Abstract
The eukaryotic origin recognition complex (ORC) is made up of six subunits and functions in nuclear DNA replication, chromatin structure, and gene silencing in both fungi and metazoans. We demonstrate that disruption of a plant ORC subunit homolog, AtORC2 of Arabidopsis (Arabidopsis thaliana), causes a zygotic lethal mutant phenotype (orc2). Seeds of orc2 abort early, typically producing embryos with up to eight cells. Nuclear division in the endosperm is arrested at an earlier developmental stage: only approximately four nuclei are detected in orc2 endosperm. The endosperm nuclei in orc2 are dramatically enlarged, a phenotype that is most similar to class B titan mutants, which include mutants in structural maintenance of chromosomes (SMC) cohesins. The highest levels of ORC2 gene expression were found in preglobular embryos, coinciding with the stage at which homozygous orc2 mutant seeds arrest. The homologs of the other five Arabidopsis ORC subunits are also expressed at this developmental stage. The orc2 mutant phenotype is partly suppressed by a mutation in the Polycomb group gene MEDEA. In double mutants between orc2 and medea (mea), orc2 homozygotes arrest later with a phenotype intermediate between those of mea and orc2 single mutants. Either alterations in chromatin structure or the release of cell cycle checkpoints by the mea mutation may allow more cell and nuclear divisions to occur in orc2 homozygous seeds.
INTRODUCTION
The origin recognition complex (ORC) is involved in nuclear DNA replication, chromatin structure, and transcriptional silencing (reviewed in Bell, 2002). The six-subunit complex was first isolated and defined by its binding to origins of replication in Saccharomyces cerevisiae. The ORC initially recruits CDC6 to the origin of replication. Thereafter, a series of other proteins, CDT1 and the minichromosome maintenance (MCM) helicase complex, are added to form the complete prereplication complex. The replication machinery attaches to this complex at the onset of S-phase (reviewed in Bell and Dutta, 2002; de Pamphilis, 2003).
Mutations in ORC genes typically result in cells arresting at the G1/S transition of the cell cycle. But in yeast (Bell et al., 1993) and Drosophila melanogaster (Loupart et al., 2000; Pflumm and Botchan, 2001), many cells can progress through S-phase to mitosis (M-phase), where they arrest and die. M-phase–arrested cells of D. melanogaster have severe chromosomal abnormalities that are probably caused by incomplete or defective replication. The effects of lack of ORC complex subunits can differ across chromosomal regions, with euchromatic regions replicating late, after heterochromatin instead of before, and being more severely damaged. In this light, it seems significant that DmOrc2 is enriched in heterochromatin and is involved in localization of heterochromatin protein 1 (HP1) to heterochromatin (Huang et al., 1998). McNairn and Gilbert (2003) recently reviewed evidence that certain chromatin remodeling proteins localize specifically to sites of late DNA replication. The modifications of histones that mark DNA for late replication are targeted to nucleosomes close to late-replicating origins. This targeting may be achieved by binding of the histone modifying enzymes to ORC1 and MCM2 at such origins. The modification of the histones on newly replicated DNA is hypothesized to ensure the inheritance of origin replication timing. The M-phase arrest and cell death characteristic of ORC mutants also may be connected to another emerging function of the ORC, in chromosome stability. ORC3 and ORC5 were identified recently as chromosome integrity determinants in a screen for mutants with elevated rates of gross chromosomal rearrangements (Huang and Koshland, 2003). The interplay between ORC and chromatin structure, chromosome integrity, and cell cycle dynamics is still poorly understood but of considerable interest and is also relevant to the third known function of the ORC in gene regulation.
In addition to the prereplication complex, the ORC is also a central constituent of heterochromatin-mediated silencing complexes. Haploid yeast cells express only one of two possible mating types, and genes for the opposite mating type are maintained as transcriptionally silent loci called HMR and HML. Silencer sites flanking these loci contain autonomous replication consensus sites. The ORC binds to the autonomous replication consensus site elements and recruits the first member, SIR1, of the silencing protein complex that then modifies chromatin structure at the HM loci (Fox et al., 1997). The ORC plays a similar role in D. melanogaster, where it binds HP1, which is essential for heterochromatic silencing. Accordingly, localization of HP1 to heterochromatin is disrupted in DmOrc2 mutants (Pak et al., 1997; Huang et al., 1998). Furthermore, flies heterozygous for mutations in DmOrc2 show suppression of position effect variegation, a heterochromatin-mediated silencing phenomenon (Pak et al., 1997).
Individual ORC subunits can have additional functions. D. melanogaster ORC3 is postulated to be involved in transport at neuromuscular junctions (Rohrbough et al., 1999), and additional roles for HsORC6 in cytokinesis and chromosome segregation were reported recently (Prasanth et al., 2002). In humans, the six ORC subunits are differentially expressed in different tissues and were shown to be present in complexes with proteins other than ORC subunits (Thome et al., 2000). As investigations continue, it is likely that the ORC, or individual ORC subunits, will be shown to be in more protein complexes and to have more functions in the cell.
Surprisingly little is known about the role of ORC in the plant kingdom, despite AtORC2 being among the earliest ORC subunits cloned (Gavin et al., 1995). More recently, cloning of ORC1 of rice (Oryza sativa) was reported together with its upregulation after starved cultured cells were supplied with sucrose (Kimura et al., 2000). Transcript levels of Arabidopsis (Arabidopsis thaliana) ORC1 were high in plants overexpressing the E2F-DP transcription factor (De Veylder et al., 2002). These results are consistent with ORC1 transcription being upregulated in proliferating cells via E2F-DP similarly to the regulation of ORC1 in metazoans (Ohtani et al., 1996). Homologs of all six ORC subunits from Arabidopsis and rice are reported in the databases, and cloning of the maize (Zea mays) cDNAs of ORC1-5 has been reported recently (Witmer et al., 2003). Witmer et al. (2003) postulated that Arabidopsis ORC may lack an ORC4 subunit because only a short polypeptide with homology to the extreme C terminus of other ORC4 proteins is encoded in the Arabidopsis genome.
Here, we demonstrate that a Dissociation (Ds) transposon insertional mutant of ORC2 exon 6 causes a zygotic-lethal mutant phenotype (orc2) in which embryos and endosperm of mutant seeds abort at an early developmental stage. The endosperm nuclei of orc2 mutants are dramatically enlarged, similar to those of titan mutants. The orc2 phenotype could be partially suppressed by mutation of the MEDEA gene, an imprinted gene that acts to suppress proliferation in the embryo and endosperm of developing seeds. MEDEA belongs to the Polycomb group of proteins that are involved in chromatin remodeling and epigenetic regulation of gene expression. We also show that AtORC4 encodes an mRNA for a longer protein that comprises all conserved ORC4 domains. Thus, in contrast with Witmer et al. (2003), we demonstrate that genes encoding homologous proteins to all six ORC subunits exist in Arabidopsis and that all of them are transcribed during early seed development and/or flower development.
RESULTS
Isolation and Complementation of the orc2 Mutant
The Arabidopsis orc2 mutant described here was identified in a systematic screen for mutants affecting megagametogenesis (Moore et al., 1997; Page and Grossniklaus, 2001). The mutagenesis screen was performed on a collection of insertional mutants created using the Cold Spring Harbor Laboratory system of enhancer trap and gene trap transposable elements (Sundaresan et al., 1995). These transposable elements are derived from the maize Ds transposons and carry the NPT2 gene for kanamycin resistance. Mutant lines were screened for reduced fertility and then (using the kanamycin resistance marker) for non-Mendelian segregation indicative of gametophytic mutations. Enhancer trap line ET3723 had reduced fertility, but the 2:1 segregation of kanamycin resistance to sensitivity was consistent with the segregation of a recessive lethal mutation.
Analysis of developing seeds in the ET3723 line showed that ∼25% aborted, also consistent with a zygotic lethal phenotype (Figure 1A, Table 1). Most aborting seeds arrested at an early developmental stage and subsequently turned brown and shriveled. Some mutant seeds, ∼10%, were white and translucent, rather like seeds of titan (ttn) mutants (Liu and Meinke, 1998), when compared with their green phenotypically normal siblings. But these seed also turned brown and shriveled before the phenotypically normal seeds reached their full size.
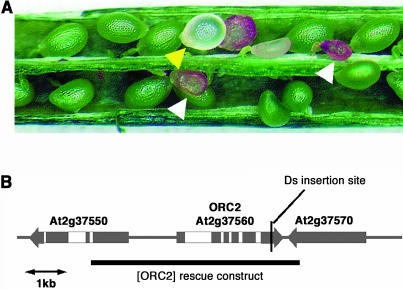
Figure 1.
The orc2 Mutant.
(A) Developing seeds of orc2/+ plants showing the typical seed abortion phenotype (white arrowheads) and the less common translucent phenotype (yellow arrowhead).
(B) Predicted gene structure of AtORC2 and flanking genes. The insertion site of the Ds transposable element in the orc2 mutant allele is indicated. The genomic fragment used to complement the orc2 mutant is shown by a black bar.
Table 1.
Rescue of the orc2 Phenotype in ORC2 Transformants of ET3723
| Seed Phenotype
|
|||
|---|---|---|---|
| Plant | Percent Normal | Percent Aborted | Total Seeds |
| Ler | 100 | 0 | 800 |
| ET3723 | 78 | 22 | 785 |
| MC5.4 | 96 | 4 | 313 |
| MC6.1 | 91 | 9 | 421 |
| MC6.4 | 94 | 6 | 191 |
| MC6.5 | 98 | 2 | 135 |
| MC6.3 | 76 | 24 | 258 |
In F1 seeds derived from reciprocal crosses with the wild-type accession Landsberg erecta (Ler), the 25% seed abortion phenotype was not observed. Kanamycin resistance strictly segregated in all cases with the seed abortion phenotype, and DNA gel blot analysis using a probe specific for the Ds transposon showed that there was only one Ds element present in the line (data not shown). PCR analyses showed that all kanamycin-resistant progeny of ET3723 were hemizygous for the Ds element. We performed thermal asymmetric interlaced PCR (Liu et al., 1995) to determine the location of the Ds element in line ET3723 and found that it was in the coding region of At2g37560 (Figure 1B). This gene was cloned and named AtOrc2 by Gavin et al. (1995) and encodes a protein that is similar to ORC2 from other organisms.
To confirm that the insertion of the Ds element in the AtORC2 coding region caused the zygotic lethal phenotype of line ET3723, we introduced a genomic fragment spanning from 2638 bp upstream to 400 bp downstream of the AtORC2 open reading frame (ORF) (Figure 1B) into the ET3723 mutant background. In primary transformants of ET3723 carrying the rescue construct at a single locus that is unlinked to ORC2, we expected the abortion frequency in self-pollinated siliques to be 6.25% (cf. 25% in the parental line). Table 1 shows the seed abortion ratios in five independent transformants (prefix MC) together with Ler and ET3723. The abortion ratio in MC5.4, MC6.1, MC6.4, and MC6.5 was significantly reduced from 25% and close to the expected 6.25%, which shows that the AtORC2 gene complementation construct could rescue the seed abortion phenotype. We found T2 individuals from three transformants whose progeny were all kanamycin resistant (n > 200), showing that they were homozygous for the Ds insertion in AtORC2. This established that the AtORC2 complementation construct was sufficient in trans to fully rescue mutant embryos.
Because the mutant phenotype was caused by the Ds insertion in the AtORC2 gene, we refer to the ET3723 insertion line from now on as the orc2 mutant.
orc2 Embryos Die at the Early Globular Stage
The seed abortion phenotype was further analyzed by microscopic observation of semithin sections and cleared, whole-mounted siliques at stages before aborting seeds turned brown (i.e., up to late heart stage embryo in normal seeds) (Figure 2). The seed abortion phenotypes observed in cleared siliques of Ler and orc2 are summarized in Table 2, and examples are shown in Figures 2F to 2H. The early aborted seeds in Ler were very small but had the distinct shape of fertilized ovules (data not shown) and were similar in appearance to the early class in orc2. Therefore, this phenotype is most likely caused by environmental factors and is not specifically an orc2 mutant phenotype. The homozygous orc2 mutant seeds are disrupted in both embryo and endosperm development. Embryos mostly arrested around the four-cell stage and did not have the normal smooth shape with regular cell division planes. Instead, the cells had enlarged vacuoles and bulged (Figures 2D and 2F) in a manner reminiscent of the (much later arresting) raspberry phenotype (Yadegari et al., 1994) and of ttn4 (Tzafrir et al., 2002). Abnormalities were frequently seen in the suspensor in older siliques, but this is a common secondary effect of arrested embryo development (Yadegari et al., 1994).
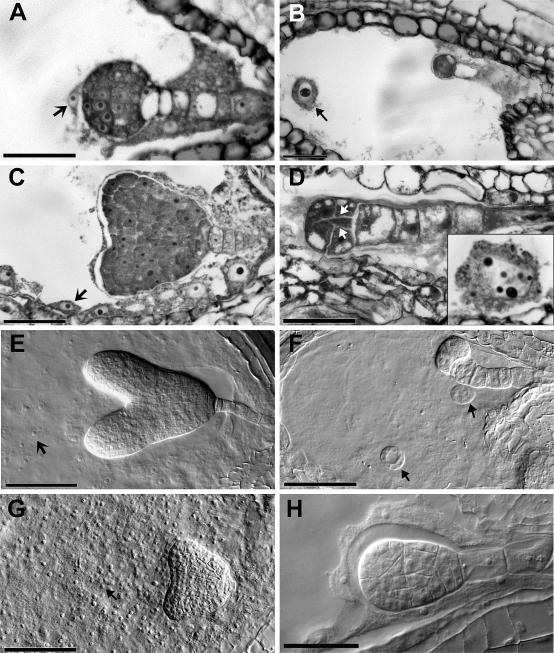
Figure 2.
Microscopic Analysis of the orc2 Zygotic Lethal Phenotype.
(A) to (D) Thin-sectioned seeds of orc2/+ plants stained with toluidine blue. Wild-type ([A] and [C]) and mutant ([B] and [D]) seeds from the same siliques. In (B), the arrow points to an enlarged endosperm nucleus. In (D), the arrows show aberrant cell divisions, and the inset shows an enlarged endosperm nucleus from the same seed.
(E) to (H) Wild-type (E) and mutant ([F] to [H]) seeds that were cleared, whole mounted, and viewed with Nomarski optics.
(F) Typical orc2 phenotype in a seed from the same silique as (E). Arrows point to enlarged endosperm nuclei.
(G) Normal embryo but abnormal endosperm (arrow).
(H) Rare almond-shaped embryo.
Arrows in (A), (C), and (E) point to normal endosperm nuclei. Bar = 10 μM.
Table 2.
Frequencies of Aborted Seed Phenotypes
| Percent
|
||
|---|---|---|
| Aborted Seed Phenotype | Ler | ET3723 |
| Early | 3.5 | 1.6 |
| Two-cell embryo; 0 to 4 endosperm nuclei | 0 | 5.5 |
| Embryo has 4 nuclei and 1 to 4 cells; 0 to 4 endosperm nuclei | 0 | 9.4 |
| Embryo has 4 or more nuclei with bulging cells; 0 to 4 endosperm nuclei | 0 | 5.7 |
| Embryo normal; endosperm abnormal | 0 | 2.0 |
| Almond-shaped globular embryo; endosperm normal | 0 | 0.2 |
| Seed abortion total | 3.5 (n = 417) | 24.4 (n = 459) |
Embryos arrested at the two-cell stage clearly had two cells, and the division was mostly symmetrical, although there were exceptions. In the cleared, whole-mounted seeds, it was not always possible to determine whether there were four cells in embryos with four nuclei because one longitudinal division plane cannot be seen. However, abnormal division planes were apparent at this stage and were clearly seen in the semithin sections (Figure 2D). In embryos with more than four nuclei, more than four cells also could be seen. Thus, it seems that cell divisions become progressively irregular in orc2 embryos, but that they do take place. We did not observe any multinucleate cells, except at late stages in the suspensor.
A small class of seeds had embryos that looked normal but in which the endosperm was degenerate (Figure 2G). The almond class, with its abnormally shaped but smooth-outlined embryo, is rare (Figure 2H) but was observed in several experiments.
In Arabidopsis, the fertilized central cell forms the endosperm. The primary endosperm nuclei divide first without cytokinesis, forming a syncytium of regularly sized nuclei that become cellularized at the heart stage of embryo development (reviewed in Berger, 2003). Endosperm development in orc2 mutant seeds was also arrested very early; mostly only two to four nuclei were detected. In contrast with the embryo nuclei that looked normal, the endosperm nuclei were dramatically enlarged, and nucleoli were often disc shaped or pitted (Figures 2B, 2D, and 2F). This feature in particular makes the orc2 phenotype very similar to the class B ttn mutants (Liu and Meinke, 1998; Tzafrir et al., 2002). The differences are that class B ttn embryos mostly arrest slightly earlier, at the two- to four-cell stage, and with a regular shape; more endosperm nuclei are observed; and mutant seeds reach normal sizes but are white and translucent.
AtORC2 Expression
ORC subunits are required for DNA replication and cell cycle progression, but unlike other prereplication complex components, CDC6, and the MCM proteins, ORC2 levels are not upregulated in proliferating cells in other eukaryotes (DePamphilis, 2003). Consistently, there was no evidence for increased AtORC2 expression in seedlings or meristems. In reverse transcriptase (RT)-PCR experiments with Ler tissue, a specific AtORC2 transcript was amplified from seedlings, leaves, and flowers (Figure 3A).
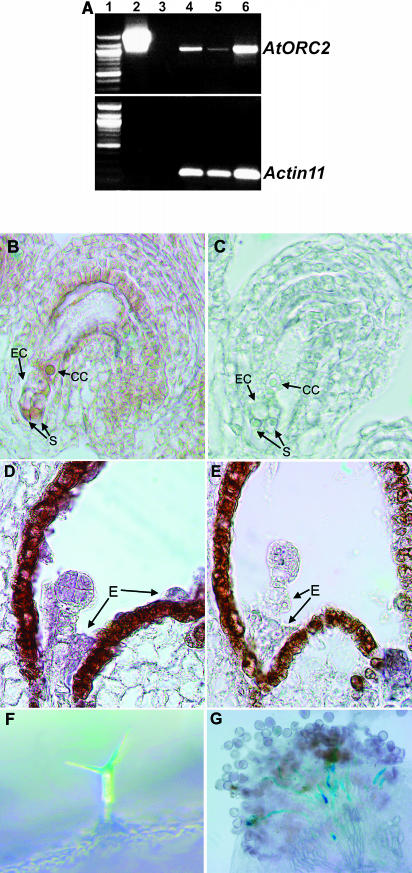
Figure 3.
Analysis of AtORC2 Expression.
(A) Assay for AtORC2 and actin11 transcripts by RT-PCR: lane 1, DNA size markers; lane 2, large PCR product from plasmid p1300-ORC2 that contains a genomic fragment spanning the complete AtORC2 gene; lane 3, control (no CDNA); lanes 4 to 6, PCR products from cDNA of seedlings (lane 4), mature leaves (lane 5), and inflorescences (lane 6).
(B) to (E) In situ hybridization experiments. CC, central cell; E, endosperm; EC, egg cell; S, synergid.
(B) Female gametophyte, antisense probe.
(C) Female gametophyte, sense probe.
(D) Preglobular embryo, antisense probe.
(E) Preglobular embryo, sense probe.
(F) and (G) GUS activity assays on tissue from orc2/+ plants.
(F) GUS activity in the trichome.
(G) GUS activity in pollen tubes from an orc2/+ plant on a wild-type stigma.
To help determine in which cells or tissues AtORC2 expression is essential to embryo and endosperm development, we performed in situ hybridization experiments on inflorescences. AtORC2 transcript was detected in the mature female gametophyte, in the synergids and central cell (Figures 3B and 3C). The strongest hybridization signals were detected in embryos (but not the suspensor) and endosperm of preglobular stage seeds (Figures 3D and 3E). This corresponds with the developmental stage when orc2 homozygous embryos arrest. By early heart stage, there was no observable difference between signal from sense and antisense probes (data not shown).
The in situ hybridization experiments revealed relative levels of spatial and temporal expression specifically during female gametophyte and seed development. We also wished to investigate whether there is any expression in other tissues and stages of development, especially in pollen, where we did not obtain conclusive results by in situ hybridization experiments. We addressed this question using the enhancer trap of the Ds transposable element causing the mutation. The enhancer trap consists of the β-glucuronidase (GUS) gene preceded by a minimal 35S promoter of Cauliflower mosaic virus (Sundaresan et al., 1995). GUS activity was detected in mature female gametophytes and in preglobular stage embryo and endosperm. GUS staining was particularly strong in some aborted seeds presumably because they arrested at a stage when the AtORC2 gene was active. We also detected weak GUS activity in trichomes (Figure 3F) and the mesophyll of leaves. These patterns were fully consistent with our RT-PCR and in situ hybridization data.
GUS activity was observed in both developing and mature pollen as well as in pollen tubes (Figure 3G). No staining was detectable in pollen tubes as they grew through the transmitting tract and along the funiculus into the micropyle, but intense staining was observed in penetrated synergids, presumably from GUS protein released into the synergid when the pollen tube opened to release the gametes (Huck et al., 2003). Further evidence that AtORC2 is expressed in pollen could be gleaned from microarray data released on the Nottingham Arabidopsis Stock Centre Web site (http://ssbdjc2.nottingham.ac.uk/narrays/static.pl?page=psp-wbubn.html). Transcript levels from AtORC2 varied little among all of the experiments except for those on pollen, where they were substantially greater, being highest in developing, mononucleate, and binucleate pollen.
We could not detect GUS activity in seedlings, even after incubation for 6 d. Therefore, we must conclude that the enhancer trap pattern reflects many but not all aspects of AtORC2 expression.
Expression of Other ORC Subunit Genes
To determine whether the zygotic lethality of orc2 might be related to the function of the complete ORC, we investigated whether the other ORC subunits are also expressed before and after fertilization. To do this, we accessed results of hybridization experiments to the Affymetrix ATH1 GeneChip with RNA from flowers before and after fertilization and in siliques containing preglobular-stage embryos. In agreement with the in situ hybridization results, AtORC2 is indeed expressed before fertilization, and transcript levels increase after fertilization (Table 3).
Table 3.
Expression of Genes Encoding ORC Subunit Homologs in Flowers
| Transcript Signala
|
||||
|---|---|---|---|---|
| Gene | Gene ID | Before Fertilization | After Fertilization | Preglobular Embryo |
| AtORC1 | At4g12620 | 354.9 | 425.0* | 425.0* |
| AtORC2 | At2g37560 | 417.6 | 515.4 | 724.2** |
| AtORC3 | At5g16690 | (204.1)b | (169.3)b | 240.0 |
| AtORC4 | At2g01120 | 517.8 | 561.8 | 602.1 |
| AtORC5 | At4g29910 | 673.6 | 640.9 | 715.4 |
| AtORC6 | At1g26840 | 452.7 | 733.9* | 343.9 |
Transcript signals are mean of two independent experiments.
Signal values in parentheses are classified as having no transcript present by the Affymetrix MAS5.0 software in one or both experiments.
The asterisks denote signals significantly higher than that before fertilization in one (*) or both (**) experiments.
Predicted genes encoding proteins with high similarity to metazoan ORC1 (At4g12620 and At4g14700), ORC3 (At5g16690), ORC4 (At2g01120), ORC5 (At4g29910), and ORC6 (At1g26840) are present in the Arabidopsis genome. All of these genes were expressed in flowers at all three stages except the AtORC3 homolog, which was only detectably expressed after fertilization. No ESTs or cDNAs exist for this gene, and we performed RT-PCR experiments to confirm that it is indeed expressed. We could not detect expression in seedlings and leaves, but a weak signal was obtained with inflorescence RNA (data not shown), confirming the microarray data.
The article by Witmer et al. (2003) noted that gene At2g01120 is currently annotated as only a peptide with homology to the C termini of ORC4s. Because the ORC subunits 1 to 6 are conserved across all known eukaryotes, an Arabidopsis ORC lacking a functional ORC4 would be an evolutionary anomaly. We therefore set out to determine whether the At2g01120 gene might be mispredicted by the genome annotation software. Performing t-BLASTX on the Arabidopsis genome using the maize ORC4 protein sequence as the query, we detected regions upstream of the predicted ORF of At2g01120 that encoded all conserved regions of ORC4 proteins. RT-PCR using primers spanning these regions indicated that they were expressed. We used 5′ rapid amplification of cDNA ends (RACE) to redefine the transcribed sequence of At2g01120 and entered it in the database as AtORC4. The AtORC4 gene and a comparison of the predicted protein sequence with ORC4 proteins are presented in the supplemental data online.
In summary, genes that encode full-length homologs of all six ORC subunits are present in the Arabidopsis genome, and all six genes are expressed at the preglobular embryo stage when orc2 mutant seeds arrest.
The medea Mutation Can Partially Rescue the orc2 Phenotype
To further elucidate the role of ORC2 in Arabidopsis during embryo and endosperm development, we investigated any possible genetic interactions between the orc2 and medea mutations. MEDEA (MEA) is a maternally expressed, imprinted gene that encodes a Polycomb group protein and has an essential function in the regulation of cell proliferation during seed development (Grossniklaus et al., 1998). Mutations in MEA cause seed abortion only when the mutant allele (meaM) originates from the mother. This gametophytic, maternal-effect, embryo lethality is completely independent of the paternal MEAP allele because imprinting results in a functionally silent MEAP allele.
In self-pollinated plants that are heterozygous for mea, 50% of seeds abort. The aborted seeds maternally inherit the mutant mea allele (i.e., meaM genotype). The typical meaM arrest phenotype is shown in Figure 4B. The meaM embryos develop slowly and overproliferate, typically arresting as large heart-stage embryos. The endosperm does not cellularize and coagulates into cysts before degenerating. Because the mea mutation causes overproliferation, we investigated whether the timing of embryo and endosperm arrest in orc2 mutants would alter in a meaM background. Conversely, because ORC2 is involved in silencing in S. cerevisiae and D. melanogaster, we looked for any modification of the mea-imprinted phenotype by orc2.
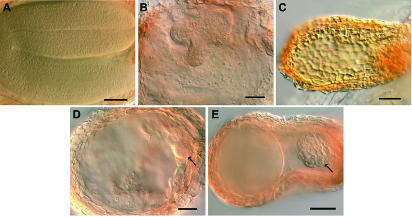
Figure 4.
Phenotypes of Developing Seeds in orc2/+; mea/+ Plants.
Cleared, whole-mounted seeds from siliques at the same developmental stage.
(A) The wild type.
(B) meaM phenotype with large heart-stage embryo and aggregated endosperm.
(C) Typical orc2/orc2 phenotype.
(D) meaM-like phenotype but with no large embryo; arrow points to small orc2/orc2-like embryo.
(E) orc2/orc2-like phenotype but bigger seed and more cells in embryo than ever seen in orc2/+ plants.
Bar = 10 μM.
We analyzed the frequencies of orc2 and mea phenotypes in the F2 progeny of a cross between orc2/+ and mea/+ plants (Table 4). The seed abortion phenotypes of orc2 and mea are readily distinguishable. As seen in Figures 1 and 4B, orc2 homozygous seeds turn brown and shriveled while they are still very small. By contrast, seeds carrying the meaM allele grow to full size but then turn yellowish-white and flaccid just before the point when their phenotypically normal siblings begin to desiccate (Grossniklaus et al., 1998). If the orc2 seed abortion phenotype was not affected by mea, we expected orc2 homozygous F2 seeds to arrest at an early stage before the meaM phenotype was apparent. If orc2 had any effect on meaM seed abortion, it could be seen in the remaining heterozygous siblings. MEA and ORC2 are unlinked genes on different chromosomes. Therefore, if there was no interaction between MEA and ORC2, the selfed seed progeny from a mea/MEA; orc2/ORC2 double heterozygote should segregate as follows: 37.5% normal, 25% small aborting, and 37.5% large aborting seeds (Tables 4 and 5, expected 1). However, what we observed was 43.0% large aborting seeds and only 18.1% small aborting seeds (Table 5, observed). Thus, our results indicate that orc2 has no effect on the meaM seed abortion ratio (i.e., on imprinting at the MEAP locus), but mea suppressed the orc2 small seed abortion phenotype, with approximately one-quarter of orc2 homozygotes aborting as large, rather than small, seeds (Table 5, expected 2). These ratios would be consistent with mea/mea suppressing the orc2 phenotype (Table 4), but because the MEAP allele is inactive in early seed development, an incompletely penetrant effect of the meaM allele is a more likely explanation.
Table 4.
Segregation of mea and orc2 and Predicted F2 Seed Phenotypes after a mea/+, orc2/+ Cross
| Paternal Gametes
|
||||
|---|---|---|---|---|
| Female Gametes | meaP orc2 | meaP ORC2 | MEAP orc2 | MEAP ORC2 |
| meaM orc2 | meaM/meaPorc2/orc2 | meaM/meaP*orc2/ORC2 | meaM/MEAPorc2/orc2 | meaM/MEAP*orc2/ORC2 |
| meaM ORC2 | meaM/meaP* ORC2/orc2 | meaM/meaP*ORC2/ORC2 | meaM/MEAP*ORC2/orc2 | meaM/MEAP*ORC2/ORC2 |
| MEAM orc2 | MEAM/meaPorc2/orc2 | MEAM/meaPorc2/ORC2 | MEAM/MEAPorc2/orc2 | MEAM/MEAPorc2/ORC2 |
| MEAM ORC2 | MEAM/meaPORC2/orc2 | MEAM/meaPORC2/ORC2 | MEAM/MEAPORC2/orc2 | MEAM/MEAPORC2/ORC2 |
The orc2 homozygotes, early abortion phenotype, are shown in bold, and the orc2 heterozygotes (meaM leads to late abortion) are marked by asterisks.
Table 5.
Predicted and Observed Seed Phenotypes in the F2 Generation of a mea/+, orc2/+ Cross
| Seed Phenotype
|
|||||
|---|---|---|---|---|---|
| Normal | Small Aborted | Large Aborted | Total | χ2 Testa | |
| Observed | 1273 (38.8%) | 595 (18.1%) | 1411 (43%) | 3279 | |
| Expected 1 | 1229 (37.5%) | 820 (25%) | 1229 (37.5%) | 3279 | χ2 = 90.26, df = 2, P < 0.0005 |
| Expected 2 | 1229 (37.5%) | 614 (18.75%) | 1434 (43.75%) | 3279 | χ2 = 2.5, df = 2, P > 0.25 |
df, degrees of freedom.
To test these alternatives, we performed embryo rescue (Vielle-Calzada et al., 1999) and obtained one mea/mea; orc2/+ plant. If orc2 homozygotes have a large seed aborting phenotype when combined with homozygous mea, then all orc2 homozygous seeds should be late aborting. However, we counted 18.5% small aborting seeds in this mea/mea; orc2/+ plant, similar to the number obtained in mea/+; orc2/+ plants. Some seeds from mea homozygous plants are viable, so we were able to test the progeny of this individual plant. Again, in the next generation, the siliques of mea/mea; orc2/+ individuals contained ∼18.5% small aborting seeds. This shows that it was not the mea homozygous condition that altered the orc2/orc2 phenotype, consistent with MEAP being inactive and thus having no effect. Because we did not see a difference in the proportion of small aborting seeds in mea/+ and mea/mea backgrounds, we propose that the phenomenon can only be explained by mea regulating one or more other loci (i.e., neither MEA nor ORC2).
Our observations of silique contents implied that some orc2 homozygotes now had a mea seed abortion phenotype. To test this, we cleared developing siliques from F2 double heterozygotes and from their mea/+ and orc2/+ siblings. The phenotypes of the developing seeds were analyzed and their frequencies compared. The results are shown in Table 6, and examples of phenotypes are shown in Figure 4. Similar to the data shown in Table 5, there was a significant reduction in the number of small aborting seeds in mea orc2 double mutants compared with orc2/+ plants. However, there was no concomitant rise in the number of seeds with the typical mea phenotype. Instead, a new class of intermediate phenotypes was seen, which were presumably orc2 homozygotes that were partially rescued by mea (Table 6, rescued orc2; Figures 4D and 4E). In all of these intermediate seeds, the embryos were larger and contained more cells than orc2/orc2 embryos (Figure 4C) but were much smaller than mea embryos (Figure 4B). The endosperm of these seeds fell into two general types. The first was mea-like, with many relatively normal-sized nuclei aggregated into cysts (Figure 4D). The second class was orc2-like in shape and with little or no detectable endosperm (Figure 4E).
Table 6.
Seed Phenotypes in the Progeny of a mea/+, orc2/+ Cross
| Genotype | Normal | orc2 | mea | Rescued orc2 | Unclassifiable | Total |
|---|---|---|---|---|---|---|
| mea/+ | 51.8% | 0.0% | 44.7% | 0.0% | 3.5% | 114 |
| orc2/+ | 74.1% | 25.9% | 0.0% | 0.0% | 0.0% | 116 |
| mea/+; orc2/+ | 36.5% | 19.5% | 33.7% | 7.1% | 3.2% | 282 |
DISCUSSION
The orc2 mutant described here is the first reported mutation in a plant ORC subunit. Our results show that AtORC2 is an essential gene in plant cells, as it is in fungi and animals. We also show that the Arabidopsis genome includes complete homologs for all six ORC subunits and that these are all expressed in flowers after fertilization when embryos are at the preglobular stage (Table 3). All six ORC homologs also are found in rice and probably in maize (from which ORC1 to ORC5 have been cloned; Witmer et al., 2003), indicating that the ORC complex and potentially its functions have been retained over long periods of evolutionary time. It is conceivable that the functional defect causing lethality in plant orc2 homozygotes could be in a process that involves an ORC, such as that characterized in yeast, D. melanogaster, and human cells (Bell, 2002).
Seeds homozygous for orc2 arrest and abort early in their development. Embryos arrest with two to eight cells and become progressively irregular in shape. Only approximately four nuclei are detectable in the endosperm, and these are dramatically enlarged. Transcript levels are below the level of detection by in situ hybridization before formation of the mature female gametophyte, and they reach maximum levels just before the stage at which homozygous mutant seeds arrest development (Figure 3). AtORC2 is actively expressed in pollen, but its expression is not required for male gametogenesis because pollen carrying the orc2 allele (monitored by GUS activity) is capable of fertilization (Figure 3). Reciprocal outcrosses of orc2 heterozygotes with the wild type resulted in F1 progeny with 100% normal seeds, and transmission frequencies of the orc2 allele are normal (data not shown). This data suggests that there are no substantial maternal reserves of ORC2 deposited in the egg cell or central cell, and that postfertilization expression of AtORC2 is essential to both embryo and endosperm development.
Possible Roles of the Plant ORC in Cell Cycle and Chromatin Dynamics
In S. cerevisiae and D. melanogaster, the phenotypes of orc mutants appear to be connected with the roles of ORC in chromosome structure as well as in DNA replication (Bell et al., 1993; Dillin and Rine, 1998; Loupart et al., 2000; Pflumm and Botchan, 2001). Our results indicate that this is the case in the plant kingdom as well.
Like the plant orc2 mutant described here, D. melanogaster mutations of Orc2, Orc3, and Orc5 also cause zygotic lethality but at a late stage (at the third instar larval/pupal boundary), and imaginal discs are lacking (Landis et al., 1997; Pinto et al., 1999; Pflumm and Botchan, 2001). Zygotic lethality at this stage of D. melanogaster development is a common feature of mutations affecting genes regulating the diploid cell cycle (Gatti and Baker, 1989) and reflects the developmental features of holometabolous insects such as D. melanogaster. Larval cells stop dividing early in embryogenesis but continue to grow and become polytenized. The larval cell divisions are thought to be driven by the large maternal stores of proteins, which exist also for ORC subunits (Gossen et al., 1995; Chesnokov et al., 1999). Diploid imaginal cells do rely on zygotic transcription of cell cycle genes because they divide later, during the larval instars. But defects in the cell cycle and, in turn, imaginal disc formation are manifested only at the early pupal stage when these cells normally differentiate.
In arresting D. melanogaster orc2 (as well as orc3 and orc5) mutants, diploid cells conduct little DNA synthesis, and as expected for a prereplication complex protein, most arrest in G1. But there is also a high mitotic index because some cells continue to a second arrest point in mitosis where they die with abnormally condensed chromatin and severe chromosome defects (Loupart et al., 2000; Pflumm and Botchan, 2001). Similarly, in S. cerevisiae, orc2 and orc5 mutants arrest at the G1/S boundary, but, with prolonged incubation at the restrictive temperature, cells completed S-phase and lost viability (Bell et al., 1993; Foss et al., 1993). Later, an additional role for ORC in M-phase was detected by Dillin and Rine (1998). Indeed, evidence is now increasing that tight links exist between DNA replication, heterochromatin formation, and chromatin condensation, and the members of the prereplication complex, including ORC, are involved in this connection (Christensen and Tye, 2003).
As noted above, lethality at the larval/pupal boundary with missing imaginal discs is symptomatic of genes required for the cell cycle in D. melanogaster. No such synchronicity or close similarity of phenotypes is observed in the arrest stage of mutants affecting cell cycle genes in plants. This could reflect differing levels of reserves and stability of the proteins as well as the differential requirements for the different cell cycle genes in the gametophyte, embryo, and endosperm (Nacry et al., 2000; Weijers et al., 2001; Sørensen et al., 2002). Even the similarity seen in the arrest phenotypes among prereplication complex mutants in D. melanogaster is not seen in Arabidopsis. The prereplication complex consists of ORC, CDC6, CDT1, and the MCM complex. Diploid cells in D. melanogaster orc2, orc5, and mcm4 mutants all arrest with condensation defects (Pflumm and Botchan, 2001), and RNA interferance depletion of Mcm10, Cdc45, Mcm2, Mcm5, and Orc2 in cell cultures results in aberrant chromosome condensation (Christensen and Tye, 2003). In Arabidopsis, a mutation of the MCM7 homolog prolifera has a gametophytic effect, resulting in diverse arrest phenotypes from female gametophyte to late embryo abortion (Springer et al., 2000). Enlarged endosperm nuclei were observed in mutant prolifera seeds, seemingly in greater numbers, and multinucleate cells in embryos were common (Holding and Springer, 2002), a phenomenon that was not observed in orc2 mutant seeds except at late stages in the suspensor.
The orc2 mutation causes a phenotype that is instead more akin to the ttn mutants. These zygotic lethal mutants arrest at the preglobular stage (except ttn6) and with enlarged endosperm nuclei. The ttn phenotypes were hypothesized to be caused by defects in the cytoskeleton, cell cycle control, or the structural mechanics of mitosis (Liu and Meinke, 1998). This theory was borne out when ttn phenotypes were correlated with mutations in genes for proteins involved in microtubule function and in structural maintenance of chromosomes (SMC) proteins (Liu et al., 2002; Tzafrir et al., 2002). The ttn mutants have been placed into different classes, and the orc2 phenotype is most similar to the class B ttn mutants (Liu and Meinke, 1998; Tzafrir et al., 2002). The differences are that in this ttn class, embryos arrest slightly earlier, at the two to four cell stage, and with a regular shape; more of the enlarged endosperm nuclei are observed; and mutant ttn seeds reach normal sizes but are white and translucent. This can be summarized as an earlier effect on embryo and a later effect on endosperm of class B ttn compared with the orc2 mutations.
The class B ttn mutants ttn7 and ttn8 are in genes coding for SMC3 and SMC1 cohesins, respectively (Liu et al., 2002). Cohesins are loaded onto the DNA after replication and are thought to form a ring sealed by Scc1 that effects sister chromatid cohesion (Nasmyth, 2002). In prophase, separase opens the ring, and most cohesin disassociates, allowing chromosome segregation. The similarity in phenotype between mutants in an ORC subunit and in cohesins implies that these mutants may have shared defects. One possibility is that incomplete or defective replication in orc2 causes a defect in sister chromatid cohesion, condensation, or both, as observed in D. melanogaster orc2 mutants (Loupart et al., 2000; Pflumm and Botchan, 2001).
Why Do Endosperm Nuclei Enlarge?
In the orc2 mutant, endosperm development arrested with approximately four detectable nuclei that were dramatically enlarged, whereas embryos had up to eight cells and their nuclei were normal in size. Thus, normal nuclear division cycles in the endosperm are arrested earlier than they are in the embryo and are presumably replaced by endoreplication cycles. Such enlarged endosperm nuclei are seen in the ttn and the pilz mutants (Mayer et al., 1999; Steinborn et al., 2002), which are related to the class A ttn mutants, as well as in prolifera (Holding and Springer, 2002). Berger (2003) proposed that this phenotype may be more common but overlooked. Clearly, the control of the cell cycle is different in the endosperm. In the embryo, there must be a cell cycle checkpoint like the one proposed to act in S. cerevisiae and D. melanogaster (Dillin and Rine, 1998; Loupart et al., 2000) that detects defective DNA replication or chromosomes and prevents the entry into S-phase. This checkpoint must be lacking in the endosperm. Nevertheless, it was surprising at first that mutation of a protein required for origin firing could allow new developmentally abnormal rounds of replication.
The most plausible explanation relates to that put forward by Landis et al. (1997) to account for the differential requirement of ORC2 for chorion gene amplification and endoreplication in D. melanogaster. Despite chorion gene amplification being completed before polyploidization of larval cells, only chorion gene amplification (not endoreplication) is prevented in orc2 mutant larvae. Initiation of chorion gene amplification, and presumably also chromosome amplification, is only twofold lower in the D. melanogaster orc2 mutant than in the wild type. It was proposed that because chorion genes have only a single major origin of replication, whereas chromosomes have multiple presumably redundant origins, an effect is seen only in the chorion genes. This is supported by the observation that the duration of S-phase is normal in cells depleted for ORC2 (Dhar et al., 2001; Christensen and Tye, 2003), and yeast cells in which most origins are inactivated can still replicate normally (Campbell and Newlon, 1991). In Arabidopsis orc2 seeds, depletion of AtORC2 by cell and nuclear divisions may cause DNA replication or chromosomal defects and trigger checkpoints that arrest the cell cycle in the embryo but only nuclear division in the endosperm. Several rounds of (endo)replication still may be initiated and completed in endosperm nuclei before AtORC2 levels fall below a level preventing origin activation.
Mutation of a Polycomb Group Protein Partially Suppresses orc2
Altered origin usage or accessibility may account for the suppression of the orc2 phenotype by mutation of MEA. In metazoans, origins of replication are not defined by sequence, and origin use changes during development. It is therefore thought that unlike in S. cerevisiae, ORC alone does not recognize origins. Chromatin structure and other factors, such as transcription factors, must modify origin activity and be involved in recruiting ORC (Gerbi and Bielinsky, 2002). MEA is a Polycomb group protein that is required for embryo and endosperm development. It may modify chromatin structure and control gene expression, including that of transcription factors (Köhler et al., 2003; Reyes and Grossniklaus, 2003). In the mea mutant, access of ORC to some origins may be increased, or ORC (or AtORC2) may be released from protein complexes or chromosome sites to which it binds with higher affinity. This may increase the number of active origins above a critical threshold level and enable the extra rounds of cell and nuclear division that are seen in mea/+, orc2/orc2 seeds. Both of these scenarios would be consistent with two recent reports regarding ORC action. In a Xenopus egg replication system, ORC did not bind to CpG-methylated DNA, and DNA replication was inhibited (Harvey and Newport, 2003). In mea mutants, a lack or reduction in DNA methylation at sites normally controlled by MEA-containing complexes may make them more accessible to dwindling levels of the ORC. The second report showed that S. cerevisiae ORC binds more strongly to the HMR-E silencer (an inefficient origin of DNA replication) than it does to nonsilencing efficient origins. This accounts for the differential effect of the yeast orc2-1 mutant on replication and silencing. This mutant lowers the concentration of the ORC in vivo, reducing replication at many origins, but it has no effect on silencing at HMR-E, whose high affinity for the ORC presumably allows it to compete successfully with other origins for the remaining ORC in the cell (Palacios DeBeer et al., 2003).
An alternative explanation is that the mea mutation affects cell cycle checkpoints that are normally activated by the orc2 mutation. It could be the lack of these checkpoints in mutant mea embryos that causes their overproliferation.
Conclusion
Chromosomal DNA sequence information must be accurately copied during DNA replication. Similarly, for endogenous epigenetic inheritance patterns to be maintained for gene regulatory purposes, chromatin and chromosome structure and the epigenetic marks that govern gene regulation must all be carried over into the newly synthesized DNA so that they can be faithfully transmitted to the next generation. In S. cerevisiae and D. melanogaster, ORC has functions in all these processes and may even be involved in coordinating them. Whether all of these functions have been conserved in plants remains to be seen. Our characterization of the orc2 mutant will allow us to address this question and opens the door to investigation of the role of AtORC2 and the ORC in plant development.
The roles for ORC in Arabidopsis postulated so far assume that the phenotype of the orc2 mutant is related to the function of AtORC2 in a complete ORC complex analogous to that described in fungi and metazoans. However, we cannot rule out a specific role for AtORC2 in postzygotic development. First, the development of the gametophytes involves mitotic divisions, but these proceed normally, and defects are seen only in post-zygotic development. Second, similar to the human ORC subunits (Thome et al., 2000), the genes for the Arabidopsis ORC subunit homologs are not coregulated (Table 3). We detected expression of AtORC2 in pollen (Figure 3), and transcripts of the maize homologs of ORC2 and ORC5, but not the other ORC subunit homologs, were found in maize sperm (Engel et al., 2003). Finally, the differential effect of the orc2 mutation on endosperm and embryo might be caused by defects in different processes. Whether the orc2 lethal defect is in a process involving the complete ORC, a subset of the subunits, or just ORC2 can be answered using mutants for the other subunits.
METHODS
Plant Material and Growth Conditions
The orc2 mutant, line ET3723, was isolated during a screen for semisterility from a collection of enhancer trap Ds transposants (Moore et al., 1997). The transposants were generated using the Ac/Ds insertional mutagenesis system of Sundaresan et al. (1995) in the Arabidopsis accession Ler. Seeds were surface sterilized using 6% sodium hypochlorite and allowed to germinate on MS medium (Duchefa, Haarlem, The Netherlands) containing 10 g/L of sucrose and 8 g/L of agar. To select for plants carrying the Ds transposon, 50 mg/L of kanamycin was added to the MS medium. Seedlings were transplanted to ED73 soil (Tränkle Einheiteserde, Kappelrodeck-Waldulm, Germany) and transferred to a growth chamber with 70% humidity and a day/night cycle of 16 h light at 21°C and 8 h dark at 18°C.
Whole-Mount Preparations and Histology
For superficial evaluation of ovule fates, siliques were opened using insulin needles (Becton Dickinson, Franklin Lakes, NJ) and observed using a Leica MZ6 binocular microscope (Leica Microsystems, Bernsheim, Germany). The contents were classed as unfertilized ovules and normal and aborted seeds. For morphological analysis of developing seeds, an incision was made in the carpel walls, and the siliques then were fixed and cleared with chloral hydrate (Yadegari et al., 1994). Siliques were dissected and mounted in clearing solution and analyzed using a Leica DMR microscope under bright-field Nomarski optics. GUS activity assays were as described by Vielle-Calzada et al. (2000).
Molecular Biology
Genomic DNA for determination of the Ds insertion site and genotype analysis was extracted as described by Meyerowitz (http://iprotocol.mit.edu/protocol/122.htm). The insertion site of the Ds enhancer trap element in the genome of line ET3723 was determined by thermal asymmetric interlaced PCR (Liu et al., 1995; as described in Grossniklaus et al., 1998). For precise analysis of the insertion site, the regions flanking the insertion site were amplified from genomic DNA using primer combinations orciF (5′-TTGTGTCCATTGGCAGTGTGGGACA-3′) and Ds3-1 (5′-CGATTACCGTATTTATCCCGTTCG-3′) and orciR (5′-ATATTCAAACACTCTTGACCATCGGA-3′) and Ds5-1 (5′-CCGTTTACCGTTTTGTATATCCCG-3′). For genotyping, diagnostic fragments of the wild type and mutant AtORC2 alleles were amplified from genomic DNA using orciF and orciR, and orciF and Ds3-1, respectively.
For RT-PCR expression analysis of AtORC2, first-strand cDNA synthesis was performed on 5 μg of total RNA extracted from Ler seedlings, mature rosette leaves, and inflorescences using SuperscriptII (Invitrogen, Carlsbad, CA). PCR amplification was performed on 1 μL (out of 20 μL) of the cDNA using primers ORC2-TF (5′-ACATAGAAGAAGATGAGTATGGGTT-3′) and orciR that span the AtORC2 mRNA and, for the actin-11 gene (At3g12110), with primers act11F (5′-AACTTTCAACACTCCTGCCATG-3′) and act11R (5′-CTGCAAGGTCCAAACGCAGA-3′).
Complementation
The AtORC2 gene (At2g37560), from 2638 bp upstream to 400 bp downstream of the coding region, was amplified from Ler with primers ORC2GF (5′-ACAGAGAAGTCACTCGCATGTCGCAT-3′) and ORC2GR (5′-CAGGTGAATTTGTCAGAGAGATCCCAT-3′) and the Expand Long Template PCR system (Roche, Indianapolis, IN) and then cloned into the pCAMBIA-1300 binary vector (Roberts et al., 1997) to produce p1300-ORC2. ET3723 (orc2/+) plants were transformed by Agrobacterium tumefaciens, and T1 plants were selected on MS medium containing 25 mg/L of hygromycin and 50 mg/L of kanamycin (the markers for pCAMBIA-1300 and the Ds element, respectively) and allowed to self-pollinate.
High-Density Oligonucleotide Array Expression Analysis
Expression analysis of Ler flowers before and at fertilization and of siliques containing preglobular-stage embryos was performed as described by Köhler et al. (2003).
5′-RACE and Definition of the AtORC4 ORF
To determine the transcript of the AtORC4 homolog At2g01120, 5′-RACE and RT-PCR was performed on RNA extracted from Ler inflorescences using Trizol (Invitrogen). One microgram of total RNA was processed using the SMART RACE cDNA amplification kit (Clontech) using ORC4-R2 (5′-TTCTCTGCATAGCATATAACTTGTCG-3′), ORC4-R3 (5′-ACTTGAAAGTGCTGCTTTGAAGTT-3′), and ORC3-R4 (5′-CTGCTGGTAGACAGCAGGTGT-3′) as the gene-specific primers. RT-PCR was used to amplify the coding region using primers ORC4-F1 (5′-TTGTCAGTTTTCCCGCCAGTCCGATG-3′) and ORC4-R1 (5′-AACACAGAGATCAATGGTCCAATAAC-3′). Clone GBGE182 (the Arabidopsis transcribed genome; GDR cDNA program, Centre Naitonal de la Recherche Scientifique) representing the currently annotated mRNA of AtORC4 was obtained from ABRC. Its sequence was used to establish the 3′ end of the transcript.
Sequence data were analyzed using Vector NTI (Informax, Frederick, MD). The mRNA sequence of the AtORC4 gene has been deposited in the database under accession number AJ586913. Intron/exon boundaries were determined using SlicePredictor (at bioinformatics.iastate.edu/cgi-bin/sp.cgi) and the gene diagram (see supplemental data online) made using Vector NTI. The protein alignment (see supplemental data online) was generated at clustalw.genome.ad.jp and presented using Boxshade at www.chembnet.org/software/BOX-form.html.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AJ586913.
Supplementary Material
Acknowledgments
J. Moore and W. Gagliano generated and screened the collection of Ds enhancer trap transposants. We thank Chantal Ebel for critical reading of the manuscript, Valeria Gagliardini and Peter Kopf for technical support, Michi Federer for making the thin sections, Norbert Huck for all his help with microscopy, and Jean-Jaques Pitet for help with rendering photographs into presentable figures. C.K. received fellowships from European Molecular Biology and Human Frontier Science Program. The project was supported by the University of Zürich and, in part, by Grant 31-64061.00 from the Swiss National Foundation to U.G.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Margaret A. Collinge (collinge@botinst.unizh.ch).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.019059.
References
- Bell, S.P. (2002). The origin recognition complex: From simple origins to complex functions. Genes Dev. 16, 659–672. [DOI] [PubMed] [Google Scholar]
- Bell, S.P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374. [DOI] [PubMed] [Google Scholar]
- Bell, S.P., Kobayashi, R., and Stillman, B. (1993). Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262, 1844–1849. [DOI] [PubMed] [Google Scholar]
- Berger, F. (2003). Endosperm: The crossroad of seed development. Curr. Opin. Plant Biol. 6, 42–50. [DOI] [PubMed] [Google Scholar]
- Campbell, J.L., and Newlon, C.S. (1991). Chromosomal DNA replication. In The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics, J.R. Broach, J.R. Pringle, and E.W. Jones, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 41–146.
- Chesnokov, I., Gossen, M., Remus, D., and Botchan, M. (1999). Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes Dev. 13, 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, T.W., and Tye, B.K. (2003). Drosophila Mcm10 interacts with members of the prereplication complex and is required for proper chromosome condensation. Mol. Biol. Cell 14, 2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis, M.L. (2003). The ‘ORC cycle’: A novel pathway for regulating eukaryotic DNA replication. Gene 310, 1–15. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., de Almeida Engler, J., Ormenese, S., Maes, S., Naudts, M., Van Der Schueren, E., Jacqmard, A., Engler, G., and Inze, D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21, 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar, S.K., Delmolino, L., and Dutta, A. (2001). Architecture of the human origin recognition complex. J. Biol. Chem. 276, 29067–29071. [DOI] [PubMed] [Google Scholar]
- Dillin, A., and Rine, J. (1998). Roles for ORC in M phase and S phase. Science 279, 1733–1737. [DOI] [PubMed] [Google Scholar]
- Engel, M.L., Chaboud, A., Dumas, C., and McCormick, S. (2003). Sperm cells of Zea mays have a complex complement of mRNAs. Plant J. 34, 697–707. [PubMed] [Google Scholar]
- Foss, M., McNally, F.J., Laurenson, P., and Rine, J. (1993). Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science 262, 1838–1844. [DOI] [PubMed] [Google Scholar]
- Fox, C.A., Ehrenhofer-Murray, A.E., Loo, S., and Rine, J. (1997). The origin recognition complex, Sir1, and the S phase requirement for silencing. Science 276, 1547–1551. [DOI] [PubMed] [Google Scholar]
- Gavin, K.A., Hidaka, M., and Stillman, B. (1995). Conserved initiator proteins in eukaryotes. Science 270, 1667–1671. [DOI] [PubMed] [Google Scholar]
- Gatti, M., and Baker, B.S. (1989). Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 3, 438–453. [DOI] [PubMed] [Google Scholar]
- Gerbi, S.A., and Bielinsky, A.K. (2002). DNA replication and chromatin. Curr. Opin. Genet. Dev. 12, 243–248. [DOI] [PubMed] [Google Scholar]
- Gossen, M., Pak, D.T., Hansen, S.K., Acharya, J.K., and Botchan, M.R. (1995). A Drosophila homolog of the yeast origin recognition complex. Science 270, 1674–1677. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Harvey, K.J., and Newport, J. (2003). CpG methylation of DNA restricts prereplication complex assembly in Xenopus egg extracts. Mol. Cell. Biol. 23, 6769–6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding, D.R., and Springer, P.S. (2002). The Arabidopsis gene PROLIFERA is required for proper cytokinesis during seed development. Planta 214, 373–382. [DOI] [PubMed] [Google Scholar]
- Huang, D., and Koshland, D. (2003). Chromosome integrity in Saccharomyces cerevisiae: The interplay of DNA replication factors, elongation factors and origins. Genes Dev. 17, 1741–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D.W., Fanti, L., Pak, D.T.S., Botchan, M.R., Pimpinelli, S., and Kellum, R. (1998). Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: Their phosphorylation levels and associations with origin recognition complex proteins. J. Cell Biol. 142, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck, N., Moore, J.M., Federer, M., and Grossniklaus, U. (2003). The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130, 2149–2159. [DOI] [PubMed] [Google Scholar]
- Kimura, S., Ishibashi, T., Hatanaka, M., Sakakibara, Y., Hashimoto, J., and Sakaguchi, K. (2000). Molecular cloning and characterization of a plant homologue of the origin replication complex 1 (ORC1). Plant Sci. 158, 33–39. [DOI] [PubMed] [Google Scholar]
- Köhler, C., Hennig, L., Spillane, C., Gruissem, W., and Grossniklaus, U. (2003). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17, 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis, G., Kelley, R., Spradling, A.C., and Tower, J. (1997). The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of the yeast origin recognition complex subunit 2. Proc. Natl. Acad. Sci. USA 94, 3888–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.-M., McElver, J., Tzafrir, I., Joosen, R., Wittich, P., Patton, D., Van Lammeren, A.A.M., and Meinke, D. (2002). Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J. 29, 405–415. [DOI] [PubMed] [Google Scholar]
- Liu, C.-M., and Meinke, D.W. (1998). The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 16, 21–31. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Loupart, M.-L., Krause, S.A., and Heck, M.M.S. (2000). Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr. Biol. 10, 1547–1556. [DOI] [PubMed] [Google Scholar]
- Mayer, U., Herzog, U., Berger, F., Inzé, D., and Jürgens, G. (1999). Mutations in the PILZ group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsis embryo and endosperm. Eur. J. Cell Biol. 78, 100–108. [DOI] [PubMed] [Google Scholar]
- McNairn, A.J., and Gilbert, D.M. (2003). Epigenomic replication: Linking epigenetics to replication. Bioessays 25, 647–656. [DOI] [PubMed] [Google Scholar]
- Moore, J.M., Vielle-Calzada, J.-P., Gagliano, W., and Grossniklaus, U. (1997). Genetic characterization of hadad, a mutant disrupting female gametogenesis in Arabidopsis thaliana. Cold Spring Harb. Symp. Quant. Biol. 62, 35–47. [PubMed] [Google Scholar]
- Nacry, P., Mayer, U., and Jürgens, G. (2000). Genetic dissection of cytokinesis. Plant Mol. Biol. 43, 719–733. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. (2002). Segregating sister genomes: The molecular biology of chromosome separation. Science 297, 559–565. [DOI] [PubMed] [Google Scholar]
- Ohtani, K., DeGregori, J., Leone, G., Herendeen, D.R., Kelly, T.J., and Nevins, J.R. (1996). Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol. Cell. Biol. 16, 6977–6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, D.R., and Grossniklaus, U. (2002). The art and design of genetic screens: Arabidopsis thaliana. Nat. Rev. Genet. 3, 124–136. [DOI] [PubMed] [Google Scholar]
- Pak, D.T.S., Pflumm, M., Chesnokov, I., Huang, D.W., Kellum, R., Marr, J., Romanowski, P., and Botchan, M.R. (1997). Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91, 311–323. [DOI] [PubMed] [Google Scholar]
- Palacios DeBeer, M.A., Müller, U., and Fox, C.A. (2003). Differential DNA affinity specifies roles for the origin recognition complex in budding yeast heterochromatin. Genes Dev. 17, 1817–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflumm, M.F., and Botchan, M.R. (2001). Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development 128, 1697–1707. [DOI] [PubMed] [Google Scholar]
- Pinto, S., et al. (1999). latheo encodes a subunit of the origin recognition complex and disrupts neuronal proliferation and adult olfactory memory when mutant. Neuron 23, 45–54. [DOI] [PubMed] [Google Scholar]
- Prasanth, S.G., Prasanth, K.V., and Stillman, B. (2002). Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science 297, 1026–1031. [DOI] [PubMed] [Google Scholar]
- Reyes, J.C., and Grossniklaus, U. (2003). Diverse functions of Polycomb group proteins during plant development. Semin. Cell Dev. Biol. 14, 77–84. [DOI] [PubMed] [Google Scholar]
- Roberts, C.S., et al. (1997). A comprehensive set of modular vectors for advanced manipulations and efficient transformation of plants by both Agrobacterium and direct DNA uptake methods: pCAMBIA vector release manual version 3.05.
- Rohrbough, J., Pinto, S., Mihalek, R., Tully, T., and Broadie, K. (1999). latheo, a Drosophila gene involved in learning, regulates functional synaptic plasticity. Neuron 23, 55–70. [DOI] [PubMed] [Google Scholar]
- Sørensen, M.B., Mayer, U., Lukowitz, W., Robert, H., Chambrier, P., Jürgens, G., Somerville, C., Lepiniec, L., and Berger, F. (2002). Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129, 5667–5676. [DOI] [PubMed] [Google Scholar]
- Springer, P., Holding, D.R., Groover, A., Yordan, C., and Martienssen, R.A. (2000). The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during G1 phase and is required maternally for early Arabidopsis development. Development 127, 1815–1822. [DOI] [PubMed] [Google Scholar]
- Steinborn, K., Maulbetsch, C., Priester, B., Trautmann, S., Pacher, T., Geiges, B., Küttner, F., Lepiniec, L., Stierhof, Y.-D., Schwarz, H., Jürgens, G., and Mayer, U. (2002). The Arabidopsis PILZ group genes encode tubulin-folding cofactor orthologs required for cell division but not cell growth. Genes Dev. 16, 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D.G., Dean, C., Ma, M., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Thome, K.C., Dhar, S.K., Quintana, D.G., Delmolino, L., Shahsafaei, A., and Dutta, A. (2000). Subsets of human origin recognition complex (ORC) subunits are expressed in non-proliferating cells and associate with non-ORC proteins. J. Biol. Chem. 275, 35233–35241. [DOI] [PubMed] [Google Scholar]
- Tzafrir, I., McElver, J.A., Liu, C.-M., Yang, L.J., Wu, J.Q., Martinez, A., Patton, D.A., and Meinke, D.W. (2002). Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol. 128, 38–51. [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Baskar, R., and Grossniklaus, U. (2000). Delayed activation of the paternal genome during seed development. Nature 404, 91–94. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Thomas, J., Spillane, C., Coluccio, A., Hoeppner, M.A., and Grossniklaus, U. (1999). Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13, 2971–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers, D., Franke-van Dijk, M., Vencken, R.-J., Quint, A., Hooykaas, P., and Offringa, R. (2001). An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128, 4289–4299. [DOI] [PubMed] [Google Scholar]
- Witmer, X., Alvarez-Venegas, R., San-Miguel, P., Danilevskaya, O., and Avramova, Z. (2003). Putative subunits of the maize origin of replication recognition complex ZmORC1-ZmORC5. Nucleic Acids Res. 31, 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari, R., de Paiva, G.R., Laux, T., Koltunow, A.M., Apuya, N., Zimmerman, J.L., Fischer, R.L., Harada, J.L., and Goldberg, R.G. (1994). Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryos. Plant Cell 6, 1713–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.