Abstract
Th9 cells protect hosts against helminthic infection but also mediate allergic disease. Here we show that nitric oxide (NO) promotes Th9 cell polarization of murine and human CD4+ T cells. NO de-represses the tumor suppressor gene p53 via nitrosylation of Mdm2. NO also increases p53-mediated IL-2 production, STAT5 phosphorylation and IRF4 expression, all essential for Th9 polarization. NO also increases the expression of TGFβR and IL-4R, pivotal to Th9 polarization. OVA-sensitized mice treated with an NO donor developed more severe airway inflammation. Transferred Th9 cells induced airway inflammation, which was exacerbated by NO and blocked by anti-IL-9 antibody. Nos2−/− mice had less Th9 cells and developed attenuated eosinophilia during OVA-induced airway inflammation compared to wild-type mice. Our data demonstrate that NO is an important endogenous inducer of Th9 cells and provide a hitherto unrecognized mechanism for NO-mediated airway inflammation via the expansion of Th9 cells.
INTRODUCTION
It is now generally accepted that CD4+ T cells are functionally divided into various subsets: Th1, Th2, Th17, Th9 and Treg (regulatory T cells). Although plasticity among these subsets are frequently reported, these cells are mainly defined by the cytokines they produce together with their signature master transcription factors (reviewed in ref. 1). Th9 is the latest subset of Th cells defined by their production of IL-9 (ref. 2, 3) and the transcription factors PU.1 (ref. 4) and IRF4 (ref. 5), although these factors are not unique to Th9.
IL-9 has been considered a classical Type 2 cytokine and is associated with the host defense against intestinal nematode infection and the development of allergic responses. Mice overexpressing IL-9 develop intestinal mastocytosis and are resistant to Trichuris muris infection6,7 and administration of neutralizing anti-IL-9 antibody abrogated the worm expulsion8. Consistent with the characteristics of Type 2 cytokines, overexpression of IL-9 in the lung led to spontaneous airway inflammation in mice9,10. Increased expression of IL-9 and its receptor were also found in the lungs of asthmatic patients11. Th9 cells as a source of IL-9 contribute to allergic inflammation4,5. In humans, allergic donors have substantially more circulating Th9 cells than non-allergic donors12.
Th9 cells are generated in vitro by culturing naïve CD4+ T cells with IL-4 and TGFβ2,3,13. This pair of cytokines forms the basic environment for driving Th9 cells, the polarizing of which can be enhanced by the presence of IL-1 (ref. 14, 15), IL-2 (ref. 13), and IL-25 (ref. 16). We report here that nitric oxide (NO), a free radical, is a potent enhancer of Th9 polarization and maintenance.
NO is a crucial mediator of a range of biological functions, including vascular relaxation, platelet aggregation, neurotransmission, tumoricidal and microbicidal activities, and immune regulation (reviewed in ref. 17-20). NO is also associated with some of the most important immune pathologies, including rheumatoid arthritis, diabetes, systemic lupus erythematosus and septic shock. NO is derived from the guanidino nitrogen atom(s) and molecular oxygen in a reaction catalyzed by three forms of nitric oxide synthase (NOS). The neuronal form (nNOS or NOS1) and endothelial form (eNOS or NOS3) produce physiological level of NO at steady state. The cytokine-inducible form (iNOS or NOS2) is activated by a number of immunological stimuli, including IFNγ, TNFα and LPS generated during infection, and catalyzes high output of NO, which can be cytotoxic and kill intracellular pathogens.
We have recently shown21 that NO can suppress the proliferation and function of polarized murine and human Th17 cells via the down regulation of the expression of aryl hydrocarbon receptor which participates in the induction of Th17. We now show that, in contrast to its effect on Th17, NO markedly enhances the polarization and function of Th9 cells. NO does so by elevating the expression of p53, which in turn increases the production of IL-2 and activates the down stream events including phosphorylation of STAT5 and the expression of IRF4. In vivo, administration of an NO donor increases airway inflammation whereas Nos2-deficiency reduces Th9 cell development and allergic reaction. Our data demonstrate a hitherto unrecognized NO function and indicate that NO is an important endogenous inducer of Th9 responses and may play a key role in infectious and allergic diseases.
RESULTS
NO inhibits Th17 but enhances Th9 differentiation
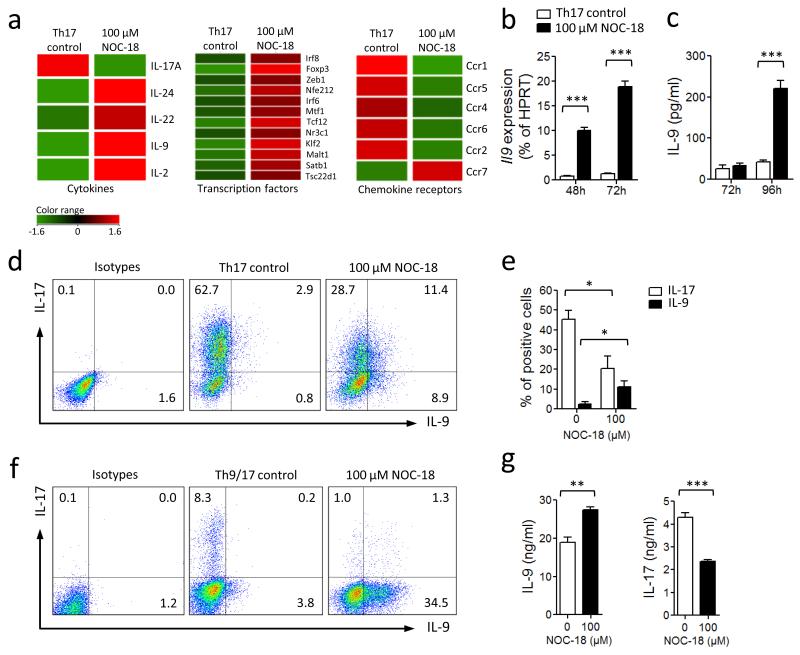
We reported recently that NO selectively inhibits the differentiation of Th17 cells21. We then investigated the effect of NO on the expression of other genes during Th17 differentiation. CD4+ T cells were purified from the spleen and lymph nodes of naïve BALB/c mice and cultured under a Th17 polarizing conditions (anti-CD3 antibodies + IL-6, TGFβ, IL-1β, IL-23, anti-IFNγ, anti-IL-4 and mitomycin-C-treated antigen-presenting cells, APC) in the presence of 100 μM of NOC-18 (an NO donor) as described previously21. The cells were harvested 72 h after the culture (when all APCs had disappeared) and total RNA extracted and subjected to microarray analysis. As expected a vast array of messages was affected by the presence of NO (Fig. 1a and supplementary Fig. 1). Focusing on cytokines, Il17a message was clearly decreased but Il24, Il22, Il9 and Il2 mRNA were markedly up regulated (Fig. 1a). In this report, we elected to focus on IL-9. The enhancement of Il9 expression by NO was confirmed by quantitative PCR (qPCR) assay (Fig. 1b), and by ELISA of the culture supernatants (Fig. 1c). FACS analysis demonstrated that decrease in the percentage of IL-17+ T cells was accompanied by an increase in the percentage of IL-9+ T cells and IL-17/IL-9 double positive T cells (Fig. 1d, e). We then determined whether NO can influence the differentiation of Th17 and Th9 cells under the mixed polarization conditions for Th17 and Th9. CD4+ T cells were cultured for 4 days with plate-bounded anti-CD3 and soluble anti-CD28 + IL-4, TGFβ, IL-6, IL-1β, IL-23, and anti-IFNγ (without APC) in the presence of NOC-18. These culture conditions resulted in a modest percentage of Th17 cells and a distinct population of Th9 cells. NO decreased the percentage of IL-17+ cells and enhanced the percentage of IL-9+ T cells, without producing significant number of IL-17/IL-9 double positive T cells (Fig. 1f). The differential effect of NO on Th17 and Th9 cell polarization was also reflected in the concentrations of IL-9 and IL-17 in the supernatants of these cultures (Fig. 1g). These results therefore demonstrate that NO inhibits Th17 development but enhances the differentiation of Th9 cells under Th17 and Th17/9 polarizing conditions.
Figure 1.
NO inhibits Th17 but enhances Th9 development. Purified BALB/c CD4+ T cells were cultured for 3 d under Th17 polarizing conditions (round-bottom 96-well plate with APC, αCD3, IL-6, TGF-β, IL-23, IL-1β, αIFNγ and αIL-4). NO donor (NOC-18) was added at the beginning of culture. (a) Microarray analysis of differential mRNA expression between cells cultured with or without NO. Full microarray information is provided in Supplementary Fig. 1 and deposited in MIAME (E-MEXP-3959). (b) Il9 mRNA was further quantified by qPCR. (c) IL-9 concentration in the supernatant determined by ELISA. (d) IL-17 and IL-9 producing cells were analyzed by FACS. Numbers in quadrants indicate percentage of cells and are summarized in (e). (f) CD4+ T cells were cultured under Th17/Th9 conditions (αCD3 + IL-4, TGFβ, IL-6, IL-1β, IL-23, anti-IFNγ) ± NO. Percentage of cells producing IL-9 and IL-17 determined by FACS. (g) IL-9 and IL-17 concentrations in the culture supernatant assayed by ELISA. Data are representative of at least 3 independent experiments; Mean ± SEM, n=3; *P<0.05, **P< 0.01, ***P< 0.001 using a 2-way ANOVA (b,c and e) or a 2-tailed unpaired Student’s t test (g).
Characterization of NO-induced Th9 cells
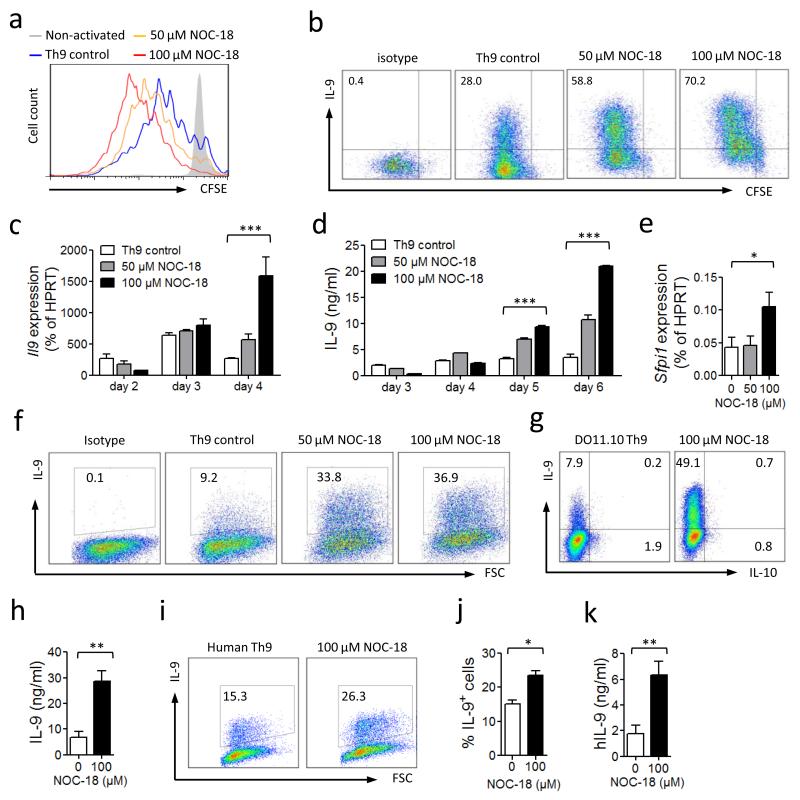
We next investigated the role of NO on Th9 cell differentiation under Th9-polarizing culture conditions. CD4+ T cells were purified from naïve BALB/c or B6 mice and cultured for up to 6 days with anti-CD3/CD28 antibodies + TGFβ, IL-4 and graded concentrations of NOC-18. NO enhanced T cell proliferation as determined by CFSE dilution (Fig. 2a). NO also markedly increased the percentage of IL-9+ T cells in a dose-dependent manner (Fig. 2b). The NO-induced enhancement of Th9 development was also evident in Il9 mRNA expression (Fig. 2c) and IL-9 protein produced in the culture supernatant (Fig. 2d). The expression PU.1 (Sfpi1), a transcription factor, which has been reported to be required for Th9 development4, was also significantly increased by NO (Fig. 2e). Next we tested if NO was able to enhance the expansion of the number of established Th9 cells. CD4+ T cells were polarized to Th9 by 2 rounds of culture, rested for 24 h and then re-cultured with anti-CD3/CD28 in the presence of graded concentrations of NOC-18. NO significantly enhanced the percentage of the polarized Th9 cells (Fig. 2f). Next we examined whether NO can promote Th9 cell polarization in an antigen-specific manner. CD4+ T cells were purified from the spleen and LN of OVA-TCR transgenic mice (DO11.10) and cultured in vitro with OVA peptide and mitomycin C-treated APC in the presence of NOC-18. NO markedly increased the percentage of IL-9+ T cells (Fig. 2g) and IL-9 in the culture supernatant (Fig. 2h). We also tested the effect of NO on human Th9 cells. CD4+ T cells were purified from the peripheral blood of 7 healthy volunteer donors and cultured with anti-hCD3/hCD28 antibodies, hTGFβ and hIL-4 in the presence of NOC-18. NO significantly increased the percentage of IL-9+ T cells (Fig. 2i, j) and IL-9 in the culture supernatant (Fig. 2k). These results therefore demonstrate that NO potently enhances the development of murine and human Th9 cells in vitro at antigen-specific and polyclonal levels.
Figure 2.
NO enhances cell proliferation and IL-9 production during Th9 polarization. CFSE-labeled BALB/c CD4+ T cells were cultured under Th9 polarizing conditions (flat-bottom 96-well plate with αCD3 and αCD28 + IL-4, TGF-β) ± NO. (a) T cell proliferation (CFSE dilution) and (b) IL-9 producing cells were analyzed by FACS on day 4. (c) Il9 mRNA expression was determined by qPCR. (d) IL-9 concentrations in culture supernatant were assessed by ELISA. (e) sfpil mRNA was determined by qPCR on day 2. Similar results were obtained with cells from C57BL/6 mice. (f) Effect of NO on polarized Th9 cells. CD4+ T cells were polarized 2x as above, washed and re-cultured ± NO for a further 4 days. Percentage of IL-9+ cells determined by FACS. (g, h) Effect of NO on antigen-specific Th9 cells. CD4+ T cells purified from DO11.10 mice were cultured for 2 rounds under Th9 polarization conditions with APC and 1 μM of OVA323-339 peptide ± NO. IL-9 and IL-10 producing cells were analyzed by FACS on day 2 of the second round (g) and IL-9 concentration in the supernatant was assessed by ELISA (h). CD4+ T cells purified from total human PBMC (n=7) were cultured with autologous APC, αhCD3, αhCD28+ hIL-4 and hTGF-β ± NO. (I, J) Percentage of IL-9+ cells analyzed by FACS on day 4, and (k) IL-9 concentrations in supernatants determined by ELISA on day 5. Data are representative of 3 independent experiments (mouse cells); mean ± SEM, n=3, *P<0.05, **P<0.01, ***P<0.001 using a 2-way ANOVA (c, d and e) or a 2-tailed unpaired Student’s t test (h, j and k).
NO-induced Th9 cell differentiation is IL-2-dependent
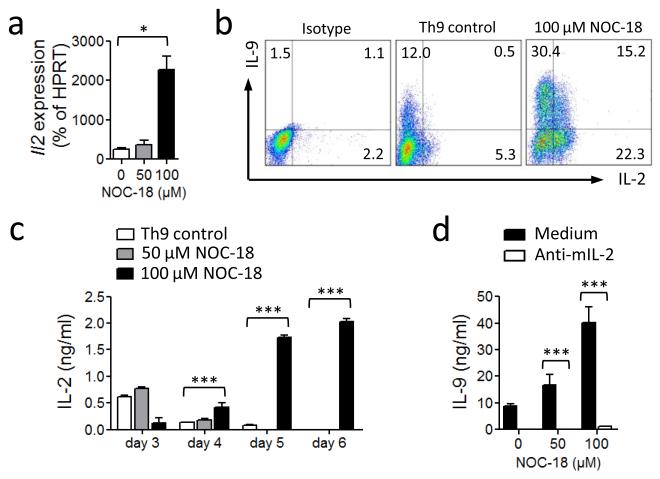
We next investigated the mechanism by which NO enhances Th9 differentiation. Since IL-2 has been shown to increase Th9 development13 and we have confirmed this in our own culture conditions (supplementary Fig. 2), we first examined the role of IL-2 in NO-induced Th9 development. CD4+ T cells were polarized under Th9 conditions in the presence of graded concentrations of NOC-18. The cells were cultured up to 6 days and the IL-2 expression analyzed. Il2 mRNA was clearly increased by the presence of NO (Fig. 3a). The percent of IL-2+, IL-9+ and IL-2/IL-9 double positive T cells were also markedly increased (Fig. 3b), together with the concentrations of IL-2 in the culture supernatant (Fig. 3c). Importantly, the enhancement of Th9 differentiation by NO was completely blocked by the presence of a neutralizing anti-IL-2 antibody (Fig. 3d). These results therefore demonstrate that the enhancement of Th9 differentiation by NO is IL-2-dependent and not via an additional pathway. CD25 expression was not affected by NO; all cells were 99% CD25+ after culture with or without NOC-18.
Figure 3.
Up-regulation of Th9 cell polarization by NO is IL-2-dependent. C57BL6 CD4+ T cells were cultured under Th9 polarizing conditions ± NO. (a) Il2 mRNA was determined by qPCR on day 2. (b) IL-9 and IL-2-producing cells were analyzed by FACS on day 4. (c) IL-2 in cell culture supernatants determined by ELISA. (d) IL-9 concentrations in the supernatant of Th9 polarized cells cultured in the presence of αIL-2 antibodies (20 μg/ml) were determined on day 5. Data are representative of at least 3 independent experiments; Mean ± SEM, * P<0.05, *** P< 0.001 using a 2-way ANOVA.
NO-induced Th9 development is STAT5- and IRF4-dependent
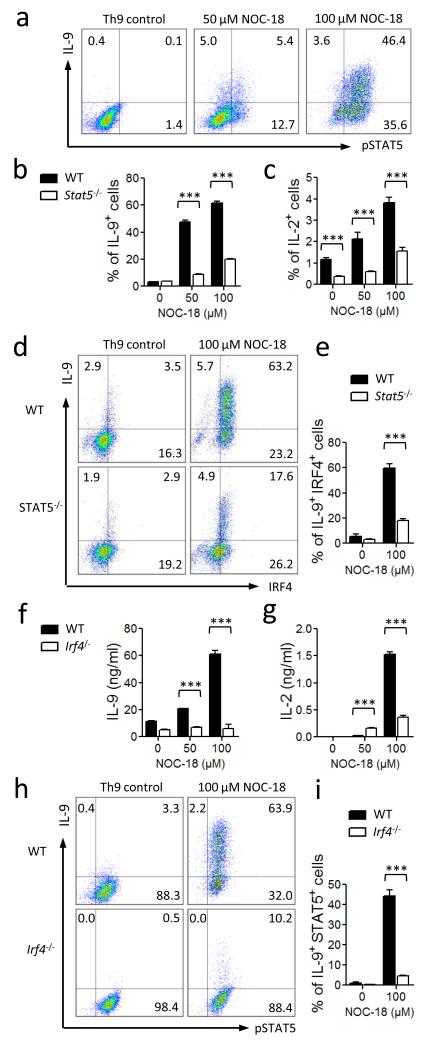
IL-2 activates the phosphorylation of the signal transducer and activator of transcription 5 (STAT5) (ref. 22-26), which has recently been shown to be important for IL-9 synthesis27. We therefore reasoned that pSTAT5 might be important in NO-induced Th9 differentiation. CD4+ T cells were cultured under Th9 polarizing conditions with NOC-18, and the percentage of pSTAT5+ cells analyzed by FACS. NO clearly increased the percentage of pSTAT5+ T cells in IL-9+ as well as IL-9− cell populations (Fig. 4a). To directly determine the role of STAT5 in NO-induced Th9 differentiation, we cultured purified CD4+ T cells from Stat5−/− mice under Th9 polarizing conditions in the presence of NOC-18. The percentage of IL-9+ cells was significantly reduced in Stat5−/− cells compared to wild-type (WT) cells (Fig. 4b). Interestingly, STAT5-deficient T cells also exhibited significantly less IL-2+ T cells when cultured under these conditions (Fig. 4c), indicating that STAT5 functions as an amplifier for the auto-induction loop of IL-2.
Figure 4.
Up-regulation of Th9 polarization by NO depends on IRF4 and STAT5. CD4+ T cells from WT, Stat5−/− or Irf4−/− mice were cultured under Th9 polarizing conditions with graded concentration of NO. (a) IL-9+ and pSTAT5+ cells were analyzed by FACS on day 4. (b, c) Percentage of IL-9+ and IL-2+ T cells from WT and Stat5−/− mice polarized under Th9 conditions with NO analyzed by FACS on day 4. (d, e) Percentage of IL-9+IRF4+ T cells from WT and Stat5−/− mice cultured ± NO for 4 days under Th9 conditions analyzed by FACS. (f, g) Concentrations of IL-9 and IL-2 in the supernatants of T cells from WT and Irf4−/− mice cultured under Th9 conditions determined by ELISA. (h, i) Percentage of IL9+pSTAT5+ T cells from WT and Irf4−/− mice cultured under Th9 conditions was analyzed by FACS. Data are representative or pool of at least 3 independent experiments; mean ± SEM, *** P<0.001 using a 2-way ANOVA.
We also investigated the role of IRF4, a key downstream molecule of STAT5 (ref.27) and an important transcription factor for the induction of IL-9 (ref. 5). CD4+ T cells from WT or Stat5−/− mice were cultured under Th9-polarizing conditions in the presence of NOC-18, and the level of intracellular IRF4 expression analyzed by FACS. NO markedly increased the differentiation of IRF4+ T cells in the WT cell population but only modestly in STAT5-deficient cells (Fig. 4d). Furthermore, NO strongly increased the frequency of IL-9/IRF4 double positive T cells in the WT cells but only marginally in the STAT5-deficient cells (Fig. 4e). To directly determine the role of IRF4 in NO-induced Th9 cell differentiation, we cultured CD4+ T cells from Irf4−/− mice under Th9 polarizing conditions in the presence of NOC-18. CD4+ T cells from Irf4−/− mice produced significantly less IL-9 compared to cells from the WT mice (Fig. 4f). Furthermore, Irf4−/− T cells also produced markedly reduced levels of IL-2 (Fig. 4g), indicating that IRF4 is also a mediator of the IL-2 self-amplification loop. The Irf4−/− cells also developed less percentage of IL-9+/pSTAT5+ T cells (Fig. 4 h, i). However, The frequency of total pSTAT5+ T cells (IL-9+/−) was the same in the WT or Irf4−/− T cells when cultured with NO under Th9 polarizing conditions (Fig. 4h), confirming that IRF4 functions downstream of STAT5 in the induction of IL-9. Together, these results demonstrate that NO-induced Th9 differentiation is also dependent on the activation of STAT5 and IRF4.
NO-induced Th9 differentiation is p53-dependent
Results obtained above so far indicate that NO induces IL-2 synthesis, which then activates STAT5 phosphorylation leading to the induction of IRF4 that promotes IL-9 synthesis. We then investigated the mechanism by which NO induces IL-2 synthesis. In a previous study, we reported that low doses of NO could activate IL-12Rβ expression via cyclic guanosine monophosphate (cGMP)28. We therefore examined the role of cGMP in the present culture system. We found that 8-Bromo-GMP, at concentrations (3-5 mM) known to activate several downstream events of cGMP, was not able to enhance Th9 differentiation or IL-2 synthesis (Supplementary Fig. 3a, b). Furthermore, ODQ (1-H-oxdiazolo-[1,2,4]-[4,3-α]quinoxalin-1-one, 5-10 μM) a potent inhibitor of cyclic guanylate cyclase (cGC), the enzyme catalyzing cGMP synthesis, also had no effect on NO-mediated enhancement of Th9 differentiation or IL-2 production (Supplementary Fig. 3c, d). Thus the effect of NO on Th9 differentiation is unlikely to be mediated by the induction of cGMP.
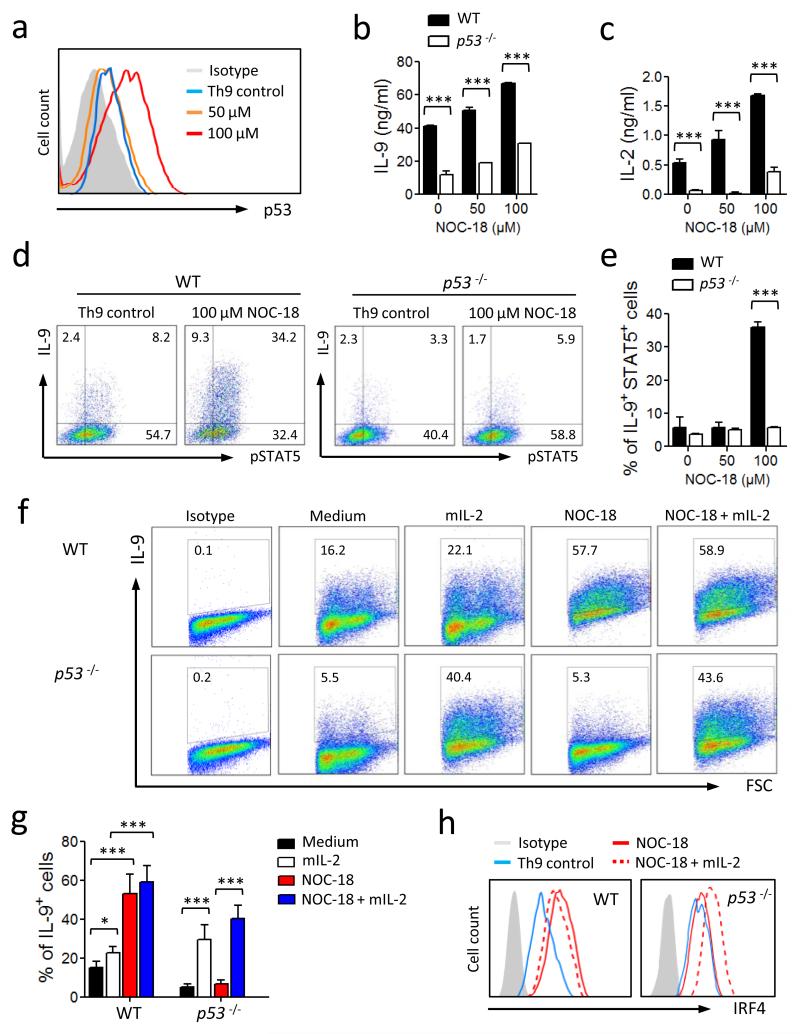
In another previous report29 we demonstrated that NO induced a population of Foxp3-negative regulatory T cells (NO-Treg) via the activation of p53, the tumor suppressor gene, which increased IL-2 synthesis. We therefore investigated the possibility that p53 may also play a role in the NO-mediated enhancement of Th9 differentiation. Naïve C57BL/6 CD4+ T cells were cultured under Th9 polarizing conditions in the presence of NOC-18, and the expression of p53 analyzed by FACS. NO induced substantial expression of p53 (Fig. 5a). To directly test the role of p53 in our culture system, CD4+ T cells from p53−/− or littermate WT mice were cultured under Th9 polarizing conditions in the presence of NOC-18. CD4+ T cells from p53−/− mice produced significantly less IL-9 (Fig. 5b) and IL-2 (Fig. 5c) compared to that of the WT mice. It should be noted that p53−/− cells retained a low capacity to polarize to Th9 cells, indicating that a p53-independent pathway may also be involved in NO-mediated enhanced Th9 polarization. The p53-deficient CD4+ T cells also generated significantly less IL-9+ as well as IL-9+pSTAT5+ double positive T cells compared to that of the WT mice (Fig. 5d, e). The frequency of total pSTAT5+ cells however remained unchanged, suggesting that the effect of p53 on pSTAT5 is confined to Th9 cells in our system. We then investigated if recombinant IL-2 can reverse the reduced ability of p53−/− cells to generate IL-9 with or without NO. As expected, NO induced a low frequency of Th9 cells from CD4+ T cells of p53−/− mice. This was markedly increased by the addition of recombinant murine IL-2 (mIL-2) in the culture in the absence of NO. This increase was even more evident in the presence of NO (Fig. 5f, g). NO induced lower levels of IRF4 in p53−/− cells compared to WT cells (Fig. 5h). However, exogenous mIL-2 was also able to reverse the reduced ability of NO to enhance IRF4 expression (Fig. 5h). These results therefore indicate that p53 is a key molecule for the NO-mediated enhancement of Th9 differentiation and p53 is likely upstream of the cascade of events leading to the enhanced Th9 development.
Figure 5.
NO-induced up-regulation of Th9 polarization is p53-dependent. CD4+ T cells from WT and p53−/− B6 mice were cultured under Th9 polarizing conditions ± NO. (a) The expression of p53 in WT Th9 cells on day 4 was analyzed by FACS. Concentrations of IL-9 (b) and IL-2 (c) in the culture supernatants of WT and p53−/− cells on day 5 were determined by ELISA. (d, e) Percent of IL-9+ and pSTAT5+ T cells from WT and p53−/− mice polarized under Th9 conditions was analyzed by FACS on day 4. Representative (f) and percent (g) of IL-9+ T cells from WT or p53−/− mice cultured under Th9 conditions in the presence of NO with or without IL-2 are shown. (h) Expression of IRF4 in the Th9 cells from WT and p53−/− mice polarized under Th9 conditions ± NO ± IL-2 was analyzed by FACS. Data are representative or pool of at least 3 experiments, mean ± SEM, n=3, *P<0.05, ***P<0.001 using a 2-way ANOVA.
NO enhances p53 expression by nitrosylation of Mdm2
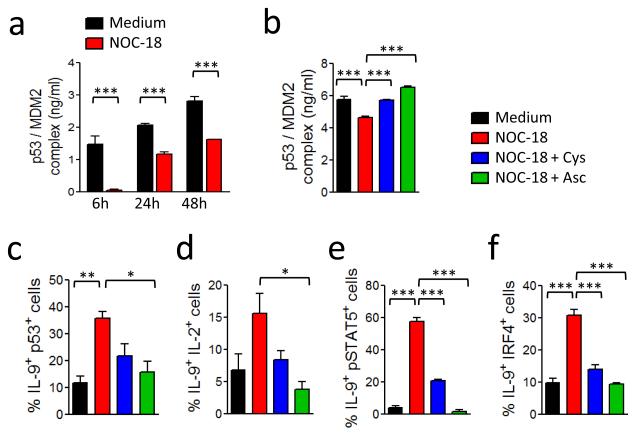
We next investigated the mechanism by which NO enhances the activity of p53. A key feature of the p53 response is p53 stabilization, leading to an increase in p53 steady state levels30,31. In non-stressed cells, the level of p53 is controlled by the mouse double minute 2 homolog (Mdm2). Mdm2, also known as E3 ubiquitin-protein ligase, is a highly conserved protein that in humans is encoded by the HDM2 gene. Mdm2 is an important negative regulator of p53 and functions both as an E3 ubiquitin ligase that recognizes the N-terminal trans-activation domain of the p53 and an inhibitor of p53 transcriptional activation32-34. Hdm2 can also regulate p53 by binding to p53 and inhibiting its transcriptional activity35. It has been demonstrated that NO nitrosylates a critical cysteine residue (C77) of Hdm2 and disrupts Hdm2-p53 binding. We therefore tested the possibility that NO enhances p53 expression by nitrosylation of Mdm2. C57BL/6 CD4+ T cells were polarized under Th9 polarizing conditions in the presence of NOC-18 and the cell lysate assayed for Mdm2-p53 binding by ELISA at various time points. NO markedly reduced Mdm2-p53 binding, evident at 6 h and sustained for at least 48 h (Fig. 6a). To ascertain that NO enhances Th9 differentiation via S-nitrosylation of Mdm2, we added excess amounts of cysteine into Th9 polarization culture. Exogenous cysteine significantly abrogated the NO-mediated inhibition of Mdm2-p53 binding (Fig. 6b, p<0.001). Cysteine also blocked the increase in the frequency of IL-9+p53+ T cells induced by NO (Fig. 6c). To confirm these results we chemically reversed the S-nitrosylation with ascorbate. This mild reductant abrogates the nitrosylation of thiols without affecting the ability to form disulfide bonds36. Ascorbate reversed NO-mediated inhibition of Mdm2-p53 binding (Fig. 6b) and limited the enhancement of the polarization of IL-9+p53+ T cells (Fig. 6c). Furthermore, cysteine and ascorbate also inhibited the increase of the frequency of IL-9+IL-2+ (Fig. 6d), IL9+pSTAT5+ (Fig. 6e) and IL-9+IRF4+ T cells (Fig. 6f). Together, these results demonstrate that NO enhances p53 expression by nitrosylation of cysteine of Mdm2 and blocking Mdm2-p53 binding. Interestingly, NO also enhanced STAT6 phosphorylation (Supplementary Fig. 4a), which was also reversed by cysteine and ascorbate (Supplementary Fig. 4a, b).
Figure 6.
NO enhances Th9 polarization via nitrosylation of MDM2-p53 complex. (a) BALB/c CD4+ T cells were polarized under Th9 conditions ± 100 μM NOC-18. p53/MDM2 complex in the cell extracts were determined by ELISA. (b) CD4+ T cells were polarized as above in the presence of cysteine (1 mM) or ascorbate (300 μM) for 24 h and the cell extracts analyzed for p53/MDM2 complex. Percentage of IL-9+p53+ T cells on day 4 (c,), IL-9+IL-2+ T cells on day 4 (d), IL-9+pSTAT5+ T cells on day 5 (e), and IL-9+IRF4+ T cells (f) on day 4 analyzed by FACS. Data are representative or pooled of at least 3 experiments; mean ± SEM, n=3-5, *P<0.05, **P<0.01, ***P<0.001 using a 2-way ANOVA.
NO enhances the expression of TGFβR and IL-4R
TGFβ and IL-4 are essential for the polarization of Th9 cells2,3,13. Moreover, STAT6 phosphorylation is a key downstream event of IL-4 signaling37-39. We therefore investigated if NO also has an effect on the expression of TGFβR and IL-4Rα on CD4+ T cells during Th9 polarization. Purified CD4+ T cells were cultured under Th9 conditions with or without NOC-18. NO markedly increased the percentage of Th9 cells expressing TGFβR2 (Supplementary Fig. 5a). It should be noted that not all Th9+ cells expressed TGFβR2. This may reflect the dynamic nature of this receptor, being internalized and re-expressed continuously40. The increase of Tgfβr2 and Il4rα mRNA expression was also evident (Supplementary Fig. 4b). These results therefore demonstrate that the direct enhancement of TGFβR and IL-4Rα expression may be an additional pathway by which NO promotes the differentiation of Th9 cells. This observation may explain the low but significant level of enhanced Th9 differentiation in response to NO in the p53−/− cells (Fig. 4b) above.
The role of NO on Th9 cells function in vivo
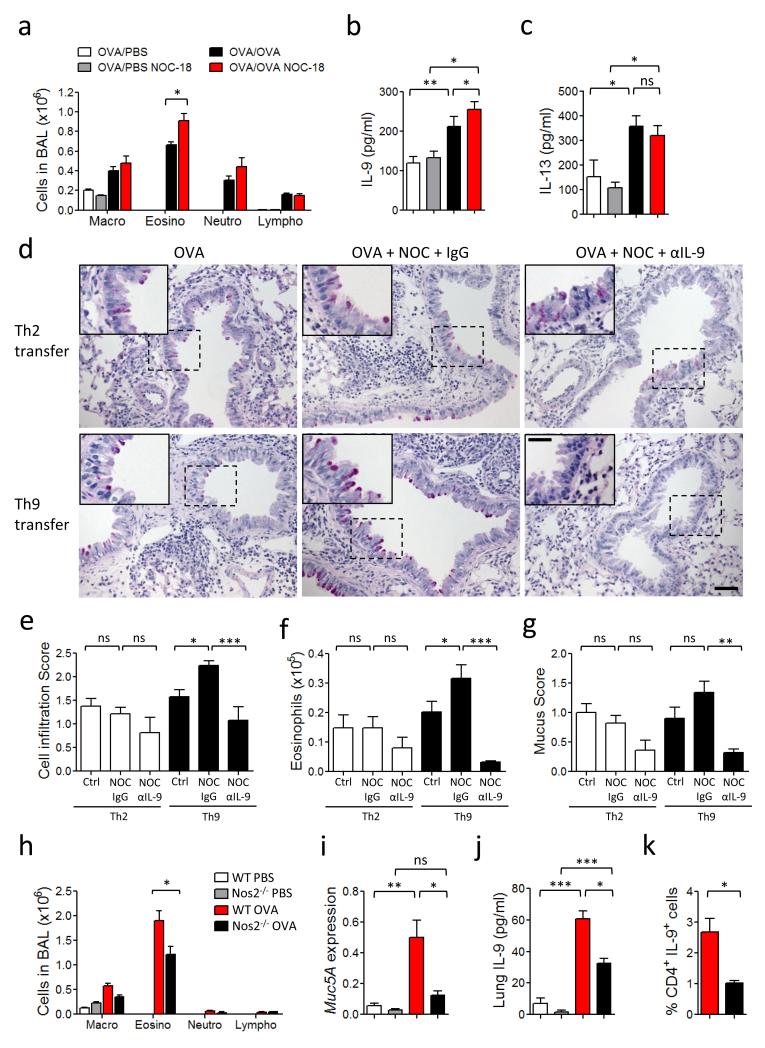
Next we investigated the in vivo relevance of our observation. Th9 cells play an important role in allergic diseases, including airway inflammation4,5,12. We therefore examined the effect of NO in an experimental model of allergic airway inflammation using the OVA priming and challenge system. C57BL/6 mice were immunized intraperitoneally (i.p.) with OVA plus Alum and challenged intranasally (i.n.) with OVA on days 7, 10, 13, 16 and 19. The mice were also injected subcutaneously every other day with NOC-18 (10 mg/kg) or PBS. Mice were sacrificed on day 20 and the bronchoalveolar lavage (BAL) collected for analysis. Mice treated with NOC-18 showed significantly increased eosinophilia in the BAL compared to PBS-treated control mice (Fig. 7a). The BAL fluid from NOC-18-treated mice also contained significantly more IL-9 compared to that of the PBS-treated control mice (Fig. 7b). IL-13 was also detected in the BAL of the OVA-immunized and challenged mice but the level of IL-13 was not affected by NOC-18 treatment indicating that NO has little effect on IL-13 synthesis.
Figure 7.
NO enhances IL-9 production and Th9 frequency and exacerbates allergic airway inflammation in mice. (a-c) OVA-immunized C57BL/6 mice were challenged with OVA and injected s.c. every 2 days with NOC-18. Differential cell counts (a), IL-9 concentration (b) and IL-13 content (c) in the BAL fluid were determined 24 h after the last challenge. (d-g) CD4+ T cells from OT-II mice were polarized to Th9 or Th2 cells and adoptively transferred i.n. to naïve recipient mice which were then challenged i.n. with OVA ± NOC-18. Some mice were injected i.p. with αIL-9 antibody or IgG. PAS staining were performed on lung sections obtained 24 h after the last challenge (d). Scale bars represent 100 μm and 400 μm for the insert. Cell infiltration in the lungs (e), Eosinophils in BAL fluids (f), and mucus score in the lung tissues (g) were measured. (h-k) WT and Nos2−/− mice were immunized s.c. with OVA and challenged i.n. with OVA or PBS. BAL was analyzed by differential cell count (h). Lungs were analyzed for Mucin5A mRNA expression by qPCR (i), and IL-9 concentration by ELISA (j). Percentage of Th9 (CD4+/IL9+) cells in the draining lymph node analyzed by FACS (k). Experiments were performed at least twice; data are mean ± SEM, n=5-6 mice per group; *P<0.05, **P<0.01, ***P<0.001 using 2-way ANOVA.
To confirm the role of NO in Th9 function in this model, we polarized OVA-specific OT-II transgenic CD4+ T cells to Th9 cells in vitro. The polarized cells were >95% IRF4+ and were negative for IL-13, IL-10, IL-4, IFNγ, IL-17, Foxp3, T-bet or RORγt. As control, we also polarized OT-II cells to Th2 cells as described previously21. The cells were adoptively transferred i.n. to naïve C57BL/6 mice, which were then challenged i.n. with OVA and treated i.n. with NOC-18 or PBS. Some mice were also injected i.p. with anti-IL-9 antibody or control IgG. OVA-challenged mice that had received Th9 cells developed symptoms of airway inflammation including increased inflammatory cell infiltration in the lungs (Fig. 7d, PAS staining and Fig. 7e), the number of eosinophils in BAL (Fig. 7f) and mucus production in the lungs (Fig 7g), which were all exacerbated by NO-treatment. Th2 cells also induced eosinophilia, inflammatory cells infiltration and mucus production in the lungs. However, these parameters were not affected by the NOC-18 treatment (Fig. 7d-g). Treatment of recipient mice with a neutralizing anti-IL-9 antibody strongly ameliorated eosinophilia, lung inflammation, mucus production induced by Th9 cells; but this antibody had a only minor effect on the Th2-induced airway inflammation (Fig 7d-g).
To demonstrate an endogenous role of NO in this model, we immunized and challenged Nos2−/− or WT mice with OVA. Nos2−/− mice showed significantly less eosinophilia in the BAL (Fig. 7h) and Mucin 5A expression in the lungs (Fig. 7i) than that of the WT mice. The attenuated airway inflammation in the Nos2−/− mice was accompanied by a significant reduction in the level of IL-9 in the lungs of Nos2−/− mice compared to the WT mice (Fig. 7j). Importantly, OVA-immunized Nos2−/− mice also produced significantly less CD4+IL-9+ T cells in the draining lymph nodes compared to WT mice (Fig. 7k and supplementary Fig. 6a). There was, however, no difference in the level of Nos3 expression between the OVA-immunized and challenged Nos2−/− and WT mice (Supplementary Fig. 6b) showing that Nos2-deficiency did not lead to an elevated Nos3 expression. CD4+ T cells from Nos2−/− and WT mice produced similar percentage of Th9 cells and secreted equivalent amounts of IL-9 when polarized under Th9 conditions in vitro (Supplementary Fig. 6c). These results are consistent with the relatively low level of NO production by T cells, and the notion that in vivo NO is likely provided by other cell types including endothelial and myeloid cells.
Together these result demonstrate that NO plays an important endogenous role in the induction of airway inflammation and that this effect is associated with the enhanced Th9 polarization in vivo.
DISCUSSION
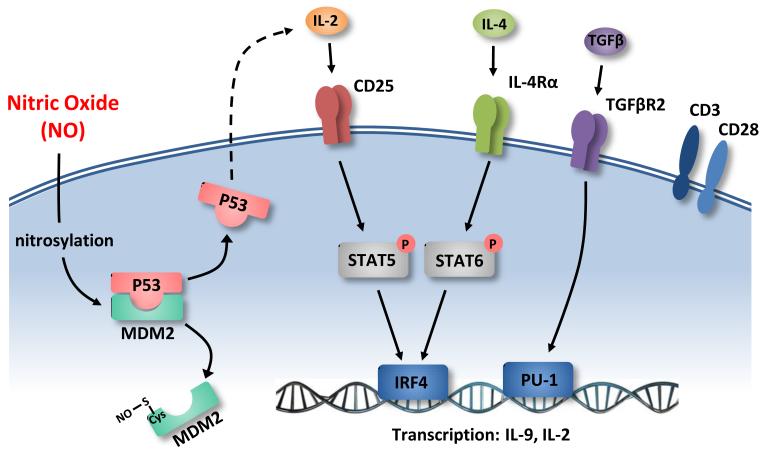
Data presented in this report demonstrate a hitherto unrecognized role of NO in the induction of Th9 cells. This finding expands the regulatory role of NO in the immune system, linking NO to the differentiation and function of the latest subset of T-helper cells and the endogenous effect of NO in airway inflammation. We also provide the molecular mechanism by which NO potentiates the expansion and maintenance of Th9 (Fig. 8). NO nitrosylates cysteine residues of Mdm2, a repressor of p53, thereby reduces the binding of Mdm2 to p53 and thus stabilizes and increases the level of p53, leading to the increased production of IL-2. IL-2, via its receptor CD25, induces the phosphorylation of STAT5, which increases the expression of IRF4 that can enhance IL-9 synthesis by binding to the promoter of I19. pSTAT5 may also bind directly to the promoter of I19 and enhance IL-9 synthesis42.
Figure 8.
Schematic representation of the mechanism of NO-mediated enhancement of Th9 polarization. NO nitrosylates cysteine of MDM2 leading to the release of p53 from the MDM2-p53 complex. p53 induces IL-2 synthesis, which binds to CD25 (IL-2R α-chain) leading to STAT5 phosphorylation that activates IRF4 which binds to the promoter of Il9 for enhanced IL-9 transcription. NO also promotes IL-4Rα expression thus stimulates STAT6 phosphorylation that can also induce IRF4 expression. NO can also enhance TGFβR2 expression leading to the activation of PU.1 that can promote IL-9 transcription by binding to Il9 promoter. NO-mediated Th9 promotion is dependent on conventional TCR and CD28 activation.
Although we have focused on the NO→Mdm2→p53→IL-2-STAT5→IRF4→Th9 axis, other contributing signaling pathways are not excluded. For example, we have shown that NO enhances the expression of TGFβR and I14rα, Sfpi1 (PU.1) and pSTAT6 in Th9 cells. It is thus likely that PU.1 activated by TGFβ could contribute to the promotion of IL-9 synthesis43. Furthermore, the increased IL-4Rα expression could lead to the phosphorylation of STAT6 that enhances IL-9 synthesis by binding directly to the promoter of Il9 (ref. 42) or indirectly via IRF4 (ref. 42, 43). IL-2 has also been reported to activate STAT6 (ref. 44). OX40 signaling has recently been reported to favor the induction of Th9 and airway inflammation45. However, in our experimental system we did not use antigen-presenting cells and thus precluded an effect on OX40. In a previous report29 we showed that NO inhibits Th17 induction by down regulating the aryl hydrocarbon receptor. We show here that NO can elevate the phosphorylation of STAT5 which has been shown to be an inhibitor of IL-17 synthesis46. Thus the induction of pSTAT5 under the Th9 polarizing conditions could be an additional mechanism by which NO suppresses Th17 differentiation.
In an acute (7 days) papain-induced lung inflammation model in mice, Wilhelm et al. reported that IL-9 production was largely restricted to innate lymphoid cells (ILCs)40. IL-9 production by ILCs depended on IL-2 and was rapidly lost in favor of IL-13 and IL-5. In a murine model of allergic inflammation, Jones et al. reported that Th9 cells were rapidly generated in vivo after exposure to HDM12. Interestingly, Th9 cell recruitment to the lung peaked one week after initial allergen administration, before Th2 cell recruitment. In contrast, in a chronic allergen exposure mouse model using the clinically relevant antigen Aspergillus fumigatus to determine the kinetics of Th9 development in vivo, Kerzerho et al. reported that Th9 began to appear in the lungs from day 39 and sustained till at least day 46 (ref. 47). These IL-9-producing CD4+ T cells did not co-express IL-4, IL-13 or IL-10, a phenotype consistent with the murine and human Th9 cells we observed here. Our data show that NO enhances not only the differentiation of Th9 but also the maintenance of Th9 cells.
Although NOS2 is not readily detected in human peripheral blood monocytes, the overall role of NOS2 in humans is not in doubt. NOC-18 at a concentration of 100-200 μM consistently releases 200-400 nM of NO with a half-life of 20 h48. This dose of NO occurs in vivo in sites of infection and inflammation49 and has been used routinely in experiments in vitro29,49. The expression of NOS2 in the site of inflammation has also been extensively reported50,51. Bronchial biopsy specimens from human asthmatic patients show increased expression of Il9 and its receptor compared to healthy controls11,52-54. A humanized monoclonal anti-IL-9 antibody has recently been used in two phase II clinical trials in asthmatics with evidence of clinical activity55. In the present report we demonstrate an endogenous enhancing role of NO in Th9 differentiation and airway inflammation.
NO is detected in the exhaled air of animals and humans 1. A marked increase in the concentration of exhaled NO has been reported in asthmatic patients 2, 3 and an association of NO with asthmatic inflammation has long been recognized 4. Although not widely implemented, fraction of exhaled NO (FeNO) has emerged in recent years as a potential useful, rapid and noninvasive biomarker for assessing airway inflammation, particularly for patients who would be responsive to inhaled corticosteroid treatment 5-7. The mechanism of NO causes severe asthma is hitherto unclear. This is counter-intuitive since NO itself is a bronchodilator 8. Our data indicates that NO may act via promoting the differentiation of Th9 cells. This finding may, at least in part, explain the lack of effectiveness of anti-IL-5 monoclonal antibody (mepolizumab) in treating asthma. Mepolizumab significantly reduces blood and sputum eosinophils without affecting FeNO levels 9.
Data reported here therefore not only reveal a hitherto unrecognized mechanism of Th9 regulation by NO, but also provide a long-sought explanation by which NO exacerbates airway inflammation.
METHODS
Mice
BALB/c and C57BL/6 (B6) mice were obtained from Harlan UK. Nos2−/− mice, DO11.10 mice (BALB/c background) and OT-II mice (B6 background) were originally from the Jackson Laboratory. Irf4−/− mice (B6 background) were as described previously56. Cells from the p53−/− mice (B6 background) were provided by Dr. Ewen McGregor (University of Glasgow). Cells from the Stat5−/− mice (B6 background) were provided by Dr. Arian Laurence (NIH, Bethesda). Nos2−/− mice (B6 background) were obtained from the Jackson laboratory. Male and female mice were used at the age of 6-10 weeks. Cells from BALB/c and C57BL/6 mice gave similar results. All animal experiments were performed according to institutional guidelines in the Ribeirão Preto Medical School, University of São Paulo, Brazil (USP Number. 038/2009) and in accordance with the French ethical and animal experiments regulations in the project approved by the Ethics Committee for Animal Experimentation of CNRS Campus Orleans (N° CLE CCO 2012-050, UMR7355, Orleans, France).
In vitro murine cell cultures
CD4+ T cells were purified from the pool of spleens and lymph nodes of naïve mice by negative selection (routine purity >98%) using an AutoMacs (Miltenyi Biotec). Culture medium was RPMI-1640 supplemented with 10% (vol/vol) FCS (LONZA), 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.05 M 2-mercaptoethanol. For Th17 cell differentiation, 5 × 105 cells/ml were cultured in round-bottom 96-well plates with mitomycin-C-treated spleen cells (antigen-presenting cells, 1:1 ratio APC:T cells), 1 μg/ml soluble anti-CD3, 1 ng/ml TGF-β, 10 ng/ml IL-6, 10 ng/ml IL-1β, 10 μg/ml anti-IFNy and 10 μg/ml anti-IL-4. For Th9 cell differentiation, 1 × 106 cells/ml were cultured for up to 6 days in flat-bottom 96-well plates with 3 μg/ml plate-bound anti-CD3, 1.5 μg/ml soluble anti-CD28, 5 ng/ml TGF-β, 10 ng/ml IL-4 and 10 μg/ml anti-IFNγ. In some experiments, CD4+ T cells were cultured under Th9/17 conditions in flat bottom plate with plate-bound anti-CD3 (3 μg/ml) and anti-CD28 (1.5 μg/ml), 1 ng/ml TGF-β, 10 ng/ml IL-6, 10 ng/ml IL-1β, 10 ng/ml IL-4 and 10 μg/ml anti-IFNγ. NO-donor (NOC-18) was added only once at the beginning of the culture (immediately before cells) at the indicated doses. In some experiments, IL-2 (5 – 20 ng/ml), anti-IL-2 antibody (10 μg/ml), cysteine (1 mM, Sigma) or ascorbate (100-300 μM, Sigma) was also added in the culture. Cultures were incubated at 37°C in 5% CO2 for up to 6 days. At the end of cultures, supernatants were collected for cytokine assay using ELISA and the cells were used for RNA extraction or FACS analysis. Th9 cells polarized under the conditions indicated in the legend to Fig. 6 were harvested at 6-72 h. Cell lysates were prepared for Mdm2-p53 binding assay (Enzo Life Sciences) as per instruction of the manufacturer. Cell division was determined by labeling CD4+ T cells with CFSE (Molecular Probes, Inc.) by flow cytometry at the indicated time points. All cytokines and antibodies were obtained from PeproTech and R&D Systems.
Human cell culture
Peripheral blood was obtained from healthy donors (n=7) with informed consent (WSBTS-12-10). Mononuclear cells were purified by Ficoll-Paque Plus (GE Healthcare), followed by CD4+ T cell isolation using AutoMacs. The cells were cultured (1×106/ml) for 5 days in round-bottom 96 well plates (Costar) with mitomycin C-treated autologous APC (1×106/ml) together with soluble anti-hCD3 (1.5 μg/ml, Invitrogen) and anti-hCD28 (1 μg/ml, Invitrogen) plus hIL-4 (10 ng/ml, R&D System), hTGFβ (2.5 ng/ml, eBioscience), hIL-1β (10 ng/ml, R&D System), anti-hIFNγ (10 ng/ml, R&D System) in the presence of NOC-18. In some experiments, CD4+ T cells were cultured with anti-hCD3/CD28-conjugated Dynabeads (4×107/ml, Invitrogen) instead of APC, with similar results. At the end of the culture, supernatants were harvested for ELISA and the cells were stained for intracellular cytokines.
FACS analysis
For measurement of intracellular cytokines, cells were stimulated for 4 h with PMA and ionomycin (both from Sigma) in the presence of GolgiStop (BD Bioscience). The cells were first stained for CD4, then permeabilized with Perm/Fix solution (eBioscience) and finally stained with antibodies against IL-17A, IL-9, IL-10, p53, pSTAT5, pSTAT6, IRF4, TGFβR2, (eBioscience, Biolegend or R&D System). Isotype-matched rat anti-mouse antibodies (directly conjugated) were used as controls. Antibodies were routinely used in 1/100 dilution for 1 million cells. Data were acquired using a FACSCalibur (BD Biosciences) and analyzed using FlowJo software (Tree Star).
OVA-induced allergic airway inflammation
Mice were sensitized intraperitoneally on day 0 with 50 μg of ovalbumin (OVA, grade V, Sigma-Aldrich) with 1.6 mg of alum adjuvant (Sigma-Aldrich). On days 7, 10, 13, 16 and 19 mice were challenged i.n. with 25 μg of OVA or PBS. In some experiments, NO-donor (NOC-18, 10 mg/kg freshly diluted in PBS) was injected subcutaneously every 2 days. Control mice received the same volume of PBS. In some experiments, WT and Nos2−/− mice were immunized s.c with 10 μg of OVA in PBS on days 1 and 7, and challenged i.n. with 10 μg of OVA in PBS on day 14, 15 and 16. Mice were culled 24 h after the last challenge and serum, BAL, lung and draining lymph nodes were collected for analysis.
Th9 cell transfer
CD4+ T cells purified from OT-II mice were cultured for 2 days with IL-4 (10 ng/ml), TGFβ (5 ng/m), anti-IFNγ (20 μg/ml), plate-bound anti-CD3 (3 μg/ml) and soluble anti-CD28. Th9 cells were harvested, washed with PBS and 2×106 cells were adoptively transferred i.n. into naïve C57BL/6 recipient mice. Mice were challenged i.n. with 10 μg of OVA on day 1, 2 and 3 after adoptive transfer. Simultaneously, mice received i.n. NOC-18 (10 mg/kg) or PBS. In some experiments a neutralizing anti-IL-9 antibody (Clone 222622, R&D Systems) or a control antibody (rat IgG2b, R&D Systems) was given i.p. 30 min before each challenge. Animals were culled 24 h after the last challenge and the extent of airway inflammation assessed in the BAL, lung and mLN. In some experiments, Th2 cells were also transferred and the recipients treated similarly to those given Th9 cells. Th2 cells were generated as described previously21. Briefly CD4+ T cells were cultured for 2 days with plate-bound anti-CD3 and anti-CD28 plus IL-4, anti-IL-12 and anti-IFNγ. The cells were then harvested washed and transferred.
BAL and lung analysis
BAL fluid was obtained by 4 tracheal infusions of 0.5 ml PBS. After centrifugation for 10 min at 1500 rpm, supernatant was stored at −20°C for cytokine analysis and total cells were counted in Turks solution. Cytospin preparations were stained with Wright’s stain (Diff-Quik, Dade Behring), and differential cell counts were obtained based on morphology and staining characteristics. For cytokine detection in total lung, the tissue was homogenized in 50 mM Tris·HCl, pH 7.5, 0.002% Tween 20, and protease inhibitor (Roche Applied Science). After centrifugation, supernatants were collected and stored at −80°C.
Lung histology
Mice were perfused with 10 ml of PBS through the right ventricle. For histological analysis, a piece of lung was fixed overnight in 4% formaldehyde solution and embedded in paraffin. Lung sections (4 μm) were stained with H&E or with periodic acid Schiff reagent (PAS). Histo-pathological assessment was performed blind on randomized sections. Inflammatory changes were graded using a semi-quantitative scale of 0-3 for perivascular and peri-bronchial cell infiltration and mucus production.
Microarray analysis
Total RNA was extracted using Qiagen RNeasy Mini kit (Qiagen), reverse-transcribed, followed by cDNA extraction with a PhaseLock gel (Eppendorf), and precipitated with ethanol and ammonium acetate. cRNA was transcribed with the MEGAscript high yield transcription kit (Ambion). The hybridization cocktail was prepared according to Affymetrix protocols (15 μg fragmented biotin-labeled cRNA spiked with Eukaryotic Hybridization control). The Murine Genome 430A version 2 GeneChip arrays (Affymetrix) were hybridized at 45°C for 16 h, stained with streptavidin-phycoerythrin using the Affymetrix GeneChip Fluidics Workstation 400 and scanned on an Affymetrix GeneChip Scanner 3000 (MG430Av2 arrays). Data were analyzed using the Microarray Suite 5.0 software (Affymetrix) and deposited under the accession code E-MEXP-3959. Microarrays were globally normalized and scaled to a trimmed mean expression value of 200. Fold induction of individual gene between untreated and NO-treated Th17 cells was represented by the ratio of normalized gene expression value in NO-treated cells to value in untreated Th17 cells.
RNA isolation and qPCR
mRNA was isolated 48, 72 and 96 h after Th17 or Th9 differentiation using the RNeasy Micro kit (Qiagen). cDNA was synthesized using 200 ng of total RNA through a reverse transcription reaction kit (Applied Biosystems). qPCR was performed with the Sybr Green Gene Expression Assay using the ABI Prism 7900 Sequence Detection System instrument (Applied Biosystems) according to the manufacturer’s protocol. Target gene expression was calculated using the comparative method for relative quantification after normalization to Hprt gene expression. The sequences for primers are as follows: Hprt: forward, 5′-GCA GTA CAG CCC CAA AAT GG-3′, reverse, 5′-AAC AAA GTC TGG CCT GTA TCC AA-3′; I19: forward, 5′ - CTG ATG ATT GTA CCA CAC CGT GC-3′, reverse, 5′- GCC TTT GCA TCT CTG TCT TCT GG- 3′; I14ra: forward, 5′- GAG TGA GTG GAG TCC TAG CAT C-3′, reverse, 5′- GCT GAA GTA ACA GAA CAG GC-3′; I12: forward, 5′- CCT GAG CAG GAT GGA GAA TTA CA -3′, reverse, 5′- TCC AGA ACA TGC CGC AGA G-3′; TgfbrII: forward, 5′- CCG CTG CAT ATC GTC CTG TG- 3′, reverse, 5′- AGT GGA TGG ATG GTC CTA TTA CA-3′; Sfpi1: forward, 5′-GAT GGA GAA GCT GAT GGC TTG G-3′, reverse, 5′-TTC TTC ACC TCG CCT GTC TTG C-3′. In some experiments, Nos3 and Mucin5A expression in the lungs of WT and Nos2−/− mice immunised with OVA and challenged with OVA or PBS (described above) were analyzed by qPCR. Total RNA from the lungs was extracted using Trizol reagent and analysed with the Syber green method as above. Primers sequences are: Gapdh: forward 5, GGG TGT GAA CCA CGA GAA AT 3′, reverse 5′ CCT TCC ACA ATG CCA AAG TT 3′; Mucin5A: forward 5′ AAA GAC ATA GTC ACT CAG CAA 3′, reverse 5′ CTG GGA AGT TCA GTG TCA AAC CA 3′; Nos3: forward 5′ GGG AAA GCT GCA GGT ATT TGA T 3′, reverse 5′ CAC TGT GAT GGC TGA ACG AAG A 3′.
ELISA
ELISA was carried out with paired antibodies according to the manufacturer’s instructions: mouse IL-17A, IL-9, IL-2, and human IL-9 (all from BD Bioscience). The sensitivity was <50 pg/ml. For the Mdm2-p53 assay, ELISA kit (Enzo Life Sciences) was used according to the manufacturer’s instruction. Murine IL-9 assay was also performed using a mouse Single-plex Cytokine Detection System (Biorad) according to the manufacturer’s instructions. The plates were analyzed on a Bio-Rad Bio-Plex Luminex 100 plate reader.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 5.0. Comparisons between 2 groups were performed using a 2-tailed unpaired Student’s t test. Multiple groups were compared using a 2-way ANOVA followed by a Bonferroni’s post-test. Values for all measurements are expressed as mean ± SEM, p<0.05 was considered statistically significant. All experiments were performed at least two times.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Medical Research Council, UK and the Wellcome Trust (to FYL), and the Deutsche Forschungsgemeinschaft Grant SCHM 1014/5-1 (to T.B., and E.S.).
Footnotes
CONFLICT OF INTEREST
The authors declared no conflict of interest.
REFERENCES
- 1.O’Shea JJ, Hunter CA, Germain RN. T cell heterogeneity: firmly fixed, predominantly plastic or merely malleable? Nat Immunol. 2008;9:450–453. doi: 10.1038/ni0508-450. [DOI] [PubMed] [Google Scholar]
- 2.Dardalhon V, et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(−) effector T cells. Nat Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veldhoen M, et al. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 4.Chang HC, et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol. 2010;11:527–534. doi: 10.1038/ni.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staudt V, et al. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity. 2010;33:192–202. doi: 10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Faulkner H, Humphreys N, Renauld JC, Van Snick J, Grencis R. Interleukin-9 is involved in host protective immunity to intestinal nematode infection. Eur J Immunol. 1997;27:2536–2540. doi: 10.1002/eji.1830271011. [DOI] [PubMed] [Google Scholar]
- 7.Faulkner H, Renauld JC, Van Snick J, Grencis RK. Interleukin-9 enhances resistance to the intestinal nematode Trichuris muris. Infect Immun. 1998;66:3832–3840. doi: 10.1128/iai.66.8.3832-3840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan WI, et al. Modulation of intestinal muscle contraction by interleukin-9 (IL-9) or IL-9 neutralization: correlation with worm expulsion in murine nematode infections. Infect Immun. 2003;71:2430–2438. doi: 10.1128/IAI.71.5.2430-2438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Temann UA, Geba GP, Rankin JA, Flavell RA. Expression of interleukin 9 in the lungs of transgenic mice causes airway inflammation, mast cell hyperplasia, and bronchial hyperresponsiveness. J Exp Med. 1998;188:1307–1320. doi: 10.1084/jem.188.7.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Temann UA, Ray P, Flavell RA. Pulmonary overexpression of IL-9 induces Th2 cytokine expression, leading to immune pathology. J Clin Invest. 2002;109:29–39. doi: 10.1172/JCI13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimbara A, et al. IL-9 and its receptor in allergic and nonallergic lung disease: increased expression in asthma. J Allergy Clin Immunol. 2000;105:108–115. doi: 10.1016/s0091-6749(00)90185-4. [DOI] [PubMed] [Google Scholar]
- 12.Jones CP, Gregory LG, Causton B, Campbell GA, Lloyd CM. Activin A and TGF-beta promote T(H)9 cell-mediated pulmonary allergic pathology. J Allergy Clin Immunol. 2012;129:1000–1010. e1003. doi: 10.1016/j.jaci.2011.12.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitt E, et al. IL-9 production of naïve CD4+ T cells depends on IL-2, is synergistically enhanced by a combination of TGF-beta and IL-4, and is inhibited by IFN-gamma. J Immunol. 1994;153:3989–3996. [PubMed] [Google Scholar]
- 14.Helmby H, Grencis RK. Interleukin 1 plays a major role in the development of Th2-mediated immunity. Eur J Immunol. 2004;34:3674–3681. doi: 10.1002/eji.200425452. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt E, et al. IL-1 serves as a secondary signal for IL-9 expression. J Immunol. 1991;147:3848–3854. [PubMed] [Google Scholar]
- 16.Angkasekwinai P, Chang SH, Thapa M, Watarai H, Dong C. Regulation of IL-9 expression by IL-25 signaling. Nat Immunol. 2010;11:250–256. doi: 10.1038/ni.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogdan C, Rollinghoff M, Diefenbach A. The role of nitric oxide in innate immunity. Immunol Rev. 2000;173:17–26. doi: 10.1034/j.1600-065x.2000.917307.x. [DOI] [PubMed] [Google Scholar]
- 18.Liew FY. Regulation of lymphocyte functions by nitric oxide. Curr Opin Immunol. 1995;7:396–399. doi: 10.1016/0952-7915(95)80116-2. [DOI] [PubMed] [Google Scholar]
- 19.Moncada S, Higgs A. The L-arginine-nitric oxide pathway. N Engl J Med. 1993;329:2002–2012. doi: 10.1056/NEJM199312303292706. [DOI] [PubMed] [Google Scholar]
- 20.Nathan CF, Hibbs JB., Jr. Role of nitric oxide synthesis in macrophage antimicrobial activity. Curr Opin Immunol. 1991;3:65–70. doi: 10.1016/0952-7915(91)90079-g. [DOI] [PubMed] [Google Scholar]
- 21.Niedbala W, et al. Regulation of type 17 helper T-cell function by nitric oxide during inflammation. Proc Natl Acad Sci U S A. 2011;108:9220–9225. doi: 10.1073/pnas.1100667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank DA, Robertson MJ, Bonni A, Ritz J, Greenberg ME. Interleukin 2 signaling involves the phosphorylation of Stat proteins. Proc Natl Acad Sci U S A. 1995;92:7779–7783. doi: 10.1073/pnas.92.17.7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaffen SL, et al. Signaling through the interleukin 2 receptor beta chain activates a STAT-5-like DNA-binding activity. Proc Natl Acad Sci US A. 1995;92:7192–7196. doi: 10.1073/pnas.92.16.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilmour KC, Pine R, Reich NC. Interleukin 2 activates STAT5 transcription factor (mammary gland factor) and specific gene expression in T lymphocytes. Proc Natl Acad Sci U SA. 1995;92:10772–10776. doi: 10.1073/pnas.92.23.10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou J, Schindler U, Henzel WJ, Wong SC, McKnight SL. Identification and purification of human Stat proteins activated in response to interleukin-2. Immunity. 1995;2:321–329. doi: 10.1016/1074-7613(95)90140-x. [DOI] [PubMed] [Google Scholar]
- 26.Lin JX, et al. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity. 1995;2:331–339. doi: 10.1016/1074-7613(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 27.Yao W, et al. Interleukin-9 is required for allergic airway inflammation mediated by the cytokine TSLP. Immunity. 2013;38:360–372. doi: 10.1016/j.immuni.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niedbala W, et al. Nitric oxide preferentially induces type 1 T cell differentiation by selectively up-regulating IL-12 receptor beta 2 expression via cGMP. Proc Natl Acad Sci U S A. 2002;99:16186–16191. doi: 10.1073/pnas.252464599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niedbala W, et al. Nitric oxide induces CD4+CD25+ Foxp3 regulatory T cells from CD4+CD25 T cells via p53, IL-2, and OX40. Proc Natl Acad Sci U S A. 2007;104:15478–15483. doi: 10.1073/pnas.0703725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oren M. Regulation of the p53 tumor suppressor protein. J Biol Chem. 1999;274:36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- 31.Woods DB, Vousden KH. Regulation of p53 function. Exp Cell Res. 2001;264:56–66. doi: 10.1006/excr.2000.5141. [DOI] [PubMed] [Google Scholar]
- 32.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 33.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 34.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 35.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 36.Forrester MT, Foster MW, Stamler JS. Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J Biol Chem. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 38.Shimoda K, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 39.Takeda K, et al. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 40.Wilhelm C, et al. An IL-9 fate reporter demonstrates the induction of an innate IL-9 response in lung inflammation. Nat Immuno1. 2011;12:1071–1077. doi: 10.1038/ni.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salmond RJ, et al. IL-33 induces innate lymphoid cell-mediated airway inflammation by activating mammalian target of rapamycin. J Allergy Clin Immunol. 2012;130:1159–1166. e1156. doi: 10.1016/j.jaci.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang XO, et al. The signaling suppressor CIS controls proallergic T cell development and allergic airway inflammation. Nat Immunol. 2013;14:732–740. doi: 10.1038/ni.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goswami R, Kaplan MH. Gcn5 is required for PU.1-dependent IL-9 induction in Th9 cells. J Immunol. 2012;189:3026–3033. doi: 10.4049/jimmunol.1201496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bessoles S, et al. IL-2 triggers specific signaling pathways in human NKT cells leading to the production of pro- and anti-inflammatory cytokines. J Leukoc Biol. 2008;84:224–233. doi: 10.1189/jlb.1007669. [DOI] [PubMed] [Google Scholar]
- 45.Xiao X, et al. OX40 signaling favors the induction of T(H)9 cells and airway inflammation. Nat Immunol. 2012;13:981–990. doi: 10.1038/ni.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laurence A, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 47.Kerzerho J, et al. Programmed cell death ligand 2 regulates TH9 differentiation and induction of chronic airway hyperreactivity. J Allergy Clin Immunol. 2013;131:1048–1057. e1041–1042. doi: 10.1016/j.jaci.2012.09.027. 1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keefer LK, Nims RW, Davies KM, Wink DA. “NONOates” (1-substituted diazen-1-ium-1,2-diolates) as nitric oxide donors: convenient nitric oxide dosage forms. Methods Enzymol. 1996;268:281–293. doi: 10.1016/s0076-6879(96)68030-6. [DOI] [PubMed] [Google Scholar]
- 49.Macphail SE, et al. Nitric oxide regulation of human peripheral blood mononuclear cells: critical time dependence and selectivity for cytokine versus chemokine expression. J Immuno1. 2003;171:4809–4815. doi: 10.4049/jimmunol.171.9.4809. [DOI] [PubMed] [Google Scholar]
- 50.Okuda Y, Sakoda S, Fujimura H, Yanagihara T. Nitric oxide via an inducible isoform of nitric oxide synthase is a possible factor to eliminate inflammatory cells from the central nervous system of mice with experimental allergic encephalomyelitis. J Neuroimmunol. 1997;73:107–116. doi: 10.1016/s0165-5728(96)00194-4. [DOI] [PubMed] [Google Scholar]
- 51.Staykova MA, Paridaen JT, Cowden WB, Willenborg DO. Nitric oxide contributes to resistance of the Brown Norway rat to experimental autoimmune encephalomyelitis. Am J Pathol. 2005;166:147–157. doi: 10.1016/S0002-9440(10)62240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hohlfeld JM, Schmiedl A, Erpenbeck VJ, Venge P, Krug N. Eosinophil cationic protein alters pulmonary surfactant structure and function in asthma. J A1lergy Clin Immunol. 2004;113:496–502. doi: 10.1016/j.jaci.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Toda M, Tulic MK, Levitt RC, Hamid Q. A calcium-activated chloride channel (HCLCA1) is strongly related to IL-9 expression and mucus production in bronchial epithelium of patients with asthma. J Allergy Clin Immunol. 2002;109:246–250. doi: 10.1067/mai.2002.121555. [DOI] [PubMed] [Google Scholar]
- 54.Ying S, Meng Q, Kay AB, Robinson DS. Elevated expression of interleukin-9 mRNA in the bronchial mucosa of atopic asthmatics and allergen-induced cutaneous late-phase reaction: relationships to eosinophils, mast cells and T lymphocytes. Clin Exp Allergy. 2002;32:866–871. doi: 10.1046/j.1365-2222.2002.01376.x. [DOI] [PubMed] [Google Scholar]
- 55.Parker JM, et al. Safety profile and clinical activity of multiple subcutaneous doses of MEDI-528, a humanized anti-interleukin-9 monoclonal antibody, in two randomized phase 2a studies in subjects with asthma. BMC Pulm Med. 2011;11:14. doi: 10.1186/1471-2466-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robbins RA, et al. Expression of inducible nitric oxide in human lung epithelial cells. Biochem Biophys Res Commun. 1994;203:209–218. doi: 10.1006/bbrc.1994.2169. [DOI] [PubMed] [Google Scholar]
- 57.Kharitonov SA, et al. Increased nitric oxide in exhaled air of asthmatic patients. Lancet. 1994;343:133–135. doi: 10.1016/s0140-6736(94)90931-8. [DOI] [PubMed] [Google Scholar]
- 58.Persson MG, Zetterstrom O, Agrenius V, Ihre E, Gustafsson LE. Single-breath nitric oxide measurements in asthmatic patients and smokers. Lancet. 1994;343:146–147. doi: 10.1016/s0140-6736(94)90935-0. [DOI] [PubMed] [Google Scholar]
- 59.Barnes PJ, Liew FY. Nitric oxide and asthmatic inflammation. Immunol Today. 1995;16:128–130. doi: 10.1016/0167-5699(95)80128-6. [DOI] [PubMed] [Google Scholar]
- 60.Hahn PY, Morgenthaler TY, Lim KG. Use of exhaled nitric oxide in predicting response to inhaled corticosteroids for chronic cough. Mayo Clin Proc. 2007;82:1350–1355. doi: 10.4065/82.11.1350. [DOI] [PubMed] [Google Scholar]
- 61.Smith AD, et al. Exhaled nitric oxide: a predictor of steroid response. Am J Respir Crit Care Med. 2005;172:453–459. doi: 10.1164/rccm.200411-1498OC. [DOI] [PubMed] [Google Scholar]
- 62.Smith AD, et al. Diagnosing asthma: comparisons between exhaled nitric oxide measurements and conventional tests. Am J Respir Crit Care Med. 2004;169:473–478. doi: 10.1164/rccm.200310-1376OC. [DOI] [PubMed] [Google Scholar]
- 63.Hogman M, Frostell CG, Hedenstrom H, Hedenstierna G. Inhalation of nitric oxide modulates adult human bronchial tone. Am Rev Respir Dis. 1993;148:1474–1478. doi: 10.1164/ajrccm/148.6_Pt_1.1474. [DOI] [PubMed] [Google Scholar]
- 64.Haldar P, et al. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med. 2009;360:973–984. doi: 10.1056/NEJMoa0808991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.