Highlights
-
•
Ornithine cyclodeaminase homolog from an archeon was characterized biochemically.
-
•
This protein functions as a novel Δ1-pyrroline-2-carboxylate reductase.
-
•
This enzyme is probably involved in trans-3-hydroxy-l-proline metabolism as in bacteria and mammals.
Abbreviations: OCD, ornithine cyclodeaminase; CRYM, μ-crystallin; AlaDH, l-alanine dehydrogenase; Pyr2C, Δ1-pyrroline-2-carboxylate; T3LHyp, trans-3-hydroxy-l-proline; NMAlaDH, N-methyl-l-alanine dehydrogenase
Keywords: Ornithine cyclodeaminase, Δ1-pyrroline-2-carboxylate reductase, Molecular evolution, trans-3-Hydroxy-l-proline metabolism
Abstract
l-Ornithine cyclodeaminase (OCD) is involved in l-proline biosynthesis and catalyzes the unique deaminating cyclization of l-ornithine to l-proline via a Δ1-pyrroline-2-carboxyrate (Pyr2C) intermediate. Although this pathway functions in only a few bacteria, many archaea possess OCD-like genes (proteins), among which only AF1665 protein (gene) from Archaeoglobus fulgidus has been characterized as an NAD+-dependent l-alanine dehydrogenase (AfAlaDH). However, the physiological role of OCD-like proteins from archaea has been unclear. Recently, we revealed that Pyr2C reductase, involved in trans-3-hydroxy-l-proline (T3LHyp) metabolism of bacteria, belongs to the OCD protein superfamily and catalyzes only the reduction of Pyr2C to l-proline (no OCD activity) [FEBS Open Bio (2014) 4, 240–250]. In this study, based on bioinformatics analysis, we assumed that the OCD-like gene from Thermococcus litoralis DSM 5473 is related to T3LHyp and/or proline metabolism (TlLhpI). Interestingly, TlLhpI showed three different enzymatic activities: AlaDH; N-methyl-l-alanine dehydrogenase; Pyr2C reductase. Kinetic analysis suggested strongly that Pyr2C is the preferred substrate. In spite of their similar activity, TlLhpI had a poor phylogenetic relationship to the bacterial and mammalian reductases for Pyr2C and formed a close but distinct subfamily to AfAlaDH, indicating convergent evolution. Introduction of several specific amino acid residues for OCD and/or AfAlaDH by site-directed mutagenesis had marked effects on both AlaDH and Pyr2C reductase activities. The OCC_00387 gene, clustered with the TlLhpI gene on the genome, encoded T3LHyp dehydratase, homologous to the bacterial and mammalian enzymes. To our knowledge, this is the first report of T3LHyp metabolism from archaea.
1. Introduction
l-Proline plays many important roles in the protein structure and cell signaling and is synthesized within cells and organisms from an intermediate, glutamate γ-semialdehyde, commonly involved in biosynthesis pathways from l-glutamate and l-arginine: l-glutamate → γ-glutamyl phosphate → glutamate γ-semialdehyde (Route 1); l-arginine → l-ornithine → glutamate γ-semialdehyde (Route 2) (Fig. 1A). The glutamate γ-semialdehyde then undergoes “non-enzymatic cyclization” to give the imine, Δ1-pyrroline-5-carboxyrate (Pyr5C), which is subsequently converted to l-proline by NAD(P)H-dependent Pyr5C reductase (EC 1.5.1.2) [1]. Route 1 is the main mechanism of l-proline biosynthesis in bacteria, while eukaryotes use this route predominantly under stress and limited nitrogen conditions. Under high nitrogen input, Route 2 appears to be prominent and is mainly found in higher plants.
Fig. 1.
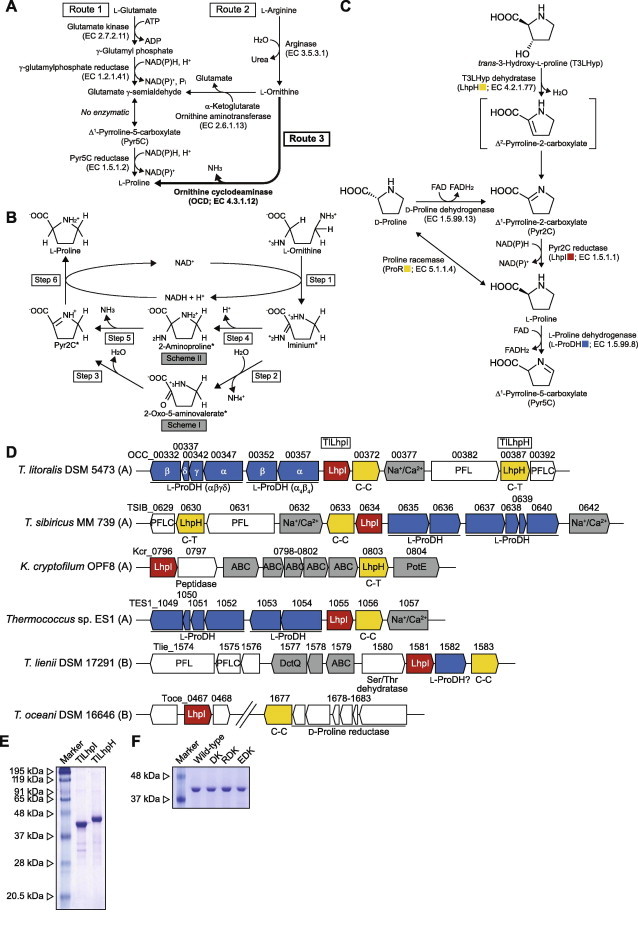
(A) Pathways of l-proline biosynthesis. (B) Proposed hydrolytic (Scheme I) and non-hydrolytic schemes (Scheme II) for the conversion of l-ornithine to l-proline by OCD [9]. Step 6 corresponds to the reaction by Pyr2C reductase. Asterisks indicate putative intermediates. (C) T3LHyp pathway and the metabolic network with d-proline. (D) Schematic gene clusters related to T3LHyp and/or proline metabolism of archaea (A) and bacteria (B). Putative genes in the box were purified and characterized in this study (see (E)). TlLhpI gene was fused with OCC_00362 and OCC_00367 genes (Supplementary Methods). C–C and C–T indicate a pair of catalytic amino acid residues of proline racemase superfamily enzymes (see Fig. S1B). Gray putative genes are sequentially similar to other (amino acid) transporters. (E) Purification of recombinant His6 tag proteins. Five micrograms each of purified protein were applied to 12% (w/v) gel (also (F)). (F) Purification of recombinant His6 tag TlLhpI mutants.
Alternatively, l-proline is also directly synthesized from the amino acid l-ornithine, itself an intermediate along the above Route 2, by l-ornithine cyclodeaminase (OCD; EC 4.3.1.12) (Route 3) [2]. This unique enzyme initially catalyzes NAD+-dependent oxidative deamination from the α-amino group of l-ornithine, and subsequently cyclization to form Δ1-pyrroline-2-carboxyrate (Pyr2C) (Fig. 1B). The actual cyclization step in this pathway occurs with the participation of enzymes, in contrast to glutamate γ-semialdehyde → Pyr5C in Routes 1 and 2, described above. The final step by OCD is NADH-dependent reduction of Pyr2C to l-proline. OCD belongs to the ornithine cyclodeaminase/μ-crystallin (OCD/CRYM) superfamily (see Fig. 4). It was recently reported that μ-crystallin (CRYM) from mammalians, known as an NADPH-dependent T3 thyroid hormone (triiodothyronin)-binding protein without enzymatic catalysis, functions as an NAD(P)H-dependent ketimine reductase (EC 1.5.1.25) [3]. Pyr2C is one of the substrates of this enzyme.
Fig. 4.
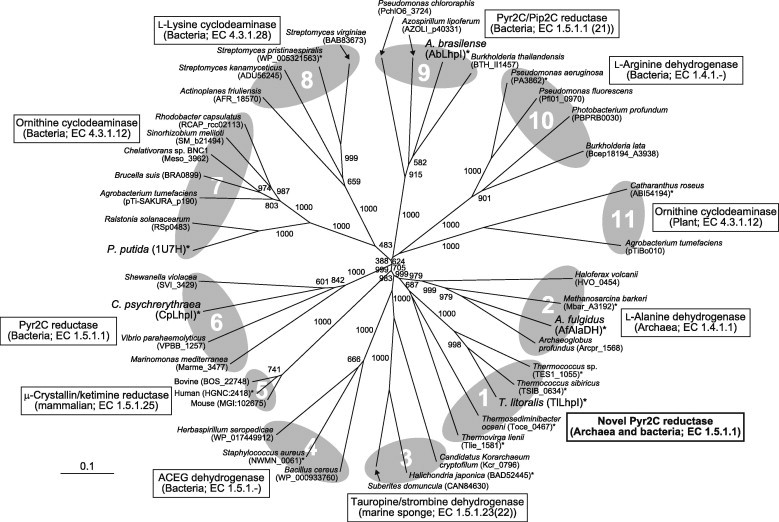
Phylogenetic tree of OCD/CRYM protein superfamily consisting of eleven subfamilies. The number on each branch indicates the bootstrap value. Proteins with asterisks were used for Fig. 5A.
Although Route 3 of l-proline biosynthesis is found in several bacteria, including Pseudomonas putida [2], Agrobacterium tumefaciens [4], Clostridium sporogenes [5], and Sinorhizobium meliloti [6], many archaea possess OCD/CRYM family genes (proteins) (20–40% sequence identity with bacterial OCD). Among them, only AF1665 protein (gene) from the hyperthermophilic archaeon Archaeoglobus fulgidus has been characterized at the molecular level, and the protein functions as an l-alanine dehydrogenase (AlaDH; EC 1.4.1.1) but not OCD: l-alanine + NAD+ + H2O ↔ pyruvate + NH3 + NADH + H+ [7,8]. It is proposed that the enzyme (AfAlaDH) is involved in l-alanine metabolism (degradation), whereas the catalytic efficiency value for pyruvate is much higher than that for l-alanine. Although OCD activity is detected in a cell-free extract of Methanococcus jannaschii [9], no OCD/CRYM family gene is found on the genome. Up to now, the physiological role of OCD-like protein including AfAlaDH has been unclear in archaea.
There are two different mechanisms for OCD (Fig. 1B), in which l-ornithine is initially oxidized to an iminium (Cα = NH2+ containing) intermediate (step 1 in Fig. 1B). In scheme I, the iminium undergoes hydrolysis to 2-oxo-5-aminovalerate by water before cyclization (step 2), and elimination of water produces Pyr2C (step 3). This scheme is analogous to the oxidative deamination of l-amino acid to 2-oxo acids by AlaDH [8], and has been preliminarily proposed based on biochemical experiments [10]. However, the crystal structure and molecular dynamic analysis of OCD [2,11] indicate an alternative mechanism (scheme II) that no attack on the iminium by water occurs: intramolecular cyclic addition of C-5 nitrogen of the iminium to the C-2 carbon to form 2-aminoproline (step 4), loss of ammonia, and reduction of the resulting imine, Pyr2C (step 5).
Recently, we characterized novel bacterial pathways of trans-4-hydroxy-l-proline (T4LHyp) and trans-3-hydroxy-l-proline (T3LHyp) metabolism [12,13]. In mammalian systems, T4LHyp and T3LHyp occur by post-translational hydroxylation of l-proline residue in certain proteins, in particular, collagen [14]. In contrast to T4LHyp, the metabolic pathways of T3LHyp from bacteria and mammalians may be similar [13,15] and consist of two enzymes, T3LHyp dehydratase (encoded by LhpH gene; EC 4.2.1.77) and NAD(P)H-dependent Pyr2C reductase (encoded by LhpI gene; EC 1.5.1.1), by which T3LHyp is converted to l-proline via Pyr2C (Fig. 1C). Interestingly, LhpI protein is a novel member of the OCD/CRYM superfamily, and catalyzes only the reduction of Pyr2C to l-proline (no OCD activity) [11]. Furthermore, there is no sequence similarity to DpkA protein as is known for the reductase of Pyr2C from bacteria [16], strongly suggesting their convergent evolution.
Metabolism of T3LHyp (and T4LHyp) by archaea has been unclear until now, and it is impossible to further assess the physiological functions of OCD-like proteins in archaea based on a simple homology search. In this study, we selected OCD-like protein (gene) from a hyperthermophilic archaeon, Thermococcus litoralis DSM 5473, as a target, and this protein functions as an NAD(P)H-dependent Pyr2C reductase rather than AlaDH. This gene was clustered with T3LHyp dehydratase and proline racemase genes involved in putative T3LHyp and/or proline metabolism. Molecular evolution of the OCD/CRYM family is also discussed.
2. Results
2.1. Candidates of target genes
Although it is unclear whether there are archaea capable of metabolizing T3LHyp as a sole carbon source, a homology search by the Protein-BLAST program was carried out against genome sequences of archaea using bacterial Pyr2C reductases (LhpI from Azospirillum brasilense (AbLhpI), GenBank: BAO21622; LhpI from Colwellia psychrerythraea 34H (CpLhpI), GenBank: YP_268197) as the probe protein sequences. As described below, bacterial Pyr2C reductases are classified into two different subfamilies, in which CpLhpI and AbLhpI are contained, respectively (see Fig. 4). However, over 900 homologous genes (proteins) with sequence identities of 20–40% were found (data not shown). Similar results was also obtained when other OCD/CRYM family proteins including AfAlaDH was used instead of LhpI protein as the probe.
On the other hand, when bacterial T3LHyp dehydratases (LhpH from A. brasilense (AbLhpH), GenBank: BAO21621; LhpH from C. psychrerythraea 34H (CpLhpH), GenBank: YP_268195) was used as the probe, 31 homologous genes were found. T3LHyp dehydratase belongs to the proline racemase superfamily, which also includes the archetypical proline racemase (EC 5.1.1.4) and T4LHyp epimerase (EC 5.1.1.8) (Fig. S1) [15,17]. In contrast to bacterial Pyr2C reductases, T3LHyp dehydratase form a single subclass in this superfamily. On the basis of two specific residues at the active sites, the members have been classified into three types: Cys-Cys type (proline racemase and T4LHyp epimerase); Cys-Thr type (T3LHyp dehydratase); Ser-Cys type (function unknown). Furthermore, two Cys-Cys type enzymes can be clearly distinguished in the phylogenetic tree. Based on this insight, we could select only three proline racemase-like genes as putative T3LHyp dehydratases, although it has been believed that the enzyme is found only in animals and fungi (and bacteria) [13,15]: T. litoralis DSM 5473 (OCC_00387, GenBank: YP_008428263; 45.6% identity with AbLhpH), Thermococcus sibiricus MM 739 (TSIB_0630, GenBank: YP_002994044; 47.1%), and Candidatus Korarchaeum cryptofilum OPF8 (OPF8 Kcr_0803, GenBank: YP_001737233; 44.1%) (Fig. 1). Interestingly, these genes were clustered with OCD-like genes: T. litoralis (OCC_00362→YP_008428257 OCC_00367, GenBank: YP_008428258); T. sibiricus (TSIB_0634, GenBank: YP_002994048); K. cryptofilum (Kcr_0796, GenBank: YP_001737226) (Fig. 1D). In the cases of T. litoralis and T. sibiricus, another proline racemase-like gene (Cys-Cys type) and genes encoding two different types of putative heterooligomeric l-proline dehydrogenase (l-ProDH) [18,19] were also contained in this gene cluster. This analysis strongly indicated that the gene cluster is related to T3LHyp and/or proline metabolism. Therefore, in this study, we selected GenBank: OCC_00362, GenBank: OCC_00367, and GenBank: OCC_00387 genes from T. litoralis as target genes (referred to as TlLhpI and TlLhpH genes, respectively).
2.2. Preparation of recombinant His6-tag proteins
TlLhpI gene was fused with OCC_00362 and OCC_00367 genes by PCR (Supplementary Methods). After cloning all target genes into the vector pETDuet-1, the recombinant enzymes with attached His6-tags at their N-termini were expressed in Escherichia coli and purified with an Ni2+-chelating affinity column (Fig. 1E). Apparent molecular masses of TlLhpI and TlLhpH, estimated by SDS–PAGE, were 40 (36.9) and 43 (38.1) kDa (values in parentheses indicate the calculated molecular mass of the enzyme with His6-tag), and those estimated by analytic gel filtration were 72 and 75 kDa, respectively (data not shown). Therefore, the two enzymes appear to be dimeric.
2.3. Characterization of TlLhpI as AlaDH
Although TlLhpI shows only 44.3% sequence identity with AfAlaDH, we could detect both activities of the NAD+-dependent oxidative deamination of l-alanine and NADH-dependent reductive amination of pyruvate (in presence of 700 mM NH4+): specific activities of 0.0265 and 4.43 (unit/mg protein), respectively. NADP+ was not used as a coenzyme. The optimum pH values of each reaction were 10–12 and 6–10.5, respectively (Fig. 2A). The kcat/Km value for pyruvate was ∼230-fold higher than that for l-alanine, caused by the ∼120-fold lower Km value: namely, the reaction equilibrium favors the direction toward reductive amination (Tables 1 and 2). Activity was also observed with several l-amino acids and 2-oxo acids with a hydrophobic side chain (see below for l-proline). Among them, l-2-aminovalerate and 2-oxovalerate with a C3 aliphatic side chain were the best substrates, and the kcat/Km values were 146- and 4.7-fold higher than those for l-alanine and pyruvate, respectively. When pyruvate and NADH were alternatively incubated together with methylamine (50 mM) instead of NH4+ as the nitrogen donor, reduction activity was detected. This result indicated that TlLhpI functions not only as AlaDH but also N-methyl-l-alanine dehydrogenase (NMAlaDH; EC 1.4.1.17). Activities of AlaDH and NMAlaDH were also detected in HPLC analysis using the reaction products and zymogram staining analysis (Figs. 2B and 3A, B).kcat/Km values for l-alanine and pyruvate of TlLhpI were over 1000-fold lower than those of AfAlaDH [7]. Furthermore, the Km value for NH4+ of TlLhpI was clearly beyond the physiological level (Table 2). AfAlaDH shows similar optimum pH of the oxidative and reductive reactions (∼7.0), and no significant activity for hydrophobic amino acids including l-2-aminovalerate (2-oxovalerate). This indicated strongly that TlLhpI possesses largely different properties from AfAlaDH, and that the physiological function is not the metabolism of l-alanine and pyruvate.
Fig. 2.
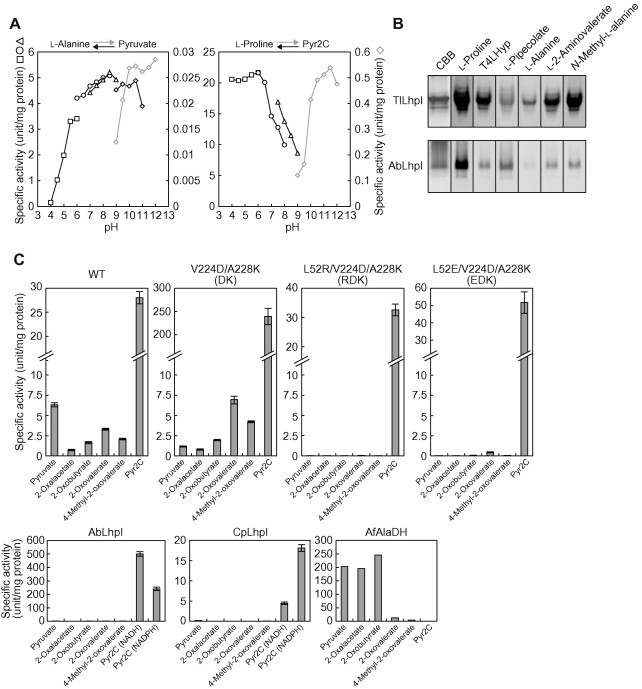
Enzymatic properties of TlLhpI Pyr2C. (A) Effect of pH on the activity as AlaDH left panel) and Pyr2C reductase (right panel). 50 mM acetate-NaOH (pH 4.0–6.0) (square), 50 mM potassium phosphate (pH 6.0–8.5) (circle), 50 mM Tris–HCl (pH 7.0–9.0) (triangle) for the reduction of pyruvate or Pyr2C (left axis and black symbol), and 50 mM Bis–tris propane (diamond) for the oxidization of l-alanine or l-proline (right axis and gray symbol), instead of 50 mM buffer under each standard assay condition. (B) Zymogram staining. Five micrograms each of purified protein was applied on 10% (w/v) gel. After electrophoresis, the gel was soaked at 50 °C (for TlLhpI) or 30 °C (for AbLhpI) in staining solution in the presence of the indicated substrate and NAD+ (each of 10 mM). Protein stained with Coomassie Brilliant Blue (CBB) was used as a loading control. (C) Comparison of NADH-dependent AlaDH and Pyr2C reductase activities of wild-type and mutated TlLhpI proteins. The assay was performed with standard assay solution containing the indicated substrate (10 mM). AbLhpI and CpLhpI were assayed at 30 °C under the standard assay conditions for TlLhpI. Data for AfAlaDH are from Ref. [7].
Table 1.
Kinetic parameters of NAD+-dependent oxidation reaction.
| Enzymes | Substrates | Specific activitya (units/mg protein) | Km (mM) | kcat (min−1) | kcat/Km (min−1 mM−1) |
|---|---|---|---|---|---|
| Wild type | l-Alanine | 0.0265 ± 0.0015 | 18.2 ± 5.0 | 2.64 ± 0.55 | 0.147 ± 0.012 |
| l-2-Aminobutyrate | 0.136 ± 0.010 | 6.00 ± 0.29 | 6.71 ± 0.16 | 1.12 ± 0.03 | |
| l-2-Aminovalerate | 0.396 ± 0.030 | 0.306 ± 0.009 | 6.56 ± 0.08 | 21.5 ± 0.4 | |
| l-Leucine | 0.248 ± 0.010 | 1.32 ± 0.10 | 10.5 ± 0.4 | 8.00 ± 0.28 | |
| N-Methyl-l-alanine | 0.526 ± 0.025 | 0.406 ± 0.044 | 11.6 ± 0.7 | 28.6 ± 2.1 | |
| l-Proline | 0.384 ± 0.015 (0.0376 ± 0.0005)b | 1.12 ± 0.12 (13.1 ± 1.2)b | 13.8 ± 1.0 (3.27 ± 0.24)b | 12.3 ± 0.4 (0.250 ± 0.006)b | |
| l-Pipecolate | 0.219 ± 0.020 | 0.608 ± 0.115 | 12.2 ± 1.3 | 20.3 ± 1.8 | |
| V224D/A228K | l-Alanine | 0.0133 ± 0.0010 | 36.9 ± 0.9 | 2.12 ± 0.02 | 0.0575 ± 0.0008 |
| l-2-Aminovalerate | 0.666 ± 0.050 | 2.13 ± 0.30 | 7.72 ± 0.82 | 3.64 ± 0.12 | |
| l-Proline | 1.51 ± 0.10 | 0.444 ± 0.008 | 36.5 ± 0.9 | 82.2 ± 1.1 | |
Under standard assay conditions in “Experimental procedures”. All substrate concentrations were 10 mM.
NADP+-dependent activity.
Table 2.
Kinetic parameters of NADH-dependent reductive reaction.
| Enzymes | Substrates | Specific activitya (units/mg protein) | Km (mM) | kcat (min−1) | kcat/Km (min−1 mM−1) |
|---|---|---|---|---|---|
| Wild type | Pyruvate | 4.43 ± 0.50 | 9.37 ± 0.23 | 315 ± 4 | 33.6 ± 0.4 |
| Oxalacetate | 1.78 ± 0.18 | 8.86 ± 0.91 | 192 ± 15 | 21.7 ± 0.5 | |
| 2-Oxobutyrate | 2.46 ± 0.25 | 0.701 ± 0.020 | 92.0 ± 1.8 | 131 ± 1 | |
| 2-Oxovalerate | 2.59 ± 0.21 | 0.969 ± 0.093 | 152 ± 11 | 157 ± 3 | |
| 4-Methyl-2-oxovalerate | 1.02 ± 0.19 | 2.52 ± 0.45 | 129 ± 18 | 51.4 ± 2.0 | |
| NH4+ (+2-oxovalerate)b | 2.45 ± 0.22 | 1450 ± 240 | 267 ± 33 | 0.183 ± 0.007 | |
| Pyr2C | 23.9 ± 2.5 | 0.944 ± 0.090 | 995 ± 33 | 1058 ± 66 | |
| V224D/A228K | Pyruvate | 1.60 ± 0.15 | 62.0 ± 12.2 | 413 ± 75 | 6.68 ± 0.11 |
| 2-Oxovalerate | 7.00 ± 0.95 | 1.82 ± 0.14 | 212 ± 16 | 116 ± 0 | |
| Pyr2C | 239 ± 35 | 2.77 ± 0.44 | 33500 ± 4900 | 12900 ± 300 | |
Under standard assay conditions in “Experimental procedures”. All substrate concentrations were 10 mM.
The NH4+ concentrations were changed with a fixed concentration of 2-oxovalerate (10 mM).
Fig. 3.
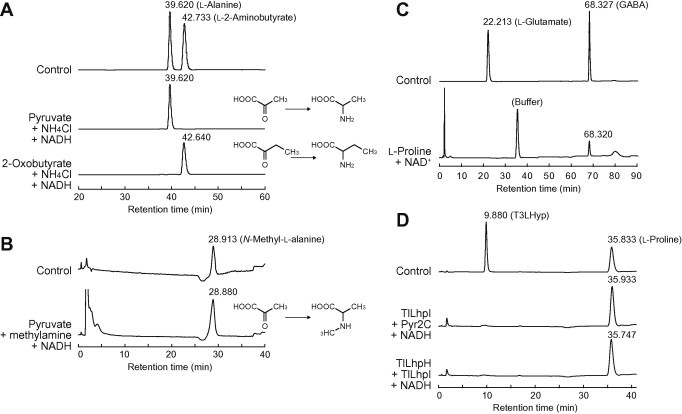
HPLC analysis of the reaction products. (A) Reaction product from pyruvate or 2-oxobutyrate by TlLhpI (as AlaDH) in presence of NADH and NH4Cl. Control indicates authentic l-alanine and l-2-aminobutyrate. (B) Reaction product from pyruvate by TlLhpI (as NMAlaDH) in the presence of NADH and methylamine. Control indicates authentic N-methyl-l-alanine. (C) Reaction product from l-proline by TlLhpI (as l-ProDH) in the presence of NAD+. After the enzymatic reaction, the product was treated by H2O2, and then analyzed. Control indicates authentic l-glutamate and γ-aminobutyrate (GABA), which are potentially yielded from Pyr5C and Pyr2C, respectively. (D) Reaction products from Pyr2C by TlLhpI and from T3LHyp by TlLhpH and TlLhpI, in the presence of NADH. Control indicates authentic T3LHyp and l-proline. No peak corresponding to Pyr2C was observed, probably caused by no reaction with labeling reagent for amino acid analysis (data not shown).
2.4. Characterization of TlLhpI as novel Pyr2C reductase in OCD/CRYM superfamily
In contrast to AfAlaDH [7], TlLhpI showed NAD+-dependent oxidization activity for l-proline. The kcat/Km value in the presence of NADP+ was ∼50-fold lower than that in the presence of NAD+ (Table 1): the specificity for NAD+ is described above. In addition to l-proline, several l-proline analogs, including cis-4-hydroxy-l-proline (85%), trans-4-hydroxy-l-proline (70%), and l-pipecolate (49%), were active substrates (relative activity to l-proline in parentheses), whereas it is known that bacterial LhpI proteins are strictly specific to l-proline [13]. These activities were also detected in zymogram staining analysis (Fig. 2B). When the reaction product of l-proline was further oxidized by H2O2, 4-aminobutyrate (GABA) was identified in HPLC analysis (Fig. 3C), indicating that TlLhpI catalyzes the conversion of l-proline to Pyr2C. Generally, l-ProDH (EC 1.5.99.8) including archaeal enzymes [18,19] is a flavoprotein, and the reaction product is Pyr5C (if so, l-glutamate is yielded by the above H2O2 treatment).
Therefore, we compared the dehydrogenation activity toward l-proline and other l-amino acids (Tables 1 and 2). The kcat/Km value for Pyr2C (1058 min−1 mM−1) was 6.7-fold higher than that for 2-oxovalerate, caused by the high kcat value. Furthermore, the ratio of Pyr2C to l-proline in kcat/Km was 86, suggesting the preference of the reaction equilibrium in the direction toward NADH-dependent reduction. Interestingly, the optimum pH of Pyr2C reductase was 4.0–6.0, clearly different from that of the reductive deamination of AlaDH (pH 6–10.5) (Fig. 2A). These results strongly indicated that Pyr2C is the preferred substrate for TlLhpI. Although AbLhpI and CpLhpI showed only 25.3% and 31.6% sequence identity to TlLhpI, respectively, we unexpectedly detected AlaDH activity in both enzymes (Fig. 3C). Commonly, these bacterial enzymes showed significantly high ratios of Pyr2C to pyruvate in specific activity: 176 and 28, respectively (∼5 for TlLhpI). Namely, TlLhpI possessed “vestigial” properties to AlaDH, probably due to the close phylogenetic relationship between them, as described in the main section.
Using the reductase activity toward Pyr2C, several properties of TlLhpI as an enzyme from “hyperthermophiles” were identified: T. litoralis grows between 55 and 98 °C with an optimal growth temperature of 88 °C [20]. Although the optimum temperature was above 75 °C, and clearly higher than that of (mesophilic) AbLhpI, the assay could not be performed above 75 °C due to the thermolability of NADH (Fig. S2A). Thus, TlLhpI appears to be a typical (hyper)thermophilic enzyme.
2.5. Phylogenetic analysis of TlLhpI
As expected from the preliminary annotation, TlLhpI belongs to the OCD/CRYM superfamily, including the archetype OCD (subfamilies 7 and 11) [2,21], CRYM/ketimine reductase (subfamily 5) [3], AlaDH (subfamily 2) [7,8], l-arginine dehydrogenase (subfamily 10) [22], l-lysine cyclodeaminase (EC 4.3.1.28; subfamily 8) [23], tauropine and strombine dehydrogenases (EC 1.5.1.23(22); subfamily 3) [24,25], l-2,3-diaminopropionate synthase (subfamily 4) [26], and Pyr2C reductase (from bacteria) (subfamilies 6 and 9) [13] (Fig. 4). TlLhpI had a poor relationship to the bacterial LhpI and mammalian CRYM proteins, in spite of their similar activity, and formed a close but distinct subfamily to many archaeal OCD-like proteins, including AfAlaDH (novel subfamily 1). Surprisingly, two OCD-like proteins from “thermophilic bacteria”, Thermosediminibacter oceani (Toce_0467) and Thermovirga lienii (Tlie_1581), were closely related to TlLhpI rather than (bacterial) AbLhpI and CpLhpI, suggesting the possibility of horizontal gene transfer between bacteria and archaea.
Putative amino acid sequences of TlLhpI contained essential amino acid residues for coenzyme binding (Rossmann-fold motif consisting of Gly-X-Gly-X2-[Ala/Ser], where X indicates any amino acid) and the catalytic tetrad for interacting (and/or binding) with carboxyl and amino groups of substrate ([Arg/Lys]-Lys-Arg-Asp): Gly134-Ala-Gly-Val-Gln-Ala139 and Arg39-Lys67-Arg110-Asp303, respectively [2,8] (Fig. 5). Furthermore, TlLhpI possessed a specific aspartate residue (Asp159) for NAD+(H)-dependent enzymes of the OCD/CRYM superfamily [2,7,23–26], conforming to the coenzyme specificity, described above.
Fig. 5.
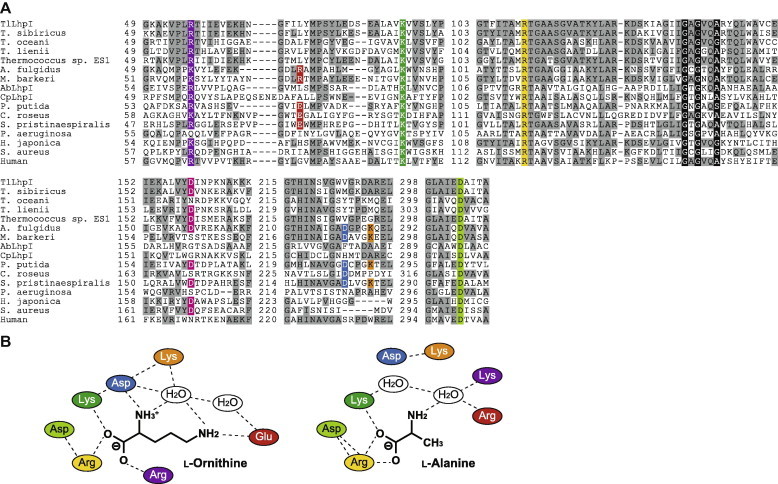
(A) Partial multiple sequence alignment of deduced amino acid sequences of TlLhpI. Binding sites for a carboxyl group of substrate are shaded in violet, green, yellow and light-green. Aspartate, lysine and glutamate residues, related to the cyclization by OCD, are shaded in blue, orange, and red, respectively. A specific aspartate residue for NAD+(H)-dependent enzymes, potentially interacting with 2′- and 3′-hydroxyl groups in the ribosyl moiety of NAD+(H), are shaded in pink. Amino acid residues Coenzyme-binding motif of Rossmann-fold are shown as white letters in black boxes. Gray-shaded letters indicate highly conserved amino acid residues. (B) Schematic diagram showing the interactions of l-ornithine (left panel) and l-alanine (right panel), and nearby residues in OCD and AlaDH, respectively. Color of residues correspond to Fig. 5A. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.6. Identification of catalytic amino acid residues
It is proposed that three amino acid residues of OCD, E56-D228-K232 (numbering from P. putida), are related to the unique cyclization of l-ornithine [2] (Fig. 5). In TlLhpI and AfAlaDH, these amino acid residues correspond to L52-V224-A228 and R52-D219-K223, respectively (conserved amino acid residues are underlined). To obtain insight into the catalytic mechanism of OCD/CRYM superfamily enzymes, we constructed three mutants of TlLhpI, V224D/A228K (DK mutant), L52E/V224D/A228K (EDK mutant), and L52R/V224D/A228K (RDK mutant). They were overexpressed in E. coli cells as a His6-tagged enzyme and purified with the same procedures as for the wild-type enzyme (Fig. 1F). Approximately 12-fold enhancement of Pyr2C reductase activity (6.7-fold for the oxidization activity for l-proline), but not AlaDH activity, was found in the DK mutant, caused by the increasing kcat value (Tables 1 and 2). On the other hand, when the mutations were combined with L52E and L52R, AlaDH activities for the 2-oxo acids were completely eliminated, whereas Pyr2C reductase activity was the same as in the wild type (Fig. 2C).
2.7. Characterization of TlLhpH
Potential T3LHyp dehydratase activity in TlLhpH was initially assayed spectrophotometrically in the coupling system with TlLhpI (in the presence of NADH). A significant decrease in absorbance at 340 nm was observed as a result of cooperative reactions by TlLhpH (T3LHyp → Pyr2C) and TlLhpI (Pyr2C → l-proline): 44.6 unit/mg protein of specific activity with T3LHyp. Km and kcat values for T3LHyp were 0.288 mM and 1710 min−1, respectively, comparable with those of AbLhpH (data not shown). On the other hand, optimum temperature for the activity was 80–100 °C, which was significantly higher than that of the (mesophilic) AbLhpH (40–45 °C) (Fig. S2B). These results indicated that the TlLhpH gene encodes T3LHyp dehydratase with enzymatic properties as a typical (hyper)thermophilic enzyme: this is the first example for archaea.
2.8. Physiological meaning of TlLhpI and TlLhpH
Although a minimal medium for the cultivation of T. litoralis is not avairable, we found that T. litoralis can grow in the complex medium [27] supplemented with l-proline, d-proline or T3LHyp as a sole carbon source, instead of maltose and peptone (data not shown). On the other hand, Pyr2C reductase and T3LHyp dehydratase activities in cell-free extract prepared from T. litoralis cells grown on such as carbon source were similar to those on maltose and peptone. These suggested one possibility that similar metabolic pathway of T3LHyp to bacteria and mammalian is operative in T. litoralis.
3. Discussion
The discovery of archaeal reductase for Pyr2C revealed that OCD/CRYM superfamily enzymes with this activity are large: subfamilies of 1, 5, 6, 7, 9, and 11 at least (Fig. 4). Furthermore, TlLhpI (and AbLhpI and CpLhpI) possesses not only Pyr2C reductase activity but also (slight but significantly detectable) AlaDH and NMAlaDH activities. Among OCD/CRYM members, enzymatic reactions by subfamilies 2 and 10 are homologous to AlaDH, and enzymes of subfamilies 3 and 4 catalyze similar reactions to NMAlaDH, in which taurine, glycine, and l-2,3-diaminopropionate are used as the nitrogen donor instead of NH4+, respectively [24–26]. Furthermore, cyclodeaminases for l-ornithine and l-lysine possess the same initial step of the catalytic reaction as AlaDH and NMAlaDH (step 1 in Fig. 1B). This suggests that the origin of l-ProDH (Pyr2C reductase) activity (step 6) is the same as step 1. A common ancestor possessed NAD(P)+-dependent dehydrogenation activity toward broad primary and secondary amines, and (strict) substrate specificity convergently evolved in bacteria, mammalians, and archaea. If this hypothesis is true, TlLhpI acquired catalytic specificity as Pyr2C reductase later than AbLhpI and CpLhpI, because the relative high activity of AlaDH and broad substrate specificity of l-ProDH remain (Fig. 2B and C).
Although it is unclear whether OCD possesses (remaining) oxidative deamination activity for l-amino acid(s) except l-ornithine, the “potential” mechanism to produce Pyr2C (from l-ornithine) by TlLhpI corresponds to scheme I, because of the AlaDH activity. Therefore, to obtain evolutional insight into Pyr2C reductase and OCD, we introduced several specific amino acid residues for OCD in TlLhpI by site-directed mutagenesis. Although Asp228 of OCD (numbering from P. putida) plays an important role in all steps of catalysis [2,11], Asp219 conserved in AfAlaDH is not involved in catalysis (steps 1 and 2) [8]. Therefore, it may be reasonable that the V244D (and A228K) mutant of TlLhpI has no significant effect on AlaDH activity (Tables 1 and 2, and Fig. 3C), because of the similar mechanism to AfAlaDH, as described above. On the other hand, Glu56 of OCD favorably interacts with the δ-amino group of l-ornithine. In CRYM, it is also proposed that Gly60 at the equivalent position (numbering from mouse) is important for the binding of a large ligand, T3, because of the small side chain [28]. These insights explain why the substitution of Leu52 in hydrophilic glutamate or arginine in TlLhpI leads to a complete loss of reductive amination activity for hydrophobic 2-oxo acid substrates (Fig. 3C).
As in our previous proposal [13], dehydratase activity with T3LHyp in the proline racemase superfamily was acquired once at an early evolutional stage, because TlLhpH also forms a single subfamily together with enzymes from bacteria and mammalians (Fig. S1). Several (thermophilic) archaea and bacteria possess another proline racemase-like gene instead of the LhpH gene within the gene cluster containing the LhpI gene (Fig. 1D). The enzymatic function may be interconversion of l- and d-isomer of proline (and/or hydroxyproline), but not dehydration of T3LHyp: Pyr2C is potentially produced from d-proline by FAD-dependent d-ProDH (Fig. 1C). TlLhpI (this study; Fig. 2B) and l-ProDH (α4β4) [29] show activities not only for Pyr2C and l-proline but also Δ1-piperidine-2-carboxylate and l-pipecolate, respectively, which are involved in the so-called “l-pipecolate pathway” of d-lysine metabolism [16]. Putative l-ProDH genes are also found within the gene cluster. Furthermore, (putative) pyruvate-formate lyase (PFL) and serine/threonine dehydratase genes, producing pyruvate and/or 2-oxobutyrate, are often located within the flanking region of the LhpI gene (Fig. 1D). These indicate the possibility that the TlLhpI gene (and the gene cluster) is also involved not only in T3LHyp but also in d-proline, l-proline and d-lysine metabolism, and several 2-oxo acids. Although T. litoralis can grow on T3LHyp as a sole carbon source, Pyr2C reductase (and T3LHyp dehydratase) activity is not significantly induced in cell-free extract, in contrast of bacteria [13], conforming to the putative functions in multiple metabolism described above. Development of gene disruption of T. litoralis would be useful for understanding more detailed physiological role(s) of TlLhpI gene.
4. Experimental procedures
4.1. General procedures
Basic recombinant DNA techniques were performed as described by Sambrook et al. [30]. PCR was carried out using a GeneAmp PCR System 2700 (Applied Biosystems) for 30 cycles in a 50 μl reaction mixture containing 1 U of KOD FX DNA polymerase (TOYOBO), appropriate primers (15 pmol) and template DNA under the following conditions: denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s and extension at 68 °C for the time periods calculated at an extension rate of 1 kbp min−1. DNA sequencing was carried out using the BigDye Cycle Sequencing Kit ver.3.1 (Applied Biosystems) and appropriate primers with the Genetic Analyzer 3130 (Applied Biosystems). Protein concentrations were determined by the method of Lowry et al. [31] with bovine serum albumin as the standard. SDS–PAGE was performed as described by Laemmli [32].
4.2. Strain and growth conditions
T. litoralis DSM5473 were grown under anaerobic conditions at 80 °C in a complex medium, as described previously [27], in the absence of sulfur. If necessary, maltose, peptone and yeast extract were omitted, and l-proline, d-proline or T3LHyp (30 mM) was added alternatively.
4.3. Plasmid construction for expression of recombinant proteins
Primer sequences used in this study are shown in Table S1. In this report, the prefixes Tl (T. litoralis), Af (A. fulgidus), Ab (A. brasilense) and Cp (C. psychrerythraea) have been added to gene symbols or protein designations when required for clarity. TlLhpI (fusion of OCC_00362 and OCC_00367; see Supplementary Methods) and TlLhpH genes (OCC_00387) were amplified by PCR using primers containing appropriate restriction enzyme sites at the 5′ and 3′ ends and genome DNA of T. litoralis as a template. Each amplified DNA fragment was introduced into BamHI-PstI sites in pETDuet-1 (Novagen), a plasmid vector for conferring N-terminal His6-tag on expressed proteins, to obtain pET/TlLhpIWT and pET/TlLhpH.
4.4. Expression and purification of His6-tagged recombinant proteins
E. coli BL21(DE3)-RIL (Novagen) harboring the expression plasmid for His6-tagged proteins was grown at 37 °C to a turbidity of 0.6 at 600 nm in Super broth medium (pH 7.0, 12 g tryptone, 24 g yeast extract, 5 ml glycerol, 3.81 g KH2PO4, and 12.5 g K2HPO4 per liter) containing 50 mg/liter ampicillin. After the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), the culture was further grown for 6 h at 37 °C to induce the expression of His6-tagged protein. Cells were harvested and resuspended in Buffer A (50 mM sodium phosphate buffer (pH 8.0) containing 300 mM NaCl and 10 mM imidazole). The cells were then disrupted by sonication for 20 min at appropriate intervals on ice using the Ultra Sonic Disruptor Model UR-200P (TOMY SEIKO Co., Ltd., Tokyo, Japan), and the solution was centrifuged at 108,000×g for 20 min at 4 °C. The supernatant was loaded onto a Ni–NTA Superflow column (Qiagen) equilibrated with Buffer A linked to the BioAssist eZ system (TOSOH). The column was washed with Buffer B (50 mM sodium phosphate buffer (pH 8.0) containing 300 mM NaCl, 10% (v/v) glycerol, and 50 mM imidazole). The enzymes were then eluted with Buffer C (pH 8.0, Buffer B containing 250 mM imidazole instead of 50 mM imidazole), concentrated by ultrafiltration with Centriplus YM-30 (Millipore), dialyzed against 50 mM Tris–HCl buffer (pH 8.0) containing 50% (v/v) glycerol, and stored at −35 °C until use. If necessary, AbLhpI and CpLhpI proteins were prepared, as described previously [13].
The native molecular mass of recombinant proteins was estimated by gel filtration, which was carried out using HPLC with a Multi-Station LC-8020 model II system (TOSOH) at a flow rate of 1 mL min−1. The purified enzyme (∼10 mg mL−1) was loaded onto a TSKgel G3000SWXL column (TOSOH) equilibrated with 50 mM Tris–HCl buffer (pH 8.0). A high molecular weight gel filtration calibration kit (GE Healthcare) was used as a molecular marker.
4.5. Enzyme assay
All enzyme assays were performed at 50 °C unless otherwise indicated.
Pyr2C reductase was assayed routinely in the direction of Pyr2C reduction by measuring the oxidization of NADH at 340 nm, using a Shimadzu UV-1800 spectrophotometer (Shimadzu GLC Ltd., Tokyo, Japan). The standard assay mixture contained 10 mM Pyr2C in 50 mM potassium phosphate (pH 6.0) buffer. The reactions were started by the addition of 100 μl of a 1.5 mM NADH solution to a final volume of 1 m1. To assay the reverse reaction, the reaction mixture consisted of 50 mM Bis–tris propane (pH 10.5) and 10 mM l-proline. The reaction was started by the addition of 15 mM NAD+ (100 μl) with a final reaction volume of 1 ml. One unit of enzyme activity refers to 1 μmol NADH produced/min. Pyr2C was enzymatically synthesized from T3LHyp, described previously [13].
AlaDH activity was measured as oxidative deamination of l-alanine and as reductive amination of pyruvate. For the oxidative deamination assay, the standard assay mixture contained 50 mM Bis–tris propane (pH 10.5), 10 mM l-alanine, and 1.5 mM NAD+. The reductive amination assay mixture consisted of 50 mM Tris–HCl (pH 8.0), 700 mM NH4Cl, 10 mM pyruvate, and 0.15 mM NADH. For the assay of NMAlaDH activity, 50 mM methylamine was used instead of 700 mM NH4Cl.
T3LHyp dehydratase was assayed spectrophotometrically in the coupling system with TlLhpI. The reaction mixture consisted of 50 mM Tris–HCl (pH 8.0), 0.15 mM NADH, and 10 μg purified TlLhpI. The reaction was started by the addition of 100 mM T3LHyp (100 μl) with a final reaction volume of 1 ml. The enzyme was alternatively assayed by the colorimetric method based on the reaction of 2-aminobenzaldehyde with Pyr2C, which yields a yellow reaction product [15]. This method was used to determine the optimum temperature for activity.
4.6. Reaction product analysis
Purified TlLhpI (10 μg) was added to the following four mixtures (1 ml): for AlaDH activity, 50 mM Tris–HCl (pH 8.0) buffer containing 10 mM pyruvate (or 2-oxobutyrate), 500 mM NH4Cl, and 10 mM NADH; for NMAlaDH activity, 50 mM Tris–HCl (pH 8.0) buffer containing 10 mM pyruvate, 100 mM methylamine, and 10 mM NADH; for l-ProDH activity; 50 mM Bis–tris propane (pH 10.5) buffer containing 10 mM l-proline and 10 mM NAD+; for T3LHyp dehydratase activity of TlLhpH; 50 mM Tris–HCl (pH 8.0) buffer containing 10 mM T3LHyp, 10 mM NADH, and TlLhpH (10 μg). After incubation at 50 °C overnight, each enzyme product was then analyzed by a Hitachi L-8900 amino acid analyzer (Tokyo, Japan), using ion exchange chromatography followed by post-column derivatization with ninhydrin. In the case of l-ProDH activity, the product was further oxidized by the addition of 30% H2O2 (20 μl) to the mixture before HPLC analysis [33].
4.7. Zymogram staining analysis
Purified TlLhpI was separated at 4 °C on non-denaturing PAGE with 10% (w/v) gel, which was performed by omitting SDS and 2-mercaptoethanol from the solution used in SDS–PAGE. The gels were then soaked in 10 ml staining solution consisting of 50 mM Bis–tris propane (pH 10), 0.25 mM nitroblue tetrazolium (NBT), 0.06 mM phenazine methosulfate (PMS), 10 mM substrate, and 10 mM NAD+ at 50 °C for 5 min in the dark. Dehydrogenase activity appeared as a dark blue band.
4.8. Site-directed mutagenesis
The mutation in TlLhpI was introduced by sequential steps of PCR [34] using sense and antisense primers (Table S1) and pET/TlLhpIWT (for V224D/A228K (DK) mutant) or pET/TlLhpIDK (for L52E/V224D/A228K (EDK) and L52R/V224D/A228K (RDK) mutants) as a template. Briefly, the codons used for single mutants were as follows: L52E, CTG → AGG; L52R, CTG → GAG; V224D, GTG → GAC; A228K, GCC → AAG. The coding region of the mutated genes was confirmed by subsequent sequencing in both directions. Mutant proteins were expressed and purified by the same procedures as for the wild-type enzyme.
4.9. Amino acid sequence alignment and phylogenetic analysis
Protein sequences were analyzed using the Protein-BLAST and Clustal W program distributed by DDBJ (DNA Data Bank of Japan) (www.ddbj.nig.ac.jp). Multiple sequence alignment was performed with the following default parameters: Protein Weight Matrix, Gonnet; GAP OPEN, 10; GAP EXTENSION, 0.20; GAP DISTANCES, 5; NO END GAPS, no; ITERATION, none; NUMITER, 1 and CLUSTERING, NJ. The phylogenetic tree was constructed by the neighbor-joining method [35]. Bootstrap resampling was performed 1000 times.
Acknowledgments
This work was partially supported by KAKENHI (25440049) (to S.W.) and HOKTO Research Foundation (to S.W.). We thank Prof. Haruhiko Sakuraba (Faculty of Agriculture, Kagawa University) and Dr. Ryushi Kawakami (Institute of Socio-Arts and Sciences, University of Tokushima) for the gift of T. litoralis DSM 5473. Our thanks are extended especially to Dr. Miyuki Kawano-Kawada (Integrated Center for Sciences (INCS), Ehime University) for amino acid analysis.
Appendix A. Supplementary data
Primers used in this study.
Phylogenetic analysis of TlLhpH.
Temperature dependence of Pyr2C reductase and T3LHyp dehydratase activities.
References
- 1.Nocek B., Chang C., Li H., Lezondra L., Holzle D., Collart F., Joachimiak A. Crystal structures of Δ1-pyrroline-5-carboxylate reductase from human pathogens Neisseria meningitides and Streptococcus pyogenes. J. Mol. Biol. 2005;354:91–106. doi: 10.1016/j.jmb.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodman J.L., Wang S., Alam S., Ruzicka F.J., Frey P.A., Wedekind J.E. Ornithine cyclodeaminase: structure, mechanism of action, and implications for the mu-crystallin family. Biochemistry. 2004;43:13883–13891. doi: 10.1021/bi048207i. [DOI] [PubMed] [Google Scholar]
- 3.Hallen A., Cooper A.J., Jamie J.F., Haynes P.A., Willows R.D. Mammalian forebrain ketimine reductase identified as μ-crystallin; potential regulation by thyroid hormones. J. Neurochem. 2011;118:379–387. doi: 10.1111/j.1471-4159.2011.07220.x. [DOI] [PubMed] [Google Scholar]
- 4.Cho K., Fuqua C., Martin B.S., Winans S.C. Identification of Agrobacterium tumefaciens genes that direct the complete catabolism of octopine. J. Bacteriol. 1996;178:1872–1880. doi: 10.1128/jb.178.7.1872-1880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costilow R.N., Laycock L. Ornithine cyclase (deaminating): purification of a protein that converts ornithine to proline and definition of the optimal assay conditions. J. Biol. Chem. 1971;246:6655–6660. [PubMed] [Google Scholar]
- 6.Soto M.J., van Dillewijn P., Olivares J., Toro N. Ornithine cyclodeaminase activity in Rhizobium meliloti. FEMS Microbiol. Lett. 1994;11:209–214. [Google Scholar]
- 7.Schröder I., Vadas A., Johnson E., Lim S., Monbouquette H.G. A novel archaeal alanine dehydrogenase homologous to ornithine cyclodeaminase and mu-crystallin. J. Bacteriol. 2004;186:7680–7689. doi: 10.1128/JB.186.22.7680-7689.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallagher D.T., Monbouquette H.G., Schröder I., Robinson H., Holden M.J., Smith N.N. Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin. J. Mol. Biol. 2004;342:119–130. doi: 10.1016/j.jmb.2004.06.090. [DOI] [PubMed] [Google Scholar]
- 9.Graupner M., White R.H. Methanococcus jannaschii generates l-proline by cyclization of l-ornithine. J. Bacteriol. 2001;183:5203–5205. doi: 10.1128/JB.183.17.5203-5205.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muth W.L., Costilow R.N. Ornithine cyclase (deaminating). III. Mechanism of the conversion of ornithine to proline. J. Biol. Chem. 1974;249:7463–7467. [PubMed] [Google Scholar]
- 11.Ion B.F., Bushnell E.A., Luna P.D., Gauld J.W. A molecular dynamics (MD) and quantum mechanics/molecular mechanics (QM/MM) study on ornithine cyclodeaminase (OCD): a tale of two iminiums. Int. J. Mol. Sci. 2012;13:12994–13011. doi: 10.3390/ijms131012994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe S., Morimoto D., Fukumori F., Shinomiya H., Nishiwaki H., Kawano-Kawada M., Sasai Y., Tozawa Y., Watanabe Y. Identification and characterization of d-hydroxyproline dehydrogenase and Δ1-pyrroline-4-hydroxy-2-carboxylate deaminase involved in novel l-hydroxyproline metabolism of bacteria: metabolic convergent evolution. J. Biol. Chem. 2012;287:32674–32688. doi: 10.1074/jbc.M112.374272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe S., Tanimoto Y., Yamauchi S., Tozawa Y., Sawayama S., Watanabe Y. Identification and characterization of trans-3-hydroxy-l-proline dehydratase and Δ1-pyrroline-2-carboxylate reductase involved in trans-3-hydroxy-l-proline metabolism of bacteria. FEBS Open Bio. 2014;4:240–250. doi: 10.1016/j.fob.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu G., Bazer F.W., Burghardt R.C., Johnson G.A., Kim S.W., Knabe D.A., Li P., Li X., McKnight J.R., Satterfield M.C., Spencer T.E. Proline and hydroxyproline metabolism: implications for animal and human nutrition. Amino Acids. 2011;40:1053–1063. doi: 10.1007/s00726-010-0715-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visser W.F., Verhoeven-Duif N.M., de Koning T.J. Identification of a human trans-3-hydroxy-l-proline dehydratase, the first characterized member of a novel family of proline racemase-like enzymes. J. Biol. Chem. 2012;287:21654–21662. doi: 10.1074/jbc.M112.363218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muramatsu H., Mihara H., Kakutani R., Yasuda M., Ueda M., Kurihara T., Esaki N. The putative malate/lactate dehydrogenase from Pseudomonas putida is an NADPH-dependent Δ1-piperideine-2-carboxylate/Δ1-pyrroline-2-carboxylate reductase involved in the catabolism of d-lysine and d-proline. J. Biol. Chem. 2005;280:5329–5335. doi: 10.1074/jbc.M411918200. [DOI] [PubMed] [Google Scholar]
- 17.Goytia M., Chamond N., Cosson A., Coatnoan N., Hermant D., Berneman A., Minoprio P. Molecular and structural discrimination of proline racemase and hydroxyproline-2-epimerase from nosocomial and bacterial pathogens. PLoS One. 2007;2:e885. doi: 10.1371/journal.pone.0000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawakami R., Sakuraba H., Ohshima T. Gene and primary structures of dye-linked l-proline dehydrogenase from the hyperthermophilic archaeon Thermococcus profundus show the presence of a novel heterotetrameric amino acid dehydrogenase complex. Extremophiles. 2004;8:99–108. doi: 10.1007/s00792-003-0368-x. [DOI] [PubMed] [Google Scholar]
- 19.Kawakami R., Sakuraba H., Tsuge H., Goda S., Katunuma N., Ohshima T. A second novel dye-linked l-proline dehydrogenase complex is present in the hyperthermophilic archaeon Pyrococcus horikoshii OT-3. FEBS J. 2005;272:4044–4054. doi: 10.1111/j.1742-4658.2005.04810.x. [DOI] [PubMed] [Google Scholar]
- 20.Neuner A., Jannasch H.W., Belkin S., Stetter K.O. Thermococcus litoralis sp. nov.: a new species of extremely thermophilic marine archaebacteria. Arch. Microbiol. 1990;153:205–207. [Google Scholar]
- 21.Trovato M., Maras B., Linhares F., Costantino P. The plant oncogene rolD encodes a functional ornithine cyclodeaminase. Proc. Natl. Acad. Sci. USA. 2001;98:13449–13453. doi: 10.1073/pnas.231320398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C., Lu C.D. Arginine racemization by coupled catabolic and anabolic dehydrogenases. Proc. Natl. Acad. Sci. USA. 2009;106:906–911. doi: 10.1073/pnas.0808269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gatto G.J., Jr, Boyne M.T., 2nd, Kelleher N.L., Walsh C.T. Biosynthesis of pipecolic acid by RapL, a lysine cyclodeaminase encoded in the rapamycin gene cluster. J. Am. Chem. Soc. 2006;128:3838–3847. doi: 10.1021/ja0587603. [DOI] [PubMed] [Google Scholar]
- 24.Kan-No N., Matsu-Ura H., Jikihara S., Yamamoto T., Endo N., Moriyama S., Nagahisa E., Sato M. Tauropine dehydrogenase from the marine sponge Halichondria japonica is a homolog of ornithine cyclodeaminase/mu-crystallin. Comp. Biochem. Physiol. 2005;B141:331–339. doi: 10.1016/j.cbpc.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Plese B., Schröder H.C., Grebenjuk V.A., Wegener G., Brandt D., Natalio F., Müller W.E. Strombine dehydrogenase in the demosponge Suberites domuncula: characterization and kinetic properties of the enzyme crucial for anaerobic metabolism. Comp. Biochem. Physiol. 2009;B154:102–107. doi: 10.1016/j.cbpb.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Kobylarz M.J., Grigg J.C., Takayama S.J., Rai D.K., Heinrichs D.E., Murphy M.E. Synthesis of l-2,3-diaminopropionic acid, a siderophore and antibiotic precursor. Chem. Biol. 2014;21:379–388. doi: 10.1016/j.chembiol.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 27.Sakuraba H., Kawakami R., Takahashi H., Ohshima T. Novel archaeal alanine:glyoxylate aminotransferase from Thermococcus litoralis. J. Bacteriol. 2004;186:5513–5518. doi: 10.1128/JB.186.16.5513-5518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borel F., Hachi I., Palencia A., Gaillard M.C., Ferrer J.L. Crystal structure of mouse mu-crystallin complexed with NADPH and the T3 thyroid hormone. FEBS J. 2014;281:1598–1612. doi: 10.1111/febs.12726. [DOI] [PubMed] [Google Scholar]
- 29.Monaghan P.J., Leys D., Scrutton N.S. Mechanistic aspects and redox properties of hyperthermophilic l-proline dehydrogenase from Pyrococcus furiosus related to dimethylglycine dehydrogenase/oxidase. FEBS J. 2007;274:2070–2087. doi: 10.1111/j.1742-4658.2007.05750.x. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J., Fritsch E.F., Maniatis T. 3rd edn. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 31.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 32.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Satomura T., Kawakami R., Sakuraba H., Ohshima T. Dye-linked d-proline dehydrogenase from hyperthermophilic archaeon Pyrobaculum islandicum is a novel FAD-dependent amino acid dehydrogenase. J. Biol. Chem. 2002;277:12861–12867. doi: 10.1074/jbc.M112272200. [DOI] [PubMed] [Google Scholar]
- 34.Penning T.M., Jez J.M. Enzyme redesign. Chem. Rev. 2001;101:3027–3046. doi: 10.1021/cr000049n. [DOI] [PubMed] [Google Scholar]
- 35.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this study.
Phylogenetic analysis of TlLhpH.
Temperature dependence of Pyr2C reductase and T3LHyp dehydratase activities.


