Abstract
Saccharomyces cerevisiae has been a key experimental organism for the study of infectious diseases, including dsRNA viruses, ssRNA viruses and prions. Studies of the mechanisms of virus and prion replication, virus structure and structure of the amyloid filaments that are the basis of yeast prions have been at the forefront of such studies in these classes of infectious entities. Yeast has been particularly useful in defining the interactions of the infectious elements with cellular components: chromosomally encoded proteins necessary for or blocking the propagation of the viruses and prions, and proteins involved in expression of viral components. Here we emphasize the L-A dsRNA virus and its killer-toxin-encoding satellites, the 20S and 23S ssRNA naked viruses, and the several infectious proteins (prions) of yeast.
Keywords: narnavirus, totivirus, killer yeast, Sup35p, [PSI+], Ure2p, [URE3]
I. Introduction
Here we review the viruses and prions of Saccharomyces cerevisiae, including the dsRNA viruses that resemble the cores of mammalian dsRNA viruses, the 'naked RNA' single-stranded RNA viruses that are closest in sequence to some RNA bacteriophage, and the yeast prions that are self-propagating amyloids of various chromosomally encoded proteins. These infectious elements of yeast share a mode of transmission by cell-cell mating; none are known to include an extracellular phase. The dsRNA and ssRNA viruses lack the outer virion layer typical of the corresponding viruses of other organisms that use an extracellular route as the primary mode of transmission. The retrotransposons of yeast, Ty1, Ty2,.., Ty5, (not reviewed here, but see: Maxwell and Curcio 2007; Beauregard et al. 2008) likewise resemble the cores of retroviruses, with no env gene and no extracellular phase, but with a similar intracellular life cycle.
Biologically, the yeast viruses resemble other virus groups that do not have a known extracellular phase. The Endornaviruses (Fukuhara et al. 2006; Roossinck et al. 2011) are large (~14 kb) dsRNA unencapsidated replicons in plants and fungi. Partitiviruses are encapsidated dsRNA viruses of plants and fungi, also without an extracellular phase (Ghabrial et al. 2005). An array of Hypoviruses, moderating the pathogenicity of Cryphonectria parasitica for Chestnut trees, are strictly intracellular RNA viruses (Nuss 2005). Moreover, endogenous retroviruses and some Herpesviruses are unable to spread via a cell-free intermediate.
II. L-A virus and the killer satellite dsRNAs
L-A, as the type species of Totiviridae, is a 4.6 kb single-segment dsRNA virus with a icosahedral coat comprised of a single major coat protein (called Gag, for reasons that will be evident), and a Gag-Pol fusion protein formed by ribosomal frameshifting and including the RNA-dependent RNA polymerase. L-A is stably maintained in cells, without apparent slowing of growth, but its presence in only 15 of 70 wild S. cerevisiae (Nakayashiki et al. 2005) indicates it must have a net detrimental effect.
Several satellite dsRNAs are found in some strains, called M1, M2, M28 or Mlus, each encoding a secreted protein toxin and immunity to the toxin (Schmitt and Breinig 2006). M dsRNAs encode a preprotoxin which is processed by cleavage to produce the mature toxin. Preprotoxin ORFs are not related with host-encoded genes, with the exception of klus prepotoxin, which shows a high degree of conservation with S. cerevisiae YFR020W ORF, of unknown function, suggesting an evolutionary relationship (Rodriguez-Cousino et al. 2011)
These toxins kill other cells by different mechanisms. Among them, K1 and K28 have been studied in detail. They interact with sensitive cells through receptors in the cell wall or the plasma membrane. Ionophoric K1 disrupt cytoplasmic membrane function by forming cation-selective ion channels (Martinac et al. 1990). K28 toxin, in contrast, blocks DNA synthesis and arrests cells in early S phase of the cell cycle (Schmitt et al. 1996).
A. Virion structure
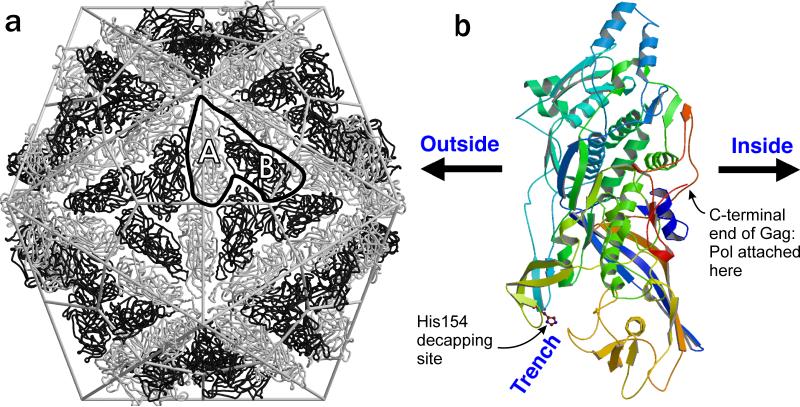
The X-ray structure of the L-A virus (Naitow et al. 2002; Tang et al. 2005) shows an icosahedral structure with 'T=2' symmetry, a supposedly forbidden mode (Fig. 1a). In fact, the structure is a T=1 structure (sixty units) with an assymetric dimer of Gag as the unit (Esteban and Wickner 1986; Caston et al. 1997; Naitow et al. 2002). The dsRNA is packed in layers inside the virion, and pores at the 5-fold axes allow access of nucleotides to the interior for RNA synthesis and the escape of the new viral (+) strands to the cytoplasm. A groove in the virion surface includes residue His154, the site of attachment of 7mGMP removed from cellular mRNAs (Fig. 1b; see below).
Fig. 1.
L-A virion structure. a. “T=2” architecture with two non-identical forms of the Gag major coat protein (A and B). b. Structure of a single monomer. Note that the trench with His154, the site of cap attachment, is outside the particles, while the C-terminus of Gag, which is extended as Gag-Pol in one or two subunits per particle, is internal (modified from (Naitow et al. 2002)).
B. L-A and M encoded proteins
L-A has two long ORFs, called Gag and Pol for their analogy with retroviruses (Fig. 2). The 5' Gag ORF encodes the major coat protein and Pol, overlapping with the 3' end of the Gag ORF, encodes the RNA dependent RNA polymerase, expressed only as a fusion protein with Gag. Virus particles contain only 1-2 copies of Gag-Pol, made by a -1 ribosomal frameshift similar in mechanism to that of many retroviruses (Dinman et al. 1991).
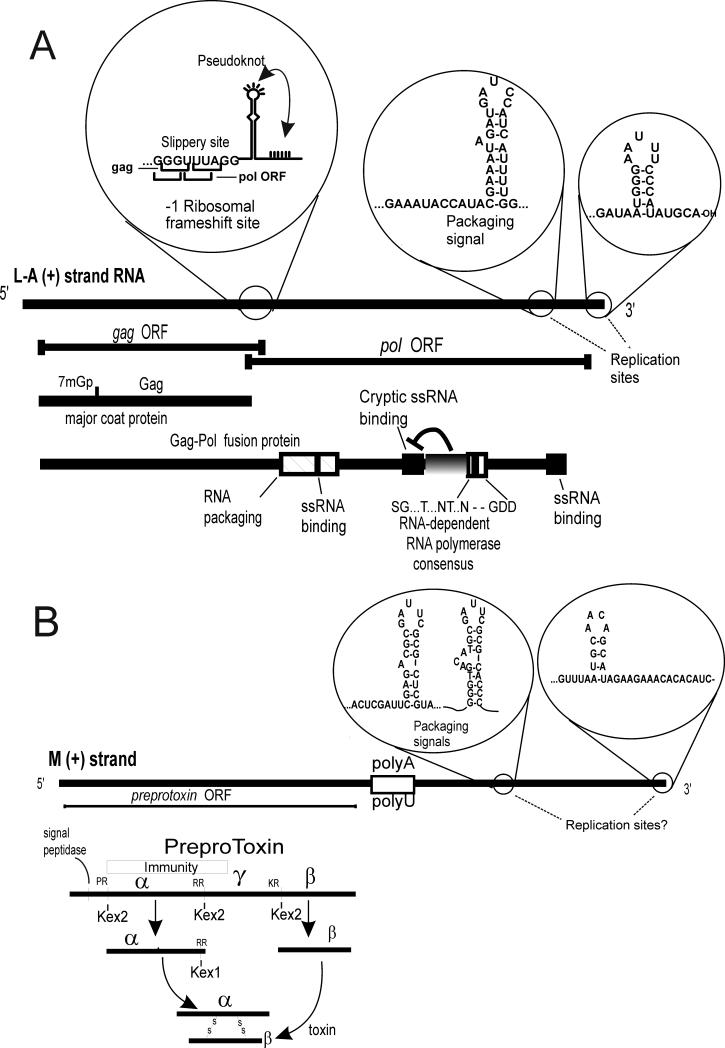
Fig. 2.
L-A and M dsRNA cis sites and encoded proteins. a. The L-A genome is shown with the sites of ribosomal frameshifting, the packaging site (and the overlapping internal replication enhancer), and the 3' replication site. The His154 cap attachment site is shown as 7mGp.
M dsRNA encodes a preprotoxin which is processed by cleavage after dibasic residues by Kex2p followed by removal of the dibasic residues by Kex1p (Leibowitz and Wickner 1976; Bostian et al. 1983; Julius et al. 1984; Cooper and Bussey 1989) (Fig. 2). This processing pattern was immediately recognized to resemble that of precursors of insulin and several other human hormones, and led to the discovery of the corresponding (homologous) processing enzymes (Steiner et al. 1992).
C. The replication cycle
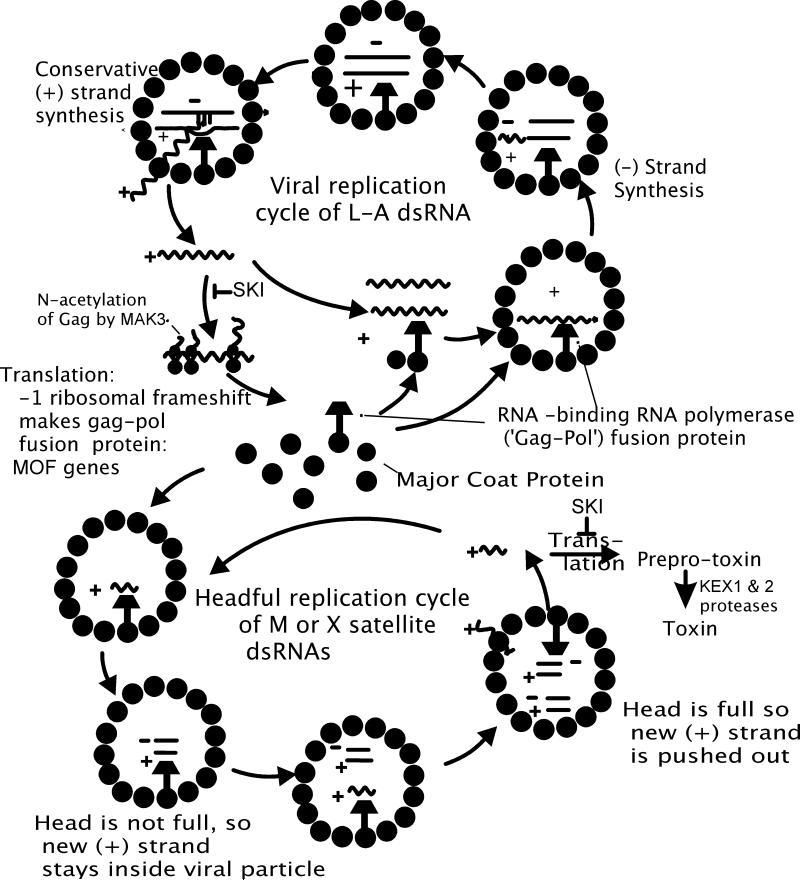
The L-A virus replicates entirely inside the coat. New (+) strands are made inside the coat from the dsRNA template by a conservative mechanism and are extruded from the particles where they are translated and/or encapsidated to form new particles (Fig. 3). Newly encapsidated (+) strands are converted to dsRNA form within the particles to complete the cycle. Note that the (+) and (-) strands are made at different points of the replication cycle.
Fig. 3.
L-A and M replication cycles.
Examination of virus particles carrying a deletion mutant of L-A (called X) and of the M1 satellite dsRNA suggested a 'head-full replication' model, in which a single (+) strand is encapsidated, and it replicates within the virion until the particle is full of dsRNA (Esteban and Wickner 1986; Esteban and Wickner 1988) (Fig. 3). Then new (+) strands are extruded from the particles. Indeed, M1 particles with a single dsRNA molecule often retain newly made (+) strands, while those with two M1 dsRNA molecules per particle export all of their new (+) strand transcripts (Esteban and Wickner 1986). Moreover, it has been directly shown that a single (+) strand is initially encapsidated (Fujimura et al. 1990).
D. Transcription
The (+) strand synthesis process goes on inside the particles and is a conservative reaction, meaning parental strands stay together (Herring and Bevan 1974; Fujimura et al. 1986). Detailed studies of the transcription, replication and packaging of the L-A virus employed an in vitro system in which the enzymes were supplied by empty particles, produced by exposure of virions to low salt conditions (Fujimura and Wickner 1988). These opened empty particles specifically bind viral (+) ssRNA in a reaction whose specificity was shown to be the same as the packaging reaction (see below), can convert these (+) strands to dsRNA form (the replication reaction, see below) and can transcribe these dsRNA molecules or dsRNA added initially to the particles.
E. Translation. Ribosomal frameshifting
The 5' Gag and 3' Pol open reading frames overlap by 130 nt. Most ribosomes translating Gag terminate at the Gag termination codon, but about 1% of ribosomes translating Gag perform a -1 ribosomal frameshift at a special sequence into the Pol open reading frame and synthesize a Gag-Pol fusion protein (Fujimura and Wickner 1988; Icho and Wickner 1989; Dinman et al. 1991). The frameshift signal consists of a 'slippery site' whose sequence is ...G GGU UUA GG... with the Gag reading frame indicated, and a pseudoknot just 3' to this slippery site. The pseudoknot makes the ribosomes pause with the underlined codons in the ribosomal A and P sites. The slippery site is such that pairing of the non-wobble bases is still correct if the ribosome shifts back one base. This happens to about 1% of the ribosomes, and when they resume translation in the new frame, they extend the polypeptide to make the Gag-Pol fusion protein (Dinman et al. 1991). This mechanism serves to a) encode two proteins from one RNA segment, conserving space, b) fixes the ratio of Gag to Gag-Pol proteins by adjusting the structure of the frameshift site, a ratio which is critical for the efficient propagation of the virus (Dinman and Wickner 1992), c) strongly suggests that the Pol domain is properly incorporated on the inside of the viral particle when the Gag domain is incorporated into the particles [note that the C-terminus of the Gag domain faces the inside of the particles (Naitow et al. 2002)].
F. 60S ribosomal subunits and SKI2, SKI3, etc
Mutants in any of a wide array of genes that are partially deficient in 60S subunits result in loss of M dsRNA and decreased copy number of L-A dsRNA (e.g. Ohtake and Wickner 1995; Edskes et al. 1998). Because the viral mRNAs lack a 3' polyA, and polyA may have a special role in the 60S subunit joining step (Munroe and Jacobson 1990; Searfoss et al. 2001; Kahvejian et al. 2005), mutants deficient in 60S subunits may tip the balance of translation away from the viral transcripts and toward the cellular mRNAs (Edskes et al. 1998).
The SKI (superkiller) genes (Toh-e et al. 1978; Ridley et al. 1984; Benard et al. 1998) include SKI1/XRN1, a 5' exoribonuclease specific for uncapped RNAs with a prominent role in mRNA degradation (Larimer et al. 1992), while SKI4 and SKI6 are subunits of the 'exosome' complex, originally named because it was believed to be composed of exoribonucleases (Mitchell et al. 1997), but now known to be devoid of such activities (Chlebowski et al. 2011). SKI2, SKI3 and SKI8, encode a cytoplasmic complex (Brown et al. 2000). SKI2, 3, 6, 7 and 8 together block the expression of nonpolyA mRNAs, as judged by electroporation experiments (Masison et al. 1995; Benard et al. 1998; Benard et al. 1999). While all are involved in the 3' exonuclease degradation of mRNA (Jacobs-Anderson and Parker 1998), kinetic experiments suggest that translation effects may also be involved. SLH1 is a paralog of SKI2, and the ski2 slh1 double mutant expresses Cap+ poly(A)- mRNAs with the same kinetics as it does Cap+ polyA+ mRNAs (Searfoss and Wickner 2000). This result indicates that the translation apparatus does not inherently need polyA.
G. N-acetylation of Gag is needed for viral assembly
MAK3, MAK10 and MAK31 are each necessary for L-A and M propagation, and together form an N-acetyltransferase complex with Mak3p the catalytic subunit (Sommer and Wickner 1982; Toh-e and Sahashi 1985; Tercero and Wickner 1992; Rigaut et al. 1999). Mak3p recognizes the four N-terminal residues of L-A Gag, and acetylates the initiator methionine, a modification necessary for viral assembly (Tercero et al. 1992; Tercero and Wickner 1992; Tercero et al. 1993). Homologs of the Mak3-Mak10-Mak31 N-acetyltransferase complex are now known in many organisms including a human complex (e.g., (Starheim et al. 2009)).
H. L-A cap-snatching
The prominent feature of eukaryotic mRNA is the presence of the cap structure (m7GpppX-) at the 5’ end. The structure is crucial for the efficient translation and stability of mRNA. In the cell and for most viruses the cap is installed on mRNA co-transcriptionally by three enzymatic reactions (Venkatesan et al. 1980; Shuman 1995). RNA triphosphatase eliminates the 5’ γ phosphate from a nascent PolII transcript. Guanylyltransferase forms a Gp-enzyme intermediate with GTP and transfers Gp to the diphosphorylated 5’ end of the transcript to form a 5’-5’ triphosphate linkage. Finally, methyltransferase methylates the guanine base at N7. Influenza virus installs its mRNA with a cap structure in a non-conventional mechanism (cap snatching) (Plotch et al. 1981; Boivin et al. 2010). The trimeric viral polymerase binds to the cap structure of host mRNA, cleaves the RNA endonucleolytically 10-13 nt downstream and utilizes the capped fragment as a primer to transcribe the viral genome. Recently, L-A virus has been found to furnish its transcript with a cap structure by a novel cap-snatching mechanism (Fujimura and Esteban 2011). L-A only transfers the m7Gp moiety from host mRNA to the diphosphorylated 5’ end of the viral transcript. Furthermore, unlike influenza virus, L-A utilizes Gag to catalyze the reaction.
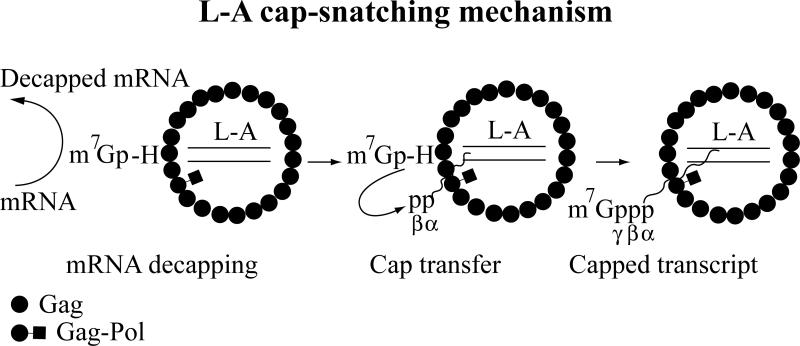
In 1992, Sonenberg and coworkers found that Gag of L-A virus covalently binds to the cap structure of mRNA (Blanc et al. 1992). Subsequent studies showed that m7Gp derived from mRNA decapping is attached covalently to His-154 of Gag (Blanc et al. 1994). Gag with a mutation at His-154, when expressed from a vector, could support replication of M1, interestingly however, its expression (killer toxin production) was severely affected by the mutation (Blanc et al. 1994). L-A and M1 transcripts have diphosphate at their 5’ ends (Fujimura and Esteban 2010). Isolated L-A virions can transfer the m7Gp moiety from mRNA to the 5’ end of the L-A transcript, thus forming the authentic cap structure found in the yeast (Fig. 4) (Fujimura and Esteban 2011). The α and β phosphates at the 5’ end of the transcripts are conserved in the triphosphate linkage of the product. A mutation at His-154 abolished the cap transfer reaction. Because the toxin production is severely affected by the mutation, it indicates that capping is essential for the efficient expression of the viral transcript. The N7 methylation of the 5’ terminal G is essential for cap donor activity, and the smallest molecule with donor activity is the dinucleotide cap analog m7GpppG (Fujimura and Esteban 2012). The cap acceptor needs to be 5’ diphosphorylated. A 5’ tri- or monophosphorylated viral transcript does not function as cap acceptor. Although L-A virions can utilize exogenously added viral transcripts as cap acceptors, the capping reaction requires the viral polymerase actively engaging in transcription (Fujimura and Esteban 2012). Because the polymerase is confined inside the virion, whereas the cap-snatching site is located on the cytoplasmic surface of the virion (see below), it indicates coordination between the transcription and cap-snatching sites. This coordination may minimize the risk of accidental capping of non-viral RNA when the polymerase is dormant. The physical separation of the cap-snatching site from the transcription site may ensure that not all of transcripts are capped. This will allow the virus to synthesize two types of transcripts: one, non-capped transcripts presumably destined for encapsidation (genomic ssRNA) and the other, capped transcripts for translation (mRNA).
Fig. 4.
Schematic diagram of L-A cap-snatching mechanism. Gag of an L-A virion decaps mRNA and forms an intermediate with m7Gp through His-154. Then m7Gp is transferred to the diphosphorylated 5’ end of the viral transcript to form a 5’-5’ triphosphate linkage.
Structural studies of L-A virions have revealed a trench on the outer surface of Gag that includes His-154 (Naitow et al. 2002; Tang et al. 2005). The trenches are located in the Gag asymmetric dimmer close to the icosahedral 2-and 3-fold symmetric axes. L-A transcripts are made inside the virion and presumably released to the cytoplasm through one of the pores located at the 5-fold axes (Naitow et al. 2002). His-154 is located at the tip of a loop (residues 144-163) that is part of the upper rim of the trench. Upon m7GDP binding, the rim moves inwardly and forms a closed conformation (Tang et al. 2005). Guanylyltransferase also contains a trench that can adopt either open or closed conformation during the mRNA capping reaction (Hakansson et al. 1997; Chu et al. 2011). Furthermore, secondary structure elements around the trench of Gag resemble those of guanylyltransferase (Naitow et al. 2002). The latter enzyme forms a Gp-enzyme intermediate with GTP and transfers Gp to the diphosphorylated 5’ terminus of PolII transcript. Thus the capping reaction of L-A resembles that of guanylyltransferase. These similarities suggest a convergent evolution.
In the trench, Tyr-150, Asp-152, Tyr-452 and Tyr-538 are located close to the bound m7GDP, suggesting their involvement in cap recognition. Mutagenesis analyses indicate that these residues are crucial for decapping activity (Tang et al. 2005). The eminent feature of cap binding proteins is the presence of two aromatic amino acids in their cap-binding pockets (Hakansson et al. 1997; Chu et al. 2011). The aromatic rings of these amino acids sandwiched the m7G aromatic ring of the cap structure by stacking. The delocalization of the positive charge arising from the N7 methylation of the guanine contributes to strong interactions between the πelectrons of the stacked aromatic rings (Hu et al. 1999). Tyr-452 and Tyr-538 of L-A Gag sandwich m7GDP by a potential stacking (Tang et al. 2005). Furthermore, the cap-snatching reaction requires N7 methylation of the cap for donor activity (Fujimura and Esteban 2012). It is likely that L-A virus utilizes a mechanism similar to those of cap binding proteins to recognize mRNA for cap snatching. Fungal totiviruses (members of the genus Totivirus), including L-BC, share the same (or similar) four amino acids aforesaid as well as His-154 at comparable positions in their capsid proteins (Fujimura and Esteban 2011). Gag of LBC has been observed to possess decapping activity (Blanc et al. 1992). Therefore, it is likely that the cap-snatching mechanism of L-A is widespread among totiviruses of fungi.
I. Encapsidation
Viral (+) strands, made in the transcription reaction, are extruded from the particles where they may serve as mRNAs and/or be encapsidated by Gag and Gag-Pol to make new viral particles. The RNA encapsidation signal is located 400 nt from the 3' end of the L-A (+) strand and consists of a stem-loop with a critical A residue protruding from the 5' side of the stem (Esteban et al. 1988; Fujimura et al. 1990). The encapsidation signal is recognized by the N-proximal part of the Pol part of Gag-Pol, resulting in packaging of a single (+) strand in each virus particle (Fujimura et al. 1992; Ribas et al. 1994).
J. Replication
The viral (+) strands are converted to dsRNA form by the RNA dependent RNA polymerase in a reaction involving recognition of an internal site, largely overlapping with the packaging site, and synthesis beginning at the 3' end of the template (Esteban et al. 1989). The polymerase apparently binds first to the internal site, and then begins synthesis at the 3' end, without sliding along the template (Fujimura and Wickner 1992).
III. Biology of the S. cerevisiae dsRNA viruses
L-BC is a 4.6 kb dsRNA virus with sequence similarity to L-A, but clearly constituting a distinct replicon (Sommer and Wickner 1982; Park et al. 1996). L-BC and L-A replicate stably in the same cells, and while L-A supports the M dsRNA satellites, L-BC does not. L-A requires MAK3, MAK10 and MAK31, the N-acetyltransferase encoding genes, but L-BC does not, and the clo1 mutation that results in loss of L-BC does not affect L-A (Wickner 1980; Wickner and Toh-e 1982; Wesolowski and Wickner 1984).
Wild isolates of L-A show a variety of genetic activities, defined by their ability to support the propagation of M dsRNAs, and their interaction with each other (Wickner 1980; Wickner and Toh-e 1982). Although cDNA clones have been used extensively in defining the activities of L-A encoded functions, these activities resisted study because it has not yet been possible to launch the L-A virus from transcripts of the cDNA clone.
Mutants in (at least) ski2, 3, and 8 are cold-sensitive for growth, and actually die at low temperature if they carry M dsRNA (Ridley et al. 1984). Although if carrying only L-A, these ski mutants have a great deal more dsRNA than when they also carry M, they are not cold-sensitive (Ball et al. 1984; Ridley et al. 1984). The molecular basis of this phenotype has not been found, but it suggests that the presence of the toxin-immunity-encoding M dsRNAs is not an undiluted advantage. It has recently been shown that S. cerevisiae can support an RNAi system if only Dicer and Argonaut are imported from S. castetllii (Drinnenberg et al. 2009), and shown that the M and L-A dsRNAs are lost from the constructed strains (but not L-BC) (Drinnenberg et al. 2011). However, M dsRNAs are rather rare in wild strains (Young and Yagiu 1978; Nakayashiki et al. 2005), indicating that there must be a significant disadvantage resulting from carrying it that more than makes up for the obvious advantage. This remains an unsolved problem.
IV. Yeast Narnaviruses: 20S and 23S RNAs
Most laboratory strains of S. cerevisiae harbor 20S RNA and fewer strains 23S RNA. These viruses are small positive-stranded RNA viruses belonging to the family Narnaviridae and encode only their RNA dependent RNA polymerases. The RNA genomes are not packed into a massive protein capsid, but form ribonucleoprotein complexes with their cognate RNA polymerases in the host cytoplasm. 20S RNA was first found in 1971 by Kadowaki and Halvorson as an RNA species accumulated under sporulation conditions (Kadowaki and Halvorson 1971). The name was based on its mobility relative to those of 18S and 25S rRNAs in a gel. Garvik and Haber established that 20S RNA is a cytoplasmically inherited genetic element (Garvik and Haber 1978) and that the accumulation of 20S RNA is independent of the sporulation process of the host. In 1984, Wesolowski and Wickner reported two heat-inducible, cytoplasmically inherited dsRNAs, W and T (Wesolowski and Wickner 1984). When 20S RNA (Matsumoto and Wickner 1991) and W (Rodriguez et al. 1991) were independently cloned and sequenced, it was evident that they were identical but in two different molecular forms, ssRNA and dsRNA. Subsequently, T dsRNA was also characterized by sequencing and its ssRNA form (23S RNA) was identified (Esteban et al. 1992) .
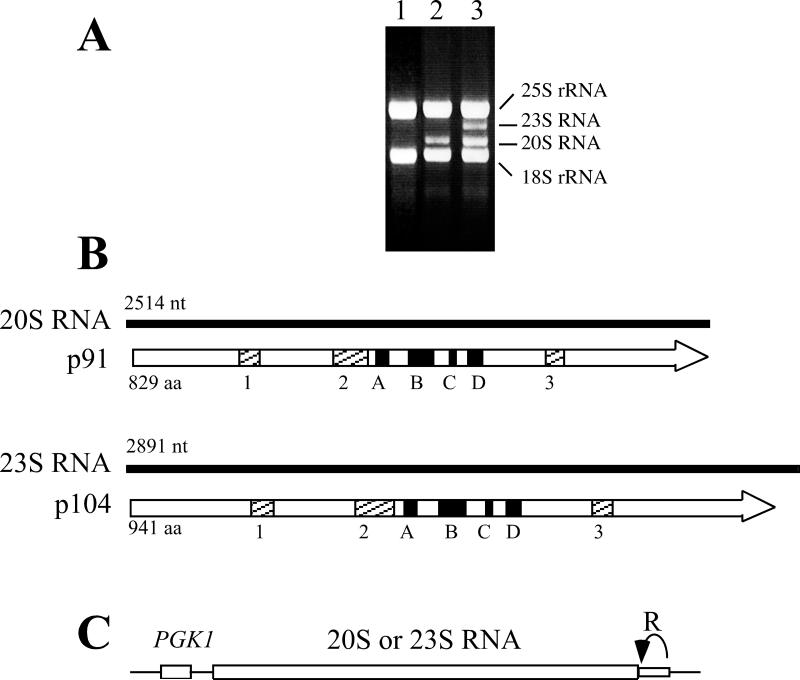
The 20S RNA and 23S RNA genomes are small (2514 and 2891 nt long, respectively) (Rodriguez-Cousino et al. 1998) and rich in GC. Both RNAs possess 5 nt inverted repeats (5’ GGGGC...GCCCC-OH) at the 5’ and 3’ termini. Each RNA has a single open reading frame that spans almost the entire viral genome (Fig. 5A). The 5’ non-translating regions are only 12 nt in 20S RNA and 6 nt in 23S RNA. 20S RNA and 23S RNA encode 91 kDa (p91) and 104 kDa proteins (p104), respectively (Matsumoto and Wickner 1991; Rodriguez et al. 1991; Esteban et al. 1992), both with consensus sequences for RNA-dependent RNA polymerases and most closely related to those of the positive stranded RNA coliphages such as Qβ.
Fig. 5.
20S and 23S RNA viral genomes. A. Total RNA extracted from induced cells containing no viruses (lane 1), 20S RNA alone (lane 2), or 20S and 23S RNAs (lane 3). Ethidium bromide staining of an agarose gel is shown. 18S and 25S rRNAs are indicated. B. 20S and 23S RNA genomes and the encoded proteins, p91 and p104. P91 and p104 contain the consensus sequences (A-D) for RNA-dependent RNA polymerases, which are most closely related to those of RNA coliphages. 1-3 indicates amino acid stretches conserved between p91 and p104. C. Diagram of a launching plasmid. The complete sequence of the 20S or 23S RNA genome is inserted downstream of the constitutive PGK1 promoter (PGK1). The hepatitis delta virus ribozyme (R) is fused directly to the 3’ end of the viral genome. Thin lines indicate sequences derived from the vector.
20S and 23S RNAs migrate in sucrose gradients almost as naked RNAs, and phenol treatment hardly changes their mobility (Widner et al. 1991; Esteban et al. 1994; Garcia-Cuellar et al. 1995). The mass of most viruses consists of capsids and lipid layers, indicating that 20S and 23S RNAs are not encapsidated. However, p91 and p104 are associated with 20S RNA and 23S RNA, respectively (Esteban et al. 1994; Garcia-Cuellar et al. 1995) in a 1:1 protein:RNA stoichiometry in the host cytoplasm (Solorzano et al. 2000). Preliminary data indicate that the resting complexes of 20S RNA contain no host proteins. Pull-down experiments detected no proteins other than p91 in metabolically labeled resting complexes. Furthermore, p91 can form a resting complex with 20S RNA in E. coli in the absence of yeast proteins (Vega 2010).
A. Launching systems of narnaviruses
20S and 23S RNA viruses can be generated from vectors (Esteban and Fujimura 2003; Esteban et al. 2005). In both cases, the complete sequences of the viral genomes are placed downstream of the constitutive PGK1 promoter (Fig. 5B). The efficiency of virus launching is high. 20S RNA can be generated in more than 70% of colonies of the yeast transformed with the vector. It is critical to directly fuse the hepatitis delta virus ribozyme to the 3’ end of the viral genome. In the absence of the ribozyme the vectors failed to generate the viruses, and insertion of a few Gs between the viral 3’ end and the ribozyme greatly reduced viral generation. In the latter case, the virus, once generated, is indistinguishable from the authentic one because the extra Gs at the 3’ end were eliminated during the launching process. The launching vectors also contained non-viral sequences (about 40 nt) between the major transcription initiation site of the promoter and the viral 5’ end. These sequences were also eliminated during the launching process and, as will be discussed later, the Ski1 5’ exonuclease plays the major role in their elimination.
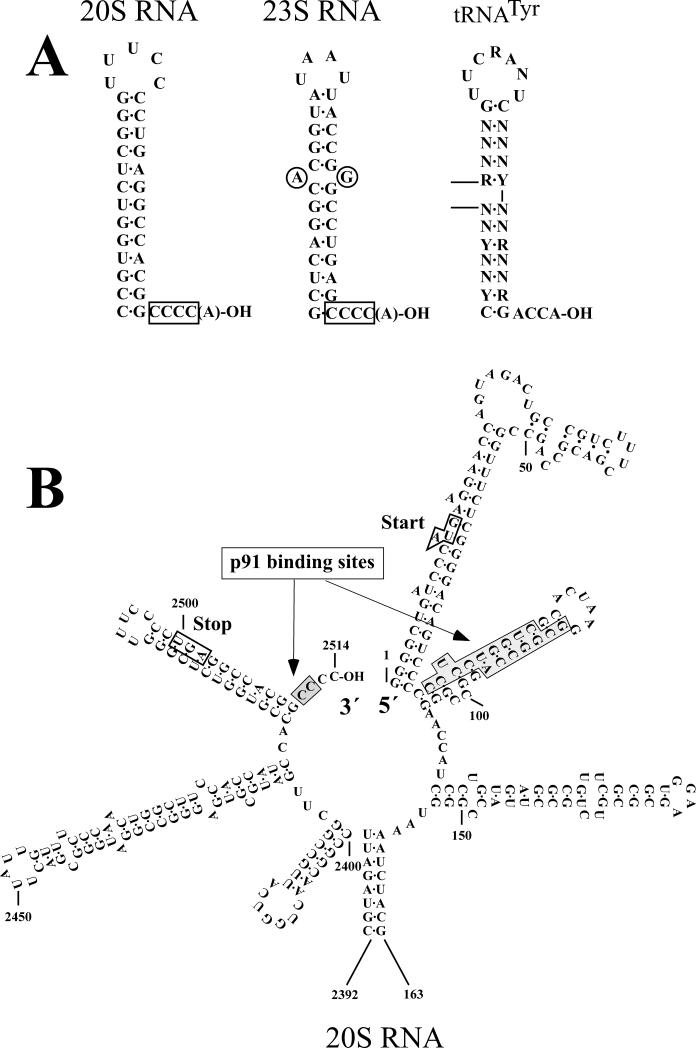
The narnaviruses have a terminal repair system (Fujimura and Esteban 2004; Esteban et al. 2005). The elimination of the last and penultimate Cs at the viral 3’ end in the vector, or changing them to the other nucleotides did not affect virus generation. However, the viruses generated were found to have recovered these terminal Cs. It indicates that these terminal Cs are essential for replication, but that the presence of a repair system makes them dispensable for virus launching (Fig. 6A). By contrast, modifications at the 3rd or 4th C from the 3’ end abolished virus generation. Although the 5th G is part of the 5 nt inverted repeat, this G is not essential for replication and a modified nucleotide at this position can be retained in the generated viruses. The terminal repair could be done by host enzymes, or by the viral RNA polymerase. A modified nucleotide at the 3’ terminus may be removed by an exonuclease and then the correct nucleotides might be install by the CCA-adding enzyme. The positive and negative strands of 20S and 23S RNAs contain stem-loop structures adjacent to the 3’ ends, which resemble the so-called top-half domain of tRNA (Fig. 6A). The CCA-adding enzyme recognizes the top-half domain of tRNA precursors and adds CCA to their 3’ ends in a non-templated fashion. That 20-30% of the positive and negative strands of W and T dsRNAs have a non-templated A at the 3’ termini (Rodriguez-Cousino and Esteban 1992) is consistent with the possible involvement of this enzyme in the repair. Alternatively, the terminal repair could be done by the viral polymerase during replication. RdRps are also known to add a non-templated A at the 3’ end of the products. Although 20S RNA and 23S RNA genomes have 5 nt inverted repeats at both termini, it is unlikely that the 5’ complementary sequence is used as template to correct modifications at the 3’ end because both a modification at the 3’ end and the compensatory mutation at the 5’ end were corrected to the wild type sequences without negative effects on virus generation (Fujimura and Esteban 2004).
Fig. 6.
Cis-acting signals. A. The 3’ terminal sequences of 20S and 23S RNAs with cis-acting signals. For comparison, the top half domain of tRNATyr is shown. B. The 5’ and 3’ terminal regions of the 20S RNA genome. The 5’ and 3’ cis sites are indicated by lightly shaded boxes. The initiation (Start) and termination (Stop) codons of p91 are also indicated.
20S RNA can also be generated from the negative strands expressed from a vector (Esteban et al. 2005). Because p91 can not be decoded from the negative strand, virus generation required p91 expressed from a second plasmid. Since modifications or deletions at the 3’ terminal or penultimate positions did not affect virus generation, it is likely that the same repair mechanism observed on the positive strand also operates on the negative strand.
B. Cis-acting signals
In a resting complex p91 interacts with 20S RNA at least at three different sites, 5’ end, 3’ end and, to a lesser extent, internal sites (Fujimura and Esteban 2007). The 5’ end site is located in the second stem-loop structure from the 5’ end (Fig. 6B). The stem structure is important for complex formation but the loop sequence is not. It has been observed that there is a tight correlation between complex formation and virus generation among those mutants, suggesting that a stable complex formation is a prerequisite for virus generation. The 3’ end site is located at the very end of the 3’ terminus, partially overlapping with the 5 nt inverted repeat. The 3rd and 4th Cs from the 3’ end are important for complex formation but the 3’ terminal and penultimate Cs, as well as the 5th G were dispensable. Since the 3rd and 4th Cs but not the 5th G were essential for replication, there is again a good correlation between replication and complex formation at the 3’ end site. It is not known whether the 3’ end site extends further inside, as in the 23S RNA virus. The internal site is located in the middle of the 20S RNA molecule between nt 1253 and 1513. Mutations at the 5’ or 3’ end site reduced complex formation to a basal level (10-20%) compared with the unmodified RNA. However, the effects of these mutations are not additive, that is, the combination of mutations at these two sites does not reduce complex formation further down. It suggests that the internal site is responsible for the basal level of complex formation. Secondary structure prediction reveals intramolecular long distance interactions among the 5’ end, 3’ end and internal regions, which bring the three sites close together (Fujimura and Esteban 2007). This may allow a p91 molecule to simultaneously interact with these three sites in the resting complex.
In the case of the 23S RNA/p104 complex, only the 3’ end site has been analyzed in detail (Fujimura and Esteban 2004; Fujimura and Esteban 2004). The 3’ end site is bipartite, consisting of the 3rd and 4th Cs from the 3’ end and a stem-loop structure adjacent to the 3’ end (Fig. 6A). Like 20S RNA, the 5th G from the 3’ end is dispensable for complex formation and also for virus generation. The stem-loop structure contains a mismatched pair of purines in the middle of the stem. This mismatch is important for both complex formation and virus generation. Any combination of purines in the mismatch supported both complex formation and virus generation, however, eliminating one of the purines or changing them with pyrimidines abolished both activities. Because 20S RNA has no mismatched pair at the stem-loop structure adjacent to the 3’ end, this may contribute to the specificity of complex formation in these viruses.
C. SKI1 anti-viral activity
During virus launching, the 20S RNA and 23S RNA genomes in the vector are transcribed from the PGK1 promoter by PolII. The transcripts possess not only non-viral sequences (about 40 nt) from the vector but also the cap structure at the 5’ ends (Fig. 5B). The generated viruses, however, have no extra nucleotides at the 5’ ends. These extra nucleotides are eliminated during virus generation. mRNA degradation in eukaryotes usually begins with the shortening of the poly(A) tail at the 3’ end, followed by decapping at the 5’ end by the Dcp1/Dcp2 decapping enzyme (Wilusz et al. 2001; Parker and Song 2004). Because decapping is a crucial step in mRNA degradation, numerous proteins (Lsm1p-7p, Pat1p, Dhh1p, etc) are involved in this reaction. Then the decapped RNA is degraded by the SKI1/XRN1 5’ exonuclease. Alternatively, deadenylated RNA is digested by the 3’ exonuclease exosome (Mitchell et al. 1997; Jacobs-Anderson and Parker 1998). In yeast, the SKI1/XRN1 5’ exonuclease plays a major role in mRNA degradation. It was found that the launching plasmid failed to generate the 20S RNA virus in ski1Δ or dhh1Δ strains (Esteban et al. 2008), signifying the importance of the mRNA degradation pathway in virus launching. When the 5’ non-viral sequence was reduced from 47 to 9 nt, the plasmid generated the 20S RNA virus in ski1Δ strains indicating that the SKI1 5’ exonuclease is largely responsible for eliminating the long 5’ extra sequence. Interestingly, these genes did not affect replication of 20S RNA virus when the endogenous virus was introduced by a cytoplasmic mixing from a donor strain. This confirms that these genes are involved in the 5’ processing of the transcripts but not in viral replication per se.
Because of the 5’ exonuclease activity, SKI1 is involved in host defense against L-A virus (Masison et al. 1995). The copy number of L-A increases several-fold in ski1Δ strains and the virus can be easily cured from the cell by over-expression of the SKI1 gene. By contrast, endogenous 20S RNA virus is fairly insensitive to overexpression of SKI1 (Esteban et al. 2008). The positive strand of L-A has no prominent structures at its 5’ end, whereas 20S RNA (and also 23S RNA) has a strong secondary structure at the 5’ end (Fig. 6B). Furthermore, the first 4 nt at the 5’ ends of both 20S and 23S RNAs are Gs and these nucleotides are buried at the bottom of the stem. It is known that the progression of the 5’ SKI1 exonuclease is blocked by a cluster of Gs and also by a strong secondary structure. It is likely that these features confer on 20S RNA insensitivity to the antiviral activity of SKI1. Indeed, when mutations were introduced to the stem to destabilize the structure, 20S RNA became vulnerable to SKI1 activity (Esteban et al. 2008). These observations indicate that SKI1 is a potent weapon for the host to fight against RNA virus infection and suggest that 20S and 23S RNAs have evolved to develop elaborated secondary structures at the 5’ ends, partly as a countermeasure against SKI1 because the viruses have no protective capsid. The 3rd and 4th Cs from the 3’ ends are not only essential for replication but also important for resting complex formation in 20S and 23S RNA viruses. It strongly suggests that p91 and p104 directly interact with these terminal nucleotides of the cognate RNAs. These interactions may be important to protect the viral genomes from degradation by the exosome. The copy numbers of 20S and 23S RNAs increase several-fold in ski2, ski3, ski4, ski7 or ski8 mutants (Matsumoto et al. 1990; Ramirez-Garrastacho and Esteban 2011), suggesting that the viruses have some stages in their replication cycles vulnerable to the 3’ exosome. This group of Ski proteins also blocks expression specifically of non-polyA mRNAs (Masison et al. 1995; Benard et al. 1999; Brown and Johnson 2001), a feature common to L-A, 20S and 23S.
D. Replication Intermediates
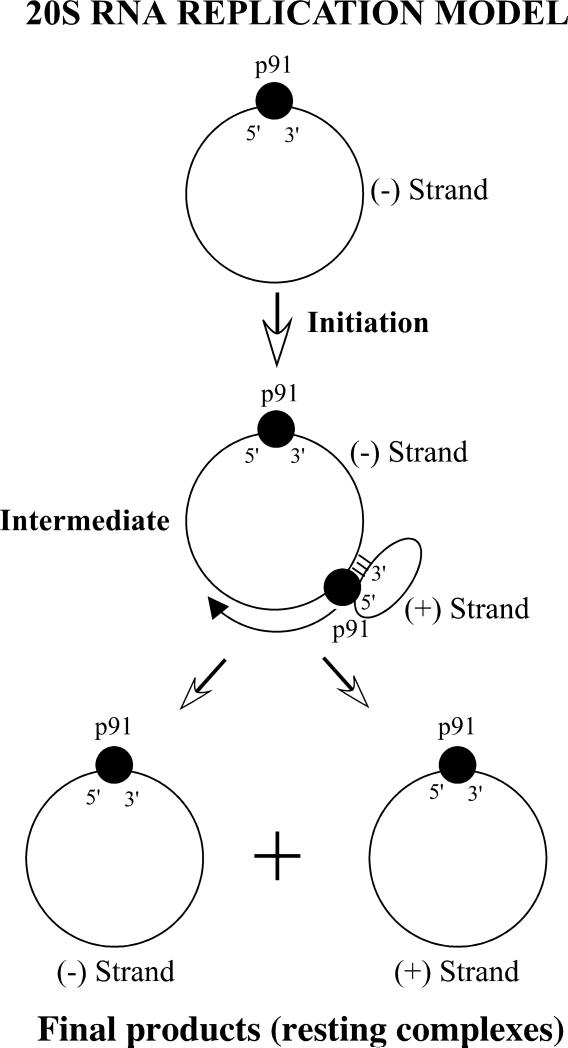
In vegetative growing cells the copy number of 20S RNA is very low (5-20 copies per cell) (Matsumoto et al. 1990). Under sporulation conditions (1% K acetate) it reaches up to 20000 copies per cell, predominantly in the form of resting complexes with the positive strands. Lysates prepared from these cells contain a minor amount of replication intermediates that possess actinomycin D- and α-ammanitin insensitive RNA polymerase activity (Garcia-Cuellar et al. 1997). The intermediate consists of a full-length negative strand template, a positive strand with less than unit length, and p91 (Fujimura et al. 2005). It is not known whether the intermediate also contains host proteins. The synthesis of RNA results from chain-elongation of the positive strand. The de novo synthesis of 20S RNA has not been observed in the lysates. Upon completion of RNA synthesis, the positive strand product and the negative strand template are released from the intermediate. The released strands can be retained on a negatively-charged membrane, suggesting that they are still associated with protein. The RNAs in the replication intermediate have largely a single-stranded RNA backbone. Deproteination with phenol converts them to a dsRNA form. Therefore, W is not a replication intermediate of 20S RNA but a byproduct. Fig. 7 shows a model of the replication intermediates of 20S RNA (Esteban and Fujimura 2006). The initiation process of intermediate formation is not known. The intermediate may contain two p91 molecules: one interacting with the negative strand at the 3’ and 5’ end regions and the other polymerizing the positive strand and also concurrently interacting with its 5’ cis site. These interactions may protect both RNAs from degradation by exonucleases. Furthermore, these interactions may prevent both RNAs from annealing in a long stretch beyond the polymerization site, thus keeping the RNA backbone largely single stranded. Because each p91 has distinct interactions with the respective strands in the intermediate, this may condition their partition when the positive and negative strands are released. In the cell, the negative strand, as soon as released, may be recruited again to form a new replication intermediate, because the majority of negative strands are found in the form of positive strand-synthesizing intermediates.
Fig. 7.
Model for 20S RNA positive strand synthesis. The intermediate may contain two p91 molecules (filled circles). For simplicity, the interaction of p91 with the internal cis site of the positive strand is omitted.
Although a resting complex of the negative strand as well as a negative strand-synthesizing intermediate apparently are also constituents of the 20S RNA replication cycle, these complexes have not been analyzed yet because of their low abundance in the cell.
VI. Yeast Prions
In addition to the nucleic acid-containing viruses, yeast can harbor any of several infectious proteins or prions, conceptually similar to the mammalian prions (Table 1). Most yeast prions are based on self-propagating amyloids, although one is a protease that can activate its own inactive precursor protein (Roberts and Wickner 2003). Amyloid is a linear polymer of a single protein species, with a largely beta-sheet structure in which the beta strands are perpendicular to the long axis of the filament. Surprisingly, a single prion protein can be the basis of any of many different prion variants that differ genetically and biologically. Based on the in-register parallel beta sheet architecture of several yeast prions, a mechanism has been proposed for the templating of amyloid conformation that must underlie this stable inheritance of prion variant.
Table 1.
Amyloid-based yeast prions.
| Prion | Protein | Protein function | Phenotype of Prion | Ref. |
|---|---|---|---|---|
| [URE3] | Ure2p | Negative regulator of catabolism of poor nitrogen sources | Inappropriate derepression of enzymes and transporters for poor N sources | (Wickner 1994) |
| [PSI+] | Sup35p | Translation termination subunit | Inappropriate readthrough of translation terimination codons | (Wickner 1994) |
| [PIN+] | Rnq1p | Unknown | Rare priming of other prion formation | (Derkatch et al. 2001) |
| [SWI+] | Swi1p | Chromatin remodeling complex subunit | Poor growth on raffinose, glycerol, or galactose | (Du et al. 2008) |
| [OCT+] | Cyc8p | Transcription repression factor subunit | Defective transcription repression | (Patel et al. 2009) |
| [MOT+] | Mot3p | Transcription repressor of aerobic-repressed genes | Inappropriate derepression of ‘anaerobic’ genes | (Alberti et al. 2009) |
| [ISP+] | Sfp1p | Positive transcription factor of translation-related genes | Antisuppression | (Rogoza et al. 2010) |
| [MOD+] | Mod5p | tRNA isopentenyltransferase | Partial inactivation of enzyme; | (Suzuki et al. 2012) |
A. The range of yeast prions
[URE3] and [PSI+] were the first yeast prions found, each long known as mysterious non-chromosomal genetic elements assumed to be nucleic acid based, but not correlated with known RNA or DNA molecules. Based on their genetic properties, [URE3] and [PSI+] were shown to be prions of Ure2p, a regulator of nitrogen catabolism, and Sup35, a subunit of the translation termination factor (Wickner 1994). These prions produce a phenotype similar to that of partial deficiency of the corresponding protein. [PIN+] is a prion of Rnq1p, a protein of no known function. The [PIN+] phenotype is the ability to rarely cross-seed other prions, originally [PSI+]. Several other new prions were discovered based on their ability to produce a Pin-like effect, including [SWI+] (Du et al. 2008), a prion of the chromatin-remodeling factor Swi1p, [OCT+], a prion of the transcription repressor Cyc8p (Patel et al. 2009), and [MOD+], a prion of the tRNA isopentenyltransferase Mod5p (Suzuki et al. 2012). Another, [MOT+], a prion of Mot3p, was found in a general screen of Q/N-rich proteins (Alberti et al. 2009).
B. Most yeast prions are amyloid filaments
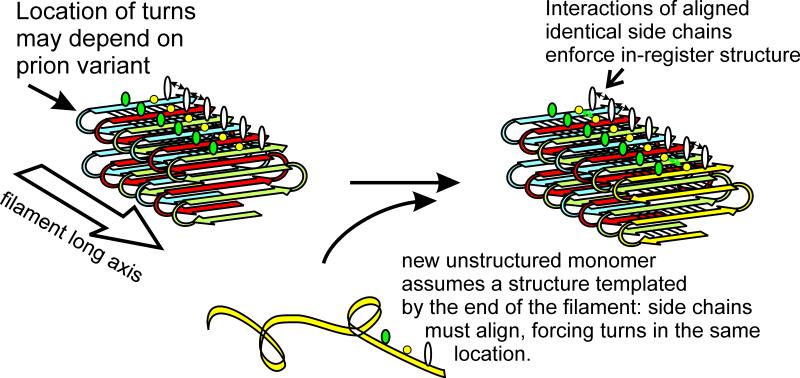
Each of the prions listed in Table 1 is an amyloid of the corresponding protein. Each prion protein has a domain (the prion domain) which is responsible for the prion properties of the whole protein, and constitutes the part of the protein that forms amyloid. The Ure2p prion domain changes from unstructured (Pierce et al. 2005) to beta sheet (Taylor et al. 1999; Baxa et al. 2005), the remainder of the molecule remaining largely unchanged in structure (Bai et al. 2004). Solid-state NMR studies combined with electron microscopic data showed that amyloids of the prion domains of Ure2p, Sup35p and Rnq1p are parallel in-register beta sheets, with the sheets folded length-wise along the long axis of the filaments (Fig. 8) (Shewmaker et al. 2006; Baxa et al. 2007; Wickner et al. 2008).
Fig. 8.
Yeast prion parallel in-register beta sheet architecture and hypothesized mechanism of variant information propagation. Parallel in-register beta sheets are characterized by lines of identical amino acid side chains extending the length of the filaments. These yeast prion beta sheets are known to be folded along the filament long axis as shown. It is proposed that different prion variants have the folds in different locations. The parallel structure is maintained by the favorable interactions of identical side chains. The energetic advantage in these interactions force a new (unstructured) monomer joining the end of the filament to become in-register, thereby insuring that the folds/turns of this monomer are in the same locations as those of previous molecules in the filament. This insures inheritance of prion variant information (conformation) by templating, analogous to DNA templating of sequence information.
C. Prion variants
Remarkably, a single protein, with a single sequence, can be the basis of several (perhaps many) different prion variants, with different biological properties and different amyloid conformations. Most variants are quite stably propagated (though some variants are unstable and are lost or change to another variant). Different prion variants are due to the same protein sequence adopting (and propagating) different amyloid conformations (Bessen and Marsh 1994; Tanaka et al. 2004). How does a prion protein adopt any of several/many different conformations and reliably transmit that conformation to new molecules joining the ends of the filaments? This has long been the central puzzle of prions, and because proteins were believed to be unable to template their conformation, it was argued that prions must have a nucleic acid component, just like other elements of heredity.
D. In-register parallel architecture of prions can explain heritable conformation
The parallel in-register architecture greatly restricts the possible structures. The beta sheet of the prion domain is known, in each case to be folded lengthwise, but the locations of the folds are not known. It is hypothesized that the folds are at different locations in different prion variants, and that the resulting different conformations have different biochemical and biological properties as a result (Wickner et al. 2007; Wickner et al. 2010) (Fig. 8). Most importantly, this hypothesis provides a means to explain how a given architecture can be stably propagated and transmitted to new molecules joining the end of the filament. The same energetically favorable interactions between aligned identical amino acid side chains that keep the structure in-register will direct a newly joining molecule to adopt the same conformation as those already in the filament, and thus to have its turns (folds) at the same locations (Wickner et al. 2007; Wickner et al. 2010) (Fig. 8). There is currently no other model to explain prion variant information propagation. The only prion for which a detailed atomic structure has been determined, that of HET-s of the filamentous fungus Podospora anserina (Saupe 2011), has a beta-helix structure (Wasmer et al. 2008), and this prion does not have prion variants.
E. Chaperones and yeast prions
Because protein conformation is such a central aspect of prions, it has been found that chaperones play several crucial roles in their generation and propagation. Hsp104, acting with Hsp70s and Hsp40s, is important in prion replication (Chernoff et al. 1995), breaking long filaments into shorter ones, perhaps by extracting protein monomers from the middle of the filaments (Paushkin et al. 1996; Ness et al. 2002; Kryndushkin et al. 2003). All of the known amyloid-based yeast prions require Hsp104 for their propagation. Overproduction of Hsp104 cures the [PSI+] prion, apparently by a different, but as yet unknown, mechanism (reviewed by Reidy and Masison 2011), The cytoplasmic Hsp70s, Ssa1p and Ssa2p, as well as the Hsp40s, Ydj1p and Sis1p, are also critical for prion propagation (Newnam et al. 1999; Jung et al. 2000; Moriyama et al. 2000; Sondheimer et al. 2001). Hsp70s are ATPases, whose ATP/ADP states determine the binding and release of substrates. These states are regulated by the Hsp40s, by nucleotide exchange factors and other components, with a clear correlation between ATP/ADP state and prion propagating ability (Sharma and Masison 2009).
F. Transmission barriers: interspecies and intraspecies
Early recognition that sheep scrapie could be transmitted to goats, but only with difficulty, gave rise to the species barrier concept, shown to be largely based on differences between donor and recipient in the prion protein sequence (Prusiner et al. 1990). A similar phenomenon is known in yeast for [PSI+] (Chen et al. 2007) and for [URE3] (Edskes et al. 2011). Indeed, there are even intra-species barriers to transmission of [PSI+] between different polymorphs of Sup35p within S. cerevisiae, suggesting that such barriers have been selected to protect cells from the ill effects of this prion (Bateman and Wickner 2012). It is notable that the species barriers are prion variant - dependent, so that with the same donor - recipient pair of prion protein sequences, transmission may be efficient or near zero, depending on the particular prion variant/amyloid conformation whose transmission is being tested (Edskes et al. 2009; Chen et al. 2010).
VII. Biology of yeast prions
Although yeast prions are proposed to benefit their host (Eaglestone et al. 1999; True and Lindquist 2000), there is as yet no reproducible evidence for this conclusion (Namy et al. 2008). Prion variants differ dramatically in their properties, with the usual mild variants of [PSI+] and [URE3] used in laboratory studies showing little discernible effect on growth, but other, probably more common variants being lethal or nearly so (McGlinchey et al. 2011). To estimate the overall benefit or detriment of yeast prions, a survey of 70 wild strains was carried out. Although the 2 micron DNA plasmid, estimated by two groups to confer a ~1-3% slowing of growth on various strains, was found in 38 of 70 wild strains, none of these strains had [URE3] or [PSI+] and only 11 had [PIN+] (Nakayashiki et al. 2005). This suggests that even the mild variants of each of these prions confers a net overall detriment of >1% on their hosts.
Other lines of evidence indicate that the [URE3] and [PSI+] prions are detrimental. Cells infected with either prion show a stress response with induction of Hsp104 and Hsp70s, indicating that the cells consider prion infection a stress (Jung et al. 2000; Schwimmer and Masison 2002). The prion domains have non-prion functions, a central role in mRNA turnover for the Sup35p prion domain and an important role in stabilization of Ure2p for its prion domain (Hoshino et al. 1999; Hosoda et al. 2003; Shewmaker et al. 2007). Thus, these domains are not conserved to allow prion formation, but for their normal function. In fact, prion formation is not conserved. Although a range of species conserve some sequence in the Ure2p prion domain, several of these are unable to form [URE3] as tested in S. cerevisiae, but some species that do not show sequence conservation can form the prion (Edskes et al. 2011; Safadi et al. 2011). Thus, prion formation appears to be a sporadic condition. Indeed, prion formation occurs stochastically, and often under conditions which make matters worse for the cell, suggesting that it is not adaptive.
VIII. Prospects
Although a large body of work has accumulated on the yeast viruses and prions, many interesting problems remain to be solved. Each of these elements are found in a limited array of wild strains, indicating that they must be a net detriment to the host, probably beyond simply use of some energy and material resources. Elucidation of the nature of these interactions would be of broad interest. The M dsRNAs use the coat supplied by L-A, and several lines of evidence suggests that L-A has first call on its own encoded coat proteins, but clear proof of this is lacking. The newly elucidated cap – stealing by L-A helps explain the ability of viral mRNAs to be translated, but they still lack the 3' polyA, a stringent requirement for translation in eukaryotes. Moreover, the ski1/xrn1 gene was first detected by its 'superkiller' phenotype (Toh-e et al. 1978), suggesting that cap – stealing by L-A is not 100% efficient. Do Ski2,3,8 affect translation or only 3'->5' decay?
How and why are 20S and 23S amplified under sporulation conditions (10,000x!), the phenomenon that led to their first detection (Kadowaki and Halvorson 1971). Can the virus-launching vectors be put to use for biotechnologic purposes? Narnaviruses exist not in the form of a conventional virion structure with a protective capsid, but as ribonucleoprotein complexes in the host cytoplasm. A fine structural analysis of these complexes is essential to understand the unique mode of narnavirus existence. Resting complexes seem to have no host proteins. But, are there any host proteins directly involved in narnavirus replication? 20S and 23S RNAs have strong secondary structures at the 5’ ends. This leads to the question of how p91 and p104 are translated, cap-dependent or –independent? In the latter case, these structures may serve as ribosome entry sites.
While yeast prions seem to have a parallel in-register architecture, the heterogeneity of amyloids made in vitro has so far precluded detailed structures, so that the hypothesis that prion variants differ in the locations of the folds in the sheet (see above) has yet to be verified. Moreover, the basis of the different properties of yeast prions is not yet understood. Yeast prions can be lethal (McGlinchey et al. 2011), but the variety of lethal prion variants and the mechanisms of the pathology they produce are just beginning to be explored. The fungal prion [Het-s] is so far unique in having demonstrated benefit to the cell (Coustou et al. 1997), but it also produces a (pathologic) meiotic drive phenomenon (Dalstra et al. 2003), clouding the issue. Suggestions that yeast prions are beneficial (Eaglestone et al. 1999; True and Lindquist 2000) have met with an enthusiastic reception not yet justified by the data. Yeast prions are important models for the increasingly common amyloidoses and promise to continue to reveal important clues to understanding and dealing with these diseases.
Acknowledgements
R.W. was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases. T.F. and R.E. were supported by Grant BFU2010-15768 from the Spanish Ministry of Education and Science.
References
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Zhou JM, Perrett S. The yeast prion protein Ure2 shows glutathione peroxidase activity in both native and fibrillar forms. J Biol Chem. 2004;279:50025–30. doi: 10.1074/jbc.M406612200. [DOI] [PubMed] [Google Scholar]
- Ball SG, Tirtiaux C, Wickner RB. Genetic control of L-A and L-BC dsRNA copy number in killer systems of Saccharomyces cerevisiae. Genetics. 1984;107:199–217. doi: 10.1093/genetics/107.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman DA, Wickner RB. [PSI+] prion transmission barriers protect Saccharomyces cerevisiae from infection: intraspecies ‘species barriers’. Genetics. 2012;190:569–579. doi: 10.1534/genetics.111.136655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U, Cheng N, Winkler DC, Chiu TK, Davies DR, Sharma D, Inouye H, Kirschner DA, Wickner RB, Steven AC. Filaments of the Ure2p prion protein have a cross-beta core structure. J Struct Biol. 2005;150:170–9. doi: 10.1016/j.jsb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Baxa U, Wickner RB, Steven AC, Anderson D, Marekov L, Yau W-M, Tycko R. Characterization of β-sheet structure in Ure2p1-89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard L, Carroll K, Valle RCP, Wickner RB. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 1998;18:2688–2696. doi: 10.1128/mcb.18.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard L, Carroll K, Valle RCP, Wickner RB. The Ski7 antiviral protein is an EF1-α homolog that blocks expression of non-poly(A) mRNA in Saccharomyces cerevisiae. J. Virol. 1999;73:2893–2900. doi: 10.1128/jvi.73.4.2893-2900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc A, Goyer C, Sonenberg N. The coat protein of the yeast double-stranded RNA virus L-A attaches covalently to the cap structure of eukaryotic mRNA. Molecular and Cellular Biology. 1992;12:3390–3398. doi: 10.1128/mcb.12.8.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc A, Ribas JC, Wickner RB, Sonenberg N. His154 is involved in the linkage of the Saccharomyces cerevisiae L-A double-stranded RNA virus gag protein to the cap structure of mRNAs and is essential for M1 satellite virus expression. Mol. Cell. Biol. 1994;14:2664–2674. doi: 10.1128/mcb.14.4.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin S, Cusack S, Ruigrok RW, Hart DJ. Influenza A virus polymerase: structural insights into replication and host adaptation mechanisms. J. Biol. Chem. 2010;285:28411–28417. doi: 10.1074/jbc.R110.117531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostian KA, Jayachandran S, Tipper DJ. A glycosylated protoxin in killer yeast: models for its structure and maturation. Cell. 1983;32:169–80. doi: 10.1016/0092-8674(83)90507-x. [DOI] [PubMed] [Google Scholar]
- Brown JT, Bai X, Johnson AW. The yeast antiviral proteins Ski2p, Ski3p and Ski8p exist as a complex in vivo. RNA. 2000;6:449–457. doi: 10.1017/s1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JT, Johnson AW. A cis-acting element known to block 3' mRNA degradation enhances expression of polyA-minus mRNA in wild-type yeast cells and phenocopies a ski mutant. RNA. 2001;7:1566–1577. [PMC free article] [PubMed] [Google Scholar]
- Caston JR, Trus BL, Booy FP, Wickner RB, Wall JS, Steven AC. Structure of L-A virus: a specialized compartment for the transcription and replication of double-stranded RNA. J. Cell Biol. 1997;138:975–985. doi: 10.1083/jcb.138.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Bruce KL, Newnam GP, Gyoneva S, Romanyuk AV, Chernoff YO. Genetic and epigenetic control of the efficiency and fidelity of cross-species prion transmission. Mol. Microbiol. 2010;76:1483–1499. doi: 10.1111/j.1365-2958.2010.07177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Newnam GP, Chernoff YO. Prion species barrier between the closely related yeast proteins is detected despite coaggregation. Proc Natl Acad Sci U S A. 2007;104:2791–6. doi: 10.1073/pnas.0611158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B-I, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Chlebowski A, Tomecki R, Lopez ME, Seraphin B, Dziembowski A. Catalytic properties of the eukaryotic exosome. Adv. Exp. Med. Biol. 2011;702:63–78. doi: 10.1007/978-1-4419-7841-7_6. [DOI] [PubMed] [Google Scholar]
- Chu C, Das K, Tyminski JR, Bauman JD, Guan R, Qiu W, Montelione GT, Arnold E, Shatkin AJ. Structure of the guanylyltransferase domain of human mRNA capping enzyme. Proc. Natl. Acad. Sci. USA. 2011;108:10104–10108. doi: 10.1073/pnas.1106610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Bussey H. Characterization of the yeast KEX1 gene product: a carboxypeptidase involved in processing secreted precursor proteins. Mol Cell Biol. 1989;9:2706–14. doi: 10.1128/mcb.9.6.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl. Acad. Sci. USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalstra HJP, Swart K, Debets AJM, Saupe SJ, Hoekstra RF. Sexual transmission of the [Het-s] prion leads to meiotic drive in Podospora anserina. Proc Natl Acad Sci U S A. 2003;100:6616–6621. doi: 10.1073/pnas.1030058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN]. Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Dinman JD, Icho T, Wickner RB. A -1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl. Acad. Sci. U S A. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman JD, Wickner RB. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333:1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Park K-W, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone SS, Cox BS, Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Engel A, McCann LM, Brachmann A, Tsai H-F, Wickner RB. Prion-forming ability of Ure2 of yeasts is not evolutionarily conserved. Genetics. 2011;188:81–90. doi: 10.1534/genetics.111.127217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, McCann LM, Hebert AM, Wickner RB. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics. 2009;181:1159–1167. doi: 10.1534/genetics.108.099929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Ohtake Y, Wickner RB. Mak21p of Saccharomyces cerevisiae, a homolog of human CAATT-binding protein, is essential for 60S ribosomal subunit biogenesis. J. Biol. Chem. 1998;273:28912–28920. doi: 10.1074/jbc.273.44.28912. [DOI] [PubMed] [Google Scholar]
- Esteban LM, Fujimura T, Garcia-Cuellar M, Esteban R. Association of yeast viral 23S RNA with its putative RNA-dependent RNA polymerase. J. Biol. Chem. 1994;269:29771–29777. [PubMed] [Google Scholar]
- Esteban LM, Rodriguez CN, Esteban R. T double-stranded RNA (dsRNA) sequence reveals that T and W dsRNAs form a new RNA family in Saccharomyces cerevisiae. Identification of 23 S RNA as the single-stranded form of T dsRNA. J Biol Chem. 1992;267:10874–81. [PubMed] [Google Scholar]
- Esteban R, Fujimura T. Launching the yeast 23S RNA Narnavirus shows 5' and 3' cis-acting signals for replication. Proc Natl Acad Sci U S A. 2003;100:2568–2573. doi: 10.1073/pnas.0530167100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban R, Fujimura T. In: Recent advances in RNA virus replication. Heffereon KL, editor. Transworld Research Network; Kerala, India: 2006. pp. 171–194. [Google Scholar]
- Esteban R, Fujimura T, Wickner RB. Site-specific binding of viral plus single-stranded RNA to replicase-containing open virus-like particles of yeast. Proc. Natl. Acad. Sci. U S A. 1988;85:4411–4415. doi: 10.1073/pnas.85.12.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban R, Fujimura T, Wickner RB. Internal and terminal cis-acting sites are necessary for in vitro replication of the L-A double-stranded RNA virus of yeast. EMBO J. 1989;8:947–954. doi: 10.1002/j.1460-2075.1989.tb03456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban R, Vega L, Fujimura T. Launching of the yeast 20S RNA narnavirus by expressing the genomic or antigenomic viral RNA in vivo. J. Biol. Chem. 2005;280:33725–33734. doi: 10.1074/jbc.M506546200. [DOI] [PubMed] [Google Scholar]
- Esteban R, Vega L, Fujimura T. 20S RNA narnavirus defies the antiviral activity of SKI1/XRN1 in Saccharomyces cerevisiae. J. Biol. Chem. 2008;283:25812–25820. doi: 10.1074/jbc.M804400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban R, Wickner RB. Three different M1 RNA-containing viruslike particle types in Saccharomyces cerevisiae: in vitro M1 double-stranded RNA synthesis. Mol Cell Biol. 1986;6:1552–61. doi: 10.1128/mcb.6.5.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban R, Wickner RB. A deletion mutant of L-A double-stranded RNA replicates like M1 double-stranded RNA. J. Virol. 1988;62:1278–1285. doi: 10.1128/jvi.62.4.1278-1285.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura T, Esteban R. Bipartite 3'-cis-acting signal for replication in yeast 23S RNA virus and its repair. J. Biol. Chem. 2004;279:13215–13223. doi: 10.1074/jbc.M313797200. [DOI] [PubMed] [Google Scholar]
- Fujimura T, Esteban R. The bipartite 3'-cis-acting signal for replication is required for formation of a ribonucleoprotein complex in vivo between the viral genome and its RNA polymerase in yeast 23 S RNA virus. J. Biol. Chem. 2004;279:44219–44228. doi: 10.1074/jbc.M408530200. [DOI] [PubMed] [Google Scholar]
- Fujimura T, Esteban R. Interactions of the RNA polymerase with the viral genome at the 5' and 3' ends contribute to 20S RNA narnavirus persistence in yeast. J. Biol. Chem. 2007;282:19011–19019. doi: 10.1074/jbc.M702432200. [DOI] [PubMed] [Google Scholar]
- Fujimura T, Esteban R. Yeast double-stranded RNA virus L-A deliberately synthesizes RNA transcripts with 5'-diphosphate. J. Biol. Chem. 2010;285:22911–22918. doi: 10.1074/jbc.M110.138982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura T, Esteban R. Cap-snatching mechanism in yeast L-A double-stranded RNA virus. Proc. Natl. Acad. Sci. USA. 2011;108:17667–17671. doi: 10.1073/pnas.1111900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura T, Esteban R. Cap snatching of yeast L-A double-stranded RNA virus can operate in trans and requires viral polymerase actively engaging in transcription. J. Biol. Chem. 2012;287:12797–12804. doi: 10.1074/jbc.M111.327676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura T, Esteban R, Esteban LM, Wickner RB. Portable encapsidation signal of the L-A double-stranded RNA virus of S. cerevisiae. Cell. 1990;62:819–828. doi: 10.1016/0092-8674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- Fujimura T, Esteban R, Wickner RB. In vitro L-A double-stranded RNA synthesis in virus-like particles from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986;83:4433–4437. doi: 10.1073/pnas.83.12.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura T, Ribas JC, Makhov AM, Wickner RB. Pol of gag-pol fusion protein required for encapsidation of viral RNA of yeast L-A virus. Nature. 1992;359:746–749. doi: 10.1038/359746a0. [DOI] [PubMed] [Google Scholar]
- Fujimura T, Solorzano A, Esteban R. Native replication intermediates of the yeast 20S RNA virus have a single-stranded RNA backbone. J. Biol. Chem. 2005;280:7398–7406. doi: 10.1074/jbc.M412048200. [DOI] [PubMed] [Google Scholar]
- Fujimura T, Wickner RB. Gene overlap results in a viral protein having an RNA binding domain and a major coat protein domain. Cell. 1988;55:663–671. doi: 10.1016/0092-8674(88)90225-5. [DOI] [PubMed] [Google Scholar]
- Fujimura T, Wickner RB. Replicase of L-A virus-like particles of Saccharomyces cerevisiae. In vitro conversion of exogenous L-A and M1 single-stranded RNAs to double-stranded form. J Biol Chem. 1988;263:454–460. [PubMed] [Google Scholar]
- Fujimura T, Wickner RB. Interaction of two cis sites with the RNA replicase of the yeast L-A virus. J Biol Chem. 1992;267:2708–2713. [PubMed] [Google Scholar]
- Fukuhara T, Koga R, Aoki N, Yuki C, Yamamoto N, Oyama N, Udagawa T, Horiuchi H, Miyazaki S, Higashi Y, Takeshita M, Ikeda K, Arakawa M, Matsumoto N, Moriyama H. The wide distribution of endornaviruses, large double-stranded RNA replicons with plasmid-like properties. Arch. Virol. 2006;151:995–1002. doi: 10.1007/s00705-005-0688-5. [DOI] [PubMed] [Google Scholar]
- Garcia-Cuellar MP, Esteban LM, Fujimura T, Rodriguez-Cousino N, Esteban R. Yeast viral 20S RNA is associated with its cognate RNA-dependent RNA polymerase. J. Biol. Chem. 1995;270:20084–20089. doi: 10.1074/jbc.270.34.20084. [DOI] [PubMed] [Google Scholar]
- Garcia-Cuellar MP, Esteban R, Fujimura T. RNA-dependent RNA polymerase activity associated with the yeast viral p91/20S RNA ribonucleoprotein complex. RNA. 1997;3:27–36. [PMC free article] [PubMed] [Google Scholar]
- Garvik B, Haber JE. New cytoplasmic genetic element that controls 20S RNA synthesis during sporulation in yeast. J. Bacteriol. 1978;134:261–269. doi: 10.1128/jb.134.1.261-269.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial SA, Buck KW, Hillman BI, Milne RG. Partitiviruses. In: Fauquet CM, Mayo M, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press; London: 2005. pp. 581–590. [Google Scholar]
- Hakansson K, Doherty AJ, Shuman S, Wigley DB. X-ray crystallography reveals a large conformation change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- Herring AJ, Bevan AE. Virus-like particles associated with the double-stranded RNA species found in killer and sensitive strains of the yeast Saccharomyces cerevisiae. J. Gen. Virol. 1974;22:387–394. doi: 10.1099/0022-1317-22-3-387. [DOI] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3'-poly(A) tail of mRNA. J. Biol. Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- Hosoda N, Kobayashii T, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S, Katada T. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 2003;278:38287–38291. doi: 10.1074/jbc.C300300200. [DOI] [PubMed] [Google Scholar]
- Hu G, Gershon PD, Hodel AE, Quiocho FA. mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains. Proc. Natl. Acad. Sci. USA. 1999;96:7149–7154. doi: 10.1073/pnas.96.13.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icho T, Wickner RB. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J. Biol. Chem. 1989;264:6716–6723. [PubMed] [Google Scholar]
- Jacobs-Anderson JS, Parker R. The 3' to 5' degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3' to 5' exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius D, Brake A, Blair L, Kunisawa R, Thorner J. Isolation of the putative structural gene for the lysine -arginine -cleaving endopeptidase required for the processing of yeast prepro-alpha factor. Cell. 1984;37:1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki K, Halvorson HO. Appearance of a new species of ribonucleic acid during sporulation in Saccharomyces cerevisiae. J. Bacteriol. 1971;105:826–830. doi: 10.1128/jb.105.3.826-830.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Larimer FW, Hsu CL, Maupin MK, Stevens A. Characterization of the XRN1 gene encoding a 5' -> 3' exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted cells. Gene. 1992;120:51–57. doi: 10.1016/0378-1119(92)90008-d. [DOI] [PubMed] [Google Scholar]
- Leibowitz MJ, Wickner RB. A chromosomal gene required for killer plasmid expression, mating, and spore maturation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1976;73:2061–5. doi: 10.1073/pnas.73.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac B, Zhu H, Kubalski A, Zhou XL, Culbertson M, Bussey H, Kung C. Yeast K1 killer toxin forms ion channels in sensitive yeast spheroplasts and in artificial liposomes. Proc Natl Acad Sci U S A. 1990;87:6228–6232. doi: 10.1073/pnas.87.16.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison DC, Blanc A, Ribas JC, Carroll K, Sonenberg N, Wickner RB. Decoying the cap- mRNA degradation system by a dsRNA virus and poly(A)- mRNA surveillance by a yeast antiviral system. Mol. Cell. Biol. 1995;15:2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Fishel R, Wickner RB. Circular single-stranded RNA replicon in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U S A. 1990;87:7628–7632. doi: 10.1073/pnas.87.19.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Wickner RB. Yeast 20 S RNA replicon. Replication intermediates and encoded putative RNA polymerase. J. Biol. Chem. 1991;266:12779–12783. [PubMed] [Google Scholar]
- Maxwell PH, Curcio MJ. Host factors that control long terminal repeat retrotransposons in Saccharomyces cerevisiae: implications for the regulation of mammalian retroviruses. Eukary. Cell. 2007;6:1069–1080. doi: 10.1128/EC.00092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey R, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. Proc. Natl. Acad. Sci. USA. 2011;108:5337–5341. doi: 10.1073/pnas.1102762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome, a conserved eukaryotic RNA processing complex containing multiple 3'->5' exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe D, Jacobson A. mRNA poly(A) tail, a 3' enhancer of translation initiation. Mol. Cell. Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naitow H, Canady MA, Wickner RB, Johnson JE. L-A dsRNA virus at 3.4 Angstroms resolution reveals particle architecture and mRNA decapping mechanism. Nature, Struct. Biol. 2002;9:725–728. doi: 10.1038/nsb844. [DOI] [PubMed] [Google Scholar]
- Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci U S A. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Galopier A, Martini C, Matsufuji S, Fabret C, Rousset C. Epigenetic control of polyamines by the prion [PSI+]. Nat. Cell. Biol. 2008;10:1069–1075. doi: 10.1038/ncb1766. [DOI] [PubMed] [Google Scholar]
- Ness F, Ferreira P, Cox BS, Tuite MF. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss DL. Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 2005;3:632–642. doi: 10.1038/nrmicro1206. [DOI] [PubMed] [Google Scholar]
- Ohtake Y, Wickner RB. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol. Cell. Biol. 1995;15:2772–2781. doi: 10.1128/mcb.15.5.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Lopinski JD, Masuda J, Tzeng T-H, Bruenn JA. A second double-stranded RNA virus from yeast. Virology. 1996;216:451–454. doi: 10.1006/viro.1996.0083. [DOI] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat. Cell Biol. 2009;11:344–349. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Pierce MM, Baxa U, Steven AC, Bax A, Wickner RB. Is the prion domain of soluble Ure2p unstructured? Biochemistry. 2005;44:321–8. doi: 10.1021/bi047964d. [DOI] [PubMed] [Google Scholar]
- Plotch SJ, Bouloy M, Ulmanen I, Krug RM. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- Prusiner SB, Scott M, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, Carlson GA, Hoppe PC, Westaway D, DeArmond SJ. Transgenic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- Ramirez-Garrastacho M, Esteban R. Yeast RNA viruses as indicators of exosome activity: human exosome hCsl4p participates in RNA degradation in Saccharomyces cerevisiae. Yeast. 2011;28:821–832. doi: 10.1002/yea.1909. [DOI] [PubMed] [Google Scholar]
- Reidy M, Masison DC. Modulation and elimination of yeast prions by protein chaperones and co-chaperones. Prion. 2011;5:245–249. doi: 10.4161/pri.5.4.17749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas JC, Fujimura T, Wickner RB. Essential RNA binding and packaging domains of the Gag-Pol fusion protein of the L-A double-stranded RNA virus of Saccharomyces cerevisiae. J. Biol. Chem. 1994;269:28420–28428. [PubMed] [Google Scholar]
- Ridley SP, Sommer SS, Wickner RB. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol. Cell. Biol. 1984;4:761–770. doi: 10.1128/mcb.4.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotech. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Roberts BT, Wickner RB. A class of prions that propagate via covalent auto-activation. Genes Dev. 2003;17:2083–2087. doi: 10.1101/gad.1115803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Cousino N, Esteban R. Both yeast W double-stranded RNA and its single-stranded form 20S RNA are linear. Nucleic Acids Res. 1992;20:2761–2766. doi: 10.1093/nar/20.11.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Cousino N, Maqueda M, Ambrona J, Zamora E, Esteban R, Ramirez M. A new wine Saccharomyces cerevisiae killer toxin (Klus), encoded by a double-stranded RNA virus, with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl. Environ. Microbiol. 2011;77:1822–1832. doi: 10.1128/AEM.02501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Cousino N, Solorzano A, Fujimura T, Esteban R. Yeast positive-strand virus-like RNA replicons. 20S and 23S RNA terminal nucleotide sequences and 3' end secondary structures resemble those of RNA coliphages. J. Biol. Chem. 1998;273:20363–20371. doi: 10.1074/jbc.273.32.20363. [DOI] [PubMed] [Google Scholar]
- Rodriguez CN, Esteban LM, Esteban R. Molecular cloning and characterization of W double-stranded RNA, a linear molecule present in Saccharomyces cerevisiae. Identification of its single-stranded RNA form as 20 S RNA. J. Biol. Chem. 1991;266:12772–12778. [PubMed] [Google Scholar]
- Rogoza T, Goginashvili A, Rodionova S, Ivanov M, Viktorovskaya O, Rubel A, Volkov K, Mironova L. Non-mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc. Natl. Acad. Sci. USA. 2010;107:10573–10577. doi: 10.1073/pnas.1005949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roossinck MJ, Sabanadzovic S, Okada R, Valverde RA. The remarkable evolutionary history of endornaviruses. J. Gen. Virol. 2011;92:2674–2678. doi: 10.1099/vir.0.034702-0. [DOI] [PubMed] [Google Scholar]
- Safadi RA, Talarek N, Jacques N, Aigle M. Yeast prions: could they be exaptations? The URE2/[URE3] system in Kluyveromyces lactis. FEMS Yeast Res. 2011;11:151–153. doi: 10.1111/j.1567-1364.2010.00700.x. [DOI] [PubMed] [Google Scholar]
- Saupe SJ. The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. Sem. Cell & Dev. Biol. 2011;22:460–468. doi: 10.1016/j.semcdb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- Schmitt MJ, Breinig F. Yeast viral killer toxins: lethality and self-protection. Nat. Rev. Microbiol. 2006;4:212–221. doi: 10.1038/nrmicro1347. [DOI] [PubMed] [Google Scholar]
- Schmitt MJ, Klavehn P, Wang J, Schonig I, Tipper DJ. Cell cycle studies on the mode of action of yeast K28 killer toxin. Microbiology. 1996;142:2655–62. doi: 10.1099/00221287-142-9-2655. [DOI] [PubMed] [Google Scholar]
- Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searfoss A, Dever TE, Wickner RB. Linking the 3' poly(A) Tail to the Subunit Joining Step of Translation Initiation: Relations of Pab1p, eIF5B (Fun12p) and Ski2p-Slh1p. Mol. Biol. Cell. 2001;21:4900–4908. doi: 10.1128/MCB.21.15.4900-4908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searfoss AM, Wickner RB. 3' poly(A) is dispensable for translation. Proc. Natl. Acad. Sci. USA. 2000;97:9133–9137. doi: 10.1073/pnas.97.16.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Masison DC. Hsp70 structure, function, regulation and influence on yeast prions. Prot. & Peptide Lett. 2009;16:571–581. doi: 10.2174/092986609788490230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F, Mull L, Nakayashiki T, Masison DC, Wickner RB. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics. 2007;176:1557–1565. doi: 10.1534/genetics.107.074153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc. Natl. Acad. Sci. USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S. Capping enzyme in eukaryotic mRNA synthesis. Prog. Nucleic Acid Res. Mol. Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- Solorzano A, Rodriguez-Cousino N, Esteban R, Fujimura T. Persistent yeast single-stranded RNA viruses exist in vivo as genomic RNA:RNA polymerase complexes in 1:1 stoichiometry. J. Biol. Chem. 2000;275:26428–26435. doi: 10.1074/jbc.M002281200. [DOI] [PubMed] [Google Scholar]
- Sommer SS, Wickner RB. Yeast L dsRNA consists of at least three distinct RNAs; evidence that the non-Mendelian genes [HOK], [NEX] and [EXL] are on one of these dsRNAs. Cell. 1982;31:429–441. doi: 10.1016/0092-8674(82)90136-2. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starheim KK, Gromyko D, Evjenth R, Ryningen A, Varhaug JE, Lillehaug JR, Arnesen T. Knockdown of human Nα-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Mol. Cell. Biol. 2009;29:3569–3581. doi: 10.1128/MCB.01909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner DF, Smeekens SP, Ohagi S, Chan SJ. The new enzymology of precursor processing endoproteases. J. Biol. Chem. 1992;267:23435–23438. [PubMed] [Google Scholar]
- Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336:355–359. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- Tang J, Naitow H, Gardner NA, Kolesar A, Tang L, Wickner RB, Johnson JE. The structural basis of recognition and removal of cellular mRNA 7-methyl G ‘caps’ by a viral capsid protein: a unique viral response to hose defense. J. Mol. Recogn. 2005;18:158–168. doi: 10.1002/jmr.724. [DOI] [PubMed] [Google Scholar]
- Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- Tercero JC, Dinman JD, Wickner RB. Yeast MAK3 N-acetyltransferase recognizes the N-terminal four amino acids of the major coat protein (gag) of the L-A double-stranded RNA virus. J. Bacteriol. 1993;175:3192–3194. doi: 10.1128/jb.175.10.3192-3194.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JC, Riles LE, Wickner RB. Localized mutagenesis and evidence for post-transcriptional regulation of MAK3, a putative N-acetyltransferase required for dsRNA virus propagation in Saccharomyces cerevisiae. J. Bio. Chem. 1992;267:20270–20276. [PubMed] [Google Scholar]
- Tercero JC, Wickner RB. MAK3 encodes an N-acetyltransferase whose modification of the L-A gag N-terminus is necessary for virus particle assembly. J. Biol. Chem. 1992;267:20277–20281. [PubMed] [Google Scholar]
- Toh-e A, Guerry P, Wickner RB. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J. Bacteriol. 1978;136:1002–1007. doi: 10.1128/jb.136.3.1002-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-e A, Sahashi Y. The PET18 locus of Saccharomyces cerevisiae: a complex locus containing multiple genes. Yeast. 1985;1:159–172. doi: 10.1002/yea.320010204. [DOI] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- Vega L. Ph.D. University of Salamanca; Salamanca, Spain: 2010. [Google Scholar]
- Venkatesan S, Gershowitz A, Moss B. Modification of the 5' end of mRNA. Association of RNA triphosphatase with the RNA guanylyltransferase-RNA (guanine-7-)methyltransferase complex from vaccinia virus. J. Biol. Chem. 1980;255:903–908. [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218-279) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- Wesolowski M, Wickner RB. Two new double-stranded RNA molecules showing non-mendelian inheritance and heat inducibility in Saccharomyces cerevisiae. Mol. Cell. Biol. 1984;4:181–187. doi: 10.1128/mcb.4.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. Plasmids controlling exclusion of the K2 killer double-stranded RNA plasmid of yeast. Cell. 1980;21:217–226. doi: 10.1016/0092-8674(80)90129-4. [DOI] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in S. cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc Natl Acad Sci U S A. 2008;105:2403–2408. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat. Rev. Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Shewmaker F, Edskes H, Kryndushkin D, Nemecek J, McGlinchey R, Bateman D, Winchester C-L. Prion amyloid structure explains templating: how proteins can be genes. FEMS Yeast Res. 2010;10:980–991. doi: 10.1111/j.1567-1364.2010.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Toh-e A. [HOK], a new yeast non-Mendelian trait, enables a replication-defective killer plasmid to be maintained. Genetics. 1982;100:159–174. doi: 10.1093/genetics/100.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widner WR, Matsumoto Y, Wickner RB. Is 20S RNA naked? Mol Cell Biol. 1991;11:2905–8. doi: 10.1128/mcb.11.5.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- Young TW, Yagiu M. A comparison of the killer character in different yeasts and its classification. Antonie Van Leeuwenhoek. 1978;44:59–77. doi: 10.1007/BF00400077. [DOI] [PubMed] [Google Scholar]