Abstract
Reconstructing the evolution and ancestral functions of synaptic proteins promises to shed light on how neurons first evolved. The postsynaptic density (PSD) protein Homer scaffolds membrane receptors and regulates Ca2+ signaling in diverse metazoan cell types (including neurons and muscle cells), yet its ancestry and core functions are poorly understood. We find that the protein domain organization and essential biochemical properties of metazoan Homer proteins, including their ability to tetramerize, are conserved in the choanoflagellate Salpingoeca rosetta, one of the closest living relatives of metazoans. Unlike in neurons, Homer localizes to the nucleoplasm in S. rosetta and interacts directly with Flotillin, a protein more commonly associated with cell membranes. Surprisingly, we found that the Homer/Flotillin interaction and its localization to the nucleus are conserved in metazoan astrocytes. These findings suggest that Homer originally interacted with Flotillin in the nucleus of the last common ancestor of metazoans and choanoflagellates and was later co-opted to function as a membrane receptor scaffold in the PSD.
Keywords: choanoflagellate, Homer, Flotillin, postsynaptic scaffold evolution, synapse, S. rosetta
Introduction
Neurons, synapses, and the proteins required for their function are critical to the biology and behavior of eumetazoans but little is known about how they first evolved. In neurons, the transmission of chemical signals (called neurotransmitters) from the presynapse to the postsynapse requires distinct sets of pre- and postsynaptic protein networks. Understanding when the proteins required for synaptic activity first evolved and how they functioned in the first metazoans promises to illuminate evolutionary processes underlying the origin of neurons.
The phylogenetic distribution of neurons in extant metazoans suggests that they likely evolved in the stem lineage leading to eumetazoans (fig. 1) (Ryan and Grant 2009). In addition to bilaterians, cnidarians and ctenophores have neurons and relatively simple nervous systems but neurons are apparently absent from sponges and placozoans. Nonetheless, some sponges contract their water canals in response to stimulation with the neurotransmitters glutamate and GABA (Elliott and Leys 2010) suggesting that sponges may contain elements of a primordial nervous system. Indeed, the sponge Amphimedon queenslandica encodes proneural atonal-related basic helix loop helix genes (Richards et al. 2008) and diverse components of the postsynaptic density (PSD), a protein complex that is essential for the morphology and function of excitatory synapses in bilaterians (Sakarya et al. 2007; Srivastava et al. 2010). Likewise, the genome of the placozoan Trichoplax adhaerens encodes genes for synapse formation and conduction of nerve impulses, as well as genes associated with neuronal migration and axon guidance in bilaterians (Srivastava et al. 2008). Thus, proteins required for synaptic transmission evolved early in metazoan evolution, perhaps during the transition to multicellularity or earlier (Kosik 2009; Emes and Grant 2011). However, it is not known how these proteins originally functioned.
Fig. 1.
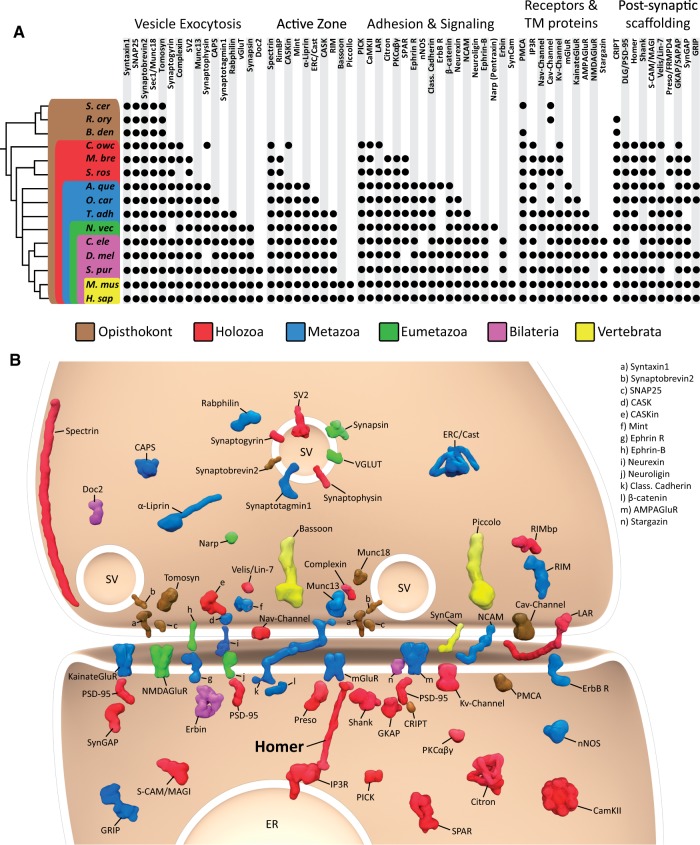
The genome of the choanoflagellate Salpingoeca rosetta encodes diverse synaptic protein homologs. (A) Metazoan synaptic proteins fall into five major functional categories: Vesicle exocytosis proteins, active zone proteins, adhesion and signaling proteins, receptors and transmembrane (TM) proteins, and postsynaptic scaffolding proteins. A black dot indicates the presence of clear homologs (also see supplementary table S1, Supplementary Material online), whereas absence of a dot indicates that a homolog was not detected in that taxon. Taxonomic groupings are indicated as follows: Brown box, Opisthokonta; red box, Holozoa; blue box, Metazoa; green box, Eumetazoa; violet box, Bilateria; yellow box, Vertebrata. The topology of the reference phylogeny was based on a consensus phylogeny (Baldauf 2003; Ruiz-Trillo et al. 2008; Philippe et al. 2009; Fairclough et al. 2013), with the nodes of the earlier branching metazoan lineages collapsed to indicate current controversy concerning their evolutionary relationships. A. que, Amphimedon queenslandica; B. den, Batrachochytrium dendrobatidis; C. ele, Caenorhabditis elegans; C. owc, Capsaspora owczarzaki; D. mel, Drosophila melanogaster; H. sap, Homo sapiens; M. bre, Monosiga brevicollis; M. mus, Mus musculus; N. vec, Nematostella vectensis; O. car, Oscarella carmela; R. ory, Rhizopus oryzae; S. cer, Saccharomyces cerevisae; S. pur, Strongylocentrotus purpuratus; S. ros, Salpingoeca rosetta; T. adh, Trichoplax adhaerens. (B) Graphic representation of an excitatory synapse indicating the subcellular contexts in which many pre- and postsynaptic proteins function. The phylogenetic distribution of each protein is indicated using the color scheme in (A). SV, synaptic vesicle; ER, endoplasmic reticulum.
The study of choanoflagellates, the closest living relatives of metazoans (Carr et al. 2008; King et al. 2008; Ruiz-Trillo et al. 2008), has begun to provide clues into the ancestry of metazoan neuronal proteins. Choanoflagellates are microbial eukaryotes that exist either as single cells or simple colonies of undifferentiated cells. Importantly, they do not produce synapses and neurons. Nonetheless, neurosecretory proteins such as SNAREs and Munc18 interact in the choanoflagellate Monosiga brevicollis by mechanisms that are conserved with metazoans, suggesting that ancestral modes of secretion may have been co-opted for neuronal functions (Burkhardt et al. 2011). Moreover, the genome of M. brevicollis encodes homologs of diverse Ca2+-signaling proteins (Cai 2008), synaptic proteins, including voltage-dependent sodium channels (Liebeskind et al. 2011), and the PSD proteins Homer, Shank, and PSD-95 (Sakarya et al. 2007; Ryan and Grant 2009; Alié and Manuel 2010; Emes and Grant 2012).
The Homer proteins are of particular interest as they are among the most abundant proteins in the PSD of metazoan excitatory synapses (Cheng et al. 2006), where they form tetramers and higher order scaffolds. Homer proteins help regulate the size of dendritic spines, the size of the PSD (Sala et al. 2001), activity-dependent changes in synaptic strength, and, potentially, higher order brain functions such as learning and memory (Xiao et al. 1998; Thomas 2002). Moreover, aberrant Homer function has been associated with several neurological diseases, including addiction, epilepsy, and schizophrenia (Szumlinski et al. 2006). In addition to their neuronal roles, Homer proteins play critical roles in diverse other metazoan-specific cell types, including acting as negative regulators of T-cell activation (Huang et al. 2008), regulating Ca2+-signaling and mechanotransduction in diverse muscle cells (Worley et al. 2007; Stiber et al. 2008), and anchoring maternal effect transcripts in Drosophila melanogaster oocytes (Babu et al. 2004). It is unclear how these diverse functions evolved or whether any of these functions reflect the function(s) of Homer in the first metazoans. We here report on our study of Homer in the colony-forming choanoflagellate Salpingoeca rosetta (Fairclough et al. 2010; Dayel et al. 2011; Fairclough et al. 2013) and our discovery that Homer interacted with Flotillin in the nucleus of the last common ancestor of choanoflagellates and metazoans.
Results
Diverse Pre- and Postsynaptic Proteins in Metazoans and Their Closest Living Relatives
The functioning of excitatory synapses in the brain is governed by a core set of synaptic proteins that fall into five major functional categories: 1) neuronal exocytosis proteins, 2) active zone proteins, 3) adhesion and signaling proteins, 4) receptors and transmembrane proteins, and 5) postsynaptic scaffolding proteins (fig. 1). We searched for their homologs in the genomes of the choanoflagellates S. rosetta and M. brevicollis; the sponges A. queenslandica and Oscarella carmela; and the placozoan T. adhaerens and the filasterean Capsaspora owczarzaki (fig. 1, supplementary table S1, Supplementary Material online). In agreement with previous studies (Sakarya et al. 2007; Srivastava et al. 2008; Ryan and Grant 2009; Srivastava et al. 2010; Conaco et al. 2012; Emes and Grant 2012; Suga et al. 2013), we find that many synaptic proteins from all five major functional categories evolved before the origin of synapses (fig. 1). Moreover, we have identified a number of additional pre- and postsynaptic proteins in sponges and T. adhaerens that were previously thought to be restricted to metazoans with synapses. For example, we have identified the ionotropic glutamate receptor in the genome of O. carmela (Nichols et al. 2012). We have also identified homologs of important active zone proteins in sponges (e.g., Erc/Cast and Liprin-alpha) and in placozoans (e.g., CASK and RIM), as well as important adhesion molecules such as Neurexin (Nichols et al. 2006) and Ephrin receptors in sponges and placozoans (fig. 1). Our analysis reveals that only a handful of synaptic proteins (e.g., the adhesion molecules Neuroligin and Ephrin) seem to be restricted to metazoans with synapses.
The genome of S. rosetta encodes homologs of diverse proteins involved in neuronal exocytosis (e.g., neurosecretory SNARE proteins, Sec1/Munc18, and Synaptogyrin), adhesion/signaling (e.g., LAR, PICK, and CamKII), and postsynaptic scaffolding (e.g., Homer, Shank, and PSD-95). Interestingly, although we identified the neuronal SNARE-binding protein Complexin in the genomes of C. owczarzaki and M. brevicollis, it is apparently missing in the genome of S. rosetta (fig. 1). Importantly, the presence of postsynaptic scaffolding protein homologs such as Homer, Shank, and PSD-95 in choanoflagellates raises the possibility that a primordial synaptic-like scaffold evolved prior to the origin of the first metazoans.
Conserved Protein Domain Composition and Biochemical Properties of Metazoan and S. rosetta Homer Proteins
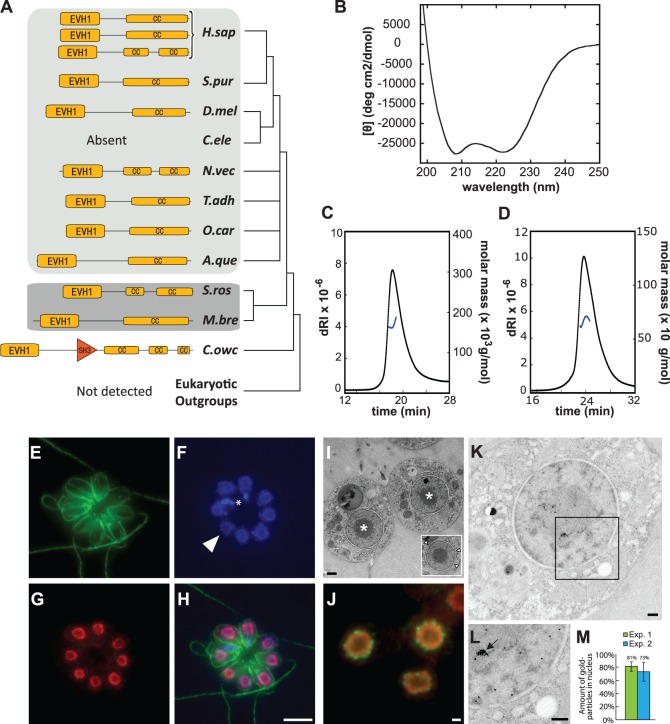
Homer proteins in metazoans contain an N-terminal-enabled/vasodilator-stimulated phosphoprotein homology 1 (EVH1) domain (PF00568) and a C-terminal coiled coil (CC) domain. We found that the protein domain composition and arrangement of Homer in metazoans is conserved in choanoflagellates (fig. 2A) (Shiraishi-Yamaguchi and Furuichi 2007). Moreover, four residues (W24, F74, Q76, and G89) that are necessary for the binding of vertebrate Homer proteins to Shank (Tu et al. 1999), metabotropic glutamate receptor (Brakeman et al. 1997), inositol trisphosphate receptor (Tu et al. 1998), and Dynamin (Tu et al. 1998; Gray et al. 2003) are conserved in S. rosetta Homer (SrHomer; supplementary fig. S1, Supplementary Material online). Finally, similar to the C-terminal CC domain of metazoan Homer proteins, the C-terminal CC domain of SrHomer is alpha-helical (fig. 2B), as revealed by circular dichroism (CD) spectroscopy of bacterially expressed SrHomer.
Fig. 2.
Conserved biochemical properties and nuclear localization of Homer in the choanoflagellate Salpingoeca rosetta. (A) The protein domain organization of Homer is conserved in S. rosetta and metazoan Homer proteins. The protein domain organizations of Homer homologs from Capsaspora owczarzaki, the choanoflagellates S. rosetta and Monosiga brevicollis, and diverse metazoan taxa were mapped onto the consensus phylogeny from figure 1(A). Yellow boxes highlight the conserved Enabled/VASP homology 1 (EVH1) domain and the CC domains of Homer. Orange triangle indicates SH3 domain in C. owczarzaki Homer protein. Although Homer was found in almost all metazoan (light gray), choanoflagellate (dark gray), and C. owczarzaki genomes analyzed in this study, it was not detected in any other lineage. (B) CD spectroscopy of the C-terminal CC domain of bacterially expressed SrHomer (SrHomerCC). The CD spectrum, with two clearly defined minima at 208 and 222 nm, reveals that SrHomerCC is alpha-helical. (C and D) Oligomerization of SrHomer in solution. Size-exclusion chromatography (SEC)-MALS data are shown for full-length SrHomer (C) and SrHomerΔCC (D). Full-length SrHomer exists predominantly as a tetramer (MW: 167 kDa), whereas the deletion of the C-terminal CC domain renders SrHomer dimeric (MW: 67 kDa). The signal from the refractive index detector (dRI; left, Y axis) is shown as solid, black lines. Molecular mass as calculated across the protein elution peak (right, Y axis) is shown as blue lines. (E–I) A rosette colony in which the apical flagellum of each cell is oriented outward. The plasma membrane and flagellum of each cell are revealed by staining with antibodies to beta-tubulin (E, green). (F) DAPI staining reveals DNA in the nucleus of each cell. Arrowhead indicates a representative nucleus from one cell. DNA from prey bacteria (indicated with asterisk) also stains with DAPI. (G) SrHomer colocalizes with DNA in the nucleus, as revealed in the merge (H). (I) In S. rosetta, the nucleus is organized with the electron-dense spherical nucleolus (asterisk) in the center surrounded by a shell of nuclear DNA and nucleoplasm, as revealed by TEM of a thin section through a S. rosetta rosette colony. Inset: Nuclear pores from a single nucleus are indicated with arrowheads. (J) Staining with antibodies to nuclear pore complexes (green) revealed that SrHomer (red) is contained within the nuclear membrane and distributed throughout the nucleoplasm. The hollow center of the nucleus suggests that SrHomer is either excluded from the nucleolus or that antibodies to SrHomer did not efficiently penetrate the nucleolus. (K) Immunogold labeling of S. rosetta cells with an antibody directed against SrHomer. Gold particles are visible as black dots that localize throughout the nucleus. (L) Higher magnification of the region of the nucleus contained within the box in (K). Homer was seen to localize to both the nucleoplasm and nucleolus. High densities of Homer localization in the nucleus (arrow) were frequently observed. (M) Two independent immunogold labeling experiments (Exp. 1 and Exp. 2) show that Homer localizes predominantly to the nucleus (73% and 81%). Nuclear versus cytosolic localization of gold particles was measured from 25 cells per experiment. Scale bars: 5 µm in (G), 500 nm in (I) and (J), 200 nm in (K) and (L). Species abbreviations as in figure 1.
Homer tetramerization in metazoans, which is dependent upon the alpha-helical C-terminal CC domain, is required for the structural integrity of dendritic spines and for recruitment of specific proteins to synapses (Hayashi et al. 2009). Deletion of the C-terminal CC domain prevents tetramerization of rat Homer, although the remaining truncated peptide still dimerizes. To investigate whether SrHomer can also tetramerize through its C-terminal CC domain, we used multiangle light scattering (MALS) to compare the behavior of full-length SrHomer with a truncated SrHomer that lacks the C-terminal CC domain (SrHomerΔCC). Full-length SrHomer (aa 1–355, MW: 41 kDa) eluted with a molecular mass of 167 ± 6 kDa, equaling 4.1× that of the monomer (fig. 2C), although SrHomerΔCC (aa 1–282, MW: 32 kDa) eluted with a molecular mass of 67 ± 10 kDa (fig. 2D), equaling only 2.1× that of the monomer. Thus, similar to metazoan Homer proteins, full-length SrHomer forms tetramers while deletion of the C-terminal CC domain results in truncated peptides that dimerize but do not tetramerize. Our results indicate that Homer’s ability to tetramerize likely evolved before the origin of metazoans and synapses.
Unexpected Localization of SrHomer to the Nucleus
In metazoan cells, Homer protein function is dependent upon its localization to a number of different subcellular regions. For example, Homer 1 in vertebrates associates with two different types of membranes to scaffold membrane proteins: The plasma membrane of the postsynapse in neurons and the ER membrane in neurons and muscle cells (Salanova et al. 2002; Hayashi et al. 2009). In addition, Homer 3 in human T cells stimulated with anti-CD3- or anti-CD28-coated beads is rapidly recruited to the area of contact with the beads and, after prolonged stimulation, translocates into the nucleus (Ishiguro and Xavier 2004). To investigate the subcellular context of SrHomer function, we used affinity-purified and validated antibodies that we raised against SrHomer (supplementary fig. S2, Supplementary Material online). In all S. rosetta cell types, including single cells and colonies, we detected Homer colocalized with DNA in the S. rosetta nucleus in a doughnut-shaped ring (fig. 2E–H). The potential nature of this ring was revealed by transmission electron microscopy (TEM) of S. rosetta thin sections, which showed that the DNA and nucleoplasm of S. rosetta are arranged around a large centrally localized nucleolus (fig. 2I). In live cells, we infer that the DNA and nucleoplasm form a continuous shell around the nucleolus. Further analysis of the localization of Homer, by simultaneously costaining S. rosetta cells with antibodies to the nuclear pore complex and to SrHomer, revealed that SrHomer is distributed throughout the nucleoplasm (fig. 2J), rather than being solely associated with the nuclear membrane. Moreover, TEM of S. rosetta thin sections following immunogold labeling of SrHomer provided independent evidence for the localization of Homer to the nucleoplasm and revealed that it is also localized to the nucleolus (fig. 2K–M). Therefore, we infer that SrHomer localizes throughout the nucleus, in contrast to expectation based on its localization in neurons.
Conserved and Novel SrHomer-Binding Proteins
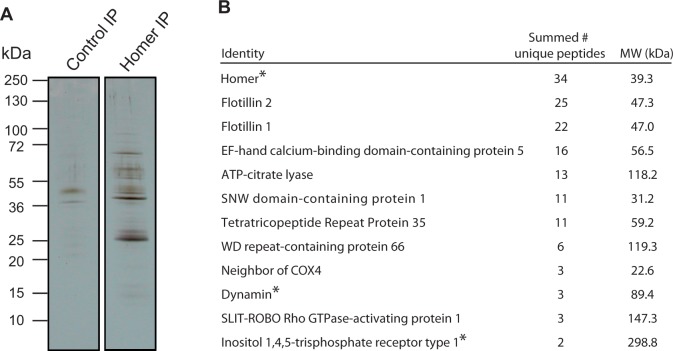
Homer function in metazoans is shaped by its context—its subcellular localization and its protein-binding partners. After finding that SrHomer localizes to the nucleus in all S. rosetta cell types, we next set out to identify its native-binding partners in S. rosetta. High-resolution mass spectrometry in combination with liquid chromatography tandem mass spectrometry (LC-MS/MS) was used to identify proteins purified selectively in a SrHomer immunoprecipitation (IP) compared with a control IP from lysates of S. rosetta colonies (fig. 3A). Based on the nature of the LC-MS/MS analysis, the number of identified peptides corresponding to any given protein can be used as a rough estimate of relative protein abundance in the SrHomer IP compared with the control IP. For example, a protein with an equivalent number of peptide hits in both the SrHomer IP and the control IP was likely to have been pulled down through nonspecific interactions. In contrast, a protein with a large number of hits in the SrHomer IP and few-to-no hits in the control IP was more likely to bind specifically to Homer (or a Homer-containing protein complex). For this study, only proteins with peptide matches detected solely in the SrHomer IP, to the exclusion of the control IP, were considered to be potential interaction partners of SrHomer.
Fig. 3.
Mass spectrometric identification of Salpingoeca rosetta Homer-binding proteins. (A) In a coimmunoprecipitation experiment, diverse proteins were pulled down by SrHomer antibodies, but not by control antibodies. Whole cell lysates from S. rosetta rosette colonies were used for coimmunoprecipitation experiments and the resulting bound proteins analyzed by SDS-PAGE, silver staining, and LC-MS/MS (supplementary table S2, Supplementary Material online). (B) Select candidate binding partners of SrHomer identified by LC-MS/MS analysis of SrHomer coimmunoprecipitates. The summed number of unique peptides represents the total number of identified peptides from two independent SrHomer IP experiments. No peptides were detected in the control IP experiments for these proteins, suggesting that they are selective binding partners of SrHomer. Previously known Homer-binding partners from metazoans are marked with an asterisk.
Following this standard, approximately 300 proteins were identified exclusively in the SrHomer IP based on LC-MS/MS analysis. This list of proteins contains both previously known Homer-binding partners and new potential binding partners (fig. 3B, supplementary table S2, Supplementary Material online). For example, as in metazoans, SrHomer likely interacts with the inositol trisphosphate (IP3) receptor, a calcium channel activated by IP3 (Tu et al. 1998), and with Dynamin, a GTPase involved in endocytosis and cytokinesis (Tu et al. 1998; Gray et al. 2003) (fig. 3B). Some other known Homer-binding partners that are encoded by the S. rosetta genome (e.g., Shank, Tu et al. 1999) were not detected, although they may interact with Homer under conditions not tested in this experiment.
The two most abundant detected putative SrHomer-binding partners, based on the number of unique peptides identified in the SrHomer IP and their complete absence in the control IP, were homologs of Flotillin 1 and Flotillin 2 (fig. 3B), neither of which have previously been shown to interact with Homer. Other previously unidentified potential binding partners included Slit-Robo Rho GTPase-activating protein (srGAP), a protein critical for neuronal migration in bilaterians (Guerrier et al. 2009) and ATP-citrate lyase (ACLY), a key enzyme in the cytosolic acetyl-CoA biosynthesis pathway that has important roles in regulating de novo lipid synthesis and histone acetylation in metazoans (Wellen et al. 2009) (fig. 3B). Moreover, we found multiple potential SrHomer-binding proteins with unknown functions, including EF-hand calcium-binding domain-containing protein 5, SNW domain-containing protein 1, Tetratricopeptide Repeat Protein 35 (TTC 35), and Neighbor of COX4 (COX4NB). Interestingly, TTC 35 was first identified as part of the mouse nuclear envelope proteome (Dreger et al. 2001) and was later shown to interact with COX4NB in a yeast two-hybrid screen (Boxem et al. 2008).
Homer Binds Directly to and Colocalizes with Flotillin 1 in the Nucleus of S. rosetta
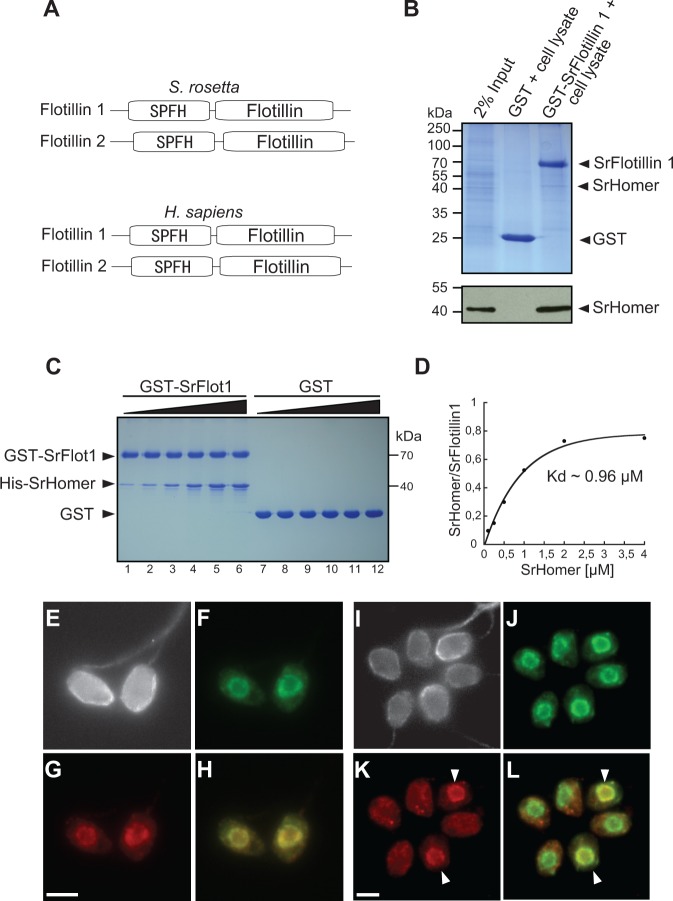
The most abundant detected SrHomer-binding partners, the Flotillins (also known as Reggies [Schulte et al. 1997]), are markers of lipid rafts (Bickel et al. 1997) and act as scaffolds for large heteromeric complexes that signal across plasma membranes in metazoans. Originally discovered in neurons during axon regeneration (Schulte et al. 1997), Flotillins were subsequently identified in diverse eukaryotes and bacteria (Rivera-Milla et al. 2006). Nonetheless, their molecular functions remain elusive (Stuermer 2010). We found that the protein domain composition and arrangement of Flotillins 1 and 2 are conserved in metazoans and in choanoflagellates (fig. 4A, supplementary fig. S3, Supplementary Material online), raising the possibility that aspects of their in vivo functions may also be conserved.
Fig. 4.
SrHomer interacts directly with SrFlotillin 1 in the Salpingoeca rosetta nucleus. (A) Salpingoeca rosetta and human Flotillins 1 and 2 contain a conserved N-terminal SPFH-domain and a C-terminal Flotillin domain. (B) Purified GST-SrFlotillin 1 (GST-SrFlot1) pulls down SrHomer from S. rosetta cell lysate. Glutathione-sepharose beads bound to GST alone or GST-SrFlotillin 1 were incubated with S. rosetta cell lysate. The input lane represents 2% of the lysate used in the pull-down experiments. The remaining lanes show elutions from the beads separated by SDS-PAGE and visualized with Coomassie Blue (upper panel) or by immunoblotting with SrHomer antibodies (lower panel). (C) Purified His-SrHomer binds directly to GST-SrFlotillin 1 but not GST alone. To measure the binding affinity between SrHomer and SrFlotillin 1, increasing concentrations of His-SrHomer were incubated with 2 µM GST-SrFlotillin 1 (lanes 1–6), or with 2 µM GST (lanes 7–12) bound to glutathione-Sepharose beads for 1 h at RT, washed and analyzed on SDS-PAGE (concentrations of His-SrHomer used for the binding assays in lanes 1–6 and lanes 7–12 were 0.1 µM, 0.25 µM, 0.5 µM, 1 µM, 2 µM, and 4 µM, respectively). Saturated binding of SrHomer to SrFlotillin 1 was reached at approximately 2 µM SrHomer. (D) Amounts of precipitated His-SrHomer were measured, normalized to GST-SrFlotillin 1, and plotted. The Kd of the reaction was determined by fitting the data to a Hill function assuming 1:1 binding stoichiometry. The apparent binding Kd is approximately 0.96 µM. (E–L) SrFlotillin 1 colocalizes with SrHomer in all solitary S. rosetta cells (E–H) and in some (indicated with white arrowhead), but not all, cells within S. rosetta rosette colonies (I–L). Subcellular localization of beta-tubulin (E and I, gray), SrHomer (F and J, green), and SrFlotillin 1 (G and K, red) in S. rosetta. (H and L). The overlay of SrHomer and SrFlotillin 1 shows colocalization of the two proteins, presumably in the nucleus. Scale bar: 2 µm. Sr, S. rosetta.
To test whether the detected Homer/Flotillin interaction is physiologically relevant in S. rosetta, we performed glutathione-S-transferase (GST) pull-down experiments against S. rosetta lysates. Bacterially expressed S. rosetta Flotillin 1 (GST-SrFlotillin1) immobilized on glutathione-sepharose beads pulled down SrHomer from S. rosetta cell lysates (fig. 4B). The interaction between SrFlotillin 1 and SrHomer is apparently through direct binding; we found that bacterially expressed SrHomer binds to GST-SrFlotillin 1 but not to GST alone. Moreover, saturated binding of SrHomer to SrFlotillin 1 was reached at approximately 2 µM SrHomer (fig. 4C), and we measured a Kd of approximately 0.96 µM (fig. 4D). This relatively high affinity compares well with affinities measured for other synaptic protein–protein interactions (Jahn and Fasshauer 2012) and is consistent with this being a physiologically relevant interaction.
The detection of a direct interaction between SrHomer and SrFlotillin predicted that SrFlotillin should localize to the nucleus in S. rosetta. Indeed, immunofluorescence microscopy reveals that SrHomer and SrFlotillin colocalize in the nucleoplasm of all solitary cells (fig. 4E–H). Interestingly, although SrHomer was found in the nucleus in all cells in colonies, SrFlotillin colocalized with SrHomer in only a subset of colony cell nuclei and was otherwise found distributed throughout the cytoplasm (fig. 4I–L).
A Conserved Homer 1/Flotillin 1 Complex in the Nucleus of Vertebrate Astrocytes
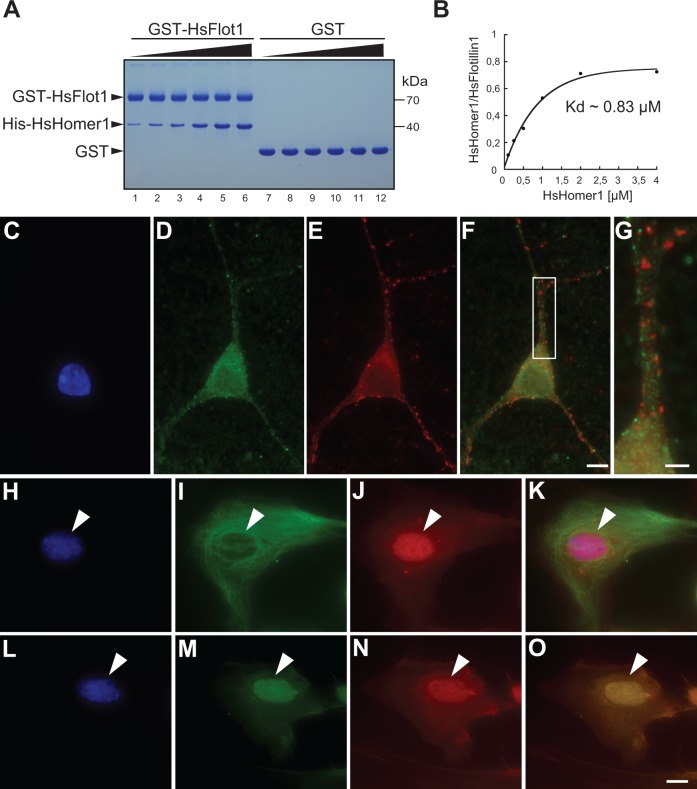
Although Homer and Flotillin are each commonly considered to be membrane-associated proteins, both have been detected in the nuclei of certain types of cells. The direct interaction between SrHomer and SrFlotillin 1 and their colocalization in the S. rosetta nucleus raised the possibility that a Homer:Flotillin interaction might also be conserved in metazoans. To test for an interaction between the two proteins, human Flotillin 1 (HsFlotillin1) protein fused to GST (GST-HsFlotillin1) was immobilized on glutathione-sepharose beads and incubated with human Homer 1 (HsHomer1) protein. We found that HsHomer 1 binds to GST-HsFlotillin 1 but not to GST alone (fig. 5A). Saturated binding of HsHomer 1 to HsFlotillin 1 was reached at approximately 2 µM HsHomer and we measured a Kd of approximately 0.83 µM (fig. 5B). This relatively high Kd, which is comparable to the Kd of the S. rosetta Homer/Flotillin 1 interaction, suggests that the interaction of HsHomer 1 to HsFlotillin 1 reported here may be physiologically relevant. Moreover, the observation that HsHomer 1 and HsFlotillin 1 are capable of interacting directly, as they do in S. rosetta, suggests that the two proteins interacted in the last common ancestor of metazoans and choano-flagellates.
Fig. 5.
Homer 1 and Flotillin 1 from vertebrates interact directly and colocalize in the nuclei of primary astrocytes. (A) Bacterially expressed His-HsHomer 1 binds to GST-HsFlotillin 1 (GST-HsFlot1) but not GST. To measure the binding affinity between HsHomer and HsFlotillin 1, increasing concentrations of His-HsHomer were incubated with 2 µM GST-HsFlotillin 1 (lanes 1–6) or with 2 µM GST (lanes 7–12) bound to glutathione-Sepharose beads for 1 h at RT, washed, and analyzed on SDS-PAGE (concentrations of His-HsHomer 1 used for the binding assays in lanes 1–6 and lanes 7–12 were 0.1 µM, 0.25 µM, 0.5 µM, 1 µM, 2 µM, and 4 µM, respectively). Saturated binding of HsHomer 1 to HsFlotillin 1 was reached at approximately 2 µM SrHomer. (B) The Kd of the reaction was determined by fitting the data to a Hill function assuming 1:1 binding stoichiometry. The apparent binding Kd is approximately 0.83 µM. (C–G) Homer 1 and Flotillin 1 do not appear to colocalize in rat hippocampal neurons. Hippocampal neurons (12 days in vitro) were stained with DAPI (C, blue), Flotillin 1 antibodies (D, green), and Homer 1 antibodies (E, red). Although Flotillin 1 and Homer 1 both form puncta throughout the dendrites and cytoplasm, both appear to be excluded from the nucleus. An overlay of Flotillin 1 and Homer 1 is shown in panel (F). (G) Segment of dendrite magnified from (F) shows little colocalization between Flotillin 1 and Homer 1. (H–K) Homer 1 localizes to the nucleus in rat astrocytes (4 days in vitro) as shown by staining with DAPI (H, blue), antibodies to the astrocyte marker GFAP (I, green), and antibodies to Homer 1 (J, red). (K) The overlay of DAPI, GFAP, and Homer 1 reveals that Homer 1 colocalizes with DNA in the nucleus. (L–O) Flotillin 1 and Homer 1 colocalize in rat astrocyte nuclei. Subcellular localization of DNA (L, blue), Flotillin 1 (M, green), and Homer 1 (N, red) are shown. (O) The overlay of DAPI-stained DNA, Flotillin1, and Homer 1 shows colocalization in the nucleus. Scale bars: 10 µm in (O) and (F), 5 µm in (G). Hs, Homo sapiens.
We next investigated circumstances under which Homer and Flotillin might colocalize in metazoans. Because the function of Homer is best understood in the context of neurons, we investigated the localization of Homer and Flotillin in two different neuronal cell types in rats: Hippocampal neurons and hippocampal astrocytes. Homer 1 and Flotillin 1 did not colocalize in dendritic spines and, consistent with prior studies, neither protein was detected in the nuclei of primary rat hippocampal neurons (fig. 5C–G). However, we detected Homer 1 in nuclei of primary rat hippocampal astrocytes (fig. 5H–K), which were identified by the astrocyte marker Glial Fibrillary Acidic Protein (GFAP; fig. 5I). Moreover, costaining with antibodies to Homer 1 and Flotillin 1 revealed that the two proteins colocalize in the nuclei of astrocytes (fig. 5L–O), just as their homologs do in the nuclei of S. rosetta. Thus, not only is the interaction of Homer and Flotillin conserved but also their colocalization in the nucleus.
Discussion
Our study of the choanoflagellate protein SrHomer has revealed new features of Homer and Flotillin biology in metazoans, suggesting that the interaction between these two proteins is conserved. Indeed, the evolution of the Homer protein family may provide an example of exaptation at the molecular level. In the last common ancestor of metazoans and choanoflagellates, long before the evolution of neurons and the PSD, an ancestral Homer protein likely functioned in the nucleus and interacted with Flotillin. If the nucleus was the sole context for Homer function in the first metazoans, the later co-option of Homer to its better known function as a scaffolding protein (e.g., at the PSD and in the ER) likely involved the evolution of new interactions with proteins that were already encoded in the genome (e.g., Shank and PSD-95). Alternatively, Homer may have been pleiotropic in the last common ancestor of metazoans and choanoflagellates, functioning both as a scaffolding protein and a nuclear protein. In this case, we infer that the scaffolding function for Homer was either lost in the choanoflagellate lineage or not apparent under the experimental conditions in this study.
Although an association between Homer and Flotillin has not previously been reported and the functions of these binding partners in the nucleus remain to be elucidated, both proteins have been observed independently in the nuclei of metazoan cells. For example, Homer in the nucleus of T cells interacts with the transcription factor C/EBPbeta to regulate serum response element activation (Ishiguro and Xavier 2004) and has been observed in nuclei of muscle fiber cells and cardiomyocytes (Volpe et al. 2004; Chiarello et al. 2013), where its functions are not well understood. Moreover, nuclear localization of Flotillin 1 can induce cell proliferation in prostate cancer cells (Santamaría et al. 2005) and regulate the abundance and activity of the mitotic regulator Aurora B kinase in cervical cancer cells (Gómez et al. 2010). We hypothesize that nuclear-localized Homer in choanoflagellates, and possibly in astrocytes, regulates nuclear Ca2+-entry and dynamics, as two Ca2+-channels—ryanodine receptor and IP3 receptor (which we detected as a potential binding partner of SrHomer, albeit with only a few unique peptide hits)—are known to be activated by Homer and have been detected in the nuclear membranes of diverse metazoan cell types (Gerasimenko O and Gerasimenko J 2004). In addition, our finding that Flotillin 1 is localized to the nucleus in all solitary S. rosetta cells (fig. 4F), while localizing to the nucleus in only a subset of colonial cells (fig. 4J), suggests that SrFlotillin may influence cell differentiation in S. rosetta colonies.
In addition to Flotillins 1 and 2, we identified other novel potential Homer-binding partners, some of which have been shown to localize to the nucleus in metazoan cells (e.g., ACLY [Wellen et al. 2009], SNW domain-containing protein 1 [also known as SKIP 1] [Folk et al. 2004], and srGAP [Guerrier et al. 2009]). For example, srGAP 1 and 3 translocate during development between the nucleus and the cytoplasm of rat neurons (Yao et al. 2008). In addition, srGAP 2 has been shown to accumulate in the head of dendritic spines within Homer 1 clusters and is detected in the PSD of mouse cortical neurons (Charrier et al. 2012). Both Homer 1 and srGAP 2 regulate axon pathfinding in the central nervous system (Foa et al. 2001; Guerrier et al. 2009) and may facilitate signal transduction from cell-surface receptors to intracellular proteins that govern the establishment of axon trajectories. Whether srGAP and other novel identified binding partners detected in this study are true Homer interacting proteins in vivo and might cooperate (e.g., in axon pathfinding) will require further study.
Despite the evolutionary distance separating choanoflagellates from vertebrates, we find that critical biochemical properties of Homer proteins are conserved in both lineages. Importantly, the ability of Homer to form tetramers, a feature that is necessary for the structural integrity of the PSD and dendritic spines in neurons, likely evolved long before the origin of eumetazoans and synapses. On the other hand, our immunoprecipitation experiments did not detect an interaction between SrHomer and Shank in S. rosetta. It is possible that the two proteins might interact under other conditions or that they interacted in the last common ancestor of choanoflagellates and metazoans but that the interaction was lost in S. rosetta. On the other hand, it is also possible that the core of the postsynaptic protein complex formed by Homer and Shank evolved after the divergence of the choanoflagellate and metazoan lineages. Consistent with the hypothesis that the protein interactions involved in the PSD are restricted to metazoans, we also did not detect other postsynaptic proteins, including GKAP, PSD-95, Cortactin, and CamKII, that are known to be part of the Homer protein interaction network in metazoans and encoded in the S. rosetta genome. Identifying Homer-binding partners in sponges and cnidarians may help to clarify when and how core postsynaptic proteins such as Homer, Shank, GKAP, and PSD-95 were integrated to form a primordial synaptic-like scaffold and contributed to the evolution of synapses and neurons.
Materials and Methods
Protein Searches and Analysis
We performed initial protein homology-based searches using the basic local alignment sequence tool (BLAST: BLASTp and tBLASTn, Altschul et al. 1997) using Homo sapiens proteins as queries in the genomes of eight metazoans (Mus musculus, Strongylocentrotus purpuratus, D. melanogaster, Caenorhabditis elegans, Nematostella vectensis, T. adhaerens, O. carmela, and A. queenslandica), two choanoflagellates (M. brevicollis and S. rosetta), one filasterean (C. owczarzaki), and three fungi (Rhizopus oryzae, Saccharomyces cerevisiae, and Batrachochytrium dendrobatidis) with the default BLAST parameters and an e-value threshold of 10 × 10−5 at the National Center for Biotechnology Information (NCBI). In addition, we searched databases at the Joint Genome Institute (genomes of M. brevicollis, T. adhaerens, and N. vectensis), databases associated with the Origins of Multicellularity Sequencing Project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/annotation/genome/multicellularity_project/MultiHome.html, last accessed June 6, 2014), and the A. queenslandica genome database (http://www.metazoa.ensembl.org/Amphimedon_queenslandica/blastview, last accessed June 6, 2014).
The stand-alone BLAST search algorithm was used to search the best predicted protein set from the draft genome of O. carmela (Nichols et al. 2012) using synaptic proteins from H. sapiens as a query. When the BLAST searches of the genome data described above returned significant “hits,” the sequences obtained were then reciprocally searched against the NCBI protein database by BLAST to confirm the validity of the sequences retrieved with the initial search. Pfam (Finn et al. 2010), SMART, and Phobius (Käll et al. 2007) domain prediction programs were then run on all proteins identified to analyze their protein domain composition, and this additional criterion was used to identify bona fide candidate orthologs. In addition, we specifically searched for, but did not find, Homer homologs in Bacteria, Chlorophyta, Dictyosteliida, Apusozoa, Fungi, and Ichthyosporea. Protein accession numbers and protein domain composition are shown in supplementary table S1, Supplementary Material online.
Protein Expression and Purification
Full-length SrHomer (amino acids 1–355) and a C-terminal deletion of SrHomer (SrHomerΔCC, amino acids 1–282), full-length SrFlotillin 1 (amino acids 1–427), and full-length HsHomer 1 (amino acids 1–354) were amplified by polymerase chain reaction (PCR) from S. rosetta cDNA or H. sapiens cDNA (Invitrogen), with the following primers:
1.) Full-length SrHomer forward: GGAATTCCATATGTTCAAGACACAGGCGCAT, which contains a NdeI restriction site (underlined).
2.) Full-length SrHomer reverse: GCGCCCTCGAGTCATGACACGCTGTGTGCTGCCTC, which contains a XhoI restriction site (underlined) and a stop codon.
3.) SrHomerΔCC forward: GGAATTCCATATGTTCAAGACACAGGCGCAT, which contains a NdeI restriction site (underlined).
4.) SrHomerΔCC reverse: GCGCCCTCGAGTCATGACGCCTCAAGCTGCTG, which contains a XhoI restriction site (underlined) and a stop codon.
5.) Full-length SrFlotillin 1 forward: CCGGAATTCATGGCCTTCTACTCCTCT, which contains an EcoRI restriction site (underlined).
6.) Full-length SrFlotillin 1 reverse CCGGCGGCCGCTCAGACCTGGGACACCTTGAT, which contains a NotI restriction site (underlined) and a stop codon.
7.) Full-length HsHomer 1 forward: GGAATTCCATATGGGGGAACAACCTATC, which contains NdeI restriction site (underlined).
8.) Full-length HsHomer 1 reverse: CCGGAATTCTCAGCTGCATTCTAGTAG, which contains an EcoRI restriction site (underlined) and a stop codon.
In addition, codon-optimized full-length HsFlotillin 1 (amino acids 1–427) was prepared by gene synthesis (GenScript) and amplified by PCR with the following primers:
9.) Full-length HsFlotillin 1 forward: CCGGAATTCATGTTTTTCACTTGTGGC, which contains an EcoRI restriction site (underlined).
10.) Full-length HsFlotillin 1 reverse: GCGCCCTCGAGTCAGGCTGTTCTCAAAGG, which contains a XhoI restriction site (underlined) and a stop codon.
We cloned SrHomer and HsHomer into the pET28a+ (Novagen) expression vector to generate proteins N-terminally tagged with hexahistidine tag (His-SrHomer and His-HsHomer). SrFlotillin 1 and HsFlotillin 1 were cloned into the pGEX-6P-1 (Amersham Biosciences) expression vector to generate proteins N-terminally tagged with GST (GST-SrFlotillin 1 and GST-HsFlotillin 1). We transformed Escherichia coli BL21 cells to express each construct. SrHomer and HsHomer were expressed at 37 °C for 3 h and SrFlotillin 1, and HsFlotillin 1 were expressed at 16 °C for 16 h. Fusion proteins were purified by Ni2+ NTA or glutathione affinity chromatography. Bacteria were mixed with 1 mM phenylmethanesulfonyl fluoride (PMSF) and 1 mg/ml lysozyme and incubated at room temperature for 20 min. This was followed by addition of 5 mg DnaseI, 1 mM MgCl2, and 1% (v/v) Triton X-100. Cells were sonicated by 4 × 30 s pulses. Immediately after, 1 mM PMSF was added, and the lysate was incubated at room temperature for 10 min again. Bacterial debris was removed by centrifugation at 14,000 rpm for 1 h. The supernatant containing the protein of interest was incubated either with Ni2+-NTA-agarose or glutathione sepharose beads for about 2 h at 4 °C. Ni2+-NTA-beads were washed with 200 ml Ni2+-washing buffer (20 mM Tris pH 7.4, 500 mM NaCl, 8 mM imidazole), and the His-tagged proteins were eluted with elution buffer (5 ml, Ni2+-washing buffer containing 400 mM imidazole). Glutathione sepharose beads were washed with 200 ml GST-washing buffer (20 mM Tris pH 7.4, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid [EDTA], and 1 mM dithiothreitol [DTT]), and the GST-tagged proteins were eluted with elution buffer (5 ml, GST-washing buffer containing 5 mM glutathione). Eluates were dialyzed overnight at 4 °C. The dialysis buffer contained 20 mM Tris pH 7.4, 200 mM NaCl, 1 mM EDTA, and 1 mM DTT. All proteins were further purified by gel filtration on a Superdex-75 10/300 or Superdex-200 10/300 column.
CD Spectroscopy
The bacterially expressed and purified SrHomer coiled-coil domain (amino acids 283–355) (SrHomerCC) was further purified by gel filtration on a Superdex 75 10/300 column. The hexahistidine tag was removed by thrombin digestion of the SrHomerCC prior to the gel filtration run. SrHomerCC was dialyzed over night at 4 °C in 20 mM NaH2PO4/Na2HPO4, 200 mM NaCl buffer. CD spectroscopy measurements were performed on an Aviv 62DS spectrometer at a protein concentration of 0.25 mg/ml SrHomerCC in 20 mM NaH2PO4/Na2HPO4, 200 mM NaCl buffer. Hellma quartz cuvettes with 1 cm path length were used. The absorption signal was collected in a wavelength range of 195–250 nm using steps of 1 nm with a scan rate of 60 nm/min and an averaging time of 2 s. The measurements were carried out at 25 °C.
MALS
For MALS studies, purified SrHomer and SrHomerΔCC (∼1 mg/ml injected concentration) were subjected to a Superdex 200 10/300 analytical SEC equilibrated overnight in gel filtration buffer (20 mM Tris, 200 mM NaCl, 1 mM EDTA, 1 mm DTT, and pH 7.4). The chromatography system was coupled to an 18-angle light scattering detector (DAWN HELEOS-II) and a refractive index detector (Optilab T-rEX) (Wyatt Technology). Data were collected every second at a flow rate of 0.5 ml/min. Data analysis was carried out using the program ASTRA, yielding the molar mass and mass distribution of the sample. For normalization of the light scattering detectors and data quality control, monomeric bovine serum albumin (BSA; Sigma) was used. The measurements were carried out at 25 °C.
SrHomer Antibody Production and Western Blot Analysis
We generated rabbit anti-SrHomer polyclonal antibodies by PCR amplifying the region of SrHomer encoding the C-terminal CC domain (amino acids 283–355) from S. rosetta cDNA using the following primers: GGAATTCCATATGCAGAACGCGGAGAAGTCC and reverse GCGCCCTCGAGTCATGACACGCTGTGTGCTGCCTC. The forward primer contained a NdeI restriction site (underlined), and the reverse primer contained a XhoI restriction site (underlined) as well as a stop codon. We cloned the synthesized DNA into pET28a+ (Novagen) and pMAL-c2X (New England BioLabs) expression vectors to generate two fusion proteins: SrHomerCC protein N-terminally tagged with hexahistidine tag (His-SrHomerCC) and the same protein tagged N-terminally with maltose-binding protein (MBP-SrHomerCC). We transformed E. coli BL21 cells and expressed His-SrHomerCC and MBP-SrHomerCC for 3 h at 37 °C. His-SrHomerCC and MBP-SrHomerCC were purified essentially as described above. For MBP-SrHomerCC, amylose resin was used. Amylose resin beads were washed with 20 ml washing buffer (20 mM Tris pH 7.4, 200 mM NaCl) and the MBP-SrHomerCC was eluted with elution buffer (5 ml, washing buffer containing 10 mM maltose).
Polyclonal antibodies were raised in rabbits against His-SrHomerCC antigen (Gensript), and we affinity purified anti-SrHomer antibodies from the rabbit serum using MBP-SrHomerCC protein. For affinity purification, 5 mg MBP-SrHomerCC protein was covalently bound to an AminoLink Plus Coupling column (Thermo Scientific) overnight at 4 °C in coupling buffer (0.1 M sodium phosphate, 150 mM NaCl, pH 10) and 40 μl of cyanoborohydride. The column was washed with 4 ml of Quenching Buffer (1 M Tris HCl, pH 7.4) and mixed gently for 30 min by end-over-end rocking with 2 ml Quenching Buffer and 40 μl of Cyanoborohydride Solution. The column was then equilibrated by adding 6 ml of Wash Buffer (1× phosphate buffered saline [PBS], pH 7.4). The rabbits serum was allowed to flow into the resin bed of the column and incubated for 1 h at 25 °C. The column was washed with 12 ml of wash buffer, and the antibody was eluted with 1 ml 0.2 M glycine HCl, pH 2.5. The pH of each fraction was adjusted to neutral by adding 50 μl of Tris HCL pH 9.3. Fractions with highest amount of antibodies were pooled and dialyzed against 1× PBS.
For the Western blot analysis, S. rosetta cells (40 mg choanoflagellate wet pellet) were resuspended in 1 ml lysis buffer (20 mM potassium phosphate buffer, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM ethyleneglycoltetraacetic acid [EGTA], 1% Triton X-100) containing protease inhibitor cocktail (Roche) for 30 min at 4 °C. The crude lysate was clarified by centrifugation at 20,000 × g, and 20 µl per lane were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 14% gel. Proteins were transferred to Immobilon-P membranes (Millipore). Membranes were blocked overnight at 4 °C in 5% nonfat dry milk in 1× PBS containing 0.05% Tween 20 (PBST) and then incubated with antibodies against SrHomer (1:1,000) in blocking solution for 1 h at room temperature and washed extensively in PBST. After 1 h incubation with the secondary antibody (goat antirabbit IgG antibody, horseradish peroxidase conjugate, Jackson Immuno Research, 1:10,000) at room temperature, membranes were washed in PBST and developed by using the SuperSignal West Pico Chemiluminescent substrate (Thermo Scientific). Antibodies against His-SrHomer recognize a single band of approximately 41 kDa (supplementary fig. S2, Supplementary Material online). In addition, preincubation of the antibody with a competitor (2 μg of maltose-binding protein-tagged SrHomer) nearly eliminated the detection of the protein, indicating that the competitor bound to the available antibody (supplementary fig. S2, Supplementary Material online).
Salpingoeca rosetta Cell Culture and Microscopy
For immunofluorescence, cultures of S. rosetta enriched in single cells or rosette colonies were grown to a density of 106 cells/ml. Cells were pelleted by spinning for 10 min at 500 × g and resuspended in a small volume of artificial seawater pH 8.0. Approximately 0.4 ml of the cells were applied to poly-l-lysine-coated coverslips and left to attach for 30 min. Cells were fixed for 5 min with 1 ml 6% acetone, for 15 min with 1 ml 4% formaldehyde, and for 10 min with 1 ml 100% ice cold methanol. Acetone and formaldehyde were diluted in artificial seawater, pH 8.0. After incubation with 100% methanol, coverslips were washed gently four times with 1 ml washing buffer (100 mM PIPES at pH 6.9, 1 mM EGTA, and 0.1 mM MgSO4) and incubated for 30 min in 1 ml blocking buffer (washing buffer with 1% BSA, 0.3% Triton X-100). Cells were incubated with primary antibodies diluted in 0.15 ml blocking buffer for 1 h, washed four times with 1 ml of blocking buffer, and incubated for 1 h in the dark with fluorescent secondary antibodies (1:100 in blocking buffer, Alexa Fluor 488 goat antimouse, and Alexa Fluor 568 goat antirabbit; Invitrogen). Coverslips were washed three times with washing buffer and mounted onto slides with Fluorescent Mounting Media (4 μl; Prolong Gold Antifade, Invitrogen). The following primary antibodies were used: Mouse monoclonal antibody against β-tubulin (E7, 1:400; Developmental Studies Hybridoma Bank) and nuclear pore complexes (1:100, Covance); rabbit polyclonal antibodies against SrHomer (1:200); and rat Flotillin 1 (1:100, Abcam). For colocalization studies of SrHomer and SrFlotillin, antibodies against SrHomer were labeled with Alexa Fluor 488 Protein Labeling Kit (Invitrogen). Images were taken with a 100× oil immersion objective on a Leica DMI6000 B inverted compound microscope and Leica DFC350 FX camera.
For TEM, choanoflagellate cells were concentrated by gentle centrifugation (3,200 g for 10 min), loaded into 100-µm deep specimen carriers, and high pressure frozen in a Bal-Tec HPM 010 high pressure freezer (Bal-Tec AG, Liechtenstein). Freeze substitution with 1% osmium tetroxide plus 0.1% uranyl acetate in acetone was performed over 2 h by the SQFS method of McDonald (McDonald 2013), then infiltrated with Eponate 12 resin and polymerized in a Pelco Biowave research microwave oven (Ted Pella, Inc., Redding, CA) over a period of 2 h. Sections were cut at 70 nm thickness, poststained with uranyl acetate and lead citrate, and viewed in a Tecnai 12 transmission EM (FEI Inc., Hillsboro, OR) operating at 120 kV. Images were recorded on a Gatan Ultrascan 1000 CCD camera. Immunogold labeling was performed essentially as described (McDonald 2013). Sections of choanoflagellate cells were incubated in primary Homer antibody diluted 1:50 for 1 h. Secondary antimouse antibodies conjugated to 10 nm gold were used to label the sections. Grids were poststained for 4 min in 2% aqueous uranyl acetate and 2 min in lead citrate.
IP and Mass Spectrometry
Two independent SrHomer IP experiments were performed: IP1 with 2.0 mg antibody-coupled beads/ml and 40 mg choanoflagellate wet pellet and IP2 with 7.5 mg antibody-coupled beads/ml and 800 mg choanoflagellate wet pellet. To prepare samples for mass spectrometry, choanoflagellate rosette colonies were dissolved in lysis buffer (20 mM potassium phosphate buffer, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 1% Triton X-100) containing protease inhibitor cocktail (Roche). The crude lysate was clarified by centrifugation at 20,000 × g for 20 min and then incubated with anti-SrHomer beads, which were prepared by conjugating affinity purified SrHomer antibodies to Dynabeads M-270 Epoxy (Invitrogen) according to the manufacturer’s instructions. Negative control beads were prepared in parallel using a purified rabbit polyclonal control antibody (Genscript). The lysate was incubated with antibody-coupled beads for 1 h at 4 °C, washed with lysis buffer, and proteins were eluted in 0.2 M Glycine pH 2.5, 1 mM EDTA. For mass spectrometry, each sample was loaded in its entirety on a 4–20% Tris-HCl Gel (BioRad) and run until the dye front was 1 cm from the bottom of the gel. The gel was cut into ten equal slices and digested in-gel by trypsin (Olsen et al. 2007). The tryptic digests were analyzed by LC-MS/MS using a nanoflow EASY-nLC system and an Orbitrap Q-Exactive instrument (Thermo Scientific, Bremen, Germany). The data were acquired in a DDA manner with a loop count of 12. The resolution in the full scan was 70,000, AGC target of 1E6, maximum IT 120 ms, and 1 microscan. The MS/MS scans were recorded with a resolution of 17,500, AGC target of 1E6, maximum IT 60 ms, isolation window 1.2 m/z, stepped NCE of 25, and underfill ratio 1.2% (intensity threshold of 2E5). The dynamic exclusion was set to 30 seconds. The peptides were separated on a 90 min gradient (8–40% acetonitrile in 0.1% formic acid) on a reverse-phase column (1.9 µm beads, Dr Maisch GmbH, Ammerbach, Germany). The raw data was searched against a S. rosetta FASTA-protein database (Origins of Multicellularity Sequencing Project, Broad Institute of Harvard and MIT; http://www.broadinstitute.org/, last accessed June 6, 2014) using Mascot as a search engine in Proteome Discoverer v1.3 (Thermo Scientific, Bremen, Germany) with a peptide false discovery rate of less than 1%, which was estimated on the number of reverse hits. Carbamidomethylation and oxidation of methionine were specified as variable modifications. A partial list of identified proteins is shown in figure 3B and the complete list in supplementary table S2, Supplementary Material online.
In Vitro Protein–Protein Interaction Assay
Salpingoeca rosetta lysate was incubated with 70 µg of bacterially expressed GST-Flotillin 1 immobilized on glutathione-Sepharose beads in binding buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 2 mM MgCl2, and 1 mM DTT). Incubations were carried out for 1 h at RT. The beads were then washed three times in 1 ml of binding buffer. Proteins bound to the beads were finally solubilized in SDS sample buffer (final concentrations: 60 mM Tris, pH 6.8, 2% SDS, 10% glycerine, 3% β-mercaptoethanol), heated for 5 min at 95 °C and subjected to SDS-polyacrylamide gel electrophoresis. Immunoblot analysis was performed using affinity purified SrHomer polyclonal antibody.
Bacterially expressed GST-SrFlotillin 1 or GST-HsFlotillin 1 (70 μg) bound to glutathione-Sepharose was incubated with 2–80 μg of bacterially expressed SrHomer or HsHomer 1 in 250 μl of binding buffer. Samples were rotated for 1 h at room temperature, washed three times with 1.0 ml of binding buffer, and 30 μl of SDS sample buffer was added to the bead bed; 15 μl of all samples were heated for 5 min at 95 °C and separated by SDS-PAGE on a 14% gel. Gels were stained with Coomassie blue and destained. For quantification, gel band intensities were measured in ImageJ, normalized to GST-Flotillin 1, and plotted. The Kd of the reaction was determined by fitting the data to a Hill function assuming 1:1 binding stoichiometry.
Immunohistochemical Staining of Rat Hippocampal Astrocytes and Neurons
Hippocampal astrocytes were isolated from the cerebral cortices of P0 rat pups. Cells were dissociated with papain and mechanical trituration. The cells were plated on poly-l-lysine-coated glass coverslips (12 mm) and cultured in high-glucose Dulbecco’s modified eagle medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C and in 5% CO2 (medium was replaced every 3–4 days). Dissociated hippocampal neurons were obtained from postnatal rats (P0-1) and plated on poly-l-lysine-coated glass coverslips (12 mm). Neurons were maintained in media containing MEM supplemented with 5% fetal bovine serum, B27 (Invitrogen), and GlutaMAX (Invitrogen). Both astrocytes and neurons were fixed with 4% PFA for 30 min at RT, rinsed three times with 1× PBS and incubated with blocking solution (10% (v/v) normal goat serum/0.2% Triton X-100 in 1× PBS) for 60 min at RT. Cells were then incubated for 1 h with primary antibodies dissolved in blocking solution, and after further washes (blocking solution), incubated for 1 h in the dark with fluorescent secondary antibodies (1:200 in blocking solution, Alexa Fluor 488 goat antimouse, and Alexa Fluor 568 goat antirabbit; Invitrogen), and washed again four times (blocking solution). After three washes (1× PBS), cells on coverslips were mounted onto slides with Fluorescent Mounting Media (4 μl; Prolong Gold Antifade, Invitrogen). Images were taken with a 100× oil immersion objective on an inverted Leica microscope. The following primary antibodies have been used: Mouse monoclonal antibodies against GFAP (1:1,500; Cell Signaling Technology) and Flotillin 1 (1:50, BD Biosciences), and rabbit polyclonal antibody against Homer 1 (1:200, Synaptic Systems).
Supplementary Material
Supplementary tables S1 and S2 and figures S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Bill Weis, Daniel Richter, Tera Levin, Rosanna Alegado, Nipam Patel, and John Kuriyan for helpful discussions and valuable comments on the manuscript; Katie Hart and Susan Marqusee for help with CD-measurements; Anna Geraghty and Daniela Kaufer for astrocytes; Josh Levitz, Ehud Isacoff, Tara Tracy, and Lu Chen for hippocampal neurons; and Emina Begovic for help with sequence alignments. They thank Burkhard Rammner (Sciloop) for his help with the graphic representation of the synapse in figure 1. This work was supported by a Deutsche Forschungsgemeinschaft postdoctoral fellowship to P.B. and NIH NIGMS GM089977 to N.K. N.K. is an Investigator in the Howard Hughes Medical Institute and a Senior Fellow in the Integrated Microbial Biodiversity program of the Canadian Institute for Advanced Research.
References
- Alié A, Manuel M. The backbone of the post-synaptic density originated in a unicellular ancestor of choanoflagellates and metazoans. BMC Evol Biol. 2010;10:34. doi: 10.1186/1471-2148-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu K, Cai Y, Bahri S, Yang X, Chia W. Roles of bifocal, Homer, and F-actin in anchoring Oskar to the posterior cortex of Drosophila oocytes. Genes Dev. 2004;18:138–143. doi: 10.1101/gad.282604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- Bickel PE, Scherer PE, Schnitzer JE, Oh P, Lisanti MP, Lodish HF. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J Biol Chem. 1997;272:13793–13802. doi: 10.1074/jbc.272.21.13793. [DOI] [PubMed] [Google Scholar]
- Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, de Lichtervelde L, Mul JD, van de Peut D, Devos M, et al. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakeman P, Lanahan A, O’Brien R. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- Burkhardt P, Stegmann CM, Cooper B, Kloepper TH, Imig C, Varoqueaux F, Wahl MC, Fasshauer D. Primordial neurosecretory apparatus identified in the choanoflagellate Monosiga brevicollis. Proc Natl Acad Sci U S A. 2011;108:15264–15269. doi: 10.1073/pnas.1106189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X. Unicellular Ca2+ signaling “toolkit” at the origin of metazoa. Mol Biol Evol. 2008;25:1357–1361. doi: 10.1093/molbev/msn077. [DOI] [PubMed] [Google Scholar]
- Carr M, Leadbeater BSC, Hassan R, Nelson M, Baldauf SL. Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc Natl Acad Sci U S A. 2008;105:16641–16646. doi: 10.1073/pnas.0801667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier C, Joshi K, Coutinho-Budd J, Kim JE, Lambert N, de Marchena J, Jin WL, Vanderhaeghen P, Ghosh A, Sassa T, et al. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012;149:923–935. doi: 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, Xu P, Wijayawardana SR, Hanfelt J, Nakagawa T, et al. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5:1158–1170. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- Chiarello C, Bortoloso E, Carpi A, Furlan S, Volpe P. Negative feedback regulation of Homer 1a on norepinephrine-dependent cardiac hypertrophy. Exp Cell Res. 2013;319:1804–1814. doi: 10.1016/j.yexcr.2013.04.019. [DOI] [PubMed] [Google Scholar]
- Conaco C, Bassett DS, Zhou H, Arcila ML, Degnan SM, Degnan BM, Kosik KS. Functionalization of a protosynaptic gene expression network. Proc Natl Acad Sci U S A. 2012;109:10612–10618. doi: 10.1073/pnas.1201890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, Alegado RA, Fairclough SR, Levin TC, Nichols SA, McDonald K, King N. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev Biol. 2011;357:73–82. doi: 10.1016/j.ydbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreger M, Bengtsson L, Schöneberg T, Otto H, Hucho F. Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc Natl Acad Sci U S A. 2001;98:11943–11948. doi: 10.1073/pnas.211201898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott GRD, Leys SP. Evidence for glutamate, GABA and NO in coordinating behaviour in the sponge, Ephydatia muelleri (Demospongiae, Spongillidae) J Exp Biol. 2010;213:2310–2321. doi: 10.1242/jeb.039859. [DOI] [PubMed] [Google Scholar]
- Emes RD, Grant SGN. The human postsynaptic density shares conserved elements with proteomes of unicellular eukaryotes and prokaryotes. Front Neurosci. 2011;5:44. doi: 10.3389/fnins.2011.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emes RD, Grant SGN. Evolution of synapse complexity and diversity. Annu Rev Neurosci. 2012;35:111–131. doi: 10.1146/annurev-neuro-062111-150433. [DOI] [PubMed] [Google Scholar]
- Fairclough SR, Chen Z, Kramer E, Zeng Q, Young S, Robertson HM, Begovic E, Richter DJ, Russ C, Westbrook MJ, et al. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 2013;14:R15. doi: 10.1186/gb-2013-14-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough SR, Dayel MJ, King N. Multicellular development in a choanoflagellate. Curr Biol. 2010;20:875–876. doi: 10.1016/j.cub.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foa L, Rajan I, Haas K, Wu GY, Brakeman P, Worley P, Cline H. The scaffold protein, Homer1b/c, regulates axon pathfinding in the central nervous system in vivo. Nat Neurosci. 2001;4:499–506. doi: 10.1038/87447. [DOI] [PubMed] [Google Scholar]
- Folk P, Půta F, Skruzný M. Transcriptional coregulator SNW/SKIP: the concealed tie of dissimilar pathways. Cell Mol Life Sci. 2004;61:629–640. doi: 10.1007/s00018-003-3215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimenko O, Gerasimenko J. New aspects of nuclear calcium signalling. J Cell Sci. 2004;117:3087–3094. doi: 10.1242/jcs.01295. [DOI] [PubMed] [Google Scholar]
- Gómez V, Sesé M, Santamaría A, Martínez JD, Castellanos E, Soler M, Thomson TM, Paciucci R. Regulation of aurora B kinase by the lipid raft protein Flotillin-1. J Biol Chem. 2010;285:20683–20690. doi: 10.1074/jbc.M110.130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NW, Fourgeaud L, Huang B, Chen J, Cao H, Oswald BJ, Hémar A, McNiven MA. Dynamin 3 is a component of the postsynapse, where it interacts with mGluR5 and Homer. Curr Biol. 2003;13:510–515. doi: 10.1016/s0960-9822(03)00136-2. [DOI] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MK, Tang C, Verpelli C, Narayanan R, Stearns MH, Xu R-M, Li H, Sala C, Hayashi Y. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell. 2009;137:159–171. doi: 10.1016/j.cell.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GN, Huso DL, Bouyain S, Tu J, McCorkell KA, May MJ, Zhu Y, Lutz M, Collins S, Dehoff M, et al. NFAT binding and regulation of T cell activation by the cytoplasmic scaffolding Homer proteins. Science. 2008;319:476–481. doi: 10.1126/science.1151227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro K, Xavier R. Homer-3 regulates activation of serum response element in T cells via its EVH1 domain. Blood. 2004;103:2248–2256. doi: 10.1182/blood-2003-08-2671. [DOI] [PubMed] [Google Scholar]
- Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. Exploring the early origins of the synapse by comparative genomics. Biol Lett. 2009;5:108–111. doi: 10.1098/rsbl.2008.0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebeskind BJ, Hillis DM, Zakon HH. Evolution of sodium channels predates the origin of nervous systems in animals. Proc Natl Acad Sci U S A. 2011;108:9154–9159. doi: 10.1073/pnas.1106363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KL. Rapid embedding methods into epoxy and LR white resins for morphological and immunological analysis of cryofixed biological specimens. Microsc Microanal. 2013;20:1–12. doi: 10.1017/S1431927613013846. [DOI] [PubMed] [Google Scholar]
- Nichols SA, Dirks W, Pearse JS, King N. Early evolution of animal cell signaling and adhesion genes. Proc Natl Acad Sci U S A. 2006;103:12451–12456. doi: 10.1073/pnas.0604065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols SA, Roberts BW, Richter DJ, Fairclough SR, King N. Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/β-catenin complex. Proc Natl Acad Sci U S A. 2012;109:13046–13051. doi: 10.1073/pnas.1120685109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Mann M, Shevchenko A, Tomas H, Havlis J. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2007;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Philippe H, Derelle R, Lopez P, Pick K, Borchiellini C, Boury-Esnault N, Vacelet J, Renard E, Houliston E, Quéinnec E, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- Richards GS, Simionato E, Perron M, Adamska M, Vervoort M, Degnan BM. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr Biol. 2008;18:1156–1161. doi: 10.1016/j.cub.2008.06.074. [DOI] [PubMed] [Google Scholar]
- Rivera-Milla E, Stuermer CAO, Málaga-Trillo E. Ancient origin of Reggie (Flotillin), Reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cell Mol Life Sci. 2006;63:343–357. doi: 10.1007/s00018-005-5434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol. 2008;25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- Ryan TJ, Grant SGN. The origin and evolution of synapses. Nat Rev Neurosci. 2009;10:701–712. doi: 10.1038/nrn2717. [DOI] [PubMed] [Google Scholar]
- Sakarya O, Armstrong KA, Adamska M, Adamski M, Wang IF, Tidor B, Degnan BM, Oakley TH, Kosik KS. A post-synaptic scaffold at the origin of the animal kingdom. PLoS One. 2007;2:e506. doi: 10.1371/journal.pone.0000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Piëch V, Wilson NR, Passafaro M, Liu G, Sheng M. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- Salanova M, Priori G, Barone V, Intravaia E, Flucher B, Ciruela F, McIlhinney RA, Parys JB, Mikoshiba K, Sorrentino V, et al. Homer proteins and InsP 3 receptors co-localise in the longitudinal sarcoplasmic reticulum of skeletal muscle fibres. Cell Calcium. 2002;32:193–200. doi: 10.1016/s0143416002001549. [DOI] [PubMed] [Google Scholar]
- Santamaría A, Castellanos E, Gómez V, Benedit P, Renau-Piqueras J, Morote J, Reventós J, Thomson TM, Paciucci R. PTOV1 enables the nuclear translocation and mitogenic activity of Flotillin-1, a major protein of lipid rafts. Mol Cell Biol. 2005;25:1900–1911. doi: 10.1128/MCB.25.5.1900-1911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte T, Paschke KA, Laessing U, Lottspeich F, Stuermer CA. Reggie-1 and Reggie-2, two cell surface proteins expressed by retinal ganglion cells during axon regeneration. Development. 1997;124:577–587. doi: 10.1242/dev.124.2.577. [DOI] [PubMed] [Google Scholar]
- Shiraishi-Yamaguchi Y, Furuichi T. The Homer family proteins. Genome Biol. 2007;8:206. doi: 10.1186/gb-2007-8-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiber JA, Zhang Z-S, Burch J, Eu JP, Zhang S, Truskey GA, Seth M, Yamaguchi N, Meissner G, Shah R, et al. Mice lacking Homer 1 exhibit a skeletal myopathy characterized by abnormal transient receptor potential channel activity. Mol Cell Biol. 2008;28:2637–2647. doi: 10.1128/MCB.01601-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuermer CAO. The reggie/flotillin connection to growth. Trends Cell Biol. 2010;20:6–13. doi: 10.1016/j.tcb.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Suga H, Chen Z, de Mendoza A, Sebé-Pedrós A, Brown MW, Kramer E, Carr M, Kerner P, Vervoort M, Sánchez-Pons N, et al. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun. 2013;4:2325. doi: 10.1038/ncomms3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumlinski KK, Kalivas PW, Worley PF. Homer proteins: implications for neuropsychiatric disorders. Curr Opin Neurobiol. 2006;16:251–257. doi: 10.1016/j.conb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Thomas U. Modulation of synaptic signalling complexes by Homer proteins. J Neurochem. 2002;81:407–413. doi: 10.1046/j.1471-4159.2002.00869.x. [DOI] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, et al. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Volpe P, Sandri C, Bortoloso E, Valle G, Nori A. Topology of Homer 1c and Homer 1a in C2C12 myotubes and transgenic skeletal muscle fibers. Biochem Biophys Res Commun. 2004;316:884–892. doi: 10.1016/j.bbrc.2004.02.132. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley PF, Zeng W, Huang G, Kim JY, Shin DM, Kim MS, Yuan JP, Kiselyov K, Muallem S. Homer proteins in Ca2+ signaling by excitable and non-excitable cells. Cell Calcium. 2007;42:363–371. doi: 10.1016/j.ceca.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Tu JC, Petralia RS, Yuan JP, Doan A, Breder CD, Ruggiero A, Lanahan AA, Wenthold RJ, Worley PF. Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of Homer-related, synaptic proteins. Neuron. 1998;21:707–716. doi: 10.1016/s0896-6273(00)80588-7. [DOI] [PubMed] [Google Scholar]
- Yao Q, Jin W-L, Wang Y, Ju G. Regulated shuttling of Slit-Robo-GTPase activating proteins between nucleus and cytoplasm during brain development. Cell Mol Neurobiol. 2008;28:205–221. doi: 10.1007/s10571-007-9187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.