Abstract
Epithelial to mesenchymal transition (EMT) is activated during cancer invasion and metastasis, enriches for cancer stem cells (CSCs), and contributes to therapeutic resistance and disease recurrence. Signal transduction kinases play a pivotal role as chromatin-anchored proteins in eukaryotes. Here we report for the first time that protein kinase C-theta (PKC-θ) promotes EMT by acting as a critical chromatin-anchored switch for inducible genes via transforming growth factor β (TGF-β) and the key inflammatory regulatory protein NF-κB. Chromatinized PKC-θ exists as an active transcription complex and is required to establish a permissive chromatin state at signature EMT genes. Genome-wide analysis identifies a unique cohort of inducible PKC-θ-sensitive genes that are directly tethered to PKC-θ in the mesenchymal state. Collectively, we show that cross talk between signaling kinases and chromatin is critical for eliciting inducible transcriptional programs that drive mesenchymal differentiation and CSC formation, providing novel mechanisms to target using epigenetic therapy in breast cancer.
INTRODUCTION
Epithelial to mesenchymal transition (EMT) is a key step in cancer progression and the process of metastasis that creates a reservoir for cancer stem cells (CSCs) and is associated with highly aggressive traits (1–4). As well as driving metastasis, these CSCs, or “precursor” metastatic cells, play a pivotal role in therapeutic resistance and relapse in breast cancer patients (5, 6). Breast CSCs are a distinct subpopulation of mesenchymal cells that possess several important features, namely, expression of key surface markers (CD44high and CD24low) (7), a distinct transcriptome (3), the ability to form spherical colonies in suspension cultures (termed mammospheres) (8), and enhanced resistance to chemotherapy (9, 10) and ionizing radiation (11–14).
Eukaryotes utilize the chromatin landscape as its epigenetic template within the nucleus of living cells in order to promote inducible gene transcription in response to environmental signals. Highly compacted chromatin structures are enriched in nucleosomes and are transcriptionally silent, and a net loss of nucleosomes from gene-specific regulatory regions increases chromatin accessibility and initiates context-specific transcriptional programs. In addition to ATP-dependent chromatin remodeling and exchange of histone variants with canonical histones, histone modifications are thought to alter gene expression, by changing chromatin structure and/or by providing a platform that promotes binding of transcriptional regulators (15–23).
Novel classes of chromatin-associated enzymes that play critical roles in modulating chromatin structure within the human genome have recently been discovered. In particular, signaling kinases can act as chromatin regulators of inducible gene transcription in both higher and lower eukaryotes by two distinct mechanisms: relaying signals from the cytoplasm to the nucleus and direct association with chromatin-bound transcription complexes at activated target genes in the nucleus (24–28). For example, the evolutionarily conserved protein kinase C (PKC) family protein PKC-θ translocates to the nucleus to directly influence both inducible immune responsive gene transcription and the microRNAs essential for an effective immune response in T cells (29).
Dysregulation of PKC-θ has been directly implicated in inflammatory disorders (30–32), tumor progression, and metastasis and has been recently linked to aggressive breast cancers (33–35). Elucidating the chromatin-associated role of PKC-θ in EMT and CSC processes will be a crucial step to unraveling the molecular events that contribute to cancer metastasis. To date, there is no information regarding the cross talk between signaling kinases and chromatin for eliciting transcriptional programs that drive mesenchymal differentiation and CSC formation. Here we report that PKC-θ is essential for EMT and the formation of CSCs in breast cancer. Specifically, we show that chromatinized PKC-θ acts as a critical molecular switch for inducible genes unique to mesenchymal cells and whose expression is also elevated in CSCs. Our findings show that the PKC pathway cooperates with the transforming growth factor β (TGF-β) pathway and collaborates with the NF-κB pathway to promote a distinct transcriptional program of inducible EMT and CSC signature genes. Using genome-wide analysis, a distinct cohort of inducible PKC-θ-sensitive genes that are directly tethered to chromatinized PKC-θ in the mesenchymal state is identified. Several of the genes within this cluster are known regulators of EMT and cancer progression, suggesting that chromatin-anchored PKC-θ is a novel upstream regulator of genes that drive EMT and cancer. This represents a novel chromatin-regulatory mechanism that operates in EMT and CSCs and shows that cross talk between the signaling kinases and chromatin axis in the mesenchymal state exists.
MATERIALS AND METHODS
Cell culture and separation of CSCs from NCSCs.
The adherent human mammary adenocarcinoma cell lines MCF-7 and MDA-MB-231 were cultured in low-glucose Dulbecco's modified Eagle medium (DMEM; Gibco) and supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, and 0.1% penicillin-streptomycin-neomycin (PSN) antibiotics. Cells were stimulated with 0.65 ng/ml of phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich) or 20 ng/ml of TGF-β (R&D systems) for the times indicated. Fluorescence-activated cell sorting (FACS) was performed on single-cell suspensions stained with anti-CD44-APC (559942; BD Biosciences) and anti-CD24-PE antibodies (555428; BD Biosciences) along with Hoechst 33258 (561908; BD Biosciences) to monitor cell viability as previously detailed (3). The FACS gating strategy used in this study is adopted and modified from the pioneer breast CSC publications (7), which sorted cells based on the CD44high/CD24low phenotype, shown to be associated with human breast CSCs (7, 36, 37), and are commonly used as CSC surface markers. A flow cytometry gating strategy was always included in the analysis to exclude dead cells (Hoechst-negative cells).
Immortalized human mammary epithelial (HMLE) cells were obtained from Robert Weinberg (Whitehead Institute, MIT) and cultured in serum-free HuMEC ready medium (Gibco). EMT was induced by culturing cells in DMEM–F-12 (1:1) supplemented with 5% fetal calf serum, 20 ng/ml epidermal growth factor (EGF; R&D systems), 0.5 mg/ml hydrocortisone (Sigma), 10 μg/ml insulin (Novo Nordisk Pharmaceuticals), and 2 ng/ml TGF-β (R&D systems) for 15 days, with cells passaged every 3 to 4 days. Affymetrix human gene arrays were performed on parental HMLE and TGF-β-induced HMLE cells that had undergone a full EMT (termed HMLE inducible model [HMLE-IM] cells).
Mammosphere assay.
Mammosphere culture medium components were purchased from StemCell Technologies, and mammosphere assays were performed as recommended by the manufacturer. Briefly, 40,000 cells/2 ml cell dilutions were prepared, and 2 ml of cells was seeded in 6-well ultralow-attachment, flat-bottomed plates (Costar). Mammospheres larger than 60 μm were counted in each well on day 7, and images were taken.
Transfection conditions.
Transfection was performed as previously described (29, 38). Human PKC-θ small interfering RNA (siRNA) (sc-36252), p50 siRNA (sc-44211), p65 siRNA (sc-44212), and mock siRNA A (fluorescein conjugate) (sc-36869) were purchased from Santa Cruz Biotechnologies, and forward transfections were performed with 20 nM siRNA using Lipofectamine 2000 (Invitrogen).
Patients and tumor samples.
Whole sections of formalin-fixed paraffin-embedded (FFPE) postsurgical breast specimens and clinical data were used in accordance with ethics approval ETHLR 13.041 (ACT Health Human Research Ethics Committee). Normal breast tissues were breast reduction mammoplasty specimens from women with no history of breast cancer. Patients' disease status, hormone receptor status, and treatment information were obtained from the Australian Capital Territory and South East New South Wales Breast Cancer Treatment Group (ACT & SE NSW BCTG).
Immunohistochemistry.
Immunohistochemistry was performed using the Bond automated system (Vision Biosystems) according to a standard protocol as previously described (39). Intracellular staining of cell lines was performed as detailed previously (29, 38).
Antibodies used for intracellular staining.
The primary antibodies used were anti-laminin-5 (MAB 1949; Millipore) and antifibronectin (MAB 1926; Millipore) followed by secondary antibodies conjugated to Alexa Fluor 488 (A11001; Life Technology) or Alexa Fluor 568 (A10037; Life Technology).
PKC activity assay.
PKC-θ and PKC-β activity assays (Enzo Life Sciences; DI-EKS-420A) were performed according to the manufacturer's protocol and as previously described (38).
NF-κB activity assay.
The TransAM NF-κB family kit was used for NF-κB activity assays (43296; Active Motif). Assays were performed according to the manufacturer's guidelines. Five micrograms of protein from MDA-MB231- or MCF-IM-derived whole-cell, nuclear, and cytoplasmic extracts was used in triplicate for the assays. Raji nuclear extract was used as a positive control. Wild-type and mutated consensus oligonucleotides were also used as a negative and a positive control, respectively.
Immunoblot analysis.
Immunoblot analysis was performed according to the manufacturer's protocol and as previously described (19) with anti-PKC-θ (sc-212; Santa Cruz), anti-p65 (ab7970; Abcam), anti-phospho (ser-468)-p65 (3039; Cell Signaling), anti-phospho (ser-536)-p65 (3031; Cell Signaling), and RNA polymerase (Pol) II(c-21) (ab817; Abcam) antibodies.
GSEA.
Gene set enrichment analysis (GSEA) was performed using default parameters, except that gene set perturbation was used, and the gene ranking was performed based on differences between the classes. The gene sets used were from the chemical and genetic permutations group (c2 cgp version 4) MSigDB (40). Sets were considered enriched if the false-discovery rate (FDR) was less than 25% with a nominal P value of less than 0.01.
Total RNA isolation and qRT-PCR.
Total RNA was extracted, and reverse transcription-quantitative PCR (qRT-PCR) was performed as previously described (22). The TaqMan microRNA reverse transcription kit (ABI 4366596) was used for microRNAs, and the data were normalized to RNU6B as previously described (29).
ChIP, sequential ChIP, and FAIRE assays.
Chromatin immunoprecipitation (ChIP) assays were performed according to the protocol supplied by Upstate Biotechnology, as previously detailed (29). In all ChIP assays, a no-antibody IP control and an isotype-specific control antibody were included to ensure that only specific enrichment was being monitored. ChIP samples were subsequently used for SYBR green real-time PCR (Sigma). Standard curves were generated for each primer set to correct for differences in primer efficiency. The human interleukin-2 (IL-2) promoter region, a gene not expressed in our EMT/CSC models, was routinely included as a negative control. ChIP enrichment ratios were calculated such that only enrichment above the negative control was considered specific protein binding, as described previously (22). Briefly, cycle threshold (CT) values from the real-time PCR amplification plots were first converted to arbitrary copies. Sample data were next normalized to the corresponding total input. Fold changes above the no-antibody control were then calculated to generate the ChIP enrichment ratio. Sequential ChIP assays were performed as described previously (29). Formaldehyde-associated isolation of regulatory elements (FAIRE) assays were performed as previously described, and the DNA was subsequently utilized for SYBR green real-time PCR (41). To compare the relative FAIRE-enriched DNA copy number for each amplicon across samples, the CT value was converted to arbitrary copies and then normalized to the reference gene, PPIA, as an endogenous control for chromatin accessibility. All assay results represent means ± standard errors (SE) from three independent experiments.
Antibodies and promoter primers used for ChIP, sequential ChIP, and FAIRE assays.
The antibodies used for ChIP, sequential ChIP, and FAIRE assays were anti-PKC-θ (sc-212; Santa Cruz), anti-PKC-θ phospho s695 (ab75658; Abcam), anti-Pol II (c-21) (ab817; Abcam), anti-Zeb-1 (sc-10573; Santa Cruz), anti-p50 (sc-1191; Santa Cruz), and anti-p65 (ab7970; Abcam). The promoter primers used for RT-PCR were human CD44 (Fwd, TGAGCTCTCCCTCTTTCCAC; Rev, TTGGATATCCTGGGAGAGGA), PLAUR (Fwd, GGGAAGCAAAGCAAGGGTTA; Rev, GTTTTGTCAGGAGGGATACTGG), and IL-6 (Fwd, CTCACCCTCCAACAAAGATTT; Rev, CAGAATGAGCCTCAGACATC).
ChIP-seq.
Ten nanograms of PKC-θ-immunoprecipitated ChIP-DNA and the corresponding total input (TI) were used to prepare the ChIP-sequencing (ChIP-seq) DNA library with the NEBNext ChIP-Seq Library Prep master mix set for Illumina (NEB number E6240L; New England BioLabs Inc.) according to the manufacturer's instructions. ChIP-DNA size was selected with AMPure XP beads (Beckman Coulter, Inc., part number A63881). The selected DNA was repaired with NEBNext end repair enzyme mix and buffer, followed by treatment with Klenow fragment (3′→5′ exo−) to generate “A” base overhangs. Subsequent adaptor ligation of the dA-tailed DNA was performed with 1.5 μM NEBNext adaptor primers, Quick Ligation reaction buffer, and Quick T4 DNA ligase. AMPure beads were used to isolate library fragments ranging between 175 and 225 bp in size, and the purified DNA was combined with the NEBNext High-Fidelity 2X PCR master mix, 25 μM Universal PCR primer, and 25 μM index primer for multiplexing purposes (NEBNext multiplex oligonucleotides for Illumina, NEB number E7335L; New England BioLabs Inc.). A total of 15 cycles were used for PCR amplification of the adaptor-ligated DNA. The resultant ChIP-DNA library was subjected to DNA purification with AMPure beads. The quality of the ChIP-DNA library was assessed on a Bioanalyzer. A 10-fold serial dilution of the library was prepared for RT-PCR with qPCR primers 1.1 and 1.2 to check the amplification efficiency of the ChIP DNA library. A total of 2 nM was captured on the Illumina flow cell for cluster generation. The ChIP DNA library was sequenced on the Illumina HiSeq2000 using a single-read 50-bp run at the Ramaciotti Centre for Gene Function Analysis, University of New South Wales, Sydney, Australia.
Bioinformatics for ChIP sequencing.
DNA sequences from bp −1000 to +500 relative to the transcription start site (TSS) of each gene (promoter analysis) or of 1,000 bp centered on the ChIP-seq peak were obtained from the University of California—Santa Cruz (UCSC) genome browser (Hg19). The MatInspector and Match programs were used to scan the sequences for position weight matrices (PWM) for differential transcription factor binding sites (42). Both the core and matrix similarity score cutoffs for the PWMs were set to minimize false-positive and -negative rates. For promoter analysis, Fisher's exact test was used to determine statistically overrepresented sites in the gene cohorts compared to a random set of 974 promoters. For ChIP-seq, Z-scores were used to determine which motifs were overrepresented compared to promoter regions and genomic regions.
Reads were first trimmed of adapter sequences using Cutadapt and then mapped to the human genome (Hg19) using local alignment in Bowtie 2 (43). Only reads that uniquely mapped to the genome were used for subsequent analysis. Duplicate reads were removed using Picard (http://picard.sourceforge.net), and enriched regions were called against the relevant total input samples using a P value cutoff of 0.05 for the zero-inflated negative binomial algorithm (ZINBA) (44) and a posterior probability cutoff of 0.999 for BayesPeak (45). For representation in UCSC, reads were extended by 200 bp, and HTSeq was used to create wig files. The number of reads per peak was normalized to the reads per million (uniquely) mapped reads. The nearest Ensembl transcript to the peaks was determined using ChIPpeakAnno (46).
Microarray analysis.
Microarray studies were performed on MCF-IM and HMLE-IM cells. Briefly, MCF-7 cells were either nonstimulated or stimulated with PMA for 60 h; stimulated cells were sorted by FACS for CSCs and NCSCs. HMLE cells were stimulated with TGF-β as previously described (3). Additional studies were carried out on MCF-7 cells nonstimulated or stimulated with PMA for 60 h in the presence of mock siRNA or PKC-θ siRNA.
Hugene 1.0 microarrays were normalized using the robust multiarray average (RMA) and then loess in R utilizing only the main probes, with small RNAs and uncharacterized probe sets removed. Hugene 2.0 arrays were RMA normalized using Affymetrix Powertools, and only “main” and “rescue” probe sets were used for analysis. Differences of log2 (0.5) (1.4-fold) were used as cutoff for expression changes unless otherwise stated. Heat maps were generated using the Heatplus library in R and clustered with complete linkage and Euclidean distances.
Gene ontology (GO) enrichment analysis was performed using DAVID (47) with a Benjamini P value cutoff of 0.05 and a requirement for the GO group to have at least 10 genes in the query list. The query list was compared to either all genes on the relevant microarray (13,261 genes) or a sample of human genes (13,528 genes). Replicate arrays were not performed, but extensive validation was undertaken by quantitative RT-PCR.
Statistical analysis.
All comparisons between treated and untreated samples were performed using the two-tailed nonparametric Mann-Whitney test. For multiple comparisons, the Friedman test with Dunn's multiple-comparisons posttest was used. Data are expressed as means ± standard errors (SE).
Microarray data accession number.
Microarray and ChIP sequencing data are deposited in GEO under accession number GSE53335.
RESULTS
The PKC pathway orchestrates a distinct inducible gene expression program in EMT and CSCs.
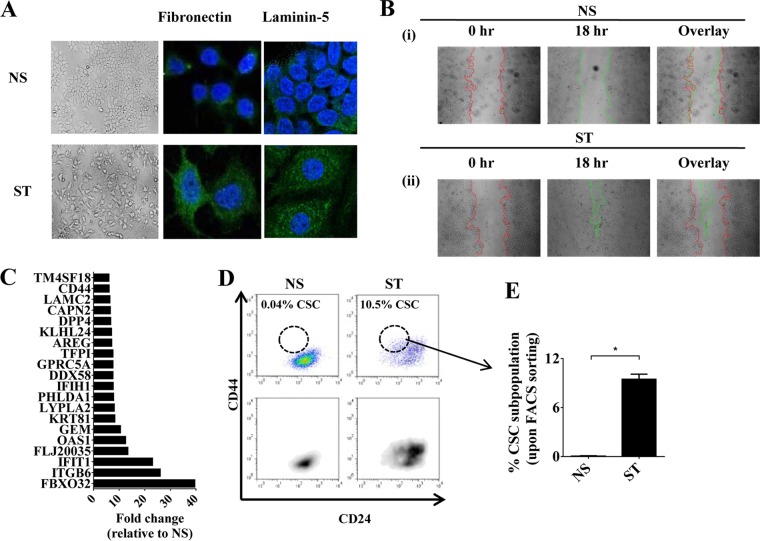
To determine to what extent the PKC pathway contributes to the induction of EMT in breast cancer, phorbol 12-myristate 13-acetate (PMA), a known PKC pathway inducer (48, 49), was used to induce EMT in the human epithelial MCF-7 breast cancer cell line. Consistent with previous reports in a prostate cancer cell line (50), treatment of MCF-7 cells with PMA induced morphological changes of EMT, with loss of cell-cell contacts, spindle cell morphology, increased laminin-5 and fibronectin staining (Fig. 1A), and increased motility (Fig. 1B). There was rapid induction of the EMT-related genes PLAUR, LAMC2, ZEB1, ITGB6, FN1, SNAI1, and SNAI2 (see Fig. S1A and B in the supplemental material) and a reduction in CDH1 and microRNAs MIR200B and MIR200C within 24 to 60 h poststimulation (see Fig. S1A and C in the supplemental material). An optimized dose of PMA was utilized, such that it resulted in maximal EMT with minimal cell death (see Fig. S1D in the supplemental material). It should be noted that the PMA concentration used in this study is much lower than commonly used PMA concentrations either in T cells (29) or in previous studies that showed EMT induction by PMA (20 ng/ml) (50). Here, the PMA-induced EMT in MCF-7 cells is referred to as the MCF-7-inducible model (MCF-IM). In this model, epithelial cells are denoted as nonstimulated (NS) and mesenchymal cells induced by PMA denoted as stimulated (ST).
FIG 1.
The PKC pathway results in a distinct inducible gene expression program in EMT and CSCs. (A) Phase-contrast and confocal microscopy with antibody staining for antifibronectin or anti-laminin-5 (green) and DAPI (4′,6-diamidino-2-phenylindole; nuclear stain; blue) of MCF-7 IM cells, nonstimulated (NS) or PMA stimulated (ST). (B) Wound healing assay at 0 h (red line) and 18 h (green line) of MCF-IM cells as indicated above. (C) Top 20 genes in MCF-IM cells via microarray analysis. Data depicted as fold changes relative to NS. (D) FACS analysis of CD44high/CD24low CSCs. (E) Percent CD44high/CD24low CSCs from panel D. (F) Mammosphere assay of MCF-IM. (G) mRNA expression measured by real-time PCR. mRNA levels are expressed as arbitrary copy numbers; fold changes relative to NS are shown above error bars. (H) H3K4me3 ChIP across the CD44 promoter. Data represented as ChIP enrichment ratios relative to no-antibody control. (I) Top 60 genes expressed in CD44high/CD24low CSCs. Blue, low expression; red, high expression. (J) Percentages of CD44high/CD24low CSCs induced with PMA, TGF-β, or both, determined by by FACS analysis. (K) mRNA expression by real-time PCR analysis of samples as described for panel J. (L) Enrichment of genes upregulated by TGF-β in embryonic fibroblasts in the list of genes ranked from higher (red) to lower (blue) in MCF-IM. All results either represent the means ± the standard errors from three independent experiments or (for panels A and B) are the individual results from a representative experiment of three replicates (n = 3). **, P < 0.01; *, P < 0.05; ns, not significant.
MCF-IM cells treated with PMA for 60 h induced a distinct gene expression microarray profile, with the top 20 most highly induced genes including the EMT genes LAMC2 and ITGB6 and the key CSC marker gene, CD44 (Fig. 1C; see Table S1 in the supplemental material). The microarray data were successfully validated for a subset of transcripts by quantitative real-time PCR (see Fig. S1E and F in the supplemental material). Formation of a CD44high and CD24low CSC subpopulation (here referred to as CD44high/CD24low CSCs) was confirmed by FACS (Fig. 1D and E) and mammosphere assays (Fig. 1F). PMA stimulation generated three other subpopulations, namely, CD44low/CD24low, CD44low/CD24high, and CD44high/CD24high, which we collectively termed non-CSCs (or NCSCs) (see Fig. S1G in the supplemental material). In addition, data suggested that continuous induction of the PKC pathway by PMA is required for the CD44high/CD24low CSC phenotype (see Fig. S1H to J in the supplemental material), and the FACS-sorted population of NCSCs could not be converted into CSCs (see Fig. S1K in the supplemental material), which suggested that non-CSCs in this model do not have the ability to gain CSC phenotype despite prolonged stimulation.
To investigate the transcriptional characteristics of MCF-IM cells, induced CD44high/CD24low CSCs and NCSCs were FACS sorted to high purity (>95%) (see Fig. S1L and M in the supplemental material) and a subset of EMT genes were analyzed by quantitative real-time PCR. EMT-associated genes, such as CD44, LAMC2, and PLAUR, displayed increased abundance in CSCs compared to NCSCs, while others, such as FN1 and ITGB6, did not (Fig. 1G). Consistent with increased expression of CD44 in the CSC subset, the active chromatin mark H3K4me3 was selectively enriched on the CD44 promoter in CSCs (Fig. 1H). CD24 transcript expression remained as low in CD44high/CD24low CSCs as in nonstimulated cells, while in the NCSC population CD24 expression was higher than in nonstimulated cells (see Fig. S1N in the supplemental material). Therefore, a cohort of EMT-associated genes are preferentially expressed in the CD44high/CD24low CSCs; the top 60 genes displayed increased abundance in CSCs compared to NCSCs, and nonstimulated (NS) MCF-7 cells are depicted in Fig. 1I. In addition, the data also showed that CSCs, but not NCSCs, express key “stemness” factors such as MYC and KLF4 (see Fig. S1O in the supplemental material). Together, these data suggest that the PKC pathway is involved in the induction of two distinct inducible gene expression programs: (i) the mesenchymal-enriched gene program, which is induced in both CSCs and NCSCs; and (ii) the CD44high/CD24low CSC-enriched gene program, which is preferentially induced in CSCs.
The TGF-β pathway has a well-established role in EMT and CD44high/CD24low CSC formation, so we examined whether the TGF-β and PKC pathways cooperate to induce CSCs. TGF-β enhanced PMA-induced CD44high/CD24low CSC formation in MCF-7 cells but did not induce EMT or CSC generation alone (Fig. 1J); this was confirmed by transcript analysis, which showed increased expression of CD44, ALDH, and IL-6 (Fig. 1K), suggesting cooperative gene regulation by the two pathways. To ascertain a global picture of the cooperation between the PKC and TGF-β pathways, we compared the genes induced by PMA to those known to be associated with EMT and/or TGF-β induction. Genes induced by PMA include COL6A1, SLCO2A1, PALM, DCHS1, SCARB2, LIMS2, LTBP3, COL6A2, SERPINE1, SERPINE2, ITGAV, CRIP2, FLNC, and LOXL2, which are associated with EMT (51), and NRP1, TUBA1A, OLFML3, BIN1, NEBL, THY1, UGDH, C10orf56, LTBP2, TPM1, RGL1, MYL9, and PPAP2B, which are associated with TGF-β induction (52) (see Table S1 in the supplemental material). Gene set enrichment analysis (GSEA) (40) showed that 651 and 592 gene sets were enriched in the PMA-treated cells or CSCs, respectively, compared to the untreated cells, with a P value of <0.01 and an FDR of <25% (see Tables S2 and S3 in the supplemental material). These included several lists of TGF-β targets such as that from Plasari et al. (53) and Verrecchia et al. (54), an EMT-upregulated gene list (55), and genes upregulated in mammary stem cells (56) (Fig. 1L; see also Fig. S1P and Q in the supplemental material). Furthermore, GSEA also showed that many of the top enriched sets were genes upregulated during inflammatory processes (see Tables S2 and S3 and Fig. S2R and S in the supplemental material), such as the interferon pathway (57–59), the tumor necrosis factor (TNF) pathway (60), and growth factors such as the EGF pathway (61). Thus, PMA induction of the PKCs operates via key inflammatory processes previously shown to induce EMT (62–65) and cooperates with the TGF-β pathway.
Taken together, our findings illustrate that induction of the PKC pathway elicits EMT and CD44high/CD24low CSC formation to enhance a distinct program of inducible EMT signature genes.
PKC-θ, a member of a novel subfamily of PKCs, is essential for the induction of key EMT and CD44high/CD24lowCSC signature genes.
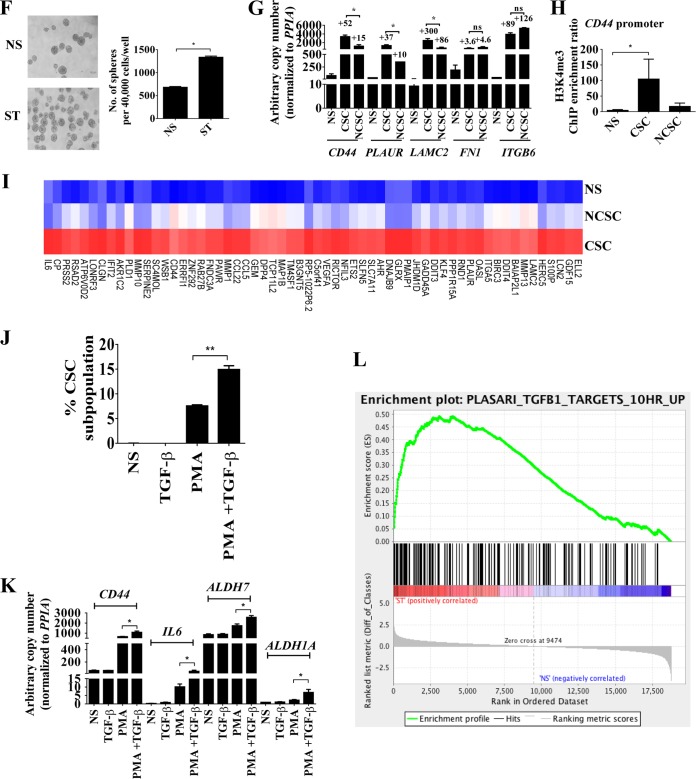
Our results thus far indicate that PKC family members may play a role in eliciting inducible gene expression programs during EMT and CSC formation. Furthermore, our previous T cell study hypothesized a role for PKC-θ in EMT, since ChIP-on-ChIP promoter analysis in Jurkat T cells suggested direct tethering of this kinase to genes involved with migration (29). To address the extent to which the PKC-θ pathway contributes to PKC-driven inducible gene expression during EMT, we performed global transcript analysis of mock- and PKC-θ siRNA-treated MCF-IM cells using a previously validated pool of five PKC-θ-specific 19- to 25-nucleotide (nt) siRNAs (Santa Cruz) (29). Effective PKC-θ knockdown was confirmed by Western blotting with anti-PKC-θ antibody (see Fig. S2A in the supplemental material) and ChIP (see Fig. S2B in the supplemental material). One thousand three hundred fifty-six (40%) of the genes that were induced by PMA-mediated activation of PKC were inhibited by PKC-θ knockdown (Fig. 2A). The microarray data were successfully validated for a subset of transcripts by quantitative real-time PCR (see Fig. S2C and D in the supplemental material). The “inducible PKC-θ-sensitive gene cluster” (Fig. 2A; see also Table S4 in the supplemental material) included EMT signature genes such as SERPINE1, FN1, HSPG2, ILIRL1, TM4SF1, ITGAV, SERPINE2, SLCO2A1, EMP1, LOXL2, IFI27, EDN1, TNFRSF21, CD59, AFRN, and ANXA2 (52). Approximately 15% of the genes in this cluster also displayed increased abundance in HMLE-IM cells (see Table S4 in the supplemental material). Additionally, microarray profiling showed that 17.8% of genes from this cohort were enriched in CSCs compared to NCSCs (see Table S4 in the supplemental material), and quantitative real-time PCR confirmed that the key CSC signature genes CD44, PLAUR, IL-6, and ALDH were highly dependent on PKC-θ (Fig. 2B). In further support of the key role of PKC-θ in CSCs, knockdown of PKC-θ using siRNAs in MCF-IM showed reduction in the CSC subpopulation (Fig. 2C), mammosphere formation (Fig. 2D), and increased MIR200C expression (see Fig. S2E in the supplemental material). Furthermore, a PKC-θ-specific peptide inhibitor, which effectively inhibited PKC-θ activity (Fig. 2E), consistent with previous reports (66), also abolished EMT and CD44high/CD24low CSC formation as assessed by morphology (Fig. 2F), FACS (Fig. 2G), and transcript analysis (Fig. 2H) in MCF-IM. This was further verified using PKC-θ short hairpin RNA (shRNA) in the MDA-MB-231 model, which predominantly contains CD44high/CD24low CSCs (67) (see Fig. S2F and G in the supplemental material).
FIG 2.
Nuclear PKC-θ is required for the induction of key EMT and CSC signature genes. (A) Venn diagram of microarray analysis depicting “inducible PKC-θ-sensitive gene cluster” by comparison of “PKC pathway induced genes” with “PKC-θ siRNA-sensitive genes.” Genes with a log2 (0.5)-fold difference were considered induced. (B) mRNA expression by real-time PCR in MCF-IM following transfection with either mock siRNA or PKC-θ siRNA and subsequently either nonstimulated (NS) or PMA stimulated (ST) for 60 h. mRNA expression changes are presented as fold changes (FC) relative to NS. (C) FACS analysis of CD44high/CD24low CSCs as described for panel B. (D) Mammosphere assay of mRNA under conditions as described for panel B. (E) PKC enzyme-linked immunosorbent assay (ELISA)-based kinase assays for PKC-θ peptide inhibitor. Relative kinase activity was calculated after subtracting blank wells. (F) Phase-contrast microscopy images of MCF-7 cells captured with either pretreatment with vehicle control (−) or PKC-θ-specific peptide (+) and subsequently nonstimulated (NS) or PMA stimulated (ST). (G) CD44high/CD24low CSC FACS analysis of samples as described for panel F. (H) mRNA expression by real-time PCR of CD44 and IL-6 as described for panel F. (I) Fluorescence microscopy images of MCF-7 cells following overexpression of PKC-θ WT (wild type) or PKC-θ NLS. (J) CD44high/CD24low CSCs FACS analysis of samples as described for panel H. Data are plotted as percent CSC increase above results for mock-treated samples. (K) mRNA expression by real-time PCR of samples as described for panel H. Data are plotted as percent change in transcript above results for mock-treated samples. (L) Immunoblotting for PKC-θ in cytoplasmic (C) and nuclear (N) fractions of indicated human breast cancer cell lines. (M) Confocal microscopy of MCF-IM with PKC-θ-P (phospho-T538) antibody (green) and nuclei stained with DAPI (blue). (N) Ratio of nuclear to cytoplasmic fluorescence (Fn/c) values for confocal microscopy images of MCF-IM as described for panel M. Image analysis was performed on confocal laser scanning microscopy (CLSM) files to determine Fn/c after subtraction of the fluorescence due to background. (O) Photomicrographs of nuclear and cytoplasmic staining of normal and triple negative invasive human breast cancer tissues using antibodies as above. All results represent either the means ± the standard errors from three independent experiments or, for panels F, I, L, M, and O, the individual results from a representative experiment of three replicates (n = 3). *, P < 0.05.
To ascertain the contribution of nuclear PKC-θ in CSC-inducible-gene regulation, we utilized previously validated PKC-θ wild-type (WT) and nuclear localization sequence (NLS) constructs (38). We observed reduced nuclear localization of PKC-θ in NLS mutant-transfected MCF-7 cells (Fig. 2I; see also Fig. S2H in the supplemental material). Transfection of PKC-θ NLS mutants inhibited generation of CD44high/CD24low CSCs compared to PKC-θ WT constructs in MCF-IM cells (Fig. 2J). This inhibitory effect of NLS mutation on the formation of CD44high/CD24low CSCs was also confirmed by reduced transcription of the CSC-inducible genes CD44, PLAUR, and IL-6 in MCF-IM (Fig. 2K). Overall, these data show that nuclear PKC-θ is required for the transcription of CSC-enriched inducible genes.
To establish whether nuclear PKC-θ has clinical relevance in breast cancer, we initially analyzed the subcellular distribution of PKC-θ in the nuclear and cytoplasmic compartments of luminal and basal breast cancer cell lines by immunoblotting. PKC-θ was predominantly located in the nuclei of a panel of human basal cell lines (Fig. 2L; see also Fig. S2I in the supplemental material) with little detection in the cytoplasmic fractions of these cell lines. The purity of the nuclear extracts was confirmed by performing immunoblotting against lamin A/C (nuclear protein control) and β-adaptin (cytoplasmic protein control), which was negative (Fig. 2L; see also Fig. S2J in the supplemental material). In addition, PKC-θ fluorescence microscopy of MCF-IM showed minimal detection of PKC-θ in the epithelial state but increased presence of predominantly nuclear PKC-θ (PKC-θ phosphorylated at T538 [PKC-θ-P]) in the mesenchymal state following activation with PMA (Fig. 2M and N). Consistent with these observations, PKC-θ mRNA expression was the most induced in the mesenchymal state compared to cPKC-α, cPKC-β, and PKC-δ family members in the MCF-IM (see Fig. S2K in the supplemental material). The MDA-MB231 metastatic cell line (>90% CD44high/CD24low CSCs) also expressed high levels of PKC-θ transcripts (see Fig. S2K).
Next, we analyzed the presence of PKC-θ protein in normal breast tissue and invasive breast cancers. Triple negative (TN) breast cancers showed immunoreactivity for active PKC-θ (PKC-θ-P) within the nuclear compartment of mitotic cells (Fig. 2O). TN subtypes also showed cytoplasmic staining of PKC-θ compared to normal breast ducts, which showed only weak cytoplasmic and patchy nuclear immunoreactivity (Fig. 2O). Additionally, strong circumferential membranous staining of CD44 was observed in TN subtypes. For all the antibodies used in the immunohistochemistry studies, a section of breast cancer and normal breast tissue was run as a negative control by omitting the primary antibody (see Fig. S2L in the supplemental material). Together with in vitro data, these clinical data implicate both nuclear and cytoplasmic PKC-θ in the formation and maintenance of CSCs in human breast cancer.
Chromatinized PKC-θ tethers as an active transcription complex to EMT- and CSC-inducible genes and is required for chromatin accessibility at these loci.
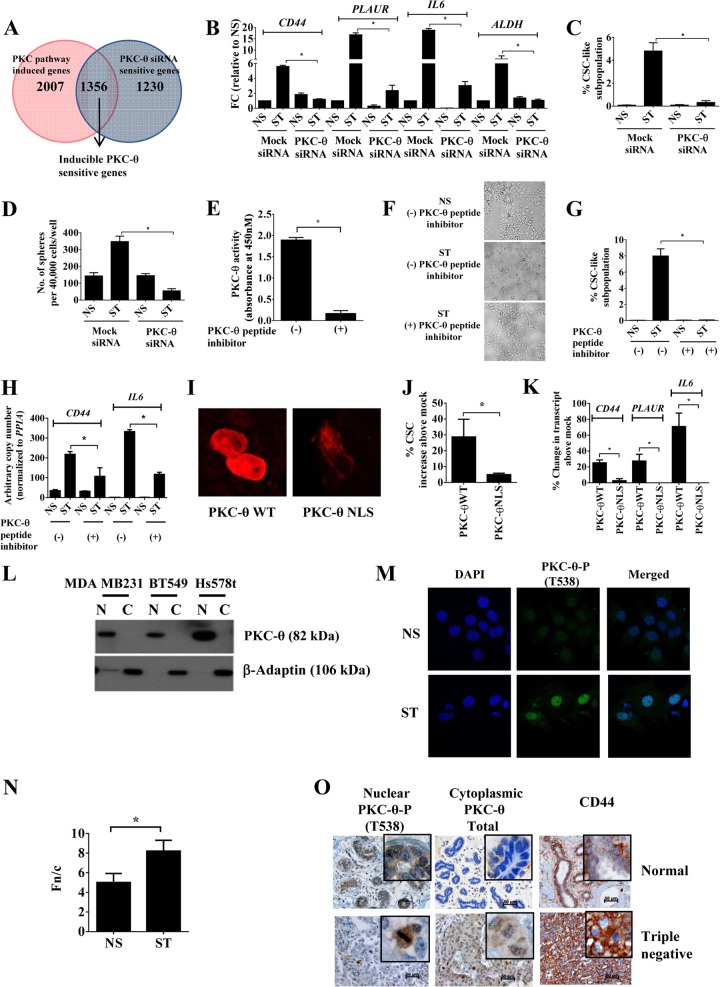
Nuclear PKC-θ therefore contributes to inducible gene expression programs in the mesenchymal state. Given that this kinase has previously been implicated as a chromatin-anchored protein (29), we queried whether PKC-θ has the ability to directly tether to mesenchymal inducible genes. Chromatin immunoprecipitation (ChIP) using a pan-PKC-θ-specific antibody shows that PKC-θ directly tethers to the proximal promoter of CD44 in three human EMT models but not the IL-2 gene promoter, a gene not transcribed in these EMT models (Fig. 3A). In addition, the active form of PKC-θ (PKC-θ-P) and the active transcription mark, RNA polymerase II (Pol II), were significantly enriched across the proximal promoters of the CSC-inducible genes PLAUR and CD44 in the mesenchymal state in the MCF-IM model (Fig. 3B and C). Furthermore, sequential ChIP analysis demonstrated that Pol II and PKC-θ coexist on the PLAUR gene promoter in stimulated cells (Fig. 3D), suggesting that PKC-θ exists as part of an active transcription complex in the mesenchymal state. PKC-θ knockdown decreased chromatin association of Pol II (Fig. 3E) and the active chromatin mark H3K4me2 (see Fig. S3A in the supplemental material) on the PLAUR gene promoter in MCF-IM cells, suggesting that PKC-θ is required for the association of Pol II onto the chromatin template in the mesenchymal state.
FIG 3.
Chromatinized PKC-θ exists as an active transcription complex on EMT- and CSC-inducible genes and is required for chromatin accessibility. (A) PKC-θ ChIP in MCF-IM, HMLE-IM, and MDA-MB-231 breast cancer cell lines across the CD44 and IL-2 promoter region for MCF-IM. A no-antibody (NA) control was used as a negative control. (B and C) PKC-θ-P ChIP (B) or RNA polymerase II (Pol II) ChIP (C) in MCF-IM across PLAUR and CD44 promoter regions. (D) Enrichment of DNA of the PLAUR promoter seen when sequential ChIP was performed in MCF-IM first with anti-Pol II antibody and then with anti-PKC-θ. (E) Pol II ChIP in MCF-IM transfected with either mock siRNA or PKC-θ siRNA followed by PMA stimulation (ST) or no stimulation (NS) across the CD44 promoter region. (F) FAIRE across CD44, PLAUR, IL-6, and IL-2 promoter regions in MCF-IM transfected with either mock siRNA or PKC-θ siRNA. All results represent the means ± the standard errors from three independent experiments (n = 3). **, P < 0.01; *, P < 0.05.
Given that our data thus far implicate nuclear PKC-θ in the transcription of inducible CSC genes and that PKC-θ directly tethers to chromatin at these loci, we tested the function of PKC-θ in mediating chromatin accessibility. Formaldehyde-associated isolation of regulatory elements (FAIRE) assay of mock siRNA-transfected cells showed increased chromatin accessibility across the promoters of CSC-inducible genes CD44, PLAUR, and IL-6 in the mesenchymal state in MCF-IM cells (Fig. 3F). In contrast, PKC-θ knockdown resulted in decreased accessibility across the proximal promoter region of all three of these genes (Fig. 3F).
Overall, these data suggest that chromatinized PKC-θ exists as an active transcription complex of key inducible genes in the mesenchymal state. Our data also support the notion that PKC-θ is essential for establishing chromatin accessibility of these inducible genes.
PKC-θ activity is required for chromatin accessibility of key inducible EMT and CD44high/CD24low-CSC signature genes.
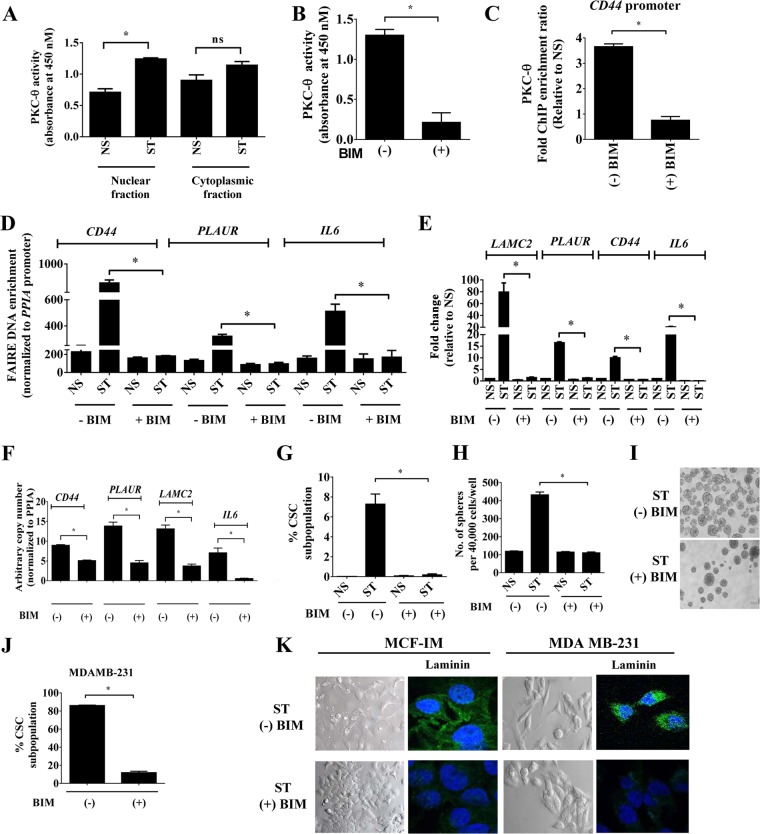
PKC-θ activity profiling of cytoplasmic and nuclear extracts from the MCF-IM revealed kinase activity in both compartments, with significant nuclear PKC-θ activity induction in the mesenchymal state (Fig. 4A). PKC activity was also detected in both cytoplasmic and nuclear fractions of the MDA-MB231 metastatic model (>90% CSCs) (see Fig. S4A in the supplemental material). Since active PKC-θ predominates on the chromatin template of inducible genes in the mesenchymal state, we next addressed the importance of PKC-θ catalytic activity in its ability to tether to chromatin with a selective inhibitor of PKC catalytic activity, bisindolylmaleimide I (BIM) (68, 69), which also successfully inhibited PKC-θ activity (Fig. 4B).
FIG 4.
PKC activity is required for transcription of inducible EMT signature genes. (A) PKC ELISA-based kinase assays of nuclei and whole cells of MCF-IM using anti-PKC-θ antibody for immunoprecipitation. Kinase assays were performed on these PKC-θ fractions, and absorbance was measured at 450 nM. Kinase activity was calculated after subtracting blank wells. Recombinant active PKC-θ was used as a positive control; secondary antibody alone was used as a negative control. (B) PKC ELISA-based kinase assays as described for panel A, using PKC activity inhibitor bisindolylmaleimide-I (BIM). (C) PKC-θ ChIP across the CD44 promoter in MCF-IM either without (−BIM) or with (+BIM) PKC inhibitor bisindolylmaleimide-I pretreatment and subsequent stimulation. (D) FAIRE assays in MCF-IM as described for panel B. (E) mRNA expression by real-time PCR with samples as in panel C. (F) mRNA expression by real-time PCR in MDA-MB-231 cells either without (−) or with (+) BIM. (G) FACS analysis of CD44high/CD24low CSCs as in panel D. (H and I) Mammosphere assays of MCF-IM cells preincubated with (+) or without (−) BIM. (J) FACS analysis of CD44high/CD24low CSCs either without (−) or with (+) BIM in MDA-MB-231 cells. (K) Representative images of MCF-IM or MDA-MB-231 cells either with pretreatment (ST + BIM) or without pretreatment (ST − BIM). Images were either taken by phase-contrast microscope or stained with anti-laminin-5 (green) and DAPI (nuclear stain; blue). All results either represent the means ± the standard errors from three independent experiments or are, for panels I and K, the individual results from a representative experiment of three replicates (n = 3). *, P < 0.05; ns, not significant.
Pretreatment of MCF-IM with BIM (Fig. 4B) substantially prevented PKC-θ from associating with the CD44 promoter (Fig. 4C). Furthermore, PKC-θ catalytic activity was also required for chromatin accessibility (Fig. 4D) and corresponding transcription of the mesenchymal enriched genes LAMC2, CD44, PLAUR, and IL-6 (MCF-IM [Fig. 4E] and MDA-MB-231 [Fig. 4F]).
Consistent with these transcriptional data, CSC formation was abrogated in BIM-treated MCF-IM cells as measured by FACS analysis (Fig. 4G) and mammosphere assays (Fig. 4H and I). Similar effects were observed in basal/metastatic MDA-MB-231 cells, in which BIM treatment reduced the CD44high/CD24low CSC population (Fig. 4J). Interestingly, incubation of MDA-MB-231 cells with BIM visually appeared to prevent morphological changes of EMT, with the appearance of an increase in cell-cell contacts, decrease in spindle cell morphology, and decreased laminin-5 staining (Fig. 4K).
Overall, these results suggest that catalytic activity of this kinase is required for chromatin accessibility for EMT and the formation of CSCs in human breast cancer cells.
Signaling via PKC-θ is a prerequisite for NF-κB activity.
We next sought to establish which transcription factors enable PKC-θ to tether to the chromatin template of inducible genes in the mesenchymal state. The promoter regions (kb +1 kb to −0.5) of the 420 most highly expressed transcripts in CSCs versus NCSCs and NS cells (see Table S1 in the supplemental material) were analyzed for overrepresented transcription factor binding motifs compared to 974 random promoters. The NF-κB matrix occurred in significantly more CSC gene promoters (Fisher's exact test, P = 0.0008) (Fig. 5A); indeed, the NF-κB family has an established role as a partner to PKC signaling (70–72), and it has recently been implicated in both pancreatic and breast CSCs (73, 74).
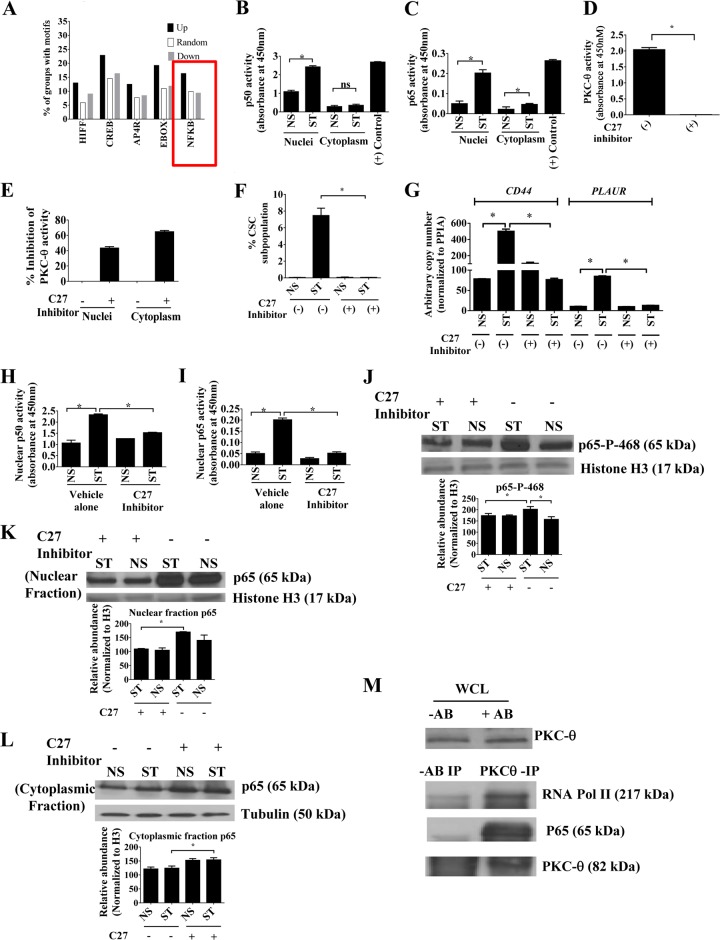
FIG 5.
Active PKC-θ is required for activity of the NF-κB family. (A) Transcription factor motifs significantly overrepresented in the promoters of the genes increased and decreased in expression in CSCs (compared to both NCSC and NS) compared to a randomly chosen set of 974 promoters. (B and C) ELISA-based NF-κB activity assays using p50 antibody or p65 antibody, respectively. Raji cell extract was used as a positive control. Nuclear and cytoplasmic extracts were prepared from MCF-IM cells, and 5 μg/well protein was used in triplicate wells. (D) PKC ELISA-based kinase assays for PKC-θ specific activity inhibitor compound 27 (C27). Kinase activity was calculated after subtracting blank wells. (E) PKC ELISA-based kinase assays of nuclear extract or cytoplasmic extract from MCF-IM cells without (−) or with (+) C27 inhibitor. Assays were performed without specific immunoprecipitates for global PKC activity. (F) FACS analysis of CD44high/CD24low CSC subpopulation in MCF-IM. MCF-7 cells were stimulated with PMA (0.65 ng/ml for 60 h) either without pretreatment of PKC-θ specific activity inhibitor compound 27 (−C27) or with pretreatment with compound 27 (+C27) (1 μM for 24 h). (G) mRNA expression by real-time PCR with samples as described for panel F. (H and I) Nuclear p50 or nuclear p65 activity in samples as described for panel B after treatment of MCF-IM cells either with vehicle alone or with PKC-θ-specific inhibitor compound 27 (C27). (J) Immunoblotting of MCF-IM nuclear extracts for phosphorylated NF-κB p65 at serine 468 (p468) after treatment with (+) or without (−) compound 27. Fifteen micrograms of the protein was used for Western blots, and histone H3 antibody was used as a nuclear control. (K and L) Immunoblotting of MCF-IM cell nuclear fraction (K) and cytoplasmic fraction (L) for global NF-κB p65 after treatment with (+) or without (−) compound 27. Densitometric analyses using Image J software are provided below Western blots in panels J, K, and L. (M) Halfway ChIP of stimulated MCF-IM cells with anti-PKC-θ antibody. WCL, whole-cell lysate utilized for the immunoprecipitation (IP) experiments. WCL is immunoblotted with anti-PKC-θ antibody. −AB or +AB, absence or presence, respectively, of antibody. Immunoblotting was done with either anti-RNA Pol II, anti-p65 antibodies, or anti-PKC-θ antibody. All results either represent the means ± the standard errors from three independent experiments or, for panels J, K, L, and M, are the individual results from a representative experiment of three replicates (n = 3). *, P < 0.05; ns, not significant.
To further investigate the nature of NF-κB family members in EMT, we carried out NF-κB activity assays, which suggested that both nuclear and cytoplasmic p50 and p65 activities were overall increased upon stimulation in MCF-IM cells (Fig. 5B and C). In addition, activity assays in MDA-MB-231 cells also displayed p65 and p50 activity, with nuclear fractions detecting greater activity than the cytoplasmic fractions (see Fig. S5A and S5B in the supplemental material). Given that previous studies in the context of the immune system have established that PKC-θ signals to and activates NF-κB family members (31, 72), we next addressed the impact of PKC-θ on NF-κB activity in our system by utilizing a novel, highly selective ATP-competitive PKC-θ specific inhibitor, compound 27 (C27) (75). As anticipated, C27 abrogated PKC-θ activity both in our in vitro PKC-θ activity assay (Fig. 5D) and in nuclear and cellular extracts generated from MCF-IM cells pretreated with C27 (Fig. 5E). Consistent with the BIM data (Fig. 4G), C27 also abolished CSC formation as measured by FACS (Fig. 5F; see also Fig. S5C in the supplemental material) and transcription of CSC-inducible genes (Fig. 5G). Furthermore, C27 significantly decreased p50 and p65 nuclear activity (Fig. 5H and I), suggesting that PKC-θ also signals to NF-κB proteins in the mesenchymal state.
Previously published data suggest that stimulus-induced phosphorylation of the p65 subunit, including at serine 468 and serine 536, plays a key role in the transcriptional activation following nuclear translocation (76–80). To address whether PKC-θ has an impact on phosphorylation at serine 468 and serine 536 in the EMT/CSC process, immunoblot analysis of nuclear extracts from MCF-IM was carried out with anti-phospho p65 (serine 536) and anti-phospho p65 (serine 468) antibodies. p65 phosphorylated at serine 468 (p65-P-468) was induced following stimulation, and this phosphorylation was inhibited in the presence of C27 (Fig. 5J). Interestingly, p65 phosphorylated at serine 536 (p65-P-536) showed no effect with the C27 inhibitor (see Fig. S5D in the supplemental material), suggesting that phosphorylation of this residue on p65 occurs independently of PKC-θ. Furthermore, immunoblotting with pan-p65 antibody revealed that C27 also inhibited p65 abundance in the nuclei of both epithelial and mesenchymal states (Fig. 5K). On the other hand, global accumulation of p65 was observed in the cytoplasmic fraction of C27-treated epithelial and mesenchymal cells derived from the MCF-IM (Fig. 5L). Collectively, this suggests that PKC-θ activity is possibly required globally both for maintaining p65 in the nucleus and for its phosphorylation at a key active site (serine 468) of this transcription factor.
Finally, we addressed whether PKC-θ and NF-κB directly associate in the MCF-IM. PKC-θ halfway ChIP on MCF-IM nuclear extracts showed an association between endogenous PKC-θ and p65 or RNA Pol II (Fig. 5M). Hence, nuclear PKC-θ exists in the proximity of NF-κB and the active transcription complex in a mesenchymal state.
Overall, these data suggest that active PKC-θ collaborates with active NF-κB family members.
NF-κB family members are part of the chromatinized PKC-θ-containing active transcription complex on inducible EMT and CD44high/CD24low-CSC signature genes.
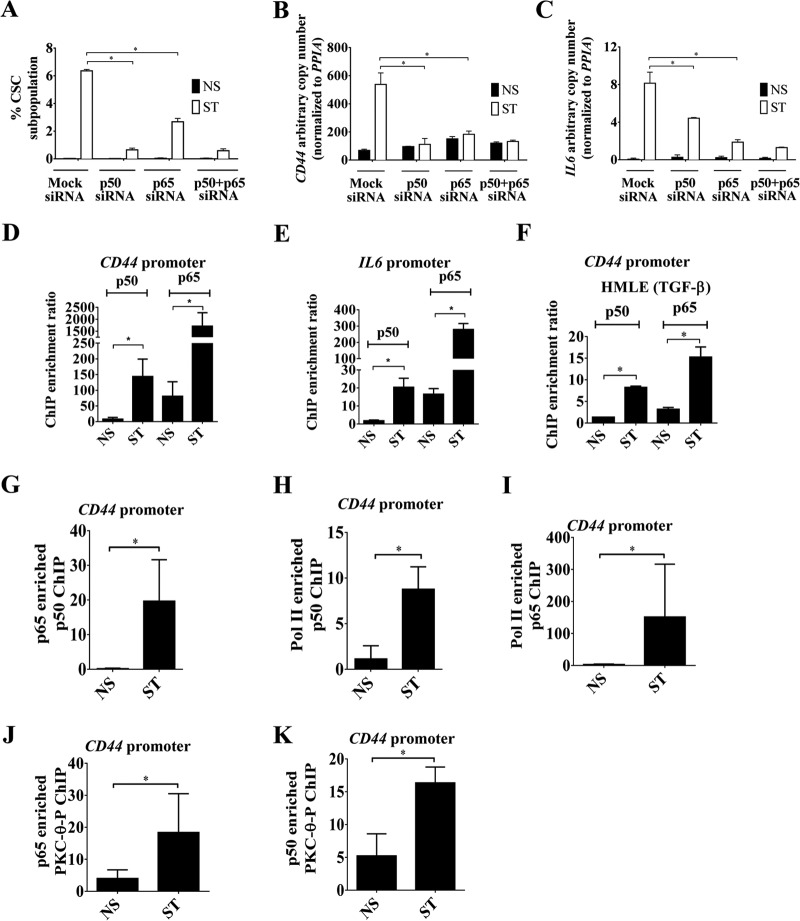
Our results thus far suggest that there is cross talk between the NF-κB family members and PKC-θ. However, the identity of the NF-κB family members that participate in the regulation of inducible EMT genes remains elusive. We therefore embarked on a systematic analysis of which NF-κB family members have an impact on CD44high/CD24low CSC formation using FACS. Knockdown of p50 and p65 alone (see Fig. S6A in the supplemental material) or in combination reduced CD44high/CD24lowCSC formation compared to mock-treated cells (Fig. 6A) and reduced transcription of two CSC-enriched genes, CD44 and IL-6 (Fig. 6B and C); c-Rel knockdown had no effect on CSC formation (see Fig. S6B and C in the supplemental material). ChIP and sequential ChIP confirmed that both p50 and p65 were recruited on the promoters of inducible genes in both the MCF-IM (Fig. 6D and E) and HMLE-IM (Fig. 6F) in the mesenchymal state and that they coexist on these promoters (Fig. 6G) with Pol II (Fig. 6H and I). Both p65 and p50 demonstrated increased cooccupancy with PKC-θ-P across the CD44 promoter in the stimulated state (Fig. 6J and K). Taken together, our results suggest that the NF-κB family members p50 and p65 associate with chromatinized PKC-θ on the CSC-enriched mesenchymal genes CD44 and IL-6.
FIG 6.
NF-κB family members partner with chromatinized PKC-θ on inducible EMT and CSC genes. (A) CD44high/CD24low CSCs FACS analysis of MCF-IM cells transfected with either mock siRNA, p50 siRNA, or p65 siRNA and subsequently either nonstimulated (NS) or stimulated (ST). (B and C) mRNA expression by real-time PCR of CD44 and IL-6, respectively, of samples as described for panel A. (D and E) p50 and p65 ChIP assays in MCF-IM cells across the CD44 and IL-6 promoters, respectively. (F) p50 and p65 ChIP assays in HMLE-IM cells across the CD44 promoter region. (G) Enrichment of DNA on the CD44 promoter seen when sequential ChIP was performed first with anti-p65 antibody and then with anti-p50 antibody. (H and I) Sequential ChIP as described for panel G, except first with anti-Pol II and then with either anti-p50 (H) or anti-p65 (I) antibody. (J and K) Sequential ChIP as for panel H, except first with either anti-p65 antibody (J) or anti-p50 antibody (K) and then with anti-PKC-θ antibody. All results represent the means ± the standard errors from three independent experiments (n = 3). *, P < 0.05.
NF-κB family members p50 and p65 exhibit differential effects in the tethering of chromatinized PKC-θ on inducible genes.
We next addressed to what extent p50 and p65 were required to tether the PKC-θ-containing active transcription complex onto chromatin. Knockdown of p65 completely abolished the recruitment of Pol II, PKC-θ-P, and p50 proteins across both the CD44 (Fig. 7A to C) and IL-6 promoters (Fig. 7D to G) in the mesenchymal state, suggesting that p65 is required for the assembly of the PKC-θ active transcription complex at inducible genes enriched in the mesenchymal state.
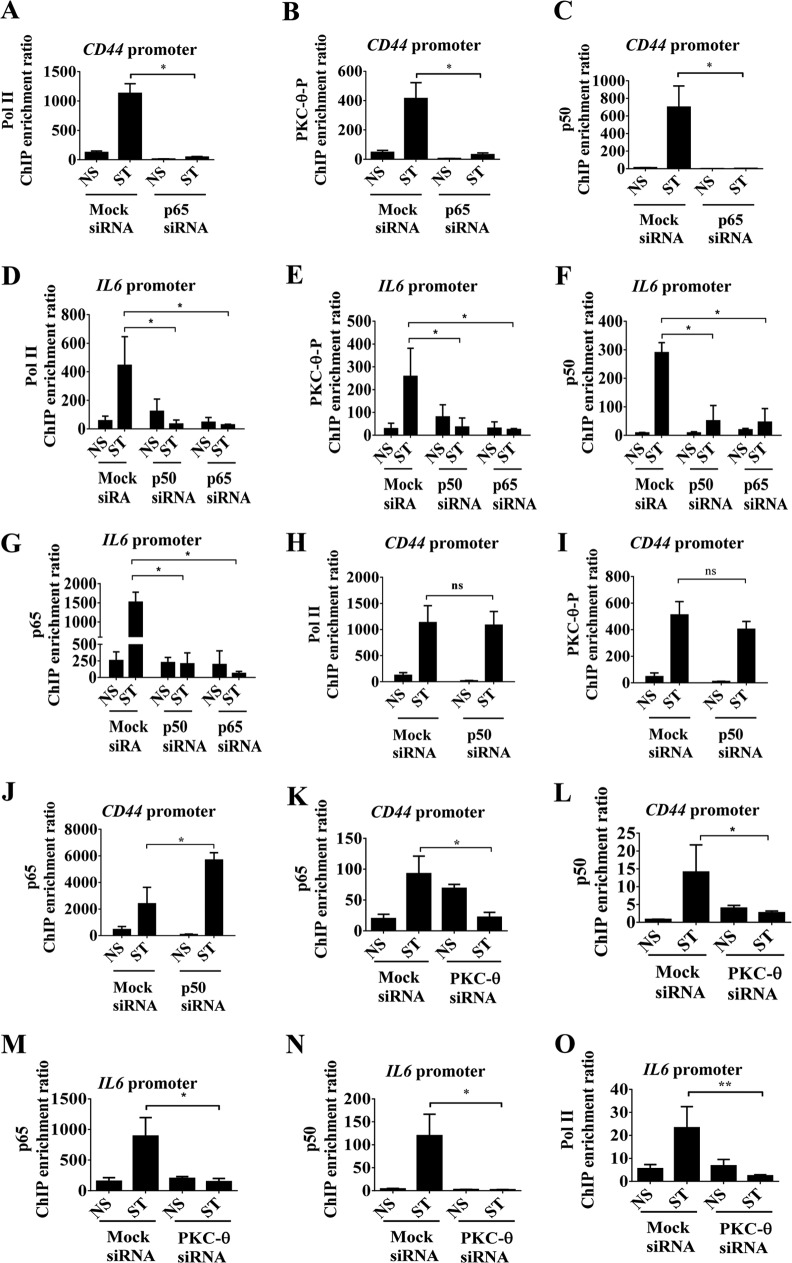
FIG 7.
Differential effects of p50 and p65 in the anchoring of chromatinized PKC-θ on inducible genes. (A to C) ChIP on mock and p65 siRNA-pretreated MCF-IM cells, nonstimulated (NS) and PMA stimulated for 60 h (ST) across the CD44 promoter using antibodies against Pol II (A), PKC-θ-P (B), and p50 (C). Results are from 3 independent experiments. (D to G) ChIP on mock and p65 siRNA-pretreated MCF-IM cells, nonstimulated (NS) and PMA stimulated for 60 h (ST) across the IL-6 promoter using antibodies against Pol II (D), PKC-theta-P (E), p50 (F), and p65 (G). (H to J) ChIP on either mock or p50 siRNA-treated MCF-IM cells, nonstimulated (NS) and PMA stimulated for 60 h (ST) across the CD44 promoter using antibodies against Pol II (H), PKC-θ-P (I), and p65 (J). (K and L) ChIP on either mock or PKC-θ siRNA-treated MCF-IM, nonstimulated (NS) and PMA stimulated for 60 h (ST) across the CD44 promoter using antibodies against p65 (K) and p50 (L). (M to O) ChIP similar to those described for panels K and L, but across the IL-6 promoter. All results represent the means ± standard errors from three independent experiments (n = 3). **, P < 0.01; *, P < 0.05; ns, not significant.
In comparison, knockdown of p50 displayed minimal effects on the tethering of Pol II and PKC-θ-P (Fig. 7H and I) but increased p65 association (Fig. 7J) on the CD44 gene promoter in mesenchymal cells, suggesting that in the absence of p50 a compensatory mechanism may occur. In contrast, knockdown of p50 reduced Pol II, PKC-θ-P, and p65 across the IL-6 promoter in mesenchymal cells, comparable to that of p65 (Fig. 7D, E, and G). Taken together, knockdown of p50 had surprisingly different effects across the CD44 and IL-6 promoters. p50 appears to be indispensable for the tethering of the PKC-θ active transcription complex on the IL-6 promoter but is dispensable on the CD44 gene promoter.
We next determined whether PKC-θ could affect the recruitment of p50 and p65 onto these inducible CSC-enriched genes. We observed that PKC-θ knockdown reduced the association of both p65 and p50 on the CD44 (Fig. 7K and L) and IL-6 (Fig. 7M and N) promoters in the MCF-IM. As anticipated, knockdown of PKC-θ prevented Pol II recruitment (Fig. 7O).
Overall, these results suggest that the NF-κB family members p50 and p65 are critical for association of chromatinized PKC-θ onto mesenchymally enriched gene promoters such as CD44 and IL-6.
Genome-wide analysis reveals that chromatinized PKC-θ preferentially binds to the 5′ untranslated region (5′UTR) of a distinct cohort of inducible PKC-θ-sensitive genes.
Chromatinized PKC-θ is therefore targeted to a key set of inducible-gene promoters whose expression is induced in the mesenchymal state. It was important to ascertain the genome-wide distribution of PKC-θ in EMT and CSC processes. PKC-θ ChIP-seq analysis was undertaken in the MCF-IM. A total of 14,108,692 and 4,645,493 reads from nonstimulated and stimulated MCF7–PKC-θ ChIP samples, respectively, were uniquely mapped to the Hg19 genome; 23,370,927 and 21,886,666 reads were uniquely mapped from the respective total input samples. In stimulated MCF-7 cells, 9,175 PKC-θ-enriched peaks were detected in PKC-θ versus genomic samples using ZINBA (P < 0.05), while 10,867 peaks were detected using BayesPeaks (with a posterior probability of >0.9990); 2,945 peaks were detected by both programs.
PKC-θ peaks were relatively broad compared to those seen for many transcription factors in ChIP-seq, compared by three peak caller programs: BayesPeaks, ZINBA, and MACS2 (see Fig. S7A to F in the supplemental material). MACS2 failed to call many of the smaller, broader peaks (see Table S5 in the supplemental material), such as those at the start of the BHLHE40 and TRIB1 transcripts, but enrichment of PKC-θ at this region was confirmed by real time-PCR (see Fig. S8 in the supplemental material).
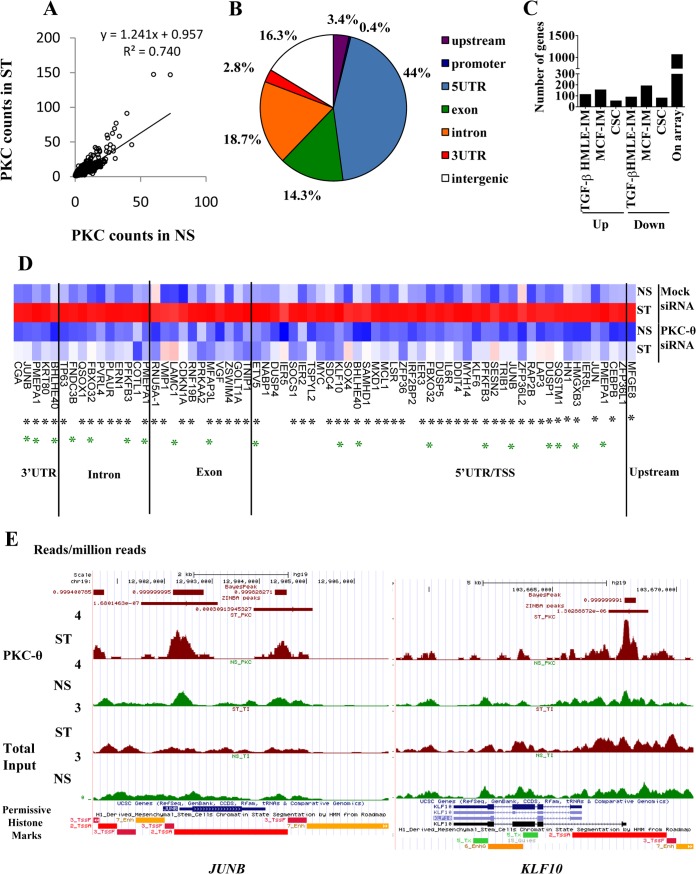
When nonstimulated and stimulated read counts for the ZINBA/BayesPEAK peaks were regressed (y = 0.9575 + 1.241x [Fig. 8A]), a subset of peaks were clearly shown to have increased PKC-θ presence in the stimulated cells. Of 1,721 peaks with at least 1.5-fold-greater normalized sequencing tag in the stimulated sample, the majority were within 10 kb of an Ensembl transcript (Fig. 8B). Nearly one-half (44%) of these peaks occurred in the 5′UTR (Fig. 8B). Peaks occurring at the TSS (transcription start site) were annotated as 5′UTR. Gene ontology analysis indicated that the genes bound to PKC-θ were enriched for genes associated with transcription, cell cycle, and endosomal transport (Benjamini value, <0.05).
FIG 8.
Genome-wide analysis identifies the direct tethering of PKC-θ to a distinct cohort of inducible PKC-θ-sensitive genes. (A) PKC-θ binding in NS and ST MCF-7 cells as measured by normalized tag counts for the PKC peaks. Tag counts were normalized to per million uniquely mapped reads. The regression line is shown. (B) PKC-θ peaks associated with stimulated MCF7 cells bind different regions of the genome relative to the transcription start site (TSS) of the nearest Ensembl transcript. Peaks were classified as 5′UTR, 3′UTR, exon, intron, promoter (<1 kb from TSS), upstream (<10 kb from TSS), or intergenic. Only peaks with >1.5-fold normalized sequencing reads in the stimulated MCF7 sample were considered. (C) Numbers of PKC-θ bound genes that displayed increased or decreased abundance by TGF-β treatment of HMLE cells, PMA treatment of MCF-7 cells, or CSCs compared to NCSCs. Genes in which expression went up or down were determined using Hugene 1.0 Affymetrix microarrays with a log2 (0.5) cutoff. (D) Expression of the PKC-dependent genes with PKC-θ binding as detected by ChIP-seq (Hugene 2.0 Affymetrix arrays). The heat map shows log2 expression values scaled across the gene and clustered by Euclidean distance using complete linkage. Black asterisks mark PKC-θ binding regions that contain the NF-κB binding motif. Green asterisks mark genes that are induced by TGF-β in HMLE. Blue, low expression; red, high expression. (E) PKC-θ binding at JUNB and KLF10 in nonstimulated (NS) and PMA-stimulated (ST) MCF-7 cells. ChIP-seq read counts (number of reads per million mapped reads) are shown for PKC-θ and total input samples in the UCSC genome browser. The RoadMap data for H1-derived mesenchymal stem cells show permissive histone environments (red, high H3K4me3 and histone acetylation) near the PKC-θ binding peaks. Peaks called by Zinba are shown with their P values, while peaks called by BayesPeak are shown with posterior probabilities.
We next determined the extent to which PKC-θ directly associates at the loci of PKC-θ-dependent inducible genes in the mesenchymal state. Of the PKC-θ-bound gene set, 1,064 genes were represented on the microarray, and a subset was differentially expressed in both the inducible HMLE-IM and MCF-IM. As shown in Fig. 8C, 110, 151, and 52 genes were induced in the HMLE-IM, in the MCF-IM, and in CSCs, respectively. We identified 62 inducible PKC-θ-sensitive genes that also showed increased PKC-θ chromatin association in mesenchymal cells (Fig. 8D; see also Table S5 in the supplemental material), termed “direct PKC-θ-dependent genes” (DPDs). Several of these PKC-θ bound target genes identified from the ChIP-seq data were further validated for a subset of genes by a quantitative real-time PCR on ChIP NS and ST samples to confirm (i) that they are indeed enriched for PKC-θ and (ii) how the observed presence of PKC-θ in the NS and ST samples compared to the presence predicted from the ChIP-seq experiments (see Fig. S7 in the supplemental material).
The size of the PKC-θ binding regions for the DPDs was adjusted to 1 kb before they were analyzed for overrepresented transcription factor binding sites using Genomatix. Twenty-eight motif families had Z-scores of >5 when their frequency was compared to both promoter regions (which have similar GC content) and background genomic regions (see Table S6 in the supplemental material). One of the significant motifs was the NF-κB family, with the majority of regions possessing NF-κB motifs (Fig. 8D; see also Table S6). In addition, several DPDs, such as JUNB, had PKC-θ bound to different parts of the gene, such as both the 5′UTR and 3′UTR regions, while others, such as KLF10, had only 5′UTR binding (Fig. 8E). Interestingly, the 3′UTR PKC-θ binding domains of KLF10 and JUNB (Fig. 8E) were enriched in permissive chromatin marks, such as H3K4me3 and histone acetylation, based on the Encode Roadmap project of H1-derived mesenchymal cells.
Overall, our genome-wide analysis unveils a distinct cohort of PKC-θ-dependent inducible genes, termed DPDs. At such loci, PKC-θ directly tethers to the chromatin template in mesenchymal cells.
DISCUSSION
In this study, we show that the signal transduction kinase PKC-θ functions as a critical molecular switch to induce EMT and formation of breast CSCs. Specifically, for the first time, we (i) demonstrated the contribution of PKC-θ to the EMT/CSC process; (ii) demonstrated the essential nature of this signaling kinase in the EMT/CSC process; (iii) demonstrated a nuclear role for PKC-θ in the context of breast cancer; (iv) addressed the essential nature of nuclear PKCs in transcription regulation of inducible genes; (v) demonstrated the interplay between the nuclear PKC-θ and NF-κB in the context of EMT/CSCs; (vi) addressed the essential nature of individual NF-κB family members in EMT/CSCs and the interplay between chromatinized PKC-θ and NF-κB family members. Collectively, this study shows the cross talk between signaling kinases, inflammation transcription factor drivers (NF-κB family members), and chromatin for eliciting transcriptional programs that drive mesenchymal differentiation and CSC formation.
The requirement of the PKC-θ pathway for induction of the mesenchymal state is a new finding that paves the way for therapeutic targeting of aggressive mesenchymal cells and CSCs. The effects of pathway inhibition are apparent in both the phenotypic and the molecular states. Both the PKC-specific inhibitor BIM and a PKC-θ selective peptide inhibitor abrogate EMT and CSC formation in both the MCF-IM and the MDA-MB-231 basal-like breast cancer model. A combination of strategies were used to test the stemness of the CSCs: (i) CSCs were enriched in cells with the CD44high/CD24low CSC phenotype; (ii) CSCs, but not the non-CSCs, have the ability to form spherical colonies in suspension cultures (mammospheres); (iii) CSCs express a distinct inducible gene transcriptome program compared to non-CSCs; (iv) in the context of chromatin, CSC-enriched gene transcripts display differential histone modification patterns; (v) CSCs show a distinct microRNA profile compared to non-CSCs; and (vi) CSCs, but not non-CSCs, express key stemness factors, such as MYC and KLF4 (see Fig. S1O in the supplemental material).
Our data suggest that there is interplay between CSC formation, transcription, and chromatin-tethered PKC-θ. ChIP analysis in three EMT models, i.e., the epithelial MCF-IM, the HMLE-IM, and the mesenchymal MDA-MB231 model, shows that PKC-θ tethers to the epigenetic template on a panel of CSC-inducible genes. At the CD44 gene promoter, recruitment of PKC-θ is impeded in PKC-θ siRNA-treated cells compared to the mock siRNA cells, which is associated with a loss of a key active transcription mark, Pol II, and the key inflammatory transcription factor p65, as well as impaired CD44 transcription. Furthermore, in functional experiments we observed abrogation of the CSC subpopulation, as measured by CD44high/CD24low FACS analysis and mammosphere assays. Additionally, the chromatin accessibility assay (FAIRE) (41) shows inhibition of chromatin accessibility of the CD44 gene in PKC-θ siRNA-treated cells. In further support of the importance of chromatinized PKC-θ in the regulation of CSC gene transcription, our PKC-θ NLS mutant analysis shows that nuclear translocation of PKC-θ is essential for CSC formation and transcription of key CSC genes such as CD44. Overall, our data consistently show that nuclear PKC-θ plays an integral part in the EMT process by promoting chromatin accessibility and enhancing expression of key CSC genes such as CD44. In the global context, our ChIP-seq data show that chromatinized PKC-θ tethers to a distinct cohort of mesenchymal enriched genes, including key EMT master regulators.
Microarray profiling shows that a distinct cluster of PKC-θ-sensitive genes are in the TGF-β EMT pathway and include key CSC-inducible genes such as CD44, IL-6, and PLAUR and others defined as EMT signature genes (51, 52). The PKC-θ-dependent inducible gene expression program that occurs during mesenchymal differentiation requires collaborative signaling from NF-κB, which is usually elicited via TNF-α-mediated NOTCH and Src signaling pathways (74, 81).
The physiological or pathological signals that mediate the PKC-θ pathway in EMT and CSCs remain to be established. Indeed, our gene set enrichment analysis (GSEA) suggested that the PKC-θ pathway in the mesenchymal/CSC state operates via central inflammatory pathways such as TNF-α, interferons, and EGF pathways as well as the TGF-β pathway. Thus, our data suggest that PKC-θ acts as a central molecular node that is critical for the integration of the TGF-β and inflammatory signals via the key inflammatory transcription factor, NF-κB, for inducible gene transcription during the EMT/CSC process. In this context, we propose a model whereby PKC-theta functions as a dual signaling and chromatin-tethered kinase for the induction of a distinct program of inducible CSC-enriched genes in close collaboration with the NF-κB family members (see Fig. S9 in the supplemental material). In addition, given that PKC and TGF-β-dependent inducible genes that are PKC-θ insensitive in the mesenchymal state were detected, it is likely that PKC isoforms have distinct but critical roles in EMT and CSC formation. In support of this, Tam et al. recently showed that the PKC-α signaling node was required for CSC formation (82). It is also tempting to speculate that both conventional PKC-α and PKC-θ isoforms cooperate in the induction of mesenchymal-state-specific genes. Indeed, PKC-α activity has been shown to be an activator of PKC-θ in human T cells (83). Thus, we suggest that distinct PKC isoform-dependent nuclear and signaling nodes are likely to converge to drive EMT and breast CSC formation.
A recent report highlighted that PKC-θ is present mainly in estrogen receptor (ER)-negative basal-like breast cancer lines (34). Here we show that PKC-θ is predominantly nuclear in localization in the basal-like breast cancer lines and EMT results in an increased nuclear PKC-θ presence in mesenchymal cells. The nuclear distribution of PKC-θ was also clearly demonstrable in primary breast cancers, in contrast to what is seen in normal breast epithelium. This is consistent with active PKC-θ acting within the nucleus as part of an active transcription complex on inducible genes enriched in the mesenchymal state and elevated in CSCs. The data are also consistent with our recent report showing that PKC-θ exists as a chromatin-tethered adapter protein on immune responsive genes and is important for inflammatory responses (38). An accessible chromatin configuration prevails in the mesenchymal state across the proximal promoter of key inducible genes such as CD44 and is enriched in an active chromatin complex consisting of NF-κB, PKC-θ, and Pol II-associated complex, which is conducive to enhanced CD44 transcription (see Fig. S9 in the supplemental material). Given that PKC is essential for chromatin accessibility at such loci, we also propose that PKC-θ tethered to the chromatin template via p50 and p65 is required for the recruitment of a chromatin remodeling complex (see Fig. S9 in the supplemental material). Future proteomics-based studies will ultimately provide novel insights into other chromatin-associated proteins, such as chromatin remodelers, that form the PKC-θ active transcription complex. It remains also to be determined which repressive epigenetic mechanisms decorate the loci of inducible genes such as CD44 and IL-6 in NCSCs and impede the engagement of the active PKC-θ complex.
Our findings in this study demonstrate that PKC-θ acts as a dual cytoplasm- and nucleus-anchored kinase in cancer, as it does in T cells (29, 35). We show that PKC-θ activity is required for nuclear p65 phosphorylation at serine 468 in the mesenchymal/CSC state. As depicted in the model (see Fig. S9 in the supplemental material), we propose that this event occurs at the level of the chromatin template, akin to that previously documented for IκB kinase ε (IKKε) in response to TNF signaling (84). Given that IKKε has been identified as a breast cancer oncogene amplified and overexpressed in >30% of breast tumors (85, 86), it will be interesting to establish in the future the precise interplay between these two chromatin-tethered kinases in breast mesenchymal cells (see Fig. S9).
PKC-θ activity has been previously documented to also stimulate the regulation of cell cycle genes such as cMYC via FOXO-mediated effects on cRel in ER-negative breast cancer cells (35). Given that we show here that PKC-θ also directly tethers to cMYC, this raises the possibility that cross talk between the signaling and nuclear PKC-θ nodes exists.
Our genome-wide analysis suggested that PKC-θ predominantly anchors to the 5′ ends of genes in the mesenchymal state, not the coding regions as seen in T cells (29), further supporting the notion that chromatinized PKC-θ is acting as a transcriptional regulator of the mesenchymal state in cancer. Specifically, we identified a distinct cohort of PKC-θ-sensitive genes that showed increased PKC-θ chromatin association in mesenchymal cells, termed direct PKC-θ-dependent genes (DPDs). Several of the genes within this cluster have previously been identified as regulators of EMT and cancer such as MYC (87), SOX4 (88), TP63 (89, 90), and JUNB (91). This raises the intriguing possibility that chromatin-anchored PKC-θ is a novel upstream regulator that directly participates in regulating major EMT and cancer drivers.
Knockdown of PKC-θ using siRNA shows that it is crucial for maintaining a permissive chromatin state for CSC-inducible genes such as IL-6 and CD44, and indeed PKC-θ bound chromatin domains appear to be engulfed in key active epigenetic marks such as H3K4me3 and H3K9 acetylation. Since PKC-θ mediates chromatin accessibility, we hypothesize that chromatin-anchored PKC-θ engages with pioneering factors that are important for facilitating the opening of local chromatin in the context of EMT. In support of this hypothesis, transcription factor binding analysis of the PKC-θ-sensitive gene set (see Table S6 in the supplemental material) identifies enrichment of motifs for the pioneering transcription factor FOXO2 (92–94). Given that PKC-θ itself cannot bind DNA, one scenario would be that PKC-θ works in association with these pioneering factors to maintain permissive chromatin states. PKC-θ is likely to stabilize the pioneering factor complex on the chromatin template by locally phosphorylating proteins within the complex or functioning as an adapter protein, as previously shown for the yeast chromatin kinase Hog1 (27).
Critical to understanding the link between inflammation, cancer progression, and the epigenetic state, a chromatinized PKC-θ-NF-κB switch operates during EMT, which is essential for the induced transcription of signature mesenchymal genes. Indeed, NF-κB is known to play a role in the formation of CSCs, but here we establish that there is context dependency (74, 81). The NF-κB family members p50 and p65 are indispensable for induction of CSCs in the mesenchymal state, and in contrast, cRel appears to be redundant in this context, suggesting that cRel's role is confined to inducible immune responsive genes (19, 95). Intriguingly, the majority of DPD genes possess high-affinity binding sites for NF-κB. Critically, both p50 and p65 engage with PKC-θ on the chromatin template of CD44 and IL-6. Our data therefore suggest that the active PKC-θ transcription complex anchors to the chromatin via the p50 and p65 heterodimer on certain inducible genes critical for the mesenchymal state. Although p65 is essential for the recruitment of the PKC-θ active transcription complex to both CD44 and IL-6 promoters, p50 appears to be indispensable for the tethering of the PKC-θ active transcription complex on the IL-6 promoter but not the CD44 promoter. Two possible scenarios are likely explanations for p50 gene specific effect. (i) Given that p50 has previously been implicated in transcriptional elongation (96, 97), we propose that in the absence of p50, the Pol II–PKC-θ complex is effectively recruited onto the CD44 promoter but remains nonprocessive/stalls, preventing elongation. (ii) Our previous study in T cells suggested that chromatinized PKC-θ functions via both positive (coding genes) and negative (microRNA genes) regulatory mechanisms (29). Thus, the final output for inducible-gene expression is a consequence of a balancing act between these positive and negative signals. With this in mind, one possibility for the discrepancy in the IL-6 and CD44 p50 knockdown data could be that PKC-θ and p50 also regulate microRNAs in CSCs and, as a consequence, expression of these two CSC signature gene transcripts is ultimately a balancing act between the resulting positive and negative effects. However, substantive further studies will be required to solidify this concept and unravel the interplay between PKC-θ, p50, p65, and microRNA gene regulation.
Future genome-wide studies will ultimately establish the extent to which p50 and p65 are required for the tethering of PKC-θ to the epigenome in the mesenchymal state. Ultimately, the need for each family member may be stimulus and gene context dependent.
Overall, our data provide a new dimension to the contribution of PKCs in chromatin regulation and EMT/CSC process. Given that PKCs are considered key therapeutic targets in many cancers, such information will not only provide crucial basic mechanistic insights into the contribution of PKC signaling family members in different cell types but also pave the way for novel epitherapeutic strategies. Targeting CSCs remains an underdeveloped area of cancer therapy, but here we identify a novel epigenetic mechanism to target in the notoriously chemo- and radiotherapy-resistant CSC population, using specific inhibitors that are already in development.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elissa Sutcliffe for optimization of the ChIP library protocol in the Rao Lab. We acknowledge Harpreet Vohra and Michael Devoy for extensive support on the FACS experiments detailed in this study. We thank Cathy Gillespie and Reena Ghildyal for their help with microscopy. We also acknowledge Chris Parish for allowing us to utilize his laboratory facilities and for discussions on the project during the early phase of this work. We also thank Pek Siew Lim for critical reading of the manuscript.
We thank the support of Endeavor Postgraduate Award granted to Anjum Zafar (641_2008) and the University of Canberra PDF fellowship scheme.
Footnotes
Published ahead of print 2 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01693-13.
REFERENCES
- 1.Boyer B, Valles AM, Edme N. 2000. Induction and regulation of epithelial-mesenchymal transitions. Biochem. Pharmacol. 60:1091–1099. 10.1016/S0006-2952(00)00427-5 [DOI] [PubMed] [Google Scholar]
- 2.Huber MA, Kraut N, Beug H. 2005. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 17:548–558. 10.1016/j.ceb.2005.08.001 [DOI] [PubMed] [Google Scholar]
- 3.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133:704–715. 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiery JP, Acloque H, Huang RY, Nieto MA. 2009. Epithelial-mesenchymal transitions in development and disease. Cell 139:871–890. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 5.Glinsky GV, Berezovska O, Glinskii AB. 2005. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 115:1503–1521. 10.1172/JCI23412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee CW, Simin K, Liu Q, Plescia J, Guha M, Khan A, Hsieh CC, Altieri DC. 2008. A functional Notch-survivin gene signature in basal breast cancer. Breast Cancer Res. 10:R97. 10.1186/bcr2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. 2003. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 100:3983–3988. 10.1073/pnas.0530291100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. 2003. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17:1253–1270. 10.1101/gad.1061803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Ejeh F, Smart CE, Morrison BJ, Chenevix-Trench G, Lopez JA, Lakhani SR, Brown MP, Khanna KK. 2011. Breast cancer stem cells: treatment resistance and therapeutic opportunities. Carcinogenesis 32:650–658. 10.1093/carcin/bgr028 [DOI] [PubMed] [Google Scholar]
- 10.Dean M. 2005. Cancer stem cells: implications for cancer causation and therapy resistance. Discov. Med. 5:278–282 [PubMed] [Google Scholar]
- 11.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. 2006. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444:756–760. 10.1038/nature05236 [DOI] [PubMed] [Google Scholar]
- 12.Diehn M, Clarke MF. 2006. Cancer stem cells and radiotherapy: new insights into tumor radioresistance. J. Natl. Cancer Inst. 98:1755–1757. 10.1093/jnci/djj505 [DOI] [PubMed] [Google Scholar]
- 13.Phillips TM, McBride WH, Pajonk F. 2006. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J. Natl. Cancer Inst. 98:1777–1785. 10.1093/jnci/djj495 [DOI] [PubMed] [Google Scholar]
- 14.Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM. 2007. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc. Natl. Acad. Sci. U. S. A. 104:618–623. 10.1073/pnas.0606599104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barski A, Jothi R, Cuddapah S, Cui K, Roh TY, Schones DE, Zhao K. 2009. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 19:1742–1751. 10.1101/gr.090951.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, Carey M, Workman JL. 2007. The role of chromatin during transcription. Cell 128:707–719. 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- 17.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. 2006. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell 24:309–316. 10.1016/j.molcel.2006.08.019 [DOI] [PubMed] [Google Scholar]
- 18.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437:436–439 [DOI] [PubMed] [Google Scholar]
- 19.Rao S, Gerondakis S, Woltring D, Shannon MF. 2003. c-Rel is required for chromatin remodeling across the IL-2 gene promoter. J. Immunol. 170:3724–3731. 10.4049/jimmunol.170.7.3724 [DOI] [PubMed] [Google Scholar]
- 20.Rao S, Procko E, Shannon MF. 2001. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J. Immunol. 167:4494–4503. 10.4049/jimmunol.167.8.4494 [DOI] [PubMed] [Google Scholar]
- 21.Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403:41–45. 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 22.Sutcliffe EL, Parish IA, He YQ, Juelich T, Tierney ML, Rangasamy D, Milburn PJ, Parish CR, Tremethick DJ, Rao S. 2009. Dynamic histone variant exchange accompanies gene induction in T cells. Mol. Cell. Biol. 29:1972–1986. 10.1128/MCB.01590-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O'Shea JJ, Zhao K. 2009. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30:155–167. 10.1016/j.immuni.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chow CW, Davis RJ. 2006. Proteins kinases: chromatin-associated enzymes? Cell 127:887–890. 10.1016/j.cell.2006.11.015 [DOI] [PubMed] [Google Scholar]
- 25.Pascual-Ahuir A, Struhl K, Proft M. 2006. Genome-wide location analysis of the stress-activated MAP kinase Hog1 in yeast. Methods 40:272–278. 10.1016/j.ymeth.2006.06.007 [DOI] [PubMed] [Google Scholar]
- 26.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. 2006. Activated signal transduction kinases frequently occupy target genes. Science 313:533–536. 10.1126/science.1127677 [DOI] [PubMed] [Google Scholar]
- 27.Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, Struhl K, Posas F. 2006. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol. Cell 23:241–250. 10.1016/j.molcel.2006.05.031 [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB. 2003. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature 423:655–659. 10.1038/nature01576 [DOI] [PubMed] [Google Scholar]
- 29.Sutcliffe EL, Bunting KL, He YQ, Li J, Phetsouphanh C, Seddiki N, Zafar A, Hindmarsh EJ, Parish CR, Kelleher AD, McInnes RL, Taya T, Milburn PJ, Rao S. 2011. Chromatin-associated protein kinase C-θ regulates an inducible gene expression program and microRNAs in human T lymphocytes. Mol. Cell 41:704–719. 10.1016/j.molcel.2011.02.030 [DOI] [PubMed] [Google Scholar]
- 30.Chand S, Mehta N, Bahia MS, Dixit A, Silakari O. 2012. Protein kinase C-theta inhibitors: a novel therapy for inflammatory disorders. Curr. Pharm. Des. 18:4725–4746. 10.2174/138161212802651625 [DOI] [PubMed] [Google Scholar]
- 31.Isakov N, Altman A. 2012. PKC-theta-mediated signal delivery from the TCR/CD28 surface receptors. Front. Immunol. 3:273. 10.3389/fimmu.2012.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zanin-Zhorov A, Ding Y, Kumari S, Attur M, Hippen KL, Brown M, Blazar BR, Abramson SB, Lafaille JJ, Dustin ML. 2010. Protein kinase C-theta mediates negative feedback on regulatory T cell function. Science 328:372–376. 10.1126/science.1186068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aguilo JI, Garaude J, Pardo J, Villalba M, Anel A. 2009. Protein kinase C-theta is required for NK cell activation and in vivo control of tumor progression. J. Immunol. 182:1972–1981. 10.4049/jimmunol.0801820 [DOI] [PubMed] [Google Scholar]
- 34.Belguise K, Milord S, Galtier F, Moquet-Torcy G, Piechaczyk M, Chalbos D. 2012. The PKCtheta pathway participates in the aberrant accumulation of Fra-1 protein in invasive ER-negative breast cancer cells. Oncogene 31:4889–4897. 10.1038/onc.2011.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belguise K, Sonenshein GE. 2007. PKCtheta promotes c-Rel-driven mammary tumorigenesis in mice and humans by repressing estrogen receptor alpha synthesis. J. Clin. Invest. 117:4009–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mani SA, Guo W, Liao M-J, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133:704–715. 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sleeman JP, Thiery JP. 2011. SnapShot: the epithelial-mesenchymal transition. Cell 145:162.e1. 10.1016/j.cell.2011.03.029 [DOI] [PubMed] [Google Scholar]
- 38.Sutcliffe EL, Li J, Zafar A, Hardy K, Ghildyal R, McCuaig R, Norris NC, Lim PS, Milburn PJ, Casarotto MG, Denyer G, Rao S. 2012. Chromatinized protein kinase C-theta: can it escape the clutches of NF-kappaB? Front. Immunol. 3:260. 10.3389/fimmu.2012.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L, Dahlstrom JE, Chandra A, Board P, Rangasamy D. 2012. Prognostic value of LINE-1 retrotransposon expression and its subcellular localization in breast cancer. Breast Cancer Res. Treat. 136:129–142. 10.1007/s10549-012-2246-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon JM, Giresi PG, Davis IJ, Lieb JD. 2012. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 7:256–267. 10.1038/nprot.2011.444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21:2933–2942. 10.1093/bioinformatics/bti473 [DOI] [PubMed] [Google Scholar]
- 43.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rashid NU, Giresi PG, Ibrahim JG, Sun W, Lieb JD. 2011. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 12:R67. 10.1186/gb-2011-12-7-r67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cairns J, Spyrou C, Stark R, Smith ML, Lynch AG, Tavare S. 2011. BayesPeak—an R package for analysing ChIP-seq data. Bioinformatics 27:713–714. 10.1093/bioinformatics/btq685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu LJ, Gazin C, Lawson ND, Pages H, Lin SM, Lapointe DS, Green MR. 2010. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11:237. 10.1186/1471-2105-11-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang W-L. 2012. Ranking gene ontology terms for predicting non-classical secretory proteins in eukaryotes and prokaryotes. J. Theor. Biol. 312:105–113. 10.1016/j.jtbi.2012.07.027 [DOI] [PubMed] [Google Scholar]
- 48.Hwang YP, Yun HJ, Kim HG, Han EH, Lee GW, Jeong HG. 2010. Suppression of PMA-induced tumor cell invasion by dihydroartemisinin via inhibition of PKCalpha/Raf/MAPKs and NF-kappaB/AP-1-dependent mechanisms. Biochem. Pharmacol. 79:1714–1726. 10.1016/j.bcp.2010.02.003 [DOI] [PubMed] [Google Scholar]
- 49.Moolten FL, Schreiber B, Rizzone A, Weiss AJ, Boger E. 1981. Protection of mice against 7,12-dimethylbenz (a) anthracene-induced skin tumors by immunization with a fluorinated analog of the carcinogen. Cancer Res. 41:425–429 [PubMed] [Google Scholar]
- 50.He H, Davidson AJ, Wu D, Marshall FF, Chung LW, Zhau HE, He D, Wang R. 2010. Phorbol ester phorbol-12-myristate-13-acetate induces epithelial to mesenchymal transition in human prostate cancer ARCaPE cells. Prostate 70:1119–1126. 10.1002/pros.21146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. 2013. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339:580–584. 10.1126/science.1228522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, Hartwell K, Onder TT, Gupta PB, Evans KW, Hollier BG, Ram PT, Lander ES, Rosen JM, Weinberg RA, Mani SA. 2010. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. U. S. A. 107:15449–15454. 10.1073/pnas.1004900107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plasari G, Calabrese A, Dusserre Y, Gronostajski RM, McNair A, Michalik L, Mermod N. 2009. Nuclear factor I-C links platelet-derived growth factor and transforming growth factor beta1 signaling to skin wound healing progression. Mol. Cell. Biol. 29:6006–6017. 10.1128/MCB.01921-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verrecchia F, Chu ML, Mauviel A. 2001. Identification of novel TGF-beta/Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J. Biol. Chem. 276:17058–17062. 10.1074/jbc.M100754200 [DOI] [PubMed] [Google Scholar]
- 55.Jechlinger M, Grunert S, Tamir IH, Janda E, Ludemann S, Waerner T, Seither P, Weith A, Beug H, Kraut N. 2003. Expression profiling of epithelial plasticity in tumor progression. Oncogene 22:7155–7169. 10.1038/sj.onc.1206887 [DOI] [PubMed] [Google Scholar]
- 56.Lim E, Wu D, Pal B, Bouras T, Asselin-Labat M-L, Vaillant F, Yagita H, Lindeman GJ, Smyth GK, Visvader JE. 2010. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 12:R21. 10.1186/bcr2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Browne EP, Wing B, Coleman D, Shenk T. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319–12330. 10.1128/JVI.75.24.12319-12330.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hecker M, Hartmann C, Kandulski O, Paap BK, Koczan D, Thiesen H-J, Zettl UK. 2013. Interferon-beta therapy in multiple sclerosis: the short-term and long-term effects on the patients' individual gene expression in peripheral blood. Mol. Neurobiol. 48:737–756. 10.1007/s12035-013-8463-1 [DOI] [PubMed] [Google Scholar]
- 59.Sana TR, Janatpour MJ, Sathe M, McEvoy LM, McClanahan TK. 2005. Microarray analysis of primary endothelial cells challenged with different inflammatory and immune cytokines. Cytokine 29:256–269. 10.1016/j.cyto.2004.11.003 [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Gavriil M, Lucas J, Mandiyan S, Follettie M, Diesl V, Sum F-W, Powell D, Haney S, Abraham R, Arndt K. 2008. IkappaBalpha kinase inhibitor IKI-1 conferred tumor necrosis factor alpha sensitivity to pancreatic cancer cells and a xenograft tumor model. Cancer Res. 68:9519–9524. 10.1158/0008-5472.CAN-08-1549 [DOI] [PubMed] [Google Scholar]
- 61.Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, Amariglio N, Vaisman N, Segal E, Rechavi G, Alon U, Mills GB, Domany E, Yarden Y. 2007. A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 39:503–512. 10.1038/ng1987 [DOI] [PubMed] [Google Scholar]
- 62.Cheng J-C, Auersperg N, Leung PCK. 2012. EGF-induced EMT and invasiveness in serous borderline ovarian tumor cells: a possible step in the transition to low-grade serous carcinoma cells? PLoS One 7:e34071. 10.1371/journal.pone.0034071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ho M-Y, Tang S-J, Chuang M-J, Cha T-L, Li J-Y, Sun G-H, Sun K-H. 2012. TNF-alpha induces epithelial-mesenchymal transition of renal cell carcinoma cells via a GSK3beta-dependent mechanism. Mol. Cancer Res. 10:1109–1119. 10.1158/1541-7786.MCR-12-0160 [DOI] [PubMed] [Google Scholar]
- 64.Li C-W, Xia W, Huo L, Lim S-O, Wu Y, Hsu JL, Chao C-H, Yamaguchi H, Yang N-K, Ding Q, Wang Y, Lai Y-J, LaBaff AM, Wu T-J, Lin B-R, Yang M-H, Hortobagyi GN, Hung M-C. 2012. Epithelial-mesenchymal transition induced by TNF-alpha requires NF-kappaB-mediated transcriptional upregulation of Twist1. Cancer Res. 72:1290–1300. 10.1158/1538-7445.AM2012-1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang S, Wang X, Iqbal S, Wang Y, Osunkoya AO, Chen Z, Chen Z, Shin DM, Yuan H, Wang YA, Zhau HE, Chung LWK, Ritenour C, Kucuk O, Wu D. 2013. Epidermal growth factor promotes protein degradation of epithelial protein lost in neoplasm (EPLIN), a putative metastasis suppressor, during epithelial-mesenchymal transition. J. Biol. Chem. 288:1469–1479. 10.1074/jbc.M112.438341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giambra V, Jenkins CR, Wang H, Lam SH, Shevchuk OO, Nemirovsky O, Wai C, Gusscott S, Chiang MY, Aster JC, Humphries RK, Eaves C, Weng AP. 2012. NOTCH1 promotes T cell leukemia-initiating activity by RUNX-mediated regulation of PKC-theta and reactive oxygen species. Nat. Med. 18:1693–1698. 10.1038/nm.2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R, Jr, Badve S, Nakshatri H. 2006. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 8:R59. 10.1186/bcr1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martiny-Baron G, Kazanietz MG, Mischak H, Blumberg PM, Kochs G, Hug H, Marme D, Schachtele C. 1993. Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. J. Biol. Chem. 268:9194–9197 [PubMed] [Google Scholar]
- 69.Toullec D, Pianetti P, Coste H, Bellevergue P, Grand-Perret T, Ajakane M, Baudet V, Boissin P, Boursier E, Loriolle F. 1991. The bisindolylmaleimide GF109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 266:15771–15781 [PubMed] [Google Scholar]
- 70.Coudronniere N, Villalba M, Englund N, Altman A. 2000. NF-kappa B activation induced by T cell receptor/CD28 costimulation is mediated by protein kinase C-theta. Proc. Natl. Acad. Sci. U. S. A. 97:3394–3399. 10.1073/pnas.97.7.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hayden MS, Ghosh S. 2008. Shared principles in NF-kappaB signaling. Cell 132:344–362. 10.1016/j.cell.2008.01.020 [DOI] [PubMed] [Google Scholar]
- 72.Lin X, O'Mahony A, Mu Y, Geleziunas R, Greene WC. 2000. Protein kinase C-theta participates in NF-kappaB activation induced by CD3-CD28 costimulation through selective activation of IkappaB kinase beta. Mol. Cell. Biol. 20:2933–2940. 10.1128/MCB.20.8.2933-2940.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun L, Mathews LA, Cabarcas SM, Zhang X, Yang A, Zhang Y, Young MR, Klarmann KD, Keller JR, Farrar WL. 2013. Epigenetic regulation of SOX9 by the NF-kappaB signaling pathway in pancreatic cancer stem cells. Stem Cells 31:1454–1466. 10.1002/stem.1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamamoto M, Taguchi Y, Ito-Kureha T, Semba K, Yamaguchi N, Inoue J-I. 2013. NF-kappaB non-cell-autonomously regulates cancer stem cell populations in the basal-like breast cancer subtype. Nat. Commun. 4:2299. 10.1038/ncomms3299 [DOI] [PubMed] [Google Scholar]
- 75.Jimenez J-M, Boyall D, Brenchley G, Collier PN, Davis CJ, Fraysse D, Keily SB, Henderson J, Miller A, Pierard F, Settimo L, Twin HC, Bolton CM, Curnock AP, Chiu P, Tanner AJ, Young S. 2013. Design and optimization of selective protein kinase C theta (PKCtheta) inhibitors for the treatment of autoimmune diseases. J. Med. Chem. 56:1799–1810. 10.1021/jm301465a [DOI] [PubMed] [Google Scholar]
- 76.Geng H, Wittwer T, Dittrich-Breiholz O, Kracht M, Schmitz ML. 2009. Phosphorylation of NF-kappaB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep. 10:381–386. 10.1038/embor.2009.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mao X, Gluck N, Li D, Maine GN, Li H, Zaidi IW, Repaka A, Mayo MW, Burstein E. 2009. GCN5 is a required cofactor for a ubiquitin ligase that targets NF-kappaB/RelA. Genes Dev. 23:849–861. 10.1101/gad.1748409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mattioli I, Geng H, Sebald A, Hodel M, Bucher C, Kracht M, Schmitz ML. 2006. Inducible phosphorylation of NF-kappa B p65 at serine 468 by T cell costimulation is mediated by IKK epsilon. J. Biol. Chem. 281:6175–6183. 10.1074/jbc.M508045200 [DOI] [PubMed] [Google Scholar]
- 79.Neumann M, Naumann M. 2007. Beyond IkappaBs: alternative regulation of NF-kappaB activity. FASEB J. 21:2642–2654. 10.1096/fj.06-7615rev [DOI] [PubMed] [Google Scholar]
- 80.Schmitz ML, Mattioli I, Buss H, Kracht M. 2004. NF-kappaB: a multifaceted transcription factor regulated at several levels. Chembiochem 5:1348–1358. 10.1002/cbic.200400144 [DOI] [PubMed] [Google Scholar]
- 81.Iliopoulos D, Hirsch HA, Struhl K. 2009. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139:693–706. 10.1016/j.cell.2009.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F, Wu ZJ, Krall JA, Bierie B, Guo W, Chen X, Liu XS, Brown M, Lim B, Weinberg RA. 2013. Protein kinase C alpha is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell 24:347–364. 10.1016/j.ccr.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trushin SA, Pennington KN, Carmona EM, Asin S, Savoy DN, Billadeau DD, Paya CV. 2003. Protein kinase Calpha (PKCalpha) acts upstream of PKCtheta to activate IkappaB kinase and NF-kappaB in T lymphocytes. Mol. Cell. Biol. 23:7068–7081. 10.1128/MCB.23.19.7068-7081.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moreno R, Sobotzik J-M, Schultz C, Schmitz ML. 2010. Specification of the NF-kappaB transcriptional response by p65 phosphorylation and TNF-induced nuclear translocation of IKK epsilon. Nucleic Acids Res. 38:6029–6044. 10.1093/nar/gkq439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adli M, Baldwin AS. 2006. IKK-i/IKKepsilon controls constitutive, cancer cell-associated NF-kappaB activity via regulation of Ser-536 p65/RelA phosphorylation. J. Biol. Chem. 281:26976–26984. 10.1074/jbc.M603133200 [DOI] [PubMed] [Google Scholar]
- 86.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S, Richardson AL, Greulich H, Stewart CJ, Mulvey LA, Shen RR, Ambrogio L, Hirozane-Kishikawa T, Hill DE, Vidal M, Meyerson M, Grenier JK, Hinkle G, Root DE, Roberts TM, Lander ES, Polyak K, Hahn WC. 2007. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 129:1065–1079. 10.1016/j.cell.2007.03.052 [DOI] [PubMed] [Google Scholar]
- 87.Cho KB, Cho MK, Lee WY, Kang KW. 2010. Overexpression of c-myc induces epithelial mesenchymal transition in mammary epithelial cells. Cancer Lett. 293:230–239. 10.1016/j.canlet.2010.01.013 [DOI] [PubMed] [Google Scholar]
- 88.Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, Meyer-Schaller N, Schubeler D, van Nimwegen E, Christofori G. 2013. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell 23:768–783. 10.1016/j.ccr.2013.04.020 [DOI] [PubMed] [Google Scholar]
- 89.Crook T, Nicholls JM, Brooks L, O'Nions J, Allday MJ. 2000. High level expression of deltaN-p63: a mechanism for the inactivation of p53 in undifferentiated nasopharyngeal carcinoma (NPC)? Oncogene 19:3439–3444. 10.1038/sj.onc.1203656 [DOI] [PubMed] [Google Scholar]
- 90.Lindsay J, McDade SS, Pickard A, McCloskey KD, McCance DJ. 2011. Role of DeltaNp63gamma in epithelial to mesenchymal transition. J. Biol. Chem. 286:3915–3924. 10.1074/jbc.M110.162511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gervasi M, Bianchi-Smiraglia A, Cummings M, Zheng Q, Wang D, Liu S, Bakin AV. 2012. JunB contributes to Id2 repression and the epithelial-mesenchymal transition in response to transforming growth factor-beta. J. Cell Biol. 196:589–603. 10.1083/jcb.201109045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, Lashkari D, Shalon D, Brown PO, Botstein D. 1999. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc. Natl. Acad. Sci. U. S. A. 96:9212–9217. 10.1073/pnas.96.16.9212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale A-L, Botstein D. 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. U. S. A. 100:8418–8423. 10.1073/pnas.0932692100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25:2227–2241. 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kontgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. 1995. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 9:1965–1977. 10.1101/gad.9.16.1965 [DOI] [PubMed] [Google Scholar]
- 96.Suhasini M, Pilz RB. 1999. Transcriptional elongation of c-myb is regulated by NF-kappaB (p50/RelB). Oncogene 18:7360–7369. 10.1038/sj.onc.1203158 [DOI] [PubMed] [Google Scholar]
- 97.Williams SA, Chen L-F, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. 2006. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 25:139–149. 10.1038/sj.emboj.7600900 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.