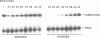Abstract
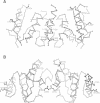
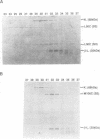
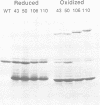
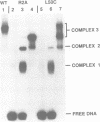
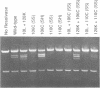
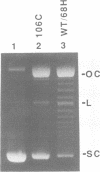
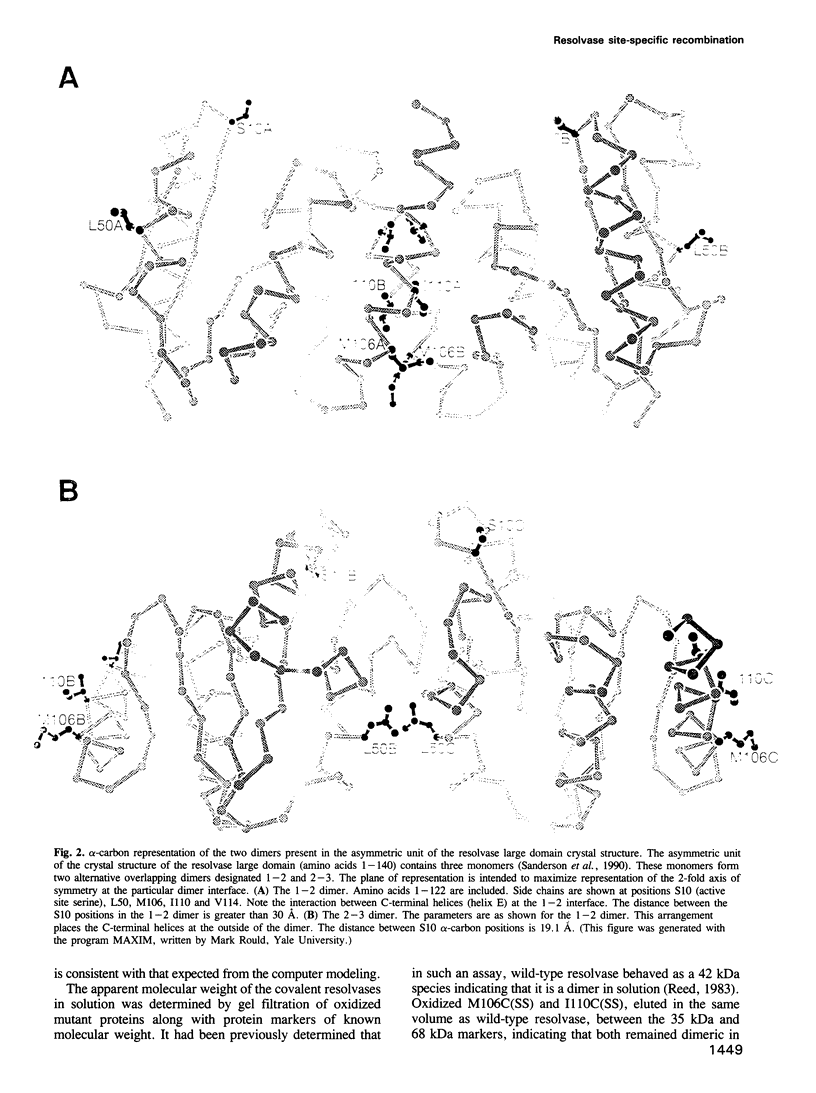
Recombination catalyzed by the gamma delta resolvase requires assembly of a nucleo-protein complex, the synaptosome, whose structure is determined by resolvase-res and resolvase-resolvase interactions. In crystals of the resolvase catalytic domain, monomers of resolvase were closely associated with one another across three different dyad axes; one of these subunit contacts was shown to be an essential inter-dimer interaction. To investigate the relevance of the remaining two interfaces, we have made site-directed mutations at positions suggested by the structure. Cysteine substitutions were designed to link the interfaces covalently, mutations to arginine were used to disrupt intersubunit contacts, and mutations to tryptophan were used to study the hydrophobicity and solvent accessibility of potential interfaces by fluorescence quenching. Characterization of the mutant proteins has allowed us to identify the dimer interface of resolvase and to assign a structural role to a second intersubunit contact. The data presented here, together with our previous results, suggest that all three of the dyad-related intersubunit interactions observed in the crystal play specific roles in synapsis and recombination.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdel-Meguid S. S., Grindley N. D., Templeton N. S., Steitz T. A. Cleavage of the site-specific recombination protein gamma delta resolvase: the smaller of two fragments binds DNA specifically. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2001–2005. doi: 10.1073/pnas.81.7.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981 Apr 30;290(5809):754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Bednarz A. L., Boocock M. R., Sherratt D. J. Determinants of correct res site alignment in site-specific recombination by Tn3 resolvase. Genes Dev. 1990 Dec;4(12B):2366–2375. doi: 10.1101/gad.4.12b.2366. [DOI] [PubMed] [Google Scholar]
- Benevides J. M., Weiss M. A., Thomas G. J., Jr Design of the helix-turn-helix motif: nonlocal effects of quaternary structure in DNA recognition investigated by laser Raman spectroscopy. Biochemistry. 1991 May 7;30(18):4381–4388. doi: 10.1021/bi00232a003. [DOI] [PubMed] [Google Scholar]
- Benjamin H. W., Cozzarelli N. R. Geometric arrangements of Tn3 resolvase sites. J Biol Chem. 1990 Apr 15;265(11):6441–6447. [PubMed] [Google Scholar]
- Benjamin H. W., Cozzarelli N. R. Isolation and characterization of the Tn3 resolvase synaptic intermediate. EMBO J. 1988 Jun;7(6):1897–1905. doi: 10.1002/j.1460-2075.1988.tb03023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R. G., Roderick S. L., Takeda Y., Matthews B. W. Protein-DNA conformational changes in the crystal structure of a lambda Cro-operator complex. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8165–8169. doi: 10.1073/pnas.87.20.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Carra J. H., Schleif R. F. Variation of half-site organization and DNA looping by AraC protein. EMBO J. 1993 Jan;12(1):35–44. doi: 10.1002/j.1460-2075.1993.tb05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröge P., Hatfull G. F., Grindley N. D., Cozzarelli N. R. The two functional domains of gamma delta resolvase act on the same recombination site: implications for the mechanism of strand exchange. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5336–5340. doi: 10.1073/pnas.87.14.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman E. H., Stasiak A. Structure of helical RecA-DNA complexes. Complexes formed in the presence of ATP-gamma-S or ATP. J Mol Biol. 1986 Oct 20;191(4):677–697. doi: 10.1016/0022-2836(86)90453-5. [DOI] [PubMed] [Google Scholar]
- Falvey E., Hatfull G. F., Grindley N. D. Uncoupling of the recombination and topoisomerase activities of the gamma delta resolvase by a mutation at the crossover point. Nature. 1988 Apr 28;332(6167):861–863. doi: 10.1038/332861a0. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Lauth M. R., Wells R. G., Wityk R. J., Salvo J. J., Reed R. R. Transposon-mediated site-specific recombination: identification of three binding sites for resolvase at the res sites of gamma delta and Tn3. Cell. 1982 Aug;30(1):19–27. doi: 10.1016/0092-8674(82)90007-1. [DOI] [PubMed] [Google Scholar]
- Haffter P., Bickle T. A. Enhancer-independent mutants of the Cin recombinase have a relaxed topological specificity. EMBO J. 1988 Dec 1;7(12):3991–3996. doi: 10.1002/j.1460-2075.1988.tb03287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G. F., Grindley N. D. Analysis of gamma delta resolvase mutants in vitro: evidence for an interaction between serine-10 of resolvase and site I of res. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5429–5433. doi: 10.1073/pnas.83.15.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G. F., Noble S. M., Grindley N. D. The gamma delta resolvase induces an unusual DNA structure at the recombinational crossover point. Cell. 1987 Apr 10;49(1):103–110. doi: 10.1016/0092-8674(87)90760-4. [DOI] [PubMed] [Google Scholar]
- Hubbard A. J., Bracco L. P., Eisenbeis S. J., Gayle R. B., Beaton G., Caruthers M. H. Role of the Cro repressor carboxy-terminal domain and flexible dimer linkage in operator and nonspecific DNA binding. Biochemistry. 1990 Oct 2;29(39):9241–9249. doi: 10.1021/bi00491a019. [DOI] [PubMed] [Google Scholar]
- Hughes R. E., Hatfull G. F., Rice P., Steitz T. A., Grindley N. D. Cooperativity mutants of the gamma delta resolvase identify an essential interdimer interaction. Cell. 1990 Dec 21;63(6):1331–1338. doi: 10.1016/0092-8674(90)90428-h. [DOI] [PubMed] [Google Scholar]
- Kanaar R., Klippel A., Shekhtman E., Dungan J. M., Kahmann R., Cozzarelli N. R. Processive recombination by the phage Mu Gin system: implications for the mechanisms of DNA strand exchange, DNA site alignment, and enhancer action. Cell. 1990 Jul 27;62(2):353–366. doi: 10.1016/0092-8674(90)90372-l. [DOI] [PubMed] [Google Scholar]
- Klippel A., Cloppenborg K., Kahmann R. Isolation and characterization of unusual gin mutants. EMBO J. 1988 Dec 1;7(12):3983–3989. doi: 10.1002/j.1460-2075.1988.tb03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnow M. A., Cozzarelli N. R. Site-specific relaxation and recombination by the Tn3 resolvase: recognition of the DNA path between oriented res sites. Cell. 1983 Apr;32(4):1313–1324. doi: 10.1016/0092-8674(83)90312-4. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Parker C. N., Halford S. E. Dynamics of long-range interactions on DNA: the speed of synapsis during site-specific recombination by resolvase. Cell. 1991 Aug 23;66(4):781–791. doi: 10.1016/0092-8674(91)90121-e. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Grindley N. D. Transposon-mediated site-specific recombination in vitro: DNA cleavage and protein-DNA linkage at the recombination site. Cell. 1981 Sep;25(3):721–728. doi: 10.1016/0092-8674(81)90179-3. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Moser C. D. Resolvase-mediated recombination intermediates contain a serine residue covalently linked to DNA. Cold Spring Harb Symp Quant Biol. 1984;49:245–249. doi: 10.1101/sqb.1984.049.01.028. [DOI] [PubMed] [Google Scholar]
- Reed R. R. The resolvase protein of the transposon gamma delta. Methods Enzymol. 1983;100:191–196. doi: 10.1016/0076-6879(83)00055-5. [DOI] [PubMed] [Google Scholar]
- Reed R. R. Transposon-mediated site-specific recombination: a defined in vitro system. Cell. 1981 Sep;25(3):713–719. doi: 10.1016/0092-8674(81)90178-1. [DOI] [PubMed] [Google Scholar]
- Rimphanitchayakit V., Grindley N. D. Saturation mutagenesis of the DNA site bound by the small carboxy-terminal domain of gamma delta resolvase. EMBO J. 1990 Mar;9(3):719–725. doi: 10.1002/j.1460-2075.1990.tb08165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Ho Y. S., Shatzman A. The use of pKc30 and its derivatives for controlled expression of genes. Methods Enzymol. 1983;101:123–138. doi: 10.1016/0076-6879(83)01009-5. [DOI] [PubMed] [Google Scholar]
- Salvo J. J., Grindley N. D. The gamma delta resolvase bends the res site into a recombinogenic complex. EMBO J. 1988 Nov;7(11):3609–3616. doi: 10.1002/j.1460-2075.1988.tb03239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson M. R., Freemont P. S., Rice P. A., Goldman A., Hatfull G. F., Grindley N. D., Steitz T. A. The crystal structure of the catalytic domain of the site-specific recombination enzyme gamma delta resolvase at 2.7 A resolution. Cell. 1990 Dec 21;63(6):1323–1329. doi: 10.1016/0092-8674(90)90427-g. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Hehir K., Stearman R. S., Weiss M. A., Jeitler-Nilsson A., Suchanek E. G., Pabo C. O. An engineered intersubunit disulfide enhances the stability and DNA binding of the N-terminal domain of lambda repressor. Biochemistry. 1986 Oct 7;25(20):5992–5998. doi: 10.1021/bi00368a024. [DOI] [PubMed] [Google Scholar]
- Shirakawa M., Matsuo H., Kyogoku Y. Intersubunit disulfide-bonded lambda-Cro protein. Protein Eng. 1991 Jun;4(5):545–552. doi: 10.1093/protein/4.5.545. [DOI] [PubMed] [Google Scholar]
- Smith D. L., Johnson A. D. A molecular mechanism for combinatorial control in yeast: MCM1 protein sets the spacing and orientation of the homeodomains of an alpha 2 dimer. Cell. 1992 Jan 10;68(1):133–142. doi: 10.1016/0092-8674(92)90212-u. [DOI] [PubMed] [Google Scholar]
- Stark W. M., Sherratt D. J., Boocock M. R. Site-specific recombination by Tn3 resolvase: topological changes in the forward and reverse reactions. Cell. 1989 Aug 25;58(4):779–790. doi: 10.1016/0092-8674(89)90111-6. [DOI] [PubMed] [Google Scholar]
- Story R. M., Weber I. T., Steitz T. A. The structure of the E. coli recA protein monomer and polymer. Nature. 1992 Jan 23;355(6358):318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- Wasserman S. A., Dungan J. M., Cozzarelli N. R. Discovery of a predicted DNA knot substantiates a model for site-specific recombination. Science. 1985 Jul 12;229(4709):171–174. doi: 10.1126/science.2990045. [DOI] [PubMed] [Google Scholar]
- Wells R. G., Grindley N. D. Analysis of the gamma delta res site. Sites required for site-specific recombination and gene expression. J Mol Biol. 1984 Nov 15;179(4):667–687. doi: 10.1016/0022-2836(84)90161-x. [DOI] [PubMed] [Google Scholar]