This work reports that the expression of HEN1, a small regulatory RNA methyltransferase essential for microRNA biogenesis, is activated by light signaling pathways to regulate Arabidopsis photomorphogenesis. The expression of positive (HY5) and negative (TCPs) regulators is tuned by miR157d and miR319, respectively, in deetiolating seedlings, demonstrating a posttranscriptional control in the photomorphogenic development.
Abstract
Light regulates growth and developmental processes in plants via global transcriptome adjustment, translational control, and multilayered posttranslational modification of proteins. The transcriptional activation and repression of light-responsive genes has been well documented; however, the impact of posttranscriptional regulation on conveying light signals has been less addressed. Here, we examined whether optimal photomorphogenesis in Arabidopsis thaliana requires the proper biogenesis of small regulatory RNAs that play pivotal roles in the posttranscriptional regulation of gene expression. Arabidopsis carrying a mutation in HUA ENHANCER1 (HEN1), required for stabilization of small regulatory RNAs, showed defects in multiple aspects of photomorphogenic and skotomorphogenic development. HEN1 negatively regulated Arabidopsis photomorphogenesis. Light-activated HEN1 expression depended on the photoreceptors phytochrome A (phyA), phyB, cryptochrome 1 (cry1), and cry2 and key transcriptional regulators ELONGATED HYPOCOTYL5 (HY5) and HY5-HOMOLOG. We also demonstrate the involvement of the small regulatory RNAs miR157d and miR319 in modulating the expression of a positive regulator, HY5, and negative regulators TEOSINTE BRANCHED1, CYCLOIDEA AND PCF family proteins, respectively, for optimal photomorphogenic development in Arabidopsis.
INTRODUCTION
Light is an important and dynamic environmental cue for many programmed growth and developmental processes in plants. The process of photomorphogenesis has been widely used for studies of light sensing and signaling pathways regulated at multiple levels. First, with light treatment, the photoreceptors or key light signaling molecules redistribute within different subcellular localizations to execute their functions (reviewed in Lorrain et al., 2006; Van Buskirk et al., 2012). Second, selective protein degradation modulates the protein abundance of both positive and negative regulators of light signaling pathways (reviewed in Yi and Deng, 2005; Henriques et al., 2009). Third, light-regulated protein phosphorylation involves the activation of both photoreceptors and downstream signaling molecules (Yeh et al., 1997; Ahmad et al., 1998; Yeh and Lagarias, 1998; Fankhauser and Chory, 1999; Colón-Carmona et al., 2000; Medzihradszky et al., 2013; Nito et al., 2013). Fourth, widespread translational regulation occurs in response to light or dark treatments (Juntawong and Bailey-Serres, 2012; Liu et al., 2012, 2013). Also, by interacting with PENTA1, phytochrome negatively regulates the translation of protochlorophyllide reductase mRNAs (Paik et al., 2012). Fifth, and likely the most well studied, is transcriptome adjustments in plants with light treatment (reviewed in Jiao et al., 2007). Recently, the splicing factor REDUCED RED-LIGHT RESPONSES IN CRY1CRY2 BACKGROUND1 was found to be a phyB-dependent signaling component (Shikata et al., 2012), suggesting alternative splicing could further increase the transcriptomic diversity.
Although light-modulated alteration in transcriptomes has been documented, most efforts have focused on the transcriptional activation or repression of light-regulated genes. Posttranscriptional regulation of gene expression in response to light signals has been less studied. A recent report showed that a key positive regulator in Arabidopsis thaliana photomorphogenesis, ELONGATED HYPOCOTYL5 (HY5), could activate the expression of MIR genes encoding microRNAs (miRNAs) by binding directly to their promoters (Zhang et al., 2011). Thus, small regulatory RNAs may participate in modulating the transcriptome shift during photomorphogenesis. Interestingly, phenotypic examination of Arabidopsis mutants carrying weak alleles of ago1 revealed a light-hypersensitive phenotype (Sorin et al., 2005), so small regulatory RNAs may act as negative regulators of the light signaling pathways. However, additional studies are needed to clarify whether light regulates the biogenesis and/or actions of small regulatory RNAs in photomorphogenic Arabidopsis.
In many eukaryotic organisms, small regulatory RNAs play a pivotal role in the transcriptional, posttranscriptional, or translational regulation of gene expression (reviewed in Chen, 2010). After being processed from single-stranded RNA hairpin or double-stranded RNAs, miRNAs and small interfering RNAs are stabilized by the action of HUA ENHANCER1 (HEN1), a small RNA methyltransferase that adds a methyl group to the 3′-most nucleotide of small RNAs, thus protecting against 3′ uridylation and truncation (Ji and Chen, 2012).
Here, we report on a light-hypersensitive phenotype in an Arabidopsis mutant defective in HEN1. Our data reveal that light upregulated the expression of HEN1, a process requiring red (R), far-red (FR), or blue (B) light photoreceptors and key transcriptional regulators, HY5 and HY5-HOMOLOG (HYH). Among the miRNAs stabilized by HEN1, miR157d and miR319 could target transcripts of HY5 and those of TEOSINTE BRANCHED1, CYCLOIDEA AND PCF24 (TCP24), respectively, for cleavage during Arabidopsis photomorphogenesis. Light may upregulate HEN1 expression for the proper biogenesis of small regulatory RNAs, and selected miRNAs could fine-tune the temporal expression patterns of key transcriptional regulators in Arabidopsis photomorphogenesis.
RESULTS
HYL1, HEN1, and HST Are Negative Regulators of Arabidopsis Photomorphogenesis
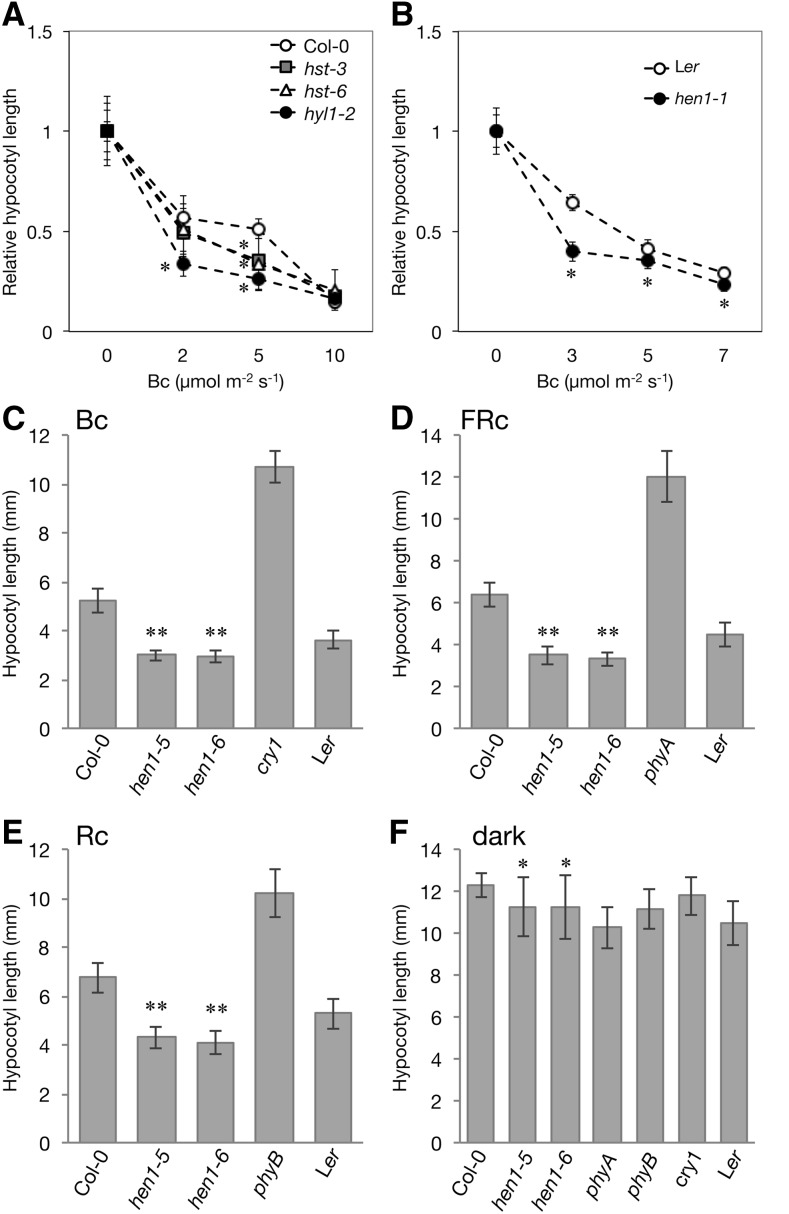
We evaluated whether small RNAs are involved in the light-regulated deetiolation process in Arabidopsis by examining the photomorphogenic phenotype of mutants defective in miRNA biogenesis genes, including HYPONASTIC LEAVES1 (HYL1) (Lu and Fedoroff, 2000), HEN1 (Park et al., 2002), and HASTY (HST) (Bollman et al., 2003). These genes are responsible for processing primary miRNA transcripts, stabilization, and export of mature miRNA duplexes (reviewed in Rogers and Chen, 2013; a simplified biogenesis scheme is shown in Supplemental Figure 1). The hyl1-2, hst-3, and hst-6 mutants have shorter hypocotyls than the wild-type Columbia-0 (Col-0) under both dark and light conditions (Supplemental Figure 2), as described previously (Lu and Fedoroff, 2000; Bollman et al., 2003). Treatment with various fluences of continuous blue (Bc) light clearly exaggerated the light-induced inhibition of hypocotyl elongation in hyl1 and hst mutants (Figure 1A). We also observed a similar light-dependent phenotype for hen1-1 mutant as compared with its corresponding wild-type Landsberg erecta (Ler) (Figure 1B; Supplemental Figure 2). The light hypersensitivity in these mutants implied that HYL1, HST, and HEN1 function as negative regulators in the deetiolation process in Arabidopsis.
Figure 1.
HEN1 Plays Multiple Roles in Skotomorphogenesis and Photomorphogenesis.
(A) and (B) Hypocotyl fluence rate responses of wild type and hyl1-2, hst3, and hst6 mutants in the Col-0 (A) and hen1-1 in the Ler background (B) under Bc light at the indicated fluence rates. Hypocotyl lengths under each indicated fluence rate were normalized to that under the dark. Data are mean ± sd. *Significantly different from the corresponding wild type (P < 0.01, Student’s t test; n = 16 to 32).
(C) to (F) Hypocotyl lengths of 4-d-old seedlings of wild types and mutants hen1-5 and hen1-6 in the Col-0, phyA, phyB, and cry1 in the Ler background grown under Bc (5 μmol m−2 s−1) (C), FRc (0.125 Wm−2) (D), Rc (5 μmol m−2 s−1) (E), and dark (F). Data are mean ± sd. ** and *, significantly different from the corresponding wild type (Col-0) (P < 10−14 and P < 0.05, Student’s t test; n = 14 to 20).
Negative Regulatory Roles of HEN1 in Multiple Aspects of Skotomorphogenic and Photomorphogenic Development
We surveyed light-regulated expression patterns of genes in miRNA biogenesis pathways, including DICER LIKE1, SERRATE, HYL1, HEN1, HST, and ARGONAUTE1 in the public transcriptome data repository Genevestigator (Zimmermann et al., 2004). Only HEN1 showed clear light responsiveness (Supplemental Figure 3). Therefore, we focused on HEN1 and its roles in deetiolating Arabidopsis.
We next examined the photomorphogenic phenotype of two independent hen1 alleles, hen1-5 (Vazquez et al., 2004) and hen1-6 (Li et al., 2005), in a Col-0 background (Supplemental Figure 4A). Neither mutant expressed the full-length HEN1 transcript (Supplemental Figure 4B). Similar to hen1-1, both hen1-5 and hen1-6 mutants were hypersensitive to Bc (Figure 1C; Supplemental Figure 5A). Under continuous far-red (FRc) and red (Rc) light, hen1-5 and hen1-6 both had a short hypocotyl as compared with the wild type (Figures 1D and 1E; Supplemental Figures 5B and 5C). The appropriate hypocotyl elongation of hen1-6 could be restored by introducing a genomic fragment of HEN1 into hen1-6 plants (Supplemental Figure 6), so loss of functional HEN1 is indeed responsible for the light hypersensitivity in hen1. Consistent with a shorter hypocotyl in hen1-1 compared with its corresponding wild-type Ler in the dark, the hypocotyl lengths of hen1-5 and hen1-6 were also slightly shorter than that of their corresponding wild-type Col-0 (Figure 1F; Supplemental Figure 5D).
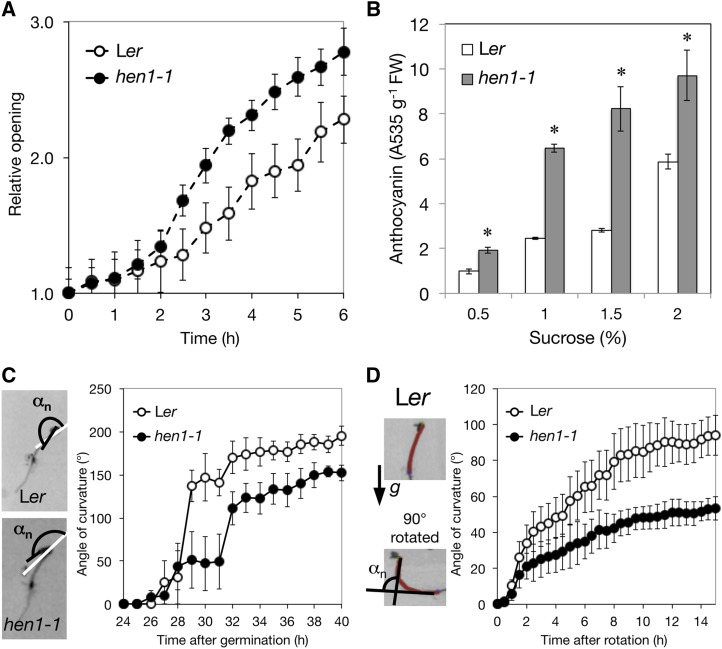
We also observed additional light-hypersensitive responses in hen1-1. Time-lapse imaging revealed that the cotyledons of the hen1-1 mutant opened faster than the Ler wild type (Figure 2A). Light also triggers the accumulation of anthocyanin in wild-type seedlings (Ahmad et al., 1995; Maier et al., 2013; Shin et al., 2013). Light-grown hen1-1 showed high anthocyanin levels (Figure 2B), which is consistent with hen1-1 being light hypersensitive.
Figure 2.
The hen1-1 Mutant Shows Broad Skotomorphogenic and Photomorphogenic Defects.
(A) Cotyledon opening of 4-d-old etiolated seedlings under continuous white light (Wc; 100 μmol m−2 s−1) analyzed by measuring cotyledon tip-to-tip distances for the wild type and hen1-1 mutant. Data are mean ± se from six biological replicates.
(B) Anthocyanin level in 4-d-old Ler and hen1-1 seedlings grown under 16-h W light (100 μmol m−2 s−1)/8-h dark cycles in the presence of sucrose with indicated concentrations. Data are mean ± sd from two biological replicates (each with two technical replicates). *Significantly different from the wild type (P < 0.05, Student’s t test; n = 4).
(C) Kinetics of hook formation in hen1-1 and wild-type seedlings under the dark. The images show apical hook angles (αn) at 1-h intervals after 24-h germination at 22°C. Data are mean ± se from three biological replicates.
(D) The hypocotyl turning angles at 0.5-h intervals after 90° change in seedling direction for the wild type and hen1-1 were measured. Images show a representative Ler seedling at time 0 and 15 h. The arrow indicates the gravity direction. The hypocotyl was traced in red to determine the angle of hypocotyl curvature. Data are mean ± se from four biological replicates.
[See online article for color version of this figure.]
Mutations in negative regulators of photomorphogenesis, such as CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and PHYTOCHROME INTERACTING FACTORs (PIFs), also resulted in aberrant skotomorphogenic development, including short hypocotyl (Leivar and Quail, 2011), opened hook (Mazzella et al., 2014) and reduced antigravity responses (Kim et al., 2011). In the dark, hypocotyl lengths were shorter for the hen1-1 mutant than Ler (Figure 1B). Also, in the dark, the apical hooks of hen1-1 seedlings were partially opened and reached a plateau at an angle of 136°, much smaller than the 181° for wild-type seedlings (Figure 2C). Additionally, hen1-1 showed reduced response to a change in gravity orientation (Figure 2D).
Therefore, consistent with it playing negative roles in light responses as do COP1 and PIFs, HEN1 has broad effects on both skotomorphogenesis and photomorphogenesis.
phyA, phyB, cry1, HY5, and HYH Regulate the Light-Responsive Expression of HEN1
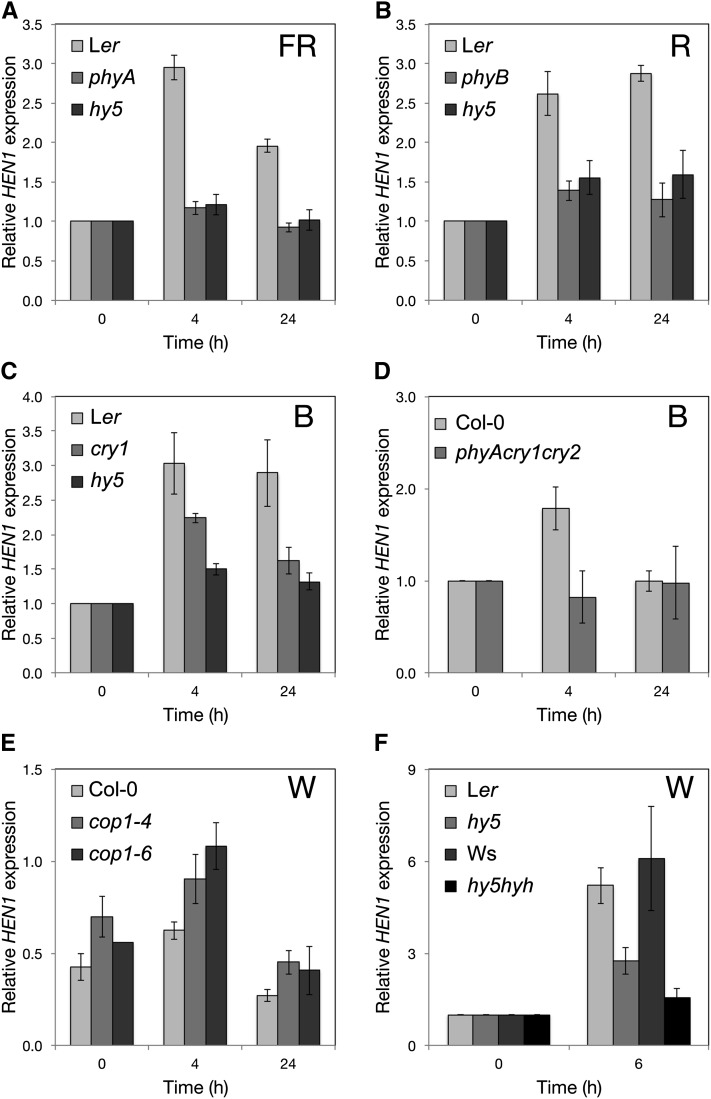
We investigated whether a broad spectrum or specific wavelengths of light could induce the expression of HEN1. R, FR, and B could all induce HEN1 expression (Figures 3A to 3C). We also examined whether the induction of HEN1 by R, FR, and B light depended on functional photoreceptors in the Arabidopsis photoreceptor mutant phyA under FR light, phyB under R light, and cry1 under B light. The light responsiveness of HEN1 was largely compromised in phyA and phyB mutants under FRc and Rc light, respectively (Figures 3A and 3B), so the FR and R light responsiveness of HEN1 depended on functional phyA and phyB, respectively. However, under B light, the expression of HEN1 could still be induced but to a lesser extent in the cry1 mutant as compared with the wild type (Figure 3C). This result suggested that cry1 was not the sole photoreceptor contributing to the B-light-inducible expression of HEN1. We therefore examined whether additional B-light photoreceptors, including phyA (Poppe et al., 1998) and cry2, could function in parallel to cry1 in inducing HEN1 expression under B light. The B light induction of HEN1 was abolished in the phyA cry1 cry2 triple mutant (Figure 3D), which indicates that phyA and cry2 could be accessory B light photoreceptors for HEN1 induction.
Figure 3.
The Expression of HEN1 Is Upregulated by Light and Requires Photoreceptors and HY5.
Real-time qRT-PCR analysis of the mRNA level of HEN1 in etiolated wild type (Ler, Wassilewskija, and Col-0), hy5, and the light signaling mutants phyA (A), phyB (B), cry1 (C), phyA cry1 cry2 (D), cop1-4, cop1-6 (E), and hy5 hyh (F) on exposure to FR (1.13 Wm−2), R (18 μmol m−2 s−1), B (15 μmol m−2 s−1), and W (100 μmol m−2 s−1), respectively, at the indicated times. The expression of UBQ10 was an internal control. Except for the relative HEN1 level in (E) being 1000-multiplied mean values of 2−ΔCT (ΔCT = CT, HEN1 – CT, UBQ10) for clarity, the level of HEN1 at each time is normalized to that of etiolated seedlings (0 h). Data are mean ± sd from three biological replicates (each with three technical replicates).
HY5 encodes a key transcription factor and is upregulated by various qualities of light signals, including R, FR, and B light (Chang et al., 2008). Therefore, we examined whether the light induction of HEN1 was regulated by HY5. The phyB- and phyA-induced expression of HEN1 primarily depended on HY5 (Figures 3A and 3B). The induction of HEN1 by B light also depended on the expression of HY5 (Figure 3C). Thus, HY5 acts downstream of multiple photoreceptors for the B light induction of HEN1. We examined the HY5-dependent induction of HEN1 in cop1 mutants that can accumulate a high level of HY5 protein in the dark (Osterlund et al., 2000). As expected, we detected increased HEN1 transcripts in cop1-4 and cop1-6 mutants (McNellis et al., 1994) in the dark and subsequent deetiolation (Figure 3E).
We noted the residual upregulation of HEN1 in the hy5 mutant under various light conditions (Figures 3A to 3C), which indicates that additional factors may function in addition to or in parallel with HY5 to induce HEN1. Because the function of HYH is partially redundant to that of HY5 in promoting photomorphogenesis (Holm et al., 2002), HYH may also contribute to the light-upregulated expression of HEN1. Indeed, the light-induced expression of HEN1 was further compromised in the hy5 hyh double mutant under white light (W) (Figure 3F). Therefore, HY5 and HYH are the primary activators of HEN1 expression under light.
Small Regulatory RNAs and Arabidopsis Photomorphogenesis
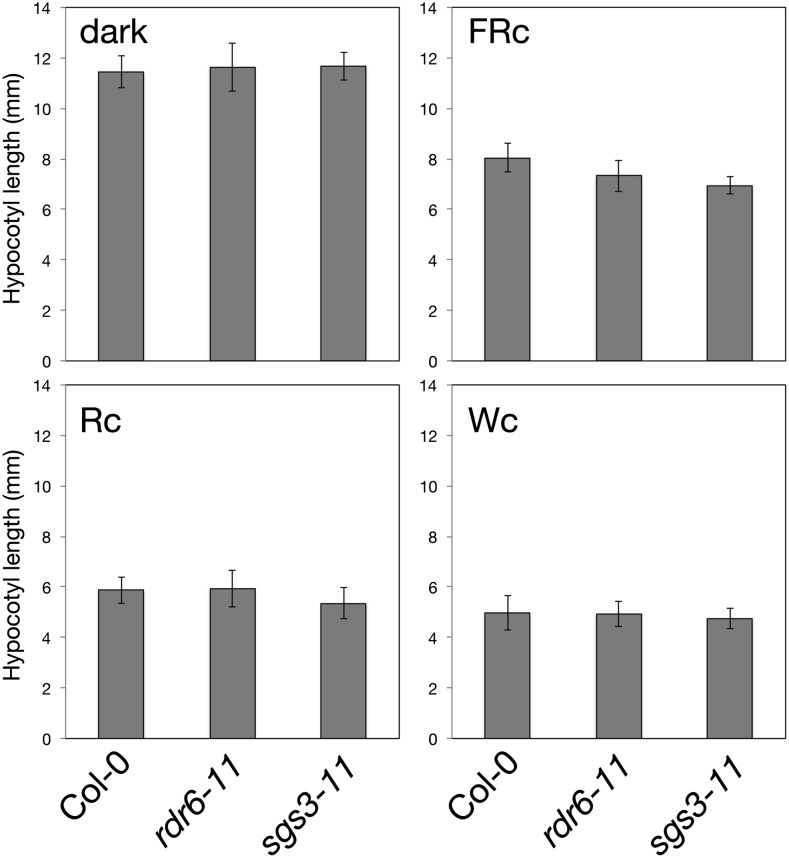
Light upregulates HEN1 to attenuate photomorphogenesis possibly by stabilizing small regulatory RNAs to silence the expression of key regulators in this important physiological process. HEN1 methylates small regulatory RNAs generated from various biogenesis pathways. To narrow down the classes of small regulatory RNAs that may be involved, we first examined the photomorphogenic development of Arabidopsis mutants defective in key genes for the biogenesis of trans-acting small interfering RNAs (ta-siRNAs). Arabidopsis mutants defective in RNA-DEPENDENT RNA POLYMERASE6 (RDR6) and SUPPRESSOR OF GENE SILENCING3 (SGS3) (Peragine et al., 2004; Vazquez et al., 2004; Xie et al., 2005; Yoshikawa et al., 2005) showed a normal photomorphogenic phenotype under the light qualities examined (Figure 4). Despite a slightly shorter hypocotyl in sgs3 under FR light (Figure 4), fluence response analysis revealed a comparable hypocotyl length for sgs3 and the wild type across a broad range of FR intensities (Supplemental Figure 7). Thus, reduced expression of ta-siRNAs alone is insufficient to confer the light hypersensitivity seen in hen1. Accordingly, we focused on characterizing miRNAs for their roles downstream of HEN1 in modulating the posttranscriptional expression of light-responsive genes.
Figure 4.
Arabidopsis Mutants Defective in ta-siRNA Biogenesis Show Normal Photomorphogenic Development.
Hypocotyl lengths of seedlings of the wild type and mutants defective in RDR6 and SGS3 measured after growth in the dark, FRc (0.125 Wm−2), Rc (5 μmol m−2 s−1), and Wc (15 μmol m−2 s−1) for 4 d. Data are mean ± sd.
Increased HEN1 Levels Lead to Increased miRNA Abundance
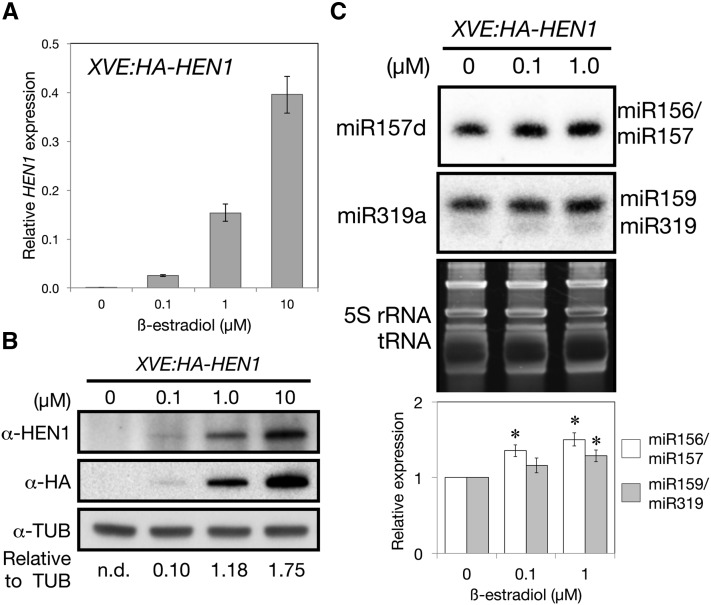
HEN1 is a methyltransferase functioning to stabilize small RNAs (Li et al., 2005). Our results showed that light could upregulate the expression of HEN1. We next examined whether the increase in HEN1 mRNAs leads to increased accumulation of HEN1 protein and consequently more small regulatory RNAs. We generated transgenic Arabidopsis carrying a HA-tagged HEN1 (HA-HEN1) under the control of a β-estradiol inducible promoter (XVE:HA-HEN1) (Zuo et al., 2006). HA-HEN1 was previously shown to be a biologically active protein (Yang et al., 2007). We first induced HA-HEN1 expression in etiolated seedlings of transgenic plants under dark conditions to eliminate the effect of light-upregulated HEN1. HEN1 transcript expression was increased with β-estradiol treatment (Figure 5A). Although too little endogenous HEN1 protein was present to be detected with anti-HEN1 antibody in etiolated seedlings, a specific protein with the expected size of HA-HEN1 (109 kD) recognized both by anti-HEN1 and anti-HA antibodies was induced to accumulate with β-estradiol treatment (Figure 5B). Etiolated seedlings treated with 0.1 and 1 μM β-estradiol showed proportionally increased HA-HEN1 transcript and protein levels (Figures 5A and 5B). The increased HA-HEN1 protein level was accompanied by increased expressions of the miR157 and miR159/319 in dark-grown seedlings (Figure 5C; Supplemental Figure 8). This suggested that, in etiolated seedlings, the level of endogenous miRNA precursors exceeded the functional capacity of endogenous HEN1 protein.
Figure 5.
HEN1 Plays a Rate-Limiting Role for miRNA Accumulation in Etiolated Seedlings.
The 3-d-old etiolated T2 seedlings of XVE:HA-HEN1 were treated with 0 to 10 μM β-estradiol for 24 h in the dark.
(A) qRT-PCR analysis of the mRNA level of HEN1 relative to that of UBQ10. Data are mean ± sd from two biological replicates (each with three technical replicates).
(B) Immunoblot analysis of protein level of HA-HEN1 with the antibodies pig anti-HEN1 (α-HEN1), mouse anti-HA (α-HA), and mouse anti-tubulin (α-TUB). Numbers below the α-TUB blot indicate the HA-HEN1 protein levels in the α-HA blot relative to TUB signals. n.d., nondetectable.
(C) Small RNA analyses of miR156/157 and miR159/319. DNA oligos used for each blot are indicated to the left of each blot. The bar graph shows 5S rRNA- and tRNA-normalized miRNA fold increase in HA-HEN1-induced seedlings. Data are mean ± sd quantitated using ImageJ on three biological replicates, shown in Supplemental Figure 8. *Significantly different from the noninduced sample (0 μM) (P < 0.05, Student’s t test; n = 3).
The results in Figures 3 and 5 imply that when wild-type etiolated seedlings were exposed to light, the increased HEN1 transcripts likely resulted in increased protein level of HEN1 to promote miRNA biogenesis/accumulation. Thus, the light-induced increase of HEN1 plays a regulatory role in increasing miRNAs in the deetiolation process.
HY5 Transcript Is a Target of miR157d
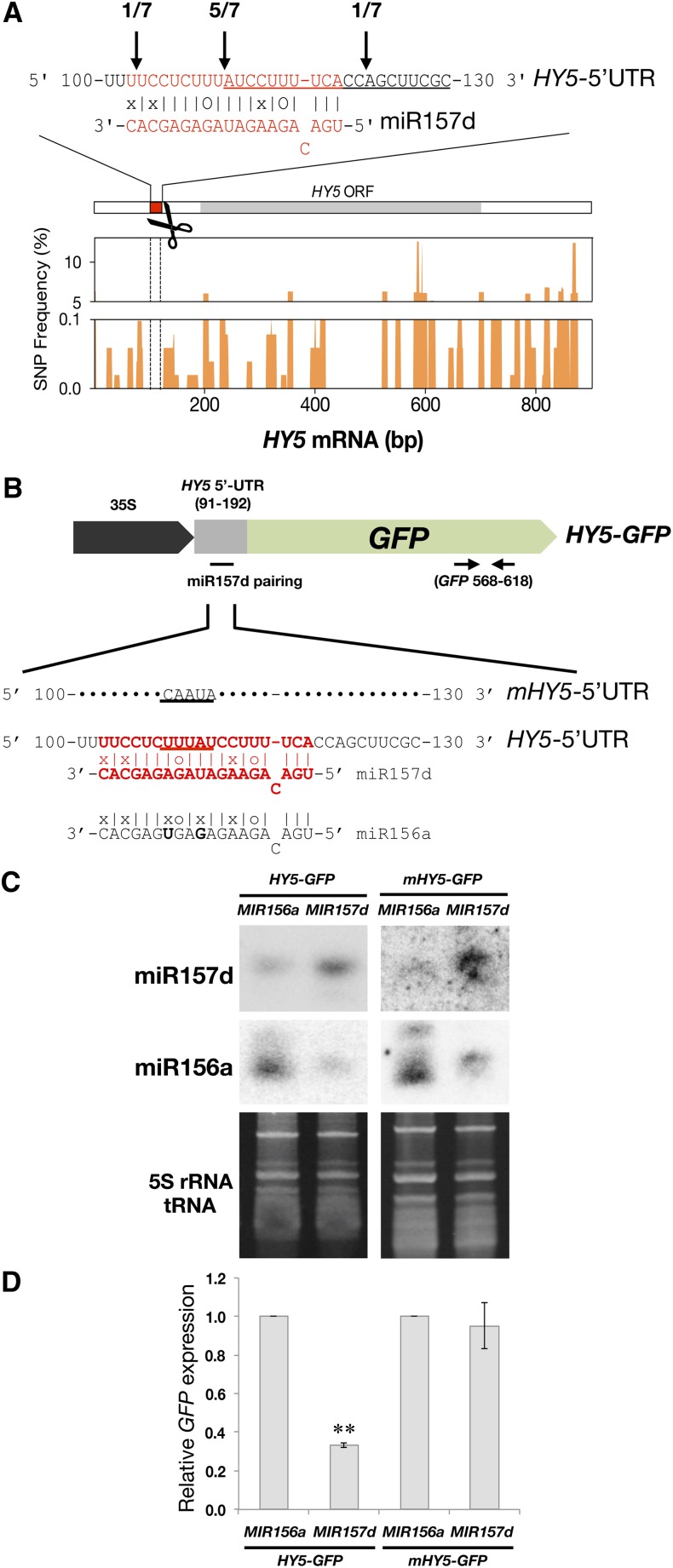
The results in Figures 1 and 2 indicate that HEN1 is a negative regulator of photomorphogenesis. Light upregulates the expression of HEN1 (Figure 3), and increased levels of HEN1 will result in increased levels of miRNAs (Figure 5). Therefore, in the deetiolating process, the increased miRNAs may target transcripts of positive regulators of photomorphogenesis for cleavage. Such cleavage may result in the accumulation of site-specific sequence signatures seen in the degradome database of 5′-end sequences of transcripts after miRNA-mediated cleavage (PARE, https://mpss.udel.edu/dbs/index.php?SITE=at_pare) (German et al., 2008). A search of such signatures revealed an enriched signature within the 5′-untranslated region (UTR) of the HY5 transcript. This putative cleavage site was located at a position opposite to nucleotides 11-12 of miR157d, rather than the canonical cleavage position 10-11, possibly because of the presence of a single nucleotide bulge on miR157d when pairing with the HY5 transcript (Figure 6A).
Figure 6.
HY5 Transcript Is Targeted by miR157d.
(A) The sequence in red represents the pairing region between miR157d and the 5′-UTR of HY5 mRNA. Numbers and vertical arrows indicate HY5 cleaved ends validated by 5′ RACE. Straight lines and “o” symbols represent perfect matches and G-U matches, respectively. The enriched signature for HY5 in the PARE database is underlined. SNP frequency for HY5 in 513 Arabidopsis tracks summed using a 10-bp sliding window across the HY5 cDNA.
(B) The HY5 5′-UTR (91 to 192 nucleotides; gray box) containing the miR157d pairing region (102 to 120 nucleotides) was fused with GFP cDNA to generate the 35S:HY5-GFP construct for N. benthamiana coinfiltration assays with 35S:MIR156a or 35S:MIR157d. The pairing region (sequences in red) of HY5-GFP was mutated (nucleotides underlined) to generate 35S:mHY5-GFP. Horizontal arrows indicate primer binding sites for GFP qRT-PCR analysis.
(C) RNA gel blot analyses of miR156a and miR157d transcripts in coinfiltration assays. DNA oligos used for each blot are indicated to the left of each blot. 5S rRNA and tRNA levels are loading controls. Each sample contained 2 μg total RNA.
(D) qRT-PCR analysis of HY5-GFP and mHY5-GFP transcripts. **HY5-GFP level reduced only when coinfiltrated with 35S:MIR157d as compared with 35S:MIR156a (P < 0.01, Student’s t test; n = 6). Data are mean ± se from two biological replicates (each with three technical replicates).
We confirmed the presence of this cleaved product of HY5 mRNA in photomorphogenic Arabidopsis by 5′ rapid amplification of cDNA ends (RACE) (Figure 6A). Because HY5 was not previously predicted as a target of miR157d, we investigated the authenticity of the targeted regulation of HY5 by miR157d by examining the sequences of both MIR157d and HY5 from 513 sequenced Arabidopsis tracks from the Arabidopsis 1001 Genomes database (http://signal.salk.edu/atg1001/3.0/gebrowser.php). The sequence encoding mature miR157d was fully conserved among the 513 Arabidopsis tracks. In addition, analysis of single nucleotide polymorphisms (SNPs) revealed 100% conservation at the miR157d target region on the HY5 transcript among the 513 genome tracks (see Methods), whereas frequent SNPs were observed across the 5′-UTR and the rest of the HY5 cDNA (Figure 6A). Therefore, sequences for complementary regions between miR157d and HY5 were preferentially retained, possibly because of functional constraint.
We next demonstrated that HY5 is an authentic target of miR157d using Agrobacterium tumefaciens-mediated transient expression assays. Tobacco (Nicotiana tabacum) leaves were coinfiltrated with agrobacteria harboring binary vectors constitutively expressing constructs of MIR157d and HY5 5′-UTR (91 to 192 bp) fused with GFP cDNA (HY5-GFP) (Figures 6B and 6C). The miR156a, which shares high sequence identity with miR157d, with only two additional mismatches within the targeting region on the HY5 5′-UTR, and mHY5-GFP carrying mutations in miR157d target site were used as controls (Figures 6B and 6C). The HY5-GFP transcript level was specifically suppressed by miR157d but not miR156a (Figure 6D). Moreover, mHY5-GFP was resistant to miR157d (Figure 6D), which suggests that there is specific targeting of HY5 5′-UTR by miR157d.
HEN1 and HY5 Form a Negative Feedback Regulatory Loop in Photomorphogenic Arabidopsis
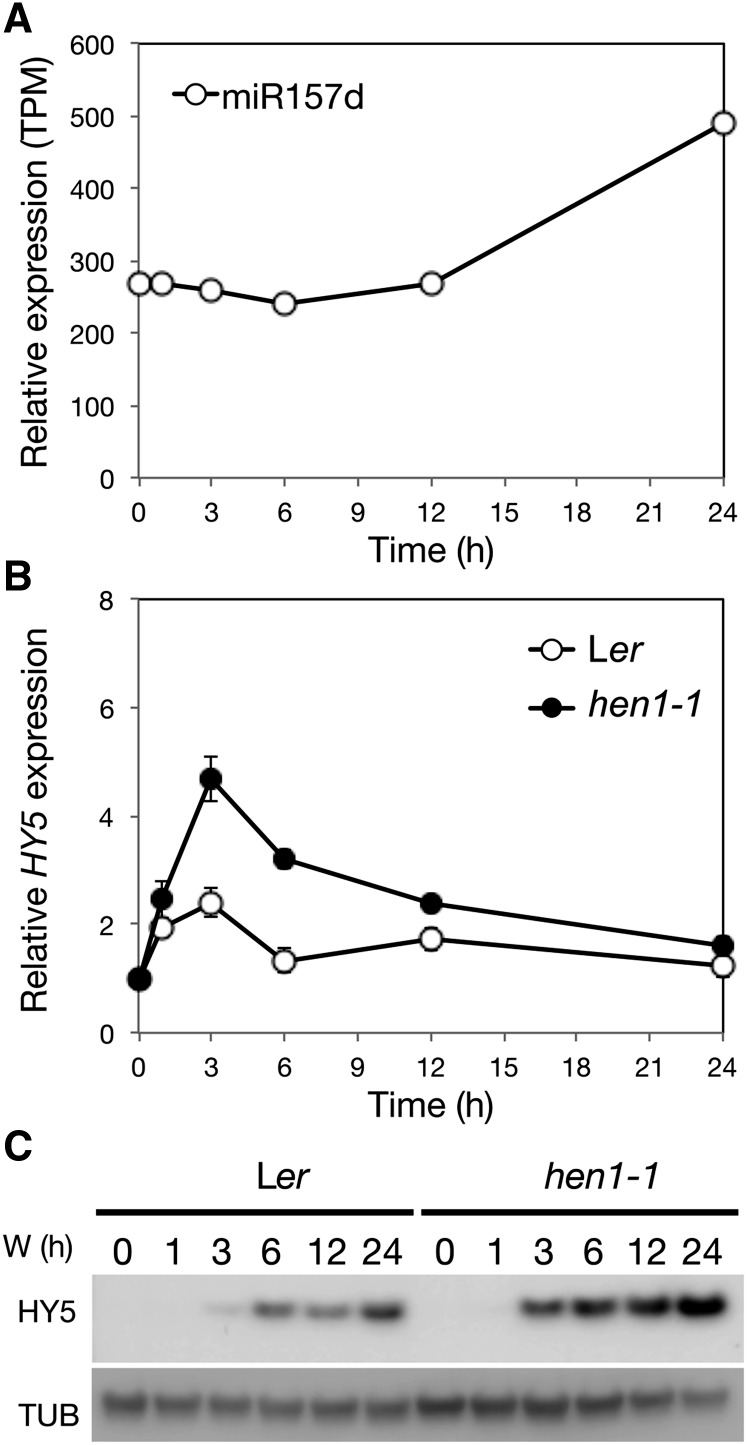
To examine whether HEN1 executes its negative roles in photomorphogenesis at least in part by regulating the level of miR157d, we first examined the accumulation of miR157d in the hen1-1 mutant. Consistent with previously investigated miRNAs in hen1 mutants (Li et al., 2005), miR157d showed only a trace uridylated signal, which confirms that the accumulation of miR157d depended on functional HEN1 (Supplemental Figure 9). Furthermore, miRNA-specific profiling of the small RNAs in deetiolating seedlings indicated that light enhanced the accumulation of miR157d in photomorphogenic Arabidopsis (Figure 7A).
Figure 7.
Light Responsiveness and Regulatory Roles of miR157d on HY5 during Photomorphogenesis.
(A) The light responsiveness of miR157d. TPM, transcripts per million.
(B) qRT-PCR analysis of the mRNA level of HY5 in wild-type (Ler) and hen1-1 mutant seedlings with UBQ10 as an internal control. The expression was normalized to that of the corresponding genotype at 0 h. Data are mean ± sd from three biological replicates (each with three technical replicates).
(C) Representative immunoblot analysis of HY5 protein level in the wild type (Ler) and hen1-1 during photomorphogenesis with tubulin (TUB) as a loading control.
With HY5 as a direct target of miR157d (Figure 6), its transcript level is expected to be increased in the hen1 mutant, which has a reduced level of miR157d. Indeed, the light-induced expression of HY5 was further elevated in the hen1-1 mutant as compared with the wild type (Ler) under photomorphogenesis (Figure 7B). The increased HY5 mRNA level also resulted in higher HY5 protein level in hen1-1 than Ler seedlings throughout the photomorphogenic process (Figure 7C; Supplemental Figure 10). The increased level of the positive regulator HY5 could enhance the light sensitivity of the hen1 mutant, which is consistent with the light-hypersensitive phenotype of hen1 (Figures 1 and 2).
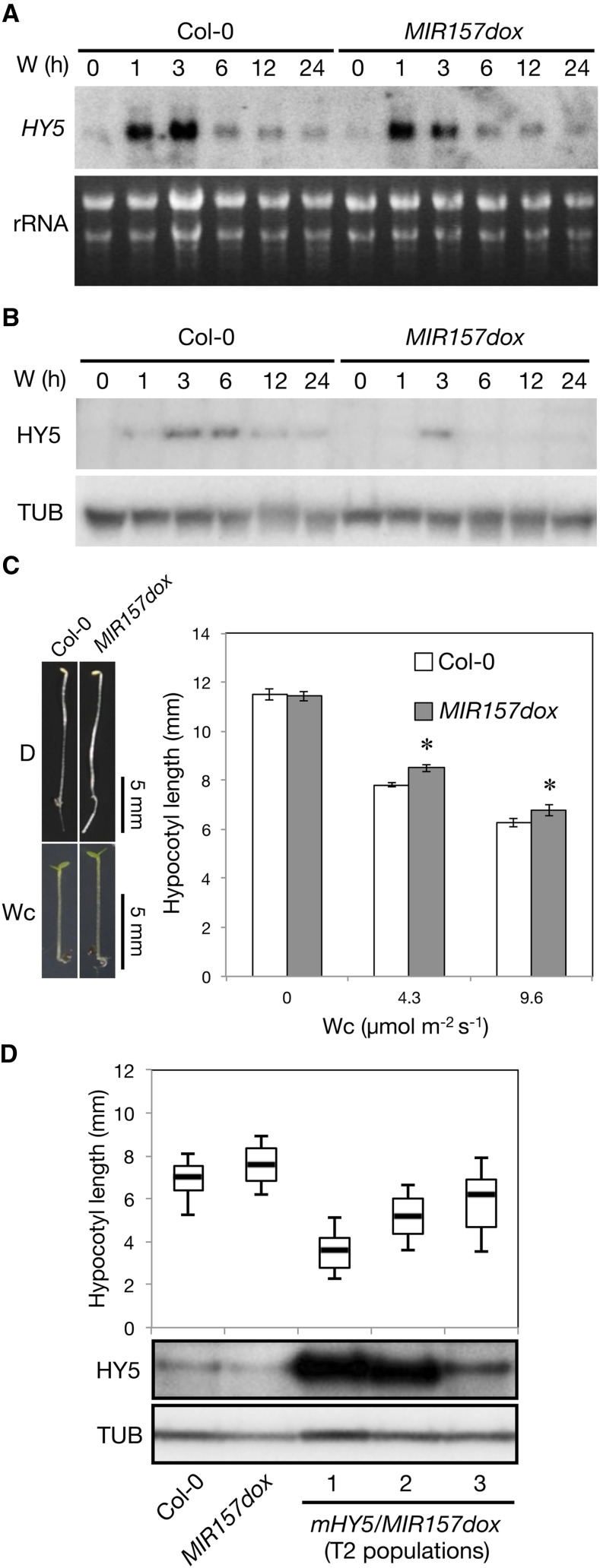
By contrast, in the MIR157d-overexpression line (MIR157dox; Supplemental Figure 11), HY5 transcript and protein levels were reduced (Figures 8A and 8B; Supplemental Figure 12). As expected, MIR157dox plants showed light hyposensitivity as compared with the wild type (Col-0) (Figure 8C). To further examine whether the light hyposensitivity of MIR157dox was due to reduced accumulation of HY5, we introduced a miR157d-resistant HY5 overexpression construct (mHY5 as in Figure 6B) into MIR157dox (denoted mHY5/MIR157dox). As expected, HY5 was overaccumulated in deetiolating mHY5/MIR157dox seedlings (Figure 8D). In contrast to the parental line (MIR157dox), mHY5/MIR157dox lines were light hypersensitive, and the degree of light hypersensitivity was proportional to HY5 protein levels in three independent mHY5/MIR157dox lines (Figure 8D). We cannot entirely rule out that miR157d may target additional positive regulators of photomorphogenesis. However, our results clearly indicated that the light hyposensitivity of MIR157dox seedlings could be rescued if HY5 protein levels were manipulated to increase (Figure 8D).
Figure 8.
The Effect of MIR157d Overexpression on Photomorphogenesis.
(A) RNA gel blot analysis of HY5 with 5 μg total RNA from 4-d-old etiolated Col-0 and MIR157dox seedlings exposed to W (50 μmol m−2 s−1) for the times indicated. The level of rRNA was a loading control. Numbers below the blot indicate HY5 level relative to the rRNA in each sample. One representative blot from three independent biological replicates is shown.
(B) Representative immunoblot analysis of HY5 in wild type (Col-0) and MIR157d overexpression line (MIR157dox) during photomorphogenesis with TUB as a loading control.
(C) Hypocotyl lengths of MIR157dox seedlings under Wc of indicated fluence rates. Data are mean ± se. *Significantly different from wild-type Arabidopsis (Col-0) (P < 0.01, Student’s t test; n = 13 to 15).
(D) Box plots (n = 65 to 98) show hypocotyl lengths of the wild type (Col-0), MIR157dox, and three independent mHY5/MIR157dox lines grown for 4 d under Wc (5 μmol m−2 s−1). The top, middle, and bottom of the box represent the 25, 50, and 75 percentiles, respectively, and the top and bottom black lines are the 10th and 90th percentiles, respectively. HY5 levels in etiolated seedlings exposed to Wc (50 μmol m−2 s−1) for 6 h were determined by immunoblot analysis with TUB as a loading control.
[See online article for color version of this figure.]
Taken together, HEN1 has a negative regulatory role on HY5 expression via the action of miR157d. Together with our findings of HY5-dependent upregulation of HEN1 (Figure 3), this demonstrates that HY5 and HEN1 form a negative feedback regulatory loop in photomorphogenesis.
miR319 Regulates the Expression of Negative Regulators of Arabidopsis Photomorphogenesis
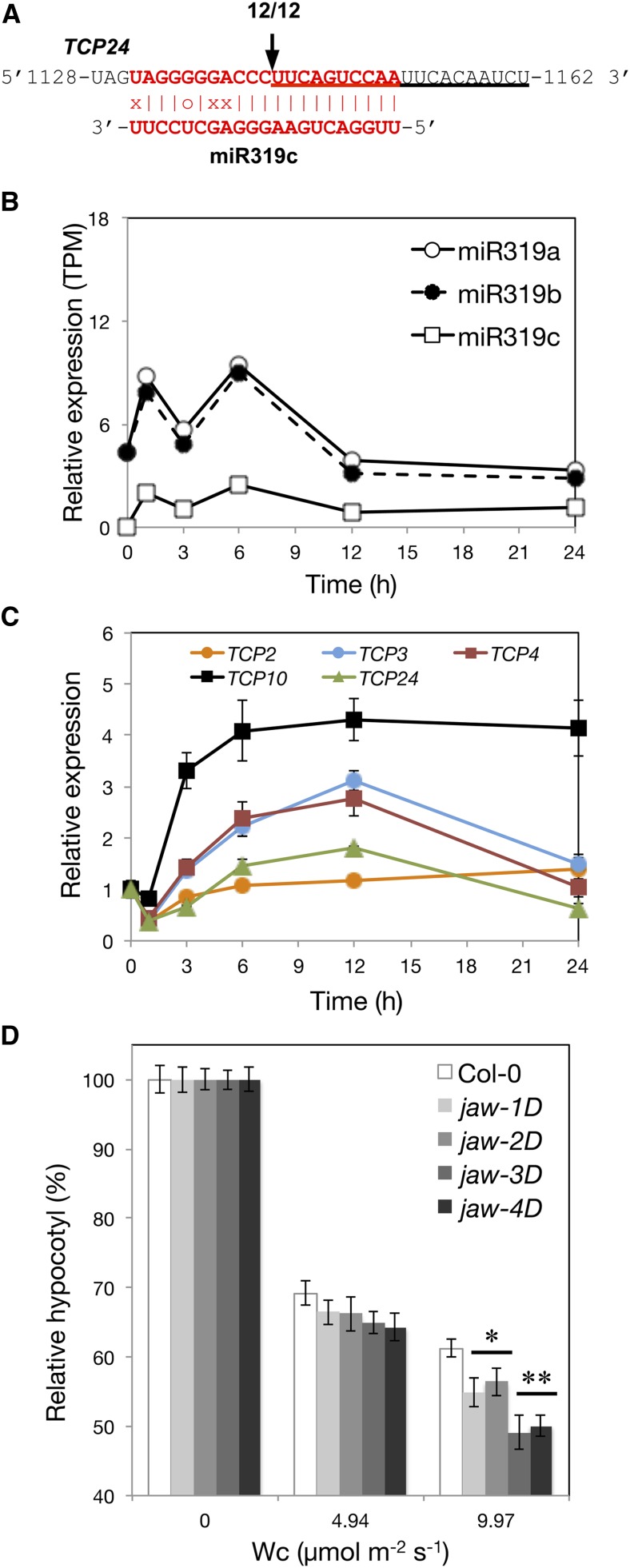
Because HEN1 is required for general miRNA biogenesis, the light hypersensitivity observed in the hen1 mutants could be a combined result of the actions of multiple miRNA-target pairs affecting photomorphogenesis. We wondered whether additional miRNAs or miRNA target genes played roles in photomorphogenesis regulation. Five TCPII subfamily transcription factor genes, TCP2, 3, 4, 10, and 24, are known targets of miR319 (Jones-Rhoades et al., 2006). Ectopic expression of TCP2/3/4 with mutated miR319 target sites results in light-hyposensitive phenotypes, so TCP2/3/4 are hypothesized to be negative regulators of hypocotyl elongation (Palatnik et al., 2003; Koyama et al., 2007; Zhao et al., 2013). We confirmed the presence of a miR319-mediated cleaved signature for transcripts of a selected TCPII gene, TCP24, in photomorphogenic Arabidopsis (Figure 9A). The expression of both MIR319 and TCPII family members (TCP2/3/4/10/24) was transiently upregulated by light (Figures 9B and 9C).
Figure 9.
Light Responsiveness and Regulatory Role of miR319 in TCP Levels in Photomorphogenic Arabidopsis.
(A) 5′-RACE analysis of the cleavage site of TCP24 mRNA within the TCP24-miR319c duplex.
(B) The light responsiveness of miR319 members.
(C) qRT-PCR analysis of the mRNA level of TCP genes. The expression of each gene under W light at times indicated was normalized to that of UBQ10 and was relative to that of etiolated seedlings (0 h). Data are mean ± se from two biological replicates (each with two technical replicates).
(D) The hypocotyl lengths of different allelic jaw-D mutant seedlings under Wc of the indicated fluence rates normalized to the length under the dark. Data are mean ± se. * and **, significantly different from wild-type Arabidopsis (Col-0) (P < 0.05 and P < 0.01, Student’s t test; n = 24 to 27).
The induction of MIR319 by light has the potential to reduce the expression of TCPs. To assess the role of miR319 on photomorphogenesis, we analyzed MIR319a overexpression lines, jaw-D mutants (Palatnik et al., 2003), known to show reduced expression of TCPII members (Nag et al., 2009). Because TCPs are negative regulators in hypocotyl elongation, their reduced levels may confer Arabidopsis seedlings with light hypersensitivity. Indeed, multiple alleles of jaw-D mutants showed a light hypersensitive phenotype (Figure 9D).
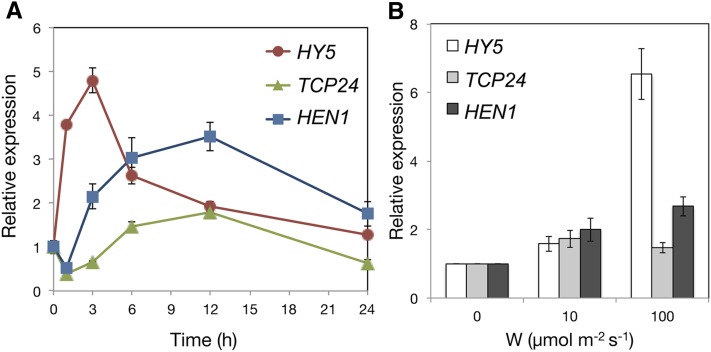
Temporal Expression Adjustment of HEN1, HY5, and TCP24
To elucidate the regulatory relationship between HEN1, HY5, and TCP24 in deetiolating Arabidopsis seedlings, we examined their expression by time-course analysis. The positive regulator HY5 quickly responded to light signals, followed by the two negative regulators, HEN1 and TCP24 (Figure 10A). This finding suggested that in deetiolating seedlings, positive regulators such as HY5 could efficiently transmit the light signals in a timely manner. With time, negative regulators such as HEN1 and TCP24 are expressed to attenuate the actions of the positive regulator(s), so the light responses could be properly fine-tuned and/or optimized.
Figure 10.
Responsiveness of HY5, TCP24, and HEN1 to Light over Time and Fluence Rates during Deetiolation.
(A) qRT-PCR analysis of the mRNA levels of HY5, TCP24, and HEN1 in deetiolating seedlings of the wild type (Col-0) at the indicated times of exposure to W (100 μmol m−2 s−1). Relative expression of each gene was normalized to that of UBQ10 and was relative to that of etiolated seedlings.
(B) qRT-PCR analysis of HY5, TCP24, and HEN1 expression in deetiolating Col-0 seedlings exposed to the indicated fluence rates for 6 h.
We also examined the expression behaviors of HEN1, HY5, and TCP24 in response to various light fluence rates. Light at lower fluence rate could trigger the expression of all three genes (Figure 10B). High light fluence rate further boosted the expression of HY5 but not TCP24 (Figure 10B). The higher expression ratio of positive to negative regulators may contribute to the exaggerated light responses under high light intensity.
DISCUSSION
HEN1 Is a Negative Regulator That Functions to Attenuate Light Signals during Photomorphogenesis in Arabidopsis
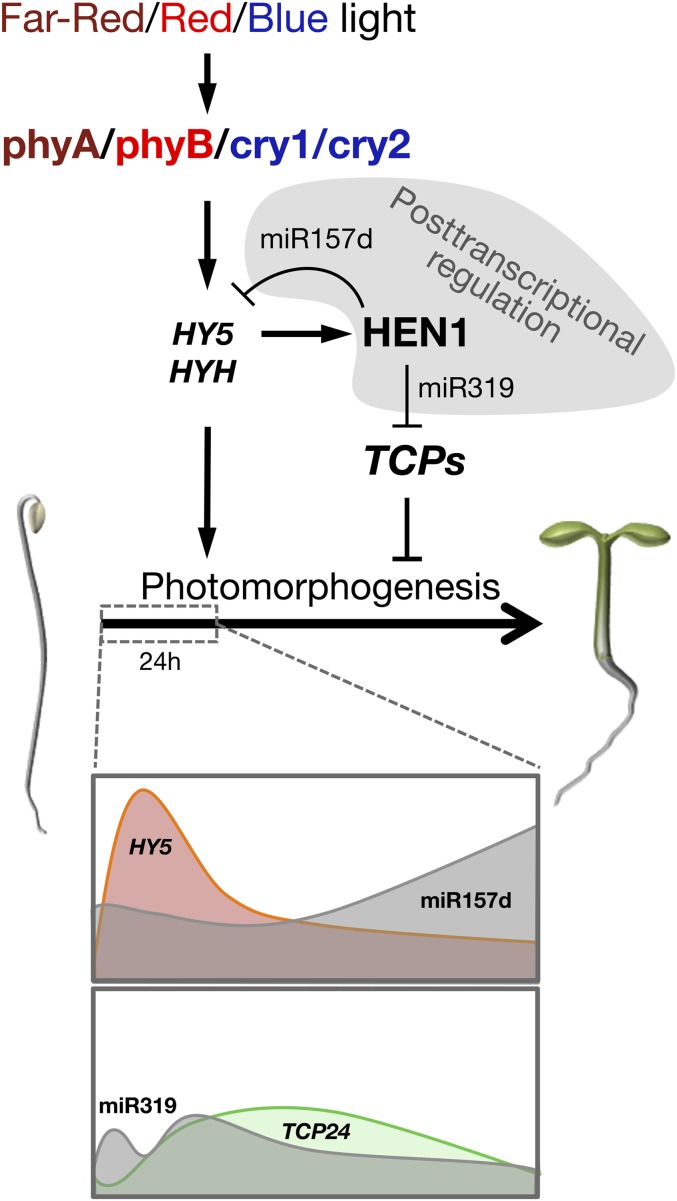
As a key enzyme in miRNA/small interfering RNA biogenesis, HEN1 likely functions to stabilize small RNA species that target the mRNAs of key regulators for degradation in light signaling pathways. From our observations, we postulate a light signaling pathway involving HEN1 in regulating Arabidopsis photomorphogenesis (Figure 11). Briefly, in wild-type Arabidopsis, environmental B, R, and FR light signals could be perceived by the photoreceptors cry1/cry2/phyA, phyB, and phyA, respectively. The light signals primarily converge at HY5 and HYH to induce the expression of HEN1, which is needed to stabilize and increase the levels of miRNAs, including but not limited to miR157d and miR319. With the progression of deetiolation, miR157d and miR319 could trigger miRNA-mediated cleavage of HY5 and TCP mRNAs. This process helps to reduce or shape the temporal expression kinetics of these genes to properly attenuate the actions of both the positive (HY5) and the negative (TCPs) regulators of photomorphogenesis (Figure 11).
Figure 11.
Diagram Depicting the Regulation and Function of HEN1 in Light Signal Transduction in Early Photomorphogenesis in Arabidopsis
The light-induced expression of HEN1 depends on the photoreceptors and positive regulators HY5/HYH. The increased HEN1 in photomorphogenic seedlings leads to the accumulation of miR157d and miR319 to fine-tune the expression patterns of both positive and negative regulators of photomorphogenesis. During early photomorphogenic development, miR157d targets HY5 transcript for degradation, resulting in a negative feedback regulatory loop between HEN1 and HY5. miR319 negatively regulates the expression of TCP24, a negative regulator of photomorphogenesis.
Our observations also reveal the complex nature of photomorphogenic development driven by a regulatory cascade or network of intertwined positive and negative regulators. HEN1 may stabilize miRNAs to prevent the overproduction of these regulators, for an antagonistic balance between positive and negative regulators. Nevertheless, the light-hypersensitive phenotype of hen1 suggests that the more predominant role of HEN1 is to desensitize positive regulators in the light signaling pathway.
Accumulation of HEN1 Transcripts in Early Photomorphogenesis
Our data indicate that the full light responsiveness of HEN1 depends on the photoreceptors phyA, phyB, cry1, and cry2 (Figures 3A to 3D). HY5 and HYH are the primary positive regulators that convey the light signal to induce HEN1 expression during early photomorphogenesis (Figure 3F). However, the promoter region of HEN1 contains no putative HY5 binding cis-element such as a G-box (CACGTG) or ACE box (ACGT-containing element) (Chattopadhyay et al., 1998; Shin et al., 2007). A recent HY5 ChIP-chip experiment also did not reveal HEN1 (Lee et al., 2007). Thus, HEN1 is likely not the immediate target of HY5. The normal light-induced expression of HEN1 in the b-box-containing protein 22/light-regulated zinc finger protein1 (bbx22/lzf1) mutant excluded BBX22/LZF1, a HY5-dependent positive regulator of photomorphogenesis (Chang et al., 2008), as responsible for HEN1 activation. Revealing the immediate transcription factor(s) governing the light responsiveness of HEN1 awaits the identification of more positive regulators in the HY5 lineage of the light-signaling pathway.
miRNA-Mediated Posttranscriptional Regulation in Photomorphogenic Arabidopsis
Etiolated hen1-1, hyl1, and hst seedlings have short hypocotyls (this study; Lu and Fedoroff, 2000; Bollman et al., 2003). However, these mutants still exhibit light-hypersensitive phenotypes by showing exaggerated light-mediated inhibition of hypocotyl elongation (Figure 1; Supplemental Figure 2). Because photomorphogenesis is normal for mutants defective in a ta-siRNA biogenesis pathway (Figure 4), miRNAs may play a more prominent role in the posttranscriptional regulation of photomorphogenesis.
We observed that the expression of miR157d and miR319 was increased by light in deetiolating seedlings (Figures 7 and 9). Such upregulation could be accomplished by increasing the transcription of primary transcripts of the MIR genes, by enhancing the capability of the processing complex to generate more precursors of miRNAs, stabilization, and/or transport of miRNAs. Among genes essential for the miRNA biogenesis pathway, only the HEN1 transcript is upregulated by light (Supplemental Figure 3). By artificially increasing the level of HA-HEN1 protein in dark-grown seedlings, miR157d/319 levels were increased (Figure 5C). This finding is indicative of a rate-limiting role for HEN1 in the biogenesis of these two miRNAs in young seedlings. Thus, the light responsiveness of HEN1 expression may serve as a signal to boost the biogenesis of miRNAs during early deetiolation. The increased miR157d and miR319 expression then imposes posttranscriptional regulation of gene expression in photomorphogenic Arabidopsis.
A future goal is to reveal additional miRNA-mRNA pairs that together achieve an optimized equilibrium of positive and negative regulators in deetiolating Arabidopsis. This investigation will require a comprehensive profiling of both mRNA and small regulatory RNAs and pairwise functional characterizations during photomorphogenic development.
METHODS
Plant Materials and Growth Conditions
Seeds of hyl1-2 (Song et al., 2007), hst-3 (Bollman et al., 2003), hst-6 (Bollman et al., 2003), rdr6-11 (Peragine et al., 2004), sgs3-11 (Peragine et al., 2004), hen1-5 (SALK_049197) (Vazquez et al., 2004), hen1-6 (SALK_090960) (Li et al., 2005), hy5-1 (Koornneef et al., 1980), cry1 (Koornneef et al., 1980), phyA-201 (Nagatani et al., 1993), phyB-5 (Reed et al., 1993), jaw-1D (CS6948), jaw-2D (CS6949), jaw-3D (CS6950), and jaw-4D (CS6951) mutants were obtained from the ABRC. The mutants hen1-1 (Park et al., 2002), phyA cry1 cry2 (Duek and Fankhauser, 2003), cop1-4/cop1-6 (McNellis et al., 1994), and hy5 hyh (Holm et al., 2002) were kindly provided by Xuemei Chen, Christian Fankhauser, Hsu-Liang Hsieh, and Christian Hardtke, respectively.
Seeds of Arabidopsis thaliana Col-0, Ler, and mutant plants were germinated on half-strength Murashige and Skoog medium with 0.8% agar and stratified at 4°C in the dark for 3 d, exposed to white light for 4 h to induce germination, and placed in the dark at 22°C for 20 h. For measuring hypocotyl length after light treatments, seedlings were moved to 22°C growth chambers for 4 d under various light conditions. Control seedlings were kept in the dark for 4 d. For HEN1 expression analysis, cold-stratified seeds were allowed to grow in the dark for 4 d at 22°C, then treated with light for the times indicated. LED light sources (Daina Electronics) were used for B (470 ± 30 nm), R (660 ± 25 nm), and FR light (730 ± 25 nm). Fluence rates used for measuring hypocotyl length under W, B, R, or FR light are in Figures 1, 4, 8, and 9 and Supplemental Figures 2, 5, 6, and 7. Hypocotyl length was analyzed using ImageJ (Schneider et al., 2012). The fluence rate was measured with use of an LI-250 radiometer (LI-COR).
Apical Hook and Antigravitropism Measurement
Seeds were germinated on a vertical plate in the dark for 24 h before seedlings were photographed every hour. The angle of curvature of the hook was measured as described (Vandenbussche et al., 2010). For the antigravitropism analysis, seedlings were grown on a vertical plate in the dark for 4 d before the plate was rotated for 90° and photographed every 30 min. The hypocotyl curvature from antigravitropism was measured using HypoPhen (Kami et al., 2012).
Anthocyanin Quantification
Ler and hen1-1 seedlings were grown for 4 d on half-strength Murashige and Skoog media containing 0.8% agar and sucrose at the indicated concentrations (Figure 2B) under 16 h light/8 h dark to enhance anthocyanin accumulation. Whole seedlings were collected and weighed for anthocyanin extraction and quantification as described (Lange et al., 1971).
RNA Isolation and Detection
Total RNA was isolated from seedlings by the pine-tree method as described (Chang et al., 1993) or with the mirVana miRNA isolation kit (Ambion). First-strand cDNA was synthesized and used for quantitative RT-PCR (qRT-PCR) as described (Wu et al., 2008) with the gene-specific primers in Supplemental Table 1. Small RNA gel blot analysis was performed as described (Chiou et al., 2006) with the miRNA-specific oligos in Supplemental Table 1. Signal intensities in Figures 5B and 5C and Supplemental Figure 12 were analyzed using ImageJ (Schneider et al., 2012).
For RNA gel blot analyses of HEN1 and HY5 transcripts, total RNA was isolated from seedlings by the PureLink Plant RNA Reagent method (Invitrogen). Five micrograms of total RNA was separated on 1% formaldehyde agarose gel and transferred to positively charged nylon membrane (GE Healthcare). HEN1 and HY5 cDNAs were amplified with specific primers (Supplemental Table 1) and used as templates for probe labeling.
Validation of miRNA-Directed Cleavage on Targets
Modified 5′ RACE with the GeneRacer Kit (Invitrogen) was used to validate cleavage sites on transcripts of HY5 and TCP24. The primers for 5′ RACE are in Supplemental Table 1.
Analysis of Frequency of SNPs
We used the Arabidopsis 1001 Genomes database (http://signal.salk.edu/atg1001/3.0/gebrowser.php) for analysis of HY5 SNPs from multiple sequence alignments of 513 Arabidopsis tracks (available on April 29, 2013). The SNP frequency was calculated for each position in HY5 exon regions by comparing HY5 sequences from different tracks to that of Col-0.
Construction of MIR Genes, HY5-GFP/mHY5-GFP Reporters, mHY5 cDNA, and XVE:HA-HEN1 System
MIR156a and MIR157d were amplified from Arabidopsis genomic DNA with corresponding forward and reverse primers (Supplemental Table 1) and inserted into the pBIN61 vector digested by XbaI and SmaI and under the control of the 35S promoter. The fragment of MIR157d driven by the 35S promoter was subcloned into the HindIII site of the pCAMBIA 1390 binary vector to generate MIR157dox line in Arabidopsis (Col-0). For the HY5-GFP reporter, HY5 5′-UTR (91 to 192 bp) containing the miR157d pairing region was amplified with HY5-5′UTR forward and reverse primers (Supplemental Table 1) and fused to the N terminus of GFP, and HY5-GFP fragment was excised by XbaI and SmaI and inserted in the pBIN61 vector to generate the HY5-GFP reporter. The miR157d-pairing region on the HY5-GFP reporter was further mutated by site-directed mutagenesis with HY5-5′UTRm forward and reverse primers (Supplemental Table 1) to generate the mHY5-GFP reporter. This mutated 5′-UTR was introduced by RT-PCR with primers HY5-5′UTRm-XbaI-F and HY5stop-EcoRI-R (Supplemental Table 1) to generate mHY5 cDNA. The XbaI-EcoRI mHY5 cDNA fragment was inserted into a Basta resistance (bar)-modified pCAMBIA 1390 under the control of the 35S promoter. The HEN1 genomic DNA fragment containing the entire coding region was amplified with primers in Supplemental Table 1. The HEN1 genomic DNA was inserted into a modified pER10-bar digested by KpnI and SpeI to generate a 3×HA epitope translationally fused HEN1 fragment in the XVE:HA-HEN1 construct for XVE-induction in Arabidopsis (Col-0) (Zuo et al., 2006).
Nicotiana benthamiana Transient Expression
Transient expression assay was performed as described (Voinnet et al., 2003). Leaves of 4-week-old N. benthamiana were coinfiltrated with cultures of Agrobacterium tumefaciens (C58C1) carrying MIR157d (or MIR156a) and HY5-GFP (or mHY5-GFP) reporter. Agrobacteria were brought to OD600 = 1.0 for input by mixing MIR and reporter cultures in a 9:1 ratio. Infiltrated tissues were collected after 3 d, and total RNA was isolated for small RNA gel blot and qRT-PCR analyses.
XVE:HA-HEN1 Induction
The T2 seed populations from independent transgenic lines of XVE:HA-HEN1 (Col-0) were germinated on the growth medium described above containing phosphinothricin (10 μg/µL) for the selection. The seeds were allowed to grow in the dark at 22°C for 3 d and transferred into 20 mL 3 mM MES buffer (pH 5.7) containing 0, 0.1, 1, or 10 μM β-estradiol dilutions carried by 20 μL DMSO for 24-h dark induction before collection for HA-HEN1 and miRNA analyses.
Sequencing of Small RNAs
Four-day-old etiolated Arabidopsis seedlings were treated with white light (100 μmol m−2 s−1) for 0, 1, 3, 6, 12, and 24 h. Total RNA was isolated with the mirVana miRNA isolation kit. The size-fractionated small RNAs were used to generate libraries for sequencing by the Illumina platform. Sequencing reads mapped to miR157d and miR319a/b/c were normalized to the total reads and extracted for profiling the light responsiveness in the deetiolation process.
Immunoblot Analyses
Total protein was extracted from 4-d-old etiolated seedlings as described (Chang et al., 2011) at different times of W light treatment. HY5 protein was detected by polyclonal anti-HY5 antibody (Liu et al., 2012). HA-tagged HEN1 protein was detected by incubation with the antibodies mouse monoclonal anti-HA (H3663; Sigma-Aldrich) or pig polyclonal anti-HEN1 (Yang et al., 2007). Endogenous α-tubulin was detected by incubation with a mouse monoclonal anti-α-tubulin antibody (T5168; Sigma-Aldrich).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative data library with the following locus identifiers: HYL1 (At1g09700), HEN1 (At4g20910), HST (At3g05040), PHYA (At1g09570), PHYB (At2g18790), CRY1 (At4g08920), CRY2 (At1g04400), HY5 (At5g11260), HYH (At3g17609), COP1 (At2g32950), RDR6 (At3g49500), SGS3 (At5g23570), TCP2 (At4g18390), TCP3 (At1g53230), TCP4 (At3g15030), TCP10 (At2g31070), TCP24 (At1g30210), MIR156A (At2g25095), MIR157D (At1g48742), MIR319A (At4g23713), MIR319B (At5g41663), and MIR319C (At2g40805).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Overview of Canonical miRNA Biosynthesis (Modified from Rogers and Chen, 2013).
Supplemental Figure 2. Skotomorphogenesis and Photomorphogenesis Are Defective in miRNA Biogenesis Mutants.
Supplemental Figure 3. The Transcript Level of HEN1 Is Light Upregulated.
Supplemental Figure 4. Molecular Characterization of hen1-5 and hen1-6 Mutants.
Supplemental Figure 5. The hen1 Mutants Are Hypersensitive to Blue (B), Far-Red (FR), and Red (R) Light.
Supplemental Figure 6. Functional Complementation of hen1 Light-Hypersensitive Phenotype.
Supplemental Figure 7. Effect of FR Fluence Rate on Hypocotyl Extension in Wild-Type and sgs3-11 Mutant Seedlings.
Supplemental Figure 8. The Levels of miR157 and miR159/319 Are Increased Accompanied with Increased Level of HA-HEN1 Protein.
Supplemental Figure 9. The Expressions of miR157d and miR319 Are Compromised in hen1-1 Mutant.
Supplemental Figure 10. HY5 Is Increased in the hen1-1 Mutant during Photomorphogenesis.
Supplemental Figure 11. Overaccumulation of miR157d Detected in the MIR157dox Line.
Supplemental Figure 12. HY5 Transcript Levels Are Decreased in the MIR157dox Transgenic Line.
Supplemental Table 1. Primers and Oligos Used in This Study.
Supplementary Material
Acknowledgments
We thank Yi-Chen Wu for constructing the HEN1 genomic fragment for the complementation experiment, Jing-Fen Wu for constructing the mHY5 overexpression and XVE:HA-HEN1 inducible constructs, Tzyy-Jen Chiou and Kuo-Chen Yeh for helpful discussion, Ho-Ming Chen for initial search of the PARE database, Mei-Jane Fang for technical assistance, Xuemei Chen for providing seeds of the hen1-1 mutant and the anti-HEN1 antibody, and Christian Frankhauser, Hsu-Liang Hsieh, and Christian Hardtke for providing seeds of the phyA cry1 cry2 triple mutant, cop1 mutant alleles, and hy5 hyh double mutant. This research was supported by a National Research Foundation of Korea grant funded by the Korean government (Ministry of Science, ICT, and Future Planning; 2008-0061988) to J.H.A., an Academia Sinica postdoctoral fellowship to H.-L.T., and the Foresight Project L20-2 and Investigator Award to S.-H.W. from Academia Sinica.
AUTHOR CONTRIBUTIONS
H.-L.T. and S.-H.W. designed the research, analyzed the data, and wrote the article. H.-L.T., Y.-H.L., W.-P.H., and M.-C.L. performed the research. J.H.A. contributed experimental materials.
Glossary
- miRNA
microRNA
- R
red
- FR
far-red
- B
blue
- Col-0
Columbia-0
- Bc
continuous blue
- Ler
Landsberg erecta
- FRc
continuous far-red
- Rc
continuous red
- W
white light
- ta-siRNA
trans-acting small interfering RNA
- UTR
untranslated region
- RACE
rapid amplification of cDNA ends
- SNP
single nucleotide polymorphism
- qRT-PCR
quantitative RT-PCR
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ahmad M., Lin C., Cashmore A.R. (1995). Mutations throughout an Arabidopsis blue-light photoreceptor impair blue-light-responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J. 8: 653–658. [DOI] [PubMed] [Google Scholar]
- Ahmad M., Jarillo J.A., Smirnova O., Cashmore A.R. (1998). The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1: 939–948. [DOI] [PubMed] [Google Scholar]
- Bollman K.M., Aukerman M.J., Park M.Y., Hunter C., Berardini T.Z., Poethig R.S. (2003). HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130: 1493–1504. [DOI] [PubMed] [Google Scholar]
- Chang C.-S.J., Maloof J.N., Wu S.-H. (2011). COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol. 156: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-S.J., Li Y.H., Chen L.T., Chen W.C., Hsieh W.P., Shin J., Jane W.N., Chou S.J., Choi G., Hu J.M., Somerville S., Wu S.-H. (2008). LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 54: 205–219. [DOI] [PubMed] [Google Scholar]
- Chang S., Puryear J., Cainey J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11: 113–116. [Google Scholar]
- Chattopadhyay S., Ang L.H., Puente P., Deng X.W., Wei N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2010). Small RNAs - secrets and surprises of the genome. Plant J. 61: 941–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T.J., Aung K., Lin S.I., Wu C.C., Chiang S.F., Su C.L. (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A., Chen D.L., Yeh K.C., Abel S. (2000). Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124: 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek P.D., Fankhauser C. (2003). HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J. 34: 827–836. [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Chory J. (1999). Light receptor kinases in plants! Curr. Biol. 9: R123–R126. [DOI] [PubMed] [Google Scholar]
- German M.A., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26: 941–946. [DOI] [PubMed] [Google Scholar]
- Henriques R., Jang I.C., Chua N.H. (2009). Regulated proteolysis in light-related signaling pathways. Curr. Opin. Plant Biol. 12: 49–56. [DOI] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J., Deng X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L., Chen X. (2012). Regulation of small RNA stability: methylation and beyond. Cell Res. 22: 624–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. (2006). MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57: 19–53. [DOI] [PubMed] [Google Scholar]
- Juntawong P., Bailey-Serres J. (2012). Dynamic light regulation of translation status in Arabidopsis thaliana. Front. Plant Sci. 3: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C., Hersch M., Trevisan M., Genoud T., Hiltbrunner A., Bergmann S., Fankhauser C. (2012). Nuclear phytochrome A signaling promotes phototropism in Arabidopsis. Plant Cell 24: 566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Shin J., Lee S.H., Kweon H.S., Maloof J.N., Choi G. (2011). Phytochromes inhibit hypocotyl negative gravitropism by regulating the development of endodermal amyloplasts through phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 108: 1729–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Rolff E., Spruit C.J.P. (1980). Genetic-control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L) Heynh. Z. Pflanzenphysiol. 100: 147–160. [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H., Shropshire W., Mohr H. (1971). An analysis of phytochrome-mediated anthocyanin synthesis. Plant Physiol. 47: 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang Z., Yu B., Liu J., Chen X. (2005). Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15: 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.-J., Wu S.-H., Chen H.-M., Wu S.-H. (2012). Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol. Syst. Biol. 8: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.-J., Wu S.-H., Wu J.-F., Lin W.-D., Wu Y.-C., Tsai T.-Y., Tsai H.-L., Wu S.-H. (2013). Translational landscape of photomorphogenic Arabidopsis. Plant Cell 25: 3699–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Genoud T., Fankhauser C. (2006). Let there be light in the nucleus! Curr. Opin. Plant Biol. 9: 509–514. [DOI] [PubMed] [Google Scholar]
- Lu C., Fedoroff N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12: 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A., Schrader A., Kokkelink L., Falke C., Welter B., Iniesto E., Rubio V., Uhrig J.F., Hülskamp M., Hoecker U. (2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 74: 638–651. [DOI] [PubMed] [Google Scholar]
- Mazzella M.A., Casal J.J., Muschietti J.P., Fox A.R. (2014). Hormonal networks involved in apical hook development in darkness and their response to light. Front. Plant Sci. 5: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzihradszky M., et al. (2013). Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell 25: 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., King S., Jack T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 22534–22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A., Reed J.W., Chory J. (1993). Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102: 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K., Wong C.C., Yates J.R., III, Chory J. (2013). Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors. Cell Reports 3: 1970–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Paik I., Yang S., Choi G. (2012). Phytochrome regulates translation of mRNA in the cytosol. Proc. Natl. Acad. Sci. USA 109: 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe C., Sweere U., Drumm-Herrel H., Schäfer E. (1998). The blue light receptor cryptochrome 1 can act independently of phytochrome A and B in Arabidopsis thaliana. Plant J. 16: 465–471. [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K., Chen X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H., Nakashima M., Matsuoka K., Matsushita T. (2012). Deletion of the RS domain of RRC1 impairs phytochrome B signaling in Arabidopsis. Plant Signal. Behav. 7: 933–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D.H., Choi M., Kim K., Bang G., Cho M., Choi S.B., Choi G., Park Y.I. (2013). HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 587: 1543–1547. [DOI] [PubMed] [Google Scholar]
- Shin J., Park E., Choi G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49: 981–994. [DOI] [PubMed] [Google Scholar]
- Song L., Han M.H., Lesicka J., Fedoroff N. (2007). Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl. Acad. Sci. USA 104: 5437–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C., Bussell J.D., Camus I., Ljung K., Kowalczyk M., Geiss G., McKhann H., Garcion C., Vaucheret H., Sandberg G., Bellini C. (2005). Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 17: 1343–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk E.K., Decker P.V., Chen M. (2012). Photobodies in light signaling. Plant Physiol. 158: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F., Petrásek J., Zádníková P., Hoyerová K., Pesek B., Raz V., Swarup R., Bennett M., Zazímalová E., Benková E., Van Der Straeten D. (2010). The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137: 597–606. [DOI] [PubMed] [Google Scholar]
- Vazquez F., Gasciolli V., Crété P., Vaucheret H. (2004). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14: 346–351. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Wu J.-F., Wang Y., Wu S.-H. (2008). Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol. 148: 948–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Vilkaitis G., Yu B., Klimasauskas S., Chen X. (2007). Approaches for studying microRNA and small interfering RNA methylation in vitro and in vivo. Methods Enzymol. 427: 139–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K.-C., Lagarias J.C. (1998). Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95: 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K.-C., Wu S.-H., Murphy J.T., Lagarias J.C. (1997). A cyanobacterial phytochrome two-component light sensory system. Science 277: 1505–1508. [DOI] [PubMed] [Google Scholar]
- Yi C., Deng X.W. (2005). COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15: 618–625. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., He H., Wang X., Wang X., Yang X., Li L., Deng X.W. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65: 346–358. [DOI] [PubMed] [Google Scholar]
- Zhao J., Favero D.S., Peng H., Neff M.M. (2013). Arabidopsis thaliana AHL family modulates hypocotyl growth redundantly by interacting with each other via the PPC/DUF296 domain. Proc. Natl. Acad. Sci. USA 110: E4688–E4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Hare P.D., Chua N.H. (2006). Applications of chemical-inducible expression systems in functional genomics and biotechnology. Methods Mol. Biol. 323: 329–342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.