Significance
Kaposi's sarcoma herpesvirus (KSHV) latently infects tumor cells, and viral episomal DNA replicates once each cell cycle. KSHV does not express DNA replication proteins during latency. Instead, KSHV latency-associated nuclear antigen (LANA) recruits host cell DNA replication machinery to the replication origin. However, the mechanism by which LANA mediates replication is uncertain. Here, we show LANA recruits replication factor C, the DNA polymerase clamp [proliferating cell nuclear antigen (PCNA)] loader, in a critical step for viral DNA replication. Our findings suggest that PCNA loading is a rate-limiting step in DNA replication that is incompatible with KSHV persistence. LANA-enhanced PCNA loading is necessary for virus replication and persistent infection. These data reveal a therapeutic target for inhibition of KSHV persistence in malignant cells.
Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) latently infects tumor cells and persists as a multiple-copy, extrachromosomal, circular episome. To persist, the viral genome must replicate with each cell cycle. The KSHV latency-associated nuclear antigen (LANA) mediates viral DNA replication and persistence, but little is known regarding the underlying mechanisms. We find that LANA recruits replication factor C (RFC), the DNA polymerase clamp [proliferating cell nuclear antigen (PCNA)] loader, to drive DNA replication efficiently. Mutated LANA lacking RFC interaction was deficient for LANA-mediated DNA replication and episome persistence. RFC depletion had a negative impact on LANA’s ability to replicate and maintain viral DNA in cells containing artificial KSHV episomes or in infected cells, leading to loss of virus. LANA substantially increased PCNA loading onto DNA in vitro and recruited RFC and PCNA to KSHV DNA in cells. These findings suggest that PCNA loading is a rate-limiting step in DNA replication that is incompatible with viral survival. LANA enhancement of PCNA loading permits efficient virus replication and persistence, revealing a previously unidentified mechanism for KSHV latency.
Kaposi's sarcoma-associated herpesvirus (KSHV or human herpesvirus 8) has a causative role in Kaposi's sarcoma, primary effusion lymphoma (PEL), and multicentric Castleman disease (1–4). KSHV infection of tumor cells is predominantly latent. During latent infection, the viral genome persists as a multicopy, circular, extrachromosomal episome (plasmid) (5, 6). To persist, the genome must replicate and segregate to progeny nuclei with each cell division. Latency-associated nuclear antigen (LANA), a 1,162-residue protein, is one of a few KSHV genes expressed in latency. LANA is necessary and sufficient for episome persistence in the absence of other viral genes (7, 8). Both N-terminal LANA (N-LANA) and C-terminal LANA (C-LANA) are essential for function. N-LANA associates with mitotic chromosomes via binding histones H2A/H2B, and C-LANA simultaneously binds KSHV terminal repeat (TR) DNA (7, 9–20). Thus, LANA tethers the viral genome to host chromosomes and distributes viral DNA to daughter nuclei during mitosis. Importantly, LANA, which lacks enzymatic function, also mediates KSHV TR DNA replication (10, 21–23) through recruitment of host cell machinery, but little is known regarding the details of this process.
We recently identified an internal 59-aa LANA region critical for efficient DNA replication and persistence (24, 25). Here, we find that LANA recruits replication factor C (RFC), the DNA polymerase clamp [proliferating cell nuclear antigen (PCNA)] loader (26), through this sequence to mediate viral DNA replication and episome persistence.
Results
LANA Associates with RFC.
To persist in replicating cells, KSHV genomes must replicate with each cell division and faithfully segregate to progeny nuclei. N-LANA and C-LANA are essential for this process. We recently showed that internal LANA residues 262–320 are also critical for efficient DNA replication and persistence.
To gain insight into the molecular mechanism underlying these effects, we sought to identify host cell protein(s) interacting with this internal sequence. We generated stable cell lines capable of doxycycline-inducible expression of LANA or LANA deletion mutants containing N-terminal ZZ and C-terminal FLAG tags (Fig. 1A). LANA expression was adjusted to levels similar to that of an infected tumor cell line (Fig. S1A). To ensure the N- and C-tags did not disrupt LANA function, we assessed LANA episome maintenance activity. Cells expressing ZZ-LANA-FLAG or control FLAG-ZZ fusion proteins were transfected with p8TR-P, which contains eight TR copies, and cells placed under puromycin selection, for which resistance is encoded on p8TR-P. Puromycin-resistant colonies were expanded and assessed for episomes by Gardella gels (27). LANA-expressing cell lines had episomal DNA in all lanes, whereas cells expressing ZZ-FLAG never contained episomes (Fig. S1B), indicating preservation of LANA function.
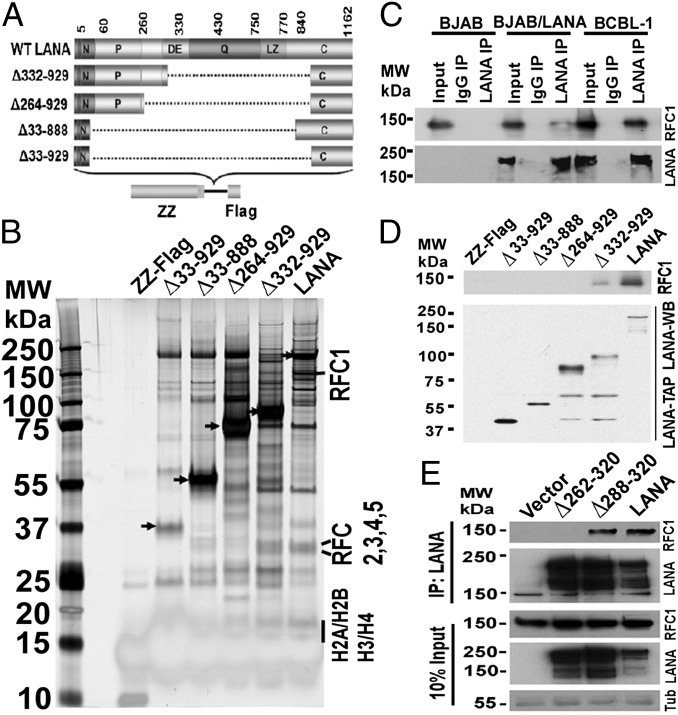
Fig. 1.
LANA interacts with RFC. (A) Schematic representation of LANA and LANA mutants with ZZ and FLAG tags. Deleted residues are indicated to the left. The N-terminal vertical bar indicates nuclear localization signal. C, C-terminal region; DE, aspartate and glutamate region; LZ, predicted leucine zipper; P, proline-rich region; Q, glutamine-rich region. (B) Silver-stained gel after affinity purification with ZZ-FLAG control, LANA, or each LANA mutant. Arrowheads indicate bait proteins. MW, molecular weight (molecular mass). (C) Western blots for RFC1 or LANA after immunoprecipitation (IP) with anti-LANA antibody from the indicated cell lines. (D) Immunoblots of LANA TAP eluants for LANA (anti-FLAG) or RFC1. WB, Western blot. (E) Western blots for LANA or RFC1 after immunoprecipitation of LANA from cell lines stably expressing LANA or LANA mutants. Tub, tubulin.
Next, LANA- or LANA mutant-associated complexes were purified from cell extracts by sequential tandem affinity purification (TAP), and the final FLAG-peptide eluate was resolved by 4–12% gradient SDS/PAGE and visualized by silver staining (Fig. 1B). Excision of bands and analysis by MS identified known LANA-associated proteins, including core histones. In addition, we identified the RFC complex, which consists of five subunits: RFC1, RFC2, RFC3, RFC4, and RFC5. Notably, RFC was only detected in the LANA and LANA∆332–929 lanes. Broad coverage of peptides corresponding to the different RFC subunits was identified (Fig. S2), suggesting a strong LANA interaction. To confirm the LANA–RFC interaction, we immunoprecipitated LANA from uninfected BJAB B lymphoma cells stably expressing LANA or from a KSHV-infected PEL cell line [body cavity-based B-cell lymphoma-1 (BCBL-1)] and found RFC1 specifically coprecipitated with LANA (Fig. 1C). To identify the region of RFC interaction, we performed an immunoblot for RFC1 on TAP products for LANA and each LANA mutant (Fig. 1D). RFC1 associated with LANA and LANA∆332–939 but not with LANA∆264–929 or other mutants, suggesting LANA residues 265–331 are critical for RFC association. LANA∆332–929 precipitated RFC1 less efficiently than did LANA, suggesting that a sequence downstream of LANA residue 332 contributes to the association. Precipitation of LANA from BJAB cell lines stably expressing LANA, LANA∆262–320, or LANA∆288–320 demonstrated RFC only precipitated with LANA and LANA∆288–320 (Fig. 1E), implicating residues 262–287 are critical for RFC interaction. Of note, RFC1 coprecipitated modestly less efficiently with LANA∆288–320 compared with LANA, as evidenced by the lack of increased RFC1 in the LANA∆288–320 lane despite enhanced LANA∆288–320 precipitation compared with that of LANA (Fig. 1E). Therefore, it is likely residues downstream of LANA 287 are required for optimal RFC1 association. Overall, these results indicate RFC associates with LANA and identify residues 262–287 as critical for interaction.
Interaction with RFC Is Crucial for LANA-Mediated DNA Replication.
We hypothesized that LANA may recruit the RFC complex to facilitate viral DNA replication. We recently showed that LANA residues 262–320 are critical for efficient replication of TR DNA in B-cell lines (25). To test this requirement in epithelial cells (which KSHV also infects), we performed a LANA-mediated DNA replication assay with LANA or LANA deletion mutants. A plasmid containing eight copies of the KSHV TR was purified from deoxyadenosine methylase (Dam) positive bacteria and transfected into 293T cells expressing LANA or LANA mutants. DpnI requires Dam methylation for digestion. After replication in mammalian cells, which lack Dam methylase, TR DNA is resistant to DpnI digestion. As expected, LANA had robust DNA replication activity (∼25-fold over control) (Fig. 2A), whereas replication was greatly diminished in the cells with LANA mutants (∆264–929 and ∆262–320) that lack RFC interaction. In contrast, DNA replication was largely preserved in cells expressing LANA∆332–929 or ∆288–320, which both retain RFC binding. The modest reductions in LANA∆332–929 and LANA∆288–320 replication are consistent with their less efficient RFC interaction compared with LANA (Fig. 1 D and E). The reduced replication activities were not due to reduced LANA expression in these mutants (Fig. 2B).
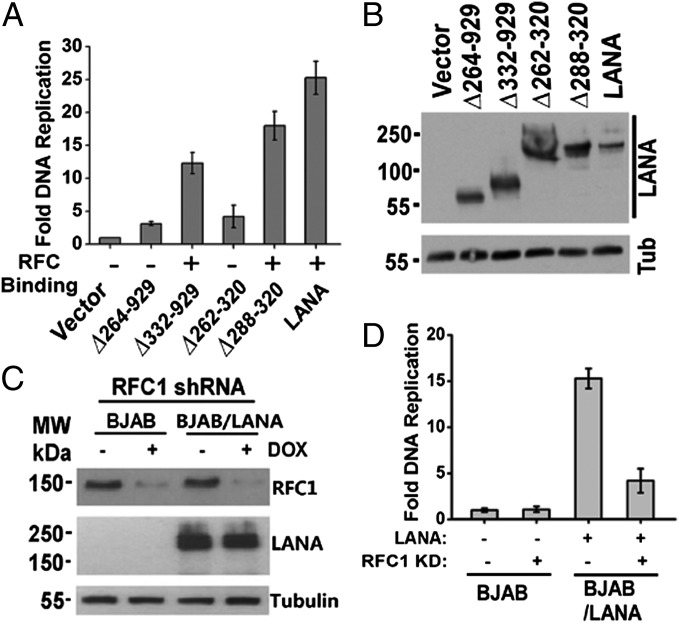
Fig. 2.
LANA interaction with RFC is critical for LANA-mediated DNA replication. (A) Vectors for LANA or LANA mutants were cotransfected with p8TR-gB, which contains eight copies of the KSHV TR element, and fold replication was determined. An average of three experiments with SD is shown. (B) Western blot analysis for LANA, LANA mutants, or Tub after transfection into 293T cells. (C) RFC knockdown in BJAB or BJAB/LANA cells. DOX, doxycycline. (D) DNA replication in BJAB cells or BJAB cells stably expressing LANA with or without RFC1 knockdown. An average of three experiments, with SD, is shown.
To assess RFC’s role in LANA-mediated DNA replication further, we constructed BJAB or BJAB/LANA cell lines with inducible knockdown for RFC1 expression and assessed LANA DNA replication in the presence or absence of ∼75% RFC1 knockdown (Fig. 2C). LANA replication was robust in the absence of RFC knockdown but was substantially reduced after RFC knockdown (Fig. 2D). Together, these results indicate that LANA’s interaction with RFC1 is critical for efficient DNA replication.
RFC Knockdown Results in Highly Deficient LANA-Mediated Episome Persistence.
We assessed the role of LANA’s interaction with RFC in episome persistence. Because we recently showed that LANA∆262–320, which cannot interact with RFC (Fig. 1E), is substantially deficient for episome persistence (25), we addressed the impact of RFC1 knockdown on LANA function. Importantly, RFC1 knockdown had little or no effect on cell proliferation as assessed by cell outgrowth after limiting dilution (Fig. 3A) or by cell cycle analysis (Fig. S3 A and B). Because RFC1 is essential for cell growth in yeast (28), this finding indicates the knockdown did not exceed a threshold necessary for normal or near-normal cell growth. After transfection of p8TR DNA (encoding G418 resistance) into BJAB or BJAB/LANA cells and seeding cells at 1,000, 100, or 10 cells per well in microtiter plates, outgrowth of G418-resistant cell lines was much more robust in BJAB/LANA cells compared with BJAB cells (Fig. 3B). This result is due to the much higher efficiency of LANA episome persistence compared with integration, which occurs infrequently, and is required for p8TR persistence in the absence of LANA. However, after induction of RFC1 knockdown, LANA cell outgrowth was substantially reduced (Fig. 3B). Further, Gardella gel analysis (27) of G418-resistant cell lines demonstrated markedly reduced levels of episomal DNA in the presence of RFC1 knockdown (Fig. 3C). This reduction was not due to reduced LANA expression (Fig. 3D). We also detected LANA by immunostaining in G418-resistant cell lines. LANA reorganizes from broad nuclear localization (Fig. 3E, arrowheads) in the absence of TR DNA to bright dots (Fig. 3E, circled cells) at sites of TR episomes (7). In RFC1 knockdown cells, the percentage of cells containing LANA dots was greatly reduced, and directly correlated with the amount of reduction in episomal DNA signal observed by Gardella gel analysis (27) (Fig. 3 E and F). For instance, RFC1 knockdown in cell line 3 had only ∼20% of cells with LANA dots (Fig. 3F) and had only a small amount of episomal DNA by Gardella gel analysis (27) (Fig. 3C). Therefore, RFC1 knockdown severely impairs LANA-mediated episome persistence.
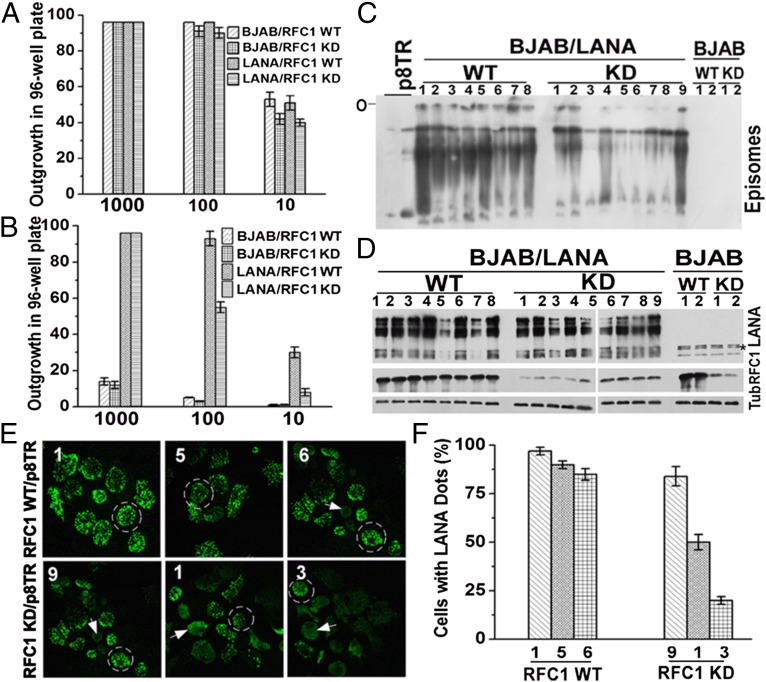
Fig. 3.
LANA interaction with RFC is critical for LANA-mediated episome persistence. (A) BJAB or BJAB/LANA outgrowth in microtiter plates after seeding at 1,000, 100, or 10 cells per well in the presence or absence of RFC1 knockdown (KD). Averages of three experiments are shown. Error bars indicate SD. (B) G418-resistant outgrowth of BJAB or BJAB/LANA cells after p8TR transfection with or without RFC1 knockdown. Averages of three experiments, with SD, are shown. (C) Gardella gel analysis (27) assessing the presence of episomal DNA in BJAB or BJAB/LANA cells with or without RFC1 KD after 20 d of G418 selection. Numbers refer to independently derived G418-resistant cell lines expanded from individual microtiter wells. The two leftmost lanes have increasing amounts of naked p8TR plasmid. O, gel origin. (D) Western blot analysis for LANA, RFC1, or Tub in cell lines used for Gardella gel analysis (27) in C. The asterisk indicates nonspecific bands. (E) LANA immunostaining in the indicated cell lines from C with or without RFC1 KD. Cell lines 1, 5, and 6 (WT, Upper) or cell lines 9, 1, and 3 (RFC1 KD, Lower) contain successively lower levels of episomal DNA as observed in C. Broad nuclear LANA staining indicates episome loss (arrowheads), whereas LANA dots (circled cells) indicate sites of episomes. (Magnification: 630×.) (F) Quantification of average percentage of cells containing LANA dots. Averages of three experiments, with SD, are shown.
RFC1 Is Critical for Persistence of KSHV Infection.
Because the prior experiments used artificial episomes containing TR DNA, we also assessed the effect of RFC1 knockdown on BJAB B lymphoma cells latently infected with KSHV expressing GFP. Latent infection requires puromycin selection (encoded by the recombinant KSHV) for stable persistence. In the absence of selection, there is gradual loss of KSHV episomes because BJAB cells are not dependent on KSHV for viability. RFC1 knockdown (Fig. S4) accelerated GFP loss from infected cells (Fig. 4A). Immunostaining at day 9 after removal of selection demonstrated loss of infection as evidenced by absence of LANA staining (Fig. 4B, arrowhead indicates no LANA staining; compare with presence of red LANA dots in circled cells) in ∼45% of cells with RFC1 knockdown compared with only ∼13% without knockdown (Fig. 4 B and C). Therefore, RFC deficiency accelerates loss of KSHV infection from BJAB cells.
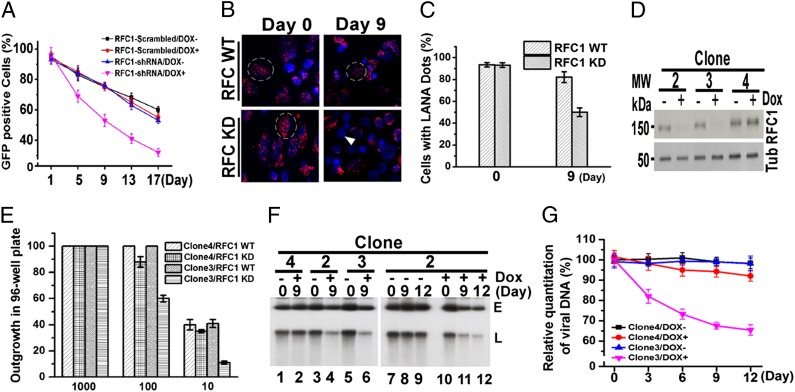
Fig. 4.
RFC deficiency results in loss of KSHV infection. (A) Loss of KSHV GFP expression from BJAB/KSHV cells was monitored in the absence or presence of RFC knockdown. Averages of three experiments at each time point, with error bars indicating SD, are shown. (B) LANA immunostaining (red) of BJAB/KSHV cells in the absence or presence of RFC knockdown. DAPI (blue) stains DNA in cell nuclei. Dashed circles indicate examples of cells with LANA dots, and the arrowhead indicates a cell lacking LANA. (Magnification: 630×.) (C) Quantification of cells containing LANA dots. (D) Immunoblot for RFC1 or tubulin in BCBL-1 cells with or without doxycycline induction of RFC1 knockdown. (E) BCBL-1 outgrowth in microtiter plates after seeding at 1,000, 100, or 10 cells per well in the presence or absence of RFC1 knockdown. Averages of three experiments, with error bars indicating SD, are shown. (F) Gardella gel analysis (27) of BCBL-1 cells in the presence or absence of RFC knockdown. E, episomal KSHV; L, linear KSHV DNA due to lytic replication. (G) Real-time PCR for KSHV DNA in BCBL-1 cells with or without RFC knockdown. Averages of three experiments are shown, with error bars indicating SD.
We also assessed the effect of RFC1 knockdown in BCBL-1 PEL cells, which are naturally infected with KSHV. BCBL-1 cell outgrowth was reduced in cell line 3 with RFC1 knockdown (Fig. 4D) after seeding at 1,000, 100, or 10 cells per well in microtiter plates (Fig. 4E), consistent with loss of viral episomes, because BCBL-1 cells are dependent on latent KSHV infection for viability and proliferation. Further, Gardella gel analysis (27) demonstrated loss of both episomal and linear (from lytic replication) DNA only in cell lines 2 and 3 with RFC1 knockdown (Fig. 4 D and F). This observation was further verified by real-time PCR of KSHV DNA (Fig. 4G). Taken together, these results demonstrate that diminished RFC levels substantially compromise the persistence of KSHV infection.
LANA Promotes PCNA Loading onto DNA.
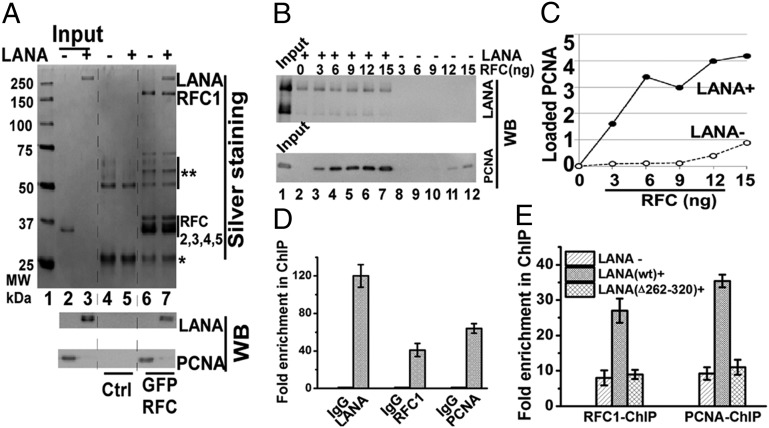
Because RFC loads PCNA onto DNA, we wished to investigate the effect of LANA on PCNA loading. Purified LANA (Fig. S5) or PCNA was incubated with RFC bound to beads. As expected, RFC precipitated PCNA (Fig. 5A, lane 6). RFC also efficiently precipitated LANA (Fig. 5A, lane 7), including in the presence of nuclease (Fig. S6), indicating a direct interaction between LANA and the RFC complex. Interaction in the presence of nuclease excludes the possibility of indirect association of LANA and RFC through simultaneously binding common DNA molecules. We assessed RFC loading of PCNA onto a nicked DNA template containing TR DNA, which has the LANA binding site (LBS) and allows for specific LANA binding (Fig. S7). PCNA loading onto DNA was dramatically enhanced in the presence of LANA compared with the absence of LANA, especially at lower RFC levels (Fig. 5 B and C). Therefore, LANA greatly enhances PCNA loading onto DNA by RFC.
Fig. 5.
LANA enhances RFC loading of PCNA onto DNA. (A) Assessment of direct RFC binding to LANA or PCNA after incubation with anti-GFP beads with or without bound GFP-RFC. PCNA is loaded in lanes 2, 4, and 6 and migrates at ∼35 kDa. The double asterisk indicates heavy chain (lanes 4–7) and degraded GFP-RFC1 (lanes 6 and 7); the single asterisk indicates a light chain. Ctrl, control. (B) PCNA loading onto DNA in the presence or absence of LANA. (C) Signal of PCNA bound to DNA from B was quantified and plotted against RFC input. A value of 1 corresponds to ∼1 ng. (D) ChIP assay for LANA, RFC1, or PCNA bound to TR DNA in BCBL-1 cells. (E) ChIP assay for RFC1 or PCNA binding to TR DNA after transfection of p8TR into BJAB, BJAB/LANA, or BJAB/LANA∆262–320 cells. Averages of three experiments, with SDs, are shown in D and E.
Next, we assessed LANA’s ability to recruit PCNA to TR DNA in cells. First, we used ChIP to assay for the presence of LANA, RFC1, and PCNA at TR DNA in KSHV-infected BCBL-1 cells. In addition to the expected presence of LANA, both RFC1 and PCNA were enriched at TR DNA (Fig. 5D). To test whether PCNA enrichment is dependent on RFC binding to LANA, we performed RFC1 or PCNA ChIP after transfection of p8TR into BJAB cells, BJAB cells stably expressing LANA, or BJAB cells expressing LANA∆262–320. RFC1 and PCNA were both substantially enriched in the presence of LANA. In contrast, LANA∆262–320, which is deficient for RFC binding (Fig. 1E), was unable to recruit either RFC or PCNA to TR DNA (Fig. 5E). To test whether LANA, RFC, or PCNA enrichment at TR DNA is dependent on the presence of the LBS, we performed LANA, RFC1, or PCNA ChIP after transfection of DNA containing a single TR element, or DNA containing a TR element in which the two adjacent LBSs are deleted, into BJAB cells or BJAB cells stably expressing LANA. As expected, LANA was enriched at TR DNA but not at TR DNA deleted for the LBSs (Fig. S8A). In the presence of LANA, RFC1 and PCNA were each substantially enriched at WT TR DNA but not at TR DNA deleted for LBSs (Fig. S8B). Together, these data indicate that LANA recruits RFC to drive PCNA loading onto viral DNA and that the LBS is required for this function.
Discussion
This work defines LANA’s interaction with the RFC complex as critical for KSHV DNA replication and viral persistence. Host and viral DNA replication is contingent on rapid and processive DNA synthesis. DNA polymerase tethering to the PCNA sliding clamp permits processive replication (29). PCNA is a closed circle and requires RFC enzymatic activity to load onto and encircle DNA. RFC, an AAA+ clamp-loading ATPase, consists of five subunits (RFC1–RFC5) (26, 29, 30). In alternative RFC complexes, other subunits substitute for RFC1 to function in specialized pathways, such as for DNA damage (26).
Remarkably, ∼70% depletion of RFC1 had negligible effects on host cell growth but severely affected LANA’s ability to mediate viral episomal replication and persistence. LANA∆262–320, which does not interact with RFC (Fig. 1E), is similarly deficient (25). After origin recognition complex (ORC) binding and formation of the prereplicative complex, transition to DNA replication occurs as a progressive cascade of events. Loading of PCNA by RFC occurs in the latter stages of replication after DNA unwinding (31). Why, then, is it critical for LANA to recruit RFC if this step normally occurs sequentially at host cell replication origins?
We suggest PCNA loading is a rate-limiting step in DNA synthesis that, although compatible with cellular replication, is incompatible with viral survival, and that LANA accelerates this step, driving KSHV DNA replication forward to enable viral persistence. Consistent with this model, LANA greatly enhanced RFC loading of PCNA onto DNA (Fig. 5). Redundancy of replication origins (32) may allow the cell to tolerate substantial inefficiencies, such as those occurring in the setting of reduced RFC. In contrast, after infection, virus survival may be contingent on efficient progression of a single or limited number of prereplication complexes to active replication at TR DNA. The ∼180-kb KSHV genome is large compared with the typical cellular 50- to 150-kb DNA distances between origins (33), further reducing any margin for inefficiency. In addition, the guanine and cytosine-rich TRs comprise ∼20% of the KSHV genome and are regions of slower DNA replication and pausing (34, 35), further contributing to the need for maximal replication efficiency. LANA interacts with and recruits ORCs to TR DNA, presumably to initiate a prereplicative complex (23, 36, 37). Notably, LANA also interacts with replication protein As (38) and topoisomerase IIβ (39), raising the possibility that these proteins may also function in downstream, rate-limiting steps. Additional, non-TR replication origins also occur in latent KSHV (34). The observed virus loss with RFC deficiency suggests that these sites may be similarly sensitive to RFC deficiency, perhaps through LANA effects. Other episomal tumor viruses, including EBV, papillomaviruses, and polyomaviruses, may face similar needs to accelerate replication (40–43). Whereas EBV nuclear antigen 1 interacts with ORCs and other licensing factors (44), Merkel cell polyomavirus (MCV) large T antigen recruits RFC to sites of MCV replication (45), suggesting that acceleration of PCNA loading may also occur with MCV.
This work implicates LANA recruitment of RFC as an attractive target for disruption. LANA’s enhancement of PCNA loading is critical for efficient viral replication and persistence. Therefore, strategies that inhibit LANA’s interaction with RFC may be effective for virus eradication.
Materials and Methods
Cell Lines.
Cell lines were maintained under standard conditions. The generation of LANA-expressing cell lines is described in SI Materials and Methods.
TAP and MS.
TAP of LANA complexes from human cells was performed using cell lines stably expressing ZZ-LANA-FLAG or ZZ-LANA-FLAG mutants. MS identified coprecipitating proteins.
Fluorescence Microscopy.
LANA was detected by immunofluorescent microscopy using a Zeiss microscope and magnification of 630×.
DNA Replication Assay.
KSHV DNA replication assays were performed as described (46), with minor modifications described in SI Materials and Methods.
Episome Maintenance Assays.
Episome maintenance was assessed in the presence or absence of RFC1 knockdown. Gardella gels (27) were used to assess the presence of episomal DNA. Loss of infection from BJAB/KSHV cells (47) was assessed by monitoring loss of GFP expression from the recombinant KSHV.
PCNA Loading Assay.
RFC loading of PCNA onto nicked DNA containing the KSHV TR was assessed in the presence or absence of purified LANA as described in SI Materials and Methods.
ChIP Assays.
Formaldehyde cross-linking and ChIP assays were performed as described (48, 49), with some modifications as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Chantal Beauchemin for helpful advice, Rolf Renne for TR DNA deleted for the LBS, and Michael Lagunoff for BJAB/KSHV cells. This work was supported by National Institutes of Health National Cancer Institute Grant CA082036 (to K.K.) and Japan Society for the Promotion of Science KAKENHI Grants 25440011 and 2513171 (to T.T.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404219111/-/DCSupplemental.
References
- 1.Chang Y, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266(5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332(18):1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Moore PS, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi’s sarcoma in patients with and without HIV infection. N Engl J Med. 1995;332(18):1181–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 4.Soulier J, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86(4):1276–1280. [PubMed] [Google Scholar]
- 5.Cesarman E, et al. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86(7):2708–2714. [PubMed] [Google Scholar]
- 6.Decker LL, et al. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J Exp Med. 1996;184(1):283–288. doi: 10.1084/jem.184.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284(5414):641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 8.Ballestas ME, Kaye KM. Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J Virol. 2001;75(7):3250–3258. doi: 10.1128/JVI.75.7.3250-3258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbera AJ, et al. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science. 2006;311(5762):856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 10.Hu J, Garber AC, Renne R. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J Virol. 2002;76(22):11677–11687. doi: 10.1128/JVI.76.22.11677-11687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley-Clarke B, Ballestas ME, Komatsu T, Kaye KM. Kaposi’s sarcoma herpesvirus C-terminal LANA concentrates at pericentromeric and peri-telomeric regions of a subset of mitotic chromosomes. Virology. 2007;357(2):149–157. doi: 10.1016/j.virol.2006.07.052. [DOI] [PubMed] [Google Scholar]
- 12.Kelley-Clarke B, et al. Determination of Kaposi’s sarcoma-associated herpesvirus C-terminal latency-associated nuclear antigen residues mediating chromosome association and DNA binding. J Virol. 2007;81(8):4348–4356. doi: 10.1128/JVI.01289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbera AJ, Ballestas ME, Kaye KM. The Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J Virol. 2004;78(1):294–301. doi: 10.1128/JVI.78.1.294-301.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krithivas A, Fujimuro M, Weidner M, Young DB, Hayward SD. Protein interactions targeting the latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus to cell chromosomes. J Virol. 2002;76(22):11596–11604. doi: 10.1128/JVI.76.22.11596-11604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim C, Choi C, Choe J. Mitotic chromosome-binding activity of latency-associated nuclear antigen 1 is required for DNA replication from terminal repeat sequence of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2004;78(13):7248–7256. doi: 10.1128/JVI.78.13.7248-7256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piolot T, Tramier M, Coppey M, Nicolas JC, Marechal V. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J Virol. 2001;75(8):3948–3959. doi: 10.1128/JVI.75.8.3948-3959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szekely L, et al. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin-associated nuclear bodies. J Gen Virol. 1999;80(Pt 11):2889–2900. doi: 10.1099/0022-1317-80-11-2889. [DOI] [PubMed] [Google Scholar]
- 18.Wong LY, Matchett GA, Wilson AC. Transcriptional activation by the Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen is facilitated by an N-terminal chromatin-binding motif. J Virol. 2004;78(18):10074–10085. doi: 10.1128/JVI.78.18.10074-10085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotter MA, 2nd, Robertson ES. The latency-associated nuclear antigen tethers the Kaposi’s sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999;264(2):254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- 20.Jeong JH, et al. Regulation and autoregulation of the promoter for the latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus. J Biol Chem. 2004;279(16):16822–16831. doi: 10.1074/jbc.M312801200. [DOI] [PubMed] [Google Scholar]
- 21.Fejér G, et al. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus interacts preferentially with the terminal repeats of the genome in vivo and this complex is sufficient for episomal DNA replication. J Gen Virol. 2003;84(Pt 6):1451–1462. doi: 10.1099/vir.0.18940-0. [DOI] [PubMed] [Google Scholar]
- 22.Grundhoff A, Ganem D. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J Virol. 2003;77(4):2779–2783. doi: 10.1128/JVI.77.4.2779-2783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim C, Sohn H, Lee D, Gwack Y, Choe J. Functional dissection of latency-associated nuclear antigen 1 of Kaposi’s sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J Virol. 2002;76(20):10320–10331. doi: 10.1128/JVI.76.20.10320-10331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Leon Vazquez E, Carey VJ, Kaye KM. Identification of Kaposi’s sarcoma-associated herpesvirus LANA regions important for episome segregation, replication and persistence. J Virol. 2013;87:12270–12283. doi: 10.1128/JVI.01243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De León Vázquez E, Juillard F, Rosner B, Kaye KM. A short sequence immediately upstream of the internal repeat elements is critical for KSHV LANA mediated DNA replication and impacts episome persistence. Virology. 2014;448:344–355. doi: 10.1016/j.virol.2013.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Indiani C, O’Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat Rev Mol Cell Biol. 2006;7(10):751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- 27.Gardella T, Medveczky P, Sairenji T, Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J Virol. 1984;50(1):248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cullmann G, Fien K, Kobayashi R, Stillman B. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15(9):4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelch BA, Makino DL, O’Donnell M, Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011;334(6063):1675–1680. doi: 10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsurimoto T, Stillman B. Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol Cell Biol. 1989;9(2):609–619. doi: 10.1128/mcb.9.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 32.Méchali M. Eukaryotic DNA replication origins: Many choices for appropriate answers. Nat Rev Mol Cell Biol. 2010;11(10):728–738. doi: 10.1038/nrm2976. [DOI] [PubMed] [Google Scholar]
- 33.Aladjem MI, Falaschi A, Kowalski D. Eukaryotic DNA replication origins. In: DePamphilis ML, editor. DNA Replication and Human Disease. Plainview, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 31–61. [Google Scholar]
- 34.Verma SC, et al. Single molecule analysis of replicated DNA reveals the usage of multiple KSHV genome regions for latent replication. PLoS Pathog. 2011;7(11):e1002365. doi: 10.1371/journal.ppat.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dheekollu J, Chen HS, Kaye KM, Lieberman PM. Timeless-dependent DNA replication-coupled recombination promotes Kaposi’s Sarcoma-associated herpesvirus episome maintenance and terminal repeat stability. J Virol. 2013;87(7):3699–3709. doi: 10.1128/JVI.02211-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stedman W, Deng Z, Lu F, Lieberman PM. ORC, MCM, and histone hyperacetylation at the Kaposi’s sarcoma-associated herpesvirus latent replication origin. J Virol. 2004;78(22):12566–12575. doi: 10.1128/JVI.78.22.12566-12575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verma SC, Choudhuri T, Kaul R, Robertson ES. Latency-associated nuclear antigen (LANA) of Kaposi’s sarcoma-associated herpesvirus interacts with origin recognition complexes at the LANA binding sequence within the terminal repeats. J Virol. 2006;80(5):2243–2256. doi: 10.1128/JVI.80.5.2243-2256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shamay M, et al. A Protein array screen for Kaposi’s sarcoma-associated herpesvirus LANA interactors links LANA to TIP60, PP2A activity and telomere shortening. J Virol. 2012;86:5179–5191. doi: 10.1128/JVI.00169-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purushothaman P, et al. Kaposi’s sarcoma-associated herpesvirus-encoded LANA recruits topoisomerase IIβ for latent DNA replication of the terminal repeats. J Virol. 2012;86(18):9983–9994. doi: 10.1128/JVI.00839-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ilves I, Kivi S, Ustav M. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J Virol. 1999;73(5):4404–4412. doi: 10.1128/jvi.73.5.4404-4412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehman CW, Botchan MR. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc Natl Acad Sci USA. 1998;95(8):4338–4343. doi: 10.1073/pnas.95.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skiadopoulos MH, McBride AA. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J Virol. 1998;72(3):2079–2088. doi: 10.1128/jvi.72.3.2079-2088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81(12):3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frappier L. EBNA1 and host factors in Epstein-Barr virus latent DNA replication. Curr Opin Virol. 2012;2(6):733–739. doi: 10.1016/j.coviro.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, et al. Bromodomain protein Brd4 plays a key role in Merkel cell polyomavirus DNA replication. PLoS Pathog. 2012;8(11):e1003021. doi: 10.1371/journal.ppat.1003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De León Vázquez E, Kaye KM. Rapid and quantitative assessment of KSHV LANA-mediated DNA replication. Arch Virol. 2011;156(8):1323–1333. doi: 10.1007/s00705-011-0985-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L, Lagunoff M. Establishment and maintenance of Kaposi’s sarcoma-associated herpesvirus latency in B cells. J Virol. 2005;79(22):14383–14391. doi: 10.1128/JVI.79.22.14383-14391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyd KE, Farnham PJ. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol Cell Biol. 1999;19(12):8393–8399. doi: 10.1128/mcb.19.12.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103(6):843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.