Abstract
A characteristic feature of asthma is the aberrant accumulation, differentiation or function of memory CD4+ T cells that produce type 2 cytokines (TH2 cells). By mapping genome-wide histone modification profiles for subsets of T cells isolated from peripheral blood of healthy and asthmatic individuals, we identified enhancers with known and potential roles in the normal differentiation of human TH1 cells and TH2 cells. We discovered disease-specific enhancers in T cells that differ between healthy and asthmatic individuals. Enhancers that gained the histone H3 Lys4 dimethyl (H3K4me2) mark during TH2 cell development showed the highest enrichment for asthma-associated single nucleotide polymorphisms (SNPs), which supported a pathogenic role for TH2 cells in asthma. In silico analysis of cell-specific enhancers revealed transcription factors, microRNAs and genes potentially linked to human TH2 cell differentiation. Our results establish the feasibility and utility of enhancer profiling in well-defined populations of specialized cell types involved in disease pathogenesis.
The acquisition of immunological memory is the hallmark of a protective immune response1. During this evolutionarily conserved process, naive T cells and B cells that have not previously encountered antigen differentiate during primary infection into memory cells that have specialized functions in immune system defense, thus permitting the organism to effectively respond to a later infection with the same pathogen. As expected for a process of cell-lineage specification, differentiation of memory T cells and B cells involves extensive epigenetic changes that are required to initiate and maintain a heritable program of gene expression2. Adaptive immunity is not without risks: some genetically susceptible individuals develop abnormal memory responses to potentially harmless antigens, which results in a multitude of immunological diseases ranging from autoimmunity to allergies and asthma3–5. A clear understanding of the molecular and epigenetic mechanisms underlying normal as well as aberrant differentiation of human memory cell types will pave the way to develop new approaches to tackle immune system–mediated diseases.
Asthma is a disease characterized by airway inflammation that is mediated by excessive memory responses to inhaled allergens, such as grass pollen3. The alarming rise in asthma incidence is a major global health concern, not only in the western world but also in large developing countries such as India, China and Brazil6. Over 200 million people suffer from asthma world-wide, which causes an economic burden that exceeds that of tuberculosis and HIV-AIDS combined6. At present, there is no cure for asthma, and most patients require long-term, daily nonspecific medication such as corticosteroids to control the underlying inflammation and prevent symptoms and life-threatening asthma attacks7. Therapies targeting specific type 2 cytokines are only efficacious in certain types of asthma8, which raises the possibility that there are unclassified molecular subtypes of asthma for which different therapies may prove beneficial.
A molecular feature of asthma and other allergic diseases is the excessive differentiation of a subset of CD4+ T helper cells known as TH2 cells, which produce a characteristic spectrum of type 2 cytokines, including the interleukins IL-4, IL-5 and IL-13 (ref. 3). Genes encoding these three cytokines are localized on human chromosome 5, in a conserved grouping known as the TH2 cytokine locus in which the IL5 gene is separated from the IL4 and IL13 genes by the RAD50 gene, which encodes a conserved DNA repair protein9. The last few introns of the RAD50 gene contain four conserved enhancers that together constitute a locus control region (LCR) for the cytokine genes; in addition, the TH2 cell cytokine locus contains many evolutionarily conserved enhancers, silencers and other cis-regulatory regions whose functions have been comprehensively analyzed in mice, by monitoring changes in histone modifications and DNase I hypersensitivity during TH2 cell differentiation as well as by gene disruption9,10. Many of these enhancers show sequence conservation in humans, and there is evidence for functional conservation as well11.
Profiling DNA and histone modifications (epigenomic marks) layered over the genome sequence provides detailed information about the location and activity of cis-regulatory elements and the likely transcriptional potential of their target genes12. For instance, chromatin immunoprecipitation (ChIP) of DNA regions associated with the H3K4me1 modification followed by high-throughput sequencing (ChIP-seq) identifies cis-regulatory regions of genes that are poised for expression but may not actually be expressed until an appropriate environmental signal is received (for example, stimulation of the T cell antigen receptor by peptide–major histocompatibility complex binding); H3K27 acetylation (H3K27Ac) is associated with enhancers of genes that are actively being expressed13. Analysis of the H3K4 dimethyl (H3K4me2) modification has a dual advantage: it reveals both active and poised enhancers and provides a more precise localization of enhancers than analyses of H3K4me1 and H3K27Ac14,15. Epigenetic modifications tend to be more stable than RNA transcripts from the associated gene and provide a better measure of transcriptional status than RNA sequencing (RNA-seq) alone14,15. However, the broader applicability of epigenomic approaches for studying primary human cells is hampered by technical and practical constraints, including the small amount of blood or tissue that can be obtained from human subjects for research purposes, the variability of genome-wide ChIP assays when performed with small numbers of cells, the fact that the cell populations available for genome-wide analysis are typically mixtures of many different cell types, the genetic heterogeneity of humans and the complex nature of most human diseases.
Our goal in this study was to develop methods to profile active and poised enhancers in naive and memory CD4+ T cells from healthy individuals and asthmatic patients. Because we wished to extend these methods eventually to very small cell populations isolated from diseased tissues, we chose to profile cis-regulatory regions using a single histone modification, H3K4me2. By generating maps of H3K4me2-marked cis-regulatory regions, we identified a large number of cell type–specific and disease-specific enhancers. Some of these are linked to genes already implicated in TH2 cell differentiation, but for many more enhancers our analysis provides the first such indication. Several of the identified enhancers also harbor asthma-associated SNPs, validating our premise that the study of TH2 subtype–enriched cells from peripheral blood of asthmatic patients would yield information relevant to disease associated with TH2 cells. We conclude that enhancer profiling for H3K4me2 is a promising approach for studying the functions of well-defined cell populations in health and disease, with particular value when only limiting cell numbers are available.
RESULTS
Mapping cis-regulatory DNA in primary human CD4+ T cells
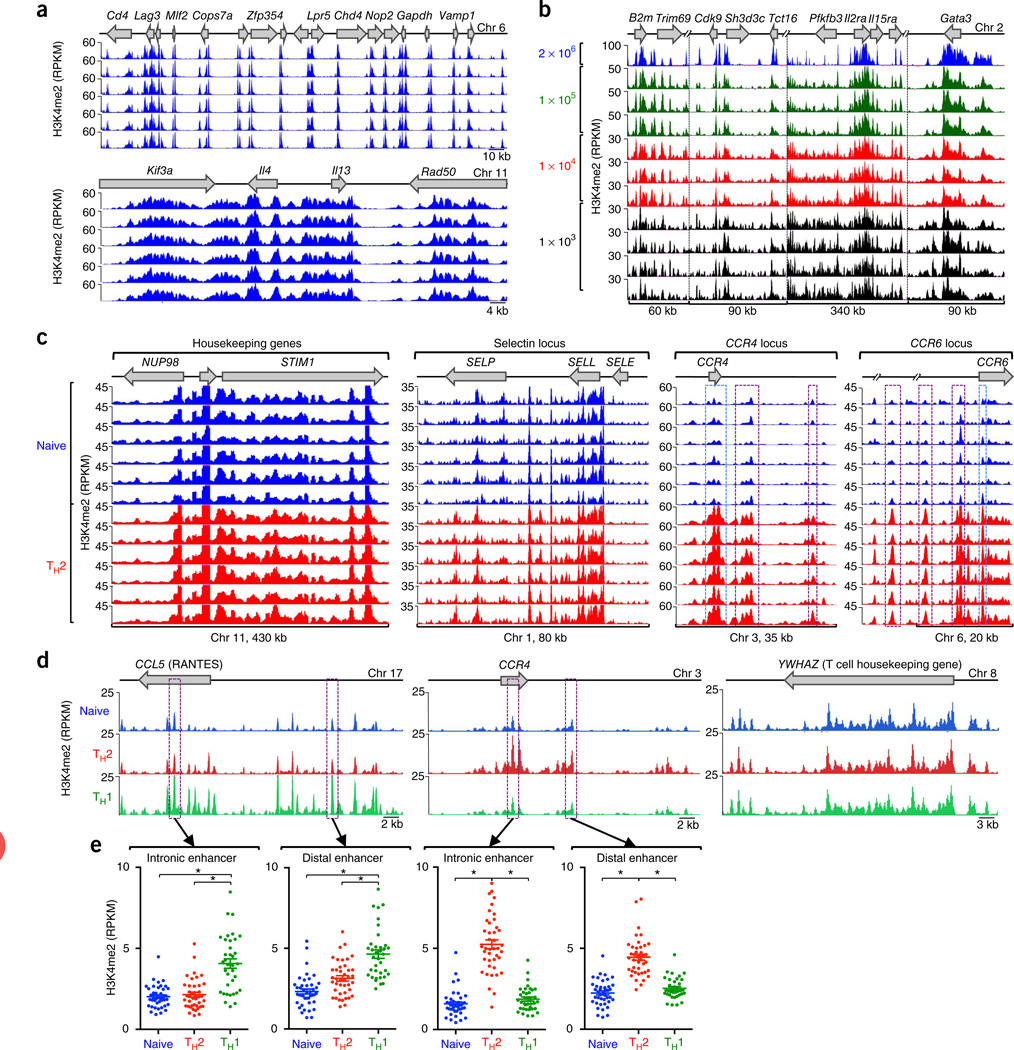
To capture the cis-regulatory landscape of human T cells, we isolated circulating naive and memory CD4+ T cells from peripheral blood of 24 subjects (12 healthy controls and 12 asthma patients; Online Methods). We subdivided CD4+ memory T cells on the basis of their surface expression of the chemokine receptor CCR4 (Supplementary Fig. 1). The CCR4+ T cells were enriched for TH2 cells, which are known to have a pathogenic role in asthma, whereas the CCR4− T cells were depleted for TH2 cells but enriched for TH1 cells, which produce interferon-γ (IFN-γ)16–18; for simplicity, hereafter we refer to CCR4+ T cells as ‘TH2 cells’ and CCR4− T cells as ‘TH1 cells’. Using ChIP-seq, we identified DNA regions associated with H3K4me2. To ensure that comparisons between sample groups were not affected by assay variability, we optimized the H3K4me2 ChIP-seq assay to provide highly reproducible results in multiple replicate assays (Fig. 1a and Supplementary Fig. 2) and also microscaled the assay (Supplementary Figs. 3 and 4) to enable reproducible detection of cis-regulatory DNA regions in as few as 104 cells (Fig. 1b). We performed 120 high-throughput whole-genome ChIP-seq assays, which generated ~2.8 × 109 reads covering ~1 × 1011 bases across three cell types (Supplementary Table 1), and computationally analyzed this data set to identify enhancers associated with CD4+ memory T cell differentiation as well as asthma.
Figure 1.
Reproducibility, microscaling and sensitivity of the H3K4me2 ChIP-seq assay. (a) Standard ChIP-seq assays (2 × 106 cells; six replicates) showing H3K4me2 enrichment patterns of the gene loci (top) in D10 cells. (b) Standard ChIP-seq assay (2 × 106 cells) and micro-scaled ChIP-seq assay (105, 104 and 103 cell samples; 3–4 replicates) showing H3K4me2 enrichment patterns in D10 cells. (c) ChIP-seq analysis showing H3K4me2 enrichment patterns, for control regions STIM1, NUP98, SELP and SELL loci, nonexpressed SELE locus, and TH2 cell–type specific CCR4 and CCR6 loci, in the naive cells and TH2 cells of six healthy subjects. Significant H3K4me2 enrichment (exact test for negative binomial distribution, using edgeR integrated in Bioconductor package MEDIPS) across distal cis-regulatory elements and promoters in these loci are highlighted by purple and blue dashed-line boxes, respectively. (d) ChIP-seq analysis showing cell-specific H3K4me2 enrichment patterns, for CCL5 (TH1 cell–specific), CCR4 (TH2 cell–specific) and control region YWHAZ (no change), in naive, TH1 cells and TH2 cells. For each cell type, data were merged from all donors, including assay duplicates. (e) H3K4me2 enrichment values for a specific 500-bp window (highlighted in purple dashed line boxes in d). Each dot represents data from a single assay; error bars indicate mean ± s.e.m. *P < 1 × 10−6, exact test for negative binomial distribution (using edgeR integrated in Bioconductor package MEDIPS).
H3K4me2 enrichment at most genes was highly comparable in the three cell types across all study subjects, which emphasizes the value of our multisample ChIP-seq assay in reducing technical variation (STIM1, NUP98, SELP and SELL genes; Fig. 1c and Supplementary Fig. 5). As expected, the H3K4me2 mark was depleted in the cis-regulatory regions of genes not expressed in T cells (e.g., SELE; Fig. 1c). We found clear evidence of cell type–specific cis-regulatory DNA elements in noncoding sequences: for instance, compared to naive T cells, the TH2 cell subset displayed extensive enrichment of H3K4me2 at promoters and cis-regulatory regions in the extended CCR4 and CCR6 loci (Fig. 1c and Supplementary Fig. 5). Merging multiple samples from the same cell types and disease categories reduced the background variability (that is, nonspecifically enriched regions) observed in individual assays, thus improving the sensitivity of our assay for detecting genomic regions with true H3K4me2 enrichment (Fig. 1d,e and Supplementary Fig. 6). Comparing enrichment values between these large sets of samples from the three different cell types, we detected highly significant differences at loci known to be regulated in a cell type–specific fashion (for example, CCL5 and CCR4 loci; Fig. 1e).
Enhancers linked to CD4+ T cell differentiation in vivo
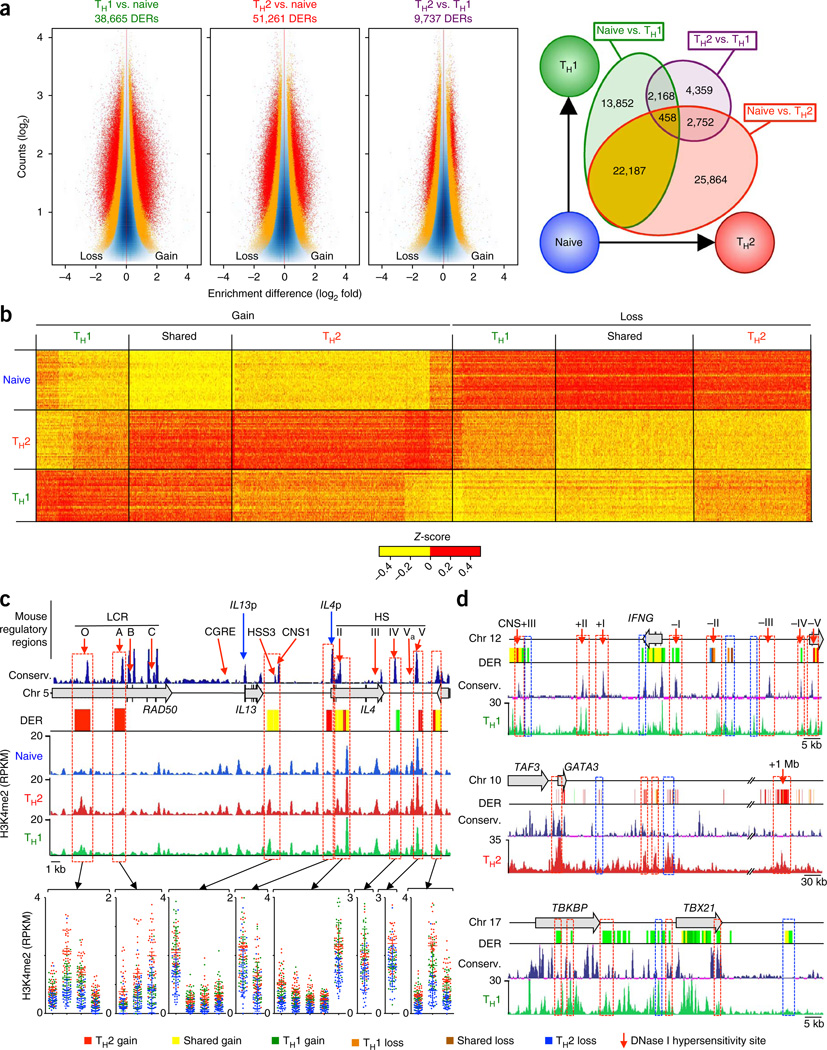
To identify cis-regulatory DNA regions differentially enriched for H3K4me2 in pairwise comparisons between cell types (differentially enriched regions; DERs), we calculated H3K4me2 enrichment based on reads in non-overlapping, consecutive 500-base-pair (bp) windows across the whole genome19,20. To identify cell-specific enhancers present in the general human population independent of disease status, we grouped each cell type from all 24 subjects and calculated differential enrichment for each group by modeling a negative binomial distribution for the data dispersion (Online Methods, Supplementary Note, and Supplementary Figs. 7 and 8). Analogous to gene-expression values in RNA-seq data, enrichment values, expressed as reads per kilobase per million mapped (RPKM), were calculated for each of the ~6 × 106 500-bp windows spanning the whole genome. 71,640 unique genomic windows (500-bp size), representing ~1% of the human genome, showed differential enrichment of H3K4me2 in the three cell types compared (Bonferroni-adjusted P < 0.05; Fig. 2a and Supplementary Table 2). We detected most of these differences (~90% of DERs) in the naive to memory T cell comparison (Fig. 2a and Supplementary Table 3) rather than between the TH2 and TH1 memory cell types, which confirmed that the activation and consequent differentiation of naive T cells into memory T cells results in major changes in the cis-regulatory landscape of chromatin21.
Figure 2.
Changes in enhancer strength among TH cell subsets. (a) ‘Minus-average’ (MA) plots for genomic regions with differences in H3K4me2 enrichment (DERs) for pairwise comparisons of indicated cell types; total numbers of DERs identified are listed (left; red and orange dots indicate windows with an adjusted P < 0.05 and raw P < 0.005, respectively; exact test for negative binomial distribution, using edgeR integrated in Bioconductor package MEDIPS). Overlap among the DERs identified for each pairwise comparison (right; Supplementary Table 3). (b) Z-scores of normalized read counts for each unique enhancer DER (columns) obtained from any of the three pairwise comparisons of naive, TH2 and TH1 cells; data are shown from each independent ChIP-seq assay (n = 120 total assays) (rows). (c) H3K4me2 enrichment tracks for each cell type were merged from all assays and illustrated along with location of mouse DNase I hypersensitivity sites (HS) and locus control regions (LCR) (red arrows), IL13 and IL4 promoter (IL13 p and IL4 p; blue arrows), University of California Santa Cruz (UCSC) multispecies conservation tracks, human TH2 cell cytokine locus and cell type–specific enhancer DERs. H3K4me2 enrichment values for specific 500-bp windows (red dashed-line boxes) are shown below. Each dot represents data from a single assay; error bars indicate mean ± s.e.m. (d) Tracks similar to those in c, for IFNG, GATA3 and TBX21. Enhancer DERs that overlap evolutionarily conserved and putative human-specific enhancers are highlighted by red and blue dashed-line boxes, respectively.
Over 90% of the ~71,000 cell-specific DERs were localized to intergenic and intronic regions; only 4% were present in promoter regions (defined as transcription start site (TSS) ± 1 kilobase (kb)) of annotated RefSeq genes (Supplementary Fig. 9). We termed all DERs outside promoter regions ‘putative differentiation-related enhancers’14,15 and classified them into three subgroups (Fig. 2b and Supplementary Table 3): ~30,263 ‘TH2 enhancers’ that exhibited the strongest H3K4me2 gain or loss in memory T cells enriched for the TH2 cell subset; ~17,483 ‘TH1 enhancers’ that exhibited the strongest H3K4me2 gain or loss in memory T cells enriched for the TH1 cell subset; and ~21,659 ‘shared memory enhancers’ that displayed equivalent H3K4me2 gain or loss in both TH2 and TH1 memory cells when compared to naive T cells. Several of these putative human enhancer DERs corresponded to well-characterized DNase I hypersensitive (HS) enhancers in the mouse TH2 locus (LCR-O, LCR-A, HS II, HS V; Fig. 2c). Moreover, a human TH1 DER (green bar) enriched in both TH2 cells and TH1 cells relative to naive T cells corresponded to a silencer region (HS IV in the mouse) that is DNase I hypersensitive in both TH1 cells and TH2 cells and restrains type 2 cytokine production in TH1 cells22. Likewise, the region corresponding to mouse hypersensitivity site S3 (HSS3), which contains a binding site for the chromatin insulator binding protein CTCF and exhibits constitutive DNase I hypersensitivity in naive, TH1 and TH2 memory cells9, appeared as a shared memory enhancer region in the human TH2 (IL5–RAD50–IL13–IL4) locus. Further, a number of evolutionarily conserved enhancers in other relevant gene loci (e.g., IFNG, GATA3 and TBX21, encoding the signature TH1 cell cytokine IFN-γ and the lineage-defining TH1 and TH2 cell transcription factors GATA-3 and T-bet, respectively) also exhibited cell type–specific enhancer activity as judged by H3K4me2 enrichment (Fig. 2d)23. However, five evolutionarily conserved regions identified as enhancers in the mouse (LCR-B, LCR-C, CGRE, CNS-1 and HS III) did not display cell-specific enhancer profiles in human T cells. In summary, we identified a number of putative lineage-specific enhancers in human T cells and showed that many but not all of these are conserved between human and mouse (Fig. 2c,d). Our findings indicate that at least some of the diverse differentiation-related enhancers identified in our screen may be important in shaping human CD4+ memory T cell fate.
Genes and molecular pathways linked to TH cell differentiation
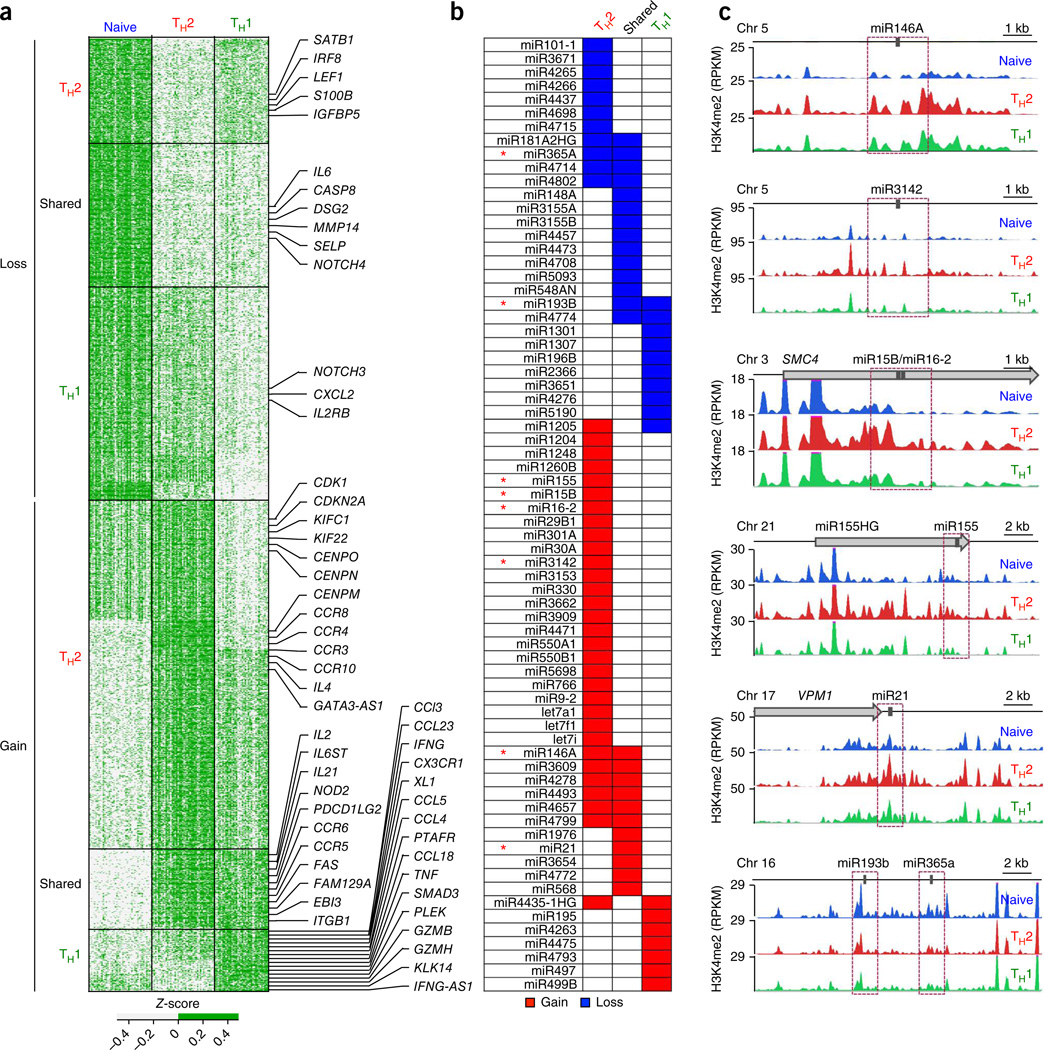
To connect our list of DERs to target genes, we focused on H3K4me2 at gene promoters (TSS ± 1 kb). Enrichment of H3K4me2 at promoters reliably marks genes that are either active or poised for expression in response to extrinsic stimuli14,15. We identified ~1,400 proteincoding genes (~7% of all annotated genes), 70 microRNAs (miRNAs) and 78 long noncoding RNAs that showed differential enrichment of H3K4me2 at their promoters (Bonferroni-adjusted P < 0.05; Fig. 3a and Supplementary Table 4). We observed concordant changes in gene expression and H3K4me2 enrichment patterns in many potential enhancers located in introns and within ± 20 kb of TSSs or transcription end sites of these genes (Supplementary Fig. 10).
Figure 3.
Genes and pathways linked to differentiation of CD4+ memory cells. (a) Z-scores of normalized read counts for each unique promoter-localized DER (rows) obtained from any of the three pairwise comparisons of naive, TH2 and TH1 cells; to illustrate samples that are above or below the average across all samples per window (row), a two-color scale (see key) was used for Z-scores; data are from each independent ChIP-seq assay (n = 120) (columns). Promoter-localized DERs were classified into six groups based on gain or loss of H3K4me2 enrichment during differentiation of naive T cells into the TH2 cells or TH1 cells (Supplementary Table 3); in each group, the promoter-localized DERs were arranged by their chromosomal location (from the start of chromosome 1 to the end of chromosome Y); ‘shared’ denotes ‘shared memory enhancers’ that displayed equivalent H3K4me2 gain or loss in both TH2 cells and TH1 cells when compared to naive cells. Target genes of some promoter-localized DERs are shown to the right. (b) miRNAs that showed the strongest gain or loss of H3K4me2 enrichment in their promoter regions for the various DER subgroups. (c) miRNAs for which the H3K4me2 enrichment tracks for each cell type are shown, for indicated miRNAs (* in b). Purple dashed-line boxes indicate the promoter regions (TSS ± 1 kb) for each miRNA.
We classified target genes of promoter-localized DERs into six subgroups based on preferential gain or loss of H3K4me2 enrichment during memory differentiation (Fig. 3a). Genes encoding many cytokines (IL4, IFNG, IL2 and IL21), chemokines (CCL3, CCL4 and CCL5), chemokine receptors (CCR3, CCR4, CCR8 and CCR10) and miRNAs (miR-146a, miR-21, miR-155, miR-29b, miR-15b and miR-193b)24 known to be differentially expressed in memory cells partitioned as expected into these subgroups (Fig. 3a–c and Supplementary Table 4). In addition, we discovered unique miRNAs and genes not previously linked to helper T cell differentiation (Fig. 3b and Supplementary Table 4).
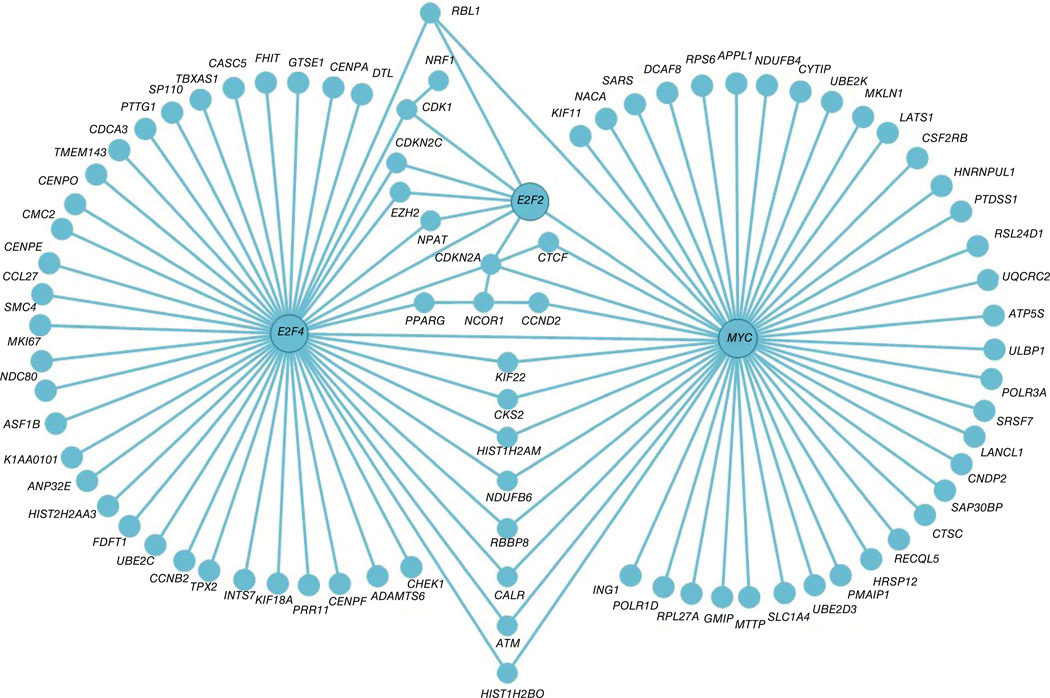
Analysis of biological process enrichment for genes that gain H3K4me2 during memory differentiation showed overrepresentation of pathways linked to positive regulation of T cell proliferation and activation, positive regulation of tyrosine phosphorylation of STAT proteins, regulation of adaptive immune response, activity of chemokine receptors, homeostasis of cellular calcium ions and regulation of immunoglobulin production (Supplementary Table 5). We observed significant enrichment for genes involved in regulation of apoptosis and mitosis (cell cycle) (Table 1) for the ‘TH2 gain’ subgroup of genes that gained promoter H3K4me2 enrichment in TH2 cells. Bioinformatic analysis of regulatory interactions among genes in the ‘TH2 gain’ subgroup pointed to three genes, MYC, E2F2 and E2F4, as potential major regulators of TH2 cell growth and survival (Fig. 4) MYC, E2F2 and E2F4 are known to regulate cell cycle, proliferation, apoptosis and metabolic reprogramming of T cells after activation25–27, and their increased expression likely confers a survival (growth) advantage to TH2 cells over TH1 cells, as previously described for mouse TH2 cells28. Thus, beside the well-known differences in cytokine production and tissue homing between human TH1 cells and TH2 cells, our data will provide information on genes that regulate other important functional differences between these cell types.
Table 1.
Enrichment for genes involved in regulation of apoptosis and mitosis
| Category | GO term | P value | Enrichment score |
Genes | Count (% of total) |
|---|---|---|---|---|---|
| TH2 cell gain | Mitosis | 3.45 × 10−7 | 6.4 | CDK1, KIFC1, KIF22, KIF11, ANAPC4, KIF18A, TPX2, CENPF, NEDD9, NDC80, CENPE, PTTG1, CEP55, UBE2C, LATS1, WEE1, SMC4, CCNB2, CDCA2, SKA2, SKA1, NEK6, TUBB3 and CDCA3 | 24 (4) |
| TH2 cell gain | Regulation of apoptosis | 6.14 × 10−4 | 3.1 | DLC1, CSF2, NUAK2, CLU, TNFSF15, TP63, FASLG, TNFRSF8, AKAP13, PMAIP1, TNFSF12, CALR, MCF2L, NLRC4, G2E3, CHD8, NOD2, CDKN2A, CDKN2C, CD2, HSPA5, BMF, MYC, CASP2, TXNIP, IL4, CFLAR, CDK1, IL2RA, IL7, ACTN3, SAP30BP, RPS6, NFKBIL1, ATM, NCSTN, CD38, NTRK1, CASP12, TNFAIP8, FAIM3, ACVR1 and IL2 | 43 (7) |
Functional biological process enrichment analysis of genes linked to promoter-localized DERs in the TH2 cell gain category. The Gene Ontology (GO) term of the biological process (BP), enrichment index and P value (calculated using database for annotation, visualization and integrated discovery (DAVID) software; Online Methods), names and number of genes present in the TH2 cell gain category (including the percentage of all genes in that biological process) are shown (full list in Supplementary Table 5).
Figure 4.
Upstream regulators of TH2 cell genes. Induced gene-regulatory network analysis (performed using version 27 software from the ConsensusPathDB interaction database; Online Methods) of genes in the TH2 cell gain category (listed in Supplementary Table 4) shows that MYC, E2F2, E2F4 are key upstream regulators of this subgroup of genes.
To identify target genes for the vast majority of DERs (~69,000) located outside promoter regions (putative enhancers), we determined the location of the nearest CTCF binding sites, potential insulator elements occupied by CTCF across many different cell types29. On the assumption that genes located in CTCF-bounded regions are controlled by enhancers located in the same region, we connected a total of ~7,600 target genes to the ~69,000 differentiation-related enhancers, which included nearly 50% of genes identified in a recent study of enhancers in in vitro–derived TH1 cells and TH2 cells30 (Supplementary Table 6). We assigned several candidate genes (median = 1.7 genes) to each enhancer and linked many enhancers (median = 4.9 enhancers) to each gene. To determine the functional significance of this large list of candidate genes, we performed a gene set enrichment analysis after classifying the genes into subgroups (as for the promoter analysis). Analysis of the enhancer-linked genes revealed many pathways important for TH2 and TH1 memory cell differentiation, and included many genes linked to promoter-localized DERs, Supplementary Table 7). An important caveat of pathway analysis performed with enhancer-linked genes is that the results can be confounded by correlation (or clustering) of genes with similar function throughout the genome. Additional experimental and computational analysis will be required to provide definitive evidence for the proposed assignments of genes to enhancers and vice versa.
Comparison of gene expression profiles with H3K4me2 enrichment profiles between cell types showed that differentially expressed genes display concordant changes in H3K4me2 enrichment patterns in the extended gene loci (Supplementary Fig. 11a and Supplementary Table 8). As expected, some of the genes that showed differential H3K4me2 enrichment did not show a significant change in gene expression (negative binomial test, false discovery rate ≤1%; Supplementary Fig. 11a). At least a fraction of these genes are likely to be poised for expression after an appropriate environmental signal, as these include the cytokine genes IL4 and IL21, which are expressed only by stimulated T cells (Supplementary Fig. 11b). Overall, comparison of H3K4me2 profiles as opposed to gene expression profiles between TH cell subtypes allowed us to define genes poised for expression in a cell-specific manner as well as identify enhancers involved in cell-specific gene regulation.
Transcription factor binding sites and motifs at TH cell enhancers
We took two approaches to identify trans-acting factors that might drive human TH cell differentiation in vivo. First, we identified known transcription factor binding motifs within the differentiation-related enhancers shown in Figure 2b (Fig. 5a). As expected, binding motifs for GATA-3 and T-bet were enriched in enhancers that gained H3K4me2 in TH2 cells and TH1 cells, respectively (Fig. 5a and Supplementary Table 9). In addition, motifs for a number of transcription factors not previously implicated in T cell differentiation were markedly enriched in ‘TH2 enhancers’ (Fig. 5a). Second, using publicly available ChIP-seq data from cell lines and primary T cells, we confirmed that some of these distinct transcription factors actually bound certain TH2 cell enhancers (Fig. 5b,c and Supplementary Table 10). NRF2 (also known as nuclear factor (erythroid-derived 2)-like 2; NFE2L2), and the structurally related factor NFE2 that binds antioxidant response elements, were strong hits in our in silico analysis (Fig. 5b). Activation of NRF2 has been shown to skew differentiation of mouse naive cells to TH2 cells and increase TH2 cell cytokine production31. Our results suggest that NRF2 may be a determining factor in human TH2 cell differentiation; experimental validation of its role in TH2 cell–driven diseases will be important because NRF2 signaling is activated by synthetic antioxidants that are commonly used as food preservatives in the United States31.
Figure 5.
Enrichment of transcription factor binding motifs and sites in enhancers linked to CD4 memory differentiation. (a) Heat map shows known transcription factor (TF) binding motifs that were significantly enriched in each DER subgroup (analysis performed using HOMER accessing its motif database; Online Methods and Supplementary Note). Motifs with P ≤ 1.00 × 10−3 and ratio of target sequences with motif versus background sequences with motif >1.1 were defined as significantly enriched (Supplementary Table 9). (b) Selected TFs (independent analysis performed for 161 TFs profiled by the ENCODE project and 18 other CD4+ T cell–related TFs, Online Methods) that showed significant enrichment of their binding sites at genomic locations of cell-specific enhancers in the TH2 cell gain subgroup (Supplementary Table 10). Shown are percentage of binding sites that overlap different cell-specific enhancer DER subgroups, the absolute number of binding sites that overlap all DERs and their percentage of the total genome-wide binding sites (in parentheses next to the name). Also shown is an example of a TF (SUZ12) whose binding sites are depleted in TH2 cell gain DERs (last row). (c) ChIP-seq peak tracks (black bars) of transcription factors (TH2 cell gain category) for human TH2 cell cytokine locus (IL4, IL13 and RAD50), IL12A and NF2L2 (encoding NRF2) along with UCSC gene tracks (top), multispecies gene conservation tracks (dark blue tracks), cell type–specific enhancer DERs and H3K27Ac track from ENCODE.
Finally, we observed a highly significant overlap between our list of cell-specific and differentiation-related enhancers with binding sites for the 161 transcription factors profiled by the encyclopedia of DNA elements (ENCODE) project32 as well as 18 other CD4-related transcription factors (odds ratio > 2.5 and P < 0.01 empirical test, Online Methods; Supplementary Table 10). Over one-third (38%) of all our identified enhancers contained at least one known transcription factor binding site and ~17% of the enhancers had > 3 such sites (Supplementary Table 10), which supports the notion that a substantial fraction of the enhancers identified in our study are likely to be functionally important.
Asthma-associated SNPs are enriched in TH2 cell enhancers
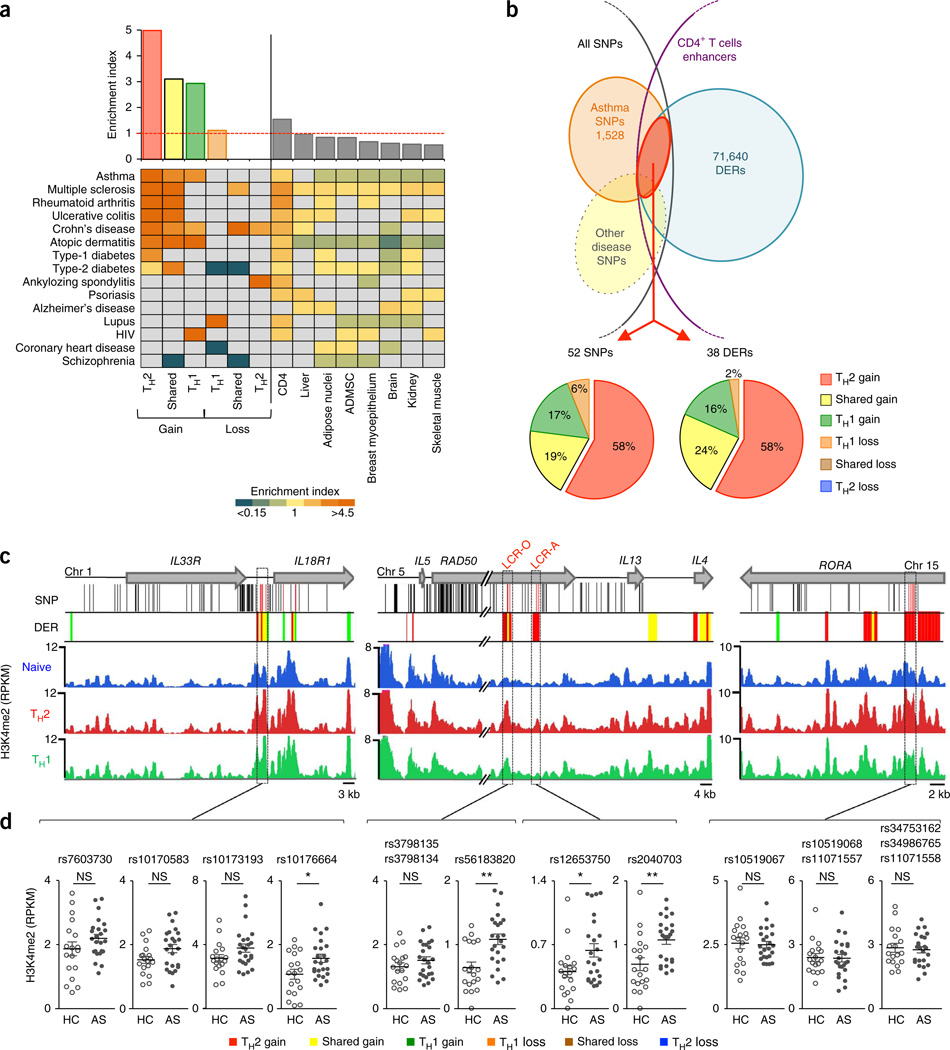
Disease-associated SNPs are enriched in regulatory regions that are active in cell types that contribute directly to disease33; in this context, we have shown previously that asthma-associated SNPs are enriched in regulatory regions that are active in CD4+ T cells34. Given that aberrant accumulation of TH2 memory cells is a key event in asthma pathogenesis, we asked whether asthma-associated SNPs are more enriched in enhancers linked to differentiation of TH2 as opposed to TH1 memory cells. We observed the highest enrichment (~4.99 fold; P < 0.0001) of asthma SNPs in enhancers (Fig. 2b) that gained H3K4me2 during development of TH2 cells rather than TH1 cells (Fig. 6a and Supplementary Table 11). This enrichment held true when considering haplotype blocks instead of individual SNPs and when limiting the source of SNPs that are significant in subjects from multiple study populations35 (Supplementary Table 11). These data support a pathogenic role of TH2 cells in asthma, and imply that SNPs associated with asthma risk may perturb TH2 memory cell differentiation by modulating the activity of enhancers that change dynamically during this developmental process. SNPs associated with some other immune system–mediated diseases showed preferential enrichment in different enhancer subgroups: SNPs associated with lupus and HIV risk were enriched in enhancers that lose and gain H3K4me2 enrichment in TH1 cells, respectively, and SNPs associated with ankylosing spondylitis risk were enriched in enhancers that lose H3K4me2 enrichment in TH2 cells (Fig. 6a and Supplementary Table 11).
Figure 6.
Asthma GWAS SNPs are enriched in TH2 cell enhancers. (a) Enrichment values of asthma GWAS SNPs in TH cell enhancer subgroups (Fig. 2b) and other cell tissue–specific enhancers32 (top) and for SNPs associated with other diseases (bottom; Supplementary Table 11). Enrichment values that did not reach significance (Chi-squared test, Online Methods) are shown in gray. ADMSC, adipose-derived mesenchymal stem cells. (b) Overlap of cell-specific DERs (shown in Fig. 2b) with asthma GWAS SNPs (top) and percentages of overlapping DERs or asthma SNPs in different DER subgroups (bottom). (c) UCSC tracks of IL33–IL18R, IL5–RAD50–IL13–IL4 and RORA loci containing large haplotype blocks of asthma-associated SNPs (black lines indicate their genomic location, red lines are SNPs that overlap DERs), along with cell-specific DERs tracks and H3K4me2 tracks for each cell type (merged from all assays shown in Fig. 2). Graphs show H3K4me2 enrichment values for each asthma-SNP-associated DERs (500-bp regions harboring the asthma SNP; highlighted in purple dashed-line boxes in c) in TH2 cells from the same H3K4me2 ChIP-seq assays shown in Figure 2. Each dot represents data from an independent assay; n = 18 assays from 10 healthy (HC) subjects, n = 24 assays from 12 asthmatic (AS) subjects; error bars indicate mean ± s.e.m.; *raw P < 0.05; **raw P < 0.01; NS, nonsignificant, calculated using MEDIPS.
Of the ~1,500 SNPs that have been associated with asthma, 52 were within the ~71,000 enhancer DERs associated with the development of CD4+ T cell memory; conversely, 38 DERs harbored one or more SNPs associated with asthma risk (Fig. 6b–d). Most of the 38 enhancers that harbored SNPs associated with asthma risk, including the conserved TH2 cell locus enhancers LCR-A and LCR-O, gained the H3K4me2 modification during TH2 cell development; a few showed a trend toward differential enrichment in asthma subjects versus healthy controls (Fig. 6b–d). These data suggest that the underlying SNPs may influence enhancer strength and have a potential role in shaping the pathological gene expression patterns observed in disease.
Enhancers associated with asthma
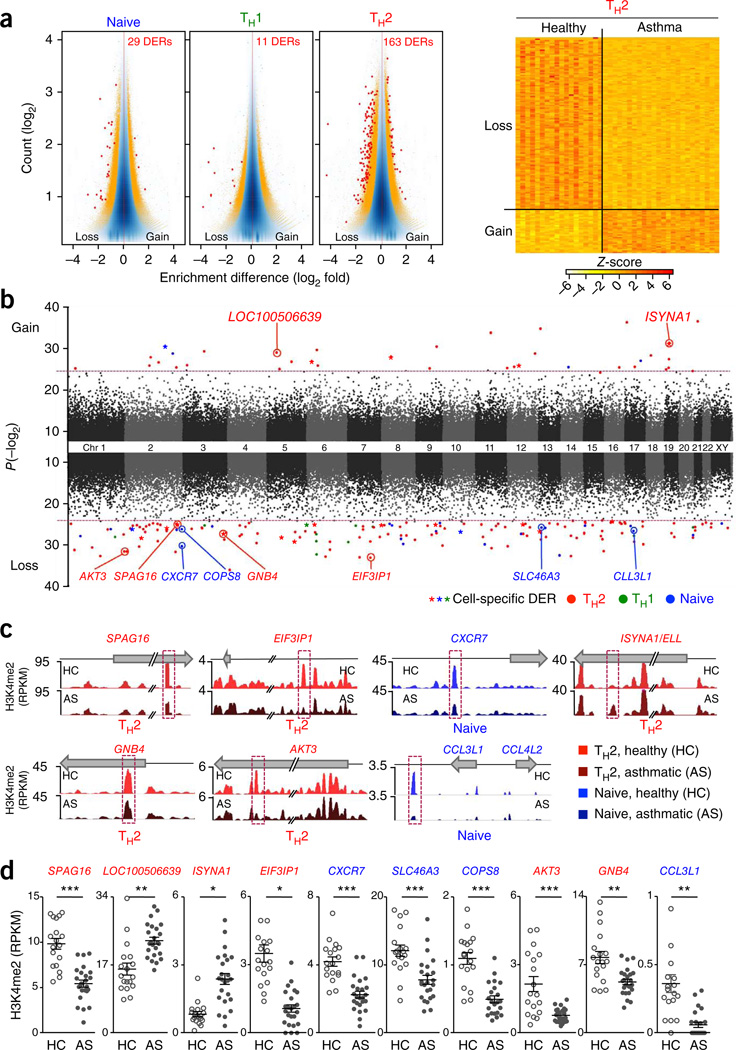
Finally, we sought to identify CD4+ T cell enhancers that differ in strength between healthy and asthmatic subjects. A total of 200 enhancer regions showed differential enrichment of H3K4me2 in the three cell types when we compared asthmatic versus healthy controls (Bonferroni-adjusted P < 0.05; Fig. 7 a–c and Supplementary Table 12). As expected, most of these regions (here termed ‘asthma-associated enhancers’) were specific to the potentially pathogenic TH2 memory cell population (163 of 200 regions, of which 33 and 130 gained and lost H3K4me2 enrichment, respectively), with smaller numbers (29 and 11 regions) specific to naive T cells and TH1 cells, respectively (Fig. 7a–d). Nearly 10% of these asthma-associated enhancers overlapped with the ~71,000 enhancer DERs associated with CD4+ T cell memory, a significant enrichment (odds ratio = 8.74; P = 1.7 × 10−12, Online Methods) that suggests an important functional role for at least a subset of the enhancers. 84 of the 200 asthma-associated enhancers (42%) contained at least one transcription factor binding site (Supplementary Table 13), with significant enrichment for binding sites of transcription factors involved in T cell differentiation (for example, GATA3, TBX21 and RUNX3; Supplementary Table 13). Enrichment analysis of candidate genes assigned to asthma-associated enhancers demonstrated significant overrepresentation of genes involved in chemokine and Toll-like receptor signaling pathways (KEGG pathway; P = 0.0009 calculated using statistical software from the ConsensusPathDB interaction database, Online Methods; Supplementary Table 14). Candidate genes enriched in this pathway include CCL3L1, CCL3L3 and CCL4L2, a cluster of genes on chromosome 17 that encodes chemokines known to bind CCR5 and prevent entry of HIV into T cells36, and AKT3 and GNB4, involved in phosphoinositide 3-kinase and G protein signaling (Fig. 7d). Further studies will clarify the role of these genes and enhancers in asthma pathogenesis.
Figure 7.
Identification of asthma-associated enhancers. (a) MA plots (vertically displayed) illustrate genomic regions with differences in H3K4me2 enrichment (DERs) between healthy and asthmatic subjects in the three different cell types (Supplementary Table 12). Red dots and orange dots indicate windows with adjusted P < 0.05, or with raw P < 0.005, respectively (exact test for negative binomial distribution, using edgeR integrated in Bioconductor package MEDIPS). Z-scores (right) of normalized read counts for each asthma-associated DER (rows) identified in the TH2 cells. (b) Manhattan plot illustrates the genome-wide distribution of asthma-associated DERs in relation to their statistical significance values (P values, MEDIPS; y-axis parameter). Red dashed line sets the threshold for an adjusted P < 0.05. (c) Comparison of H3K4me2 enrichment between healthy and asthmatic subjects in indicated cells. H3K4me2 tracks for each cell type were merged from all assays performed in healthy (HC) and asthmatic (AS) donors (same cohort as shown for the analysis above and in Fig. 2). (d) H3K4me2 enrichment values for each asthma-associated DER (highlighted in purple dashed line boxes in c from the same H3K4me2 ChIP-seq assays shown in Fig. 2). Each dot represents data from an independent assay; n = 18 assays from 10 healthy subjects (HC), n = 24 assays from 12 asthmatic patients (AS); error bars indicate mean ± s.e.m.; *P < 0.05, **P < 0.01, ***P < 0.001 (MEDIPS).
DISCUSSION
We mapped the genome-wide distribution of H3K4me2 in naive, CD4+, TH1 and TH2 cells from 24 subjects. Through computational analysis of the data, we identified putative enhancer regions that are associated with specific TH cell subsets. By comparing TH cell subsets from healthy and asthmatic individuals, we identified enhancers that reproducibly differ in H3K4me2 enrichment and thus are presumed to be differentially active in T cells from asthma patients compared to healthy controls.
Our strategy can be applied to any accessible cell type with a function relevant to human health or disease. Purified cell populations are not necessarily needed: even though we performed ChIP-seq experiments on enriched cell populations rather than on purified T cell subsets, the majority of H3K4me2-marked enhancers that we identified were clearly associated with the enriched cell type. This was particularly true for TH2 cells, which were present only in the CCR4+ population and depleted from the CCR4− population; in contrast, IFN-γ–producing TH1 cells were present in both the CCR4+ and CCR4− populations, which explains why overall we identified fewer enhancers as specifically TH1 cell–associated. To facilitate studies of rare cell populations involved in disease pathogenesis, we optimized our H3K4me2 ChIP-seq assay so that it can be applied to very small numbers of cells. This feature will be valuable not only for studies of the immune system but in cases where only limited amounts of biological materials are available for research. Our method offers a substantial advantage over transcriptional profiling by microarray or conventional RNA-seq, which identifies only those genes that are represented by steady-state transcripts at the time that the cells are lysed. In contrast, H3K4me2 ChIP-seq has the potential to identify promoters and enhancers that control the expression of genes that are not actively being transcribed but are poised for expression in response to extrinsic stimuli.
We showed that in silico analyses of enhancer profiles obtained by H3K4me2 ChIP-seq can be used to predict binding sites for transcriptional regulators that set up gene expression programs in specific cell types and disease states. Many such transcription factors are only transiently expressed at specific developmental stages that may be difficult to isolate in vivo, which implies that they cannot be identified merely through expression profiling of fully differentiated cells. However, lineage-defining or disease-initiating transcription factors often leave a footprint of their activity on the cis-regulatory landscape of a cell, by altering H3K4me2 levels at promoters and enhancers during cellular differentiation or early disease progression. Thus, H3K4me2 profiling of peripheral blood cells is particularly relevant for studies of the immune system in patients: differentiation of memory T cells occurs in relatively inaccessible compartments such as lymph nodes, spleen or thymus, and the majority of patients present in the clinic well after the first disease-initiating events have occurred.
Consistent with the idea that TH2 cell function is aberrant in asthma, we found that TH2 cell–specific enhancers were highly enriched for asthma-associated SNPs identified in genome-wide association studies (GWAS). Our approach has narrowed down the large list of SNPs in asthma-associated haplotype blocks to those that are likely to be functional in cells relevant to asthma pathogenesis by determining whether they are located in a putative enhancer region, and whether that enhancer shows evidence of altered function during memory T cell differentiation. The candidate enhancer-localized SNPs identified in our study can now be formally tested for functional importance in disease pathogenesis.
Separate from GWAS analysis, we identified enhancers associated with asthma susceptibility simply by comparing TH cell enhancer profiles of healthy and asthmatic subjects as defined by H3K4me2 enrichment patterns. By analogy with GWAS, such approaches have been termed epigenome-wide association studies37. An important consideration is the need to confirm the link between disease-associated enhancers and their target genes, for instance, by using new techniques to detect physical interactions between enhancers and gene promoters38,39; moreover, detailed testing of the candidate genes for function in human helper T cells will be necessary to attribute an important role in asthma pathogenesis. Notably, the 12 asthma patients evaluated in our study are a very small patient cohort; replication of our hits in multiple independent and larger cohorts is needed to strengthen their biological importance and to address the important question of whether epigenome-wide association studies can be used to distinguish molecular subtypes of asthma.
ONLINE METHODS
Study subjects and sample processing
The ethics committees of the Southampton University Hospitals Trust and La Jolla Institute approved the study, and written informed consent was obtained from all subjects. Twelve nonsmoking subjects with asthma (6 with mild asthma never treated with corticosteroids and 6 subjects with moderate asthma treated with inhaled corticosteroids16), meeting established diagnostic criteria40, and 12 healthy subjects were studied (Supplementary Table 15). Subjects with mild asthma had symptoms <3 times a week, with forced expiratory volume in 1 s (FEV1) > 80% of predicted, and used short-acting inhaled β2-agonists as needed for symptom relief. Subjects with moderate asthma were additionally treated with regular inhaled corticosteroids and inhaled short-acting β2-agonists. All asthmatic subjects were atopic and allergic to house dust mite (Dermatophagoides pteronyssinus). All study subjects were Caucasian adults (10 females and 14 males, age range 18–65, inclusion criterion). Healthy controls had no history of smoking or respiratory symptoms suggestive of asthma.
For isolating T cell subtypes from peripheral blood samples, peripheral blood mononuclear cells (PBMCs) were first separated into a CD4+ memory cell fraction and remaining cells by use of the memory CD4+ T cell isolation kit (Miltenyi Biotec). The CD4+ memory cells were then stained with a cocktail of fluorescently conjugated antibodies (anti-CD45RA FITC-conjugated (clone HI100), anti-CD4 APC-Cy7-conjugated (clone SK3), anti-CCR4 PE-conjugated (clone 1G1), and anti-CD25 APC-conjugated (clone M-A251) and sorted on the FACS Aria to obtain two cell populations: CD4+CD45RA−CCR4− and CD4+CD45RA−CCR4+CD25− (Supplementary Fig. 1). Naive T cells were sorted from the remaining cells after CD4+ memory cell isolation by staining with anti-CD45RA FITC-conjugated, anti-CD4 PerCP-Cy5.5-conjugated (clone SK3), anti-CD62L APC-Cy7-conjugated (clone DREG-56) antibodies and anti-CD45RO PE-conjugated (clone UCHL1). Sorted cells were washed and fixed for 10 min at 20 °C with 1:10 dilution of 11% formaldehyde (in 50 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EDTA and 0.5 mM EGTA), neutralized with 1:20 dilution of 2.5 M glycine, washed twice in ice-cold phosphate buffered saline, and cell pellets were snap frozen in liquid nitrogen before storage at −80 °C.
Microscaled multisample chromatin immunoprecipitation–sequencing
Formaldehyde-fixed cell pellets were snap-frozen in liquid nitrogen and stored at −80 °C before the ChIP assay. This storage step gave us the flexibility to batch multiple samples for ChIP-seq. Frozen cells were lysed in 120 µl of lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS, 1 mM phenyl methane sulfonylfluoride, 20 mg/ml sodium butyrate and proteinase inhibitor cocktail; Sigma), and chromatin was sheared by sonication, using BioRuptor (Diagenode), to generate 100–500 bp DNA fragments. Shearing multiple samples (n = 12) in parallel using the Bioruptor ensured equivalent shearing of chromatin among the samples that were to be compared. For the initial validation of microscaled ChIP-seq, chromatin from 105, 104 or 103 whole-cell equivalents (D10.G4.1 cells, ATCC, mycoplasma-negative) was immunoprecipitated with anti-H3K4me2 (clone Y47, lot#YH041501C; Abcam). For samples from study subjects, chromatin from 105 cells was used for immunoprecipitation. Standard H3K4me2 ChIP using 2–10 × 106 cells was performed as described previously10. For microscaled H3K4me2 ChIP-seq, chromatin was diluted in 1 ml of radioimmunoprecipitation assay (RIPA) buffer and immunoprecipitation was done overnight at 4 °C by incubating 1 µl of antibody precoated on 10 µl protein A coated magnetic beads (Invitrogen). To reduce nonspecific DNA binding (background signal in ChIP-seq assays), we used pipette tips and tubes with low DNA and protein binding (Axygen Maximum recovery). Immunocomplexes were captured and washed for 5 min each with RIPA buffer, high-salt buffer (50 mM Tris-HCl, pH 8, 500 mM NaCl, 0.1% SDS, 0.5% Na-deoxycholate, 1% Nonidet-P40 and 1 mM EDTA), LiCl buffer (50 mM Tris-HCl, pH 8, 250 mM LiCl, 1 mM EDTA, 1% Nonidet-P40 and 0.5% Na-deoxycholate), and low-salt buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA, 50 mM NaCl). Beads were resuspended in TE (10 mM Tris-HCl, pH 8, 1 mM EDTA), transferred to fresh tubes, captured and then resuspended in 200 µl of elution buffer (50 mM Tris-HCl, pH 8, 10 mM EDTA and 1% SDS). Chromatin was detached from the beads by incubating for 15 min at 65 °C, transferred to fresh tubes, then treated with 2 µl of RNase A (20 mg/ml; Invitrogen) for 30 min at 37 °C and with proteinase K (0.2 mg/ml, Invitrogen) for 6 h at 55 °C, and incubated overnight at 65 °C. DNA was purified using affinity columns (Zymo Research) and eluted in 12 µl of TE (10 mM Tris-HCl, pH 8 and 0.1 mM EDTA). Selected DNA sequences from control regions (IL4 promoter, IFNG promoter, GAPDH promoter and HOX7A promoter) were quantified by real-time quantitative PCR to assess the quality of the ChIP. The amount of DNA in the ChIP samples was quantified using picogreen assay (Invitrogen). One nanogram of ChIP DNA was PCR-amplified for 18–22 cycles using whole genome amplification (WGA) primers (WGA-SEQX, Sigma) following the manufacturer’s instructions. 2 µg of this amplified DNA was treated with restriction enzyme (WGA-SEQX, Sigma), to remove the WGA primer sequences and then purified using AmpureXP beads (Beckman Coulter). Efficiency of removal of WGA primer sequences was assessed by PCR. From this step, 500 ng of purified DNA was diluted with TE to obtain a total volume of 65 µl. Diluted DNA was sonicated with E220 Covaris multiplex sonicator (Covaris) to generate 100–250 bp DNA fragments. ~250 ng of DNA was used for preparing standard SOLiD sequencing library (5500 SOLiD Fragment 48 Library Core Kit & Fragment Library Barcode Adaptors 1–96). Following emulsion PCR, samples were sequenced on the SOLiD 5500 sequencer to obtain 35-bp single-end reads (SOLiD EZ Bead E120 System kits). Both whole-genome amplification and sequencing-library preparation were performed in a 96-well format, which reduced hands-on time, beside reducing technical and assay-to-assay variability. Multiple quality control steps (Supplementary Fig. 3) were included to determine optimal shearing of DNA, ChIP efficiency and number of PCR amplification cycles, to assess removal of whole-genome PCR amplification adaptors, efficiency of ligation of sequencing adaptors and to determine library complexity. Samples that failed quality control were eliminated from downstream steps. Reproducibility and correlation between standard and microscaled ChIP-seq, performed using various cell numbers, is shown in Figures 1a–c and Supplementary Figures 4 and 5.
RNA sequencing
Total RNA was purified using miRNAeasy kit (Qiagen) and quantified as described previously24. 10–15 ng of purified total RNA was used for poly(A) section (Poly(A)Purist Mag kit, Life Technologies). Poly(A)-selected RNA was amplified with the Whole Transcriptome Amplification Sequencing Technology kit (SEQR, Sigma) as per manufacturer’s recommendation. 1 µg of this amplified cDNA was treated with restriction enzyme (SEQR, Sigma), to remove the primer sequences and then purified using AmpureXP beads (Beckman Coulter). Efficiency of removal of SEQR primer sequences was assessed by PCR. From this step, 250 ng of purified DNA was sonicated with E220 Covaris multiplex sonicator (Covaris) to generate 100–250 bp DNA fragments. Sonicated cDNA was used for preparing standard SOLiD sequencing library as described in the previous section for ChIP-seq.
Mapping and primary data processing of sequencing reads
Human ChIP-seq data (csfasta and qual files) were mapped against the human reference genome hg19 (downloaded from the Genome Bioinformatics database of the University of California Santa Cruz) using bowtie v.0.12.7 (-n 2 -m 1 -C)41. 120 different ChIP-seq assays were performed and the obtained sam files were converted and merged into separated bam files using samtools v.0.1.18 (ref. 42). For visualization of the ChIP-seq data in public genome browsers, sequencing coverage was calculated at genome-wide 50-bp windows after extending each read to a length of 120 bp along the sequencing direction using MEDIPS v.1.10.0 (extend = 120, uniq = F, window_size = 50, BSgenome = “BSgenome. Hsapiens.UCSC.hg19”)19,20, and the resulting coverage profiles (RPKM) were exported as wiggle files. Mouse ChIP-seq data were mapped against the mouse reference genome mm10 as described for the human ChIP-seq data (bowtie v.0.12.7 -n 2 -m 1 -C). Human RNA-seq data was mapped against hg19 using tophat43 (v1.4.1.,–library-type fr-secondstrand -C) and the RefSeq gene annotation downloaded from the UCSC genome Bioinformatics site. Sequencing read coverage per gene was counted using HTSeq-count (-m union -s yes -t exon -i gene_id, http://www-huber.embl.de/users/anders/HTSeq/).
Correlation between ChIP-sequencing assays and samples
Pairwise Spearman correlation between samples were calculated by comparing the sequencing coverage at genome-wide 500-bp windows using MEDIPS v.1.10.0 (extend = 120, uniq = F, window_size = 500, BSgenome = “= “BSgenome. Mmusculus.UCSC.mm10”) (Supplementary Figs. 2 and 4).
Identification of differentially enriched cis-regulatory regions comparing cell types
To identify cell type–specific DERs, H3K4me2 ChIP-seq data were grouped according to their cell types: naive CD4+ T cells, (n = 42), TH2 cells (n = 42) and TH1 cells (n = 36). All technical and biological replicates were independently processed ChIP assays with separate ChIP, whole-genome amplification and sequencing, and each ChIP replicate was sequenced to a targeted depth of ~20 × 106 mapped reads. Additional details are available in Supplementary Note.
Sample sizes for ChIP-seq were chosen to achieve power to detect consistent differences between asthmatic and healthy donors in the presence of variability inherent to the assay. Assay variability was assessed based on biological replicates of mouse cell line data, and differences were introduced computationally at different sites. Recovery of these sites using MEDIPS was used to estimate error rates depending on sample size. To estimate a false discovery rate for our chosen significance threshold to identify DERs (Bonferroni-adjusted P < 0.05), we performed a random approach by permuting the 42 naive CD4+ T cells and the 42 TH2 cell ChIP-seq assays resulting in two groups of intermixed samples. Subsequently, we applied the same statistical framework used for our original approach. In ten independent iterations of such random sample permutations, we observed a very small number of DERs ranging from 4 to 11 (data not shown) compared to 51,261 DERs detected for the correct assignment of samples at the same significance level of a Bonferroni adjusted P < 0.05. This translates to a very low false discovery rate (FDR) of 0.02%, suggesting that the P-value threshold used for our analysis is stringent and does not result in overcalling of DERs. Further, in our approach, the detectable effect size is dependent on enhancer strength (see MA plots where the ‘enhancer strength’ is represented on the x axis). With our sample size (naive = 42, TH2 = 42) and the inherent variation between samples, we can detect differences between cell types with a ratio (‘effect size’) of at least 1.6 in the lowest 25% quantile and of at least 1.2 in the upper 25% quantile of signal-enhancer strengths.
Assignment of target genes to DERs
To assign DERs to transcripts and genes, we downloaded the hg19 table refGene (track RefSeq Genes via the Table browser at the UCSC Genome Bioinformatics site (17 October 2013), and assigned DERs to promoters if their midpoint fell within ± 1 kb of a transcription start site. This identified 2,235 promoter DERs associated with 1,710 transcripts of 1,529 genes. We classified the remaining 69,405 non-promoter–localized DERs as enhancers. To associate enhancer DERs to genes, we considered CTCF binding sites previously identified in 19 diverse human cell types44 as boundaries for defining extended transcript loci. For each transcript, their extended locus was defined as the upstream and downstream region around their start sites limited by the occurrence of a CTCF binding site or by a maximal distance of 200 kb. We observed 25,888 DERs that fell into the extended loci of 8,983 transcripts of 7,647 unique genes.
Correlation of H3K4me2 enrichment patterns at promoters and enhancers
To examine putative concordant changes of H3K4me2 enrichment at promoters of a transcript and enhancers located within or close to that transcript, we considered the following two groups of transcripts: (i) transcripts containing a DER in their promoter region with gain of H3K4me2 in TH2 cells compared to naive CD4+ T cells (TH2 gain, n = 534), (ii) transcripts containing a DER in their promoter region with loss of H3K4me2 in TH2 cells compared to naive CD4+ T cells (n = 405, TH2 cell loss). In addition, we defined the following four groups of genomic regions: upstream: −20 kb to a transcript start site, promoter: ± 1 kb around a transcript start site, transcript body: between a transcript start and end site, and downstream: +20 kb from a transcript end site. We divided the upstream and downstream regions into ten equally spaced windows of 2 kb but considered the promoter as one window and the transcript body as another window. Subsequently, we calculated sequencing read coverage at the predefined windows of the selected transcripts for each of the 42 naive CD4+ T cell and for each of the 42 TH2 cell samples using MEDIPS v.1.12.0 (function MEDIPS.createROIset, extend = 120, uniq = F, bn = 10 (for upstream and downstream) or bn = 1 (for promoter and transcript bodies), BSgenome = “BSgenome.Hsapiens.UCSC.hg19”). For each transcript and for each tested window, mean counts over all samples per group were calculated. In order to avoid division by 0, we added 1 to each tested window in both groups, before calculation of fold change between groups at individual transcripts and windows. For each transcript, we calculated the mean fold change over all 2-kb windows in the upstream or downstream regions, respectively. The heat map in Supplementary Figure 10 shows the log2 of fold changes at the four tested groups of genomic regions from left to right and at individual transcripts of groups i and ii from top to bottom. Transcripts in groups i and ii are ordered by the sum of log2 fold changes across the four tested genomic regions, where group i is sorted from highest (top) to lowest (bottom), and group ii is sorted from lowest (top) to highest (bottom).
Principal component analysis of the RNA-sequencing data and differential gene expression analysis
The principal component analysis (Supplementary Fig. 11) has been performed by calculating singular value decomposition of the centered, variance-stabilized and library-normalized counts of the 5,000 most variable genes, using the R functions svd (base package, http://www.r-project.org/) and variance Stabilizing Transformation (Bioconductor package DESeq45. To identify differential gene expression between cell types, we performed negative binomial tests for pairwise comparisons of the naive CD4+, TH2 and TH1 cells employing the Bioconductor package DESeq45 and allowing for a false discovery rate of 1%.
Correlation of gene expression with H3K4me2 enrichment patterns at promoters and enhancers
This is described in Supplementary Note.
Functional gene-set enrichment analysis
Gene sets obtained for each promoter-localized DER category (classified based on preferential gain or loss of H3K4me2 enrichment during memory differentiation; Supplementary Table 3) were analyzed using DAVID (version 6.750; refs. 46,47). Biological process and pathways enriched for genes present in our data set are shown in Supplementary Table 5. As some of the gene sets obtained for enhancer-DER groups exceeded the maximum limit of 3,000 genes for the DAVID software, we additionally used the software from the ConsensusPathDB interaction database (CPDB; version 27; refs. 48,49) for the overrepresentation analysis. Biological pathways and processes enriched for genes present in our data set are shown in Supplementary Tables 5, 7 and 14). To determine upstream regulators of genes in the TH2 cell (CCR4+) gain category, we performed induced gene-regulatory network analysis using the software from CPDB48,49. Analysis was performed using default settings on the software and allowing for intermediate nodes with a Z-score threshold of 20 (Fig. 4).
Detecting enrichment of transcription factor motifs and binding at cell-specific enhancers
This is described in Supplementary Note.
Identification of asthma-associated DERs
To identify differentially enriched regions (DERs) between healthy and asthmatic donors, we grouped the ChIP-seq data of samples into the following six groups: (i) naive CD4+ T cells, healthy (n = 18), (ii) naive CD4+ T cells, asthma (n = 24), (iii) TH2 cells, healthy (n = 18), (iv) TH2 cells, asthma (n = 24), (v) TH1 cells, healthy (n = 13), and (vi) TH1 cells, asthma (n = 23). Differential sequence coverage was calculated for each cell type between healthy and asthma samples as described above to identify cell type–specific DERs. This resulted in 29 DERs for naive CD4+ T cells (4 asthma > healthy, and 25 asthma < healthy), 163 DERs for TH2 cells (33 asthma > healthy, and 130 asthma < healthy), and 11 DERs for TH1 cells (0 asthma > healthy, and 11 asthma < healthy) (Bonferroni-adjusted P < 0.05). In total, there were 203 significant windows in the three comparisons falling into 200 distinct asthma DERs. As above, we divided DERs into those that fall into promoters and enhancers. There were 13 promoter-localized DERs associated with 15 transcripts of 14 genes. A total of 86 nonpromoter DERs were located in the extended loci of 129 transcripts of 111 unique genes. Transcription factor binding site enrichment analysis for the asthma-associated DERs has been calculated as described above for the cell-specific enhancers. Owing to the relatively small number of DERs, we considered the entire 500-bp DER and according random regions for calculating overlaps to transcription factor binding sites.
SNPs data sets and enrichment calculation
62 significant lead SNPs (P < 1.0 × 10−5) from GWAS studies of asthma were obtained by querying the HaploReg database50. Cross-referencing the P values of these lead SNPs using the GWAS Integrator database51 and the original articles confirmed that all SNPs had P < 1.0 × 10−5. An additional 20 lead SNPs associated with asthma-related traits (plasma eosinophil counts, serum IgE and YKL-40 concentrations) were obtained from the GWAS Integrator database, after manual review of the originating studies. Details of all 82 lead SNPs, P values and source study are provided in Supplementary Table 11a. For each lead SNP, SNPs in tight genetic linkage (r2 > 0.8) were retrieved based on data from the phase I of the 1,000 genome project using European (EUR) as the reference population52 (calculations performed using HaploReg v2.0, Supplementary Table 11b). The total number of asthma-associated SNPs in the combined set (lead SNPs + linked SNPs) was 1,528. For all other diseases, significant lead SNPs (P < 1.0 × 10−5) were obtained from the HaploReg50 database, and no additional traits were considered. Linked SNPs were calculated as described for the asthma SNPs above. More detailed description is available in Supplementary Note.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff at the Wellcome Trust Clinical Research Facility (University of Southampton) where samples were acquired from volunteers; M. North for assisting in patient recruitment, assessment and sample collection; R. Jewel and C. McGuire for providing assistance in the flow cytometry facility (University of Southampton; J. Day for assistance with high-throughput sequencing at the La Jolla Institute for Allergy and Immunology sequencing facility, and A. Moghaddas Gholami at the La Jolla Institute for Allergy and Immunology bioinformatics core for help with the SNP enrichment analysis. L.C. is funded by a Feodor Lynen Research Fellowship from the Alexander von Humboldt Foundation. This work was supported by the Dana Foundation (K.M.A.), GlaxoSmithKline National Clinician Scientist Fellowship Award and Peel Travel Fellowship Award (P.V.), R01 HL114093 (to B.P., A.R. and P.V.) and U19 AI100275 (to B.P., A.R. and P.V.).
Footnotes
Accession codes. Gene Expression Omnibus: GSE53646 (sequencing data).
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
G.S., K.M.A., B.P., A.R. and P.V. conceived the work, designed, performed and analyzed experiments, and wrote the paper; N.O., L.K., M.V. and A.P.V.G. assisted in the performing some of the experiments under the supervision of G.S. and P.V.; R.D. provided support and direction for obtaining and processing clinical specimens; L.C. identified DERs; and A.G., M.L. and A.C. performed the bioinformatic analysis under the supervision of L.C. and B.P.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 2.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat. Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 3.Kay AB. Allergy and allergic diseases. Second of two parts. N. Engl. J. Med. 2001;344:109–113. doi: 10.1056/NEJM200101113440206. [DOI] [PubMed] [Google Scholar]
- 4.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol. Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu. Rev. Immunol. 2009;27:363–391. doi: 10.1146/annurev.immunol.021908.132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO fact sheets N206 and N307. [accessed November 2013]; http://www.who.int/mediacentre/factsheets/fs206/en/; http://www.who.int/mediacentre/factsheets/fs307/en/index.html. [Google Scholar]
- 7.Holgate ST, Polosa R. Treatment strategies for allergy and asthma. Nat. Rev. Immunol. 2008;8:218–230. doi: 10.1038/nri2262. [DOI] [PubMed] [Google Scholar]
- 8.Wenzel SE, Wang L, Pirozzi G. Dupilumab in persistent asthma. N. Engl. J. Med. 2013;369:1276. doi: 10.1056/NEJMc1309809. [DOI] [PubMed] [Google Scholar]
- 9.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of TH2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 10.Vijayanand P, et al. Interleukin-4 production by follicular helper T cells requires the conserved Il4 enhancer hypersensitivity site V. Immunity. 2012;36:175–187. doi: 10.1016/j.immuni.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loots GG, et al. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 12.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koche RP, et al. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vijayanand P, et al. Chemokine receptor 4 plays a key role in T cell recruitment into the airways of asthmatic patients. J. Immunol. 2010;184:4568–4574. doi: 10.4049/jimmunol.0901342. [DOI] [PubMed] [Google Scholar]
- 17.Mikhak Z, Strassner JP, Luster AD. Lung dendritic cells imprint T cell lung homing and promote lung immunity through the chemokine receptor CCR4. J. Exp. Med. 2013;210:1855–1869. doi: 10.1084/jem.20130091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zielinski CE, et al. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature. 2012;484:514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
- 19.Chavez L, et al. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 2010;20:1441–1450. doi: 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L. MEDIPS: genome wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics. 2014;30:284–286. doi: 10.1093/bioinformatics/btt650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ansel KM, et al. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nat. Immunol. 2004;5:1251–1259. doi: 10.1038/ni1135. [DOI] [PubMed] [Google Scholar]
- 23.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 24.Seumois G, et al. An integrated nano-scale approach to profile miRNAs in limited clinical samples. Am. J. Clin. Exp. Immunol. 2012;1:70–89. [PMC free article] [PubMed] [Google Scholar]
- 25.Douglas NC, Jacobs H, Bothwell AL, Hayday AC. Defining the specific physiological requirements for c-Myc in T cell development. Nat. Immunol. 2001;2:307–315. doi: 10.1038/86308. [DOI] [PubMed] [Google Scholar]
- 26.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu JW, et al. E2F1 and E2F2 determine thresholds for antigen-induced T-cell proliferation and suppress tumorigenesis. Mol. Cell. Biol. 2001;21:8547–8564. doi: 10.1128/MCB.21.24.8547-8564.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandiyan P, et al. CD152 (CTLA-4) determines the unequal resistance of Th1 and Th2 cells against activation-induced cell death by a mechanism requiring PI3 kinase function. J. Exp. Med. 2004;199:831–842. doi: 10.1084/jem.20031058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hawkins RD, et al. Global chromatin state analysis reveals lineage-specific enhancers during the initiation of human T helper 1 and T helper 2 cell polarization. Immunity. 2013;38:1271–1284. doi: 10.1016/j.immuni.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rockwell CE, Zhang M, Fields PE, Klaassen CD. Th2 skewing by activation of Nrf2 in CD4(+) T cells. J. Immunol. 2012;188:1630–1637. doi: 10.4049/jimmunol.1101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerasimova A, et al. Predicting cell types and genetic variations contributing to disease by combining GWAS and epigenetic data. PLoS ONE. 2013;8:e54359. doi: 10.1371/journal.pone.0054359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moffatt MF, et al. A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolan MJ, et al. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat. Immunol. 2007;8:1324–1336. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- 37.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bousquet J. Global initiative for asthma (GINA) and its objectives. Clin. Exp. Allergy. 2000;30(suppl. 1):2–5. doi: 10.1046/j.1365-2222.2000.00088.x. [DOI] [PubMed] [Google Scholar]
- 41.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 47.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamburov A, Stelzl U, Lehrach H, Herwig R. The Consensus Path DB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–D800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamburov A, Wierling C, Lehrach H, Herwig R. Consensus Path DB–a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu W, et al. GWAS Integrator: a bioinformatics tool to explore human genetic associations reported in published genome-wide association studies. Eur. J. Hum. Genet. 2011;19:1095–1099. doi: 10.1038/ejhg.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.