 20 intergenic region between SNX5 and BANF2 that contains a SE and other enhancer sequences, which would then be located ~100kb downstream of MYC. A copy number gain of chr 8 sequences between these two breakpoints suggests, but does not prove, that both rearrangements occur in the same tumor cell |
| MMRC0425 | the interchromosomal breakpoint also occurs on chr 20 between SNX5 and BANF2 so that the intergenic SE and other enhancers are predicted to be located ~700 kb downstream of MYC |
| MMRC0417 | there were two different interchromosomal breakpoints involving MYC. The first has a chr 6 breakpoint ~100 kb downstream of FOXO3, which encompasses a strong SE, and ~15 kb downstream of LINC00222, which also is associated with several enhancer elements; the chr 8 breakpoint is ~25 kb upstream of MYC. The second has a chr 7 breakpoint within the 5’ end of GLCCI1, which has enhancer elements located within and upstream of its promoter region; the chr 8 breakpoint is ~80 kb downstream of MYC. There is a copy number gain of chr 8 sequences between these two breakpoints, consistent with - not proving - that they are on the same derivative chromosome in the same tumor cell |
| MMRC0387 | this apparent reciprocal translocation has a chr8 breakpoint ~370 kb downstream of MYC and a chr4 breakpoint at the 5’ end of C4orf45, suggesting that MYC could be dysregulated by the enhancer sequences in the C4orf45 promoter |
| MMRC0343 | there is a chr8 gain (including MYC) between two breakpoints. The chr 21 breakpoint is immediately downstream of HSF2BP, with an ENCODE LCL enhancer predicted to be located ~65 downstream of MYC. The chr5 breakpoint is ~8 kb upstream of HNRNPAB and ENCODE LCL enhancer elements associated with this gene but also an ENCODE LCL insulator separating the enhancer elements from the breakpoint |
| MMRC0319 | there are two MYC rearrangements. The first is a reciprocal translocation with a chr8 breakpoint ~33kb downstream of MYC and a chr13 breakpoint ~240 kb upstream of NBEA1, but only a combination of ENCODE LCL weak enhancers and insulators in the promoter regions of NBEA transcripts. The second is a chr 8 deletion that repositions MYC near enhancers in the PVT-1 locus |
| MMRC0376 | this is a reciprocal t(8;13) translocation that is similar to the one in MMRC0319, but with the chr8 breakpoint located ~726 kb downstream of MYC and the chr13 breakpoint located ~248 kb upstream of NBEA |
The molecular characterization of MYC locus breakpoints not involving IGH, IGL, or IGK in 14 MMCL and 11 MM tumors revealed the involvement of: MM.1S SEs (TXNDC5 [6p24.3], FAM46C [1p12], ANKRD55 [5q11.2], FAM188A [10p13], IGJ [4q13.3], FOXO3 [6q21], PRDM1 [6q21], NSMCE2 [8q24.13], CTLA4 [2q33.2], SNX5 [20p11-12]); MM.1S enhancers (AHNAK [11q12.3], PVT1 [8q24.21], MTSS1 [8q24.13], GLCCl1 [7p21.3], C4orf45 [4q32.1]); and GM12878 lymphoblastoid Stretch enhancers (AHNAK, MTSS1, TRIB1), Encode enhancers (LINC00313/HSF2PB [21q22.3], and other regions near the MM.1S enhancers or SEs).34-36 Several of these regions were recurrent (NSMCE2, TXNDC5, FAM46C, SNX5, NBEA) in our samples, or found in one of our samples but twice in the first 29 samples analyzed and publically reported for the CoMMpass project (http://research.themmrf.org) (FOXO3/LINC00313). The JIM3 MMCL has a MYC rearrangement for which there is no obvious enhancer. In addition, two MM tumors have rearrangements near NBEA with no evidence for enhancers near the breakpoints, but others reported a breakpoint that juxtaposed PVT1 and NBEA in a MMCL.15 It is possible that there are undetected rearrangements or polymorphisms that might explain these results. Curiously, there are three MMCL and five MM tumors with two different rearrangements and associated enhancers near MYC. This could reflect passenger events occurring during a complex rearrangement process, or heterogeneity for tumors.
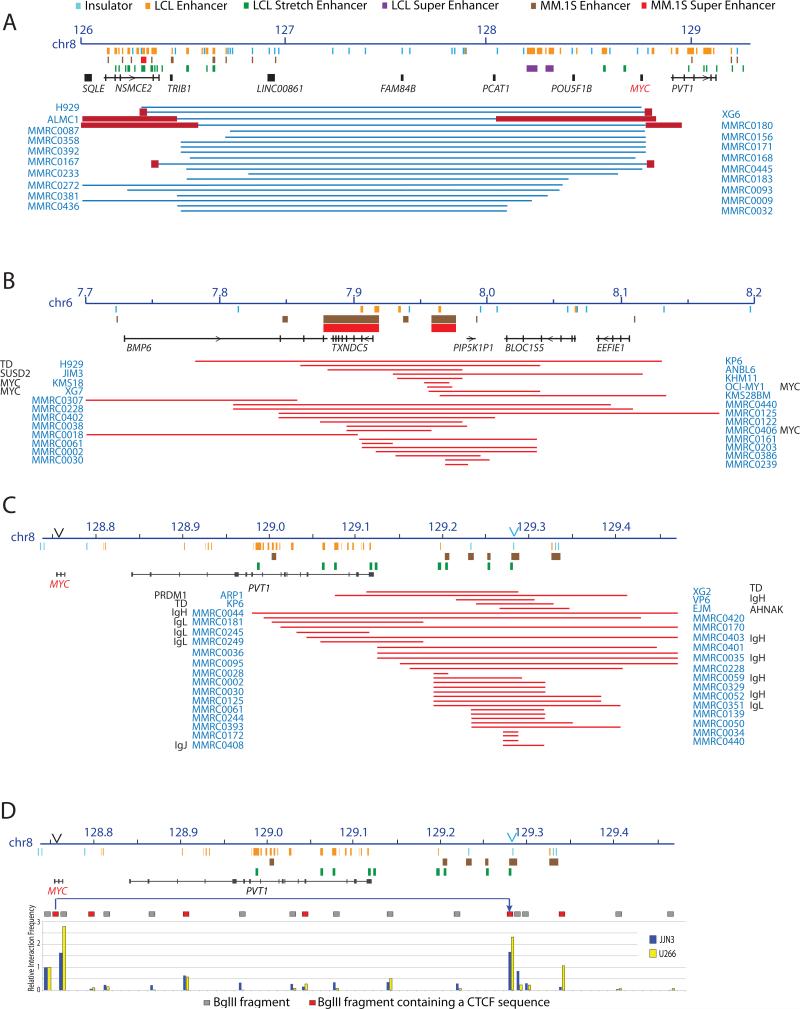
Figure 4 summarizes CGH CNA and breakpoints in two recurrent non-IG partner loci. First, it is not surprising that centromeric losses – and perhaps some gains (not shown) - that juxtapose MYC and NSMCE2/TRIB1 enhancers were the most common MYC:nonIG rearrangement since intra-chromosomal rearrangements are more common than inter-chromosomal rearrangements39 (Figure 4A). Second, in addition to the three MMCL and two MM tumors with MYC:TXNDC5 rearrangements, Figure 4B shows other MMCL and MM tumors with gains involving TXNDC5, which are clustered among tumors with CGH detected MYC rearrangements not involving IGH or IGL compared to tumors that have a MYC:IGH or MYC:IGL rearrangement or no rearrangement (P=0.0001).
Figure 4. Three chromosomal regions involved in recurrent MYC rearrangements.
Symbols at the top apply to Panels A-D; the boxed symbols at the bottom to Panel D.
A. Large deletions (blue lines) identified by Agilent 244K CGH results in 53 MMCL and 218 MMRC tumors indicates a repositioning of MYC near enhancers associated with NSMCE2 and TRIB1, which are located about 2.5 Mb centromeric to MYC. The dark red boxes indicate regions of copy number gains that flank the deleted regions. Breakpoints have been identified by sequencing or mate pairs in two MMCL (H929, XG6) and in one of the tumors (MMRC0392) shown in this figure, but also in two other tumors (MMRC0375, 0421) with complex CGH patterns not shown in this Figure.
B. Super enhancers associated with TXNDC5 on chr6. The red lines indicate regions of copy number gains determined from 244K CGH data on MMCL and 218 MMRC tumors. The copy number gain is juxtaposed near MYC in three MMCL (KMS18, OCIMY1, XG7) and one tumor (MMRC406) and near SUSD2 in the JIM3 MMCL. The copy number gains shown are significantly enriched in MMRC tumors that have MYC rearrangements not involving IGH or IGL loci (see DISCUSSION).
C. Segmental copy number gains localized >200 kb telomeric to MYC. MMCL and MMRC tumors with segmental copy number gains (red lines) identified from Agilent 244K CGH data are shown. Many of these gains flank insertions of enhancer elements (see Results and Discussion), or occur in tumors or MMCL that have MYC.IGH or MYC.IGL fusion signals. Most of these gains are centered in two regions that are about 350 kb and 500 kb downstream of MYC. The former occurs within the distal end of the PVT1 locus, near one MM.1S enhancer and multiple LCL enhancers. The latter is centered in a region that preferentially associates with the MYC promoter, but also contains a cluster of MM.1S enhancer sequences and GM12878 stretch enhancers.
D. Interaction of MYC promoter with distal telomeric sequences. 3C experiments show a preferential interaction of a MYC promoter fragment that contains a CTCF site with a fragment, which is located ~500 kb telomeric of MYC, that also contains a CTCF sequence. The JJN3 MMCL has a MYC:IGH rearrangement, but the U266 MMCL has a MYCL rearrangement and expresses virtually no MYC.
Evidence for dysregulation of MYC by rearrangements not involving an IG locus
Excluding MYC rearrangements that involve an IGH or IGL locus, allele specific expression of MYC still is associated significantly with MYC rearrangements for the combined MMCL+MM tumor samples (P=0.01) (Figure 2C). Additional evidence that rearrangements are able to dysregulate MYC includes the following. First, a t(6;22) translocation [chr6: 7932054(-) | chr22: 24572284(-)], which juxtaposes SUSD2 near a TXNDC5 SE, results in ectopic expression of SUSD2 in the JIM3 MMCL. Second, DNA fragments from putative enhancer regions in intron 4 of NSMCE2 and within ANKRD55 had enhancer activity in transient transfection assays (Figure 2D). Finally, somatic cell hybrids between several MMCL and the SP2/0 murine plasmacytoma line showed expression of MYC from EJM and L363 chromosomes having MYC rearrangements, but virtually no expression from chromosomes with germline MYC loci (Supplemental Methods).
DISCUSSION
We identified MYC rearrangements in nearly half of treated or untreated MM tumors. Avet-Loiseau et al used interphase FISH to identify MYC rearrangements in 16% of 529 newly diagnosed MM tumors and 10% of 58 relapsed MM tumors.12 Given the different methodologies, a comparison of the overall prevalence of MYC rearrangements is problematic. However, a direct comparison of the prevalence of MYC fusions with IGH or IGL loci should be informative. We found fusions of MYC with IGH or IGL loci in ~15% of treated or untreated MM tumors. By contrast, they found fusions of MYC with IGH or IGL sequences in ~4% of either newly diagnosed or relapsed MM tumors. One possible explanation is that the MMRC samples require enough material for analyses and are therefore biased for a larger tumor mass with better ex vivo viability (see Methods), but we also note the use of different IGH and IGL FISH probes.
It is likely that a significant number of MM tumors with rearrangements in the MYC locus have not been detected by the combination of CGH and interphase FISH assays that we used. First, 11 of 33 MM tumors with MYC:IG fusions did not have CGH abnormalities (Table 1). Second, a MYC:IG fusion was found in only 1 of 25 tumors with centromeric losses or gains, but was present in 21 (30%) of the 69 MM tumors in the other three CGH categories (Table 1). Assuming that other partner loci of MYC would mimic MYC:IG fusions, we would predict that about 24 (11%) of the 218 MM tumors would have MYC rearrangements not detected by CGH, e.g., the MMRC0387 tumor has a MYC rearrangement detected by whole genome sequencing but not by CGH (Table 3, Table S11).
Primary IGH translocations involving 7 recurrent partners mostly have breakpoints consistent with errors in three B cell specific DNA modification processes (IGH switch recombination, somatic hypermutation, VDJ recombination) that are active during early stages of MM pathogenesis.6, 10, 40-42 By contrast, MYC:IGH rearrangements, including all three reported here (Table S10, Table S11), usually do not have IGH breakpoints consistent with errors in the three B cell specific DNA modification processes that are thought to be inactive in plasma cells or plasma cell tumors. We identified a number of MMCL and MM tumors with variant MYC:IGH translocations for which the IGH locus, including at least one 3’ IGH enhancer and the most telomeric VH sequences are positioned telomeric to MYC.10, 11, 43 Given that there are insulator sequences centromeric to 3’ IGH enhancers44, 45, we predicted that these translocations would be associated with a deletion of the insulator, or have an inversion so that the insulator was no longer located between MYC and the 3’ IGH enhancer.43 Our analysis of the variant MYC:IGH translocation in the LP1 MMCL showed that there is an inversion, confirming our prediction (Figure S2B).
MYC:IGH (45%), MYC:IGL (11%), and MYC:IGK (4%) accounted for ~60% of MYC rearrangements in MMCL, whereas MYC:IGH (24%) and MYC:IGL (8%) accounted for ~32% of MYC rearrangements in the MMRC tumors (Table 1, Table S1, Table S2). The fraction of MYC rearrangements involving IGH or IGL is significantly higher (P=0.007) in MMCL than in the MM tumors. This is somewhat surprising given that the mean level of MYC RNA expression (and the GEP70 for tumors) is not significantly different for MMCL or MM tumors with MYC:IG vs. MYC:non-IG rearrangements. Although there is evidence for recurrent non-IG partners (see above), there is not yet enough information to reveal the full repertoire of partner loci involved in MYC rearrangements in MM. However, there are at least four (NSMCE2/TRIB1, TXNDC5, FAM46C, FOXO3) -perhaps six (SNX5, NBEA) - loci that are frequent partners. Significantly, MYC rearrangements involving 1p13 , 6p21, 6q21 and 13q14 are recurrent in previous karyotypic analyses.10, 46-49
One of the most striking findings from the CGH analyses was that 30 MM and 4 MMCL had a segmental telomeric gain that was located about 350-500 kb telomeric to MYC (Figure 1, 4C). Ten of the 30 MM tumors had a MYC:IG fusion or insertion of an IGJ SE (Figure 3C), and one (VP6) of the 4 MMCL tumors had an insertion of 3’ IGH enhancer sequences between the duplicated chr8 sequences that were identified by CGH. The EJM MMCL had a 58 kb region and AHNAK enhancer from chr11 inserted between the duplicated sequences on chr8. However, two HMCL (KP6, XG2) had only a tandem duplication of sequences in this region. The targeting of this region for duplication is consistent with chromosome conformation capture (3C) experiments, which showed that a MYC promoter fragment containing a CTCF site interacts preferentially with a fragment containing a CTCF site that is located near several enhancers, including a stretch-enhancer (Figure 4D). It seems possible that this interaction facilitates the interaction of MYC with enhancer sequences repositioned near this region. Regarding the two MMCL with tandem duplications in this region without insertion of an exogenous enhancer, we can only speculate that the duplication somehow unmasks or creates a functional enhancer element in this region.
Most MYC rearrangements in MMCL and tumors reposition MYC near a putative SE or stretch enhancer34, 35, 37, i.e., for MMCL, 11/14 not having a MYC:IG rearrangement (Table 2) but 39/42 including MMCL with MYC:IG rearrangements (Table S2); and for MM tumors, 6/11 not having a MYC:IG rearrangement (Table 3), but 39/44 including tumors with MYC:IG rearrangements (Table 1). Most rearrangements not involving an MM.1S SE appeared to involve credible MM.1S or LCL enhancer elements that might have properties of SE in specific MMCL or MM tumors, possibly as a consequence of tumor specific chromatin marks, and perhaps related to tumor specific MYC locus rearrangement. It is not clear why there is markedly non-random targeting of MYC rearrangements to a limited number of partner loci, most of which contain SE regions. Although we are unable to answer this question, it is possible that the recurrent involvement of the FAM46C SE is selected because this rearrangement can inactivate one copy of the FAM46C tumor suppressor gene while also dysregulating MYC.38, 50 Nonetheless, the identification of these partner loci suggests that they will help us to identify other genes (e.g., SUSD2:TXNDC5 SE) that are dysregulated when repositioned near these regions in plasma cell tumors. The activation of MYC by rearrangements that mostly reposition it near a SE provides clear evidence that genomic rearrangements in other kinds of tumors are likely to hijack a subset of cell and tumor specific SE that will dysregulate expression of oncogenes34, 35.
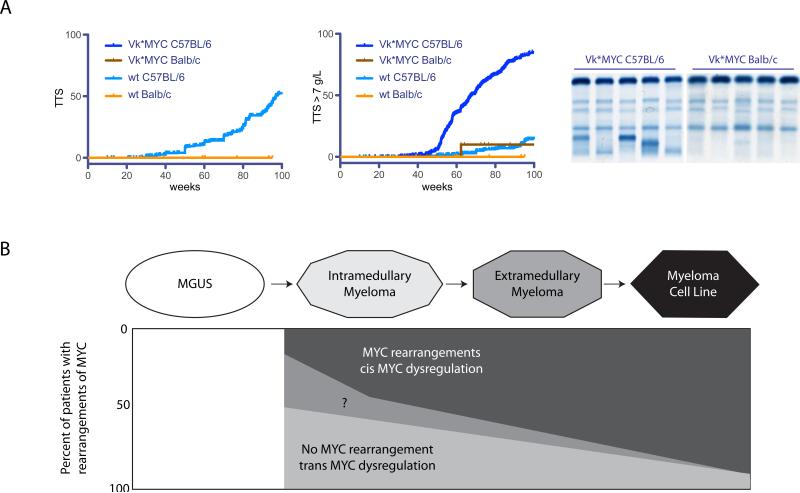
Increasingly, it has become apparent that dysregulation of MYC (less often MYCN or MYCL) is an essential event in MM.19, 51-53. Previous results indicated that germinal center activation of a MYC transgene caused progression of MGUS to MM in an MGUS prone mouse strain, a conclusion that is supported by new results showing that the same transgene does not cause MM in a mouse strain that rarely develops MGUS (Figure 5A). The increased MYC expression at the MGUS/MM transition can be bi-allelic, or mono-allelic if there is a MYC rearrangement. Given the evidence for heterogeneity of MYC rearrangements in about 15% of treated or untreated MM tumors with MYC:IGH or MYC: IGL rearrangements, it is clear that the prevalence of MYC rearrangements increases during tumor progression.10, 12, 54 Initially, we had proposed that most MYC rearrangements were a late progression event. However, in view of a high and similar prevalence of MYC rearrangements in untreated and treated MM tumors, and in SMM tumors, it now seems possible that some of these rearrangements occur early (Figure 5B). One caveat is that the MMRC tumors analyzed may be skewed towards more advanced stages of disease (Methods and Table S14).
Figure 5. Dysregulation of MYC causes the progression of MGUS to MM.
A. Wild type C57Bl/6, but not Balb/c, mice develop faint (<4g/L), non-progressive M-spikes detectable by serum protein electrophoresis, with a median Time To Spike (TTS) detection of 97 weeks (left panel). With the introduction of the Vk*MYC transgene, C57Bl/6 mice develop progressively increasing M-spikes with a median Time To Spike >7g/L of 67 weeks. In contrast wild type C57Bl/6 or Vk*MYC Balb/c mice rarely develop an M-spike >7g/L, and it is never progressive (middle panel). Representative serum protein electrophoresis is shown for Vk*MYC C56BL/6 and Vk*MYC Balb/c mice (right panel).
B. Proposed model summarizing the role of MYC in the progression of MM. The height of the boxed region indicates the fraction of tumors with MYC rearrangements at different stages. MGUS: white, low MYC expression. MM: light gray, increased MYC expression without a MYC rearrangement; dark gray, substantially increased MYC expression with a MYC rearrangement; intermediate gray, unknown. Increased MYC expression is due to unknown trans- mechanisms that result in bi-allelic expression in at least 50% of early MM tumors. Mono-allelic expression as a result of a MYC locus rearrangement might occur with this transition in some (? ~15%) MM tumors. Most MYC rearrangements probably occur during progression of MM, with >50% in advanced MM and 90% in MMCL.
Regardless of whether MYC rearrangements occur at early or late stages of pathogenesis, MYC rearrangements may provide one of several critical events contributing to increased autonomy (perhaps one manifestation being decreased expression from the non-rearranged MYC allele) and a more aggressive phenotype. Although we don't have sufficient survival data for individuals from whom the MMRC tumors were derived, we determined that the GEP70, which is correlated strongly with low patient survival25, is significantly increased in tumors with MYC rearrangements (P<0.0001), but only in some molecular types of MM (Results, Figure S1, Table S9). However, Avet-Loiseau et al55 reported that MYC rearrangements were not associated with survival, so that the prognostic significance of MYC rearrangements remains to be clarified. Given the uncertainty of the prevalence of MYC rearrangements at early vs. late stages, it is important to achieve a better understanding of when MYC rearrangements occur in different kinds of MM tumors, and how they impact MM pathogenesis and prognosis.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the Multiple Myeloma Research Consortium (MMRC) for their help in processing samples for this project and making available cytospin slides for FISH, the RNA for selective expression, and with the help of Mike Chapman, the list of structural variations in the MYC locus from the MM Genomics Initiative 38. This work is supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (WMK); grants CA83724 (RF), ECOG CA21115T(RF), CA136671 (PLB), CA133966 (PLB), Predolin Foundation, Mayo Clinic Cancer Center and the Mayo Foundation. Rafael Fonseca is a Clinical Investigator of the Damon Runyon Cancer Research Fund. Supplementary information is available at Leukemia's website.
This work is supported by grants CA83724 (RF), ECOG CA21115T (RF), CA136671 (PLB), CA133966 (PLB), Predolin Foundation, Mayo Clinic Cancer Center and the Mayo Foundation. Rafael Fonseca is a Clinical Investigator of the Damon Runyon Cancer Research Fund.
This manuscript is dedicated to the memory of Oleg K. Glebov, who played a major role during the early phases of this work.
ABBREVIATIONS
- MM
multiple myeloma
- IG
immunoglobulin genes
- IGH, IGL, IGK
immunoglobulin heavy chain, lambda, kappa genes
- BMPC
normal bone marrow plasma cell
- MGUS
monoclonal gammopathy of undetermined significance
- MMCL
MM cell lines
- PCL
plasma cell leukemia
- SMM
smoldering MM
- GEP70
UAMS 70 gene prognostic index
- CNA
copy number abnormalities
- SE
super enhancer
Footnotes
Conflict of Interest: The authors have no conflict of interest related to this work
Supplementary information is available at Leukemia's website
REFERENCES
- 1.Dalla-Favera R, Martinotti S, Gallo RC, Erikson J, Croce CM. Translocation and rearrangements of the c-myc oncogene locus in human undifferentiated B-cell lymphomas. Science. 1983 Feb 25;219(4587):963–967. doi: 10.1126/science.6401867. [DOI] [PubMed] [Google Scholar]
- 2.Potter M. Neoplastic development in plasma cells. Immunol Rev. 2003 Aug;194:177–195. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- 3.Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005 Apr;5(4):251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 4.Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr., Miljkovic V, et al. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci U S A. 2003 Mar 4;100(5):2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhan F, Tian E, Bumm K, Smith R, Barlogie B, Shaughnessy J., Jr. Gene expression profiling of human plasma cell differentiation and classification of multiple myeloma 23 based on similarities to distinct stages of late-stage B-cell development. Blood. 2003 Feb 1;101(3):1128–1140. doi: 10.1182/blood-2002-06-1737. [DOI] [PubMed] [Google Scholar]
- 6.Kuehl WM, Bergsagel PL. Molecular pathogenesis of multiple myeloma and its premalignant precursor. J Clin Invest. 2012 Oct 1;122(10):3456–3463. doi: 10.1172/JCI61188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malpas JS, Bergsagel DE, Kyle R, Anderson K. Multiple Myeloma: Biology and Management. Oxford University Press; Oxford: 2004. [Google Scholar]
- 8.Drexler HG, Matsuo Y. Malignant hematopoietic cell lines: in vitro models for the study of multiple myeloma and plasma cell leukemia. Leuk Res. 2000 Aug;24(8):681–703. doi: 10.1016/s0145-2126(99)00195-2. [DOI] [PubMed] [Google Scholar]
- 9.Dib A, Gabrea A, Glebov OK, Bergsagel PL, Kuehl WM. Characterization of MYC translocations in multiple myeloma cell lines. J Natl Cancer Inst Monogr. 2008;(39):25–31. doi: 10.1093/jncimonographs/lgn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabrea A, Martelli ML, Qi Y, Roschke A, Barlogie B, Shaughnessy JD, Jr., et al. Secondary genomic rearrangements involving immunoglobulin or MYC loci show similar prevalences in hyperdiploid and nonhyperdiploid myeloma tumors. Genes Chromosomes Cancer. 2008 Jul;47(7):573–590. doi: 10.1002/gcc.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shou Y, Martelli ML, Gabrea A, Qi Y, Brents LA, Roschke A, et al. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci U S A. 2000 Jan 4;97(1):228–233. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avet-Loiseau H, Gerson F, Magrangeas F, Minvielle S, Harousseau JL, Bataille R. Rearrangements of the c-myc oncogene are present in 15% of primary human multiple myeloma tumors. Blood. 2001 Nov 15;98(10):3082–3086. doi: 10.1182/blood.v98.10.3082. [DOI] [PubMed] [Google Scholar]
- 13.Chiecchio L, Dagrada GP, White HE, Towsend MR, Protheroe RK, Cheung KL, et al. Frequent upregulation of MYC in plasma cell leukemia. Genes Chromosomes Cancer. 2009 Jul;48(7):624–636. doi: 10.1002/gcc.20670. [DOI] [PubMed] [Google Scholar]
- 14.Fabris S, Storlazzi CT, Baldini L, Nobili L, Lombardi L, Maiolo AT, et al. Heterogeneous pattern of chromosomal breakpoints involving the MYC locus in multiple myeloma. Genes Chromosomes Cancer. 2003 Jul;37(3):261–269. doi: 10.1002/gcc.10211. [DOI] [PubMed] [Google Scholar]
- 15.Nagoshi H, Taki T, Hanamura I, Nitta M, Otsuki T, Nishida K, et al. Frequent PVT1 rearrangement and novel chimeric genes PVT1-NBEA and PVT1-WWOX occur in multiple myeloma with 8q24 abnormality. Cancer Res. 2012 Oct 1;72(19):4954–4962. doi: 10.1158/0008-5472.CAN-12-0213. [DOI] [PubMed] [Google Scholar]
- 16.Au WY, Horsman DE, Gascoyne RD, Viswanatha DS, Klasa RJ, Connors JM. The spectrum of lymphoma with 8q24 aberrations: a clinical, pathological and 24 cytogenetic study of 87 consecutive cases. Leuk Lymphoma. 2004 Mar;45(3):519–528. doi: 10.1080/10428190310001593120. [DOI] [PubMed] [Google Scholar]
- 17.Bertrand P, Bastard C, Maingonnat C, Jardin F, Maisonneuve C, Courel MN, et al. Mapping of MYC breakpoints in 8q24 rearrangements involving non544 immunoglobulin partners in B-cell lymphomas. Leukemia. 2007 Mar;21(3):515–523. doi: 10.1038/sj.leu.2404529. [DOI] [PubMed] [Google Scholar]
- 18.Lossos IS, Alizadeh AA, Diehn M, Warnke R, Thorstenson Y, Oefner PJ, et al. Transformation of follicular lymphoma to diffuse large-cell lymphoma: alternative patterns with increased or decreased expression of c-myc and its regulated genes. Proc Natl Acad Sci U S A. 2002 Jun 25;99(13):8886–8891. doi: 10.1073/pnas.132253599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chesi M, Robbiani DF, Sebag M, Chng WJ, Affer M, Tiedemann R, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008 Feb;13(2):167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T, et al. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia. 2011 Jun;25(6):1026–1035. doi: 10.1038/leu.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhan F, Hardin J, Kordsmeier B, Bumm K, Zheng M, Tian E, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002 Mar 1;99(5):1745–1757. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- 22.International Myeloma Working G. Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol. 2003 Jun;121(5):749–757. [PubMed] [Google Scholar]
- 23.Ahmann GJ, Jalal SM, Juneau AL, Christensen ER, Hanson CA, Dewald GW, et al. A novel three-color, clone-specific fluorescence in situ hybridization procedure for monoclonal gammopathies. Cancer Genet Cytogenet. 1998 Feb;101(1):7–11. doi: 10.1016/s0165-4608(97)00058-7. [DOI] [PubMed] [Google Scholar]
- 24.Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy Jr J. Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005 Mar 8;106(1):296–303. doi: 10.1182/blood-2005-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaughnessy JD, Jr., Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007 Mar 15;109(6):2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 26.Dispenzieri A, Kyle RA, Katzmann JA, Therneau TM, Larson D, Benson J, et al. Immunoglobulin free light chain ratio is an independent risk factor for progression of smoldering (asymptomatic) multiple myeloma. Blood. 2008 Jan 15;111(2):785–789. doi: 10.1182/blood-2007-08-108357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kyle RA, Durie BG, Rajkumar SV, Landgren O, Blade J, Merlini G, et al. Monoclonal gammopathy of undetermined significance (MGUS) and smoldering (asymptomatic) multiple myeloma: IMWG consensus perspectives risk factors for progression and guidelines for monitoring and management. Leukemia. 2010 Jun;24(6):1121–1127. doi: 10.1038/leu.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Persona E, Mateo G, Garcia-Sanz R, Mateos MV, de Las Heras N, de Coca AG, et al. Risk of progression in smouldering myeloma and monoclonal gammopathies of unknown significance: comparative analysis of the evolution of monoclonal component and multiparameter flow cytometry of bone marrow plasma cells. Br J Haematol. 2010 Jan;148(1):110–114. doi: 10.1111/j.1365-2141.2009.07929.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhan F, Barlogie B, Arzoumanian V, Huang Y, Williams DR, Hollmig K, et al. Gene599 expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007 Feb 15;109(4):1692–1700. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005 Sep 10;23(26):6333–6338. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 31.Chng WJ, Glebov O, Bergsagel PL, Kuehl WM. Genetic events in the pathogenesis of multiple myeloma. Best Pract Res Clin Haematol. 2007 Dec;20(4):571–596. doi: 10.1016/j.beha.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006 Sep 15;108(6):2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hose D, Reme T, Hielscher T, Moreaux J, Messner T, Seckinger A, et al. Proliferation is a central independent prognostic factor and target for personalized and risk613 adapted treatment in multiple myeloma. Haematologica. 2011 Jan;96(1):87–95. doi: 10.3324/haematol.2010.030296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013 Apr 11;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013 Oct 8; doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011 May 5;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013 Oct 29;110(44):17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011 Apr 15;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, Luquette LJ, Gehlenborg N, Xi R, Haseley PS, Hsieh CH, et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013 May 9;153(4):919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001 Sep 10;20(40):5611–5622. doi: 10.1038/sj.onc.1204641. [DOI] [PubMed] [Google Scholar]
- 41.Walker BA, Wardell CP, Johnson DC, Kaiser MF, Begum DB, Dahir NB, et al. Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood. 2013 Apr 25;121(17):3413–3419. doi: 10.1182/blood-2012-12-471888. [DOI] [PubMed] [Google Scholar]
- 42.Bergsagel PL, Chesi M, Nardini E, Brents LA, Kirby SL, Kuehl WM. Promiscuous translocations into immunoglobulin heavy chain switch regions in multiple myeloma. Proc Natl Acad Sci U S A. 1996 Nov 26;93(24):13931–13936. doi: 10.1073/pnas.93.24.13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gabrea A, Leif Bergsagel P, Michael Kuehl W. Distinguishing primary and secondary translocations in multiple myeloma. DNA Repair (Amst) 2006 Sep 8;5(9-10):1225–1233. doi: 10.1016/j.dnarep.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 44.Birshtein BK. The role of CTCF binding sites in the 3′ immunoglobulin heavy chain regulatory region. Front Genet. 2012;3:251. doi: 10.3389/fgene.2012.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volpi SA, Verma-Gaur J, Hassan R, Ju Z, Roa S, Chatterjee S, et al. Germline deletion of Igh 3′ regulatory region elements hs 5, 6, 7 (hs5-7) affects B cell-specific regulation, rearrangement, and insulation of the Igh locus. J Immunol. 2012 Mar 15;188(6):2556–2566. doi: 10.4049/jimmunol.1102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis JP, MacKenzie MR. Non-random chromosomal aberrations associated with multiple myeloma. Hematol Oncol. 1984 Oct-Dec;2(4):307–317. doi: 10.1002/hon.2900020402. [DOI] [PubMed] [Google Scholar]
- 47.Sawyer JR, Lukacs JL, Munshi N, Desikan KR, Singhal S, Mehta J, et al. Identification of new nonrandom translocations in multiple myeloma with multicolor spectral karyotyping. Blood. 1998 Dec 1;92(11):4269–4278. [PubMed] [Google Scholar]
- 48.Sawyer JR, Lukacs JL, Thomas EL, Swanson CM, Goosen LS, Sammartino G, et al. Multicolour spectral karyotyping identifies new translocations and a recurring pathway for chromosome loss in multiple myeloma. Br J Haematol. 2001 Jan;112(1):167–174. doi: 10.1046/j.1365-2141.2001.02546.x. [DOI] [PubMed] [Google Scholar]
- 49.Smadja NV, Fruchart C, Isnard F, Louvet C, Dutel JL, Cheron N, et al. Chromosomal analysis in multiple myeloma: cytogenetic evidence of two different diseases. Leukemia. 1998 Jun;12(6):960–969. doi: 10.1038/sj.leu.2401041. [DOI] [PubMed] [Google Scholar]
- 50.Boyd KD, Ross FM, Walker BA, Wardell CP, Tapper WJ, Chiecchio L, et al. Mapping of chromosome 1p deletions in myeloma identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being genes in regions associated with adverse survival. Clin Cancer Res. 2011 Dec 15;17(24):7776–7784. doi: 10.1158/1078-0432.CCR-11-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011 Sep 16;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holien T, Vatsveen TK, Hella H, Waage A, Sundan A. Addiction to c-Myc in multiple myeloma. Blood. 2012;120(12):2450–2453. doi: 10.1182/blood-2011-08-371567. [DOI] [PubMed] [Google Scholar]
- 53.Kuehl WM, Bergsagel PL. MYC addiction: a potential therapeutic target in MM. Blood. 2012 Sep 20;120(12):2351–2352. doi: 10.1182/blood-2012-08-445262. [DOI] [PubMed] [Google Scholar]
- 54.Chiecchio L, Dagrada GP, Protheroe RK, Stockley DM, Smith AG, Orchard KH, et al. Loss of 1p and rearrangement of MYC are associated with progression of smouldering myeloma to myeloma: sequential analysis of a single case. Haematologica. 2009 Jul;94(7):1024–1028. doi: 10.3324/haematol.2008.004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood. 2007 Apr 15;109(8):3489–3495. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.