Abstract
Embryonic Stem Cells (ESCs) and Epiblast Stem Cells (EpiSCs) are the in vitro representatives of naïve and primed pluripotency, respectively. It is currently unclear how their epigenomes underpin the phenotypic and molecular characteristics of these distinct pluripotent states. Here, we performed a genome-wide comparison of DNA methylation between ESCs and EpiSCs by MethylCap-Seq. We observe that promoters are preferential targets for methylation in EpiSC compared to ESCs, in particular high CpG island promoters. This is in line with upregulation of the de novo methyltransferases Dnmt3a1 and Dnmt3b in EpiSC, and downregulation of the demethylases Tet1 and Tet2. Remarkably, the observed DNA methylation signature is specific to EpiSCs and differs from that of their in vivo counterpart, the postimplantation epiblast. Using a subset of promoters that are differentially methylated, we show that DNA methylation is established within a few days during in vitro outgrowth of the epiblast, and also occurs when ESCs are converted to EpiSCs in vitro. Once established, this methylation is stable, as ES-like cells obtained by in vitro reversion of EpiSCs display an epigenetic memory that only extensive passaging and sub-cloning are able to almost completely erase.
Introduction
Two kinds of pluripotent stem cells can be captured ex vivo from the mouse embryo: embryonic stem cells (ESCs) are derived from the inner cell mass (ICM) of the blastocyst, whereas epiblast stem cells (EpiSCs) are isolated from the late epiblast of postimplantation embryos. Although both express the core triad of transcription factors Oct4/Sox2/Nanog, some other pluripotency factors identified in ESCs are absent in EpiSCs, such as Esrrb, Klf4, Rex1/Zfp42, and Dppa3/Stella [1,2]. Moreover, transcriptome comparisons have indicated that EpiSCs may be closer to the postimplantation epiblast, whereas ESCs share more characteristics with ICM cells [3]. Indeed, EpiSCs express late epiblast markers such as Nodal, Fgf5, Brachyury (T), or Cer1, which are low or absent in ESCs [4]. These contrasting signatures suggest these cell types represent two different states of pluripotency, naïve for ESCs and primed for EpiSCs [5]. In contrast to ESCs, EpiSCs are unable to form chimeras following injection into blastocysts [3,6]. However, they can contribute, at least to some extent, to embryo development if injected in the postimplantation epiblast [7].
Different signaling pathways control self-renewal of these pluripotent states: ESCs require LIF and BMP4, while EpiSCs are dependent on FGF2 and Activin/Nodal [3,8]. By switching between the appropriate culture conditions for each cell type, ESCs can be readily converted into EpiSCs, whereas EpiSCs can also be reverted into naïve ES-like cells in vitro albeit with low efficiencies [9–11]. Such interconversion abilities provide new avenues to study the relationships between the two states of pluripotency and have elicited the notion of an “epigenetic barrier” separating ESCs from EpiSCs, since reprogramming EpiSCs into naïve ESCs is an inefficient and long process. In particular, de novo DNA methylation, which takes place during epiblast development [12,13], may be a constituent of this barrier as it is in somatic cell reprogramming [14]. De novo DNA methylation is catalyzed by the de novo methyltransferase 3 (Dnmt3) enzymes [15]. Three have been isolated in mammals: Dnmt3a and Dnmt3b are catalytically active, while Dnmt3l, is a cofactor for both. In the epiblast, Dnmt3b is the first to be expressed at E3.5 while Dnmt3a expression starts 1 day later [16–18]. Dnmt3l is expressed transitorily in the epiblast, between E4.5 and E6.5 [16,18]. The ICM of E3.5 blastocyst is globally hypomethylated, while a massive wave of de novo methylation occurs between the epiblast stages E3.5 and E6.5 [13]. Interestingly, the methylome of ESC is shaped by their culture conditions: ESCs cultured in the presence of the two kinase inhibitors, inhibiting Gsk3 and FGF signaling, respectively, are globally hypomethylated and resemble ICM cells. ESCs cultured using serum conditions display a methylation profile more similar to that of the early postimplantation epiblast [12,19,20]. In contrast, little is known about the methylome of EpiSCs. Quantitatively, the global level of 5-methylcytosine in EpiSCs was found to be similar to that of ESC cultured in serum [20,21]. A recent study by Senner et al. [21] showed that four types of stem cells derived from the mouse embryo, either extra-embryonic or embryonic, contained a unique DNA methylation signature. However, an in-depth comparison of the methylome of ESCs and EpiSCs is currently lacking.
Here, we compare the patterns of DNA methylation in EpiSCs and ESCs and observed a clear bias toward promoter-associated hypermethylation in EpiSCs. By following the kinetics of methylation during ESC to EpiSC conversion, we show that de novo methylation seems to occur very rapidly and concomitant to the molecular switch. Conversely, reversion of EpiSC into ES-like cells shows that the reprogramming of methylation at the promoters is very slow and incomplete, suggesting the persistence of an epigenetic memory. Finally, a comparison of EpiSCs and late epiblast cells reveals that the in vitro and “in embryo” cells show a remarkably different promoter methylation profile.
Materials and Methods
Preparation of samples for sequencing
MethylCap
Genomic DNA was sonicated to generate 300 bp fragments on average. MethylCap was performed using the IP-STAR robot (Diagenode) as described before [22]. In short, 1 μg DNA was incubated with paramagnetic beads coated with the MBD domain of MeCP2 fused to GST. After washing with 200, 400, and 500 mM NaCl, the bound methylated DNA was eluted in two fractions using 600 and 800 mM NaCl, respectively. Twenty nonogram of DNA eluates was prepared for sequencing.
Double-stranded cDNA synthesis
Total RNA was isolated with TRIzol (Invitrogen) according to the manufacturer's recommendations. One hundred microgram total RNA was subjected to two rounds of poly(A) selection (Oligotex mRNA Mini Kit; QIAGEN), followed by DNaseI treatment (QIAGEN). About 100–200 ng mRNA was fragmented by hydrolysis (5× fragmentation buffer: 200 mM Tris acetate, pH8.2, 500 mM potassium acetate, and 150 mM magnesium acetate) at 94°C for 90 s and purified (RNAeasy Minelute Kit; QIAGEN). cDNA was synthesized using 5 μg random hexamers by Superscript III Reverse Transcriptase (Invitrogen). Double-strand cDNA synthesis was performed in second strand buffer (Invitrogen) according to the manufacturer's recommendations and purified (Minelute Reaction Cleanup Kit; QIAGEN).
Sequencing
DNA or cDNA samples were prepared for sequencing by end repair of 20 ng DNA as measured by Qubit (Invitrogen). Adaptors were ligated to DNA fragments, followed by size selection (∼300 bp) and 14 cycles of PCR amplification. Integrity of DNA libraries was confirmed by running the products on a Bioanalyzer (BioRad). Cluster generation and sequencing (36 bp) was performed with the Illumina Genome Analyzer IIx (GAIIx) (MethylCap-seq) or HiSeq (RNA-seq) platform according to standard Illumina protocols. Initial data processing, base calling, and alignment to the mouse reference genome was performed using the Illumina Analysis Pipeline allowing one mismatch. Only tags aligning to one position on the genome were considered for further analysis. For RNA-seq, further analysis was performed with the 36 bp aligned sequence. For MethylCap-seq, the uniquely mapped sequence reads were directionally extended to 300 bp, the estimated median length of the original DNA library. If multiple tags were mapped on the same genomic position, only one was included for further analysis. Mapped reads from the initial 600 and 800 mM NaCl eluate libraries were combined. For both RNA-seq and MethylCap-seq data were converted to Browser Extensible Data files for downstream analysis. To compensate for differences in sequencing depth and mapping efficiency among samples, the total number of unique reads of each sample was uniformly equalized relative to the sample with the lowest number of sequence reads, allowing quantitative comparisons. Wiggle (WIG) files for viewing the data in the UCSC Genome Browser were generated from the normalized files. All sequencing analyses were conducted based on the Mus musculus NCBI m37 genome assembly (MM9; assembly July 2007). Supplementary Table S1 summarizes the sequencing output.
MethylCap-seq analyses
Peak calling
data analysis was performed using in-house generated scripts written in LINUX shell, Perl, and R. Enriched regions (peaks) were called on the basis of a Poisson distribution of overlapping sequence reads within a dynamic window. A false discovery rate (FDR) was calculated relative to the total covered sequence, and peaks with an FDR of ≤1×10−6 were selected. All peaks from the four samples (three EpiSCs and one ESC) were merged. Per sample, the number of normalized sequence reads overlapping each region (peak) of interest was calculated, which is referred to as read density and used as a measure of DNA methylation.
Annotation
methylated peaks were annotated according to their localization in the genome, as intron, exon, promoter (−900 to +400 bp around the gene start), or intergenic based on the Ensembl release 66 (MM9 assembly). As peaks often span different genomic regions, we used the summit genomic coordinate of each peak as representative. Due to the presence of overlapping transcripts in the mouse genome, about 5% of the peaks were annotated to multiple genes. Therefore, peaks were annotated with (at least one) gene name and sequence type (exon, intron, and promoter). Nonassigned peaks were considered as intergenic.
Gene Ontology and KEGG analysis was performed using DAVID (http://david.abcc.ncifcrf.gov) [23,24]. Further data analysis was performed using IPA (Ingenuity® Systems, www.ingenuity.com).
RNA-seq analysis
To obtain RNA-seq gene expression values (RPKM), we used Genomatix (www.genomatix.de).
Statistical analysis of MethylCap seq data
Read counts for all peaks and transcripts were normalized using the normalization procedure used in the Bioconductor package “DESeq” [25]. We used these normalized counts to perform a hierarchical clustering analysis of methylome samples using the distance function 1-c, where c is the correlation coefficient, and the Ward linkage method.
To study the relation between DNA methylation and expression, we built a reduced dataset where methylation peaks corresponding to promoters were combined with read counts for the corresponding transcripts. The following thresholds were used for including genes: methylation read density >0 for either the ESC sample or for at least 2 EpiSC samples.
After log2 transformation of normalized counts, we used the R package “flexclust” [26] to group methylation and expression profiles in clusters. Only clusters having at least 10 profiles and a Pearson correlation coefficient greater than 0.9 (with the center profile of the cluster; radius–that is maximum distance to the cluster center profile=0.1), were included.
Cell lines and culture conditions
Derivation of the 129S2 ESC line was performed as previously described [27]. The rESCs (129S2 rESCs) and ESCs (129S2 ESCs, 129B6 ESCs, and R1) were grown on irradiated mouse embryonic fibroblasts in medium containing DMEM with either 15% serum or (for 129S2 ESCs) 20% KSR (Invitrogen), 0.1 mM β-mercaptoethanol and 1,000 U/mL LIF (ESGRO; Millipore) and plated feeder-free on gelatin-coated dishes for two passages before collection. EpiSCs (129S2 EpiSCs, EpiSC1, 2, and 3 described in Maruotti, 2010 and 129B6 EpiSCs) and cEpiSCs (129S2 cEpiSCs) were grown in serum-free medium (CDM) with FGF2 (12 ng/mL; R&D) and Activin A (20 ng/mL; R&D) on serum-coated dishes as previously described [3]. Conversion of 129S2 ESCs was performed as previously described [9,28]. In summary, the ESCs were trypsinized and 1.5×106–3×106 cells were seeded in 35 mm, serum-coated dishes in CDM+FGF2 and Activin. At day 4, cells were detached with collagenase-II (Sigma) and replated without dilution. This first passage promotes the appearance of flat colonies with typical EpiSC morphology. Converted cells were then cultured for several passages before harvesting. For reversion, EpiSCs were passaged onto mouse irradiated feeders in the presence of ESC medium as described. After 7 days, cells were trypsinized and passaged as ESCs for at least five passages.
Epiblast dissection
E6.5 and E7 epiblasts were dissected from CDI mouse embryos in Flushing and Handling Medium (FHM). The embryonic region was cut out from extra-embryonic tissue and incubated for 10 min in FHM containing 0.1% Trypsin (Type II, Sigma) and 2.5% Pancreatin (Sigma). The epiblast was then isolated using glass needles and either snap-frozen or plated in four-well plate in CDM supplemented with Activin and Fgf2 for EpiSC derivation as described [29].
Real-time PCR analysis
Total RNA was extracted and reverse-transcribed with Superscript III (Invitrogen). Real-Time PCR were carried out using SybrGreen mix (Qiagen) on a Step One Plus thermal cycler (Applied Biosystem) and repeated thrice on independent experiments and/or cell lines. Data were normalized using the geometric mean of Hprt and Pbgd using Qbase software (Biogazelle). Primers used are listed in Supplementary Table S2.
Western-blot analysis
Cellular samples were lysed in 3×Laemli-SDS buffer. The polypeptides were separated through 4%–12% Bis- Tris Gel NuPage electrophoresis and transferred onto a polyvinylidene difluoride membrane (Hybond-P PVDF; Amersham). After blocking with 1/1,000 Tween 20-PBS (PBS-T) containing 4% (w/v) nonfat dried milk, the membranes were incubated with primary antibodies overnight at 4°C. Antibodies used were as follows: mouse monoclonal anti-Dnmt3b (Abcam; 1/1,500 dilution) or anti-Dnmt3a (Active Motif; 1/1,000 dilution). The membranes were washed thrice with PBS-T, incubated with a peroxidase-conjugated anti-mouse antibody and washed again. Peroxidase activity was measured using the ECL-Plus Western Blot detection system (Amersham) and a LAS 1000 camera (Fuji). The membranes were incubated with an anti-Actin antibody for loading control. Band intensities were quantified using ImageJ software (http://imagej.nih.gov/ij/index.html) and normalized using Actin.
DNA methylation analysis by bisulfite sequencing
Genomic DNA was purified using DNA extraction kit (Promega) according to the manufacturer's instructions. For epiblast DNA, pools of seven (E6.5) or three (E7) epiblasts were digested by proteinase K in lysis saline buffer and DNA was extracted using NaCl/EtOH precipitation.
Bisulfite conversion was performed as previously described [30] on 1 μg of genomic DNA for the cells, or all DNA obtained from epiblasts. Regions of interest were then amplified by PCR using the KAPA HiFi HotStart Uracil+ mix (Clinisciences) using primers listed in Supplementary Table S2. The PCR program was as follows: 5 min at 95°C followed by 40 cycles of 20 s at 98°C, 30 s at 60°C, and 15 s at 72°C, with a final extension of 5 min at 72°C. PCR products were directly sequenced or subcloned into pGEMTeasy vector (Promega). Clones were amplified by PCR using the Platinum taq polymerase (Invitrogen) with 5% DMSO at 95°C 15 min followed by 35 cycles of 30 s at 95°C and 3 min at 64°C with a final extension 10 min at 64°C. PCR products of the expected size and quantity were sequenced and analyzed using BiQ Analyzer software [31].
Processing of publicly available datasets for methylation and expression in the epiblast
For comparisons with epiblast methylation, we used profiles of GEO series GSE22831 [12]. Oligo sequences of NimbleGen Mouse Promoter Array (GPL9485) were mapped to the MM9 genome assembly using bowtie2 with a maximum of three mismatches. Sequences mapping on multiple loci in the genome were discarded. For the methylation profiles of the three epiblast (E6.5) replicates within GSE22831 we computed the average log-ratios per NimbleGen probe. For each promoter regions covered by our Methylcap dataset and by the NimbleGen probes (only regions containing ≥5 probes were included), we computed the number of probes, the percentage of probes having an average log-ratio larger than 0.5 and the average log-ratios of all probes within the promoter. The classification of promoters was according to Borgel et al. [12]: low or no methylation (log2ratio <0.3), or highly methylated (log2ratio >0.4).
To determine expression levels for genes showing hypermethylated promoters in EpiSCs, we used profiles within GEO series GSE4622 [32], that is, the microarray data for epiblast at prestreak (two replicates) and mid-streak (three replicates) stages (same stages as used for EpiSC derivation, bisulfite sequencing, and RT-qPCR in this study). A gene was considered to be expressed in one sample in case of a detection P-value<0.05, and considered to be expressed in the epiblast if detected in at least four out of five samples.
Accession numbers
The GEO accession number for the MethylCaq-seq and RNA-seq data for EpiSCs reported in this article is GSE47793. The GEO accession numbers for the previously generated MethylCap-seq and RNA-seq profiles of ESCs are GSE31343 and GSE23943, respectively [33,34]. Raw sequencing data of MeDIP-Seq from Senner et al. [21] were downloaded from the EBI European Nucleotide Archive (ENA) accession number PRJEB4263.
Results
DNA methylation profiles of ESCs and EpiSCs
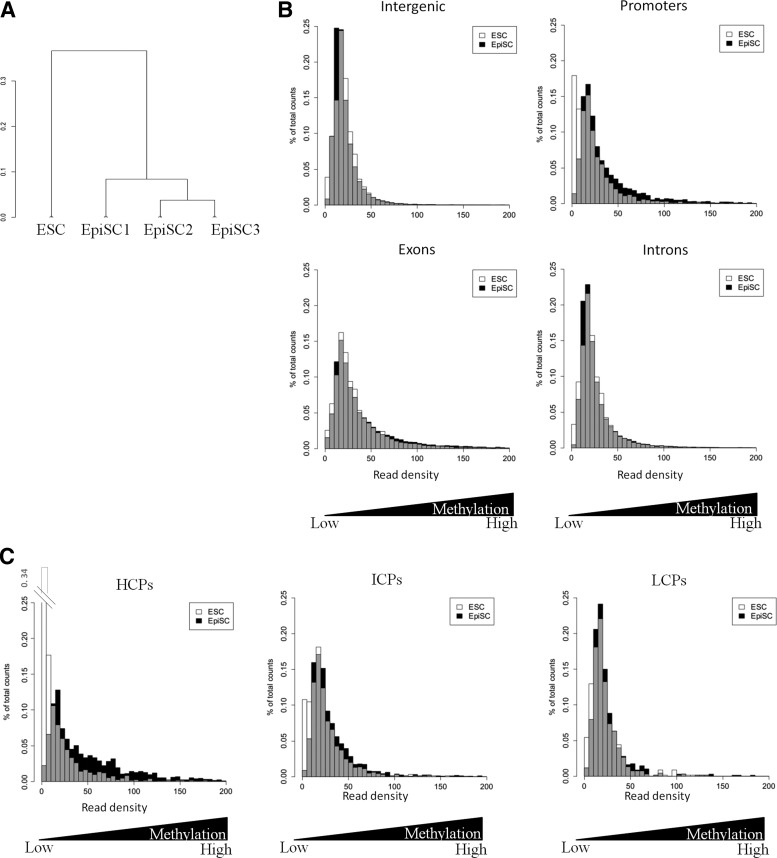
To investigate the DNA methylome of ESCs and EpiSCs at a genome-wide scale, we applied MethylCap-sequencing. This method involves capture of methylated DNA using the MBD domain of MeCP2, followed by parallel sequencing of the captured DNA. In comparison to other genome-wide DNA methylation profiling methods, MethylCap-seq stands out for its robustness, sensitivity, and costeffectiveness [22]. MethylCap-seq was performed on three EpiSC lines (EpiSC1 and 3, male; EpiSC2, female) derived from fertilized B6D2F1 embryos and characterized in [29]. We compared these new profiles with an ESC line (male, E14Tg2a) [33]. Overall a total of 90,474 methylated regions were identified, with a median length of 2,034 bp. These regions are distributed across all chromosomes (Supplementary Fig. S1A). Furthermore, the read density distribution plots of all enriched regions, representative for the level of methylation at the individual loci, in either ESCs or EpiSCs were well overlaid (Supplementary Fig. S1B). This suggests that, overall, DNA methylation is similar in ESCs and EpiSCs at a global level in accordance with Senner et al. [21].
Methylated regions were annotated according to their genomic localization: intergenic, exonic, intronic, and promoter regions. The partition of the methylated regions into these categories was the same in both pluripotent cell types, with an overrepresentation of intragenic methylation compared with the genomic background distribution within the nonrepetitive portion of the genome (Supplementary Fig. S1C). Despite these similarities between ESC and EpiSC methylation, hierarchical clustering clearly shows that the ESC methylome is distinct from that of EpiSCs (Fig. 1A). To get insight into this distinction, we plotted the read density distribution of the regions according to their genomic localization and observed that the read distribution of regions annotated as promoters was shifted toward higher read densities (ie, higher methylation) in EpiSCs compared with ESCs (Fig. 1B). Classifying promoters according to their CpG density [35], high-, intermediate-, and low-CpG content promoters (HCPs, ICPs, and LCPs, respectively), the shift revealed that the higher methylation in EpiSCs is mainly present in HCPs (Fig. 1C), and not so much in ICPs and LCPs.
FIG. 1.
Global analysis of DNA methylation in embryonic stem cell (ESC) and epiblast stem cells (EpiSC) lines. (A) Hierarchical clustering of methylation profiles, based on a Pearson's correlation distance matrix. (B, C) Distribution of read density for ESCs and EpiSCs (mean of the three cell lines) in each genomic category (B) and in promoters annotated as high (HCP), intermediate (ICP), or low (LCP) CpG content (C). Bars for EpiSC are in white, bars for ESC in black, the overlay in gray.
Relation of promoter methylation to gene expression
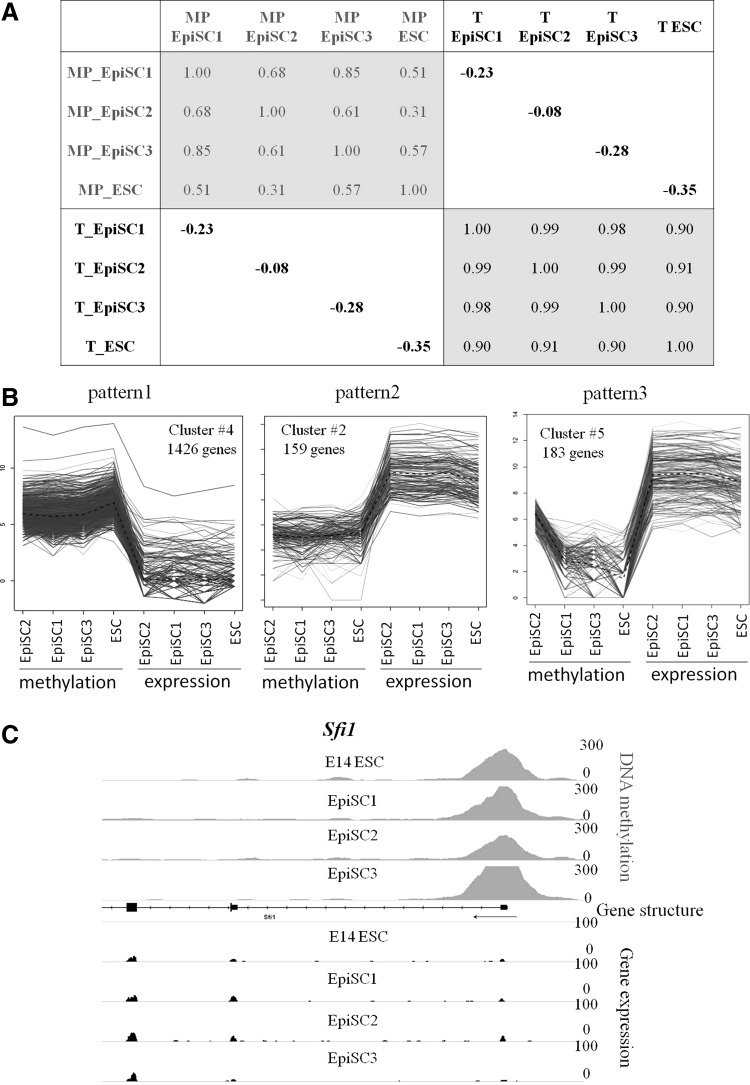
To understand the functional importance of methylation differences in the promoters, we first examined the statistic correlation between DNA methylation at promoters and gene expression in ESCs and EpiSCs. We generated transcriptome profiles of the same three EpiSC lines by RNA-seq and used previously generated RNA-Seq data for the E14 ESC line used in this study [34]. We determined the correlation coefficients between promoter methylation and expression of the corresponding genes (Fig. 2A), followed by a principal component analysis performed on the matrix of these correlations (Supplementary Fig. S2A). Overall, there is a limited variability within EpiSC lines, exhibiting high positive correlations between methylation peaks (0.61–0.85). It is noteworthy that these correlation coefficients are lower between the male lines (EpiSC1 and EpiSC3) and the female one (EpiSC2), reflecting the presence of an inactivated and highly methylated X-chromosome in the female. In addition, although gene expression of the different EpiSC and ESC lines is closely related (between 0.9 and 0.99 correlation), lower correlations were observed between ESC and EpiSC methylation (0.31–0.57). Lastly, an anti-correlation between gene expression and methylation peaks was observed (between −0.08 and −0.35).
FIG. 2.
Relationships between promoter methylation and gene expression. (A) Correlation coefficients between DNA methylation on promoters (MP) and expression data (T). (B) Three main clusters (Qt-clust) illustrating the relation between promoter methylation and gene expression. (C) Example of a gene following pattern 1.
To gain more insight in the anti-correlation between gene expression and DNA methylation we performed quality threshold clustering (QT-Clust; Supplementary Fig S2B), summarized in Fig. 2B. Fifty-eight percent of the genes present in the combined datasets followed pattern 1, that is, showed both high promoter methylation and low expression. Conversely, 29% of the genes followed the opposite pattern (pattern 2), characterized by low promoter methylation and high expression. An example of a gene following pattern 1 is shown on Fig. 2C. However, a few genes seem to escape from methylation-induced repression, as illustrated with Car4 (Supplementary Fig. S2C). Interestingly, pattern 3 characterized by high methylation only in the female EpiSC line (EpiSC2) contained mostly genes located on chromosome X (90%), in agreement with the presence of an inactivated X in this female line. This specificity of the EpiSC2 line probably explains the weaker anti-correlation (−0.08) compared with the others (−0.23 to −0.35).
Functional characterization of methylated promoters
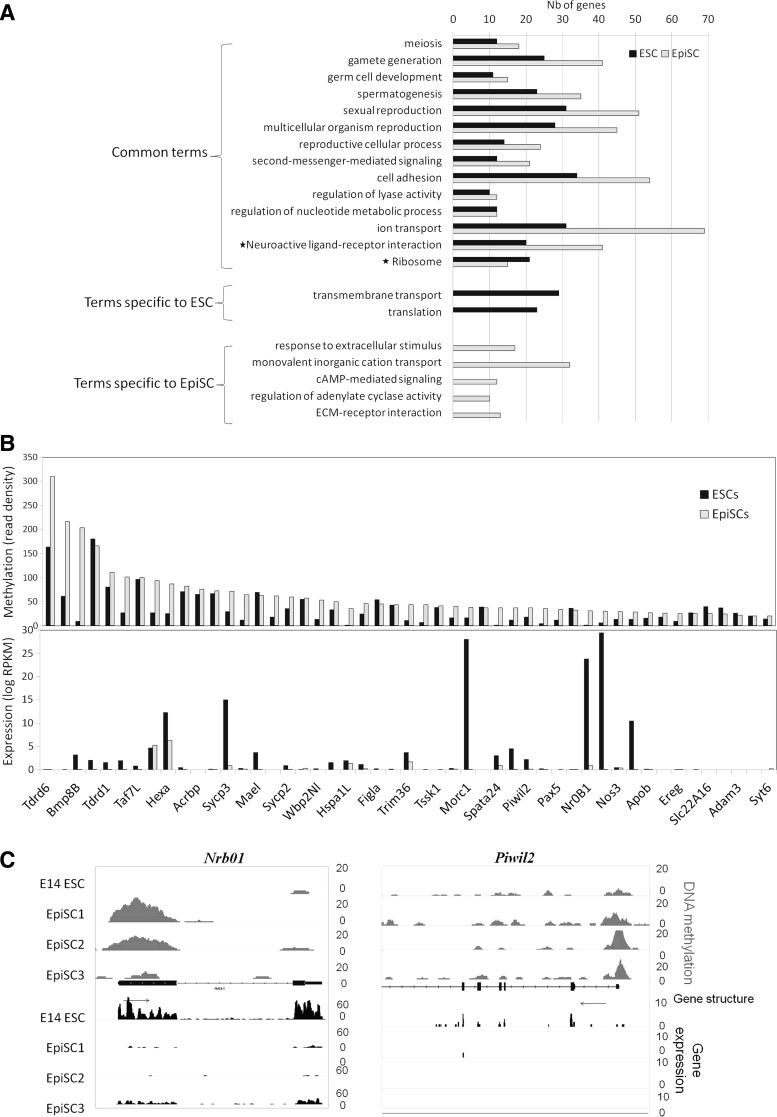
We then investigated the biological functions and pathways associated with genes having methylated promoters, and therefore likely to be repressed in ESCs or EpiSCs. We selected genes containing highly methylated promoters in EpiSCs or ESCs, that is, those with a read density of at least 20, which corresponds to the median of the read density distribution (Fig. 1C and Supplementary Table S3), resulting in 1,528 genes for ESCs and 2,151 for EpiSCs. Gene ontology analysis using DAVID showed only two terms associated with ESC specific methylation: transmembrane transport and translation (Fig. 3A). Terms showing up only in EpiSCs concerned response to endogenous or extracellular stimuli. Most terms (14/21) were common to both cell types, such as germ cell development and reproductive function, ion transport, and cell adhesion. In addition, two Kegg pathways were common to ESCs and EpiSCs: neuroactive ligand-receptor interaction and ribosome. Interestingly, there were always more genes with methylated promoters in EpiSCs belonging to these common terms than for ESCs. This suggests that the repression of certain biological processes initiated in ESCs was amplified in EpiSCs. To further document this trend, we selected genes belonging to two well-represented categories, “sexual reproduction” and “gamete generation” and compared the levels of both DNA methylation and gene expression. Most genes belonging to these categories displayed much lower expression, while higher DNA methylation, in EpiSCs as compared with ESCs (Fig. 3B). It has been reported that several germ cell markers are downregulated in EpiSCs compared with ESCs including Piwil2 and Nr0b1 [6]. We now show that both genes are hypermethylated in EpiSCs compared with ESCs (Fig. 3C).
FIG. 3.
Functional annotation of methylated promoters. (A) GO terms associated with promoters methylated (read density >20) in EpiSCs and in ESCs, according to DAVID analysis (P-value≤1%). The *indicate KEGG pathways. (B) Plots comparing methylation and expression of the 49 genes associated with the union of “gamete generation” and “sexual reproduction” GO terms. (C) IGV browser view of two germline and ESC-specific genes, Nr0b1 and Piwil2, showing the de novo methylation and loss of gene expression in EpiSCs compared to ESCs.
Among genes showing hypermethylated promoters in EpiSCs, Dppa3 (Stella) and Zfp42 (Rex1) have been shown to be methylated at their promoters and their expression repressed in EpiSCs, in contrast to ESCs ([9,36], and Supplementary Fig. S3B for Zfp42). Interestingly, also Tbx3, another “naïve” gene important for ESC maintenance [37], is specifically methylated in EpiSC, with concordant lower expression in EpiSCs as compared with ESCs (Supplementary Fig. S3C). The zygotic promoter of Dnmt3l, which controls its expression during preimplantation stages, was also methylated in EpiSCs (Supplementary Fig. S3D), as is the case in the postimplantation epiblast [16]. Expression of these genes are quickly downregulated during conversion of ESCs to EpiSCs [38]; (see below). Together, our data suggest that DNA methylation at promoters in EpiSCs contributes to the regulation of genes that are differentially expressed in naïve versus primed pluripotent cells.
Identification and characteristics of differentially methylated promoters
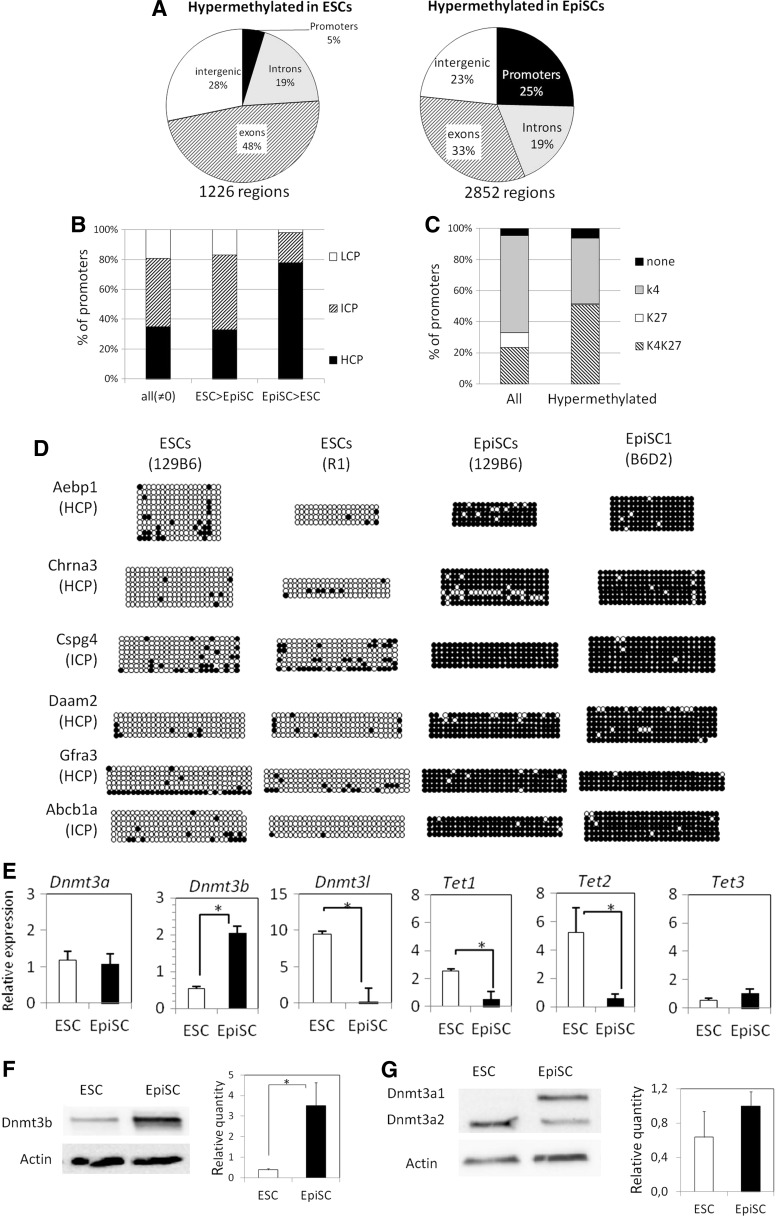
To identify differentially methylated regions (DMRs) between ESCs and EpiSCs, we selected regions displaying a read density ratio of at least three between the two cell types with a minimum of 20 reads for the category with the highest count. This analysis was performed on the whole dataset and revealed 1,226 hypermethylated regions specific for ESCs and twice more (2,852) for EpiSCs. Among these DMRs, 724 (25%) are annotated as promoters specifically hypermethylated in EpiSCs, and only 58 (5%) are promoters that are hypermethylated specifically in ESCs, again illustrating that promoters tend to become hypermethylated in EpiSCs (Fig. 4A). Promoters hypermethylated in EpiSCs were found associated with molecular transport, metabolism, signaling, and nervous system development (Supplementary Fig. S3A). The same analysis performed on the promoters that are hypermethylated in ESCs did not yield significant results because of the low number of genes.
FIG. 4.
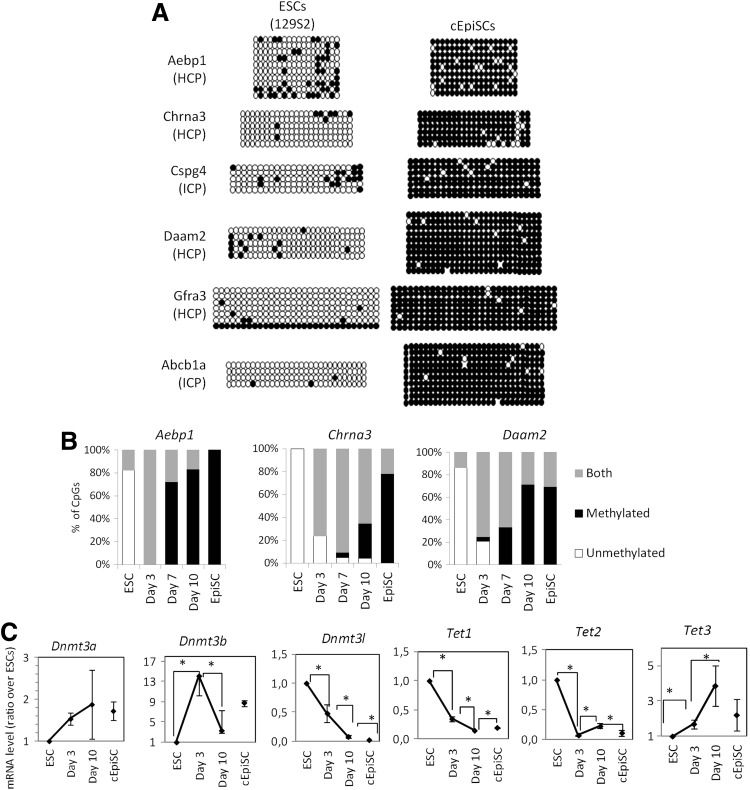
EpiSCs tend to contain hypermethylated promoters. (A) Pie charts showing the classification of hypermethylated regions in ESCs (read density ratio ESC/EpiSC >3) and hypermethylated in EpiSCs (read density ratio EpiSC/ESC >3). The number of DMRs is indicated below the pies. ESC and EpiSC distribution differ significantly (Chi-squared test, P-value<10−53). (B) Classification of all methylated (read density ≠0) and differentially methylated promoters as HCP, ICP, and LCP. Hypermethylated promoters in EpiSCs are significantly enriched in HCP (Chi-squared test, P-value<10−76). (C) Classification of all promoters (read density in EpiSC ≠0) and differentially methylated promoters in EpiSC (ratio EpiSC/ESC >3) according to their association in ESC with H3K4me3 (gray), H3K27me3 (white), bivalent (H3K4me3+H3K27me3, hatched) or neither mark (black). Hypermethylated promoters are significantly different from all promoters (Chi-squared test, P-value<10−22). (D) Validation of differential methylation between EpiSCs and ESCs. The class of each promoter according to their CpG content is indicated. Circles represent CpG nucleotides either methylated (closed) or unmethylated (open). (E) Gene expression of DNA methylation modifying enzymes in ESCs and EpiSCs determined by RT-qPCR. Error bars represent SEM of three different cell lines. (F, G) Western blots showing the protein level of Dnmt3a and Dnmt3b in ESCs and EpiSCs. The average quantity relative to Actin is shown on the right. Error bars represent SEM of two (ESCs) to three (EpiSCs) different cell lines. *in E, F: P<0.05, Mann–Whitney U test.
When CpG density was taken into account, the proportion of each category was identical for the small set of hypermethylated promoters in ESCs, compared to that of all methylated promoters (Fig. 4B). By contrast, only ICPs and HCPs were represented among promoters hypermethylated in EpiSCs, the latter being the most abundant and clearly overrepresented when compared to the population of all methylated promoters (78% compared to 35%).
Using published ChIP-seq data on histone modifications in ESCs [39,40], we determined that half of the hypermethylated promoters in EpiSCs were associated with bivalent domains (H3K4me3/H3K27me3) in ESCs (Fig. 4C), while the remaining were mainly associated with H3K4me3. Hence, these (bivalent) genes are likely stably silenced in EpiSCs by deposition of dense methylation at their promoters.
To validate the current MethylCap-seq, six DMRs in promoters were selected and assessed by bisulfite sequencing. The six genes were chosen among those with hypermethylated promoter and no expression in EpiSCs, and associated with bivalent promoters in ESCs (Supplementary Fig. S4). Of these, Abcb1a is a membrane transporter whereas the others are involved in development [41]. Chrna3, Cspg4, Daam2, and Gfra3 play roles in nervous system development [42–46], while Aebp1 is required for smooth muscle formation [47–50]. We verified the low level of DNA methylation in two different ESC lines, which contrasted with the high DNA methylation level (90%–98% methylated CpGs) in two EpiSC lines (Fig. 4D). These results highly correlated with the MethylCap-seq data.
To better understand the basis of the methylation difference between ESCs and EpiSCs, we assessed the RNA expression of the de novo methyltransferases Dnmt3a, 3b, and 3l, and the Tet enzymes that are involved in active demethylation (Fig. 4E). Dnmt3a is expressed at a similar level in the two cell types, whereas Dnmt3b expression is about four-fold higher in EpiSCs, while Dnmt3l is only expressed in ESCs. Expression of both Tet1 and Tet2 is lower in EpiSCs, while Tet3 expression is low in both cell types. The protein level of both DNMT3A and 3B was evaluated by western-blotting (Fig. 4F, G). In good correlation with transcript level, DNMT3B is greater than eightfold higher in EpiSCs compared with ESCs. As expected, the total quantity of DNMT3A was similar in both cell types, but distributed over two different isoforms: the lower band of ∼75 kDa corresponding to DNMT3A isoform 2 (DNMT3A2, [51]) is higher in ESCs, whereas the higher band (∼100 kDa; DNMT3A1) appeared only in EpiSCs and accounted for about half of the total quantity of DNMT3A in these cells. In conclusion, compared with ESCs, EpiSCs have more abundant DNMT3B and DNMT3A1, whereas Dnmt3l, Tet1, and Tet2 are much lower.
Dynamics and role of DNA methylation changes during conversion of ESCs into cEpiSCs
EpiSCs can be obtained directly in vitro from ESCs by applying EpiSC culture conditions to the ESCs (cEpiSCs, [9,10]. In cEpiSCs harvested 13–15 passages after conversion, the level of expression of the Dnmt3 and Tet genes is very similar to that of embryo-derived EpiSCs (Supplementary Fig. S5A). In line with this, the pattern of DNA methylation of embryo-derived EpiSCs was also correctly apposed in these cEpiSC: a very similar, dense methylation was observed in both EpiSCs and cEpiSCs (88%–98%) for the six promoters described above (Fig. 5A; see Fig. 4D for comparison). During conversion, colonies with an EpiSC-like morphology first appeared at day 6 (Supplementary Fig. S5B), while changes in gene expression levels of ESC markers such as Klf4 and Dppa3, or the upregulation of the EpiSC marker Fgf5, occur as early as at day 3 (Supplementary Fig. S5C). We therefore asked when the de novo methylation occurred during the conversion. To this end, bisulfite sequencing was performed on Aebp1 and Chrna3 promoters at day 3, 7, and 10 of conversion (Fig. 5B). In parallel, we also examined the dynamics of expression of the Dnmt3 and the Tet genes (Fig. 5C). At day 3, sequence polymorphism was present at most CpG loci (Fig. 5B), indicating that de novo methylation had already started, in accordance with the early upregulation of Dnmt3b and the downregulation of Tet1 and Tet2. Dnmt3l, on the other hand, was quickly downregulated and, intriguingly, Tet3 was transiently upregulated during conversion, although remaining at low level. Altogether, our results show that DNA hypermethylation at promoters occurs early in the transition from ESCs to EpiSCs.
FIG. 5.
Characterization of ESC conversion into cEpiSC. (A) Changes of methylation in the six promoters assessed by bisulfite-sequencing during conversion of ESCs into cEpiSCs. (B) Changes in the CpG level of methylation in the promoter of Aebp1, Chrna3, and Daam2 during the conversion process. Genomic DNA after bisulfite conversion was directly sequenced. Each CpG has been classified according to the presence of a C (methylated), a T (unmethylated), or a polymorphism meaning heterogeneity between the two forms in the cell population (both). (C) Changes in expression of DNA methylation modifying enzymes, during the conversion of ESCs determined by RT-qPCR. Bars represent SEM of three independent experiments. *P<0.05, Mann–Whitney U test.
Reprogramming of DNA methylation during reversion of EpiSCs to ESCs
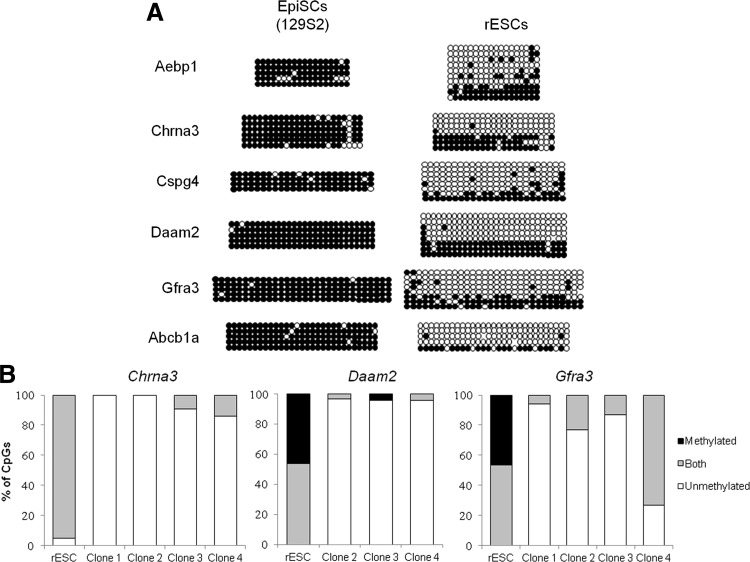
As DNA methylation is considered to be a stable epigenetic mark, we next asked whether reprogramming of EpiSCs toward ESCs would efficiently reverse methylation at promoters. The DNA methylation status at the six promoters as mentioned above was analyzed in rESCs harvested 11–13 passages after the start of reversion by transferring the EpiSCs onto feeders in LIF-containing medium [9]. Although DNA methylation was largely reduced, all six genes displayed a higher level of DNA methylation at their promoters in the rESCs as compared with the embryo-derived ESCs (Fig. 6A). Remarkably, we observed heterogeneity between clones, representing different alleles in the population: some were highly methylated while others were unmethylated as is the case for Daam2. This is in contrast with embryo-derived ESCs in which the DNA methylation level at each allele was quite similar (see Fig. 4D for comparison). We verified that the level of expression of Dnmt3s and Tets was correctly reprogrammed in rESCs compared with embryo-derived ESCs (Supplementary Fig. S6).
FIG. 6.
Characterization of EpiSC reversion into rESC. (A) Changes of methylation in the six promoters assessed by bisulfite-sequencing during reversion of EpiSCs into rESCs. (B) CpG level of methylation in the promoter of Chrna3, Daam2, and Gfra3 after clonal expansion of rESCs. As in Fig. 5B, each CpG is classified as methylated, unmethylated, or polymorphic (both) in the cell population.
Bisulfite sequencing of individual alleles after cloning does not allow distinguishing between allelic heterogeneity within cells or among the cell population, as linkage information between the different fragments is lost. Therefore, we grew four rESC clones originating from single rESCs and determined the methylation status of the three promoters showing the highest methylation heterogeneity (Chrna3, Daam2, and Gfra3; Fig. 6B). Surprisingly, clones were now mostly demethylated, with the notable exception of Gfra3 for clone 4, which still exhibited some methylated CpGs. These results indicate that DNA methylation is stable and resistant to reprogramming although further passages and severe selection by sub-cloning is able to almost, but not totally, erase this epigenetic memory.
Comparison of methylated promoters in EpiSC compared to the epiblast
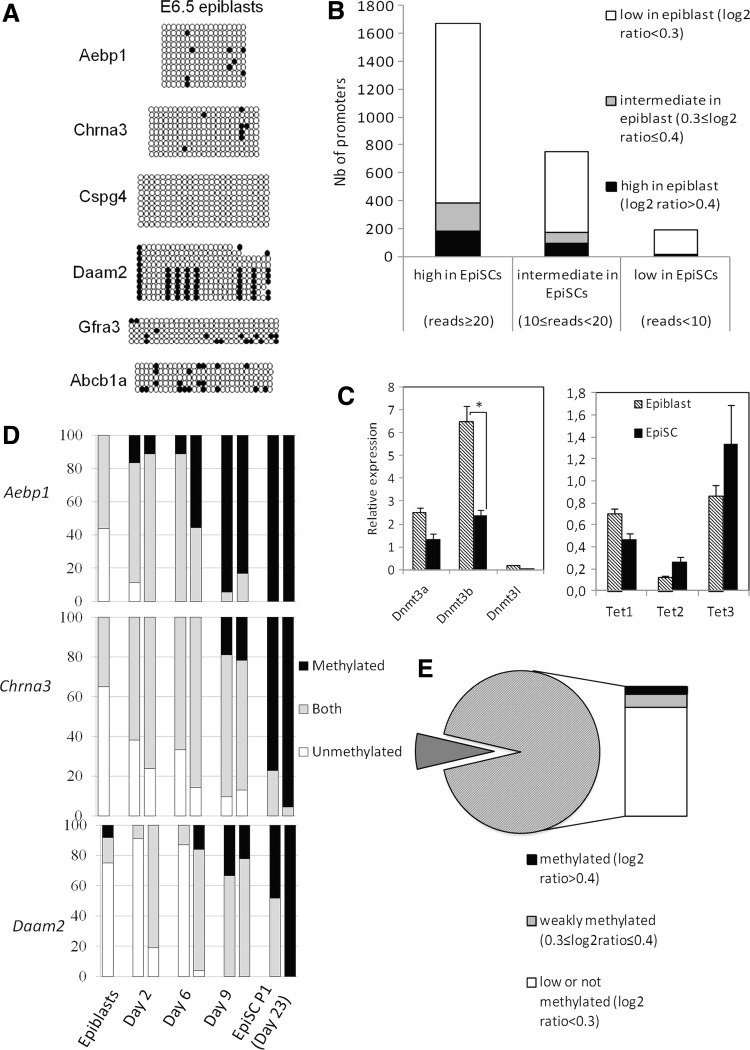
It has been reported that the DNA methylation signature at promoters in ESCs is closer to the early postimplantation epiblast than to the ICM that they are derived from [12]. We now asked how promoter methylation in EpiSCs compared with that of their in vivo counterpart. We isolated epiblasts from early (E6.5) or late (E7) gastrulating embryos and performed bisulfite sequencing on the six promoters that were strongly methylated in EpiSCs. The DNA methylation level in epiblasts was very low at the two stages and even lower than the level of methylation observed in ESCs for Aebp1, Cspg4, and Gfra3 (Fig. 7A and Supplementary Fig. S7).
FIG. 7.
Differential methylation in embryo-derived pluripotent cells and in the epiblast. (A) Methylation status in the epiblast (E6.5). (B) Comparison of methylation in promoters common to Borgel et al. [12]) and our study. Promoters were classified according to their mean methylation status in EpiSCs and each category further separated according to their methylation values in E6.5 epiblasts. (C) Gene expression of DNA methylation modifying enzymes in E6.5 epiblast and EpiSCs determined by RT-qPCR. Bars represent SEM. *P<0.05, Mann–Whitney U test. (D) Changes in the CpG level of methylation in the promoter of Aebp1, Chrna3, and Daam2 during derivation of EpiSCs. Genomic DNA after bisulfite conversion was directly sequenced and the status of the CpGs was indicated as in Figs. 5 and 6. (E) Expression status in the epiblast of genes that contain methylated promoters in EpiSCs.
To perform this analysis at a global scale, we compared our data with the publicly available dataset of promoter methylation on E6.5 epiblasts obtained by MedIP arrays [12] after selection of promoters common to the two study (2,610 promoters; Supplementary Table S4 and Fig. 7B). The majority of promoters (77%, 1,861/2,416) that were methylated in EpiSCs (at least 10 reads) had a low level of methylation in epiblast (log2ratio <0.3). Conversely, very few promoters with low level of methylation in EpiSCs were methylated in the epiblast (8%, 15/194). Methylation at these promoters is therefore unique to EpiSCs, and does not recapitulate the methylation status of the epiblast stage they are derived from. The expression level of Dnmt3s and Tets in epiblast and EpiSCs could not explain this difference in methylation deposition, as these enzymes were similarly expressed, except for Dnmt3b, which is even higher expressed in the epiblast (Fig. 7C).
To get insight into the kinetic of this methylation process during EpiSC derivation, day 6.5 epiblasts were explanted in culture and outgrowths collected at different time points. Bisulfite conversion followed by direct sequencing was performed on three representative promoters, Aebp1, Chrna3, and Daam2 (Fig. 7D). Completely methylated CpGs started to appear as early as day 2 for Aebp1 and before day 9 for Chrna3. For Aebp1, it reached the level of established EpiSCs within 9 days. Such kinetic is not in favor of a slow deposition of methylation along passages in culture but rather suggests that the removal of the epiblast from its in vivo environment and/or the culture conditions may have released constraints that prevent promoter hypermethylation within the embryo.
Lastly, to gain insight into the functional consequence of the differential DNA methylation, we asked whether the difference in promoter DNA methylation between EpiSCs and epiblast cells translated into differences in expression of the corresponding genes. Using available microarray data on expression in the epiblast [32], we determined the expression status (expressed or not expressed) of genes in the epiblast with contain hypermethylated promoters in EpiSCs (reads ≥20) (Fig. 7E and Supplementary Table S4). Most genes (93%, 554/593) were not expressed in the epiblast, although being largely unmethylated. This suggests that for a large set of genes that are repressed in both the epiblast and their in vitro counterparts (the EpiSCs), the epigenetic mechanism of repression is different.
Discussion
We have compared the DNA methylome of EpiSCs to that of ESCs and to their tissue of origin, the postimplantation epiblast. ESCs and EpiSCs have similar methylation levels and a similar distribution of methylation within the different genomic regions. However, several features distinguish the two cell types, in particular the fact that there are significantly more regions specifically methylated in EpiSCs as compared to ESCs, a large part of those being hypermethylated HCP promoters. These promoters are mostly associated with either a bivalent signature (H3K4me3 and H3K27me3) or an active H3K4me2/me3 mark in the ESCs [39,40]. Furthermore, our study suggests that the promoter methylation pattern is quickly established during the in vitro conversion of ESCs into EpiSCs.
To further validate our findings on an independent dataset, we re-examined the MeDIP-seq data generated by Senner et al. [21]. This analysis yielded the same results as with our dataset: a larger set of hypermethylated, high-CpG content promoters is present in EpiSCs compared with ESCs, and these promoters are associated with a bivalent signature in ESCs (Supplementary Fig S8A, B). GO term analysis of methylated promoters in both cell types also shows overlap with our analysis (see Fig. 3A), with a prominent targeting of methylation toward germline associated promoters (Supplementary Fig. S8C). In addition, a recent report by Hackett et al. shows an increase of methylcytosine at promoters in EpiSC [52]. Together, these two independent analyses performed on ESC and EpiSCs of different origins with different profiling techniques show that the epigenome of the primed EpiSCs clearly differ from that of the naïve ESCs.
The differences in promoter methylation in the two pluripotent cell types reported here are well correlated with the differences in expression of the enzymes involved in the control of DNA methylation. We show that Dnmt3b is more abundant in EpiSCs compared with ESCs, which in particular could explain the increased deposition of methylation at promoters of germline genes [53]. The expression of the two main Tet enzymes involved in active demethylation and present in ESCs, Tet1 and Tet2, are largely downregulated in EpiSCs. Interestingly, TET1 has been suggested to help in maintaining a demethylated state at bivalent promoters [54,55]. Dnmt3l, a cofactor of Dnmt3a/3b, also rapidly decreases during ESC-EpiSC conversion and remains low in EpiSCs. A recent study have shown that DNMT3L interacts with PRC2, the polycomb complex that tri-methylates H3K27, which helps in maintaining the bivalent domains free of DNA methylation by preventing their access by DNMT3A/3B [56]. Although the total quantity of DNMT3A remains constant in EpiSCs and ESCs, an additional splicing isoform, DNMT3A1, is present in EpiSCs [51]. Interestingly, this isoform is also upregulated upon retinoic acid ESC differentiation and exhibits distinct gene targets [57]. Altogether these data provide a functional explanation for the preferential deposition of DNA methylation in EpiSC at sites that show bivalent histone marks in ESCs.
Our study indicates that ESCs obtained by reversion of EpiSCs show persistent methylation at some promoters, which is very difficult to erase. Even after extended culture and stringent selection, a residual methylation apparently persists at some alleles and some CpGs. Culture of the revertant cells at clonal density excludes the presence of any residual EpiSCs that would die in our assay when passaged as single cells [3]. Persistence of residual methylation has been previously observed in somatic cells reprogrammed using defined factors (iPSCs). It was recognized as an epigenetic memory, which could be erased at high passages [58–60]. Reverted ESCs generated using the same protocol as ours have been shown to be transcriptionally similar to ESCs, even after a few passages, and able to give rise to germline-competent chimeras [9], suggesting that the residual methylation does not impair the establishment of naïve pluripotency. Reversion of EpiSC to ESCs has been successfully used as a model system to seek for factors that facilitate reprogramming [61]. We propose that a quick and complete erasure of DNA methylation at promoters would be a good criterion for reprogramming efficiency.
The comparison of our dataset of methylated promoters in EpiSCs with the dataset on epiblast cells [12] revealed that many methylated promoters in EpiSCs are poorly methylated in the epiblast. Hence, hypermethylation at promoters appears also to be a distinguishing feature of EpiSCs compared to their in vivo counterpart. During epiblast culture, we observed a rapid de novo methylation at the tested promoters, within 3 days, similar to deposition of methylation during conversion of ESCs into EpiSCs. Although the difference in promoter methylation in epiblast and EpiSC does not have an immediate consequence in terms of gene expression differences, it nevertheless implies that the epigenetic regulation of EpiSCs differs from that of their tissue of origin, the epiblast. It is possible that in vivo the deposition of methylation is controlled by external factors that are relieved when the epiblast is explanted in vitro without any surrounding tissues. Indeed, when the epiblast is dissociated into single cells and cultured on feeders with LIF and serum-containing medium, both Rex1 and Dppa3 promoters, initially demethylated in the epiblast, become transiently methylated, as they are in EpiSCs [9]. Culture conditions used for EpiSCs required two active signaling pathways, Activin/Nodal and FGF [29,62,63], which may be involved in the stimulation of the hypermethylation in EpiSCs. Interestingly, when ESCs are transferred from serum-containing medium to serum-free medium containing MEK and GSK3 inhibitors, they become extensively demethylated [19,20]. This is also the case for reverted ESCs, which become more demethylated when grown in the same serum-free conditions (data not shown). Several studies have highlighted a complex crosstalk between FGF signaling, Prdm14, Dnmt3b, and Tet that could play an inductive role in this process [19,64,65]. In vivo, FGF signaling activity as revealed by phosphorylated Erk1/2 is low in the pregastrulation epiblast cells but becomes activated upon derivation in the presence of FGF2-containing medium [62,66]. As shown in this study, the conversion of ESCs into EpiSCs provides an excellent system to further decipher the role of these various components in the regulation of DNA methylation.
In conclusion, our study shows that EpiSCs have a specific DNA methylome signature, in particular at promoters. It differs from both ESCs and from the epiblast they originate from, and cannot be easily erased by reprogramming EpiSCs to a more naïve ESC-like state through modulation of the culture conditions. The rapid molecular and epigenetic changes during the first days of ESC-to-EpiSC conversion make it an interesting system to further study the role of DNA methylation in the transition from naïve to a primed state of pluripotency.
Supplementary Material
Acknowledgments
We thank E.M. Janssen-Megens and Y. Tan for technical support with sequencing. Femke Simmer and Arjen Brinkman helped with the analysis of MethylCap-seq data. We are grateful to Hélène Jammes and Sylvie Ruffini for technical help in bisulfite sequencing and western-blotting, respectively. Many thanks to Gabriel Brons for the derivation of 129S2 EpiSC. The research leading to these results has received funding from PluriSys (FP7/2009: 223485) and by ANR Programme Investissements d'Avenir REVIVE. ACV and MT are recipients of a PhD fellowship from DIM STEM Pole and DIM Biotherapies Ile-de-France. HM is supported by a grant from The Netherlands Organization for Scientific Research (NWO-VIDI 864.12.007). ASB was part sponsored by the British Heart Foundation (FS/12/37/29516).
Author Disclosure Statement
No financial competing interest exists.
References
- 1.Hutchins AP, Choo SH, Mistri TK, Rahmani M, Woon CT, Ng CK, Jauch R. and Robson P. (2013). Co-motif discovery identifies an Esrrb-Sox2-DNA ternary complex as a mediator of transcriptional differences between mouse embryonic and epiblast stem cells. Stem Cells 31:269–281 [DOI] [PubMed] [Google Scholar]
- 2.Nichols J. and Smith A. (2012). Pluripotency in the embryo and in culture. Cold Spring Harb Perspect Biol 4:a008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brons IGM, Smithers LE, Trotter MWB, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA. and Vallier L. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448:191–195 [DOI] [PubMed] [Google Scholar]
- 4.Chenoweth JG, McKay RDG. and Tesar PJ. (2010). Epiblast stem cells contribute new insight into pluripotency and gastrulation. Dev Growth Differ 52:293–301 [DOI] [PubMed] [Google Scholar]
- 5.Nichols J. and Smith A. (2009). Naive and primed pluripotent states. Cell Stem Cell 4:487–492 [DOI] [PubMed] [Google Scholar]
- 6.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL. and McKay RDG. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448:196–199 [DOI] [PubMed] [Google Scholar]
- 7.Huang Y, Osorno R, Tsakiridis A. and Wilson V. (2012). In Vivo differentiation potential of epiblast stem cells revealed by chimeric embryo formation. Cell Reprog 2:1571–1578 [DOI] [PubMed] [Google Scholar]
- 8.Ying QL, Nichols J, Chambers I. and Smith A. (2003). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115:281–292 [DOI] [PubMed] [Google Scholar]
- 9.Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K. and Surani MA. (2009). Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 461:1292–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W. and Smith A. (2009). Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136:1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou HY, Li WL, Zhu SY, Joo JY, Do JT, Xiong W, Kim JB, Zhang K, Scholer HR. and Ding S. (2010). Conversion of mouse epiblast stem cells to an earlier pluripotency state by small molecules. J Biol Chem 285:29676–29680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T. and Weber M. (2010). Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet 42:1093–1100 [DOI] [PubMed] [Google Scholar]
- 13.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A. and Meissner A. (2012). A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484:339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES. and Meissner A. (2008). Dissecting direct reprogramming through integrative genomic analysis. Nature 454:49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okano M, Bell DW, Haber DA. and Li E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247–257 [DOI] [PubMed] [Google Scholar]
- 16.Guenatri M, Duffie R, Iranzo J, Fauque P. and Bourc'his D. (2013). Plasticity in Dnmt3L-dependent and -independent modes of de novo methylation in the developing mouse embryo. Development 140:562–572 [DOI] [PubMed] [Google Scholar]
- 17.Hirasawa R. and Sasaki H. (2009). Dynamic transition of Dnmt3b expression in mouse pre- and early post-implantation embryos. Gene Exp Patterns 9:27–30 [DOI] [PubMed] [Google Scholar]
- 18.Hu Y-G, Hirasawa R, Hu J-L, Hata K, Li C-L, Jin Y, Chen T, Li E, Rigolet M, et al. (2008). Regulation of DNA methylation activity through Dnmt3L promoter methylation by Dnmt3 enzymes in embryonic development. Hum Mol Gen 17:2654–2664 [DOI] [PubMed] [Google Scholar]
- 19.Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, et al. (2013). Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol 20:311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habibi E, Brinkman AB, Arand J, Kroeze LI, Kerstens HH, Matarese F, Lepikhov K, Gut M, Brun-Heath I, et al. (2013). Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 13:360–369 [DOI] [PubMed] [Google Scholar]
- 21.Senner CE, Krueger F, Oxley D, Andrews S. and Hemberger M. (2012). DNA methylation profiles define stem cell identity and reveal a tight embryonic-extraembryonic lineage boundary. Stem Cells 30:2732–2745 [DOI] [PubMed] [Google Scholar]
- 22.Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG. and Meissner A. (2010). Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol 28:1106–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang DW, Sherman BT. and Lempicki RA. (2008). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols 4:44–57 [DOI] [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT. and Lempicki RA. (2009). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucl Acids Res 37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anders S. and Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leisch F. (2006). A toolbox for -centroids cluster analysis. Comput Stat Data Anal 51:526–544 [Google Scholar]
- 27.Bryja V, Bonilla S, Cajanek L, Parish CL, Schwartz CM, Luo Y, Rao MS. and Arenas E. (2006). An efficient method for the derivation of mouse embryonic stem cells. Stem Cells 24:844–849 [DOI] [PubMed] [Google Scholar]
- 28.Jouneau A, Ciaudo C, Sismeiro O, Brochard V, Jouneau L, Vandormael-Pournin S, Coppee JY, Zhou Q, Heard E, Antoniewski C. and Cohen-Tannoudji M. (2012). Naive and primed murine pluripotent stem cells have distinct miRNA expression profiles. RNA 18:253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maruotti J, Dai XP, Brochard V, Jouneau L, Liu J, Bonnet-Garnier A, Jammes H, Vallier L, Brons IG, et al. (2010). Nuclear transfer-derived epiblast stem cells are transcriptionally and epigenetically distinguishable from their fertilized-derived counterparts. Stem Cells 28:743–752 [DOI] [PubMed] [Google Scholar]
- 30.Dupont JM, Tost J, Jammes H. and Gut IG. (2004). De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal Biochem 333:119–127 [DOI] [PubMed] [Google Scholar]
- 31.Bock C, Reither S, Mikeska T, Paulsen M, Walter J. and Lengauer T. (2005). BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 21:4067–4068 [DOI] [PubMed] [Google Scholar]
- 32.Kojima Y, Kaufman-Francis K, Studdert JB, Steiner KA, Power MD, Loebel DA, Jones V, Hor A, de Alencastro G, et al. (2014). The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell 14:107–120 [DOI] [PubMed] [Google Scholar]
- 33.Matarese F, Carrillo-de Santa Pau E. and Stunnenberg HG. (2011). 5-Hydroxymethylcytosine: a new kid on the epigenetic block?. Mol Syst Biol 7:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Francis Stewart A, Smith A. and Stunnenberg HG. (2012). The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149:590–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M. and Schubeler D. (2007). Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39:457–466 [DOI] [PubMed] [Google Scholar]
- 36.Hayashi K, Lopes SMCdS, Tang F. and Surani MA. (2008). Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3:391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niwa H, Miyazaki J. and Smith AG. (2000). Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24:372–376 [DOI] [PubMed] [Google Scholar]
- 38.Hayashi K, Ohta H, Kurimoto K, Aramaki S. and Saitou M. (2011). Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146:519–532 [DOI] [PubMed] [Google Scholar]
- 39.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, et al. (2008). Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454:766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawicki WT, Kujawa M, Jankowska-Steifer E, Mystkowska ET, Hyc A. and Kowalewski C. (2006). Temporal/spatial expression and efflux activity of ABC transporter, P-glycoprotein/Abcb1 isoforms and Bcrp/Abcg2 during early murine development. Gene Expr Patterns 6:738–746 [DOI] [PubMed] [Google Scholar]
- 42.Atluri P, Fleck MW, Shen Q, Mah SJ, Stadfelt D, Barnes W, Goderie SK, Temple S. and Schneider AS. (2001). Functional nicotinic acetylcholine receptor expression in stem and progenitor cells of the early embryonic mouse cerebral cortex. Dev Biol 240:143–156 [DOI] [PubMed] [Google Scholar]
- 43.Kida Y, Shiraishi T. and Ogura T. (2004). Identification of chick and mouse Daam1 and Daam2 genes and their expression patterns in the central nervous system. Brain Res Dev Brain Res 153:143–150 [DOI] [PubMed] [Google Scholar]
- 44.Nakaya MA, Habas R, Biris K, Dunty WC, Jr., Kato Y, He X. and Yamaguchi TP. (2004). Identification and comparative expression analyses of Daam genes in mouse and Xenopus. Gene Expr Patterns 5:97–105 [DOI] [PubMed] [Google Scholar]
- 45.Nishiyama A, Lin XH, Giese N, Heldin CH. and Stallcup WB. (1996). Co-localization of NG2 proteoglycan and PDGF α-receptor on O2A progenitor cells in the developing rat brain. J Neurosci Res 43:299–314 [DOI] [PubMed] [Google Scholar]
- 46.Widenfalk J, Tomac A, Lindqvist E, Hoffer B. and Olson L. (1998). GFRalpha-3, a protein related to GFRalpha-1, is expressed in developing peripheral neurons and ensheathing cells. Eur J Neurosci 10:1508–1517 [DOI] [PubMed] [Google Scholar]
- 47.Ith B, Wei J, Yet SF, Perrella MA. and Layne MD. (2005). Aortic carboxypeptidase-like protein is expressed in collagen-rich tissues during mouse embryonic development. Gene Expr Patterns 5:533–537 [DOI] [PubMed] [Google Scholar]
- 48.Layne MD, Yet SF, Maemura K, Hsieh CM, Bernfield M, Perrella MA. and Lee ME. (2001). Impaired abdominal wall development and deficient wound healing in mice lacking aortic carboxypeptidase-like protein. Mol Cell Biol 21:5256–5261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kucharova K. and Stallcup WB. (2010). The NG2 proteoglycan promotes oligodendrocyte progenitor proliferation and developmental myelination. Neuroscience 166:185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J, Klein C, Liang C, Rauch R, Kawamura K. and Hsueh AJ. (2009). Autocrine regulation of early embryonic development by the artemin-GFRA3 (GDNF family receptor-alpha 3) signaling system in mice. FEBS Lett 583:2479–2485 [DOI] [PubMed] [Google Scholar]
- 51.Chen T, Ueda Y, Xie S. and Li E. (2002). A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem 277:38746–38754 [DOI] [PubMed] [Google Scholar]
- 52.Hackett Jamie A, Dietmann S, Murakami K, Down TA, Leitch HG. and Surani MA. (2013). Synergistic mechanisms of DNA demethylation during transition to ground-state pluripotency. Stem Cell Rep 1:518–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velasco G, Hube F, Rollin J, Neuillet D, Philippe C, Bouzinba-Segard H, Galvani A, Viegas-Pequignot E. and Francastel C. (2010). Dnmt3b recruitment through E2F6 transcriptional repressor mediates germ-line gene silencing in murine somatic tissues. Proc Natl Acad Sci U S A 107:9281–9286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams K, Christensen J. and Helin K. (2012). DNA methylation: TET proteins—guardians of CpG islands?. EMBO Rep 13:28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE. and Zhang Y. (2011). Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25:679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neri F, Krepelova A, Incarnato D, Maldotti M, Parlato C, Galvagni F, Matarese F, Stunnenberg HG. and Oliviero S. (2013). Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA Methylation at gene bodies in ESCs. Cell 155:121–134 [DOI] [PubMed] [Google Scholar]
- 57.Kotini AG, Mpakali A. and Agalioti T. (2011). Dnmt3a1 upregulates transcription of distinct genes and targets chromosomal gene clusters for epigenetic silencing in mouse embryonic stem cells. Mol Cell Biol 31:1577–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, et al. (2009). Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5:111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buecker C, Chen H-H, Polo JM, Daheron L, Bu L, Barakat TS, Okwieka P, Porter A, Gribnau J, Hochedlinger K. and Geijsen N. (2010). A Murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell 6:535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, et al. (2010). Epigenetic memory in induced pluripotent stem cells. Nature 467:285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, et al. (2013). Deterministic direct reprogramming of somatic cells to pluripotency. Nature 502:65–70 [DOI] [PubMed] [Google Scholar]
- 62.Greber B, Wu G, Bernemann C, Joo JY, Han DW, Ko K, Tapia N, Sabour D, Sterneckert J, Tesar P. and Scholer HR. (2010). Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell 6:215–226 [DOI] [PubMed] [Google Scholar]
- 63.Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MW, Cho CH, Martinez A, et al. (2009). Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development 136:1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ficz G, Hore TA, Santos F, Lee HJ, Dean W, Arand J, Krueger F, Oxley D, Paul Y-L, et al. (2013). FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell 13:351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grabole N, Tischler J, Hackett JA, Kim S, Tang F, Leitch HG, Magnusdottir E. and Surani MA. (2013). Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO Rep 14:629–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Corson LB, Yamanaka Y, Lai KM. and Rossant J. (2003). Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development 130:4527–4537 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.