Abstract
The cysteine protease cathepsin B (CTSB) is frequently overexpressed in human breast cancer and correlated with a poor prognosis. Genetic deficiency or pharmacological inhibition of CTSB attenuates tumor growth, invasion and metastasis in mouse models of human cancers. CTSB is expressed in both cancer cells and cells of the tumor stroma, in particular in tumor-associated macrophages (TAM). In order to evaluate the impact of tumor- or stromal cell-derived CTSB on Polyoma Middle T (PyMT)-induced breast cancer progression, we used in vivo and in vitro approaches to induce human CTSB overexpression in PyMT cancer cells or stromal cells alone or in combination. Orthotopic transplantation experiments revealed that CTSB overexpression in cancer cells rather than in the stroma affects PyMT tumor progression. In 3D cultures, primary PyMT tumor cells showed higher extracellular matrix proteolysis and enhanced collective cell invasion when CTSB was overexpressed and proteolytically active. Coculture of PyMT cells with bone marrow-derived macrophages induced a TAM-like macrophage phenotype in vitro, and the presence of such M2-polarized macrophages in 3D cultures enhanced sprouting of tumor spheroids. We employed a doxycycline (DOX)-inducible CTSB expression system to selectively overexpress human CTSB either in cancer cells or in macrophages in 3D cocultures. Tumor spheroid invasiveness was only enhanced when CTSB was overexpressed in cancer cells, whereas CTSB expression in macrophages alone did not further promote invasiveness of tumor spheroids. We conclude that CTSB overexpression in the PyMT mouse model promotes tumor progression not by a stromal effect, but by a direct, cancer cell-inherent mode of action: CTSB overexpression renders the PyMT cancers more invasive by increasing proteolytic extracellular matrix protein degradation fostering collective cell invasion into adjacent tissue.
Keywords: CTSB, breast cancer, macrophages, invasion
INTRODUCTION
A tumor is not a homogenous entity of cancer cells but also comprises a variety of stromal cell types and extracellular matrix (ECM) components that together form the tumor microenvironment.1 The tumor stroma consists of endothelial cells, cancer-associated fibroblasts and tumor-associated immune cells, such as macrophages.2,3 Pathogenic interactions with cancer cells convert the tumor stroma into a favorable microenvironment for tumor growth and progression. A number of proteases are implicated in the pathogenic processes that occur in the coevolution of cancer cells and their microenvironment, among them are matrix metalloproteinases, urokinase-type plasminogen activator and cathepsins.4 Cysteine cathepsins are papain-like proteases (clan CA, family C1)5 with principal localization in the endolysosomal cell compartment. However, these enzymes are also secreted by lysosome exocytosis or via the secretory pathway.6,7 Over-expression and elevated enzymatic activities of cathepsins, most prominently cathepsin B (CTSB), have been linked to tumor invasiveness and poor patient prognosis in several cancer entities, including breast cancer.8–10 CTSB is also highly expressed in cells of the tumor stroma, in particular tumor-associated macrophages (TAMs).11,12 TAMs are educated toward the M2-like phenotype by cancer cell- or immune cell-derived interleukins.13,14 Such M2-polarized TAMs suppress their immune effector functions in favor of a pro-neoplastic ‘wound healing’ phenotype that promotes tumor progression by secretion of chemokines and by extracellular matrix breakdown.15,16 TAMs are often localized at the invasive front of the tumor,17 and they were shown to comigrate with cancer cells.18
In vivo, the impact of CTSB on tumor progression and metastasis has been studied almost exclusively in loss of function approaches by constitutive CTSB targeting11,19–21 and by selective genetic inactivation of CTSB either in cancer cells or in cells of the tumor stroma, particularly in TAMs.11,19,22–24 Pharmacologic inhibition of CTSB and other cysteine cathepsins showed therapeutic efficacy in several murine cancer models.20,25–28 Patient studies congruently establish an increased CTSB expression in human breast cancer cells8,10,29 caused by gene amplification, transcriptional activation, alternative splicing or further post translational processes (for review see Mohamed et al.30). Recently, a molecular link between expression of the epidermal growth factor receptor ErbB2/Her2 and CTSB expression, which promotes invasiveness of various human breast cancer cell lines, has been reported.31
In order to model the overexpression of CTSB that is often found in human breast cancer, we set out to study the in vivo effects of forced overexpression of human CTSB in the transgenic mouse mammary tumor virus (MMTV)/Polyoma Middle T (PyMT) mouse model of invasive breast cancer. In this mouse model, we found that transgenic overexpression of human CTSB accelerated tumor growth and increased metastatic burden in lungs.32 In this previous in vivo study, CTSB expression was regulated by the genuine human CTSB promoter, which results in ubiquitous CTSB expression and does not allow discrimination between cell type-specific effects. Therefore, we undertook the present experiments employing a combination of in vivo and 3D coculture approaches to discriminate between cancer cell- and stroma-mediated effects of CTSB overexpression on tumor growth and invasion.
RESULTS
CTSB overexpression in cancer cells promotes tumor growth, while CTSB overexpression in stroma has no effect
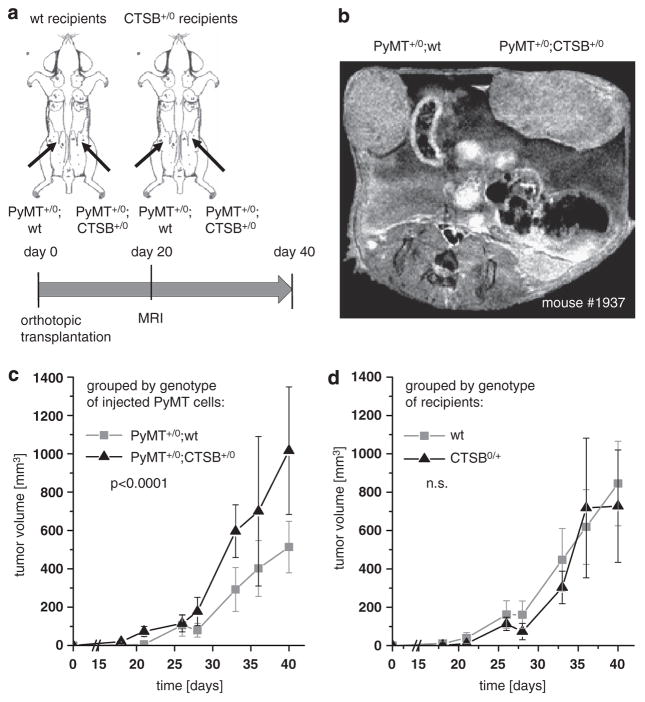
Ubiquitous overexpression of human CTSB in the transgenic PyMT model of invasive ductal mammary carcinoma resulted in enhanced tumor growth and lung metastasis in our previous study.32 Here we experimentally discriminate between cancer cell-autonomous and stromal CTSB effects by an orthotopic tumor model, for which primary PyMT breast cancer cells with human CTSB transgenic overexpression (PyMT+/0;CTSB+/0) or without the CTSB transgene (PyMT+/0;wt) were injected into a defined mammary gland of CTSB+/0 or wt recipients (Figure 1a). The recipient mice developed palpable tumors within the first week post injection, which grew to a size of 1.0 cm within 6 weeks. Correct anatomical localization of tumors in the mammary fat pad was assessed by magnetic resonance imaging (Figure 1b). Histologically, the tumors resembled primary tumors of the PyMT model and were largely undifferentiated. While encapsulated toward the skin, the tumors invaded the fat pad and the underlying breast muscle (Supplementary Figure 1a). CTSB immunohistology on orthotopic tumors showed that human CTSB is expressed in tumors derived from injection of PyMT+/0; CTSB+/0 and exhibit a very similar staining intensity and pattern as in tissue sections obtained from cancers of the primary PyMT breast cancer model with transgenic overexpression of human CTSB (Supplementary Figure 1b and Sevenich et al.32). The orthotopically growing tumors were well vascularized and showed macrophage infiltration, indicating a well-established tumor microenvironment provided by the host (Supplementary Figures 1c and d).
Figure 1.
Orthotopic transplantation of primary PyMT cells. (a) Schematic representation of experimental setup: primary tumor cells (0.5 × 106) either PyMT +/0;wt or PyMT +/0;CTSB +/0 were injected bilaterally into a defined mammary gland of female adult recipient mice of wt or CTSB+/0 genotype. Tumor volumes were determined by diameter measurement twice a week and by magnetic resonance imaging (MRI) at day 20 post injection. (b) Representative MRI image. (c) Tumor growth kinetics grouped by genotype (PyMT +/0;wt or PyMT +/0;CTSB +/0) of injected tumor cells. Bilateral tumor volumes of 9–11 animals depending on the time point. (d) Tumor growth kinetics grouped by genotype of recipient mice (wt or CTSB +/0). Bilateral tumor volumes of 9–11 animals per group depending on the time point. Data points represent mean ± s.e.m.
Tumor growth was systematically assessed over time by measurement of tumor diameters twice a week. The calculated tumor volumes were evaluated in dependance of either determinant, genotype of injected cells (that is, PyMT +/0;CTSB +/0 compared with PyMT +/0;wt) (Figure 1c) or genotype of recipient mice (that is, CTSB +/0 compared with wt) (Figure 1d). The growth curves of tumors derived from transplantation of PyMT+/0;CTSB+/0 and PyMT+/0;wt diverge significantly in the course of the experiment (P = 0.00087), whereas the growth curves of tumors in wt and CTSB+/0 recipient mice overlap and are not significantly different (P = 0.83). This reveals that the CTSB overexpression in the tumor cells is a pivotal determinant of end point tumor volume, whereas the CTSB overexpression in the recipient is not critical for tumor size. Tumors resulting from PyMT+/0;wt and from PyMT+/0; CTSB+/0 cancer cells showed similar rates of proliferating cells and only a low percentage of apoptotic cells in the tumor tissue (Supplementary Figures 2a–c). However, the orthotopic tumors had relatively large necrotic areas, but the extent of necrosis was not different in PyMT+/0;wt and PyMT+/0;CTSB +/0 tumors (Supplementary Figures 2d and e). Therefore, the observed higher tumor volume of PyMT+/0;CTSB+/0 compared with PyMT+/0;wt orthotopic tumors does not result from a shifted proliferation/cell death ratio but rather depends on other processes of tumor progression.
CTSB overexpression in tumor cells promotes collective cell invasion
Recently, the growth of tumor cells in a 3-dimensional (3D) matrix has been developed into a valuable in vitro approach to study tumor growth and invasion. In these 3D cultures, cancer cells grow as spheroids, which develop to acini-like structures that deposit basement membrane components in a collagen I matrix, thus mimicking the in vivo conditions for a growing epithelial tumor surrounded by connective tissue.33 Spheroids raised from PyMT cancer cells develop multicellular strands growing into the surrounding matrix in a collective cell invasion process, thereby degrading ECM molecules, such as collagen I (Supplementary Figures 3a and b).
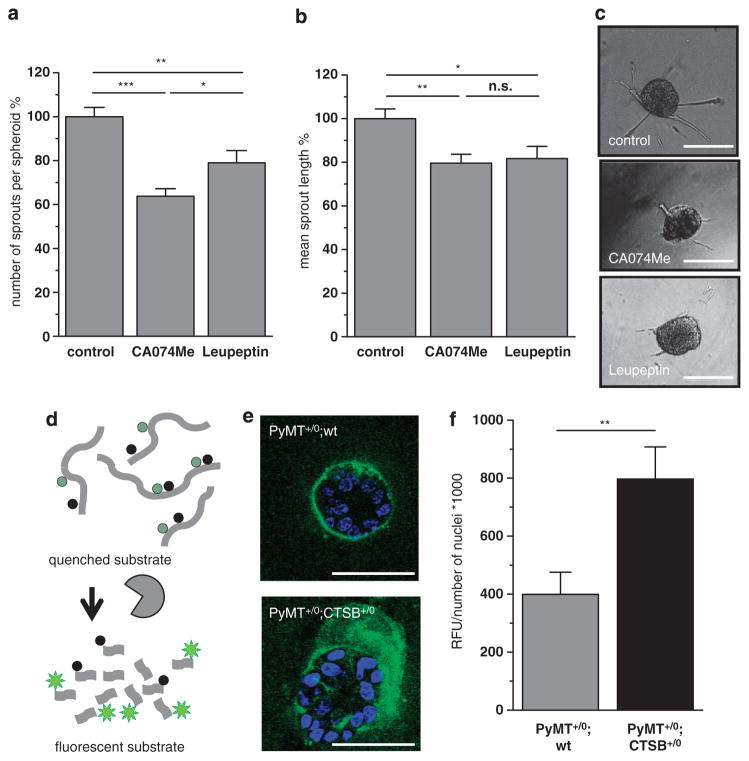
General inhibition of serine and cysteine proteases by Leupeptin significantly reduced spheroid sprouting of PyMT +/0; wt tumor cells in number of sprouts per spheroid and in sprout length. Notably, specific inhibition of CTSB by CA074Me resulted in a similar reduction in number of sprouts per spheroid and mean sprout length, indicating that enzymatic activity of CTSB, among other proteases, promotes tumor spheroid sprouting into the matrix (Figures 2a–c).
Figure 2.
Invasiveness of primary PyMT tumor spheroids and matrix degradation. (a, b) Effect of protease inhibition on average number of sprouts per spheroid and mean sprout length. PyMT +/0;wt spheroids were grown in collagen I for 24 h, inhibition of CTSB by CA074Me (10 μM), broad spectrum protease inihibiton by Leupeptin (10 μM). A total of 153, 190 and 170 spheroids, in the control-, CA074Me- and Leupeptin-group, were analyzed in 3–5 independent assays. Quantitative values represent mean ± s.e.m.; NS, not significant, *P<0.05, **P<0.01 and ***P<0.001 by ANOVA and post hoc Tukey-test. (c) Representative phase contrast images of tumor spheroids. Scale bars indicate 100 μm. (d) Schematic principle of proteolysis detection with dye-quencher (DQ)-labeled substrate collagen IV. (e) Representative pictures of tumor spheroids of primary PyMT +/0;wt or PyMT +/0;CTSB +/0 tumor cells grown in reconstructed basement membrane for 6 days. Pictures are overlays of 10 confocal planes, scale bars indicate 50 μm. (f) Quantification of fluorescence emission upon substrate cleavage detected by confocal imaging.
The enhanced sprouting of PyMT +/0;CTSB +/0 tumor spheroids could in principal be a result of several distinct processes: increased cell proliferation, enhanced motility or higher invasive capacity of the cells. Proliferation, however, was unaffected when CTSB was transgenically overexpressed in vitro (Supplementary Figures 4a–c). This is in line with the observation of similar proliferation rates in the orthotopic tumors of PyMT +/0;wt or PyMT +/0;CTSB +/0 origin (Supplementary Figures 2a and b). Motility of cancer cells is often acquired by epithelial-to-mesenchymal transition (EMT). Interestingly, PyMT tumor spheroids exhibit signs of a partial EMT, expressing epithelial E-cadherin and mesenchymal α-smooth muscle actin (αSMA) at the same time (Supplementary Figure 5a). Overexpression of CTSB did not change mRNA expression of the EMT transcription factors Snail1 and Zeb1, of the epithelial marker E-cadherin (Cdh1) or the mesenchymal markers N-cadherin (Cdh2) and Vimentin (Supplementary Figure 5b). In addition, cell morphology was not altered by CTSB overexpression (Supplementary Figure 5c). Similarly, in the orthotopic tumors, the cells expressing the EMT transcription factors Slug/Snail2 and Zeb1 were quantified, however, an CTSB overexpression had no impact on the number of cells undergoing EMT (Supplementary Figures 5d–g). In accordance with these findings, migration of PyMT cells in a boyden chamber experimental setup was not changed by CTSB overexpression (Supplementary Figures 6a and b). Invasion of tumor cells through collagen I, however, turned out to be slightly enhanced when CTSB was transgenically expressed (Supplementary Figures 6c and d). In summary, a regulatory function of CTSB in the EMT program appears unlikely, but the findings point toward a role of CTSB as an executer of tumor cell invasion.
To address the question if the higher invasiveness observed in 3D culture is caused by an enhanced proteolytic capacity of PyMT +/0;CTSB +/0 cells, we performed 3D proteolysis imaging, using dye-quenched (DQ)-labeled collagen IV, which emits fluorescence upon proteolytic cleavage (Figure 2d). Quantification of matrix-derived fluorescence of PyMT +/0;wt and PyMT +/0; CTSB+/0 tumor spheroids confirmed that CTSB overexpression promotes matrix proteolysis (Figures 2e and f).
In conclusion, PyMT tumor spheroids invade the surrounding matrix not by ameboid migration of EMT-generated single mesenchymal cells, but by a collective invasion process, most likely ‘multicellular streaming’.33 This collective invasion of the tumor cells requires proteolytic ECM degradation to which CTSB contributes significantly.
CTSB affects transcription of only few extracellular proteins in tumor cell – macrophage interaction
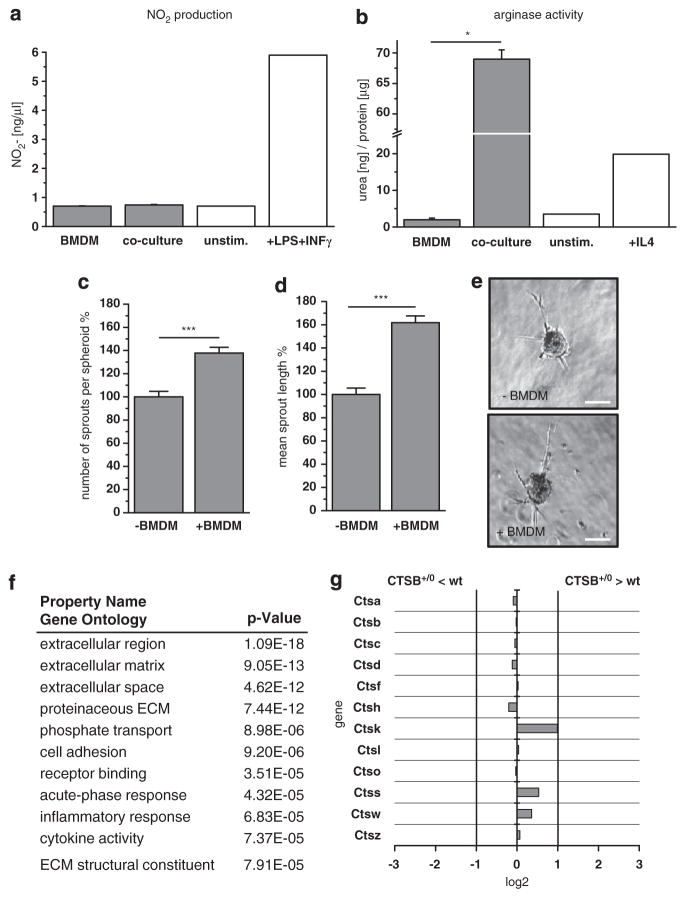
Tumor progression and invasion have often been linked to tumor–macrophage interactions, which promote comigration of invading tumor cells and tumor-associated macrophages.18 As a consequence of this heterotypic interaction, macrophages are polarized toward the tumor-promoting M2 phenotype. This does not require direct cancer cell-macrophage contact, because, in indirect cocultures of tumor cells with bone marrow-derived macrophages, in which the two cell types are separated by a porous membane, M2-polarization was detected already after 24 h, as proven by low nitric oxide production and high arginase activity (Figures 3a and b). The presence of macrophages in PyMT +/0 3D cultures promoted spheroid sprouting by about 40% in sprout number and about 60% in average sprout length (Figures 3c–e), indicating that macrophages positively affect invasiveness of PyMT breast cancer cells.
Figure 3.
Interaction of tumor cells and macrophages. (a) Nitric oxide (NO) production by bone marrow-derived macrophages after 24 h in 1:1 indirect (transwell) cocultures with primary PyMT tumor cells. (b) Arginase activity of bone marrow-derived macrophages after 24 h in 1:1 direct cocultures with primary PyMT tumor cells. (c, d) Average number of sprouts per spheroid and mean sprout length of primary tumor spheroids alone or in coculture with bone marrow-derived macrophages (ratio tumor cells: macrophages = 1:1) in collagen I matrix. A total of 161 and 251 spheroids were analyzed in three independent assays. Quantitative values represent mean ± s.e.m.; ***P<0.001 by t-test. (e) Representative phase contrast pictures of spheroids. Scale bar indicates 100 μm. (f) Gene Ontology categories of genes with changed expression in direct coculture of bone marrow-derived macrophages with primary PyMT derived from microarray gene expression analysis. Enhancement in the enlisted categories is significant by P<10–4 in Fisher’s t-test. (g) Comparison of cysteine cathepsin expression in wt and CTSB +/0 cocultures derived from microarray gene expression analysis.
Gene expression profiling studies have revealed that macrophages can induce a certain gene signature in tumor cells that primes them for invasion.34 Therefore, we investigated if CTSB transgenic overexpression influences tumor–macrophage crosstalk, resulting in sustained alterations in gene expression pattern. A microarray expression analysis of direct cocultures of primary tumor cells and bone marrow-derived macrophages (ratio 1:1) of the same CTSB genotype (that is, non-transgenic wt or transgenic CTSB+/0) was carried out in two independent experiments. Among the ~28 000 assayed genes, the analysis detected 160 genes with statistically significant, higher than twofold expression change in either direction (Supplementary Tables 1 and 2). In Gene Ontology analysis, the genes with changed expression were mainly annotated in the categories ‘extracellular region’, ‘extra-cellular matrix’ and ‘extracellular space’ (Figure 3f), indicating an influence of CTSB overexpression on transcriptional regulation of extracellularly located proteins. One of the significantly upregulated genes in CTSB transgenic cocultures was lysyl oxidase (LOX), which has been shown to be associated with invasiveness of MDA-MB-231 breast cancer cells.34 In line with the mRNA expression data, LOX activity was enhanced when CTSB was transgenically overexpressed both in PyMT tumor cells and in macrophages (Supplementary Figures 7a and b). Another known driver of tumor metastasis, the heparin receptor CD44, was not altered in expression between wt and CTSB transgenic PyMT cells and macrophages (Supplementary Figures 7c–d). Furthermore, the microarray analysis of wt and CTSB +/0 cocultures did not show compensational or synergistic regulation of other cathepsins, with the exception of cathepsin K, which was slightly overexpressed (1.9 fold) in the CTSB +/0 group (Figure 3g).
These results unravel a complex interference of CTSB overexpression with tumor–macrophage interactions. Besides extracellular proteolysis directly executed by CTSB, the effect of transgenic CTSB seems to involve transcriptional regulation of several genes. The latter effect is, however, moderate because <0.5% of the genome was affected by CTSB overexpression in the interaction of tumor cells and macrophages. Thus, the impact of CTSB on tumor progression seems to be mainly mediated by direct ECM proteolysis.
CTSB induction in tumor cells rather than macrophages increases spheroid invasion
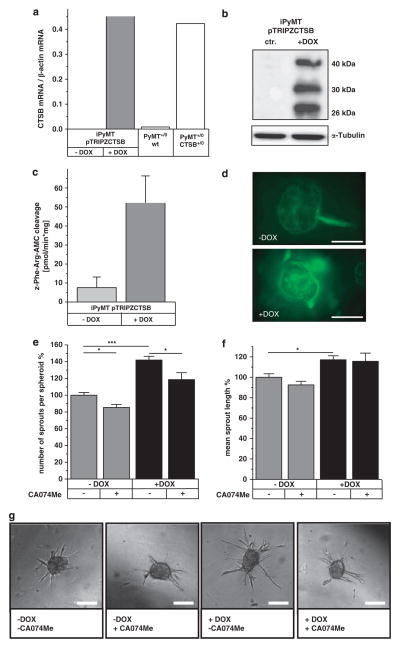
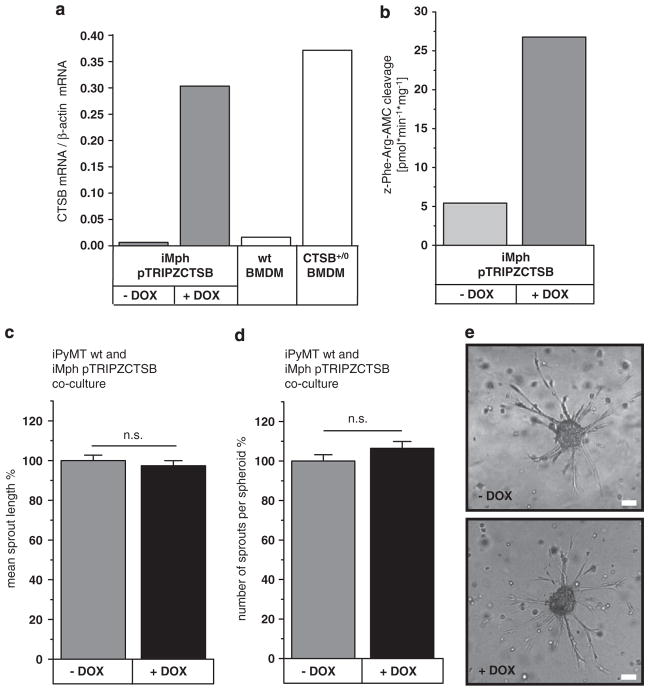
To further elucidate the role of CTSB in cocultures of tumor cells and macrophages, we induced CTSB expression in one or the other cell type. In order to minimize variation compared with primary tumor cells, we generated a PyMT +/0 cell line with inducible expression of CTSB. Spontaneously immortalized PyMT +/0 cells were transfected with the doxycycline (DOX)-inducible expression vector pTRIPZ, containing the human CTSB cDNA corresponding to mRNA variant 1, the most abundant CTSB mRNA in human cancers.35 CTSB mRNA induction reached the level of primary PyMT +/0;CTSB +/0 cells (Figure 4a), and western blots confirmed the expression and full processing of human CTSB (Figure 4b). In the time course of induction, CTSB mRNA was already detectable 6 h after DOX addition, and its expression maximum was reached after 24 h. In western blots, pro-CTSB appeared as early as 6 h after induction, and mature forms of CTSB occurred after 10 h. The mature enzyme was stable and persisted for over 4 days after DOX removal (Supplementary Figures 8a–d). Proteolytic CTSB activity upon DOX induction was shown in cell lysates by cleavage of a fluorogenic peptide, which could be inhibited by the specific CTSB inhibitor CA074Me (Figure 4c). In addition, proteolysis imaging showed a striking enhancement of matrix proteolysis when CTSB expression was induced (Figure 4d). Induction of CTSB in tumor spheroids significantly increased spheroid sprouting in number of sprouts per spheroid. CTSB inhibition in spheroids reduced the number of sprouts developed, but not the overall length of sprouts (Figures 4e–g).
Figure 4.
Induction of human CTSB expression affects spheroid sprouting and matrix degradation. (a) Induction of CTSB mRNA expression by doxycycline (1 μM) measured by qRT–PCR. (b) Human CTSB protein in western blot. (c) CTSB activity detected by proteolytic cleavage of the fluorogenic substrate Z-Phe-Arg-AMC. (d) Proteolysis of dye-quencher-labeled collagen IV in 3D culture. Scale bar indicates 100 μm. (e, f) Average number of sprouts per spheroid and mean sprout length of tumor spheroids in collagen I matrix upon induction of human CTSB by doxycycline (Dox) and under inhibition of CTSB by CA074Me (10 μM). A total of 235, 243, 167 and 93 spheroids were analyzed in 2–7 independent assays. Quantitative values represent mean ± s.e.m.; *P<0.05 and ***P<0.001 by ANOVA and post hoc Tukey-test. (g) Representative phase contrast pictures of tumor spheroids. Scale bar indicates 100 μm.
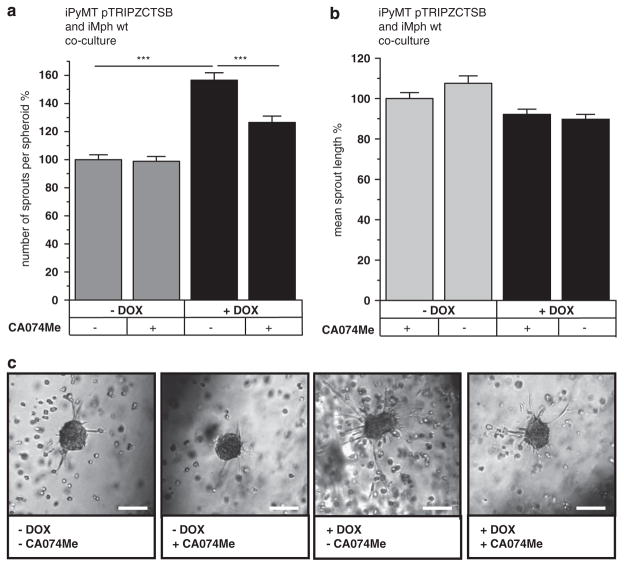
Immortalized macrophages were endowed with the same DOX-inducible expression system for CTSB as described above for tumor cells. CTSB mRNA induction in the macrophages reached the level of CTSB expression in primary bone marrow-derived macrophages from transgenic CTSB +/0 mice (Figure 5a). As in the tumor cells, the total proteolytic activity of CTSB, which is the sum of endogenous murine CTSB and the overexpressed human CTSB, was increased about fivefold upon DOX-treatment of macrophages (Figure 5b). To analyze the contribution of CTSB overexpression in macrophages to tumor cell invasiveness in vitro, DOX-inducible macrophages were cocultured with DOX-insensitive PyMT +/0 tumor spheroids. Here the addition of DOX had no effect on tumor spheroid sprouting (Figures 5c–e). In the inverse experiment, DOX-insensitive macrophages and PyMT cells with DOX-inducible CTSB were cocultured (Figure 6). In this setting, CTSB induction significantly increased the average number of sprouts per spheroid (Figure 6a). An effect on the mean sprout length could not be detected (Figure 6b), presumably because of the general trophic effect that macrophages exert on sprout length (as shown in Figure 3c).
Figure 5.
Induction of human CTSB expression in macrophages in coculture with tumor spheroids. (a) Induction of CTSB mRNA expression in macrophages iMph pTRIPZCTSB by doxycycline (1 μM) measured by qRT–PCR. (b) CTSB activity in macrophages iMph pTRIPZhCTSB upon induction of CTSB detected by proteolytic cleavage of the fluorogenic substrate Z-Phe-Arg-AMC. (c, d) Average number of spouts per spheroid and mean sprout length of DOX-insensitive tumor spheroids in cocultures with DOX-inducible macrophages (ratio 1:1). Addition of DOX (1 μM) resulting in induction of CTSB expression only in the macrophages. (e) Representative phase contrast pictures of spheroids. Scale bar indicates 100 μm. A total of 195 and 199 spheroids in five independent assays were analyzed. Quantitative values represent mean ± s.e.m.; NS, not significant, by ANOVA and post hoc Tukey-test.
Figure 6.
Induction of CTSB in PyMT tumor spheroids in coculture with macrophages. (a, b) Average number of sprouts per spheroid and mean sprout length in 3D cocultures of inducible PyMT tumor cells and Dox-insensitive macrophages. Induction with Dox (1 μM) and CTSB inhibition by CA074Me (10 μM). A total of 147, 128, 133 and 135 spheroids in three independent assays were analyzed. Quantitative values represent mean ± s.e.m.; ***P<0.001 by ANOVA and post hoc Tukey-test. (c) Representative phase contrast pictures of tumor spheroids. Scale bars indicate 100 μm.
In summary, CTSB overexpression in macrophages, even when in close proximity with tumor spheroids, could not enhance invasiveness, whereas tumor spheroid invasion was clearly fostered by CTSB overexpression in the tumor cells.
DISCUSSION
In this study, we demonstrate in vivo and in vitro that CTSB overexpression in the cancer cells increases growth of invasive ductal carcinoma in the MMTV-PyMT mouse model of breast cancer. In contrast, overexpression of CTSB in cells of the tumor microenvironment had no relevant impact on tumor progression, as shown by orthotopic transplantation of PyMT cancer cells in mouse mammary glands of wt and CTSB +/0 recipients. In 3D cocultures, macrophages enhanced the invasive phenotype of tumor spheroids, but this was not fostered by CTSB over-expression in the macrophages. Induction of CTSB transgenic overexpression in the tumor cells, in contrast, increased invasive sprouting in cocultures with macrophages. These findings indicate that the tumor-promoting effect of overexpressed CTSB in the PyMT model can be assigned to the tumor cells rather than to the microenvironment.
Our results provide new insight into the differential roles of cathepsins in cancer cells and tumor stroma, in particular TAMs, as being tumor-promoting and tumor-permissive, respectively. In the RIP1-Tag2 model of pancreatic neuroendocrine neoplasia, genetic deficiency for CTSB or cathepsin S in bone marrow-derived macrophage progenitors decreased tumor growth.12 Similarly, evidence from depletion studies in the PyMT mouse model of invasive breast cancer pointed toward a role of stroma-derived CTSB at the metastatic site. Formation of lung colonies was reduced when PyMT cells were i.v. injected into congenic CTSB-null recipient mice.11 In addition, targeting the tumor micro-environment with cathepsin inhibitor-loaded ferriliposomes reduced the growth of orthotopically transplanted PyMT cancer cells.36 Furthermore, cathepsin-expressing TAMs impaired the chemotherapy efficacy in PyMT breast cancer mice.26 Thus, TAM-derived cathepsins contribute to create a tumor micro-environment permissive for cancer growth that is not functional upon genetic ablation of cathepsins, that is, CTSB. However, our present study revealed that forced overexpression of human CTSB in the tumor stroma or particularly in macrophages did not exert an additional effect on tumor growth and invasion. Rather, CTSB overexpression in cancer cells increased matrix degradation, invasion and tumor growth. These findings are in line with the histopathological observation, for example, in prostate carcinoma, that cathepsins are highly expressed in cancer cells that are localized at the invasive front of tumors.37
A recent publication demonstrated that expression of the epidermal growth factor receptor ErbB2/Her2 is functionally linked to breast cancer cell invasion with cathepsins B and L as the effector proteases of invasiveness.31 Our data indicate that CTSB-mediated tumor spheroid sprouting in 3D cultures occurs by a collective cell invasion process, most likely by ‘multicellular streaming’,34 in which tumor cells become motile by a partial EMT while maintaining adherens junctions to neighbor cells. In this process, CTSB promoted invasion, whereas other hallmark cancer processes like proliferation, single cell motility and EMT induction were unaffected by CTSB overexpression. Biochemical investigations have identified collagen I, collagen IV, fibronectin and laminin as substrates of CTSB, whose proteolytic degradation could drive tumor cell invasion.38,39 We could confirm proteolysis of DQ-labeled collagen I and IV by confocal imaging of PyMT spheroids, but it is probable that CTSB-dependent invasion involves proteolytic degradation of further ECM proteins.
Investigating the heterotypic interaction of tumor cells and macrophages by a transcriptome analysis, we found that CTSB overexpression does not lead to alterations of general gene expression patterns, indicating that CTSB overexpression does not disturb crosstalk between the two cell types. However, transcription of 160 genes was significantly changed, among them are many extracellular proteins or ECM-modifying enzymes like lysyl oxidase, for which we could confirm higher activity upon CTSB overexpression. Other members of the cysteine cathepsin protease family were unaffected by CTSB overexpression, apart from a slight increase in cathepsin K. Macrophage-derived cathepsin K has been reported to promote bone metastasis of prostate carcinoma by degradation of collagen I.40 Therefore, a pro-invasive cooperation of CTSB, cathepsin K and other matrix modifying enzymes, such as lysyl oxidase, is possible.41
In summary, CTSB is a major factor that fosters breast cancer invasion by its proteolytic activity and should therefore be considered as a therapeutic target. In malignantly transformed cells, CTSB is tethered to the plasma membrane at specific microdomains, invadosomes and caveolae, which are associated with ECM remodeling and invasion.42–44 Despite this invasion-promoting effect of extracellular CTSB, lysosomal CTSB might also exert anti-tumor effects: cysteine cathepsins have been described as mediators of cell death by lysosomal membrane permeabilization.45–47 Lysosomal rupture for induction of cathepsin-mediated cancer cell death is investigated as a therapeutic strategy for cancer treatment.48 Thus, therapeutic approaches to prevent progression of invasive breast cancer should take the ambivalent function of cathepsins into account. A rational therapeutic strategy would selectively inhibit extracellular CTSB in order to preserve the intracellular cell death function while disrupting the extracellular pro-invasive function of CTSB in ECM degradation.
MATERIALS AND METHODS
Mouse model for breast cancer
FVB/N mice harboring the genomic human CTSB gene (CTSB+/0)49 and the oncogenic transgene MMTV-PyMT50 were generated as previously described.32 The maintenance of the animals as well as the orthotopic transplantation experiments were performed in accordance to the German law for animal protection (Tierschutzgesetz) as published on 25 May 1998.
Isolation of primary PyMT tumor cells and generation of cell line
Primary tumor cells were prepared from tumors of all 10 mammary glands of 14-week-old female PyMT mice as previously described.11,32 Vital cells were cryopreserved for further use. PyMT cell lines were generated by spontaneous immortalization of freshly isolated cells kept in culture for 6 weeks with splitting whenever 80% confluency was surpassed.
Orthotopic transplantation
Primary PyMT breast cancer cells were washed twice with PBS and passed through a 100 μm cell strainer (BD, Franklin Lakes, NJ, USA). Cell suspension was adjusted to 5 × 106/ml. With a 26-gauge needle (Braun, Melsungen, Germany), 100 μl cell suspension was injected into the fourth mammary gland of female adult recipient mice. Tumor growth was monitored by palpation twice a week for 6 weeks after tumor cell injection. Magnetic resonance imaging was performed at day 20 post injection. Mice were killed when tumors reached a diameter of 1.0 cm or after 40 days.
Magnetic resonance imaging
Magnetic resonance imaging was performed using a 9.4 tesla small bore animal scanner (BioSpec 94/21, Bruker Biospin, Ettlingen, Germany) and a dedicated mouse quadrature-resonator (Bruker Biospin) in vivo mouse body magnetic resonance imaging. Mice were anesthetized under spontaneous breathing conditions using isoflurane. Heart rate and respiration rate were continuously monitored and kept at a constant level. Gating was used to reduce motion and blood flow artefacts during the scan. The magnetic resonance imaging protocol consisted of a localizer and an axial/coronal T2-weighted spin echo RARE (Rapid Acquisition with Relaxation Enhancement) sequence in the abdomen and thorax of mice. This sequence was performed to delineate the tumor and eventual metastasis from the surrounding healthy tissue. The RARE sequence in axial and coronal orientation featured a field of view of 30 mm2, a matrix size of 256 × 256pixel2 and an in-plane resolution of 117 × 117 μm2. The slice thickness was 0.50 mm with no slice spacing to achieve contiguous image sets of the whole volume. The number of slices was adjusted to the measured volume (on average 30) to ensure complete coverage of the abdomen and lung, respectively.
Preparation of primary bone marrow-derived macrophages
Femur and tibia of killed 8- to 10-week-old mice were washed in isopropanol, opened at the ends and the bone marrow was flushed out with RPMI medium. The bone marrow was washed in DPBS and resuspended in RPMI. Cells were cultured in RPMI with 15% L292 cell supernatant containing M-CSF, 10% FCS (PAN-Biotech, Aidenbach, Germany), 5% horse serum (Invitrogen, Carlsbad, CA, USA), 1% sodium pyruvate (Invitrogen), 5% penicillin/streptavidin (Invitrogen) and 0.04% β-mercaptoethanol and cultured for 10 days for differentiation. The macrophages were collected and resuspended in DMEM with 10% FCS, 1% L-glutamine, 1% penicillin/streptavidin for use in experiments.
Tumor cell–macrophage cocultures
For assessment of macrophage polarization, primary PyMT tumor cells and bone marrow-derived macrophages were grown in a 1:1 ratio in indirect cocultures. Briefly, macrophages were plated in six-well plates, and PyMT tumor cells were plated in cell culture inserts with anapore membranes, which allow the exchange of soluble factors. Both cell types were starved overnight. The inserts containing the PyMT tumor cells were placed on top of the macrophages, and the cells were cocultured for 24 h. For gene expression analysis, a direct 1:1 coculture of primary tumor cells with bone marrow-derived macrophages was performed for 48 h.
Nitric oxide assay and arginase assay
Nitrite, a stable breakdown product of nitric oxide, was measured in cell culture supernatant by Griess reagent (1% sulfonile amide and 0.1% N-ethylenediamine in phosphoric acid solution) and subsequent determination of absorption at 520 nm. Arginase activity was determined in cell lysates by arginine hydrolysis to urea. Cell lysates were incubated with arginine solution (0.5 mM) for 1 h at 37 °C. Thereafter, hydrolysis was stopped by phosphoric acid and sulfuric acid. Produced urea reacted with α-isonitriso propiophenone 1.5% (v/w) in ethanol for 30 min at 95°, followed by 30 min at 4 °C. Absorption was measured at 540 nm.
Microarray analysis
Tumor–macrophage cocultures were washed twice with PBS, and cells were lysed directly in the dish with RLT buffer with 1% β-mercaptoethanol. Isolation of RNA was performed with the RNeasy mini kit (Qiagen, Germantown, MD, USA) and further processed with the Ambion WT Expression and Affymetrix terminal labeling kits according to the manufacturer’s instructions. Labeled fragments were hybridized to the arrays for 16 h at 45 °C. After washing and staining, the arrays were scanned with the Affymetrix GeneChip Scanner 3000 7G. Data were processed with Expressionist Refiner (Genedata, Basel, Switzerland) and Analyst software (Absiex, Framingham, MA, USA). After GC background subtraction, quantile normalization and probe summarization were performed with the Bioconductor RMA condensing algorithm as implemented in the Genedata Refiner.51 To identify differentially expressed genes between the groups, the unpaired Bayes T-test (CyberT)52 was used. To control the false discovery rate, the Benjamini-Hochberg q-value was calculated.53
Stable transfection of cell lines with a doxycycline-inducible expression system for human CTSB
The cDNA corresponding to the human CTSB mRNA variant, purchased in a TrueClone Vector (Origene, Rockville, MD, USA), was amplified by PCR with mutagenesis primers, generating two new restriction sites at the ends of the PCR product. The purified PCR product was cloned into a lentiviral pTRIPZ shRNAmir vector (Thermo Fisher Scientific, Waltham, MA, USA). Production of lentivirus containing the pTRIPZCTSB was performed in HEK293 cells, employing the pMISSION system (Sigma-Aldrich, St Louis, MO, USA) in a spinfaction with Superfect (Qiagen). Virus containing supernatant was sterile filtered (Thermo Fisher Scientific), supplemented with polybrene (5 μg/ml) and was applied to 80% confluent immortalized PyMT cells in a spinfaction. Cells were grown in DMEM with 10% FCS, 1% L-glutamine, 1% penicillin/streptavidin and selected with puromycin (5 μM) for 1 week.
Imaging collagen IV proteolysis in 3D cultures
For measurement of collagen IV proteolysis in 3D culture, a dye-quenched (DQ)-collagen IV was employed (Invitrogen; λem = 488 nm).54 Tumor cells (60 000 cells in single cell suspension) were seeded on coverslips coated with Cultrex BME PathClear (Trevigen, Gaithersberg, MD, USA) containing 25 μg/ml DQ-collagen IV and then Indicator-free DMEM with 10% FCS, 1% L-glutamine, 1% penicillin/streptavidine and 2% Cultrex was added. Cultures were maintained for 6 days to allow spheroid growth; overlay medium was changed once. Before imaging, cells were stained with Hoechst and imaged using × 20 water dipping lens on a Zeiss LSM 510 META NLO microscope. The intensity of DQ-collagen IV degradation products was determined using Metamorph 7.1.7 software (Molecular Devices, Sunyvale, CA, USA). The number of nuclei was determined using the Volocity 6.1.1 (PerkinElmer, Waltham, MA, USA), and data are represented as normalized intensity value/cell number.
PyMT tumor spheroid sprouting and proteolysis imaging in a collagen I matrix
Primary or immortalized PyMT tumor cells were collected from monolayer culture, suspended in cell culture medium (DMEM containing 10% FCS, 1% L-glutamine, 1% penicillin/streptomycin) with 0.24% (w/v) methylcellulose as previously described.32,55 Droplets containing ~500 tumor cells were applied to a plastic cell culture plate, which was subsequently turned upside down and incubated 24 h for spheroid formation. Spheroids were collected and embedded in a collagen I matrix with 0.6% methylcellulose for 24 h. Tumor spheroid invasiveness was assessed by measurement of invasive sprout length and number of sprouts per spheroid in phase contrast pictures taken with an Axiovert microscope (Zeiss, Oberkochen, Germany). The length of invasive strands was measured using the Axiovision LE 4.4 software (Zeiss). For imaging of collagen I degradation in sprouting spheroids, iPyMT cells were labeled with cell tracker red (Invitrogen) according to the manufacturer’s instructions before generation of spheroids. DQ-collagen was mixed into the collagen matrix (10 μg/ml). Cells were stained with Hoechst before imaging with the Apotome (Zeiss).
CTSB qRT-PCR, immunoblotting and detection of CTSB enzyme activity
CTSB was detected in qRT–PCR with forward primer 5′-TCT GGT GGC CTC TAT GAA TC-3′ and reverse primer 5′-GAA AGC GGA GTC AAC CTA CA-3′ and normalized to β-actin. CTSB immunoblotting was performed as previously described,32 and CTSB activity was determined by hydrolysis of the fluorogenic substrate z-Phe-Arg-7-amino-4-methylcoumarin (z-Phe-Arg-AMC) (Bachem, Bubendorf, Switzerland; 25 μmol/l) that could be inhibited by CA074 (Bachem, Bubendorf; 150 nmol/l) as previously described.32
Data presentation and statistical analyses
Quantitative data are presented as means ± s.e.m. Data from tumor spheroid sprouting assays were normalized to the mean of untreated controls. Statistics was perfomed by t-test for two group-comparisons or analysis of variance (ANOVA) for multiple groups, followed by post hoc Tukey-test for pairwise comparisons. For comparison of tumor growth kinetics, the exponential growth curves were fitted and the data sets were compared by F-test statistics (software OriginPro 8.6; OriginLab Corporation, Northampton, MA, USA).
Supplementary Material
Acknowledgments
The CTSB +/0 mice were kindly provided by Dr L. Pennacchio, Genomics Division, Lawrence Berkeley Laboratory, Berkeley, USA. The immortalized macrophage cell line from FVB/n mice was kindly provided by Eike Latz, currently University of Bonn, Germany. We thank Nicole Klemm, Anja Faulhaber and Julia Wolanski for excellent technical assistance. Proteolysis imaging was mainly performed with kind help of the Microscopy, Imaging & Cytometry Resources Core at Wayne State University, Detroit, MI, USA, directed by Dr Kamiar Moin and supported in part by NIH Center grant P30CA22453 to the Karmanos Cancer Institute, Wayne State University. This work was supported by the Excellence Initiative of the German Federal and State Governments (EXC 294 and GSC-4, Spemann Graduate School), and the Deutsche Forschungsge-meinschaft SFB 850 project B7.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 3.Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor micro-environment and breast cancer. Cancer Metastasis Rev. 2012;32:303–315. doi: 10.1007/s10555-012-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;21:228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36:D320–D325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tu C, Ortega-Cava CF, Chen G, Fernandes ND, Cavallo-Medved D, Sloane BF, et al. Lysosomal cathepsin B participates in the podosome-mediated extracellular matrix degradation and invasion via secreted lysosomes in v-Src fibroblasts. Cancer Res. 2008;68:9147–9156. doi: 10.1158/0008-5472.CAN-07-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jane DT, Morvay L, Dasilva L, Cavallo-Medved D, Sloane BF, Dufresne MJ. Cathepsin B localizes to plasma membrane caveolae of differentiating myoblasts and is secreted in an active form at physiological pH. Biol Chem. 2006;387:223–234. doi: 10.1515/BC.2006.030. [DOI] [PubMed] [Google Scholar]
- 8.Harbeck N, Alt U, Berger U, Kruger A, Thomssen C, Janicke F, et al. Prognostic impact of proteolytic factors (urokinase-type plasminogen activator, plasminogen activator inhibitor 1, and cathepsins B, D, and L) in primary breast cancer reflects effects of adjuvant systemic therapy. Clin Cancer Res. 2001;7:2757–2764. [PubMed] [Google Scholar]
- 9.Nouh MA, Mohamed MM, El-Shinawi M, Shaalan MA, Cavallo-Medved D, Khaled HM, et al. Cathepsin B: a potential prognostic marker for inflammatory breast cancer. J Transl Med. 2011;9:1. doi: 10.1186/1479-5876-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jedeszko C, Sloane BF. Cysteine cathepsins in human cancer. Biol Chem. 2004;385:1017–1027. doi: 10.1515/BC.2004.132. [DOI] [PubMed] [Google Scholar]
- 11.Vasiljeva O, Papazoglou A, Kruger A, Brodoefel H, Korovin M, Deussing J, et al. Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 2006;66:5242–5250. doi: 10.1158/0008-5472.CAN-05-4463. [DOI] [PubMed] [Google Scholar]
- 12.Gocheva V, Wang HW, Gadea BB, Shree T, Hunter KE, Garfall AL, et al. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24:241–255. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012;33:119–126. doi: 10.1016/j.it.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, et al. CD4( + ) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 17.Pollard JW. Macrophages define the invasive microenvironment in breast cancer. J Leukoc Biol. 2008;84:623–630. doi: 10.1189/jlb.1107762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 19.Gocheva V, Zeng W, Ke D, Klimstra D, Reinheckel T, Peters C, et al. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006;20:543–556. doi: 10.1101/gad.1407406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gopinathan A, Denicola GM, Frese KK, Cook N, Karreth FA, Mayerle J, et al. Cathepsin B promotes the progression of pancreatic ductal adenocarcinoma in mice. Gut. 2012;61:877–884. doi: 10.1136/gutjnl-2011-300850. [DOI] [PubMed] [Google Scholar]
- 21.Sevenich L, Schurigt U, Sachse K, Gajda M, Werner F, Müller S, et al. Synergistic anti-tumor effects of cathepsin B and cathepsin Z deficiencies on breast cancer progression and metastasis in mice. Proc Natl Acad Sci USA. 2010;107:2497–2502. doi: 10.1073/pnas.0907240107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gondi CS, Lakka SS, Dinh DH, Olivero WC, Gujrati M, Rao JS. RNAi-mediated inhibition of cathepsin B and uPAR leads to decreased cell invasion, angiogenesis and tumor growth in gliomas. Oncogene. 2004;23:8486–8496. doi: 10.1038/sj.onc.1207879. [DOI] [PubMed] [Google Scholar]
- 23.Lakka SS, Gondi CS, Yanamandra N, Olivero WC, Dinh DH, Gujrati M, et al. Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma cell line via RNA interference reduces tumor cell invasion, tumor growth and angiogenesis. Oncogene. 2004;23:4681–4689. doi: 10.1038/sj.onc.1207616. [DOI] [PubMed] [Google Scholar]
- 24.Malla RR, Gopinath S, Gondi CS, Alapati K, Dinh DH, Gujrati M, et al. Cathepsin B and uPAR knockdown inhibits tumor-induced angiogenesis by modulating VEGF expression in glioma. Cancer Gene Ther. 2011;18:419–434. doi: 10.1038/cgt.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, et al. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 26.Shree T, Olson OC, Elie BT, Kester JC, Garfall AL, Simpson K, et al. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 2011;25:2465–2479. doi: 10.1101/gad.180331.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Withana NP, Blum G, Sameni M, Slaney C, Anbalagan A, Olive MB, et al. Cathepsin B inhibition limits bone metastasis in breast cancer. Cancer Res. 2012;72:1199–1209. doi: 10.1158/0008-5472.CAN-11-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell-McGuinn KM, Garfall AL, Bogyo M, Hanahan D, Joyce JA. Inhibition of cysteine cathepsin protease activity enhances chemotherapy regimens by decreasing tumor growth and invasiveness in a mouse model of multistage cancer. Cancer Res. 2007;67:7378–7385. doi: 10.1158/0008-5472.CAN-07-0602. [DOI] [PubMed] [Google Scholar]
- 29.Gabrijelcic D, Svetic B, Spaic D, Skrk J, Budihna M, Dolenc I, et al. Cathepsins B, H and L in human breast carcinoma. Eur J Clin Chem Clin Biochem. 1992;30:69–74. [PubMed] [Google Scholar]
- 30.Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 31.Rafn B, Nielsen CF, Andersen SH, Szyniarowski P, Corcelle-Termeau E, Valo E, et al. ErbB2-driven breast cancer cell invasion depends on a complex signaling network activating myeloid zinc finger-1-dependent cathepsin B expression. Mol Cell. 2012;45:764–776. doi: 10.1016/j.molcel.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 32.Sevenich L, Werner F, Gajda M, Schurigt U, Sieber C, Muller S, et al. Transgenic expression of human cathepsin B promotes progression and metastasis of polyoma-middle-T-induced breast cancer in mice. Oncogene. 2011;30:54–64. doi: 10.1038/onc.2010.387. [DOI] [PubMed] [Google Scholar]
- 33.Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA. Multicellular tumor spheroids: an underestimated tool is catching up again. J Biotechnol. 148:3–15. doi: 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Friedl P, Locker J, Sahai E, Segall J. Classifying collective cancer cell invasion. Nat Cell Biol. 2012;14:777–783. doi: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- 35.Keppler D, Sloane BF. Cathepsin B: multiple enzyme forms from a single gene and their relation to cancer. Enzyme Protein. 1996;49:94–105. doi: 10.1159/000468619. [DOI] [PubMed] [Google Scholar]
- 36.Mikhaylov G, Mikac U, Magaeva AA, Itin VI, Naiden EP, Psakhye I, et al. Ferri-liposomes as an MRI-visible drug-delivery system for targeting tumours and their microenvironment. Nat Nanotechnol. 2011;6:594–602. doi: 10.1038/nnano.2011.112. [DOI] [PubMed] [Google Scholar]
- 37.Sinha AA, Gleason DF, Deleon OF, Wilson MJ, Sloane BF. Localization of a biotinylated cathepsin B oligonucleotide probe in human prostate including invasive cells and invasive edges by in situ hybridization. Anat Rec. 1993;235:233–240. doi: 10.1002/ar.1092350207. [DOI] [PubMed] [Google Scholar]
- 38.Maciewicz RA, Etherington DJ. A comparison of four cathepsins (B, L, N and S) with collagenolytic activity from rabbit spleen. Biochem J. 1988;256:433–440. doi: 10.1042/bj2560433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buck MR, Karustis DG, Day NA, Honn KV, Sloane BF. Degradation of extracellular-matrix proteins by human cathepsin B from normal and tumour tissues. Biochem J. 1992;282(Pt 1):273–278. doi: 10.1042/bj2820273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heroon Rajagurubandara E, Rudy DL, Chalasani A, Hardaway AL, Podgorsji I. Macrophage Cathepsin K promotes prostate tumor progression in bone. Oncogene. 2012;32:1580–1593. doi: 10.1038/onc.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Payne SL, Fogelgren B, Hess AR, Seftor EA, Wiley EL, Fong SF, Csiszar K, Hendrix MJ, Kirschmann DA. Lysyl oxidase regulates breast cancer cell migration and adhesion through a hydrogen peroxide-mediated mechanism. Cancer Res. 2005;65:11429–11436. doi: 10.1158/0008-5472.CAN-05-1274. [DOI] [PubMed] [Google Scholar]
- 42.Cavallo-Medved D, Mai J, Dosescu J, Sameni M, Sloane BF. Caveolin–1 mediates the expression and localization of cathepsin B, pro-urokinase plasminogen activator and their cell-surface receptors in human colorectal carcinoma cells. J Cell Sci. 2005;118:1493–1503. doi: 10.1242/jcs.02278. [DOI] [PubMed] [Google Scholar]
- 43.Victor BC, Sloane BF. Cysteine cathepsin non-inhibitory binding partners: modulating intracellular trafficking and function. Biol Chem. 2007;388:1131–1140. doi: 10.1515/BC.2007.150. [DOI] [PubMed] [Google Scholar]
- 44.Brisson L, Reshkin SJ, Gore J, Roger S. pH regulators in invadosomal functioning: Proton delivery for matrix tasting. Eur J Cell Biol. 2012;91:847–860. doi: 10.1016/j.ejcb.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Cesen MH, Pegan K, Spes A, Turk B. Lysosomal pathways to cell death and their therapeutic applications. Exp Cell Res. 2012;318:1245–1251. doi: 10.1016/j.yexcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 46.Kallunki T, Olsen OD, Jaattela M. Cancer-associated lysosomal changes: friends or foes? Oncogene. 2013;32:1995–2004. doi: 10.1038/onc.2012.292. [DOI] [PubMed] [Google Scholar]
- 47.Vasiljeva O, Turk B. Dual contrasting roles of cysteine cathepsins in cancer progression: apoptosis versus tumour invasion. Biochimie. 2008;90:380–386. doi: 10.1016/j.biochi.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 48.Fehrenbacher N, Jaattela M. Lysosomes as targets for cancer therapy. Cancer Res. 2005;65:2993–2995. doi: 10.1158/0008-5472.CAN-05-0476. [DOI] [PubMed] [Google Scholar]
- 49.Houseweart MK, Pennacchio LA, Vilaythong A, Peters C, Noebels JL, Myers RM. Cathepsin B but not cathepsins L or S contributes to the pathogenesis of Unverricht-Lundborg progressive myoclonus epilepsy (EPM1) J Neurobiol. 2003;56:315–327. doi: 10.1002/neu.10253. [DOI] [PubMed] [Google Scholar]
- 50.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 52.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 53.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 54.Sameni M, Cavallo-Medved D, Dosescu J, Jedeszko C, Moin K, Mullins SR, et al. Imaging and quantifying the dynamics of tumor-associated proteolysis. Clin Exp Metastasis. 2009;26:299–309. doi: 10.1007/s10585-008-9218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schurigt U, Sevenich L, Vannier C, Gajda M, Schwinde A, Werner F, et al. Trial of the cysteine cathepsin inhibitor JPM-OEt on early and advanced mammary cancer stages in the MMTV-PyMT-transgenic mouse model. Biol Chem. 2008;389:1067–1074. doi: 10.1515/BC.2008.115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.