Abstract
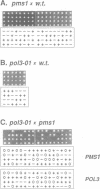
Mutation of predicted 3'-->5' exonuclease active site residues of Saccharomyces cerevisiae POL3 DNA polymerase (delta) or deletion of the PMS1 mismatch repair gene lead to relative (to wild type) spontaneous mutation rates of approximately 130 and 41, respectively, measured at a URA3 reporter gene inserted near to a defined replication origin. The POL3 exonuclease-deficient mutant pol3-01 generated most classes of single base mutation in URA3, indicating a broad specificity that generally corresponds to that of the PMS1 system. pol3-01 pms1 haploid cells ceased growth after a few divisions with no unique terminal cell morphology. A pol3-01/pol3-01 pms1/pms1 diploid was viable and displayed an estimated URA3 relative mutation rate of 2 x 10(4), which we calculate to be catastrophically high in a haploid. The relationship between the relative mutation rates of pol3-01 and pms1 was multiplicative, indicating action in series. The PMS1 transcript showed the same cell cycle periodicity as those of a set of DNA replication genes that includes POL3, suggesting PMS1 is co-regulated with these genes. We propose that the POL3 3'-->5' exonuclease and the PMS1 mismatch repair system act on a common pathway analogous to the dnaQ-->mutHLS pathway of DNA replication error correction in Escherichia coli.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Araki H., Ropp P. A., Johnson A. L., Johnston L. H., Morrison A., Sugino A. DNA polymerase II, the probable homolog of mammalian DNA polymerase epsilon, replicates chromosomal DNA in the yeast Saccharomyces cerevisiae. EMBO J. 1992 Feb;11(2):733–740. doi: 10.1002/j.1460-2075.1992.tb05106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. K., Williamson M. S., Fogel S., Kolodner R. D. The role of heteroduplex correction in gene conversion in Saccharomyces cerevisiae. Nature. 1987 Jul 23;328(6128):362–364. doi: 10.1038/328362a0. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Chung D. W., Zhang J. A., Tan C. K., Davie E. W., So A. G., Downey K. M. Primary structure of the catalytic subunit of human DNA polymerase delta and chromosomal location of the gene. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11197–11201. doi: 10.1073/pnas.88.24.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Drake J. W. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M. G., Petes T. D. Most of the yeast genomic sequences are not essential for cell growth and division. Cell. 1986 Sep 26;46(7):983–992. doi: 10.1016/0092-8674(86)90697-5. [DOI] [PubMed] [Google Scholar]
- Gordon C. B., Campbell J. L. A cell cycle-responsive transcriptional control element and a negative control element in the gene encoding DNA polymerase alpha in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6058–6062. doi: 10.1073/pnas.88.14.6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes J., Jr, Clark S., Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. H., Lowndes N. F. Cell cycle control of DNA synthesis in budding yeast. Nucleic Acids Res. 1992 May 25;20(10):2403–2410. doi: 10.1093/nar/20.10.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Williamson M. S., Fogel S. Heteroduplex DNA correction in Saccharomyces cerevisiae is mismatch specific and requires functional PMS genes. Mol Cell Biol. 1989 Oct;9(10):4432–4440. doi: 10.1128/mcb.9.10.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer W., Kramer B., Williamson M. S., Fogel S. Cloning and nucleotide sequence of DNA mismatch repair gene PMS1 from Saccharomyces cerevisiae: homology of PMS1 to procaryotic MutL and HexB. J Bacteriol. 1989 Oct;171(10):5339–5346. doi: 10.1128/jb.171.10.5339-5346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G. S., Savage E. A., Ritzel R. G., von Borstel R. C. The base-alteration spectrum of spontaneous and ultraviolet radiation-induced forward mutations in the URA3 locus of Saccharomyces cerevisiae. Mol Gen Genet. 1988 Nov;214(3):396–404. doi: 10.1007/BF00330472. [DOI] [PubMed] [Google Scholar]
- Lichten M., Goyon C., Schultes N. P., Treco D., Szostak J. W., Haber J. E., Nicolas A. Detection of heteroduplex DNA molecules among the products of Saccharomyces cerevisiae meiosis. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7653–7657. doi: 10.1073/pnas.87.19.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes N. F., Johnson A. L., Johnston L. H. Coordination of expression of DNA synthesis genes in budding yeast by a cell-cycle regulated trans factor. Nature. 1991 Mar 21;350(6315):247–250. doi: 10.1038/350247a0. [DOI] [PubMed] [Google Scholar]
- McIntosh E. M., Atkinson T., Storms R. K., Smith M. Characterization of a short, cis-acting DNA sequence which conveys cell cycle stage-dependent transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jan;11(1):329–337. doi: 10.1128/mcb.11.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Bell J. B., Kunkel T. A., Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3'----5' exonuclease activity. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Sugino A. Nucleotide sequence of the POL3 gene encoding DNA polymerase III (delta) of Saccharomyces cerevisiae. Nucleic Acids Res. 1992 Jan 25;20(2):375–375. doi: 10.1093/nar/20.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon C. S., Lipchitz L. R., Collins I., Deshpande A., Devenish R. J., Green R. P., Klein H. L., Palzkill T. G., Ren R. B., Synn S. Analysis of a circular derivative of Saccharomyces cerevisiae chromosome III: a physical map and identification and location of ARS elements. Genetics. 1991 Oct;129(2):343–357. doi: 10.1093/genetics/129.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quah S. K., von Borstel R. C., Hastings P. J. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics. 1980 Dec;96(4):819–839. doi: 10.1093/genetics/96.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. E., McCarroll R. M., Newlon C. S., Fangman W. L. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989 Oct;9(10):4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M., Grisafi P., Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M. Mechanisms of mutagenesis in the Escherichia coli mutator mutD5: role of DNA mismatch repair. Proc Natl Acad Sci U S A. 1988 Nov;85(21):8126–8130. doi: 10.1073/pnas.85.21.8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Giot L., Faye G. The 3' to 5' exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991 Aug;10(8):2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. C., Roberts J. D., Kunkel T. A. Heteroduplex repair in extracts of human HeLa cells. J Biol Chem. 1991 Feb 25;266(6):3744–3751. [PubMed] [Google Scholar]
- Varlet I., Radman M., Brooks P. DNA mismatch repair in Xenopus egg extracts: repair efficiency and DNA repair synthesis for all single base-pair mismatches. Proc Natl Acad Sci U S A. 1990 Oct;87(20):7883–7887. doi: 10.1073/pnas.87.20.7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Borstel R. C. Measuring spontaneous mutation rates in yeast. Methods Cell Biol. 1978;20:1–24. doi: 10.1016/s0091-679x(08)62005-1. [DOI] [PubMed] [Google Scholar]
- Williamson M. S., Game J. C., Fogel S. Meiotic gene conversion mutants in Saccharomyces cerevisiae. I. Isolation and characterization of pms1-1 and pms1-2. Genetics. 1985 Aug;110(4):609–646. doi: 10.1093/genetics/110.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. H., Clarke C. H., Marinus M. G. Specificity of Escherichia coli mutD and mutL mutator strains. Gene. 1990 Mar 1;87(1):1–5. doi: 10.1016/0378-1119(90)90488-d. [DOI] [PubMed] [Google Scholar]
- Zhang J., Chung D. W., Tan C. K., Downey K. M., Davie E. W., So A. G. Primary structure of the catalytic subunit of calf thymus DNA polymerase delta: sequence similarities with other DNA polymerases. Biochemistry. 1991 Dec 24;30(51):11742–11750. doi: 10.1021/bi00115a002. [DOI] [PubMed] [Google Scholar]